Abstract
Based on the identification of residues that determine receptor selectivity in arrestins and the phylogenetic analysis of the arrestin (arr) family, we introduced fifteen mutations of receptor-discriminator residues in arr-3, which were identified previously using mutagenesis, in vitro binding, and BRET-based recruitment assay in intact cells. The effects of these mutations were tested using neuropeptide Y receptors Y1R and Y2R. NPY-elicited arr-3 recruitment to Y1R was not affected by these mutations, or even alanine substitution of all ten residues (arr-3-NCA), which prevented arr-3 binding to other receptors tested so far. However, NCA and two other mutations prevented agonist-independent arr-3 pre-docking to Y1R. In contrast, eight out of 15 mutations significantly reduced agonist-dependent arr-3 recruitment to Y2R. NCA eliminated arr-3 binding to active Y2R, whereas Tyr239Thr reduced it ~7-fold. Thus, manipulation of key residues on the receptor-binding surface generates arr-3 with high preference for Y1R over Y2R. Several mutations differentially affect arr-3 pre-docking and agonist-induced recruitment. Thus, arr-3 recruitment to the receptor involves several mechanistically distinct steps. Targeted mutagenesis can fine-tune arrestins directing them to specific receptors and particular activation states of the same receptor.
Keywords: Arrestins, GPCRs, neuropeptide Y receptors, protein engineering, signal transduction, bioluminescence resonance energy transfer (BRET)
1. Introduction
G protein-coupled receptors (GPCR) are high value drug targets, with almost half of the marketed drugs acting on these cell surface proteins (1,2). The demand for specific tools for molecular intervention keeps growing, with the emphasis on minimizing side effects and expanding the repertoire of therapeutically addressable GPCRs. The identification of arrestins as key regulators in GPCR desensitization and internalization and mediators of G protein-independent signaling opened new possibilities for the manipulation of receptor function. Arrestins bind activated and phosphorylated GPCRs and block receptor-G-protein interaction, while serving as adaptors for key components of the endocytic machinery and numerous signaling proteins (3,4). The two visual subtypes, arrestin-11 (arr-1) and arrestin-4 (arr-4), are expressed exclusively in photoreceptors and pinealocytes, and bind rhodopsin and cone opsins (5,6). In contrast, two ubiquitously expressed non-visual subtypes, arrestin-2 (arr-2) and arrestin-3 (arr-3), are fairly promiscuous and interact with a huge range of GPCRs (3). Recent advances in our understanding of the molecular basis of receptor specificity of arrestin proteins pave the way to targeted construction of reengineered receptor-specific arrestins (7,8).
Here we tested the interactions of mutants of the non-visual arr-3 and two members of the neuropeptide Y (NPY) receptor family. The NPY system plays a central role in the regulation of energy homeostasis and is involved in the pathophysiology of cancer progression, obesity, mood disorders, and epilepsy (9). This makes the neuropeptide Y system a desirable drug target for future therapies. NPY receptors respond to three endogenous peptidic agonists, NPY, peptide YY (PYY) and the pancreatic polypeptide (PP). Neuropeptide Y receptor family includes four subtypes in humans: Y1R, Y2R, Y4R, and Y5R (10). Y receptors belong to class A of the GPCR superfamily, couple to Gi/o proteins (11), and are expressed in numerous tissues including brain, blood vessels, heart, and gastrointestinal tract (12–16). They have overlapping binding profiles for their endogenous ligands with Y1R, Y2R, and Y5R binding NPY and PYY with high affinity, whereas Y4R prefers PP (17).
In this complex multiligand-multireceptor system Y1R and Y2R occupy a distinct position. They are frequently expressed in the same tissues (18), where they can induce either synergistic or antagonizing effects on food intake, anxiety, and depression (19,20). Antagonism is thought to be due to the pre-synaptic expression of Y2R in NPY-ergic neurons, where it functions as an inhibitory autoreceptor blocking NPY release and thus Y1R-and Y5R-mediated effects (21), as revealed by using Y1R-and Y2R-selective agonists and antagonists. However, as Y receptor knockout models showed inconclusive results (20,22), subtype selective arrestins would be valuable tools to unravel phenotypes assigned to either Y1R-or Y2R-mediated effects.
Unlike the Y5R, both Y1R and Y2R undergo arr-3-dependent internalization, which terminates their G-protein-mediated signaling. Arr-3 binding motifs in these receptors have been recently identified. Like in many GPCRs, these motifs include C-terminal serine/threonine clusters which serve as phosphorylation substrates (23–25) for G protein-coupled receptor kinases (GRKs (26)). Even though phosphorylated serines and threonines favor the interaction of non-visual arrestins with their cognate receptors, previous studies showed that phosphorylation does not contribute to the arrestin receptor specificity and is not even mandatory for all arrestin-GPCR interactions (27–29). Although arrestins engage an extensive surface of the receptor, only a few residues on the concave sides of the arrestin N-and C-domains determine its receptor specificity (28). This finding made it possible to construct the mutants of naturally promiscuous arr-3 that are specific for GPCR groups or single receptor subtypes (30). Importantly, these studies on D1 and D2 dopamine, β2-adrenergic (β2AR) and M2 muscarinic receptors showed that mutations of arr-3 similarly affected the interactions with activated phosphorylated receptors and agonist-independent arrestin pre-docking (30). This supports the idea that arrestins pre-select their target receptors before they become active and phosphorylated, enabling the use of reengineered arrestins for the manipulation of GPCR functions. Here we report the identification of key arr-3 residues that discriminate between Y1 and Y2 receptors and their functional states.
2. Material and methods
2.1. Materials
Porcine NPY (pNPY) was produced by automated solid phase peptide synthesis using the Fmoc/tBu (9–fluorenylmethoxycarbonyl-tert-butyl) strategy, as described (31). [3H] myo-inositol was from GE Healthcare Europe GmbH (Braunschweig, Germany). Restriction endonucleases and other DNA modifying enzymes were from New England Biolabs (Ipswich, MA). Cell culture reagents and media were from Mediatech-Corning (Manassas, VA), Life-technologies (Carlsbad, CA), or PAA Laboratories GmbH (Pasching, Austria). Luciferase substrate coelenterazine-h was from NanoLight Technology (Pinetop, AZ). All other reagents were from Amresco (Solon, OH) or Sigma-Aldrich (St Louis, MO).
2.2. Mutagenesis and plasmid construction
Plasmids encoding short splice variant of bovine arr-3 (32,33) with unique restriction sites introduced by silent mutations and arr-3 mutants K2A, NCA and KNC with secondary phosphate-sensing N-terminal lysines (Lys11 and Lys12) or all key receptor-discriminator residues replaced with alanines, were described previously (27,28). All mutations were introduced on Ala87Val background, as described (7,27,30), which mimics more receptor-specific visual subtypes (34,35), reducing the flexibility of the N-domain (36,37). To generate all mutants, mutagenizing oligonucleotides were used as forward primers and an oligonucleotide downstream from the far restriction site to be used for subcloning was used as a reverse primer. Resulting fragments of various lengths and an appropriate primer upstream of the near restriction site were then used as reverse and forward primers, respectively, for the second round of PCR. Restriction sites Bam HI and Bsi WI were used for introducing mutations in the N-domain, sites Age I and Xho I were used for introducing mutations in the C-domain. The resulting fragments were purified, digested with the respective enzymes, and subcloned into the suitably digested pGEM-2 plasmid (Promega; Madison, WI) containing the sequence of wild type (WT) bovine arr-3 (38,39). For bioluminescence resonance energy transfer (BRET) assays, WT and mutant arr-3 was subcloned into pcDNA3 Venus plasmid (Life technologies), yielding arr-3 N-terminally tagged with Venus (40). Human Y1R and Y2R were C-terminally tagged with Renilla luciferase variant 8 (RLuc8 (41)), as described (27,28,30). All constructs were verified by dideoxy sequencing.
2.3. Functional characterization of RLuc-tagged Y receptors
Signaling of RLuc-tagged Y1R and Y2R was measured in transiently transfected COS-7 cells (ATCC Cat. # CRL-1651) by determining the inositol phosphate (IP) accumulation after pNPY stimulation, as described (42,43), using the method established by Berridge (44,45). Cells were treated with increasing concentrations of pNPY, from 10 pM to 1 µM. To determine EC50 values for pNPY on Y1R and Y2R, the data were fit to a three-parameter dose-response curve equation (GraphPad Prism 6.03).
2.4. Bioluminescence resonance energy transfer (BRET) assay
BRET-based (46,47) arrestin-receptor interaction assays were performed and analyzed, as described (24,27,30,48). Cells were treated with pNPY (1 µM) for 8 minutes prior to the addition of 5 µM coelenterazine-h. Absolute levels of luminescence were used as the measure of the expression levels of RLuc-tagged receptors, whereas direct fluorescence was used to gauge the expression of Venus-tagged arrestins, as described (24,27,30,37). BRET was measured 15 minutes after agonist addition, as described (24), and plotted as a function of fluorescence/luminescence ratio. The resulting curves were fit by non-linear regression to a one-site hyperbola equation (net BRET after pNPY treatment) or a three-parameter dose-response curve equation (BRET ratios vs fluorescence/luminescence (F/L) ratios for each arrestin amount) (GraphPad Prism 6.03).
2.5. Visualization of arrestin recruitment to the receptor by live-cell imaging
COS-7 cells were plated onto sterile 8-well µ-slides (ibidi GmbH, Martinsried/Munich, Germany), grown to 70% confluence and transfected with 1 µg of DNA encoding Y1R or Y2R fused to eCFP along with 0.1 µg of DNA encoding WT Venus-arr-3, or Ala87Val, Ala87Val+Tyr239Thr, and KNC mutants, using Lipofectamine™ 2000 (Life technologies). Imaging was performed 24 h after transfection using a Zeiss Axio Observer microscope with an ApoTome Imaging System equipped with a Heating Insert P Lab-Tek S1 unit. Fluorescence images from living cells were taken at 37°C on a heated microscope stage using the AxioVision Rel. 4.6 software. Arr-3 translocation was visualized in serum-starved cells for 30 min in Opti-MEM® (Life technologies) and treated with 1 µM pNPY at 37°C for 15 min. Images (EYFP/Venus channel only) were acquired before and after agonist treatment.
