Abstract
The Notch signaling pathway has been shown to be upregulated in colorectal cancer (CRC) and important for the self‐renewal of cancer stem cells. In this study, we evaluated the efficacy of PF‐03084014, a γ‐secretase inhibitor, in combination with irinotecan to identify the effects of treatment on tumor recurrence and the tumor‐initiating population in our CRC preclinical explant model. The combination of PF‐03084014 and irinotecan had the greatest effect at reducing tumor growth on four CRC tumors when compared with treatment with PF‐03084014 or irinotecan alone. The combination significantly reduced tumor recurrence in two CRC explants (CRC001 and CRC036) after treatment was discontinued. Both of these tumors exhibited elevated baseline levels of Notch pathway activation as well as an increase in NOTCH1 gene copy number when compared with the two CRC explants (CRC026 and CRC027) where tumors reappeared quickly after termination of treatment. Isolation and injection of aldehyde dehydrogenase (ALDH+ and ALDH−) cells in an in vivo explant model demonstrated that the ALDH+ cell population were tumorigenic. Evaluation of the ALDH+ cells after 28 days of treatment showed that the combination reduced the ALDH+ population in the tumors that did not regrow. Furthermore, ALDH+ cells from CRC001 and CRC027 were injected in vivo and treated immediately for 28 days. Two months after treatment, tumors were evident in the combination treatment group for CRC027 but not for CRC036. These results indicate the combination of PF‐03084014 and irinotecan may be effective in reducing tumor recurrence in CRC patients whose tumors exhibit elevated levels of the Notch pathway.
Keywords: Notch, Cancer stem cell, Gamma-secretase inhibitor, Colorectal cancer
Highlights
PF‐03084014 + irinotecan had the greatest effect at reducing tumor growth.
Combination slowed tumor recurrence in two CRC explants after treatment was stopped.
Regrowth sensitive tumors exhibited an increase in NOTCH1 gene copy number.
The combination reduced the ALDH+ TIC population in regrowth sensitive tumors.
1. Introduction
Colorectal cancer (CRC) is a common malignancy in western societies (Jemal et al., 2009). Despite high cure rates in early stages of disease, treatment modalities for advanced stages of disease are mostly ineffective. Currently, it is believed that a subset of cells within a tumor known as cancer stem cells, or tumor‐initiating cells, are responsible for tumor formation, chemotherapeutic resistance and disease recurrence (Li et al., 2007; O'Brien et al., 2007; Ricci‐Vitiani et al., 2007; Schatton et al., 2008; Singh et al., 2004). Recently, it has been shown that aberrantly expressed developmental pathways are broadly important for the self‐renewal of cancer stems cells (Zhou et al., 2009).
Notch is an evolutionarily conserved developmental pathway that is important in embryonic development and maintaining adult tissue homeostasis by influencing cell fate decisions. Dysregulation of the Notch pathway plays an integral role in the tumorigenesis of many human malignancies (Curry et al., 2005; Fan et al., 2004; Hopfer et al., 2005; Lee et al., 2004; Reedijk et al., 2005; Santagata et al., 2004). Recently, the Notch pathway has been shown to contribute to the self‐renewal or maintenance of cancer stem cells. Evidence of this was demonstrated by (van Es et al. 2005) showing in an intestinal adenomatous polyposis coli (APC) −/− adenoma mouse model that the Notch pathway was upregulated within the intestinal and colonic crypts of these tumors. Inhibition of the Notch pathway either by genetic manipulation of the RBPjκ gene or treatment with a γ‐secretase inhibitor facilitated the conversion of proliferative cells into post‐mitotic goblet cells resulting in a reduction of tumor burden. Another study involving a preclinical model of CRC showed that the Notch signaling pathway was significantly increased and activated in colon cancer initiating cells (CCIC) (Sikandar et al., 2010). Inhibition of CCIC with treatment of γ‐secretase inhibitor induced apoptosis and the formation of terminally differentiated goblet cells (Sikandar et al., 2010). These studies demonstrate that in intestinal cancers, Notch activation enhances the self‐renewal of cancer stem cells (CSCs).
Activation of the Notch (1, 2, 3, or 4) receptor through the interaction with a Notch ligand (DLL1, DLL3, DLL4, JAG‐1, or JAG2) results in the proteolytic cleavage of the Notch intracellular domain (NICD) by the γ‐secretase complex that subsequently leads to the nuclear localization and transcription of Notch target genes (Pannuti et al., 2010). In addition to maintaining the cancer stem cell population, the oncogenic role of the Notch pathway promotes growth of cells by enhancing cellular proliferation and inhibiting apoptosis. This is achieved in part through the transcriptional activation of Hes‐1, a Notch target gene, which suppresses the cyclin‐dependent kinase inhibitor p27kip1 (Murata et al., 2005) and NFκB2, a transcription factor that regulates many genes involved in augmenting cellular proliferation and inhibiting apoptosis (Oswald et al., 1998). The Notch pathway also has been shown to be important for the formation of tumor vascularity mainly through the Notch ligand DLL4 (Noguera‐Troise et al., 2006).
The Notch pathway has been shown to be upregulated and associated with resistance to chemotherapy by leading to the activation of prosurvival pathways (Meng et al., 2009). Treatment of CRC cell lines with oxaliplatin increased γ‐secretase activity evident by elevated levels of intracellular notch (ICN) (Meng et al., 2009). Use of a γ‐secretase inhibitor in addition to oxaliplatin resulted in a decrease in prosurvival pathways, thus enhancing sensitivity to oxaliplatin. It has also been shown that inhibition of DLL4 combined with irinotecan treatment resulted in a decrease in the frequency of tumor‐initiating cells and tumor recurrence after treatment in a CRC preclinical model (Hoey et al., 2009). These results suggest that combination strategies involving a chemotherapeutic and a Notch pathway inhibitor may be most effective at enhancing tumor death. Since Notch appears to be facilitator in CRC development and chemoresistance, we focused on the effects of this pathway on tumor recurrence and the tumor‐initiating population. Here we show that the combination of PF‐03084014 (a γ‐secretase inhibitor) and irinotecan may be beneficial in reducing tumor recurrence and the tumor‐initiating population in CRC patients whose tumors exhibit elevated levels of the Notch pathway.
2. Materials and methods
2.1. CRC explant xenograft model
Patient‐derived colorectal adenocarcinoma tumor specimens were obtained from consenting patients at the University of Colorado Hospital in accordance with protocols approved by the Colorado Multiple Institutional Review Board (COMIRB # 07‐0570). Tumor material not required for clinical histopathological analysis was collected and placed in excess medium consisting of RPMI supplemented with 10uM HEPES, 4.5g/L glucose, 1uM pyruvate sodium, 200units/mL penicillin, and 200ug/mL streptomycin. After washing several times in media, tumors were cut into 2–3mm3 pieces in antibiotic containing medium and coated in matrigel. Four‐to‐six week old female athymic (nu+/nu+) mice were obtained from Harlan laboratories (Washington, DC) under an approved research protocol by the Institutional Animal Care and Use Committee (IACUC # 51402007(09) 2E). The tumor pieces were then implanted in mice and expansion of the F1–F3 generations was carried out as previously described (Dangles‐Marie et al., 2007; Rubio‐Viqueira et al., 2006). Tumors were expanded in the left and right flanks of 5–6 mice (10 evaluable tumors per group). Mice were randomized into the control, PF‐03084014 (γ‐secretase), irinotecan, or PF‐03084014+irinotecan when tumor volumes reach ∼200mm3. Mice were treated daily with PF‐03084014 (125mg/kg) by oral gavage and/or weekly with irinotecan (30mg/kg) by ip for 28 days. After 28 days of treatment, tumors were monitored for an additional month to evaluate the regrowth of tumors following treatment. Of note, tumor regrowth on CRC036 was also examined at 3 months after stopping treatment. Mice were monitored daily for signs of toxicity and tumor size was evaluated twice per week by caliper measurements using the following formula: tumor volume=[length×width2]×0.52.
