Abstract
A major mechanism by which cancers escape control by the immune system is by blocking the differentiation of myeloid cells into dendritic cells (DCs), immunostimulatory cells that activate anti-tumor T cells. Tumor-dependent activation of signal transducer and activator of transcription 3 (STAT3) signaling in myeloid progenitor cells is thought to cause this block in their differentiation. In addition, a signaling pathway through protein kinase C βII (PKCβII) is essential for the differentiation of myeloid cells into DCs. Here, we found in humans and mice that breast cancer cells substantially decreased the abundance of PKCβII in myeloid progenitor cells through a mechanism involving the enhanced activation of STAT3 signaling by soluble, tumor-derived factors (TDFs). STAT3 bound to previously undescribed negative regulatory elements within the promoter of PRKCB, which encodes PKCβII. We also found a previously undescribed counter-regulatory mechanism through which the activity of PKCβII inhibited tumor-dependent STAT3 signaling by decreasing the abundance of cell-surface receptors, such as cytokine and growth factor receptors, that are activated by TDFs. Together, these data suggest that a previously unrecognized crosstalk mechanism between the STAT3 and PKCβII signaling pathways provides the molecular basis for the tumor-induced blockade in the differentiation of myeloid cells, and suggest that enhancing PKCβII activity may be a therapeutic strategy to alleviate cancer-mediated suppression of the immune system.
Introduction
In their role as professional antigen-presenting cells (APCs), conventional dendritic cells (DCs) play a central role in the induction and regulation of adaptive immune responses (1, 2). DCs are uniquely capable of presenting extracellular antigen bound to class I major histocompatibility complex (MHC) molecules through a process known as cross-presentation (1, 3). Loss of DCs prevents the cross-priming of T cells, which severely limits immune responses by CD8+ T cells, which are also known as cytotoxic T cells and which target tumor cells (3). These findings suggest that DCs play an essential role in the ability of the immune system to restrain the growth and spread of cancers (4), even in advanced disease. Indeed, studies in humans demonstrate that ongoing immune responses are an important check on tumor cell growth, even in established tumors (5, 6), and that the numbers of DCs in and around tumors is an important independent predictor of patient survival in a number of different cancers (5, 7). Furthermore, patients with genetic mutations that result in the loss of DCs (8) or that impair the function of so-called “danger receptors” on the surface of DCs (9) suffer from increased cancer incidence and early death, as well as experiencing reduced chemotherapeutic efficacy. Murine models have confirmed that intact, functional conventional DCs are required for the induction of local and systemic anti-tumor responses by CD8+T cell s (3).
Thus, the ability of cancers to progress is dependent on mechanisms that enable tumors to elude, disrupt, or suppress this immune control. One major mechanism involved is the tumor-induced blockade of the differentiation of myeloid cells into DCs. This phenomenon was first described in breast cancer patients (10), and has been widely reported in solid and hematologic malignancies (10, 11), as well as in a number of inflammatory and autoimmune conditions (11). It is now thought that this blockade in myeloid cell differentiation is one manifestation of a global disruption in normal myelopoiesis that also gives rise to the accumulation of actively immunosuppressive and pro-tumorigenic immature myeloid cells, collectively termed myeloid-derived suppressor cells (MDSCs) (10), whose presence correlates with poor overall survival in cancer patients (12). Thus, this tumor-mediated block in myeloid cell differentiation may compromise anti-tumor immunity both through the loss of mature immunostimulatory DCs (5, 7) and the accumulation of immunosuppressive MDSCs.
The signaling events that impair the generation of DCs in cancer patients are elicited by a large number of soluble, tumor-derived factors (TDFs), such as vascular endothelial growth factor (VEGF), interleukin-6 (IL-6), and granulocyte colony-stimulating factor (G-CSF), which accumulate to large amounts in the circulation, and are secreted either by the tumor itself or by stromal cells in the tumor microenvironment in response to tumor-derived signals (10, 13). The common signaling feature of these TDFs is their ability to strongly stimulate sustained activation of the signal transducer and activator of transcription 3 (STAT3) signaling pathway (10), which in turn impairs the differentiation of myeloid cells to DCs as well as DC function (14, 15).
Despite its central role in tumor-mediated immunosuppression, the molecular mechanisms by which enhanced STAT3 signaling impairs myeloid cell differentiation remain unknown, unlike the mechanisms by which it supports the survival and function of MDSCs, which have been characterized in a number of seminal studies (16–18). We previously found that signaling through the serine and threonine kinase PKCβII (protein kinase C βII) is essential for the differentiation of myeloid progenitor cells into DCs, and that subtle changes in the abundance or activity of PKCβII substantially alter the ability of the resulting DCs to activate T cells (19). This raises the possibility that decreased PKCβII protein abundance might be a potential mechanism of tumor-mediated blockade of DC generation; however, PRKCB (the gene that encodes PKCβI and PKCβII) has not been previously reported to be a STAT3 target gene.
PKCβII and PKCβI are splice variants of the PRKCB gene (20). They are fully activated by the second messengers diacylglycerol (DAG) and Ca2+, whereupon they translocate to the plasma membrane and are stabilized in an active conformation by scaffold proteins, which enables their full kinase activity (20). We and others reported that the activation of PKCβI or PKCβII specifically drives the differentiation of myeloid progenitor cells to DCs (19, 21), whereas pharmacologic inhibition of PKCβII or its spontaneous loss in human DC progenitor cell lines prevents their differentiation into DCs (19). These studies also demonstrated that PKCβII signaling positively autoregulated the PRKCB promoter, maintaining stable expression through the basal activity of PKCβII (19). Unexpectedly, however, there are myeloid progenitor cell lines that spontaneously lose PKCβII and the ability to undergo differentiation to DCs. For example, KG1 cells have readily detectable PKCβII protein, whereas the naturally arising daughter cell line KG1a does not (19, 22). These findings suggest the existence of undescribed mechanisms that inhibit the expression of PRKCB despite the positive feedback loop provided by the basal enzymatic activity of PKCβII. These observations led us to examine whether STAT3 signaling resulted in decreased PKCβII abundance, and whether this was the underlying mechanism by which tumors and TDFs blocked the differentiation of myeloid cells into DCs.
Here, we report that PKCβII abundance in myeloid cells is decreased in patients with advanced breast cancer and in tumor-bearing mice. In vitro experiments revealed that TDFs stimulated the enhanced activation of STAT3 in myeloid progenitor cells, and that STAT3 reduced the abundance of PKCβII protein and the expression of PRKCB2 by binding to previously undescribed negative regulatory elements in the PRKCB promoter. We also discovered a previously uncharacterized mechanism by which the activity of PKCβII limited the ability of TDFs to activate STAT3 signaling. This work identifies a regulatory network in which, on the one hand, STAT3 inhibits PRKCB2 expression, and on the other, PKCβII activity inhibits STAT3 activation.
Results
PKCβII abundance is decreased in myeloid cells from breast cancer patients and tumor-bearing mice
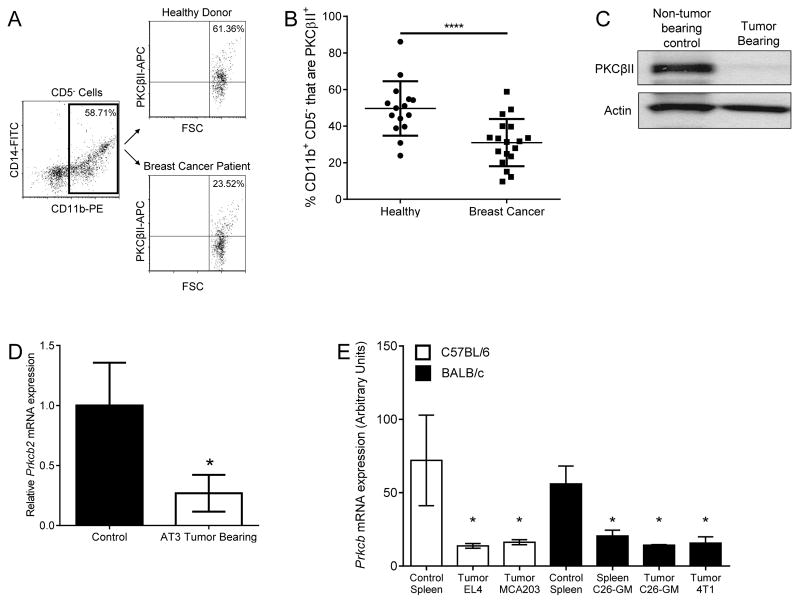
To determine whether PKCβII abundance in myeloid cells was reduced in the presence of cancer, we measured PKCβII amounts in peripheral blood myeloid cells [characterized as CD11b+CD5− (CD5 is a pan-lymphocyte marker)] from newly diagnosed patients with advanced breast cancer (table S1) and in purified splenic myeloid cells [the spleen being a major site of MDSC accumulation (10)] from tumor-free control mice or from mice bearing EL4 (thymoma) or AT3 (breast) tumors. We found that advanced breast cancer patients had significantly fewer PKCβII-containing CD11b+CD5− myeloid cells in the blood than did healthy donors (p = 0.041, Fig. 1, A and B). This was also seen in tumor-bearing mice, in which splenic myeloid cells isolated by Gr1-based positive selection from EL4 tumor–bearing mice had considerably decreased PKCβII protein abundance compared to that of non-tumor–bearing control mice (Fig. 1C). Purified CD11b+ splenic myeloid cells from AT3 tumor–bearing mice also had significantly less Prkcb2mRNA than did tumor-free control mice (p = 0.031, Fig. 1D).
Fig. 1. Decreased PKCβII abundance in myeloid cells in tumor-bearing hosts.
(A and B) Analysis of PKCβII abundance in peripheral blood myeloid cells from breast cancer patients and healthy donors. (A) Gating strategy: Myeloid cells were identified by gating on CD11b+CD5− PBMCs, and PKCβII+ cells were identified by intracellular flow cytometry. The positive population was defined in comparison to cells that were stained intracellularly with the APC-conjugated secondary antibody but without the primary antibody specific for PKCβII. (B) Percentages of CD11b+ cells identified as being PKCβII+. Data are from 15 healthy donors and 18 breast cancer patients and are presented as means ± SD. *P = 0.041 by the Mann-Whitney rank sum test. (C to E) Analysis of PKCβII abundance in myeloid cells from tumor-free and tumor-bearing mice. (C) Splenic Gr1+ myeloid cells were isolated from tumor-free mice and from mice bearing EL4 tumors and were analyzed by Western blotting with antibodies against the indicated proteins. (D) Splenic CD11b+ myeloid cells were isolated from tumor-free mice and from mice bearing AT3 tumors. The abundances of Prkcb2 and Actb mRNAs were determined by qPCR analysis. Data are means± SD from three mice of each genotype. P= 0.031 by Student’s T-test. (E) Prkcb expression was analyzed from gene expression profiles generated from CD11b+ cells isolated from the spleens or tumors of C57/BL6 mice or BALB/c mice bearing the indicated tumors, as described in (23) (GSE21927). Data are means ± SD from three mice for each condition. *P = 0.038 among the BALB/c conditions, and *P = 0.025 among the C57BL/6 conditions, as analyzed by one -way ANOVA on ranks and post-hoc test.
We further examined publically available gene expression profiles [NCBI GEO database accession GSE21927 (23)] of tumor-free mice and mice bearing a number of different tumors. In these studies, CD11b+ cells isolated from the spleens and tumors of mice bearing breast (4T1), thymoma (EL4), colon (C26-GM), and fibrosarcoma (MCA203) tumors had significantly reduced Prkcb mRNA abundance compared to that in CD11b+ splenocytes from healthy mice (p ≤ 0.038, Fig. 1E). These observations demonstrate the decreased abundance of PKCβII in myeloid cells in cancer patients and in mice bearing various solid and hematologic tumors. This reduction in PKCβII abundance would suggest a diminished capacity of these myeloid cells to undergo differentiation to DCs, because we have found that PKCβII protein abundance is a key determinant of the ability of progenitors to commit to and complete this differentiation process (19).