2.6. Data analysis and statistics
Statistical analysis was performed using Prism 6.03 (GraphPad Software, San Diego, CA). Statistical significance was determined by one-way ANOVA followed by post-hoc Dunnett’s test with correction for multiple comparisons.
3. Results
3.1. Renilla luciferase (RLuc8) tagged neuropeptide Y receptors are functional
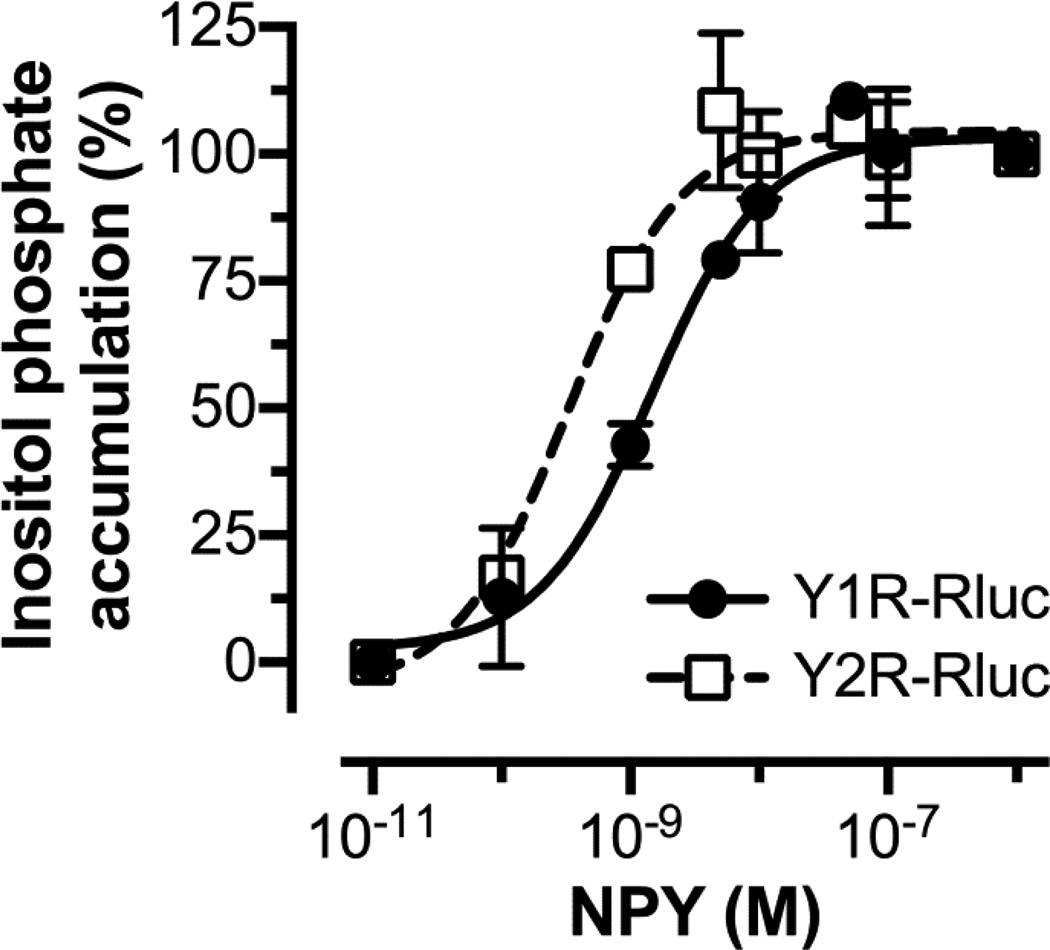
Agonist-driven arrestin recruitment studies require fluorophore or luminogen-tagged receptors, which must be functional. The functionality of fused receptors is characterized by ligand affinity, expression at the cell membrane, and agonist potency. To test the functionality of Y1R and Y2R RLuc constructs, we measured the accumulation of [3H]-inositol phosphates in cells co-transfected with a chimeric Gαq that couples to receptors that interact with Gi proteins (49). This procedure bypasses the need to pre-activate cells with forskolin to measure cAMP accumulation. Kostenis (49) described chimeric G protein α subunit (GαΔ6qi4myr) that couples to Gi-specific receptors and interacts with PLC-β, which releases inositol (1,4,5)-trisphosphate. Previous reports showed that fusing Y receptors to EYFP has little effect in G protein coupling (42). We tested RLuc tagged Y1R and Y2R and found that both effectively couple to G proteins (Fig. 1). The EC50 for NPY (1.5 nM and 0.3 nM for Y1R and Y2R respectively) for the accumulation of inositol phosphate were almost identical to those obtained for the EYFP-fused receptors (42), demonstrating receptor functionality.
Fig. 1. Functionality of RLuc-tagged Y1 and Y2 receptors.
Inositol phosphate accumulation assays where performed using COS-7 cells transfected with either Y1R (filled circles; continuous line) or Y2R (open squares; dashed line) and a chimeric GαΔ6qi4myr-protein. Cells were loaded with [3H] myo-inositol for 16 to 18 h and then treated with indicated concentrations of pNPY for 60 min. Accumulated [3H]-inositol phosphates were measured after cell lysis. The pNPY EC50 value for Y1R and Y2R were 1.5 nM (pEC50= 8.83 ± 0.12) and 0.3 nM (pEC50= 9.49 ± 0.16). These EC50 values were obtained by fitting the data to a three-parameter dose-response curve equation (GraphPad Prism 6.03). Means ± SEM from two independent experiments performed in duplicate are shown.
3.2. Arrestin recruitment by neuropeptide Y receptors is a multi-step process
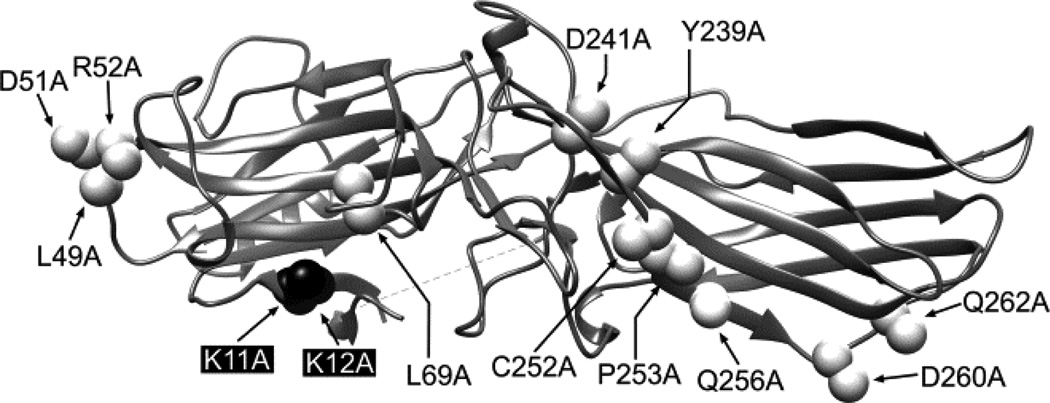
Arrestin recruitment by agonist-bound receptors can be readily evaluated in live cells using BRET between RLuc-tagged receptor and Venus-tagged arrestins (46,47). To exclude artifacts, BRET experiments must include appropriate controls to account for bystander effects. In this case, the best control would be a Venus-tagged arr-3 mutant that does not bind GPCRs. We showed that substitution of ten residues on the receptor-binding surface of arrestins with alanines completely abolishes binding to rhodopsin and significantly reduces the interaction with other receptors (28,30). In addition, alanine substitution of the two highly conserved lysines (Lys14 and 15 in bovine arr-1) significantly reduced binding to phosphorylated active rhodopsin (50). We introduced homologous mutations in bovine arr-3 (K2A, NCA and KNC mutations shown in Fig. 2) and tested by BRET their effect on arr-3 recruitment to Y1R and Y2R (Fig. 3).
Fig. 2. The structure of K2A, NCA, and KNC mutants.
Ribbon diagram of bovine arrestin-3 (PDB ID: 3P2D) (37) showing the positions of the two phosphate-binding lysines in the β-strand I (replaced with alanines in the K2A; black) and the residues replaced with alanines in the NCA (white). The KNC is the combination of K2A and NCA. The image was generated with USCF Chimera 1.8.