2.2. RT‐PCR
RT‐PCR was used to evaluate the (human and mouse) Notch pathway genes. A Notch pathway array (Qiagen # PAHS‐059) was also used to examine differences between ALDH+ cells in sensitive and resistant explants, following the manufacturer's instructions. Total RNA was extracted using the RNeasy Mini kit (Qiagen). cDNA was synthesized using the Applied Biosystems high capacity cDNA reverse transcription kit, following the manufacturer's instructions. Validated and pre‐designed primer/probes and housekeeping gene(s) were purchased from Applied Biosystems. Samples were amplified using the ABI Step One Plus RT‐PCR system. Relative expression of the mRNA analyzed was estimated using the formula: , where #CT=CT (mRNA)−CT (Housekeeper).
2.3. Immunoblotting
Tumor tissues (50–75mg/mouse) were minced on ice and homogenized using a Dounce homogenizer and centrifuged at 16,000g at 4°C for 10min. The total protein in samples was determined using the Bio‐Rad Dc Protein Assay. Forty micrograms of sample were electrophoresed on 4–12% Bis‐Tris precast gels (Bio‐Rad Laboratories, Inc.). After electrotransfer to Immobilon‐P membranes (Millipore), membranes were blocked at room temperature with TBS [10mmol/L Tris–HCl (pH 7.5), 0.5mol/L NaCl, and 0.1% (v/v) Tween 20] containing 5% nonfat milk (Bio‐Rad) for 1h. Hes‐1 primary antibody (origene) was diluted at 1:1000 in TBST containing 5% protease‐free bovine serum albumin (Sigma–Aldrich), and the membranes were incubated overnight at 4°C with rocking. After washing three times with TBST, the membranes were incubated for 1h at room temperature with anti‐mouse IgG horseradish peroxidase‐conjugated antibody at a final dilution of 1:50,000 in TBST. After washing three times with TBST, bound antibodies were detected by enhanced chemiluminescence (Millipore).
2.4. Immunohistochemistry
Tumor tissues from control and treated were fixed in formalin immediately after surgical excision and processed in to paraffin wax blocks. Sections were deparaffinized using standard histologic procedures, and an antigen retrieval method was used to ensure optimal antigen integrity and expression. The CDX2 (Cell Signaling Technology), CD34 (Becton Dickinson), & Ki‐67 (Dako) antibodies were used for IHC. IHC was assessed for overall staining of human cells. IHC staining for CDX2 and Ki‐67 was scored by eye (from a minimum of 1000 cells for the % markers) by a blinded gastrointestinal pathologist. CD34 staining was scored using the Aperio digital IHC scoring system (Vista, CA) and the % CD34 staining was calculated by moderate percent CD34 Stain=(number positive/number total)×100.
2.5. Notch pathway analysis by gene array
Total RNA (RNeasy kit) from all CRC explants were profiled using the Affymetrix Human Gene 1.0 ST Array. This gene array has 764,885 probes comprising of 28,132 genes. Sample preparation and processing procedure were done as described in the Affymetrix GeneChip Expression Analysis Manual (Affymetrix, Inc.). The gene expression levels were converted to a rank‐based matrix and standardized (mean, 0; SD, 1) for each microarray. Using this preprocessing method, we can cluster the same explants from different data sets based on their gene expression profiles. Data analyses were done on this rank‐based matrix. Gene set analysis was performed using the GSEA software Version 2.0.1 obtained from the Broad Institute (http://www.broad.mit.edu/gsea) (Subramanian et al., 2005). Gene set permutations were performed 1000 times for each analysis. We used the nominal p value and Normalized Enrichment Score (NES) to sort the pathways enriched in each phenotype. We used the pathway defined by Kyoto Encyclopedia of Genes and Genomes (KEGG) as the gene sets in this study (Kanehisa et al., 2008). The Notch human pathway annotation was downloaded from KEGG.
2.6. Evaluation of mouse and human cells by FISH
A dual‐color FISH assay using the probe mix human Cot‐1 DNA labeled with red fluorophores (SpectrumRed) and the mouse Cot‐1 DNA labeled with green fluorophores (SpectrumGreen) was developed to determine the effects of treatment on mouse stroma and human tumor tissue. The human Cot‐1 and mouse Cot‐1 probes were prepared by labelling 1μg of the commercial reagents (Invitrogen) respectively with SpectrumRed and SpectrumGreen conjugated dUTPs (Abbott Molecular) using the Nick Translation Reagent Kit (Abbott Molecular Catalog # 32‐801300), according to manufacturer's instructions. Dual‐color FISH assays were performed according to standard laboratory protocol using 50ng of each DNA per round area of 12mm diameter; 15–17min incubation in 2×SSC, and 15–17min incubation in Proteinase K/2×SSC. After post‐hybridization washes, the slides were dehydrated in ethanol and the chromatin was counterstained with DAPI (0.3μg/ml in Vectashield mounting medium, Vector Laboratories). Analysis was performed on epifluorescence microscope using single interference filters sets for green (FITC), red (Texas red), blue (DAPI), and dual (red/green) and triple (blue, red, green) band pass filters. Both red and green fluorescence ranked excellent for all samples.
2.7. Flow cytometric analysis of tumor‐initiating cells
Tumors from treated and untreated mice were excised, finely minced and placed in DMEM medium containing 10% FBS, 20U/ml collagenase, and 1μg/ml DNase. Following incubation for 60min at 37°C, any remaining intact tissue was disrupted by passage through 100μm and 70μm filters. Red blood cells were lysed using an ACK lysis buffer (Invitrogen, Carlsbad, CA). The cell suspensions were collected by centrifugation and count/cell viability was determined on the countess. Cell viability was >80% for all groups. The cell pellets were resuspended in Aldefluor® buffer at a concentration of 1×106cells/ml. Activated Aldefluor® (5μl/1×106cells/ml) was added and immediately after the addition of Aldefluor®, 500ul was placed in another tube containing DEAB buffer (negative control). All samples were incubated at 37°C for 45min. Following incubation, the samples were then centrifuged and washed with 500μl of Aldefluor® buffer. Anti‐mouse alexa‐647 H‐2Kb/H‐2Db, CD45, CD31 antibodies were added to each sample at a concentration of 1:20. The samples were rotated at room temperature for 20min and then washed 2 times with Aldefluor® assay buffer. All samples were analyzed by the University of Colorado Cancer Center flow cytometry core facility. To determine the tumorigenicity of Aldefluor® +/− cell populations in tumors of untreated and treated mice, positive and negative cells were sorted and injected subcutaneous (s.c.) in athymic nude mice. For the analysis of RT‐PCR in Aldefluor® +/− populations cells were sorted directly into RLT lysis buffer. ALDH+ cells were sorted and cytospun on slides for analysis of NOTCH1 gene copy number.
2.8. Analysis of NOTCH1 gene copy number by FISH
The initial step was to develop a FISH probe for human NOTCH1, the gene of interest. NOTCH1 gene maps at 9q34.3 [139, 388, 896 – 139, 440, 314Mb in chromosome 9, www.Ensembl.org]. The RP11‐707O3 clone, mapped at chromosome 9 139, 250, 847 – 139, 453, 805Mb, was selected for this study. The clone was ordered from CHORI (Oakland, CA), streaked on agar plates and single‐cell colonies were cultured and the presence of NOTCH1 sequences were validated via touchdown PCR. The following sets of primers were used for PCR: 5′‐ATGAGTCTTTGAAGAGGAGCTGGT‐3′ and 5′‐TCAGAACGCACTCGTTGATGT‐3′ producing a 303bp product.