When we knocked down PKCβII in the myeloid progenitor cell line K562, which we previously used to study PKC activation during the differentiation of progenitor cells to DCs (24), we observed a significant decrease in the ability of these cells to stimulate the proliferation of T cells (p < 0.05), which is a hallmark of DC function (fig. S1, A and B). In addition, we found that knockdown of PKCβII in primary human monocytes undergoing cytokine-driven differentiation to DCs resulted in a marked reduction in the extent of activation of nuclear factor κB (NF-κB) and extracellular signal–regulated kinase (ERK) pathways (fig. S1C). Both of these signaling pathways are critical mediators of DC generation (25–28), and they are activated by PKCβII in other cell types (29–31). We previously demonstrated that PKCβII mediates the activation of non-canonical NF-κB signaling in DC generation (32). Because the abundance of the NF-κB family member RelB is itself transcriptionally regulated by other NF-κB family members (33), we used increases in its abundance as a read out for NF-κB activation, whereas we used phosphorylation of ERK1/2 as a readout for the activation of ERK (fig. S1C). Unstimulated monocytes had low PKCβII protein abundance, which was markedly increased by culture in the presence of GM-CSF and IL-4. The extent of this increase was substantially reduced in monocytes transfected with plasmid encoding enhanced green fluorescent protein (EGFP) and small interfering RNA (siRNA) specific for PKCβ (pEGFP-siPKCβ) compared to that in untransfected cells and in cells transfected with a plasmid encoding EGFP and a negative control siRNA (pEGFP-siEmpty) (fig. S1C). Both the NF-κB and ERK pathways were activated in either untransfected or control-transfected monocytes in response to GM-CSF and IL-4; however, the reduced abundance of PKCβII protein in the monocytes expressing siPKCβ was reflected in a four- to eight-fold decrease in the extent of activation of both downstream pathways. These observations suggest that PKCβII is necessary for the induction of NF-κB and ERK signaling during the cytokine-dependent differentiation of myeloid cells to DCs, and suggest the possibility that tumor-induced decreases in PKCβII abundance may inhibit differentiation to DCs by inhibiting downstream NF-κB and ERK signaling.
Tumor-secreted soluble factors suppress differentiation to DCs through reduced PKCβII abundance
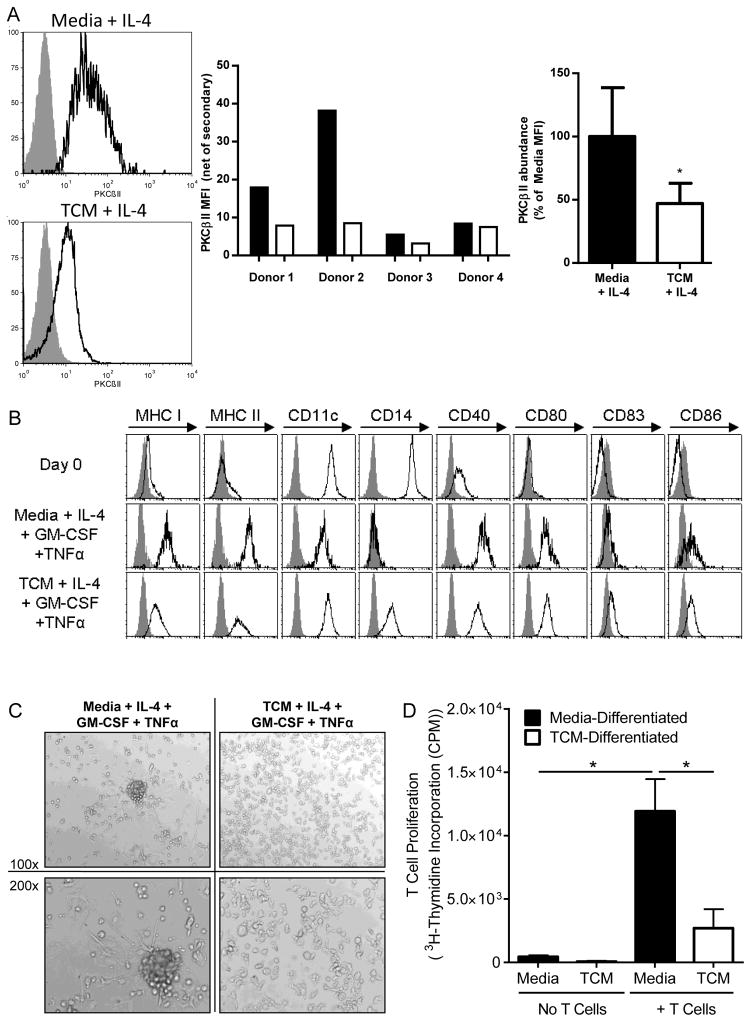
The populations of myeloid cells found in tumor-bearing individuals are likely to be qualitatively different from those found in tumor-free individuals. To distinguish between qualitative differences in cell populations versus cell-intrinsic changes in PKCβII abundance, we cultured myeloid DC progenitor cells in tumor-conditioned media (TCM) in vitro to directly test for cell-intrinsic effects. For these experiments, we used both normal human CD14+ monocytes and the KG1 cell line, both of which we previously used to model the differentiation of myeloid cells to DCs (19). KG1 cells are a human CD34+ myeloid leukemia progenitor-like cell line that undergo differentiation to DCs in response to cytokines, (including GM-CSF and tumor necrosis factor α (TNF-α), or phorbol ester [for example phorbol 12-myristate 13-acetate (PMA)], stimuli that activate PKCβII (19, 22). IL-4 was included in all monocyte cultures to suppress their differentiation to macrophages (34). Monocytes cultured in TCM for 7 days had significantly reduced amounts of PKCβII protein compared to that in cells grown in normal medium (p = 0.029, Fig. 2A). Monocytes cultured in DC-inducing cytokines (GM-CSF, IL-4, and TNF-α) for 11 additional days acquired cell-surface expression of DC markers, including MHC Class I, MHC Class II, CD11c, CD40, CD80, CD83, and CD86 (Fig. 2B), a characteristic DC morphology, based on clustering and long extended dendrites (Fig. 2C), and the ability to stimulate robust proliferation of T cells (Fig. 2D), a central functional characteristic of DCs. However, in the presence of TCM, the differentiating cells failed to acquire the classical DC morphology (Fig. 2C). These cells also had somewhat reduced cell-surface expression of MHC I, MHC II, and CD40 compared to that of cells cultured in control medium, and they had a substantially increased abundance of the monocyte marker CD14 (Fig. 2B). Furthermore, the ability of the TCM-treated cells to stimulate the proliferation of allogeneic T cells was likewise significantly reduced (p = 0.016, Fig. 2D). Together, these findings are consistent with the impaired differentiation of primary human monocytes into DCs. We observed similar results in experiments withKG1 cell s.
Fig. 2. TCM decreases PKCβII abundance.
(A to D) Monocytes from healthy donors were cultured with IL-4 and either control medium or TCM together with GM-CSF over the final 11 days and TNF-α over the final 2 days. (A) On day 7, monocytes were fixed and stained intracellularly for PKCβII and analyzed by flow cytometry. Left: Representative histograms of PKCβII staining. Gray histograms represent the APC-conjugated secondary antibody alone as a control, whereas the black line represents specific PKCβII staining. Middle: The mean fluorescence intensity (MFI) of PKCβII (less the MFI of the secondary antibody control) for each donor. Right: Combined results from the individual donors. Data are mean percentages of the PKCBII MFIs from four individual donors. *P = 0.029 by the Mann-Whitney rank sum test. (B) Monocytes and monocyte-derived DCs were incubated with antibodies specific for the indicated markers of DC differentiation on day 0 and on day 18 after incubation under the indicated conditions, and then were analyzed by flow cytometry. Gray histograms show the isotype control, whereas black lines indicate specific staining. Results are representative of three individual donors. (C) Monocytes and monocyte-derived DCs were examined under a light microscope on day 18 of culture. Images are representative of cells from three individual donors. (D) Monocytes and monocyte-derived DCs under the indicated culture conditions were tested for their ability to stimulate the proliferation of allogeneic T cells. Data are means ± SD from three individual monocyte donors. P = 0.016 by One-way ANOVA on ranks and post-hoc test for select comparisons.
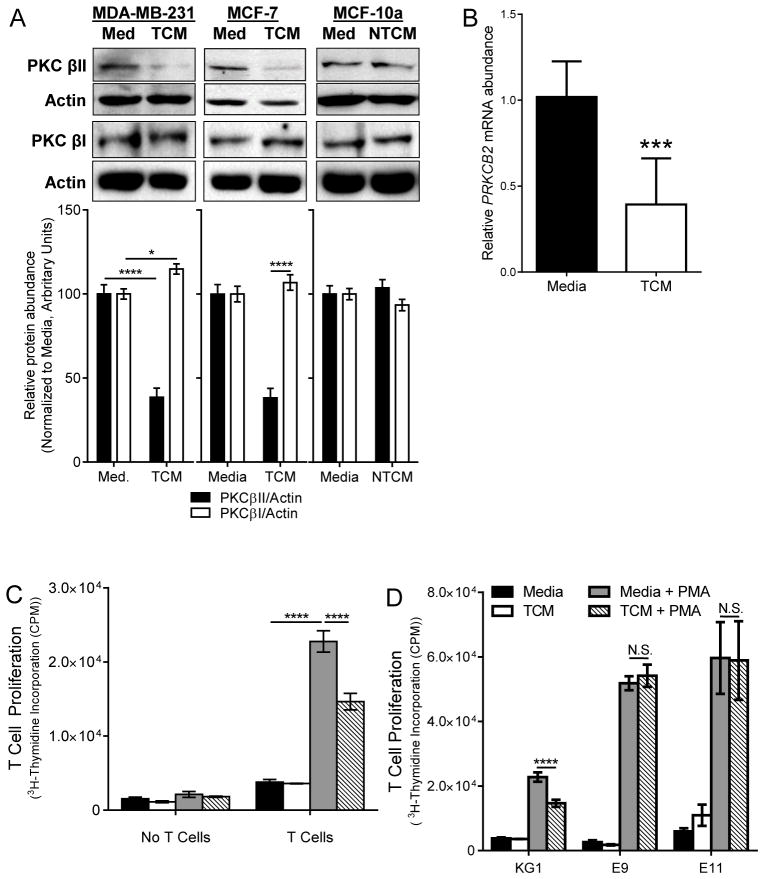
We cultured KG1 cells in TCM or in conditioned medium from non-tumorigenic, immortalized human mammary epithelial cells (non-tumor conditioned media, NTCM). We found that culture of KG1 cells in TCM significantly decreased the abundance of PKCβII protein compared to that in untreated KG1 cells (p < 0.001), whereas culture of the cells in NTCM had no effect (Fig. 3A). This finding is consistent with our observations from experiments with human monocytes. We observed small (although statistically significantly different, p = 0.026) increases in PKCβI abundance in the cells (compared with TCM from MDA-MB-231 cells, but not MCF-7 cells), but this did not offset the larger decrease in PKCβII protein abundance (Fig. 3A), suggesting that alterations in the alternative splicing to generate PKCβI or PKCβII isoforms did not play a major role in tumor-driven decreases in PKCβII abundance. Studies in other cell types have demonstrated that generation of PKCβI is favored over that of PKCβII (35), which may account for the preservation of the abundance of the former at the expense of the latter. Culture of KG1 cells in TCM likewise significantly reduced the abundance of PRKCB2 mRNA (p = 0.004, Fig. 3B) to a similar extent as that seen for the protein. KG1 cells treated with PMA for seven days undergo differentiation into DCs, which stimulated significant proliferation of allogeneic T cells (p < 0.001); however, this process was significantly reduced in the presence of TCM (p < 0.001, Fig. 3C), also similar to what we observed in experiments with primary human monocytes.
Fig. 3. TDFs suppress DC generation through reduced PKCβII abundance.
KG1 cells were cultured in complete medium (Med), TCM (derived from MCF-7 or MDA-MB-231 cells), or non-tumor conditioned medium (NTCM, derived from MCF10a cells) for 14 days, and for the last 7 days were cultured in the absence or presence of PMA. (A) Whole-cell lysates were collected on day 7 and were analyzed by Western blotting with antibodies against the indicated proteins. Top: Representative Western blots. Bottom: Densitometric analysis of combined Western blotting data. Data are means ± SEM from three (for PKCβI) or four (for PKCβII) independent experiments. ****P < 0.001 for MDA-MB-231 and MCF7 TCM PKCβII blots; *P = 0.026 for MDA-MB-231 PKCβI blots (Student’s T-test). (B) RNA was isolated on day 7 and PRKCB2 mRNA abundance was determined by qPCR analysis. Data are means ± SD from five independent experiments. ***P = 0.004 by student’s t test. (C) Cells were harvested on day 14, cultured alone or with T cells, and then examined for their ability to stimulate T cell proliferation based on [3H]thymidine incorporation. Data are means ± SD from three independent experiments. ****P < 0.001 by one-way ANOVA and post-hoc tests for select comparisons. (D) T cell stimulation by cells expressing exogenous PKCβII in the presence and absence of TCM. KG1 cells and the KG1a-PKCβII-GFP clones E9 and E11 were treated as described in (C) and were cultured alone or with T cells. Data are means ± SD from three independent experiments. ****P< 0.001 (Student’s T -test); N.S., not significant.