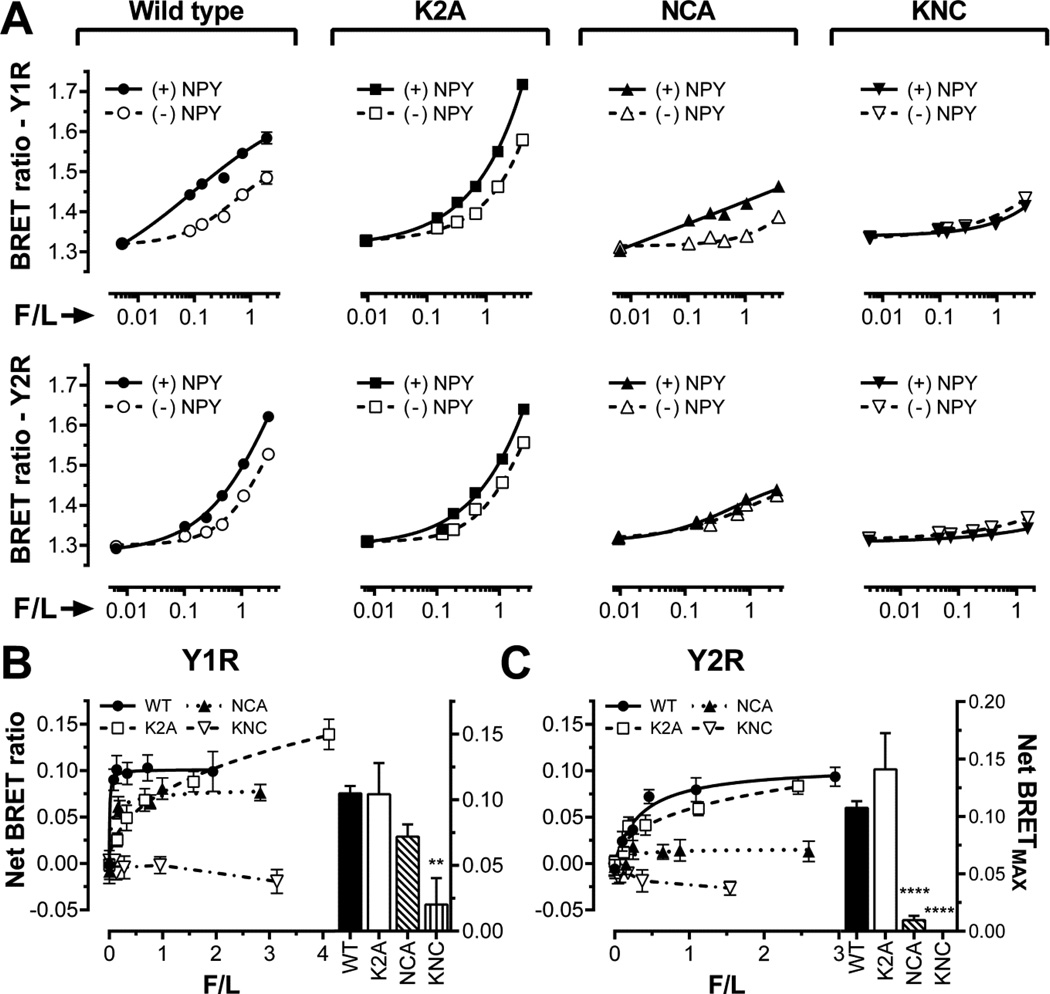
Fig. 3. The binding of K2A, NCA, and KNC mutants to Y1R and Y2R.
A–C. Raw BRET signal reflecting the interaction of WT (circles), K2A (squares), NCA (triangles), and KNC (inverted triangles) forms of Venus-arr-3 with luciferase-tagged human Y1R or Y2R. BRET ratios are plotted as a function of Venus-arr-3 fluorescence normalized by receptor-RLuc8 luminescence (F/L) in the presence of agonist (filled symbols; solid lines) or vehicle (open symbols; dashed lines) in COS-7 cells. B,C. Net BRET (agonist-induced increase in BRET signal) for the Y1R (B) or Y2R (C) receptors. The same symbols as in panel A are used. Bar graphs in B and C show net BRETMAX at saturation, determined by a fit to one-site binding hyperbola using GraphPad Prism 6.03. Means ± S.E. of six repeats of a representative experiment (out of three performed) are shown. Statistical significance was determined by one-way ANOVA followed by Dunnett's multiple comparison test. ** p< 0.01, **** p < 0.0001.
We recently found that arr-3 binding to β2AR, M2 muscarinic, D1 and D2 dopamine receptors has two components: agonist-independent pre-docking observed as progressive increase of BRET ratio with Venus-arr-3 expression, and agonist-induced increase, measured by net BRET, a difference between BRET ratio in the presence and absence of agonist (30). An increasing BRET ratio with arrestin expression (Fig 3A) and the saturable agonist-induced additional arrestin binding (Fig. 3B,C) showed that both components are also present in case of WT arr-3 interaction with Y1R and Y2R. In contrast, arr-3 KNC (a combination of K2A and NCA; Fig. 2) does not appear to pre-dock, judging by only marginal increase in BRET ratio with its expression level, which likely reflects its random encounters with receptors (bystander BRET) (Fig. 3A). There is no agonist-dependent increase in BRET signal (virtually zero net BRETMAX) (Fig 3B,C), suggesting that KNC mutant is not recruited to either Y1R or Y2R upon agonist stimulation. Thus, Venus-tagged arr-3-KNC that has exactly the same molecular weight as all other forms of Venus-arr-3, but does not bind receptors (Fig. 3) is an ideal negative control yielding only bystander BRET.
Next we used the NCA mutant, where residues engaging receptor-attached phosphates are intact, whereas other receptor-binding residues are replaced with alanines, and K2A mutant, where two key phosphate-binding lysines are replaced with alanines, whereas residues that bind non-phosphorylated parts of the receptor are intact. The comparison of these two mutants with positive (WT) and negative (KNC) controls in the presence and absence of an agonist sheds light on the interactions involved in arrestin binding to Y1R and Y2R (Fig. 3). Basal BRET ratios for NCA mutant and Y1R are similar to those obtained with KNC (Fig 3A, top row). However, in contrast to KNC, agonist significantly increases NCA binding to Y1R, as revealed by net BRETMAX (Fig 3B). Interestingly, in case of Y2R NCA mutant performs essentially like KNC, indicating that receptor-discriminator residues eliminated in NCA play key role in Y2R binding (Fig 3C). Thus, either agonist-induced arr-3 binding to Y1R is mediated solely by the phosphates attached by GRKs in response to receptor activation, or the interaction with Y1R is partially mediated by arrestin residues not mutated in NCA. The data with K2A mutant impaired in phosphate binding suggest that the latter is the case: K2A pre-docks to both receptors, and agonist-dependent increase in its binding is comparable to that observed with WT arr-3 in both cases (Fig. 3).
These data show that arr-3 binding to Y1R and Y2R is not critically dependent on receptor-attached phosphates, as K2A mutant resembles WT arr-3 (Fig. 3). The interactions with other receptor elements appear to be largely mediated by the ten residues replaced in NCA in case of Y2R, where this mutation abolishes the binding, but not in case of Y1R, where NCA mutant demonstrates reduced pre-docking, but responds to agonist stimulation almost normally (Fig. 3).
3.3. Certain mutations in arr-3 differentially affect the binding to Y1R and Y2R
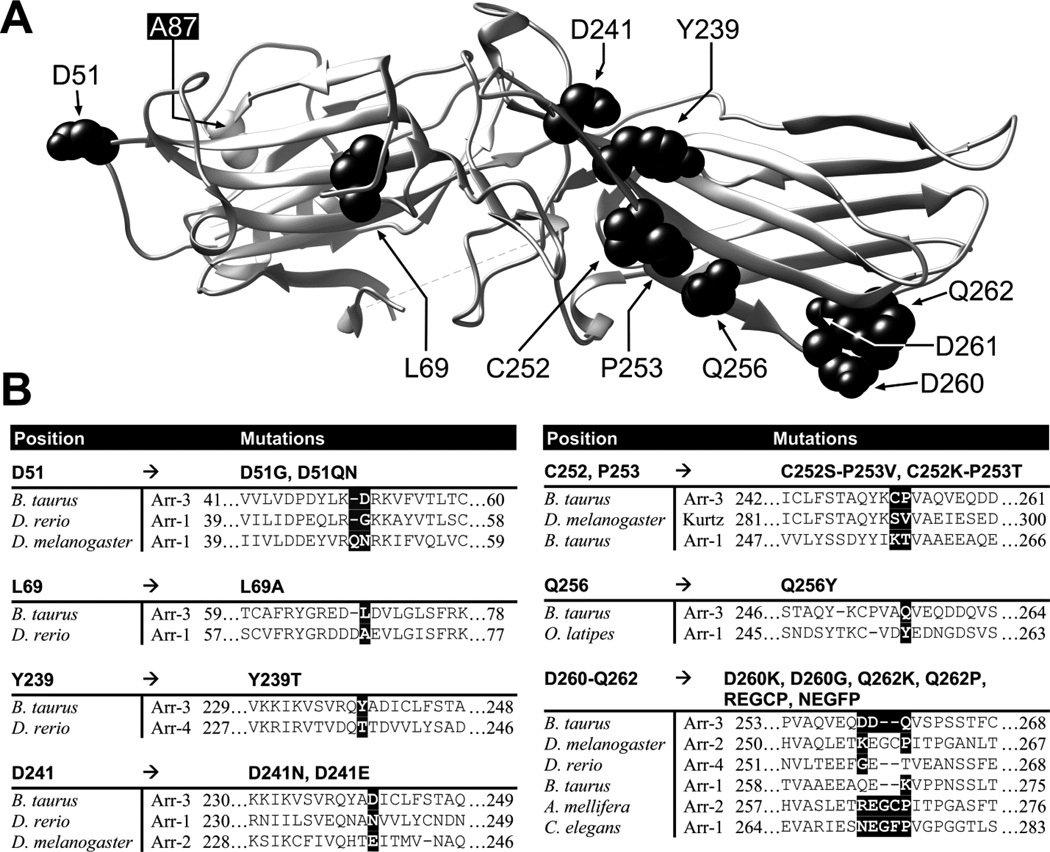
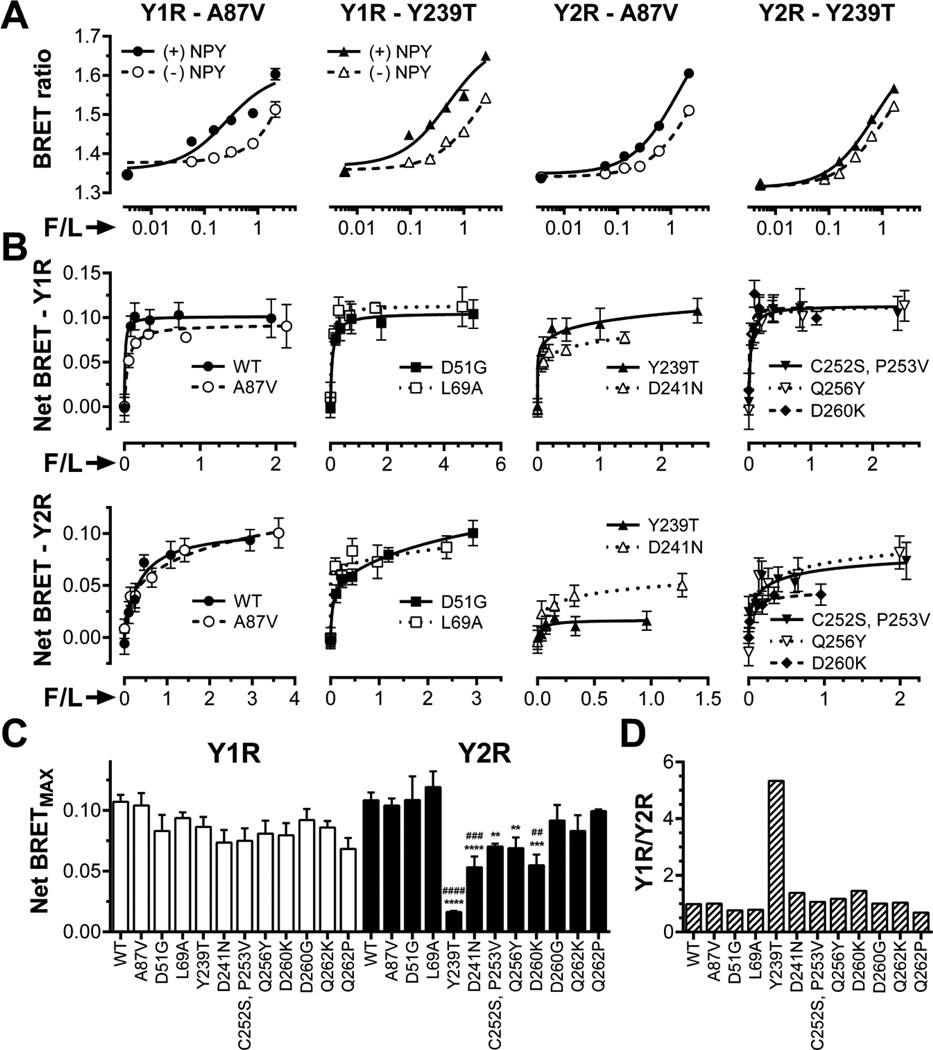