One validated single‐cell colony was expanded for DNA extraction and purification using the Qiagen QIAamp DNA Mini Kit (Valencia, CA). The NOTCH1genomic DNA was then amplified using the Qiagen Repli‐G Whole Genome Amplification Kit (Valencia, CA) per manufacturer's instructions. Amplified NOTCH1 DNA was labeled with SpectrumRed (SR) conjugated dUTPs using the Nick Translation Kit (Abbott Molecular Cat # 32‐801300), according to manufacturer's instructions. Labeled DNA was ethanol‐precipitated using herring sperm DNA as carrier and human Cot‐1 DNA for blocking of repetitive sequences, and the DNA pellet was resuspended in tDenHyb (Insitus Biotechnologies, Albuquerque, NM). The NOTCH1 probe was tested on karyotypically normal cell line AG09391 to verify mapping and quality of signal.
Formalin‐fixed, paraffin‐embedded sections from the CRC specimens were subjected to a dual‐color FISH assay using the NOTCH1 (SpectrumRed) and the commercial probe CEP9 (labeled in SpectrumGreen, from Abbott Molecular) as a control for chromosome 9 aneusomy. Initially the slides were incubated for 4h at 56°C, deparaffinized in Citri‐Solv (Fisher) and washed in 100% ethanol for 10min. The slides were sequentially incubated in 2×SSC at 75°C for 15–18min, digested in 0.6mg/ml Proteinase K/2×SSC at 45°C for 15–20min, washed in 2×SSC for 5min, and dehydrated in ethanol. For a 113mm2 hybridization area, a probe mixture was prepared using NOTCH1 (SR) (200ng), 1μl of diluted CEP9 (SG). In each specimen, the probe was applied to the selected hybridization area, which was covered with a glass coverslip and sealed with rubber cement. DNA denaturation was performed for 15min at 85°C and hybridization was allowed to occur at 37°C for 36–48h. Post‐hybridization washes were performed sequentially with 2×SSC/0.3%NP40 (pH 7.0–7.5) at 74°C for 2min and 2×SSC for 2min, and dehydrated in ethanol. Chromatin was counterstained with DAPI (0.3μg/ml in Vectashield mounting medium, Vector Laboratories). Analysis was performed on epifluorescence microscope using single interference filters sets for green (FITC), red (Texas red), blue (DAPI), dual (red/green), and triple (blue, red, green) band pass filters. A minimum of 50 tumor cells per specimen was scored for NOTCH1 and CEP9 signals.
2.9. Statistical analysis
A one‐way analysis of variance (ANOVA) was used to determine whether the means were significantly different overall at day 28 (end of treatment) and at 1 month after stopping treatment (∼Day 56). A logarithm transformation of the data was used to stabilize the variance. If the overall means were significantly different, we carried out a pair‐wise comparison. p values were adjusted using Tukey's method for multiple comparison. SE of the mean was indicated for each value by a bar. A student's t‐test was used for comparisons between two groups (ALDH+ vs. ALDH−). All analyses were carried out using GraphPad Prism version 5.0c for Windows (GraphPad Software, San Diego) and SAS statistical software (SAS, Cary, NC).
3. Results
3.1. Antitumor activity and tumor regrowth in a CRC primary tumor explant model treated with PF‐03084014 and/or irinotecan
We first examined the antitumor activity of PF‐03084014, irinotecan, and PF‐03084014+irinotecan in 4 patient‐derived CRC tumor explants. Supplementary table 1 shows the patient characteristics and common genes mutated in their tumors. As shown in Figure 1, CRC001 and CRC036 showed sensitivity to PF‐03084014, with CRC001 being the most sensitive (static effects, but no regression), while CRC026 and CRC027 were more resistant to PF‐03084014. All tumors treated showed sensitivity to irinotecan and PF‐03084014+irinotecan with the combination having the greatest effect. After 28 days, treatment was halted and tumors were observed for an additional month to evaluate tumor recurrence. The combination significantly reduced tumor recurrence in CRC001 and CRC036 (Figure 1A and B). In contrast, tumors rapidly recurred in CRC026 and CRC027 after treatment was discontinued (Figure 1C and D). Since irinotecan also prevented tumor recurrence in CRC036 at 1 month we followed the irinotecan and PF‐03084014+irinotecan groups for an additional 2 months. A time to event analysis was performed (event: time taken for a tumor to triple in size from baseline) and showed that all tumors in the irinotecan group tripled by day 108, whereas only one out of 8 tumors in the combination group tripled (log rank p<0.0001) (supplementary figure 1A). Furthermore, we evaluated maintenance therapy of PF‐03084014 immediately after treatment with irinotecan for 28 days in CRC001 (PF‐03084014 sensitive tumor). PF‐03084014 had greater antitumor activity when compared with irinotecan and control (regrowth of irinotecan) supplementary figure 1B, suggesting that sequential treatment may be effective.
Figure 1.
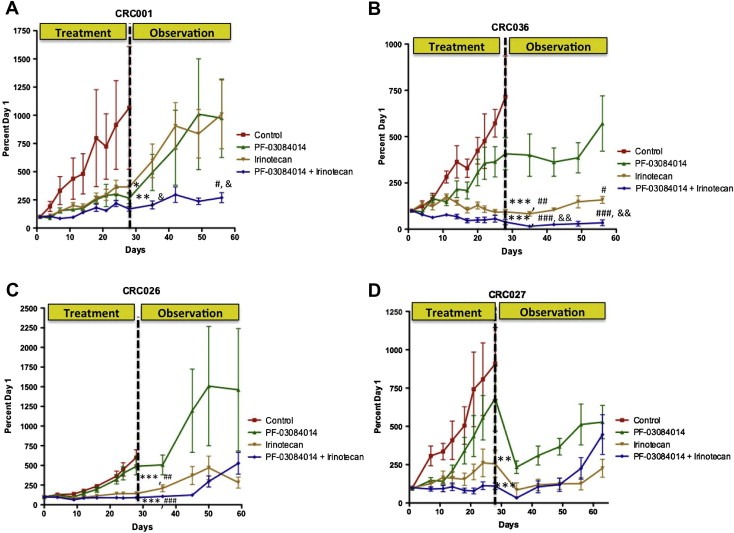
The effects of treatment on tumor growth and recurrence. Four CRC explants were treated (vehicle, PF‐03084014, irinotecan, or PF‐03084014+irinotecan) for 28 days. CRC001 and CRC036 both exhibited sensitivity to PF‐03084014, while CRC026 and CRC027 were resistant. All tumors (CRC001 (A), CRC036 (B), CRC026 (C), and CRC027 (D)) showed sensitivity to irinotecan and PF‐03084014+irinotecan. Data presented as mean±SEM, ANOVA: Tukey's adjusted p values: ∗, p<0.05, ∗∗, p<0.01, ∗∗∗, p<0.001 vs. control, ##, p<0.01, ###, p<0.001 vs. PF‐03084014, and &, p<0.05, &&, p<0.01, &&&, p<0.001 vs. IRN at day 28. Each data point represents an average of ≥10 tumors per treatment group. After 28 days, treatment was stopped and tumor growth was evaluated for an additional month. Tumor size was evaluated weekly after stopping treatment. Treatment with PF‐03084014+irinotecan significantly reduced tumor recurrence in tumors CRC001 (A) and CRC036 (B) however, tumors rapidly recurred in CRC026 (C) and CRC027 (D) after treatment was discontinued. Data presented as mean±SEM, ANOVA: Tukey's adjusted p values: #, p<0.05, ###, p<0.001 vs. PF‐03084014, and &, p<0.05, &&, p<0.01, vs. IRN at day 58.