Because TDFs impaired progenitor cell differentiation to DCs by reducing PRKCB2 mRNA and PKCβII protein abundances, we reasoned that enforced expression of PKCβII should prevent impaired differentiation. To examine this, we used KG1 cells and the KG1a-PKCβII-GFP cell line, which was stably transfected with a plasmid encoding the CMV promoter–driven PRKCB2 (19). PKCβII in these cells was thus resistant to negative regulation by TCM. We cultured these cells in normal medium or in TCM with or without PMA, and then assessed their ability to stimulate the proliferation of allogeneic T cells as a measure of DC differentiation. Undifferentiated KG1 or KG1a-PKCβII-GFP cells induced minimal T cell proliferation in all cases, whereas PMA-differentiated cells induced substantial T cell proliferation (Fig. 3D). PMA-differentiated KG1 cells cultured in TCM induced significantly less T cell proliferation than did KG1 cells cultured in control medium (p < 0.001). In contrast, the extent of T cell proliferation induced by PMA-treated differentiated KG1a-PKCβII-GFP cell clones E9 and E11 was unaffected by the presence of TCM (Fig. 3D). These findings suggest that TDFs decrease the abundance of PKCβII, which impairs the ability of myeloid progenitor cells to undergo differentiation into DCs.
STAT3 activation is associated with decreased PKCβII abundance
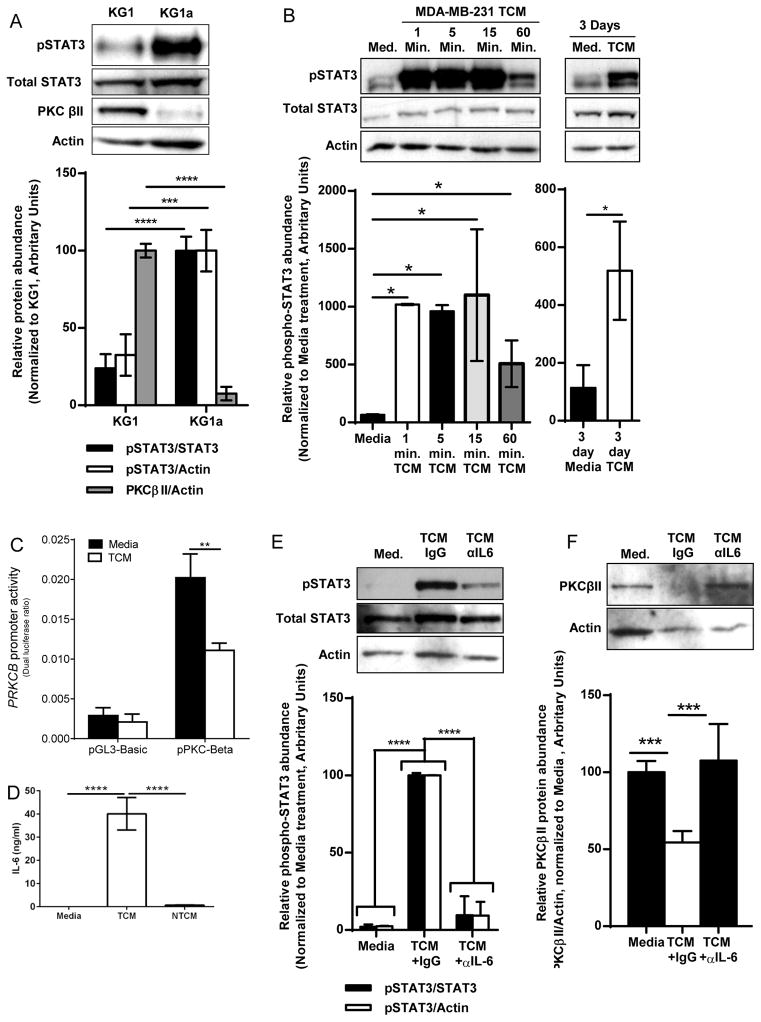
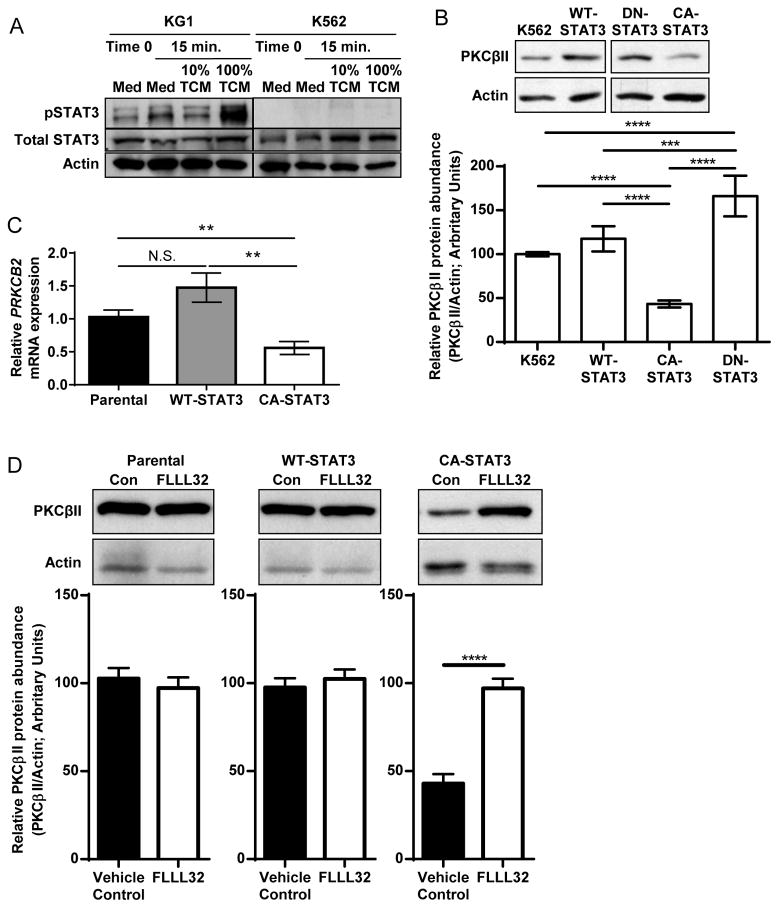
Because enhanced activation of STAT3 is a major mechanism by which TDFs inhibit the differentiation of myeloid cells into DCs (10), we next examined whether this process was occurring in our system. We began by comparing baseline STAT3 activity in KG1 cells (PKCβII+) and KG1a cells (PKCβII−), a spontaneously arising daughter cell line of KG1 cells that does not undergo differentiation into DCs because of lost expression of PRKCB2 (19, 22). Compared to the minimal amount of STAT3 activation that we observed in KG1 cells, as assessed by detection of phosphorylation of tyrosine 705, KG1a cells exhibited significantly increased STAT3 activity (p < 0.005, Fig. 4A). This negatively correlated with the pattern of PRKCB2 expression, suggesting an inverse relationship between PRKCB2 expression and basal STAT3 activity. It is unclear why KG1a cells have constitutive STAT3 activity, though it is possible that they secrete an autocrine factor that activates STAT3 or that they have an as-yet uncharacterized mutation in a cytokine receptor that results in constitutive STAT3 activity. Although KG1 cells lacked basal STAT3 activity, STAT3 phosphorylation was significantly and rapidly increased in response to TCM (p = 0.016, Fig. 4B). Although the extent of STAT3 activation peaked within 15 min, the extent of STAT3 phosphorylation was significantly greater in TCM-stimulated cells than in unstimulated cells as late as 3 days after exposure (p = 0.020, Fig. 4B), demonstrating that TCM stimulated sustained STAT3 activation.
Fig. 4. STAT3 signaling inhibits PRKCB expression.
(A) STAT3 activity in cells that do or do not have PKCβII. Whole-cell lysates of KG1 and KG1a cells were analyzed by Western blotting with antibodies against the indicated proteins. Top: Representative Western blots. Bottom: Densitometric analysis of combined Western blots. Data are means ± SD from three independent experiments. ***P< 0.005, **** P< 0.001, by Student’s t-test. (B) Kinetics of STAT3 activation by TCM. KG1 cells were cultured in complete medium or TCM, and whole-cell lysates were collected at the indicated times and analyzed by Western blotting with antibodies against the indicated proteins. Top: Representative Western blots. Bottom: Densitometric analysis of combined Western blots. Data are means ± SD from three independent experiments. Left: *P = 0.016 by one-way ANOVA on ranks and post-hoc test. Right: *P = 0.020 by Student’s T-test. (C) Analysis of PRKCB promoter activity. KG1 cells were transfected with a PRKCB promoter–based reporter construct (19) or with pGL3-basic (as a vector control) and then were cultured in the presence or absence of TCM for 24 hours before luciferase activity was determined (which was normalized to Renilla luciferase activity). Data are means ± SD from three independent experiments. **P = 0.0073 by student’s t-test. (D) Breast tumor cell lines produce IL-6. The abundances of IL-6 in fresh medium, TCM, or NTCM were determined by ELISA. Data are means ± SD from three independent experiments. ****P < 0.001 by one-way ANOVA. (E and F) Effect of IL-6 neutralization in TCM on STAT3 activation and PKCβII abundance. KG1 cells were cultured in medium or TCM (from MDA-MB-231 cells) that had been treated with either an IL-6–neutralizing antibody or an IgG isotype control antibody. Whole-cell lysates were collected after (E) 15 min or (F) 7 days and were analyzed by Western blotting. Top: Representative Western blots. Bottom: Densitometric analysis of combined Western blots. Data are means ± SD from three independent experiments. ****P < 0.001 by one-way ANOVA on ranks and post-hoc test (for E), and ***P < 0.005 by one-way ANOVA and post-hoc test (for F).
Although typically thought of as an inducer of gene transcription, STAT3 represses the transcription of certain genes, such as that encoding glucose-6-phosphatase (36). Because TCM both induced considerable STAT3 activity and significantly decreased PRKCB2 mRNA abundance (p = 0.004), and because STAT3 acts as a transcriptional repressor, we examined its effect on PRKCB promoter activity. KG1 cells transfected with a PRKCB promoter-based reporter construct exhibited a significant decrease in PRKCB promoter activity when they were cultured with TCM rather than with control medium (p = 0.0073, Fig. 4C). The TCM-induced reduction in promoter activity was similar in magnitude to the reduced abundance of PRKCB2 mRNA and PKCβII protein, suggesting that decreased transcriptional activity is the primary mechanism underlying decreased PKCβII protein abundance.
TCM contains many different TDFs that could potentially activate STAT3 and might alter PRKCB2 expression. To more precisely characterize this, we tried to determine the specific factor within the TCM that reduced PKCβII protein abundance. We found that conditioned medium from MDA-MB-231 cells contained 40.1 ± 7.0 ng/ml IL-6 (Fig. 4D), and that pretreatment of TCM with an IL-6–neutralizing antibody largely abrogated the TCM-dependent activation of STAT3 (Fig. 4E) and decrease in PKCβII protein abundance in KG1 cells (Fig. 4F). Increased serum concentrations of IL-6 correlate with poor prognosis in patients with breast cancer (37), as well as with impaired DC generation and increased MDSC accumulation in other malignancies (38). This may result from IL-6–mediated decreases in PKCβII abundance in DC progenitors. In addition to IL-6, we also examined G-CSF, which is a STAT3-activating TDF (13). We first examined additional gene expression profiles in the NCBI GEO database accession GSE21927 (23) that we interrogated earlier (Fig. 1E). We generated gene expression profiles from bone marrow cells freshly isolated from BALB/c and then treated in vitro with GM-CSF and G-CSF, and we found that Prkcb2 mRNA abundance was significantly reduced in cells treated with G -CSF and GM-CSF compared to that in control cells (p < 0.005, fig. S2).