Since NCA mutation precluded Y2R binding without significantly affecting Y1R interactions (Fig. 3), next we tested a series of point mutations in the positions substituted in NCA (Fig. 4A). Out of the fifteen mutants used, ten were previously tested with β2AR, M2 muscarinic, and D1 and D2 dopamine receptors (30) and five were generated de novo based on arrestin sequences in other organisms (Fig 4B).
Fig. 4. Mutations on the receptor-binding surface of arr-3.
A. Ribbon diagram of bovine arr-3 (PDB ID: 3P2D). The residues targeted in this study are shown as CPK models in black. UCSF Chimera 1.8 was used to generate this panel. B. Alignments of the sequences of different arrestins containing the residues introduced into bovine arr-3 (on the Ala87Val background). Substituted residues in arr-3 and corresponding amino acids in other arrestins are highlighted by black background.
Agonist-induced increase in the interaction of representative mutants with Y1R (top row) or Y2R (bottom row) (net BRET signal) clearly saturates with increased Venus-arr-3 in all cases (Fig. 5B). The mutants shown are grouped according to their binding to the Y2R. The binding of WT arr-3 and Ala87Val mutant was nearly identical. Asp51Gly and Leu69Ala had virtually no effect, whereas Cys252Ser-Pro253Val, Gln256Tyr, and Asp260Lys demonstrated a modest but significant reduction in binding. Only two mutations had dramatic effect on Y2R interaction: charge neutralization Asp241Asn reduced net BRETMAX with Y2R by about half, whereas Tyr239Thr reduced it by more than eighty percent (Fig. 5B). Interestingly, none of these mutations significantly reduced Y1R binding, in agreement with modest effect of the NCA mutation (Fig. 3). Five out of ten mutants affected arr-3 binding to Y2R (Fig. 5C). In contrast to WT arr-3 that binds both receptors comparably, arr-3-Tyr239Thr showed a more than five-fold preference for Y1R over Y2R (Fig. 5D).
Fig. 5. Mutations in arr-3 differentially affect its binding to Y1R and Y2R.
A. BRET ratio as a function of Venus-arrestin fluorescence normalized by receptor-RLuc luminescence (F/L) in the presence of agonist (filled symbols; solid lines) or vehicle (open symbols; dashed lines) for the base mutant Ala87Val (A87V, circles), Ala87V-Tyr239Thr (Y239T, triangles). B. Net BRET (agonist-induced increase in BRET signal) vs F/L. Means ± SEM of six repeats in a representative experiment (out of 3–15 performed) for indicated forms arr-3 and receptors are shown. C. Net BRETMAX for the indicated mutant-receptor combinations determined by a fit of the data shown in B to one-site binding hyperbola using GraphPad Prism 6.03. Mean BRETMAX ± SEM is shown (n=3–15). D. Ratio of Y1R/Y2R binding of indicated arr-3 mutants (the ratio for arr-3-Ala87Val base mutant was set at 1). Statistical significance was determined using one-way ANOVA followed by Dunnett's multiple comparison test. **, p < 0.01; ***, p < 0.001; ****, p<0.0001 as compared to WT arr-3. ##, p < 0.01; ###, p < 0.001; ####, p < 0.001; as compared to the Ala87Val mutant.
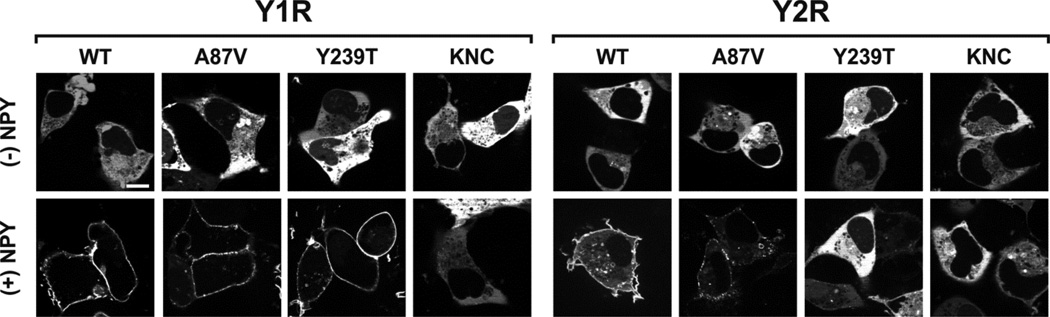
To test these findings by an independent method, we visualized the recruitment of Venus-tagged arrestins to Y1R and Y2R in COS-7 cells using live-cell imaging (Fig. 6). In the absence of agonist all forms of arr-3 were evenly distributed in the cytoplasm and virtually undetectable in the nuclei, in agreement with reported presence of functional nuclear export signal in arr-3 (38,39,51). In full agreement with BRET results (Figs. 3,5), WT arr-3 and Ala87Val base mutant were readily recruited by both receptors to the plasma membrane, whereas the distribution of KNC did not change. Arr-3-Tyr239Thr was recruited by Y1R like WT arr-3, whereas the activation of Y2R failed to change its subcellular distribution (Fig. 6). Thus, two independent lines of evidence show that in contrast to promiscuous WT arr-3 and its Ala87Val version, arr-3-Tyr239Thr has lost the ability to interact with Y2R, while retaining normal Y1R binding (Figs. 5,6).
Fig. 6. Live cell imaging of arrestin recruitment to Y1R and Y2R.
COS-7 cells were co-transfected with indicated forms of Venus-arr-3 and Y1R or Y2R. Arrestin distribution was visualized prior to (upper panels) or after 15 min of stimulation with 1 µM NPY (lower panels). Representative images from at least two independent experiments are shown. Scale bar = 10 µM.