3.2. Combination reduces notch dependent Hes‐1 protein expression
Evaluation of the Notch dependent transcription factor Hes‐1 in CRC001 (A) and CRC036 (B) by western blot confirmed that the combination had the greatest effect at reducing the Notch pathway (supplementary figure 2). Turning to the regrowth resistant tumors, there was an increase in Notch pathway with irinotecan and combination in CRC026 (C), while all treatments decreased the Notch pathway in CRC027 (D) (supplementary figure 2).
3.3. Effects of treatment on stromal and tumor cells
To assess whether treatment resulted in death to stromal cells or tumor cells, a FISH assay for mouse and human Cot‐1 was developed in order to distinguish between mouse cells and human cells within a tumor. As shown in Figure 2 and supplementary figure 3, all treatments appeared to decrease human tumor tissue (red) as a result of necrosis while the mouse stroma (green) remained intact. Necrotic cells were identified by reduced or lack of DAPI intercalation and red fluorescence as shown in CRC001 (Figure 2); initial necrosis was identified by bright red fluorescence due to extreme chromatin compaction as shown in CRC036 (supplementary figure 3). H&E staining and CDX2 (a marker of colon cancer) of the regrowth sensitive tumors (CRC001 and CRC036) showed that the combination of PF‐03084014+irinotecan exhibited a greater disintegration of tumor as a result of tumor necrosis as well as an increase in the disorganization of the tissue evident by an intermingling of stromal and tumor cells (Figure 2 and supplementary figure 3). It was intriguing to find that in the combination group of CRC036 there was a marked excess amount of foreign body giant cells and induced fibrosis that encapsulated the tumor (supplementary figure 3). These findings were not seen in CRC001 and CRC026. Of note, CRC027 had a slight increase in foreign body giant cells. Further, no reduction in gene expression of mouse Hes‐1 was seen in all explants treated (data not shown) as well as no treatment effects were observed on angiogenesis (IHC: CD34 staining of mouse endothelial cells) (supplementary figure 4) providing further evidence that treatment was only affecting tumor cells. These results suggest that PF‐03084014+irinotecan has the greatest effect at reducing tumor burden evident by an increase in necrosis and enhancing tumor/stroma disorganization.
Figure 2.
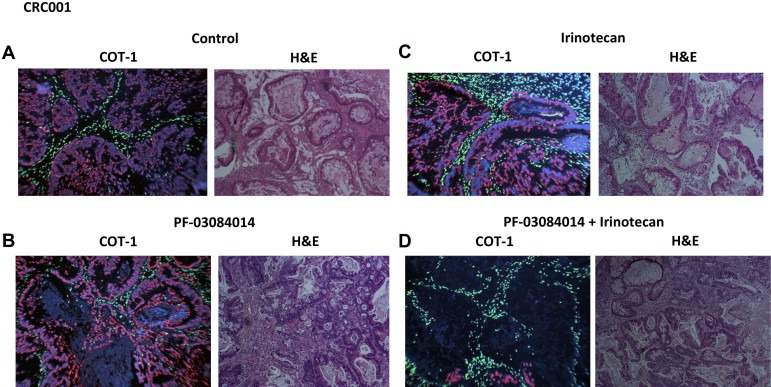
Evaluation of treatment effects (A – Control, B – PF‐03084014, C – Irinotecan, D – PF‐03084014 + Irinotecan) on mouse stromal and tumor cells. Dual‐color FISH for the human cot‐1 gene (red) and mouse cot‐1 gene (green) was used to investigate treatment effects on mouse and human cells. All treatments decreased human tumor cells as a result of tumor necrosis, while the mouse stroma remained intact. Necrotic cells were identified by reduced or lack of DAPI intercalation and red fluorescence. H&E staining of the regrowth sensitive tumor CRC001 showed that the combination of PF‐03084014+irinotecan exhibited a greater disintegration of tumor as a result of tumor necrosis as well as an increase in the disorganization of the tissue evident by an intermingling of stromal and tumor cells.
3.4. Identification and treatment effects on the tumor‐initiating population in CRC explants
Since tumor recurrence was inhibited in two of our explants (CRC001 and CRC036) with combination therapy, we next examined the effects of treatment on the tumor‐initiating population. First, we used an Aldefluor assay (a measure of aldehyde dehydrogenase (ALDH) activity) to determine if the ALDH+ cells are capable of tumorigenic growth when compared with the ALDH− population in our 4 CRC explants. As shown in Figure 3, isolation and injection of ALDH+ cells resulted in growth of tumors in CRC001, CRC026, CRC027, and CRC036. In contrast, ALDH− cells either did not form tumors (CRC001 (A) and CRC036 (B)) or had very slow growing and small tumors (CRC026 (C) and CRC027 (D)) when the same number of cells were injected. In addition, injection of 50,000–100,000 ALDH− cells also only yielded small slow growing tumors (data not shown). Morphologically, all ALDH+ tumors appeared identical to the original tumor (Figure 3).
Figure 3.
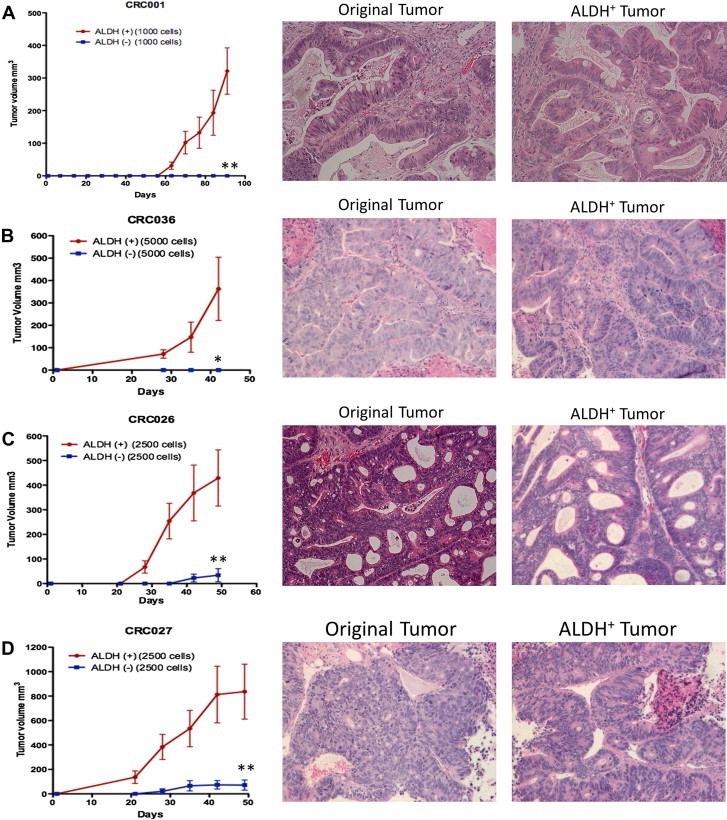
Identification of the tumorigenic population (ALDH+ cells) in CRC explants. Untreated tumors were collagenase digested and ALDH+ and ALDH− populations were sorted by flow cytometry. Injection of ALDH+ cells produced fast growing and large tumors in (A) CRC001 (1000 cells), (B) CRC036 (5000 cells), (C) CRC026 (2500 cells), and (D) CRC027 (2500 cells) when compared to ALDH− cells. Data presented as mean±SEM (T‐TEST: ∗, p<0.05, ∗∗, p<0.01). H&E staining shows that morphologically all ALDH+ tumors appeared similar to the original tumor.