To test the effect of TDF depletion in vivo, we returned to the AT3 tumor model, because AT3 tumors secrete abundant amounts of G-CSF (13). Mice bearing AT3 tumors were given daily injections for 8 days of a G-CSF–neutralizing antibody after tumors became palpable. Ten days later, spleens were harvested from the sacrificed mice. When we examined CD11b+ splenic myeloid cells taken from AT3 tumor-bearing mice, we found that Prkcb2 expression was increased in cells from mice that received the G-CSF–neutralizing antibody compared to that in cells from mice that received the isotype control antibody, although this difference did not reach statistical significance (fig. S3). This finding suggests that G-CSF plays a role in decreasing Prkcb2 expression in vivo, and we suspect that the effect of neutralization of G-CSF might have been more profound at time points closer to the completion of antibody administration or if other TDFs had been concurrently neutralized.
Activation of PKCβII inhibits the TDF-dependent activation of STAT3
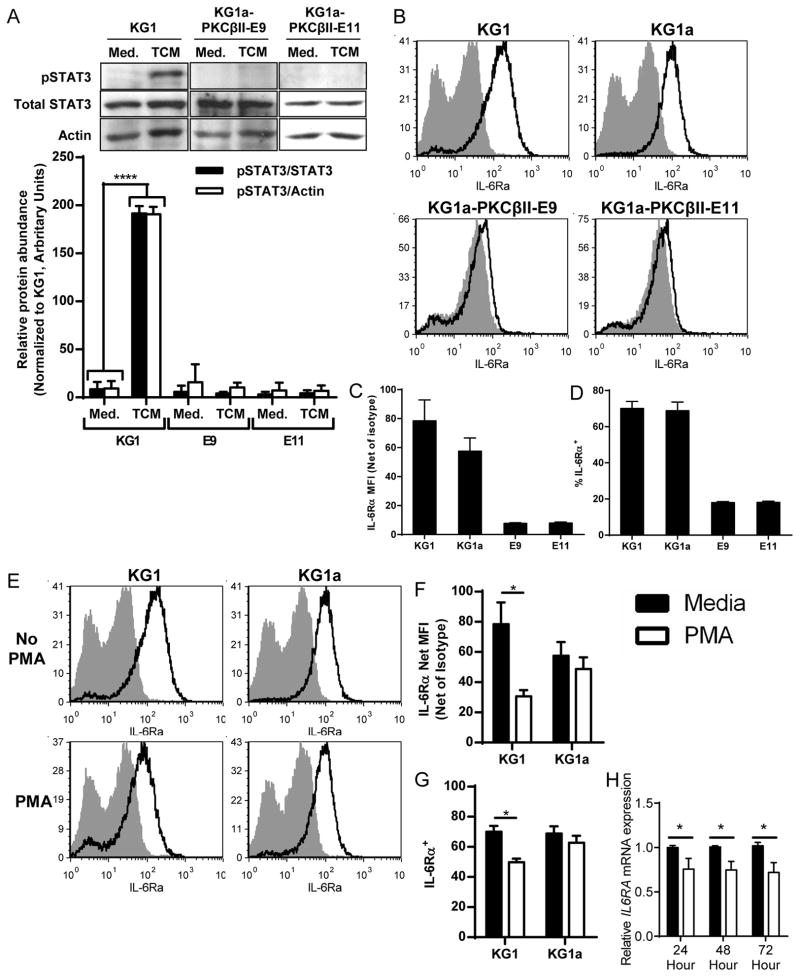
When we cultured the KG1a-PKCβII-GFP cell clones E9 and E11 in TCM to assess the effect of increased PKCβII abundance, we unexpectedly found that TCM failed to activate STAT3 in these cells (Fig. 5A). Gene expression profiling revealed that the E9 and E11 cell lines had substantially decreased abundance of the receptors for IL-6, VEGF, and G-CSF (Table 1), factors that activate STAT3 and impair the differentiation of myeloid cells to DCs (10, 13). Although both cell lines expressed transcripts for all three receptors, KG1 and KG1a cells had only IL-6Rα present on their surface, whereas the E9 and E11 cell clones had none of these receptors on their surfaces (Fig. 5, B to D and fig. S4, A to C). Although KG1 cells express PKCβII, the abundance of PKCβII in the E9 and E11 clones was increased, and thus they exhibited more basal PKCβII activity than was observed in KG1 cells, although the amount of activity was not sufficient to spontaneously induce their differentiation into DCs (19). This finding was suggestive of a previously unrecognized ability for PKCβII activity to prevent STAT3 signaling by reducing the cell-surface abundances of receptors that promote STAT3 activation. We tested this possibility directly by treating KG1 and KG1a cells with PMA to specifically activate PKC. This treatment significantly reduced the cell-surface abundance of IL-6Rα on KG1 cells (p ≤ 0.034), but not KG1a cells, indicating that this response specifically involved PKCβII (Fig. 5, E to G). We also found that PMA significantly decreased the abundance of IL-6RA mRNA (p ≤ 0.026), as well as those of CSF3R (which encodes G-CSFR, p < 0.05) and KDR (which encodes VEGFR2, p < 0.001) (Fig. 5H and fig. S4, D to E). These findings suggest the existence of counter-regulatory mechanisms by which PKCβII activity antagonizes STAT3 signaling and STAT3 activity represses PRKCB expression.
Fig. 5. PKCβII activity impairs the TCM-stimulated activation of STAT3.
(A) KG1 cells or the KG1a-PKCβII-GFP clones E9 and E11 were treated with CM or with TCM from MDA-MB-231 cells for 10 min and then were analyzed by Western blotting with antibodies against the indicated proteins. Top: Representative Western blot. Bottom: Densitometric analysis of combined Western blots. Data are means ± SD from four independent experiments. ****P = < 0.001 by student’s T-test. (B to D) KG1 and KG1a cells express IL-6Rα, whereas cell lines over-expressing PKCβII do not. The presence of IL-6Rα on the surface of KG1 and KG1a cells and of the KG1a-PKCβII-GFP clones E9 and E11 was analyzed by flow cytometry. (B) Representative histograms. Gray histograms show the isotype control, whereas the black lines show specific IL-6Rα staining. (C) The MFIs of IL-6Rα (less the MFIs of isotype antibody staining) for the indicated cells are shown. Data are means ± SEM from three independent experiments. (D) Percentages of viable cells that have cell-surface expression of IL-6Rα, as determined relative to staining with isotype control antibody. Data are means ± SEM from three independent experiments. (E to G) PKC activation decreases the cell-surface abundance of IL-6Rα on KG1 cells, but not KG1a cells. KG1 and KG1a cells were cultured in complete medium in the presence or absence of PMA for 24 hours, and the abundance of IL-6Rα on the cell surface was assessed by flow cytometric analysis. (E) Representative histograms are shown. Gray histograms show the isotype control, whereas black lines indicate specific IL-6Rα staining. (F) The MFIs of IL-6Rα (less the MFIs of isotype antibody staining) on the indicated cells cultured for 24 hours in the presence or absence of PMA. Data are means ± SEM from three independent experiments. *P = 0.034 by student’s T-test. (G) Percentages of viable cells that expressed cell-surface IL-6Rα, as determined relative to staining with the isotype control antibody. Data are means ± SEM from three independent experiments. *P = 0.011 by student’s t-test. (H) PKC activation decreases IL6RA mRNA abundance. KG1 cells were treated with CM in the presence or absence of PMA for up to 72 hours. Total RNA was then collected from the cells at the indicated times, and gene expression was analyzed by qPCR. Data are means ± SD from three independent experiments. P = 0.026 (24 hours), P = 0.01 (48 hours), and P = 0.009 (72 hours) (all determined by student’s t-test).
Table 1. Gene expression profiles of KG1 cells, KG1a cells, and the KG1a-PKCβII-GFP cell clones E9 and E11.
RNA was isolated from KG1 cells (which express PKCβII), KG1a cells (which do not express PKCβII), and the KG1a-PKCβII-GFP clones E9 and E11, and gene expression profiles were generated with Affymetrix microarrays. The raw signal intensity (Signal) returned from the microarray is expressed in arbitrary units. The expression status (Status) of the gene (present or absent) is returned from an analysis of the microarrays.
| KG1 | KG1a | KG1a-PKCBII Clone E9 | KG1a-PKCBII Clone E11 | |||||
|---|---|---|---|---|---|---|---|---|
| Signal (Arbitrary Units) | Status | Signal (Arbitrary Units) | Status | Signa (Arbitrary Units) | Status | Signal (Arbitrary Units) | Status | |
| CSF3R | 4348.3 | Present | 654.5 | Present | 114.7 | Absent | 210.9 | Absent |
| IL6RA | 3648.5 | Present | 4059.7 | Present | 47.7 | Absent | 16.5 | Absent |
| KDR | 137.2 | Present | 524.0 | Present | 5.0 | Absent | 3.4 | Absent |
STAT3 activation directly inhibits the expression of PRKCB2
To test the direct effect of STAT3 activation on PRKCB2 expression in the absence of any other signals from the TCM, we generated cell clones stably expressing wild-type STAT3, dominant negative STAT3 (DN-STAT3), or constitutively active STAT3 (CA-STAT3) in the human myeloid progenitor cell line K562 (24). We chose the K562 model because we previously demonstrated that K562 cells undergo differentiation into DCs in response to PKC activation (24), and that K562 cells had relatively less endogenous STAT3 activity compared to that of KG1 cells, and that they exhibited minimal activation of STAT3 in response to TCM (Fig. 6A). Whereas expression of wild-type STAT3 had no effect on the abundance of PKCβII protein in K562 cells, blockade of basal STAT3 signaling by the DN-STAT3 mutant significantly increased PKCβII abundance (p ≤ 0.005, Fig. 6B). Conversely, K562 clones expressing the CA-STAT3 mutant had significantly reduced abundances of PKCβII protein (p < 0.001, Fig. 6B) and PRKCB2 mRNA compared to those of untransfected K562 cells or of clones expressing wild-type STAT3 (Fig. 6C). This decrease in PKCβII abundance was dependent on continued STAT3 signaling, because the pharmacologic inhibitor of STAT3 phosphorylation FLLL-32 (39) led to a two-fold increase in PKCβII protein abundance in clones expressing the CA-STAT3 mutant, whereas it had no effect on untransfected cells or on cells expressing wild-type STAT3 (Fig. 6D).
Fig. 6. STAT3 activity inhibits the expression of PRKCB2.
(A) Comparison of STAT3 activation in KG1 and K562 cells in response to TCM from MDA-MB-231 cells. KG1 and K562 cells were cultured in CM or the indicated concentrations of TCM from MDA-MB-231 cells for 15 min, and whole-cell lysates were then analyzed by Western blotting with antibodies against the indicated proteins. Blots are representative of three independent experiments. (B to D) Constitutive STAT3 activity decreases the abundance of PKCβII. (B) The relative amounts of PKCβII and actin from parental K562 cells and from cell clones stably expressing exogenous WT-STAT3, DN-STAT3, or CA-STAT3 were quantified by Western blotting analysis. Top: Representative Western blots. Bottom: Densitometric analysis of combined Western blots. Data are means ± SD from four independent experiments. ****P = < 0.001 for all pair-wise comparisons except for DN-STAT3 vs. WT-STAT3 (***P = 0.005) and K562 vs. WT-STAT3 (P = 0.139). Statistical analysis was performed by one-way ANOVA and post-hoc test. (C) PRKCB2 mRNA abundance. Total RNA was isolated from parental K562 cells and from a panel of cell clones stably expressing exogenous WT-STAT3 or CA-STAT3, and qPCR analysis was performed to determine the relative abundance of PRKCB2 mRNA normalized to that of ACTB mRNA. Independent sample numbers were as follows: n = 6 (parental), 11 (WT-STAT3), and 16 (CA-STAT3). Data are means ± SEM. P < 0.01 by one-way ANVOA on ranks and post-hoc test for all-pairwise comparisons. (D) Parental K562 cells or cells stably expressing exogenous WT-STAT3 or CA-STAT3 were cultured with the STAT3 inhibitor FLLL32 (5 μM) or DMSO (carrier control, 0.05%) for 24 hours and then were analyzed by Western blotting. Top: Representative Western blots. Bottom: Densitometric analysis of combined Western blots. Data are means± SD from three independent experiments. ****P< 0.001 by student’s t-test.