3.4. Arr-3 mutations differentially affect pre-docking and agonist-induced increase in binding to Y1R and Y2R
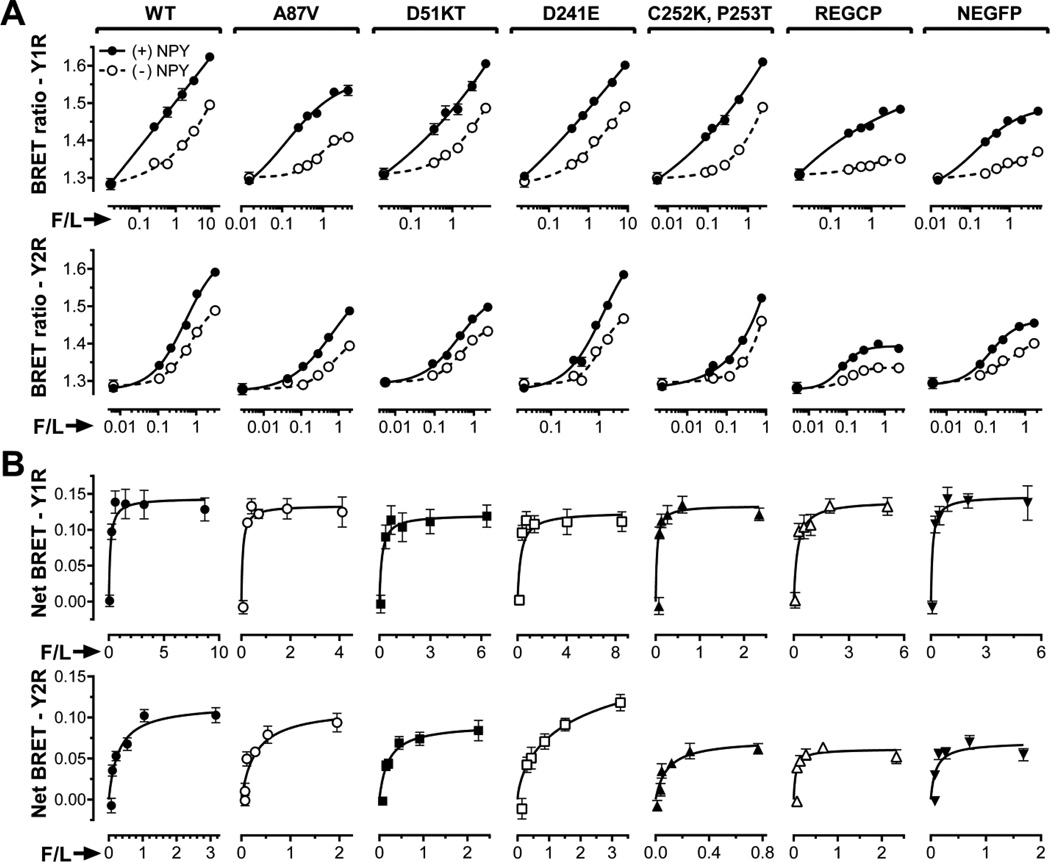
To further probe the molecular mechanisms underlying arrestin-GPCR interactions, we generated five additional mutants of arr-3-Ala87Val, targeting the same positions replaced by alanines in NCA, while introducing elements present in several evolutionary early arrestins (Fig. 4)(52). Similar to the first ten substitutions (Fig. 5), none of these new mutations significantly affected agonist-induced increase in Y1R binding reflected by net BRET (Fig. 7B). In contrast, Cys252Lys-Pro253Thr, REGCP and NEGFP reduced net BRETMAX observed with Y2R by 40–60%, whereas Asp51delinsLys (D51KT in Fig. 7) reduced it by ~15%. These data are in line with the observation that, in contrast to Y1R, Y2R interaction is sensitive to substitutions in these positions (Fig. 5).
Fig. 7. Mutations in arr-3 differentially affect pre-docking and agonist-induced receptor binding.
A. BRET ratio as a function of Venus-arr-3 fluorescence normalized by receptor-RLuc8 luminescence (F/L) in the presence of agonist (filled circles; solid lines) or vehicle (open circles; dashed lines) in COS-7 cells. B. Net BRET (agonist-induced increase in BRET signal) vs F/L. Means ± SEM of six repeats in a representative experiment (out of 3 performed) for indicated forms arr-3 and receptors are shown in both panels.
However, REGCP and NEGFP significantly reduced agonist-independent arr-3 interaction with Y1R (Fig. 7A, top row), almost to the bystander level observed with KNC (Fig. 3). Interestingly, the effects of REGCP and NEGCP mutations on agonist-independent arr-3 pre-docking to Y2R were more modest, even though these substitutions significantly reduced agonist-mediated increase in the binding (Fig. 7A,B). Mutation Tyr239Thr, which also greatly reduces agonist-dependent arr-3 binding to Y2R, does not affect pre-docking in the absence of agonist (Fig. 5A). Previously we observed a remarkable correlation between the effects of various arr-3 substitutions on basal and agonist-induced binding to β2AR, M2, D1, and D2 receptors (Fig. 5 in (30)). REGCP and NEGCP are the first mutations that show differential effects on pre-docking and agonist-induced arr-3 binding.
4. Discussion
True arrestins specifically binding active phosphorylated GPCRs were so far described in metazoans (52–54). Vertebrates express four arrestin subtypes, two of which (arr-1 and arr-4) are highly specialized and function in the visual system. The remaining two non-visual arrestins (arr-2 and arr-3) interact with hundreds of receptors belonging to the GPCR superfamily (3). Although all arrestins preferentially bind their cognate receptors in the active phosphorylated state, relative contribution of receptor-attached phosphates and non-phosphorylated receptor elements varies (27,29). The phosphates are the common theme, and their action via conserved phosphate sensing mechanism (50,55–57) to promote arrestin “activation” (58) explains how very few arrestins can “serve” hundreds of structurally different GPCRs. Significant contribution to the interaction of non-phosphorylated parts of the receptor (27,30,59), which are subtype-specific, makes the existence of receptor-specific arrestins possible. Evolution created only one of those: arrestin-1, which has high preference for rhodopsin over other GPCRs (60,61). We recently found that relatively few residues on an extensive receptor-binding surface of arrestins confer this specificity (28), and demonstrated that certain substitutions of these residues in naturally very promiscuous arr-3 can generate mutants with high preference for some GPCRs over others (30).
Here we tested the possibility of generating versions of arr-3 specific for particular NPY receptors, which play critical role in the regulation of food intake, mood disorders, and cancer (17,18). To assess the feasibility of manipulating this multireceptor-multiligand system using subtype-specific arrestins, we used ten previously characterized and five new arr-3 variants with substitutions of earlier identified key receptor-discriminator residues (28,30). We focused on Y1R and Y2R, which bind the same ligand but often play antagonistic roles in physiology (9,19,21), using two experimental approaches, BRET and arrestin recruitment assays in live cells. It has been shown that Y1R remains in complex with arrestins even after endocytosis, co-localizing with them in late endocytic vesicles (42). In contrast, arrestin is recruited to the plasma membrane after agonist stimulation of Y2R, but apparently dissociates early, as it does not co-localize with Y2R in late endocytic vesicles (24).
The residues that determine receptor preference (30) are fairly variable, even though they reside in regions with high degree of conservation across different arrestin subtypes from different species, from C. elegans to humans (52,53). The residues in bovine arr-3 here were substituted by residues found in homologous positions in other arrestins (30). For example, Asp51Gly, Leu69Ala, and Asp241Asn “borrowed” residues from zebrafish arr-1, whereas the double mutant Cys252Ser, Pro253Val was based on Kurtz from Drosophila (Fig. 4B (30)). The advantage of this approach is that resulting proteins are likely to fold properly, because they mimic existing arrestins that were apparently “approved” by evolution (52).
First, we assessed the effects of two global perturbations K2A and NCA, and their combination KNC (Fig. 2). K2A eliminates two conserved lysines (Fig. 2), which are critical for the binding of receptor-attached phosphates (50). This mutation is highly detrimental for arrestin-receptor interactions where phosphorylation plays critical role (27,50). In NCA mutant all ten known receptor-discriminator residues are replaced with alanines (Fig. 2), which blocks arrestin binding to many receptors (28). KNC, which is a combination of K2A and NCA mutations, generates an arrestin totally devoid of ability to bind GPCRs (48), which provides a perfect negative control. We found that NCA greatly reduced arr-3 pre-docking to inactive Y1R, but agonist-elicited binding of arr-3-NCA to Y1R was only ~30% lower than that of WT arr-3 (Fig. 3), which is the highest binding level observed for this mutant amongst all receptors tested so far. In contrast, NCA mutation virtually eliminated arr-3 binding to Y2R, both basal and agonist-induced (Fig. 3).
In agreement with preservation of agonist-induced arr-3-NCA binding to Y1R, none of the point mutants that significantly reduced arr-3 binding to other receptors (30) affected NPY-elicited increase in BRET signal (Fig. 5B). In contrast, Y2R binding was reduced by five out of the initial ten mutations, with Tyr239Thr showing the greatest effect, ~7-fold reduction. As a result, arr-3-Tyr239Thr demonstrated greater than 5-fold preference for Y1R over Y2R (Fig. 5D).
Although several experimental controls were used to ensure that BRET results were not artifacts, we used a different method to independently confirm our findings. To this end, we used agonist-induced arrestin recruitment to the receptor in plasma membrane in live cells (Fig. 6), a method that was validated with many GPCRs (62), including Y1R and Y2R (24,63–65) In agreement with BRET data, both WT arr-3 and Ala87Val version were readily recruited to Y1R and Y2R upon NPY stimulation in COS-7 cells, whereas arr-3-Tyr239Thr was only recruited to Y1R (Fig. 6). Receptor binding-impaired arr-3-KNC was not recruited by either receptor. Thus, two lines of evidence independently demonstrated high preference of arr-3-Tyr239Thr for Y1R over Y2R (Figs. 5,6).
Various experimental approaches used by many labs established that receptor-binding surface covers the concave sides of both arrestin domains (reviewed in (3,8)). The cavity of the N-domain of arr-1 is lined by the phosphate-binding positive charges of Lys14, Lys15, Arg171, and Arg175 (50,66,67). The two highly conserved lysines on the β-strand I, Lys14 and Lys15 in arr-1, corresponding to Lys11 and Lys12 in arr-3 (Fig. 2), are fully exposed in the basal conformation in all arrestins (34–37,50,67), and appear bind the receptor-attached phosphates first and then deliver them to the main phosphate sensor, the polar core (50) localized on the inter-domain interface (55). Alanine substitution of Lys14 and Lys15 (K2A) in arr-1 virtually eliminates its binding to phosphorylated rhodopsin (50). This is not necessarily the case for non-visual arrestins, where interaction assays in intact cells with β2AR, M2 muscarinic, or D2 dopamine receptors showed that agonist-elicited arrestin-receptor BRET was unaffected by K2A mutations in the corresponding positions of arr-2 (Lys10, Lys11) and arr-3 (Lys11 and Lys12) (27).