To evaluate the effects of treatment on the ALDH+ population, we treated mice for 28 days and assessed the percentage of positive ALDH staining cells by flow cytometry. As shown in Figure 4, a reduction of ALDH+ cells was seen in tumors CRC001 (A) and CRC036 (B) treated with PF‐03084014, while no decrease was evident in the two GSI resistant tumors CRC026 (C) or CRC027 (D). Furthermore, treatment with irinotecan resulted in a decrease in ALDH+ cells in CRC036 and CRC027 while CRC001 had increased levels. No treatment effect on the ALDH+ population was seen in CRC026. The combination of PF‐03084014+irinotecan decreased ALDH+ cells in CRC001, CRC036 and CRC027. The greatest effect seen was in the combination group of CRC036 where the ALDH+ population was reduced ∼27 fold.
Figure 4.
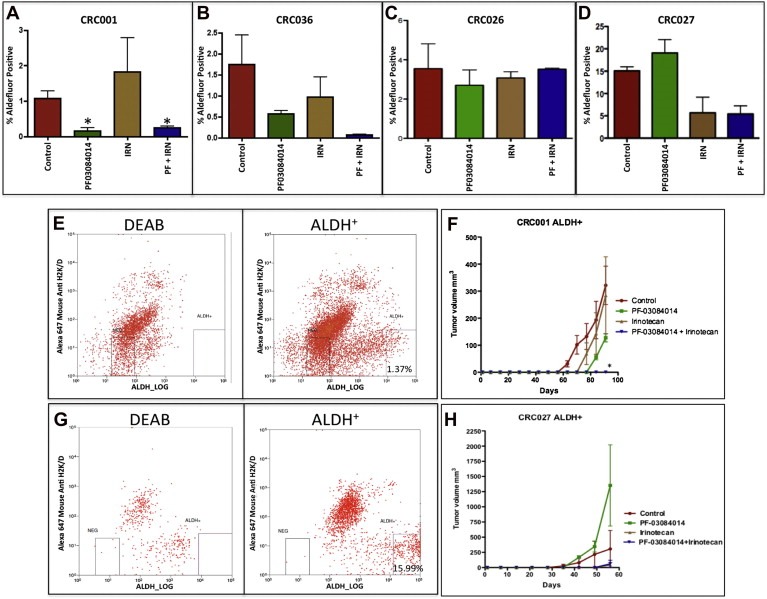
Effects of treatment on the tumor‐initiating population (ALDH+). CRC explants were treated (vehicle, PF‐03084014, irinotecan, or PF‐03084014+irinotecan) for 28 days and ALDH activity was analyzed by flow cytometry. Treatment with PF‐03084014 and PF‐03084014+irinotecan (PF + IRN) reduced the ALDH+ population in CRC001 (A) and CRC036 (B). No treatment effect on the ALDH+ population was seen in CRC026 (C). Irinotecan and PF‐03084014+irinotecan (PF + IRN) reduced the ALDH+ population in CRC027 (D). Data presented as mean±SEM (ANOVA: ∗, p<0.05, when compared with control at 28 days, n=3). E–F. ALDH+ cells from CRC001 (E) and CRC027 (G) were sorted and the same numbers of cells were injected in athymic nude mice. Mice were treated (vehicle, PF‐03084014, irinotecan, or PF‐03084014+irinotecan) for 28 days. After 28 days, treatment was stopped and tumor growth was evaluated for a period of time. Treatment with PF‐03084014 reduced tumor growth in CRC001 (F), but not in CRC027 (H). No tumors were evident in CRC001 (F) treated with PF‐03084014+irinotecan 5 months after stopping treatment, while tumors were present in CRC027 (H) at day 58. Data presented as mean±SEM (ANOVA: ∗p<0.05 when compared with control).
Treatment effects on tumorigenicity of the tumor‐initiating population (ALDH+ cells) were directly tested in vivo. ALDH+ cells were sorted from CRC001 (regrowth sensitive tumors) and CRC027 (regrowth resistant tumor) and injected into athymic nude mice and immediately followed by treatment with vehicle, PF‐03084014, irinotecan, or PF‐03084014+irinotecan for 28 days. After 28 days, treatment was stopped and tumor growth was evaluated for a period of time. As shown in Figure 4, treatment with PF‐03084014 delayed the growth of tumors in CRC001 (E–F), but not in CRC027 (G–H). No tumors were evident even 5 months following combination treatment in the regrowth sensitive tumors CRC001 (E–F) and CRC036 (data not shown). In contrast, tumors were evident in all treatment groups in the CRC027 (G–H). Together, these results indicate that the combination has a significant impact on the ALDH+ population in the regrowth sensitive tumors CRC001 and CRC036.
3.5. Notch pathway is upregulated in tumors that do not recur
Since we identified differences in tumor recurrence in CRC001 and CRC036 (regrowth sensitive tumors) vs. CRC026 and CRC027 (regrowth resistant tumors) treated with PF‐03084014+irinotecan, we explored whether differences existed in activation of the Notch pathway by gene array and pathway analysis in baseline tumors. As shown in Figure 5A and B, both CRC001 and CRC036 had increases in many components of the Notch pathway (highlighted in red). Evaluation of the Notch target transcription factor Hes‐1 by RT‐PCR confirmed that the Notch pathway had significantly greater expression in CRC001 and CRC036 in the overall tumor (Figure 5C). In addition, we performed RT‐PCR on Notch1 and showed that CRC001 and CRC036 have significantly greater expression of the Notch1 (Figure 5D) gene when compared with CRC026 and CRC027.
Figure 5.
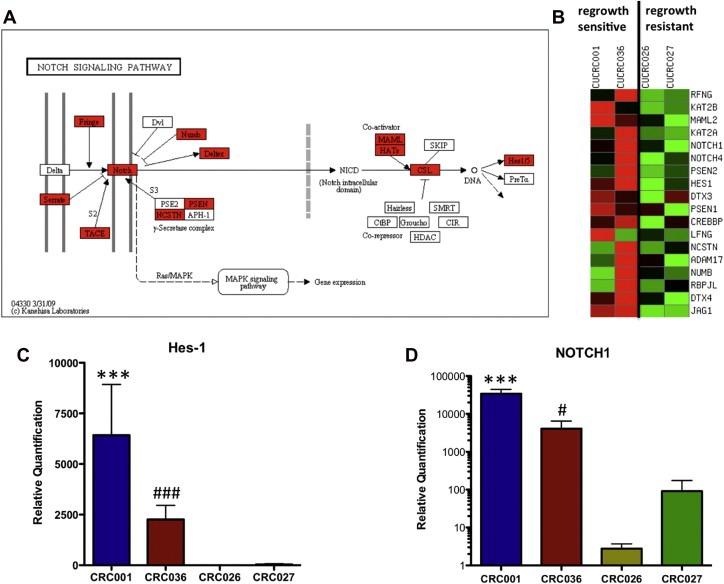
Notch pathway analysis between regrowth sensitive and regrowth resistant tumors. (A) KEGG pathway analysis of the Notch pathway between CRC001, CRC036 vs. CRC026, CRC027. The regrowth sensitive tumors CRC001 and CRC036 demonstrated an increase in the Notch pathway when compared to the regrowth resistant tumors CRC026 and CRC027. Red indicates elevated gene expression. (B) Heatmap of the relative expression of Notch genes shows an increase in many components of the Notch pathway in CRC001 and CRC036. Real‐time PCR of Hes‐1 (C) and Notch1 (D) gene expression showed a significant increase in Hes‐1 and Notch1 in the regrowth sensitive tumors when compared to the regrowth resistant tumors. Data presented as mean±SEM (ANOVA: #, p<0.05, ###, p<0.001, ∗∗∗, p<0.001, when compared with CRC026 and CRC027).