TDFs induce the binding of STAT3 to the PRKCB promoter, which reduces its activity
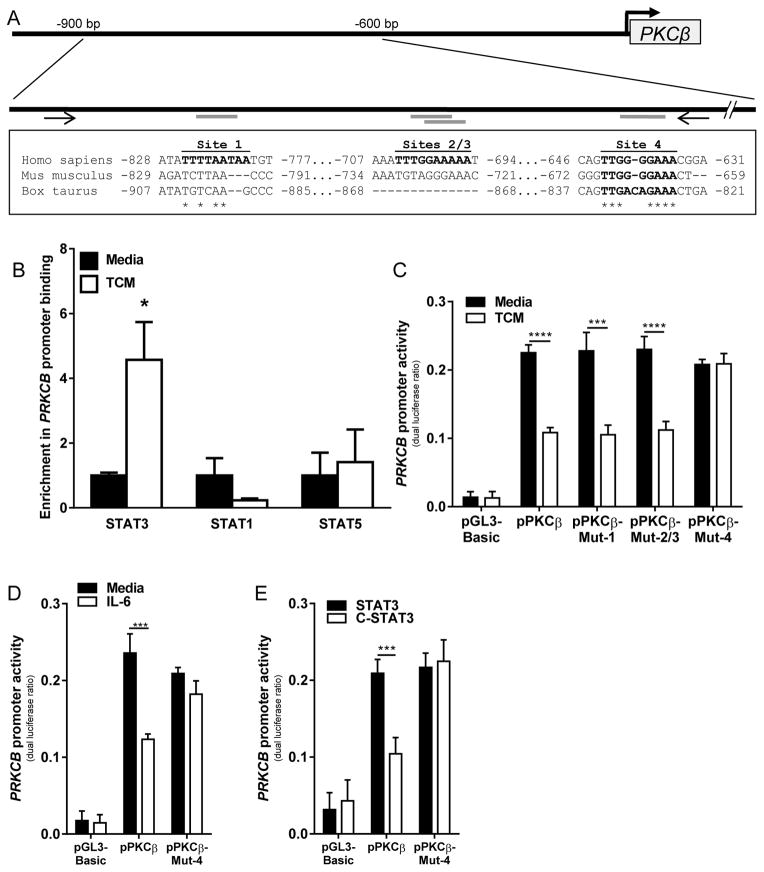
Although we found that the enhanced activation of STAT3 led to decreased PKCβII abundance, our data did not discriminate between whether STAT3 acted indirectly (for example, by altering the abundance of some other factor that is necessary for PRKCB2 expression) or bound directly to the PRKCB promoter to inhibit its activity. In silico analysis of the PRKCB promoter revealed four putative STAT3-binding sites (40–42) within a region 1000 bp upstream of the transcriptional start site and within 200 bp of each other (Fig. 7A). Further analysis revealed that of these sites, only site 4 was widely conserved among mammalian species (Fig. 7A). Chromatin immunoprecipitation (ChIP) analysis of these sites revealed that TCM significantly enhanced the binding of STAT3 to the PRKCB promoter (p = 0.029) without changing the extent of binding of either STAT1 or STAT5 (Fig. 7B). To determine whether the binding of STAT3 to the PRKCB promoter at any of these sites suppressed the expression of PRKCB2, we performed site-directed-mutagenesis of each site. We found that ablation of site 4 prevented the suppression of PRKCB promoter activity by TCM, whereas mutation of the other putative STAT3-binding sites had no effect (Fig. 7C). To further validate that the binding of STAT3 at site 4 decreased PRKCB promoter activity, we tested the activity of the PRKCB promoter mutated at site 4 in the presence of recombinant IL-6 (Fig. 7F) and the CA-STAT3 mutant (Fig. 7G). In both cases, the mutated promoter, but not the unmodified one, was unaffected by STAT3 activation. These observations reveal a previously unreported mechanism of PKCβII regulation, wherein activated STAT3 directly binds to the PRKCB promoter to repress its activity.
Fig. 7. TCM stimulates the binding of STAT3 to the PRKCB promoter.
(A) STAT3-binding sites in the PRKCB promoter. In silico analysis identified four putative STAT3-binding sites in the human PRKCB promoter, of which site 4 is conserved among mammalian species. Bolded letters indicate intact STAT3-binding motifs. Nucleotide positions are given relative to the transcription start site. An asterisk indicates sequence conservation. Arrows indicate primer-binding sites for ChIP assays. (B) KG1 cells were treated with or without TCM for 15 min and then were subjected to ChIP assays with the antibodies against the indicated targets or with isotype control antibody. Binding to the PRKCB promoter was assessed by qPCR analysis. Data are means ± SEM from three (for STAT1 and STAT5) or four (for STAT3) independent experiments. For STAT3, P = 0.029 by Mann-Whitney rank sum test, for STAT1, P = 0.227 by student’s t test, and for STAT5, P = 0.754 by student’s t test. (C to E) KG1 cells were transfected with pRL-CMV together with pGL3-basic, the unmodified PRKCB promoter reporter construct, or PRKCB promoter constructs in which site 1, sites 2 and 3, or site 4 were ablated (pPKCB-Mut-1, -Mut-2/3, and –Mut-4, respectively). Cells were cultured in the presence or absence of (C) TCM or (D) IL-6 (150 ng/ml), or (E) were co-transfected with the WT-STAT3 or CA-STAT3 constructs, and dual luciferase activity was assessed. Data are means ± SD from three independent experiments. ***P< 0.005, **** P< 0.001, by student’s t test.
Discussion
Ongoing immune responses continue to control tumor growth late into disease, and they are predictive of patient survival (5–7). Conversely, cancer induces a state of immunosuppression in patients, particularly with regard to anti-tumor immunity (10). Although there are many mechanisms by which tumors escape the immune system, the decreased number of conventional myeloid DCs, which is one manifestation of the global blockade in the differentiation of myeloid cells (10), is an important cause of tumor-mediated suppression of the immune system (10, 17). DCs are central components of the immune response, and their presence in the tumor microenvironment positively correlates with patient survival (5, 7). DCs are also necessary for the optimal efficacy of certain chemotherapeutics (9). Thus, the depletion of DCs that results from the impaired differentiation of myeloid cells would be expected to directly impair immune control of established tumors and new tumor variants, weaken immune responsiveness at potential metastatic sites, and compromise chemotherapeutic effectiveness. Thus, understanding the molecular basis behind the tumor-mediated inhibition of DC generation and preventing or reversing it are keys to improving the immune control of existing tumors.
Impaired generation of DCs in cancer is mediated by the enhanced activation of STAT3; however, the mechanism by which sustained STAT3 activation blocks the differentiation of myeloid cells remains unclear, particularly with regard to their differentiation into DCs. PKCβII signaling is activated by cytokines and phorbol esters, which drive the differentiation of myeloid cells, and its function in human myeloid progenitor cells is a prerequisite for their ability to undergo differentiation into DCs (19, 43). PKCβII activates downstream signaling pathways, including NF-κB and ERK, that play a critical role in differentiation into DCs (19), and studies are ongoing to fully elucidate this process. Basal PKCβII activation stimulates the expression of PRKCB through a stable positive feed-back loop (19), which raises the question of how PKCβII is spontaneously lost in cells, such as KG1a cells, and points to a previously undescribed negative regulatory signal. This led us to examine whether PKCβII was the target of the tumor-mediated enhanced activation of STAT3.
Here, we reported that myeloid cells from breast cancer patients and tumor-bearing mice have significantly decreased PKCβII abundance compared to that in normal controls (p ≤ 0.05). These observations are supported by our analysis of a larger, publicly available dataset (GSE21927) (23) of gene expression profiles of CD11b+ myeloid cells from tumor-free mice and two strains of mice bearing various tumors, in which the presence of a tumor led to a significant decrease in Prkcb abundance at the tumor site and in splenic myeloid cells (p ≤ 0.05). In vitro experiments with human monocytes and myeloid progenitor cell lines demonstrated that this decrease in PKCβII abundance occurred in a cell-intrinsic manner, and that it was the primary mechanism underlying the TCM-dependent impairment of the differentiation of myeloid cells into DCs. Thus, our observations suggest that tumor-dependent STAT3 signaling blocks the differentiation of myeloid cells into DCs by limiting the capacity for PKCβII to activate these downstream signals (including ERK and NF-κB signaling) in progenitor cells.
Our in vitro studies demonstrated that TDFs stimulated the sustained activation of STAT3, consistent with previously reported observations (10). This finding uncovers a previously uncharacterized mechanism of PKC regulation. Whereas PKC family members directly interact with or indirectly regulate STAT3 at the protein level (44), we have demonstrated the transcriptional control of PRKCB expression by STAT3. The PKC family is subject to tight and multi layered regulation at the protein level, including multiple required co-factors, phosphorylation steps for full activity, and differential subcellular localization (20). Moreover, the abundances of PKCβI and PKCβII are differentially regulated by alternative mRNA splicing (45); however, the transcriptional regulation of PKC isoforms has remained largely unexplored. Together, our data suggest a model in which TDFs stimulate the sustained binding of STAT3 to previously undescribed negative regulatory elements in the PRKCB promoter, impairing its transcriptional activity and blocking the differentiation of myeloid cells into DCs.
In the course of our experiments, we found that IL-6 produced by breast cancer cell lines decreased the abundance of PKCβII in KG1 myeloid progenitor–like cells. Normal and malignant breast epithelial cells produce IL-6 (46, 47), and the increased abundance of circulating IL-6 is a poor prognostic indicator in breast cancer patients (37). Our findings suggest that one potential mechanism by which IL-6 is detrimental to survival is to weaken anti-tumor immune responses by blocking the generation of DCs. IL-6 is not the only TDF that is linked to dysregulated myelopoiesis (10, 13, 17). Our model suggests that although the exact combination of TDFs may change from patient to patient, the salient feature of these TDFs is the resulting enhanced activation of STAT3, which leads to a reduction in PKCβII abundance and a blockade in differentiation to DCs. New therapies to stimulate an increase in PKCβII abundance or disrupt STAT3 signaling in myeloid progenitor cells might therefore be more effective than targeted cytokine depletion in alleviating the immunosuppressive tumor microenvironment (48).
In the course of this work, we unexpectedly found that PKCβII abundance or activation counter-regulated STAT3 signaling by inhibiting the cell-surface expression of receptors for the STAT3-activating cytokines IL-6, G-CSF, and VEGF. We observed these changes even when cells expressed the mRNAs for these receptors, without apparently making the protein. This previously unrecognized interaction suggests the existence of two negative regulatory networks that serve to positively reinforce one signal at the expense of the other: STAT3 signaling reduces PRKCB expression, which enables enhanced or sustained STAT3 signaling, whereas PKCβII signaling decreases the cell-surface expression of STAT3-activating receptors, limiting STAT3 signaling, and removing a check on PRKCB2 generation. This suggests that an increase in the abundance or activity of PKCβII might be used to disrupt the tumor-dependent increase in STAT3 activation, and thus enhance the differentiation of myeloid cells into DCs. The activation of PKCβII through cytokines, phorbol esters, or PKCβ peptide agonists (49) would induce further generation of PKCβII directly through its positive autoregulatory feedback loop (19), as well as indirectly, by reducing the ability of TDFs to transduce STAT3 signals in myeloid progenitor cells. This approach could synergize with existing attempts to develop clinically applicable pharmacologic inhibitors of STAT3. Further studies are required to determine the precise relationship between these two networks in the normal differentiation of myeloid cells into DCs.
Although the activation of PKCβII may disrupt the tumor-dependent suppression of the immune response, PKCβI and PKCβII also have pro-tumorigenic effects in epithelial tumors (50). The PKCβ inhibitor enzastaurin (50) effectively inhibits the growth of human breast cancer cell lines in vitro and in transplanted murine breast carcinomas in vivo (51). However, multiple phase II clinical trials showed that enzastaurin was ineffective in treating metastatic breast cancer alone or in combination with chemotherapeutics (52). One study found a marginal decrease in overall patient survival when enzastaurin was combined with capecitabine (52). Considering the importance of ongoing anti-tumor immune responses, this raises the possibility that any benefit derived from inhibiting PKCβ in tumor cells may be offset by the decreased generation of DCs because of impaired PKCβII activity in differentiating myeloid progenitor cells. While arguing for caution, this dichotomy also suggests that a therapy to inhibit PKCβ in tumor cells while selectively enhancing PKCβII activity in myeloid cells could yield clinical benefit.
Our findings do not directly address whether the STAT3-PKCβII mechanism leading to the blockade in DC generation also results in the accumulation of MDSCs, but they raise this possibility. The impaired differentiation of DC progenitors may directly contribute to the accumulation of the monocytic MDSC pool. At a more global level, it is possible that decreased PKCβII abundance may be a component underlying the broader dysregulation of myelopoiesis seen in cancer. In this scenario, TDF-induced loss of PKCβII in early myeloid progenitor cells inhibits their ability to fully differentiate, resulting in the accumulation of immature cells. Changes in PKCβII abundance may also alter the commitment of progenitors to specific lineages, increasing the proportion of cells that adopt the granulocytic lineage at the expense of monocytic, macrophage, or DC lineages.