Similarly, NPY induced recruitment of arr-3 by Y1R or Y2R was not affected by the K2A mutations (Fig. 3B,C). However, when combined with NCA, K2A completely eliminated arr-3 binding to both receptors, even to Y1R, where NCA mutation per se was less detrimental (Fig. 3B,C). Importantly, NCA effectively blocked agonist-independent pre-docking to both receptors, whereas K2A mutation did not (Fig. 3A). These data suggest that pre-docking does not involve the phosphates, which are attached by GRKs only to active GPCRs (reviewed in (26)). The finding that K2A mutation in the context of NCA has much stronger effect than in WT arr-3 suggests that although non-visual arrestins can rely on the interactions with non-phosphorylated receptor elements, in their absence the phosphates can drive agonist-induced binding, at least in case of Y1R (Fig. 3). An alternative interpretation is that both components play a role, and other arr-3 residues that remain intact in NCA mutant partially mediate its binding to Y1R, but not to Y2R.
An apparent increase of the extent of pre-docking of the K2A mutant, as indicated by higher than WT BRET ratios in the absence of agonist (Fig. 3A), suggests that the two lysines eliminated by the K2A mutation weaken arr-3 interactions with unphosphorylated receptors. It is tempting to speculate that arrestins with these lysines in place are repulsed by the positive charges on the cytoplasmic surface of Y1R and Y2R, which would serve to enhance their preference for the phosphorylated GPCRs. Extensive mutagenesis of the intracellular loops of these receptors and/or the crystal structure of their complexes with arr-3 are necessary to test this idea.
In addition to ten mutants previously tested with other GPCRs (30), we introduced five new arr-3 with substitutions located in both N-and C-domains (Fig. 4B). None of these 15 mutations had an appreciable effect on the agonist-induced arr-3 binding to the Y1R (Figs. 5,7). In contrast, arr-3 interaction with Y2R was much more sensitive: significant reductions in net BRETMAX were observed with eight out of fifteen mutants, including three new versions, Cys252Lys-Pro253Thr, REGCP, and NEGFP (Figs. 5,7).
In contrast to previous observations with β2AR, M2 muscarinic, or D2 dopamine receptors (30), several mutations differentially affected agonist-dependent and independent binding to Y1R (Figs. 3,7). In particular, in contrast to WT arr-3 and all the other mutants, NCA (Fig. 3), as well as REGCP and NEGCP (Fig. 7A) showed virtually no interaction with Y1R in the absence of agonist. This lack of pre-docking is similar to that observed with arr-3-KNC (Fig. 3A), whereas the Tyr239Thr mutant with severely impaired agonist-induced binding to Y2R still pre-docks normally (Fig. 5A,B). Importantly, agonist-induced increase in the interaction with Y1R of NCA, REGCP and NEGCP mutants was not significantly different from that of WT arr-3 (Figs. 3,7). While NCA prevented both agonist-independent and dependent binding of arr-3 to Y2R (Fig. 3), REGCP and NEGCP were much more detrimental for pre-docking than for Y2R binding in response to the agonist (Fig. 7). This is the first case where arrestin pre-docking to the receptor was separated from agonist-induced increase in binding. These data suggest that these two modes of the arrestin-receptor interaction are mechanistically distinct.
The physiological role of the arrestin pre-docking to GPCRs remains to be elucidated. Arr-3 mutants where this function is selectively disrupted without detrimental effect on the agonist-induced binding provide the first viable tools to investigate the biological significance of pre-docking. Our ability to eliminate pre-docking while retaining agonist-induced interactions opens new venues in arrestin engineering with the ultimate goal of avoiding possible side effects produced by more drastic interventions.
GPCRs are targeted by more clinically used drugs than any other protein family (1,2). GPCRs transduce their signals via two types of adaptors, G proteins and arrestins (3,68). There is no doubt that recent advances in developing GPCR agonists biased towards one of these pathways will significantly expand their therapeutic applications (69,70) Targeted redesign of signaling proteins holds even greater promise for channeling cell signaling in the desired direction, overcoming unavoidable limitations of small molecules (71–73). Here we showed that reengineering of the receptor-binding surface of non-visual arrestins can not only narrow down their receptor specificity, but introduce even subtler functional changes, differentially affecting basal pre-docking and agonist-induced binding to the same receptor.
5. Conclusions
Here we described two mutants of the most promiscuous non-visual subtype, arr-3, that show high preference for Y1R over Y2R. One of these, Tyr239Thr, pre-docks normally, whereas the other, NCA, demonstrates virtually exclusively agonist-induced binding. Moreover, two other mutants, REGCP and NEGCP, completely lost the ability to pre-dock to Y1R, while retaining perfectly normal agonist-induced binding. These newly described mutants expand the arsenal of molecular tools for the study of the interactions of different GPCRs with non-visual arrestins.
Highlights.
Receptor specificity of arrestins can be enhanced by targeted mutagenesis
Arrestins selective for neuropeptide Y1 over Y2 receptor were developed
Mutations differentially affect agonist-dependent and –independent recruitment
Receptor-specific non-visual arrestins are novel tools for research and therapy
Acknowledgments
We thank Mr. Denis Hüvel for help in making some of the arrestin-3 mutants. We are also grateful to Dr. J. A. Javitch (Columbia University, New York) for providing the plasmid encoding Venus and Dr. N. A. Lambert (Medical College of Georgia, Augusta, GA) for the plasmid encoding Renilla luciferase variant 8. This work was supported by NIH grants GM077561, GM081756, and EY011500 (VVG) and by the DFG (SFB1052/A3), the European Union and the Free State of Saxony ESF (AGB-S). SB is grateful for a fellowship of the Fond der Chemischen Industrie.
Abbreviations
- GPCR
G protein-coupled receptor
- arr
arrestin
- NPY
neuropeptide Y
- Y1R
neuropeptide Y1 receptor
- Y2R
neuropeptide Y2 receptor
- BRET
bioluminescence resonance energy transfer
- ANOVA
analysis of variance
- arr-3-K2A
arrestin-3-Lys11Ala, Lys12Ala
- arr-3-NCA
arrestin-3 Leu49Ala, Asp51Ala, Arg52Ala, Leu69Ala, Tyr239Ala, Asp241Ala, Cys252Ala, Pro253Ala, Asp260Ala, and Gln262Ala
- arr-3-KNC
arrestin-3 Lys11Ala, Lys12Ala, Leu49Ala, Asp51Ala, Arg52Ala, Leu69Ala, Tyr239Ala, Asp241Ala, Cys252Ala, Pro253Ala, Asp260Ala, and Gln262Ala
- REGCP
arrestin-3 Asp260-Gln262delinsArg, Glu, Gly, Cys, Pro
- NEGFP
arrestin-3 Asp260-Gln262delinsAsn, Glu, Gly, Phe, Pro
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
We use systematic names of arrestin proteins: arrestin-1 (historic names S-antigen, 48-kDa protein, visual or rod arrestin), arrestin-2 (β-arrestin or β-arrestin1), arrestin-3 (β-arrestin2 or hTHY-ARRX), and arrestin-4 (cone or X-arrestin; for unclear reasons, its gene is called “arrestin 3” in the HUGO database).
Author contribution
LEG, SB, and LW contributed new reagents, performed experiments, and analyzed data. LEG, SB, AGB-S and VVG designed the study and wrote the manuscript.
Conflict of interests
The authors declare no conflict of interest.