A Notch1 FISH assay was developed to assess whether differences in NOTCH1 gene copy existed between the regrowth sensitive and resistant explants. As displayed in supplementary figure 5, CRC001 (A) and 036 (B) had the highest NOTCH1 mean copy numbers (∼3.6 signals per cell), whereas CRC026 had the lowest NOTCH1 mean copy numbers (∼2 signals per cell). Both CRC001 and 036 exhibited doublet or triplet NOTCH1 signals on the same chromosome in numerous cells in addition to the expected “single” signal. CRC027 had a copy number gain for NOTCH1 but at a lower level than displayed by CRC001 and 036. NOTCH1 copy number increase was accompanied by CEP9 (centromere 9 probe) gain in most of the specimens, as indicated by the mean NOTCH1/mean CEP9 ratio (supplementary table 2). A lower level of imbalance was detected in CRC001 and CRC027 and a higher level of imbalance was observed in CRC036.
Analysis of the Notch pathway in the ALDH+ population by RT‐PCR showed an overall increase in many components of the Notch pathway in CRC001 and CRC036 vs. CRC026 and CRC027 (supplementary table 3). Evaluation of Hes‐1 (A) and JAG‐1 (B) gene expression by RT‐PCR confirmed this finding that Notch signaling was significantly elevated in the ALDH+ population in CRC001 and CRC036 (Figure 6). In addition, we were interested in determining whether the increase in NOTCH1 gene copy number was evident in the ALDH+ population of CRC001 and CRC036. As shown in Figure 6, both CRC001 (C) and CRC036 (D) harbored an increase in NOTCH1 gene copy number. CRC001 and CRC036 displayed doublet or triplet NOTCH1 signals in numerous cells.
Figure 6.
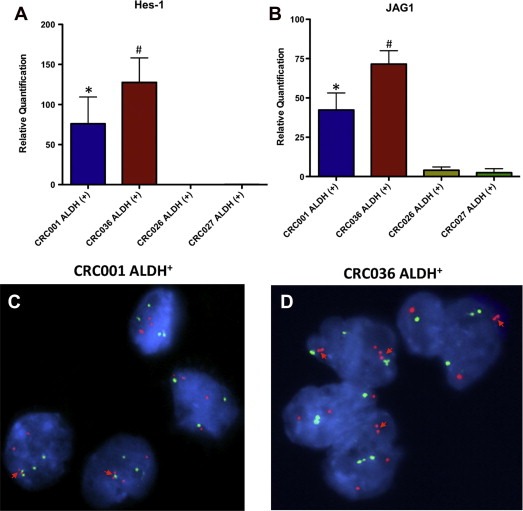
Comparison of the Notch pathway in the ALDH+ tumorigenic population between regrowth sensitive tumors and regrowth resistant tumors. Hes‐1 (A) and JAG‐1 (B) gene expression was elevated in ALDH+ cells in CRC001 and CRC036 when compared to CRC026 and CRC027. Representative graphs of interphase nuclei showing a gain in gene copies (doublets on the same chromosome) of the Notch1 gene in CRC001 (C) and CRC036 (D).
4. Discussion
In this study, we first investigated the efficacy of PF‐03084014, irinotecan and PF‐03084014+irinotecan in 4 CRC patient‐derived explants. The major advantage of this model is that tumor heterogeneity remains intact and tumors appear to retain the genetic features of the original tumor (Rubio‐Viqueira et al., 2006). Whereas single agent PF‐03084014 had efficacy on two tumors CRC001 and CRC036, irinotecan and the combination reduced tumor growth in all tumors treated. Interestingly, only the combination significantly reduced tumor recurrence in the two sensitive PF‐03084014 tumors (CRC001 and CRC036). A study by Hoey et al. (2009) showed very similar effects whereby the combination of anti‐DLL4 antibody+irinotecan prevented regrowth of a unique tumor in a similar primary xenograft model. This tumor was also shown to be sensitive to single agent DLL4. Another study demonstrated in vitro that the addition of a platinum compound with a γ‐secretase inhibitor enhanced cell death in a subset of tumors that possessed cleaved Notch (Aleksic and Feller, 2008). This same study also showed variability with respect to cleaved Notch levels in a panel of CRC cell lines. Taken together, although combination therapy was effective on all explants, prevention of tumor recurrence may only occur in tumors that exhibit dysregulation of Notch. This is plausible since it is unlikely that all tumors will possess upregulation of the Notch pathway. However, further studies are warranted in order to validate these findings.
We also examined treatment effects on Hes‐1 protein levels. In both CRC001 and CRC036, Hes‐1 protein levels were reduced the greatest with combination therapy 28 days after treatment. These results indicate that PF‐03084014+irinotecan have the greatest effect at reducing the Notch pathway. Of note, in addition to Notch, there are multiple targets of γ‐secretase that include CD44, ADAM10, and EpCAM (Pannuti et al., 2010); therefore, it is unknown whether these γ‐secretase substrates were important in the tumorigenesis of these explants.
Since stromal elements may play a role in drug sensitivity, a Cot‐1 FISH assay was developed to assess the effects of treatment on human tumor cells vs. mouse cells. Herein we show with this assay that treatment with PF‐03084014+irinotecan had the greatest effect at reducing tumor volume as a result of tumor necrosis. Furthermore, the disruption of the intricate network of tumor and mouse stroma was widely seen by the Cot‐1 assay and H&E. There was not a significant reduction in mouse stromal cells or affects on angiogenesis (CD34 staining of mouse endothelial cells). In CRC036, we did observe that treatment with the combination produced a large amount of foreign body giant cells within the tumor and it is highly likely that this inflammatory response yielded the fibrotic ring that appeared to encapsulate the tumor. This data suggest that an inflammatory component, albeit in a nude mouse, may have also been responsible for death to tumor cells in CRC036. Whether or not the loss of human stroma is strength or weakness in this model remains to be determined.
ALDH plays an important role in detoxification of aldehydes that ultimately protects these cells from oxidative damage (Vasiliou et al., 2004). This marker has been used to characterize tumor‐initiating cells in many tumor types and in CRC it has been shown to be a marker in normal colon cells and in the development and progression of CRC (Carpentino et al., 2009; Huang et al., 2009). Prior to evaluating treatment effects on the ALDH+ population, we first needed to verify that this population of cells were tumorigenic in our 4 CRC explants. As expected, we showed that ALDH+ cells produced large tumors when compared with ALDH− cells, which either grew very slowly or not at all. Importantly, we observed in CRC001 and CRC036 tumors (regrowth sensitive) that treatment with PF‐03084014+irinotecan produced a significant reduction in the ALDH+ population. These results were further validated in a separate experiment where we treated mice immediately after injection ALDH+ cells. No tumors were present in CRC001 and CRC036 even when observed 5 months after stopping treatment. Recently, it has been shown in CRC cells that the differentiated population within a tumor protects the CSC population from irinotecan (Emmink et al., 2011). They concluded that eliminating this population of cells may enhance irinotecan sensitivity to the tumorigenic population. Our findings suggest that PF‐03084014 and irinotecan resulted in marked reduction of the tumorigenic ALDH+ population in CRC001 and CRC036. Whether this reduction was a result of a decrease in the differentiated population, thereby enhancing the sensitivity of treatment to the ALDH+ population remains to be determined. Nevertheless, the reduction observed may have resulted from 1) PF‐03084014 altering the ALDH+ cells self‐renewal capability and 2) irinotecan promoting the death of these cells.