In describing a molecular basis for tumor-dependent inhibition of DC progenitor differentiation, our work suggests new avenues of immune manipulation in the treatment of disease. We predict that increasing PKCβII protein abundance in myeloid progenitor cells, or preventing its initial decrease, would alleviate a substantial proportion of tumor-associated immune suppression, improving prognosis and prospects for immunotherapy. The activation of PKCβII within myeloid progenitor cells could serve the dual purpose of abrogating STAT3 signaling and enhancing PKCβII protein abundance, thereby enhancing the differentiation of myeloid cells into DCs.
Materials and Methods
Cells and culture
K562 cells (derived from a BCR-ABL+ human CML blast), KG1 cells (derived from a CD34+ human AML blast), and KG1a cells (a spontaneously arising daughter cell line of KG1) were obtained from the American Type Culture Collection (ATCC) and were cultured at 2 to 5 × 105 cells/ml in Isocove’s modification of DMEM (IMDM) medium containing 100 mM L-glutamine, 100 U of penicillin and streptomycin (all from MediaTech Inc.), and 10% (for K562 cells) or 20% (for KG1 and KG1a cells) fetal calf serum (FCS, HyClone)(complete medium), as previously described (19, 24). Culture medium was changed every 2 to 3 days. The generation of the KG1a-PKCβII-GFP E9 and E11 clones stably expressing the PKCβII-GFP fusion protein was previously described (19). The STAT3 inhibitor FLLL32 (a gift of T. Li, Ohio State University) (39) was dissolved in DMSO and used at a final concentration of 5 μM. IL-6 (R&D Systems) was used at concentrations ranging from 1 to 150 ng/ml, with medium being changed every 2 days. KG1 cells, KG1a cells and KG1a-PKCβII-GFP clones, and K562 cells were cultured in complete medium or TCM for 14 days in the presence of PMA (10 ng/ml, Sigma-Aldrich) for days 7to 14 (for KG1, KG1a, and KG1a-PKCβII-GFP cells) or for 5 days (for K562 cells) to drive their differentiation into DCs. Monocytes were obtained from PBMCs that were isolated by Ficoll gradient centrifugation of peripheral blood from platelet donors that was obtained through IRB-approved protocols: monocytes were isolated by negative magnetic selection (StemCell Technology) and cultured in RPMI 1640 medium (GIBCO) containing 10% fetal calf serum (FCS, HyClone) 100 mM L-glutamine (MediaTech Inc.), 100 U of penicillin and streptomycin (MediaTech Inc.), and IL-4 (1000 U/ml, R&D Systems) or TCM supplemented with IL-4 (1000 U/ml). To stimulate their differentiation into DCs, monocytes were cultured in medium or TCM for 18 days, with GM-CSF (5 ng/ml, Sigma-Aldrich) for days 7 to 18, and in the absence or presence of TNF-α (10 ng/ml, R&D Systems) for the final 2 days. PBMCs from breast cancer patients or from healthy donors were obtained after informed consent through IRB-approved protocols through the Data Bank and Biorepository (DBBR) at Roswell Park Cancer Institute (53) before the patients received any treatment. Donor characteristics are detailed in table S1. K562 cells were electroporated with the Nucleofector system (Amaxa Biosystems) with pCDNA 3.0 plasmid encoding wild-type (WT) STAT3, or the similar pRc/CMV plasmid encoding a constitutively active STAT3 mutant (STAT3-CA) (54) or a dominant-negative STAT3 mutant (STAT3-DN) (55) (a gift of Drs. L. Koniaris and T. Zimmers, Miller School of Medicine, University of Miami), selected for neomycin resistance, and then single-cell clones were generated by limiting dilution.
Flow cytometry
Cells were incubated with the following antibodies: phycoerythrin (PE)-conjugated anti-CD11c, PE-conjugated anti-CD14, fluorescein isothiocyanate (FITC)-conjugated anti-CD14, PE-conjugated anti-CD40, PE-conjugated anti-CD80, PE-conjugated anti-CD83, and PE-conjugated anti-CD86 (all from Immunotech); anti-MHC Class I and anti-MHC Class II (VMDR Inc.); PerCP-conjugated anti-CD5, PE-conjugated anti-CD114 (G-CSFR), allophycocyanin (APC)-conjugated anti-CD126 (IL-6Rα), PerCP/Cy5.5-conjugated anti-CD309 (VEGFR2) (Biolegend), or the appropriate isotype controls (Immunotech and Biolegend). MHC Class I, MHC Class II, CD83, and CD86 were subsequently stained with PE-conjugated goat anti-mouse secondary antibody (Southern Biotechnology Associates). For intracellular PKCβII staining, monocytes or PBMCs were fixed with methanol and permeabilized with Fix and Perm buffers (Invitrogen) as previously described (56), and were incubated with anti-PKCβII antibody (Santa Cruz BioTech) and with APC-conjugated donkey anti-mouse secondary antibody (Jackson Immunoresearch) before cell-surface proteins were stained. Cultured monocytes were harvested through a 10- to 20-min incubation in 3 mM EDTA, washed and resuspended in FACS buffer [phosphate-buffered saline (PBS), 2% FCS, 0.1% sodium azide, 10 mM Hepes] and incubated with single-color antibodies to assess the extent of their differentiation into DCs. An LSR-Fortessa or LSRII flow cytometer (both from BD Bioscience) and DIVA (BD Bioscience) and FCS Express (De Novo Software) software were used for data acquisition and analysis.
Murine models
EL4 thymoma cells were injected subcutaneously into C57BL/6 mice, whereas AT3 breast cancer cells were injected into the mammary fat pads of BALB/c mice (13), and the mice were sacrificed when tumors reached a volume of 1500 mm3. Immature myeloid cells were isolated from the spleens of tumor-bearing or control mice by positive selection based on the cell-surface expression of Gr1 (for the C57BL/6 mice) or CD11b (Miltenyi Biotech). All murine experiments were performed under protocols approved by the Institutional Animal Care and Use Committee (IACUC). Blinding of these experiments was not possible, and randomization of age-matched mice before they received tumor cells was deemed unnecessary.
Microarray analysis
Total cellular RNA was isolated from cells with the RNeasy Mini Kit (Qiagen). Complementary DNA (cDNA) was prepared from RNA for analysis on Human Genome U133-plus 2.0 gene chips (Affymetrix) by Expression Analysis, according to the manufacturer’s instructions. Signal intensity and expression status for each transcript on the array was determined with the statistical algorithms contained within the Affymetrix GCOS software by Expression Analysis.
Preparation of conditioned media
MCF-7 and MDA-MB-231 cells (both derived from epithelial mammary carcinomas removed from patients) and MCF10a cells (an immortalized, non-malignant mammary-epithelial cell line) were obtained from ATCC and cultured at 2 × 105 cells/ml in IMDM, 10% FCS, 100 mM L-glutamine, containing 100 U each of penicillin and streptomycin. Cells were allowed to grow for 3 days, and supernatants were centrifuged at 330g for 5 min, passed through a 0.2-micron filter to remove any remaining cells or cell debris, and either frozen at −80°C or added immediately to cell culture, as required. Unless specifically noted as being generated by MCF-7 cells, TCM was generated from MDA-MB-231 cells.
Cytokine neutralization
IL-6 in TCM was neutralized with an anti -IL-6 neutralizing antibody (R&D Systems), which was added to TCM to a final concentration of 10 μg/ml for 5 min before cells were added. Depletion of G-CSF was performed as previously described (13). Briefly, AT-3 tumor–bearing mice were injected with anti–G-CSF neutralizing antibody or immunoglobulin G (IgG) isotype control (10 μg, R&D systems) on day 7, when tumors became palpable. Injections continued for 8 days, and mice were sacrificed and spleens were collected on day 30 (15 days after the final injection with anti–G-CSF).
Western blotting
Western blotting analysis was performed as previously described (19). Membranes were incubated with antibodies against PKCβI, PKCβII, STAT3, RelB, eGFP (all from Santa Cruz Biotechnology); actin (Sigma-Aldrich); phosphorylated ERK1/2 (pERK1/2), and pSTAT3-Y705 (both from Cell Signaling Technology). HRP-conjugated anti-mouse and anti-rabbit secondary antibodies were obtained from Promega. Western blots were visualized by ECL Western Blotting Substrate (Pierce) onto x-ray film (IBF) and developed in an X-OMAT 2000 film processor (Kodak). Where indicated in the figure legends, changes in protein abundance as determined by Western blot were quantified by densitometric analysis. Western blots were scanned with an HP Scanjet G3010 (Hewlett-Packard), and protein abundance was quantified with Quantity One software (BioRad). Proteins of interest were normalized against the relevant loading controls (for example, actin and STAT3).
Quantitative real -time PCR (qPCR)
Total RNA was isolated with Trizol LS reagent (Invitrogen) or RNeasy-Mini kits (Qiagen) according to the manufacturers’ instructions. Equal amounts of RNA were used to generate cDNA with the SuperScriptIII reverse transcriptase system (Invitrogen) or the iScript cDNA synthesis system (Bio-Rad) according to the manufacturers’ protocols. Gene expression was determined with gene-specific primers (table S2) through quantitative, real-time PCR (qPCR) analysis with the FastStart Universal SYBR Green system (Roche) or the iTaq Universal SYBR Green system (Bio-Rad) on AB7900HT real-time qPCR machines (Applied Biosystems) in accordance with the manufacturers’ protocols. Data are presented as the fold-change in abundance relative to control (as indicated) normalized to ACTB (human) or Actb (murine) actin.
T cell stimulation
T cells were isolated as previously described (19). Allogeneic T cell stimulation assays were performed as previously described (19). Briefly, monocytes, KG1 cells, K562 cells, or the KG1a-PKCβII-GFP clones E9 or E11 were cultured as described earlier, with KG1 and K562 lines cultured in the presence or absence of PMA, whereas monocytes were cultured in the presence of IL-4 with or without GM-CSF and TNF-α. These stimulator cells were then irradiated (for K562 cells: 12,000 R 37Co) or treated with mitomycin C (100 μg/ml), washed thoroughly, and cultured with allogeneic T cells at a DC:T cell ratio of 1:10 (for monocytes and K562 cells) or 1:3 (for KG1 cells and the KG1a-PKCβII-GFP clones E9 and E11) for 3 days in 96-well plates. KG1 cells and monocytes were neither treated with mitomycin C nor were they irradiated. Over the final 14 to 18 hours, 3H-thymidine (Perkin-Elmer) was added at a concentration of 20 μCi/ml. Cells were then harvested with a Filtermate Harvester (Perkin-Elmer) and tritium incorporation was measured with a MicroBeta Trilux (Perkin -Elmer).
ELISA
The quantity of IL-6 in culture supernatants was measured by ELISA, as previously described (57).
PRKCB promoter analysis
In silico promoter analysis was performed with the sequence analysis algorithms TRANSFAC and NEBcutter (41, 42). The region corresponding to the PRKCB promoter (drawn from the NCBI Homo sapiens genome build 37.3, chromosome 16, contig NT_010393.16) was analyzed for the consensus STAT3 (GAS)-binding sequence, TTNNNNNAA (40). Generation of the initial PRKCB promoter–based reporter construct (pPKCβ) was described previously (19). KG1 cells were cultured for 20to 24 hours at a density of 2 × 105cells/ml in fresh medium before they were electroporated with the Nucleofector system (Amaxa) and the T-016 program, according to the manufacturer’s instructions. Cells (2 × 106) were centrifuged at 90g, resuspended in 100 μl of Reagent V, and mixed with 2 μg of reporter construct (pGL3-basic, the unmodified pPKCβ construct, or pPKCβ containing site-directed mutations) together with 100 ng of the pRL-CMV transfection control plasmid (Promega) encoding Renilla luciferase. Immediately after transfection, cells were allowed to incubate in pre-warmed complete medium for 2 hours, and then were treated as indicated in the figure legends. Twenty-four hours later, cells were harvested and lysed, and luciferase activity was determined with the Dual-Luciferase Reporter Assay system (Promega) according to the manufacturer’s guidelines, with a Monolight 3010 luminometer (Pharmingen).