References
- 1.Bridges TM, Lindsley CW. G-protein-coupled receptors: from classical modes of modulation to allosteric mechanisms. ACS Chem Biol. 2008;3:530–541. doi: 10.1021/cb800116f. [DOI] [PubMed] [Google Scholar]
- 2.Lagerstrom MC, Schioth HB. Structural diversity of G protein-coupled receptors and significance for drug discovery. Nat Rev Drug Discov. 2008;7:339–357. doi: 10.1038/nrd2518. [DOI] [PubMed] [Google Scholar]
- 3.Gurevich VV, Gurevich EV. The structural basis of arrestin-mediated regulation of G-protein-coupled receptors. Pharmacol Ther. 2006;110:465–502. doi: 10.1016/j.pharmthera.2005.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shenoy SK, Lefkowitz RJ. β-Arrestin-mediated receptor trafficking and signal transduction. Trends Pharmacol Sci. 2011;32:521–533. doi: 10.1016/j.tips.2011.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Song X, Vishnivetskiy SA, Seo J, Chen J, Gurevich EV, Gurevich VV. Arrestin-1 expression level in rods: balancing functional performance and photoreceptor health. Neuroscience. 2011;174:37–49. doi: 10.1016/j.neuroscience.2010.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhu X, Brown B, Li A, Mears AJ, Swaroop A, Craft CM. GRK1-dependent phosphorylation of S and M opsins and their binding to cone arrestin during cone phototransduction in the mouse retina. J Neurosci. 2003;23:6152–6160. doi: 10.1523/JNEUROSCI.23-14-06152.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gimenez LE, Vishnivetskiy SA, Gurevich VV. Targeting Individual GPCRs with Redesigned Nonvisual Arrestins. Handb Exp Pharmacol. 2014;219:153–170. doi: 10.1007/978-3-642-41199-1_8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gurevich VV, Gurevich EV. Structural determinants of arrestin functions. Prog Mol Biol Transl Sci. 2013;118:57–92. doi: 10.1016/B978-0-12-394440-5.00003-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Babilon S, Morl K, Beck-Sickinger AG. Towards improved receptor targeting: anterograde transport, internalization and postendocytic trafficking of neuropeptide Y receptors. Biol Chem. 2013;394:921–936. doi: 10.1515/hsz-2013-0123. [DOI] [PubMed] [Google Scholar]
- 10.Larhammar D, Salaneck E. Molecular evolution of NPY receptor subtypes. Neuropeptides. 2004;38:141–151. doi: 10.1016/j.npep.2004.06.002. [DOI] [PubMed] [Google Scholar]
- 11.Michel MC, Beck-Sickinger A, Cox H, Doods HN, Herzog H, Larhammar D, Quirion R, Schwartz T, Westfall T. XVI. International Union of Pharmacology recommendations for the nomenclature of neuropeptide Y, peptide YY, and pancreatic polypeptide receptors. Pharmacol Rev. 1998;50:143–150. [PubMed] [Google Scholar]
- 12.Ferrier L, Segain JP, Bonnet C, Cherbut C, Lehur PA, Jarry A, Galmiche JP, Blottiere HM. Functional mapping of NPY/PYY receptors in rat and human gastro-intestinal tract. Peptides. 2002;23:1765–1771. doi: 10.1016/s0196-9781(02)00133-x. [DOI] [PubMed] [Google Scholar]
- 13.Rettenbacher M, Reubi JC. Localization and characterization of neuropeptide receptors in human colon. Naunyn Schmiedebergs Arch Pharmacol. 2001;364:291–304. doi: 10.1007/s002100100454. [DOI] [PubMed] [Google Scholar]
- 14.Abounader R, Elhusseiny A, Cohen Z, Olivier A, Stanimirovic D, Quirion R, Hamel E. Expression of neuropeptide Y receptors mRNA and protein in human brain vessels and cerebromicrovascular cells in culture. J Cereb Blood Flow Metab. 1999;19:155–163. doi: 10.1097/00004647-199902000-00007. [DOI] [PubMed] [Google Scholar]
- 15.Dumont Y, Jacques D, Bouchard P, Quirion R. Species differences in the expression and distribution of the neuropeptide Y Y1, Y2, Y4, and Y5 receptors in rodents, guinea pig, and primates brains. J Comp Neurol. 1998;402:372–384. [PubMed] [Google Scholar]
- 16.Gullestad L, Aass H, Ross H, Ueland T, Geiran O, Kjekshus J, Simonsen S, Fowler M, Kobilka B. Neuropeptide Y receptor 1 (NPY-Y1) expression in human heart failure and heart transplantation. J Auton Nerv Syst. 1998;70:84–91. doi: 10.1016/s0165-1838(98)00032-0. [DOI] [PubMed] [Google Scholar]
- 17.Pedragosa-Badia X, Stichel J, Beck-Sickinger AG. Neuropeptide Y receptors: how to get subtype selectivity. Front Endocrinol (Lausanne) 2013;4:5. doi: 10.3389/fendo.2013.00005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Michel MC, Beck-Sickinger AG, Colmers WF, Cox HM, Doods HN, Herzog H, Larhammar D, Quirion R, Schwartz T, Westfall T. Neuropeptide Y receptors. IUPHAR database. 2013 Last modified on: 12-03-2013. http://www.iuphardb.org/DATABASE/FamilyMenuForward?familyId=46. [PubMed] [Google Scholar]
- 19.Zhang L, Bijker MS, Herzog H. The neuropeptide Y system: pathophysiological and therapeutic implications in obesity and cancer. Pharmacol Ther. 2011;131:91–113. doi: 10.1016/j.pharmthera.2011.03.011. [DOI] [PubMed] [Google Scholar]
- 20.Morales-Medina JC, Dumont Y, Quirion R. A possible role of neuropeptide Y in depression and stress. Brain Res. 2010;1314:194–205. doi: 10.1016/j.brainres.2009.09.077. [DOI] [PubMed] [Google Scholar]
- 21.Morales-Medina JC, Dumont Y, Benoit CE, Bastianetto S, Flores G, Fournier A, Quirion R. Role of neuropeptide Y Y(1) and Y(2) receptors on behavioral despair in a rat model of depression with co-morbid anxiety. Neuropharmacology. 2012;62:200–208. doi: 10.1016/j.neuropharm.2011.06.030. [DOI] [PubMed] [Google Scholar]
- 22.Lin S, Boey D, Herzog H. NPY and Y receptors: lessons from transgenic and knockout models. Neuropeptides. 2004;38:189–200. doi: 10.1016/j.npep.2004.05.005. [DOI] [PubMed] [Google Scholar]
- 23.Kilpatrick LE, Briddon SJ, Hill SJ, Holliday ND. Quantitative analysis of neuropeptide Y receptor association with beta-arrestin2 measured by bimolecular fluorescence complementation. Br J Pharmacol. 2010;160:892–906. doi: 10.1111/j.1476-5381.2010.00676.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Walther C, Nagel S, Gimenez LE, Morl K, Gurevich VV, Beck-Sickinger AG. Ligand-induced internalization and recycling of the human neuropeptide Y2 receptor is regulated by its carboxyl-terminal tail. J Biol Chem. 2010;285:41578–41590. doi: 10.1074/jbc.M110.162156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Walther C, Morl K, Beck-Sickinger AG. Neuropeptide Y receptors: ligand binding and trafficking suggest novel approaches in drug development. J Pept Sci. 2011;17:233–246. doi: 10.1002/psc.1357. [DOI] [PubMed] [Google Scholar]
- 26.Gurevich EV, Tesmer JJ, Mushegian A, Gurevich VV. G protein-coupled receptor kinases: more than just kinases and not only for GPCRs. Pharmacol Ther. 2012;133:40–69. doi: 10.1016/j.pharmthera.2011.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gimenez LE, Kook S, Vishnivetskiy SA, Ahmed MR, Gurevich EV, Gurevich VV. Role of receptor-attached phosphates in binding of visual and non-visual arrestins to G protein-coupled receptors. J Biol Chem. 2012;287:9028–9040. doi: 10.1074/jbc.M111.311803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vishnivetskiy SA, Gimenez LE, Francis DJ, Hanson SM, Hubbell WL, Klug CS, Gurevich VV. Few residues within an extensive binding interface drive receptor interaction and determine the specificity of arrestin proteins. J Biol Chem. 2011;286:24288–24299. doi: 10.1074/jbc.M110.213835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vilardaga JP, Krasel C, Chauvin S, Bambino T, Lohse MJ, Nissenson RA. Internalization determinants of the parathyroid hormone receptor differentially regulate β-arrestin/receptor asociation. J Biol Chem. 2002;277:8121–8129. doi: 10.1074/jbc.M110433200. [DOI] [PubMed] [Google Scholar]
- 30.Gimenez LE, Vishnivetskiy SA, Baameur F, Gurevich VV. Manipulation of very few receptor discriminator residues greatly enhances receptor specificity of non-visual arrestins. J Biol Chem. 2012;287:29495–29505. doi: 10.1074/jbc.M112.366674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Beck-Sickinger AG, Wieland HA, Wittneben H, Willim KD, Rudolf K, Jung G. Complete L-alanine scan of neuropeptide Y reveals ligands binding to Y1 and Y2 receptors with distinguished conformations. Eur J Biochem. 1994;225:947–958. doi: 10.1111/j.1432-1033.1994.0947b.x. [DOI] [PubMed] [Google Scholar]
- 32.Attramadal H, Arriza JL, Aoki C, Dawson TM, Codina J, Kwatra MM, Snyder SH, Caron MG, Lefkowitz RJ. β-arrestin2, a novel member of the arrestin/bet-aarrestin gene family. J Biol Chem. 1992;267:17882–17890. [PubMed] [Google Scholar]
- 33.Sterne-Marr R, Gurevich VV, Goldsmith P, Bodine RC, Sanders C, Donoso LA, Benovic JL. Polypeptide variants of β-arrestin and arrestin3. J Biol Chem. 1993;268:15640–15648. [PubMed] [Google Scholar]
- 34.Hirsch JA, Schubert C, Gurevich VV, Sigler PB. The 2.8 A crystal structure of visual arrestin: a model for arrestin's regulation. Cell. 1999;97:257–269. doi: 10.1016/s0092-8674(00)80735-7. [DOI] [PubMed] [Google Scholar]
- 35.Sutton RB, Vishnivetskiy SA, Robert J, Hanson SM, Raman D, Knox BE, Kono M, Navarro J, Gurevich VV. Crystal structure of cone arrestin at 2.3A: evolution of receptor specificity. J Mol Biol. 2005;354:1069–1080. doi: 10.1016/j.jmb.2005.10.023. [DOI] [PubMed] [Google Scholar]
- 36.Han M, Gurevich VV, Vishnivetskiy SA, Sigler PB, Schubert C. Crystal structure of β-arrestin at 1.9 A: possible mechanism of receptor binding and membrane Translocation. Structure. 2001;9:869–880. doi: 10.1016/s0969-2126(01)00644-x. [DOI] [PubMed] [Google Scholar]