Since we identified differences in tumor recurrence in our explants, we investigated the Notch pathway between our sensitive and resistant regrowth CRC explants. Using baseline gene array, Notch pathway analysis (KEGG) showed that many components of the pathway were elevated in the sensitive tumors CRC001 and CRC036. Analysis of gene expression of the Notch dependent transcription factor Hes‐1 by RT‐PCR confirmed these findings and showed that CRC001 and CRC036 tumors had significantly elevated levels of Hes‐1 when compared with CRC026 and CRC027. Moreover, expression of the Notch1 receptor has been shown to be important for the regulation of colon cancer growth (Zhang et al., 2010) and associated with a more poorly differentiated tumor (Chu et al., 2009), disease progression and worse survival (Chu et al., 2010). Therefore, we evaluated expression of the Notch1 gene in our explants and showed that the Notch1 receptor was significantly elevated in our regrowth sensitive tumors when compared with our regrowth resistant tumors. Furthermore, we show for the first time that NOTCH1 gene copy number was the highest in sensitive tumors. Although this gain was accompanied with CEP9, CRC036 and CRC001 had a higher and lower level of imbalance respectively when compared with CRC026 and CRC027.
To determine if the Notch pathway was elevated in our ALDH+ population, we sorted ALDH+ cells from all four explants and compared the sensitive and resistant regrowth tumors using a RT‐PCR Notch pathway array. We observed that many components of the Notch pathway had greater expression in the ALDH+ population, as shown by fold increase, in our regrowth sensitive tumors CRC001 and CRC036. In addition, we investigated whether a gain in NOTCH1 gene copy existed in the ALDH+ population of cells. As shown, doublet and triplet NOTCH1 gene signals were evident within this population, suggesting that a gain in NOTCH1 gene copy as well as expression of the Notch pathway appear to be important for the development of CRC and the self‐renewal of the CSC population in CRC001 and CRC036.
This study provides substantial evidence that treatment with PF‐03084014+irinotecan was only effective at preventing tumor recurrence and decreasing the ALDH+ tumorigenic population in CRC tumors that exhibited sensitivity to PF‐03084014 and elevated levels of the Notch pathway. In addition, we show that a gain in NOTCH1 gene copy number exists in the ALDH+ cells and therefore this genetic abnormality may play a significant role in the development of colorectal cancer as well as disease recurrence in these patients. Our findings are consistent with other studies showing that Notch1 appears to be an important player in the development of CRC (Zhang et al., 2010) and chemoresistance (Meng et al., 2009). Further studies are warranted to confirm the role of Notch1 in CRC. These observations have important implications for treatment in patients. In particular, identifying and treating CRC patients that have aberrant activation of the Notch pathway with a Notch pathway inhibitor in combination with a chemotherapeutic agent may not only decrease tumor burden, but also more importantly may prevent disease recurrence. Since PF‐03084014 is in the clinic and shown to have effects in Phase I clinical trials (a complete response and several partial responses), these preclinical findings may inform further clinical development of this compound (Messersmith et al., 2011).
Supporting information
The following are the Supplementary data related to this article:
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary material 1.
1.1.
Supplementary data related to this article can be found online at doi:10.1016/j.molonc.2012.03.004.
Arcaroli John J., Powell Rebecca W., Varella-Garcia Marileila, McManus Martine, Tan Aik Choon, Quackenbush Kevin S., Pitts Todd M., Gao Dexiang, Spreafico Anna, Dasari Arvind, Touban Basel M., Messersmith Wells A., (2012), ALDH+ tumor‐initiating cells exhibiting gain in NOTCH1 gene copy number have enhanced regrowth sensitivity to a γ‐secretase inhibitor and irinotecan in colorectal cancer, Molecular Oncology, 6, doi: 10.1016/j.molonc.2012.03.004.
References
- Aleksic, T. , Feller, S.M. , 2008. Gamma-secretase inhibition combined with platinum compounds enhances cell death in a large subset of colorectal cancer cells. Cell Commun. Signal.. 6, 8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpentino, J.E. , Hynes, M.J. , Appelman, H.D. , Zheng, T. , Steindler, D.A. , Scott, E.W. , Huang, E.H. , 2009. Aldehyde dehydrogenase-expressing colon stem cells contribute to tumorigenesis in the transition from colitis to cancer. Cancer Res.. 69, 8208–8215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu, D. , Wang, W. , Xie, H. , Li, Y. , Dong, G. , Xu, C. , Chen, D. , Zheng, J. , Li, M. , Lu, Z. , Ji, G. , 2009. Notch1 expression in colorectal carcinoma determines tumor differentiation status. J. Gastrointest. Surg.. 13, 253–260. [DOI] [PubMed] [Google Scholar]
- Chu, D. , Li, Y. , Wang, W. , Zhao, Q. , Li, J. , Lu, Y. , Li, M. , Dong, G. , Zhang, H. , Xie, H. , Ji, G. , 2010. High level of Notch1 protein is associated with poor overall survival in colorectal cancer. Ann. Surg. Oncol.. 17, 1337–1342. [DOI] [PubMed] [Google Scholar]
- Curry, C.L. , Reed, L.L. , Golde, T.E. , Miele, L. , Nickoloff, B.J. , Foreman, K.E. , 2005. Gamma secretase inhibitor blocks Notch activation and induces apoptosis in Kaposi's sarcoma tumor cells. Oncogene. 24, 6333–6344. [DOI] [PubMed] [Google Scholar]
- Dangles-Marie, V. , Pocard, M. , Richon, S. , Weiswald, L.B. , Assayag, F. , Saulnier, P. , Judde, J.G. , Janneau, J.L. , Auger, N. , Validire, P. , Dutrillaux, B. , Praz, F. , Bellet, D. , Poupon, M.F. , 2007. Establishment of human colon cancer cell lines from fresh tumors versus xenografts: comparison of success rate and cell line features. Cancer Res.. 67, 398–407. [DOI] [PubMed] [Google Scholar]
- Emmink, B.L. , Van Houdt, W. , Vries, R.G. , Hoogwater, F.J. , Jimenez, C.R. , Clevers, H. , Borel Rinkes, I.H. , Kranenburg, O. , 2011. Differentiated human colorectal cancer cells protect tumor-initiating cells from irinotecan. Gastroenterology. 141, 269–278. Abstract: AACR: Colorectal Cancer: Biology to Therapy [DOI] [PubMed] [Google Scholar]
- Fan, X. , Mikolaenko, I. , Elhassan, I. , Ni, X. , Wang, Y. , Ball, D. , Brat, D.J. , Perry, A. , Eberhart, C.G. , 2004. Notch1 and notch2 have opposite effects on embryonal brain tumor growth. Cancer Res.. 64, 7787–7793. [DOI] [PubMed] [Google Scholar]
- Hoey, T. , Yen, W.C. , Axelrod, F. , Basi, J. , Donigian, L. , Dylla, S. , Fitch-Bruhns, M. , Lazetic, S. , Park, I.K. , Sato, A. , Satyal, S. , Wang, X. , Clarke, M.F. , Lewicki, J. , Gurney, A. , 2009. DLL4 blockade inhibits tumor growth and reduces tumor-initiating cell frequency. Cell Stem Cell. 5, 168–177. [DOI] [PubMed] [Google Scholar]