Chromatin immunoprecipitation assays
Following growth in tumor-conditioned medium for the time s indicated in the figure legends, DC progenitor cells were harvested, protein and DNA were cross-linked with formaldehyde, and DNA was sheared by sonication. Equal amounts of chromatin [based on protein content as determined by BCA protein assay (Pierce)] were incubated with antibodies against STAT1, STAT3, or STAT5 or with an IgG isotype control antibody (all from Santa Cruz Biotechnology), and were immunoprecipitated with recombinant-protein A/agarose beads (Repligen). Cross-linking was reversed by overnight incubation at 65°C and elution with elution buffer (Santa Cruz Biotech). DNA binding was measured by real-time SYBR green PCR (iScript cDNA synthesis, Bio-Rad) performed with promoter-specific primers for each gene of interest (see table S2) with an AB7900HT real-time qPCR machine (Applied Biosystems). Data are presented as fold change in the abundance of the promoter (based on the number of PCR cycles needed to reach the threshold of detection) immunoprecipitated by the specific antibody compared to the control IgG, normalized to KG1 cells cultured in normal medium.
Site-directed mutagenesis
Putative STAT3-binding sites in the PRKCB promoter contained in pPKCβ were eliminated individually by site-directed mutagenesis. Primers were designed to introduce the desired mutations (table S2), and were used together with pPKC-Forward and the QuikChange II XL Site-Directed Mutagenesis kit (Agilent Technologies-Genomics) according to the manufacturer’s protocol.
Knockdown of PKCβII in K562 and monocytes with specific siRNA
The plasmids pEGFP-siPKCβ and pEGFP-siEmpty were constructed as previously described (32). 5 × 106 K562 cells and 10 × 106 monocytes were transfected with nucleofector technology (Amaxa) with Kit-V and program T-16 (for K562 cells) or the Human Monocyte Nucleofector Kit and program Y-01 (for monocytes).
Primers for PCR, site-directed mutagenesis, and ChIP assasys
All of the primers used in this study were purchased from Invitrogen. Their sequences can be found in table S2.
Statistical analysis
Data were analyzed by Student’s T-test, Mann-Whitney rank sum test, one-way ANOVA, and one-way ANOVA on ranks, as appropriate. Post-hoc analysis for multiple pairwise comparisons following ANOVA was performed with the Student-Newman-Keuls method and Dunn’s method. All tests were two-tailed. Differences were considered statistically significant when P ≤ 0.05.
Supplementary Material
Fig. S1. Activation of the NF-κB and ERK pathways during the differentiation of myeloid progenitor cells into DCs is dependent on PKCβII.
Fig. S2. G-CSF decreases the extent of Prckb2 expression in freshly isolated murine bone marrow cells.
Fig. S3. Depletion of G-CSF increases the expression of Prkcb2 in myeloid cells from tumor-bearing mice.
Fig. S4. Effect of PKCβII activation on the expression of G-CSFR and VEGFR2 expression.
Table S1. Characteristics of breast cancer patients and healthy controls.
Table S2. Primers used for PCR and related applications.
Acknowledgments
We thank Roswell Park’s Databank and Biorepository, M. Nesline, and W. Davis for helping us to obtain human samples. We also thank L. Koniaris and T. Zimmers (University of Miami) for providing us with the WT-STAT3, CA-STAT3, and DN-STAT3 constructs.
Funding: This research was supported in part by grants from the NIH (R01 CA140622, R01 CA121044, R01 AI10015, and T32 CA085183), the Multiple Myeloma Research Foundation (MMRF), and the Roswell Park Cancer Institute Alliance Foundation.
Footnotes
Author contributions: M.F., D.G., S.A., and K.L. carried out the experimental design; M.F., L.C., and C.N. conducted the experiments; C.N., P.L., D.G., and S.A. provided critical experimental components and expertise; M.F. carried out data analysis; M.F. primarily authored the manuscript under the supervision of K.L.; and all authors edited and provided feedback on the manuscript.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: The data for the KG1/KG1a/KG1a-PKCβII-GFP microarray can be accessed at XXX.
References and Notes
- 1.Banchereau J, Briere F, Caux C, Davoust J, Lebecque S, Liu YJ, Pulendran B, Palucka K. Immunobiology of dendritic cells. Annu Rev Immunol. 2000;18:767–811. doi: 10.1146/annurev.immunol.18.1.767. [DOI] [PubMed] [Google Scholar]
- 2.Gallucci S, Matzinger P. Danger signals: SOS to the immune system. Curr Opin Immunol. 2001 Feb;13:114–119. doi: 10.1016/s0952-7915(00)00191-6. [DOI] [PubMed] [Google Scholar]
- 3.Heath WR, Belz GT, Behrens GM, Smith CM, Forehan SP, Parish IA, Davey GM, Wilson NS, Carbone FR, Villadangos JA. Cross-presentation, dendritic cell subsets, and the generation of immunity to cellular antigens. Immunol Rev. 2004 Jun;199:9–26. doi: 10.1111/j.0105-2896.2004.00142.x. [DOI] [PubMed] [Google Scholar]
- 4.Gabrilovich D. Mechanisms and functional significance of tumour-induced dendritic-cell defects. Nat Rev Immunol. 2004 Dec;4:941–952. doi: 10.1038/nri1498. [DOI] [PubMed] [Google Scholar]
- 5.Camus M, Tosolini M, Mlecnik B, Pages F, Kirilovsky A, Berger A, Costes A, Bindea G, Charoentong P, Bruneval P, Trajanoski Z, Fridman WH, Galon J. Coordination of intratumoral immune reaction and human colorectal cancer recurrence. Cancer Res. 2009 Mar 15;69:2685–2693. doi: 10.1158/0008-5472.CAN-08-2654. [DOI] [PubMed] [Google Scholar]
- 6.McKallip RJ, Nagarkatti M, Nagarkatti PS. Delta-9-tetrahydrocannabinol enhances breast cancer growth and metastasis by suppression of the antitumor immune response. J Immunol. 2005 Mar 15;174:3281–3289. doi: 10.4049/jimmunol.174.6.3281. [DOI] [PubMed] [Google Scholar]
- 7.Dieu-Nosjean MC, Antoine M, Danel C, Heudes D, Wislez M, Poulot V, Rabbe N, Laurans L, Tartour E, de Chaisemartin L, Lebecque S, Fridman WH, Cadranel J. Long-term survival for patients with non-small-cell lung cancer with intratumoral lymphoid structures. J Clin Oncol. 2008 Sep 20;26:4410–4417. doi: 10.1200/JCO.2007.15.0284. [DOI] [PubMed] [Google Scholar]
- 8.Collin M, Bigley V, Haniffa M, Hambleton S. Human dendritic cell deficiency: the missing ID? Nat Rev Immunol. 2011 Sep;11:575–583. doi: 10.1038/nri3046. [DOI] [PubMed] [Google Scholar]
- 9.Ghiringhelli F, Apetoh L, Tesniere A, Aymeric L, Ma Y, Ortiz C, Vermaelen K, Panaretakis T, Mignot G, Ullrich E, Perfettini JL, Schlemmer F, Tasdemir E, Uhl M, Genin P, Civas A, Ryffel B, Kanellopoulos J, Tschopp J, Andre F, Lidereau R, McLaughlin NM, Haynes NM, Smyth MJ, Kroemer G, Zitvogel L. Activation of the NLRP3 inflammasome in dendritic cells induces IL-1beta-dependent adaptive immunity against tumors. Nat Med. 2009 Oct;15:1170–1178. doi: 10.1038/nm.2028. [DOI] [PubMed] [Google Scholar]
- 10.Gabrilovich DI, Ostrand-Rosenberg S, Bronte V. Coordinated regulation of myeloid cells by tumours. Nat Rev Immunol. 2012 Apr;12:253–268. doi: 10.1038/nri3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Greten TF, Manns MP, Korangy F. Myeloid derived suppressor cells in human diseases. International immunopharmacology. 2011 Jul;11:802–807. doi: 10.1016/j.intimp.2011.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Montero AJ, Diaz-Montero CM, Kyriakopoulos CE, Bronte V, Mandruzzato S. Myeloid-derived suppressor cells in cancer patients: a clinical perspective. J Immunother. 2012 Feb-Mar;35:107–115. doi: 10.1097/CJI.0b013e318242169f. [DOI] [PubMed] [Google Scholar]
- 13.Waight JD, Hu Q, Miller A, Liu S, Abrams SI. Tumor-derived G-CSF facilitates neoplastic growth through a granulocytic myeloid-derived suppressor cell-dependent mechanism. PloS one. 2011;6:e27690. doi: 10.1371/journal.pone.0027690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kortylewski M, Jove R, Yu H. Targeting STAT3 affects melanoma on multiple fronts. Cancer Metastasis Rev. 2005 Jun;24:315–327. doi: 10.1007/s10555-005-1580-1. [DOI] [PubMed] [Google Scholar]
- 15.Kortylewski M, Kujawski M, Wang T, Wei S, Zhang S, Pilon-Thomas S, Niu G, Kay H, Mule J, Kerr WG, Jove R, Pardoll D, Yu H. Inhibiting Stat3 signaling in the hematopoietic system elicits multicomponent antitumor immunity. Nat Med. 2005 Dec;11:1314–1321. doi: 10.1038/nm1325. [DOI] [PubMed] [Google Scholar]
- 16.Corzo CA, Cotter MJ, Cheng P, Cheng F, Kusmartsev S, Sotomayor E, Padhya T, McCaffrey TV, McCaffrey JC, Gabrilovich DI. Mechanism regulating reactive oxygen species in tumor-induced myeloid-derived suppressor cells. J Immunol. 2009 May 1;182:5693–5701. doi: 10.4049/jimmunol.0900092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gabrilovich DI, Nagaraj S. Myeloid-derived suppressor cells as regulators of the immune system. Nat Rev Immunol. 2009 Mar;9:162–174. doi: 10.1038/nri2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vasquez-Dunddel D, Pan F, Zeng Q, Gorbounov M, Albesiano E, Fu J, Blosser RL, Tam AJ, Bruno T, Zhang H, Pardoll D, Kim Y. STAT3 regulates arginase-I in myeloid-derived suppressor cells from cancer patients. The Journal of clinical investigation. 2013 Mar 1; doi: 10.1172/JCI60083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cejas PJ, Carlson LM, Zhang J, Padmanabhan S, Kolonias D, Lindner I, Haley S, Boise LH, Lee KP. Protein kinase C betaII plays an essential role in dendritic cell differentiation and autoregulates its own expression. J Biol Chem. 2005 Aug 5;280:28412–28423. doi: 10.1074/jbc.M500345200. [DOI] [PubMed] [Google Scholar]
- 20.Liu WS, Heckman CA. The sevenfold way of PKC regulation. Cell Signal. 1998 Sep;10:529–542. doi: 10.1016/s0898-6568(98)00012-6. [DOI] [PubMed] [Google Scholar]
- 21.Lin YF, Lee HM, Leu SJ, Tsai YH. The essentiality of PKCalpha and PKCbetaI translocation for CD14+ monocyte differentiation towards macrophages and dendritic cells, respectively. J Cell Biochem. 2007 Oct 1;102:429–441. doi: 10.1002/jcb.21305. [DOI] [PubMed] [Google Scholar]
- 22.Hooper WC, Abraham RT, Ashendel CL, Woloschak GE. Differential responsiveness to phorbol esters correlates with differential expression of protein kinase C in KG-1 and KG-1a human myeloid leukemia cells. Biochim Biophys Acta. 1989 Sep 4;1013:47–54. doi: 10.1016/0167-4889(89)90126-2. [DOI] [PubMed] [Google Scholar]
- 23.Marigo I, Bosio E, Solito S, Mesa C, Fernandez A, Dolcetti L, Ugel S, Sonda N, Bicciato S, Falisi E, Calabrese F, Basso G, Zanovello P, Cozzi E, Mandruzzato S, Bronte V. Tumor-induced tolerance and immune suppression depend on the C/EBPbeta transcription factor. Immunity. 2010 Jun 25;32:790–802. doi: 10.1016/j.immuni.2010.05.010. [DOI] [PubMed] [Google Scholar]