- 37.Zhan X, Gimenez LE, Gurevich VV, Spiller BW. Crystal structure of arrestin-3 reveals the basis of the difference in receptor binding between two non-visual subtypes. J Mol Biol. 2011;406:467–478. doi: 10.1016/j.jmb.2010.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang P, Wu Y, Ge X, Ma L, Pei G. Subcellular localization of β-arrestins is determined by their intact N domain and the nuclear export signal at the C terminus. J Biol Chem. 2003;278:11648–11653. doi: 10.1074/jbc.M208109200. [DOI] [PubMed] [Google Scholar]
- 39.Scott MG, Le Rouzic E, Perianin A, Pierotti V, Enslen H, Benichou S, Marullo S, Benmerah A. Differential nucleocytoplasmic shuttling of β-arrestins. Characterization of a leucine-rich nuclear export signal in β-arrestin2. J Biol Chem. 2002;277:37693–37701. doi: 10.1074/jbc.M207552200. [DOI] [PubMed] [Google Scholar]
- 40.Nagai T, Ibata K, Park ES, Kubota M, Mikoshiba K, Miyawaki A. A variant of yellow fluorescent protein with fast and efficient maturation for cell-biological applications. Nat Biotechnol. 2002;20:87–90. doi: 10.1038/nbt0102-87. [DOI] [PubMed] [Google Scholar]
- 41.Loening AM, Fenn TD, Wu AM, Gambhir SS. Consensus guided mutagenesis of Renilla luciferase yields enhanced stability and light output. Protein Eng Des Sel. 2006;19:391–400. doi: 10.1093/protein/gzl023. [DOI] [PubMed] [Google Scholar]
- 42.Hofmann S, Frank R, Hey-Hawkins E, Beck-Sickinger AG, Schmidt P. Manipulating Y receptor subtype activation of short neuropeptide Y analogs by introducing carbaboranes. Neuropeptides. 2013;47:59–66. doi: 10.1016/j.npep.2012.12.001. [DOI] [PubMed] [Google Scholar]
- 43.Nordsieck K, Pichert A, Samsonov SA, Thomas L, Berger C, Pisabarro MT, Huster D, Beck-Sickinger AG. Residue 75 of interleukin-8 is crucial for its interactions with glycosaminoglycans. Chembiochem. 2012;13:2558–2566. doi: 10.1002/cbic.201200467. [DOI] [PubMed] [Google Scholar]
- 44.Berridge MJ. Rapid accumulation of inositol trisphosphate reveals that agonists hydrolyse polyphosphoinositides instead of phosphatidylinositol. Biochem J. 1983;212:849–858. doi: 10.1042/bj2120849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Berridge MJ, Dawson RM, Downes CP, Heslop JP, Irvine RF. Changes in the levels of inositol phosphates after agonist-dependent hydrolysis of membrane phosphoinositides. Biochem J. 1983;212:473–482. doi: 10.1042/bj2120473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kocan M, Pfleger KD. Detection of GPCR/β-arrestin interactions in live cells using bioluminescence resonance energy transfer technology. Methods Mol Biol. 2009;552:305–317. doi: 10.1007/978-1-60327-317-6_22. [DOI] [PubMed] [Google Scholar]
- 47.Pfleger KD, Eidne KA. Illuminating insights into protein-protein interactions using bioluminescence resonance energy transfer (BRET) Nat Methods. 2006;3:165–174. doi: 10.1038/nmeth841. [DOI] [PubMed] [Google Scholar]
- 48.Breitman M, Kook S, Gimenez LE, Lizama BN, Palazzo MC, Gurevich EV, Gurevich VV. Silent scaffolds: inhibition of c-Jun N-terminal kinase 3 activity in cell by dominant-negative arrestin-3 mutant. J Biol Chem. 2012;287:19653–19664. doi: 10.1074/jbc.M112.358192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kostenis E. Is Gα16 the optimal tool for fishing ligands of orphan G-protein-coupled receptors? Trends Pharmacol Sci. 2001;22:560–564. doi: 10.1016/s0165-6147(00)01810-1. [DOI] [PubMed] [Google Scholar]
- 50.Vishnivetskiy SA, Schubert C, Climaco GC, Gurevich YV, Velez MG, Gurevich VV. An additional phosphate-binding element in arrestin molecule. Implications for the mechanism of arrestin activation. J Biol Chem. 2000;275:41049–41057. doi: 10.1074/jbc.M007159200. [DOI] [PubMed] [Google Scholar]
- 51.Song X, Raman D, Gurevich EV, Vishnivetskiy SA, Gurevich VV. Visual and both non-visual arrestins in their "inactive" conformation bind JNK3 and Mdm2 and relocalize them from the nucleus to the cytoplasm. J Biol Chem. 2006;281:21491–21499. doi: 10.1074/jbc.M603659200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gurevich EV, Gurevich VV. Arrestins: ubiquitous regulators of cellular signaling pathways. Genome Biol. 2006;7:236. doi: 10.1186/gb-2006-7-9-236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Alvarez CE. On the origins of arrestin and rhodopsin. BMC Evol Biol. 2008;8:222. doi: 10.1186/1471-2148-8-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Aubry L, Klein G. True arrestins and arrestin-fold proteins: a structure-based appraisal. Prog Mol Biol Transl Sci. 2013;118:21–56. doi: 10.1016/B978-0-12-394440-5.00002-4. [DOI] [PubMed] [Google Scholar]
- 55.Vishnivetskiy SA, Paz CL, Schubert C, Hirsch JA, Sigler PB, Gurevich VV. How does arrestin respond to the phosphorylated state of rhodopsin? J Biol Chem. 1999;274:11451–11454. doi: 10.1074/jbc.274.17.11451. [DOI] [PubMed] [Google Scholar]
- 56.Celver J, Vishnivetskiy SA, Chavkin C, Gurevich VV. Conservation of the phosphate-sensitive elements in the arrestin family of proteins. J Biol Chem. 2002;277:9043–9048. doi: 10.1074/jbc.M107400200. [DOI] [PubMed] [Google Scholar]
- 57.Kovoor A, Celver J, Abdryashitov RI, Chavkin C, Gurevich VV. Targeted construction of phosphorylation-independent β-arrestin mutants with constitutive activity in cells. J. Biol. Chem. 1999;274:6831–6834. doi: 10.1074/jbc.274.11.6831. [DOI] [PubMed] [Google Scholar]
- 58.Gurevich VV, Gurevich EV. The molecular acrobatics of arrestin activation. Trends Pharmacol Sci. 2004;25:105–111. doi: 10.1016/j.tips.2003.12.008. [DOI] [PubMed] [Google Scholar]
- 59.Gurevich VV, Benovic JL. Visual arrestin interaction with rhodopsin. Sequential multisite binding ensures strict selectivity toward light-activated phosphorylated rhodopsin. J Biol Chem. 1993;268:11628–11638. [PubMed] [Google Scholar]
- 60.Gurevich VV, Dion SB, Onorato JJ, Ptasienski J, Kim CM, Sterne-Marr R, Hosey MM, Benovic JL. Arrestin interaction with G protein-coupled receptors. Direct binding studies of wild type and mutant arrestins with rhodopsin, b2-adrenergic, and m2 muscarinic cholinergic receptors. J Biol Chem. 1995;270:720–731. doi: 10.1074/jbc.270.2.720. [DOI] [PubMed] [Google Scholar]
- 61.Vishnivetskiy SA, Hosey MM, Benovic JL, Gurevich VV. Mapping the arrestin-receptor interface. Structural elements responsible for receptor specificity of arrestin proteins. J Biol Chem. 2004;279:1262–1268. doi: 10.1074/jbc.M308834200. [DOI] [PubMed] [Google Scholar]
- 62.Barak LS, Ferguson SS, Zhang J, Caron MG. A β-arrestin/green fluorescent protein biosensor for detecting G protein-coupled receptor activation. J Biol Chem. 1997;272:27497–27500. doi: 10.1074/jbc.272.44.27497. [DOI] [PubMed] [Google Scholar]
- 63.Walther C, Lotze J, Beck-Sickinger AG, Morl K. The anterograde transport of the human neuropeptide Y2 receptor is regulated by a subtype specific mechanism mediated by the C-terminus. Neuropeptides. 2012;46:335–343. doi: 10.1016/j.npep.2012.08.011. [DOI] [PubMed] [Google Scholar]
- 64.Bohme I, Stichel J, Walther C, Morl K, Beck-Sickinger AG. Agonist induced receptor internalization of neuropeptide Y receptor subtypes depends on third intracellular loop and C-terminus. Cell Signal. 2008;20:1740–1749. doi: 10.1016/j.cellsig.2008.05.017. [DOI] [PubMed] [Google Scholar]
- 65.Lindner D, Walther C, Tennemann A, Beck-Sickinger AG. Functional role of the extracellular N-terminal domain of neuropeptide Y subfamily receptors in membrane integration and agonist-stimulated internalization. Cell Signal. 2009;21:61–68. doi: 10.1016/j.cellsig.2008.09.007. [DOI] [PubMed] [Google Scholar]
- 66.Gurevich VV, Benovic JL. Visual arrestin binding to rhodopsin. Diverse functional roles of positively charged residues within the phosphorylation-recognition region of arrestin. J Biol Chem. 1995;270:6010–6016. doi: 10.1074/jbc.270.11.6010. [DOI] [PubMed] [Google Scholar]
- 67.Hanson SM, Gurevich VV. The differential engagement of arrestin surface charges by the various functional forms of the receptor. J Biol Chem. 2006;281:3458–3462. doi: 10.1074/jbc.M512148200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.DeWire SM, Ahn S, Lefkowitz RJ, Shenoy SK. β-arrestins and cell signaling. Annu Rev Physiol. 2007;69:483–510. doi: 10.1146/annurev.physiol.69.022405.154749. [DOI] [PubMed] [Google Scholar]
- 69.Kenakin TP. Biased signalling and allosteric machines: new vistas and challenges for drug discovery. Br J Pharmacol. 2012;165:1659–1669. doi: 10.1111/j.1476-5381.2011.01749.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Reiter E, Ahn S, Shukla AK, Lefkowitz RJ. Molecular mechanism of β-arrestin-biased agonism at seven-transmembrane receptors. Annu Rev Pharmacol Toxicol. 2012;52:179–197. doi: 10.1146/annurev.pharmtox.010909.105800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gurevich VV, Gurevich EV. Synthetic biology with surgical precision: targeted reengineering of signaling proteins. Cell Signal. 2012;24:1899–1908. doi: 10.1016/j.cellsig.2012.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gurevich VV, Gurevich EV. Custom-designed proteins as novel therapeutic tools? The case of arrestins. Expert Rev Mol Med. 2010;12:e13. doi: 10.1017/S1462399410001444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gurevich EV, Gurevich VV. Therapeutic potential of small molecules and engineered proteins. Handb Exp Pharmacol. 2014;219:1–12. doi: 10.1007/978-3-642-41199-1_1. [DOI] [PMC free article] [PubMed] [Google Scholar]