- Hopfer, O. , Zwahlen, D. , Fey, M.F. , Aebi, S. , 2005. The Notch pathway in ovarian carcinomas and adenomas. Br. J. Cancer. 93, 709–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, E.H. , Hynes, M.J. , Zhang, T. , Ginestier, C. , Dontu, G. , Appelman, H. , Fields, J.Z. , Wicha, M.S. , Boman, B.M. , 2009. Aldehyde dehydrogenase 1 is a marker for normal and malignant human colonic stem cells (SC) and tracks SC overpopulation during colon tumorigenesis. Cancer Res.. 69, 3382–3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jemal, A. , Siegel, R. , Ward, E. , Hao, Y. , Xu, J. , Thun, M.J. , 2009. Cancer statistics, 2009. CA Cancer J. Clin.. 59, 225–249. [DOI] [PubMed] [Google Scholar]
- Kanehisa, M. , Araki, M. , Goto, S. , Hattori, M. , Hirakawa, M. , Itoh, M. , Katayama, T. , Kawashima, S. , Okuda, S. , Tokimatsu, T. , Yamanishi, Y. , 2008. KEGG for linking genomes to life and the environment. Nucleic Acids Res.. 36, D480–D484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, S.F. , Shah, S. , Yu, C. , Wigley, W.C. , Li, H. , Lim, M. , Pedersen, K. , Han, W. , Thomas, P. , Lundkvist, J. , Hao, Y.H. , Yu, G. , 2004. A conserved GXXXG motif in APH-1 is critical for assembly and activity of the gamma-secretase complex. J. Biol. Chem.. 279, 4144–4152. [DOI] [PubMed] [Google Scholar]
- Li, C. , Heidt, D.G. , Dalerba, P. , Burant, C.F. , Zhang, L. , Adsay, V. , Wicha, M. , Clarke, M.F. , Simeone, D.M. , 2007. Identification of pancreatic cancer stem cells. Cancer Res.. 67, 1030–1037. [DOI] [PubMed] [Google Scholar]
- Meng, R.D. , Shelton, C.C. , Li, Y.M. , Qin, L.X. , Notterman, D. , Paty, P.B. , Schwartz, G.K. , 2009. Gamma-secretase inhibitors abrogate oxaliplatin-induced activation of the Notch-1 signaling pathway in colon cancer cells resulting in enhanced chemosensitivity. Cancer Res.. 69, 573–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messersmith, W. , LoRusso, P. , Cleary, J.M. , Dasari, A. , Zhang, X. , 2011. A phase I dose-escalation study of the novel gamma secretase inhibitor PF-03084014 in patients (pts) with advanced solid tumors. J. Clin. Oncol.. 29, suppl; abstr 3100 [Google Scholar]
- Murata, K. , Hattori, M. , Hirai, N. , Shinozuka, Y. , Hirata, H. , Kageyama, R. , Sakai, T. , Minato, N. , 2005. Hes1 directly controls cell proliferation through the transcriptional repression of p27Kip1. Mol. Cell Biol.. 25, 4262–4271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noguera-Troise, I. , Daly, C. , Papadopoulos, N.J. , Coetzee, S. , Boland, P. , Gale, N.W. , Lin, H.C. , Yancopoulos, G.D. , Thurston, G. , 2006. Blockade of Dll4 inhibits tumour growth by promoting non-productive angiogenesis. Nature. 444, 1032–1037. [DOI] [PubMed] [Google Scholar]
- O'Brien, C.A. , Pollett, A. , Gallinger, S. , Dick, J.E. , 2007. A human colon cancer cell capable of initiating tumour growth in immunodeficient mice. Nature. 445, 106–110. [DOI] [PubMed] [Google Scholar]
- Oswald, F. , Liptay, S. , Adler, G. , Schmid, R.M. , 1998. NF-kappaB2 is a putative target gene of activated Notch-1 via RBP-Jkappa. Mol. Cell Biol.. 18, 2077–2088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pannuti, A. , Foreman, K. , Rizzo, P. , Osipo, C. , Golde, T. , Osborne, B. , Miele, L. , 2010. Targeting Notch to target cancer stem cells. Clin. Cancer Res.. 16, 3141–3152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reedijk, M. , Odorcic, S. , Chang, L. , Zhang, H. , Miller, N. , McCready, D.R. , Lockwood, G. , Egan, S.E. , 2005. High-level coexpression of JAG1 and NOTCH1 is observed in human breast cancer and is associated with poor overall survival. Cancer Res.. 65, 8530–8537. [DOI] [PubMed] [Google Scholar]
- Ricci-Vitiani, L. , Lombardi, D.G. , Pilozzi, E. , Biffoni, M. , Todaro, M. , Peschle, C. , De Maria, R. , 2007. Identification and expansion of human colon-cancer-initiating cells. Nature. 445, 111–115. [DOI] [PubMed] [Google Scholar]
- Rubio-Viqueira, B. , Jimeno, A. , Cusatis, G. , Zhang, X. , Iacobuzio-Donahue, C. , Karikari, C. , Shi, C. , Danenberg, K. , Danenberg, P.V. , Kuramochi, H. , Tanaka, K. , Singh, S. , Salimi-Moosavi, H. , Bouraoud, N. , Amador, M.L. , Altiok, S. , Kulesza, P. , Yeo, C. , Messersmith, W. , Eshleman, J. , Hruban, R.H. , Maitra, A. , Hidalgo, M. , 2006. An in vivo platform for translational drug development in pancreatic cancer. Clin. Cancer Res.. 12, 4652–4661. [DOI] [PubMed] [Google Scholar]
- Santagata, S. , Demichelis, F. , Riva, A. , Varambally, S. , Hofer, M.D. , Kutok, J.L. , Kim, R. , Tang, J. , Montie, J.E. , Chinnaiyan, A.M. , Rubin, M.A. , Aster, J.C. , 2004. JAGGED1 expression is associated with prostate cancer metastasis and recurrence. Cancer Res.. 64, 6854–6857. [DOI] [PubMed] [Google Scholar]
- Schatton, T. , Murphy, G.F. , Frank, N.Y. , Yamaura, K. , Waaga-Gasser, A.M. , Gasser, M. , Zhan, Q. , Jordan, S. , Duncan, L.M. , Weishaupt, C. , Fuhlbrigge, R.C. , Kupper, T.S. , Sayegh, M.H. , Frank, M.H. , 2008. Identification of cells initiating human melanomas. Nature. 451, 345–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikandar, S.S. , Pate, K.T. , Anderson, S. , Dizon, D. , Edwards, R.A. , Waterman, M.L. , Lipkin, S.M. , 2010. NOTCH signaling is required for formation and self-renewal of tumor-initiating cells and for repression of secretory cell differentiation in colon cancer. Cancer Res.. 70, 1469–1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh, S.K. , Hawkins, C. , Clarke, I.D. , Squire, J.A. , Bayani, J. , Hide, T. , Henkelman, R.M. , Cusimano, M.D. , Dirks, P.B. , 2004. Identification of human brain tumour initiating cells. Nature. 432, 396–401. [DOI] [PubMed] [Google Scholar]
- Subramanian, A. , Tamayo, P. , Mootha, V.K. , Mukherjee, S. , Ebert, B.L. , Gillette, M.A. , Paulovich, A. , Pomeroy, S.L. , Golub, T.R. , Lander, E.S. , Mesirov, J.P. , 2005. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U.S.A.. 102, 15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Es, J.H. , van Gijn, M.E. , Riccio, O. , van den Born, M. , Vooijs, M. , Begthel, H. , Cozijnsen, M. , Robine, S. , Winton, D.J. , Radtke, F. , Clevers, H. , 2005. Notch/gamma-secretase inhibition turns proliferative cells in intestinal crypts and adenomas into goblet cells. Nature. 435, 959–963. [DOI] [PubMed] [Google Scholar]
- Vasiliou, V. , Pappa, A. , Estey, T. , 2004. Role of human aldehyde dehydrogenases in endobiotic and xenobiotic metabolism. Drug Metab. Rev.. 36, 279–299. [DOI] [PubMed] [Google Scholar]
- Zhang, Y. , Li, B. , Ji, Z.Z. , Zheng, P.S. , 2010. Notch1 regulates the growth of human colon cancers. Cancer. 116, 5207–5218. [DOI] [PubMed] [Google Scholar]
- Zhou, B.B. , Zhang, H. , Damelin, M. , Geles, K.G. , Grindley, J.C. , Dirks, P.B. , 2009. Tumour-initiating cells: challenges and opportunities for anticancer drug discovery. Nat. Rev. Drug Discov.. 8, 806–823. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The following are the Supplementary data related to this article:
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data