- 24.Lindner I, Kharfan-Dabaja MA, Ayala E, Kolonias D, Carlson LM, Beazer-Barclay Y, Scherf U, Hnatyszyn JH, Lee KP. Induced dendritic cell differentiation of chronic myeloid leukemia blasts is associated with down-regulation of BCR-ABL. J Immunol. 2003 Aug 15;171:1780–1791. doi: 10.4049/jimmunol.171.4.1780. [DOI] [PubMed] [Google Scholar]
- 25.Burkly L, Hession C, Ogata L, Reilly C, Marconi LA, Olson D, Tizard R, Cate R, Lo D. Expression of relB is required for the development of thymic medulla and dendritic cells. Nature. 1995 Feb 9;373:531–536. doi: 10.1038/373531a0. [DOI] [PubMed] [Google Scholar]
- 26.Carrasco D, Ryseck RP, Bravo R. Expression of relB transcripts during lymphoid organ development: specific expression in dendritic antigen-presenting cells. Development. 1993 Aug;118:1221–1231. doi: 10.1242/dev.118.4.1221. [DOI] [PubMed] [Google Scholar]
- 27.Pettit AR, Quinn C, MacDonald KP, Cavanagh LL, Thomas G, Townsend W, Handel M, Thomas R. Nuclear localization of RelB is associated with effective antigen-presenting cell function. J Immunol. 1997 Oct 15;159:3681–3691. [PubMed] [Google Scholar]
- 28.Xie J, Qian J, Yang J, Wang S, Freeman ME, 3rd, Yi Q. Critical roles of Raf/MEK/ERK and PI3K/AKT signaling and inactivation of p38 MAP kinase in the differentiation and survival of monocyte-derived immature dendritic cells. Exp Hematol. 2005 May;33:564–572. doi: 10.1016/j.exphem.2005.03.001. [DOI] [PubMed] [Google Scholar]
- 29.Saijo K, Mecklenbrauker I, Santana A, Leitger M, Schmedt C, Tarakhovsky A. Protein kinase C beta controls nuclear factor kappaB activation in B cells through selective regulation of the IkappaB kinase alpha. J Exp Med. 2002 Jun 17;195:1647–1652. doi: 10.1084/jem.20020408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stariha RL, Kim SU. Protein kinase C and mitogen-activated protein kinase signalling in oligodendrocytes. Microscopy research and technique. 2001 Mar 15;52:680–688. doi: 10.1002/jemt.1052. [DOI] [PubMed] [Google Scholar]
- 31.Troller U, Zeidman R, Svensson K, Larsson C. A PKCbeta isoform mediates phorbol ester-induced activation of Erk1/2 and expression of neuronal differentiation genes in neuroblastoma cells. FEBS Lett. 2001 Nov 9;508:126–130. doi: 10.1016/s0014-5793(01)03043-5. [DOI] [PubMed] [Google Scholar]
- 32.Cejas PJ, Carlson LM, Kolonias D, Zhang J, Lindner I, Billadeau DD, Boise LH, Lee KP. Regulation of RelB expression during the initiation of dendritic cell differentiation. Mol Cell Biol. 2005 Sep;25:7900–7916. doi: 10.1128/MCB.25.17.7900-7916.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bren GD, Solan NJ, Miyoshi H, Pennington KN, Pobst LJ, Paya CV. Transcription of the RelB gene is regulated by NF-kappaB. Oncogene. 2001 Nov 22;20:7722–7733. doi: 10.1038/sj.onc.1204868. [DOI] [PubMed] [Google Scholar]
- 34.Hiasa M, Abe M, Nakano A, Oda A, Amou H, Kido S, Takeuchi K, Kagawa K, Yata K, Hashimoto T, Ozaki S, Asaoka K, Tanaka E, Moriyama K, Matsumoto T. GM-CSF and IL-4 induce dendritic cell differentiation and disrupt osteoclastogenesis through M-CSF receptor shedding by up-regulation of TNF-alpha converting enzyme (TACE) Blood. 2009 Nov 12;114:4517–4526. doi: 10.1182/blood-2009-04-215020. [DOI] [PubMed] [Google Scholar]
- 35.Chalfant CE, Watson JE, Bisnauth LD, Kang JB, Patel N, Obeid LM, Eichler DC, Cooper DR. Insulin regulates protein kinase CbetaII expression through enhanced exon inclusion in L6 skeletal muscle cells. A novel mechanism of insulin- and insulin-like growth factor-i-induced 5′ splice site selection. J Biol Chem. 1998 Jan 9;273:910–916. doi: 10.1074/jbc.273.2.910. [DOI] [PubMed] [Google Scholar]
- 36.Ramadoss P, Unger-Smith NE, Lam FS, Hollenberg AN. STAT3 targets the regulatory regions of gluconeogenic genes in vivo. Mol Endocrinol. 2009 Jun;23:827–837. doi: 10.1210/me.2008-0264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ravishankaran P, Karunanithi R. Clinical significance of preoperative serum interleukin-6 and C-reactive protein level in breast cancer patients. World journal of surgical oncology. 2011;9:18. doi: 10.1186/1477-7819-9-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mundy-Bosse BL, Young GS, Bauer T, Binkley E, Bloomston M, Bill MA, Bekaii-Saab T, Carson WE, 3rd, Lesinski GB. Distinct myeloid suppressor cell subsets correlate with plasma IL-6 and IL-10 and reduced interferon-alpha signaling in CD4(+) T cells from patients with GI malignancy. Cancer Immunol Immunother. 2011 Sep;60:1269–1279. doi: 10.1007/s00262-011-1029-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bill MA, Fuchs JR, Li C, Yui J, Bakan C, Benson DM, Jr, Schwartz EB, Abdelhamid D, Lin J, Hoyt DG, Fossey SL, Young GS, Carson WE, 3rd, Li PK, Lesinski GB. The small molecule curcumin analog FLLL32 induces apoptosis in melanoma cells via STAT3 inhibition and retains the cellular response to cytokines with anti-tumor activity. Mol Cancer. 2010;9:165. doi: 10.1186/1476-4598-9-165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Darnell JE., Jr STATs and generegulation. Science. 1997 Sep 12;277:1630–1635. doi: 10.1126/science.277.5332.1630. [DOI] [PubMed] [Google Scholar]
- 41.Heinemeyer T, Wingender E, Reuter I, Hermjakob H, Kel AE, Kel OV, Ignatieva EV, Ananko EA, Podkolodnaya OA, Kolpakov FA, Podkolodny NL, Kolchanov NA. Databases on transcriptional regulation: TRANSFAC, TRRD and COMPEL. Nucleic Acids Res. 1998 Jan 1;26:362–367. doi: 10.1093/nar/26.1.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vincze T, Posfai J, Roberts RJ. NEBcutter: A program to cleave DNA with restriction enzymes. Nucleic Acids Res. 2003 Jul 1;31:3688–3691. doi: 10.1093/nar/gkg526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Geijsen N, Spaargaren M, Raaijmakers JA, Lammers JW, Koenderman L, Coffer PJ. Association of RACK1 and PKCbeta with the common beta-chain of the IL-5/IL-3/GM-CSF receptor. Oncogene. 1999 Sep 9;18:5126–5130. doi: 10.1038/sj.onc.1202896. [DOI] [PubMed] [Google Scholar]
- 44.Mattagajasingh SN, Yang XP, Irani K, Mattagajasingh I, Becker LC. Activation of Stat3 in Endothelial Cells Following Hypoxia-reoxygenation is Mediated by Rac1 and Protein Kinase C. Biochim Biophys Acta. 2012 May;1823:997–1006. doi: 10.1016/j.bbamcr.2012.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chalfant CE, Mischak H, Watson JE, Winkler BC, Goodnight J, Farese RV, Cooper DR. Regulation of alternative splicing of protein kinase C beta by insulin. J Biol Chem. 1995 Jun 2;270:13326–13332. doi: 10.1074/jbc.270.22.13326. [DOI] [PubMed] [Google Scholar]
- 46.Basolo F, Conaldi PG, Fiore L, Calvo S, Toniolo A. Normal breast epithelial cells produce interleukins 6 and 8 together with tumor-necrosis factor: defective IL6 expression in mammary carcinoma. Int J Cancer. 1993 Dec 2;55:926–930. doi: 10.1002/ijc.2910550609. [DOI] [PubMed] [Google Scholar]
- 47.Chiu JJ, Sgagias MK, Cowan KH. Interleukin 6 acts as a paracrine growth factor in human mammary carcinoma cell lines. Clin Cancer Res. 1996 Jan;2:215–221. [PubMed] [Google Scholar]
- 48.Fricke I, Mirza N, Dupont J, Lockhart C, Jackson A, Lee JH, Sosman JA, Gabrilovich DI. Vascular endothelial growth factor-trap overcomes defects in dendritic cell differentiation but does not improve antigen-specific immune responses. Clin Cancer Res. 2007 Aug 15;13:4840–4848. doi: 10.1158/1078-0432.CCR-07-0409. [DOI] [PubMed] [Google Scholar]
- 49.Ron D, Mochly-Rosen D. Agonists and antagonists of protein kinase C function, derived from its binding proteins. J Biol Chem. 1994 Aug 26;269:21395–21398. [PubMed] [Google Scholar]
- 50.Graff JR, McNulty AM, Hanna KR, Konicek BW, Lynch RL, Bailey SN, Banks C, Capen A, Goode R, Lewis JE, Sams L, Huss KL, Campbell RM, Iversen PW, Neubauer BL, Brown TJ, Musib L, Geeganage S, Thornton D. The protein kinase Cbeta-selective inhibitor, Enzastaurin (LY317615.HCl), suppresses signaling through the AKT pathway, induces apoptosis, and suppresses growth of human colon cancer and glioblastoma xenografts. Cancer Res. 2005 Aug 15;65:7462–7469. doi: 10.1158/0008-5472.CAN-05-0071. [DOI] [PubMed] [Google Scholar]
- 51.Dudek AZ, Zwolak P, Jasinski P, Terai K, Gallus NJ, Ericson ME, Farassati F. Protein kinase C-beta inhibitor enzastaurin (LY317615.HCI) enhances radiation control of murine breast cancer in an orthotopic model of bone metastasis. Investigational new drugs. 2008 Feb;26:13–24. doi: 10.1007/s10637-007-9079-y. [DOI] [PubMed] [Google Scholar]
- 52.Clemons M, Joy AA, Abdulnabi R, Kotliar M, Lynch J, Jordaan JP, Iscoe N, Gelmon K. Phase II, double-blind, randomized trial of capecitabine plus enzastaurin versus capecitabine plus placebo in patients with metastatic or recurrent breast cancer after prior anthracycline and taxane therapy. Breast Cancer Res Treat. 2010 Nov;124:177–186. doi: 10.1007/s10549-010-1152-0. [DOI] [PubMed] [Google Scholar]
- 53.Ambrosone CB, Nesline MK, Davis W. Establishing a cancer center data bank and biorepository for multidisciplinary research. Cancer Epidemiol Biomarkers Prev. 2006 Sep;15:1575–1577. doi: 10.1158/1055-9965.EPI-06-0628. [DOI] [PubMed] [Google Scholar]
- 54.Bromberg JF, Wrzeszczynska MH, Devgan G, Zhao Y, Pestell RG, Albanese C, Darnell JE., Jr Stat3 as an oncogene. Cell. 1999 Aug 6;98:295–303. doi: 10.1016/s0092-8674(00)81959-5. [DOI] [PubMed] [Google Scholar]
- 55.Kaptein A, Paillard V, Saunders M. Dominant negative stat3 mutant inhibits interleukin-6-induced Jak-STAT signal transduction. J Biol Chem. 1996 Mar 15;271:5961–5964. doi: 10.1074/jbc.271.11.5961. [DOI] [PubMed] [Google Scholar]
- 56.Lesinski GB, Kondadasula SV, Crespin T, Shen L, Kendra K, Walker M, Carson WE., 3rd Multiparametric flow cytometric analysis of inter-patient variation in STAT1 phosphorylation following interferon Alfa immunotherapy. J Natl Cancer Inst. 2004 Sep 1;96:1331–1342. doi: 10.1093/jnci/djh252. [DOI] [PubMed] [Google Scholar]
- 57.Nair JR, Carlson LM, Koorella C, Rozanski CH, Byrne GE, Bergsagel PL, Shaughnessy JP, Jr, Boise LH, Chanan-Khan A, Lee KP. CD28 expressed on malignant plasma cells induces a prosurvival and immunosuppressive microenvironment. J Immunol. 2011 Aug 1;187:1243–1253. doi: 10.4049/jimmunol.1100016. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Activation of the NF-κB and ERK pathways during the differentiation of myeloid progenitor cells into DCs is dependent on PKCβII.
Fig. S2. G-CSF decreases the extent of Prckb2 expression in freshly isolated murine bone marrow cells.
Fig. S3. Depletion of G-CSF increases the expression of Prkcb2 in myeloid cells from tumor-bearing mice.
Fig. S4. Effect of PKCβII activation on the expression of G-CSFR and VEGFR2 expression.
Table S1. Characteristics of breast cancer patients and healthy controls.
Table S2. Primers used for PCR and related applications.