Ca2+-dependent RNA–protein interactions enable activity-inducible RNA transport in dendrites.
Abstract
A key determinant of neuronal functionality and plasticity is the targeted delivery of select ribonucleic acids (RNAs) to synaptodendritic sites of protein synthesis. In this paper, we ask how dendritic RNA transport can be regulated in a manner that is informed by the cell’s activity status. We describe a molecular mechanism in which inducible interactions of noncanonical RNA motif structures with targeting factor heterogeneous nuclear ribonucleoprotein (hnRNP) A2 form the basis for activity-dependent dendritic RNA targeting. High-affinity interactions between hnRNP A2 and conditional GA-type RNA targeting motifs are critically dependent on elevated Ca2+ levels in a narrow concentration range. Dendritic transport of messenger RNAs that carry such GA motifs is inducible by influx of Ca2+ through voltage-dependent calcium channels upon β-adrenergic receptor activation. The combined data establish a functional correspondence between Ca2+-dependent RNA–protein interactions and activity-inducible RNA transport in dendrites. They also indicate a role of genomic retroposition in the phylogenetic development of RNA targeting competence.
Introduction
Neurons rely on the targeted delivery of diverse RNAs to synaptodendritic microdomains for locally controlled protein synthesis (Wells and Fallon, 2000; Job and Eberwine, 2001; Smith, 2004; Kindler et al., 2005; Darnell, 2011). RNA transport mechanisms are therefore critical underpinnings of local protein synthetic competence and, consequently, of synaptic functionality and plasticity (Miyashiro et al., 2009; Buckley et al., 2011; Darnell, 2011; Doyle and Kiebler, 2011). Dysregulated dendritic RNA transport has been implicated in the manifestation of neurological disease (Jin et al., 2007; Mus et al., 2007; Swanson and Orr, 2007; Oostra and Willemsen, 2009; Muslimov et al., 2011).
Dendritically targeted RNAs include mRNAs and regulatory RNAs (Eberwine et al., 2002; Smith, 2004; Kindler et al., 2005; Qureshi and Mehler, 2012; Iacoangeli and Tiedge, 2013). Dendritic mRNAs encode a variety of synaptodendritic proteins, including postsynaptic receptors, kinases, and cytoskeletal elements (Miyashiro et al., 2009; Darnell, 2011). Some of these mRNAs are delivered to dendrites in an activity-dependent manner (Tongiorgi et al., 1997, 2004; Dictenberg et al., 2008), suggesting that neuronal stimulation can lead to increased availability of such mRNAs for local translation at the synapse. Dendritic regulatory RNAs include miRNAs (Kim et al., 2004; Kosik and Krichevsky, 2005; Schratt et al., 2006; Muddashetty et al., 2011) and brain cytoplasmic (BC) RNAs (Muslimov et al., 1997, 1998, 2006, 2011). Regulatory BC RNAs are translational repressors that target translation initiation by interacting with eukaryotic initiation factors (Iacoangeli and Tiedge, 2013). Translational control of gene expression is thus performed in synaptodendritic domains by regulatory RNAs that are selectively delivered to such sites (Cao et al., 2006; Iacoangeli and Tiedge, 2013).
Dendritic RNA targeting enables postsynaptic microdomains to retain relevant mRNAs for local translation. The dynamic modulation of dendritic RNA delivery is therefore a key specificity determinant for local mRNA availability and thus for the local management of synaptic protein repertoires. However, our understanding of dendritic RNA delivery mechanisms is underdeveloped as important questions have remained unanswered. At the most fundamental level, we need to (a) decipher spatial targeting codes that dendritic RNAs use to specify their destinations, (b) understand how such codes are recognized and interpreted into dendritic targeting, and (c) establish how targeting codes mediate dendritic RNA transport that is induced by neuronal activity. Despite recent progress in some of these areas (Dictenberg et al., 2008; Buckley et al., 2011; Darnell, 2011), the molecular basis of activity-dependent dendritic RNA targeting has remained elusive.
Here, we describe a novel molecular mechanism for the stimulus-inducible delivery of neuronal RNAs to dendrites. Key to targeting conditionality is the organization of noncanonical motif interactions in the RNA targeting element. We show that such noncanonical conditional targeting elements specify dendritic delivery that is inducible by neuronal stimulation and receptor activation. Underlying the conditional delivery mechanism is the Ca2+-dependent recognition of targeting motifs by the RNA transport factor heterogeneous nuclear RNP (hnRNP) A2. Such conditional targeting motifs are often encoded in mobile genomic elements of the retroposon type, and our work therefore also implicates genomic retroposition as an innovative force in the evolution of plastic cellular mechanisms in neurons.
Results
Rationale
The dendritic targeting of regulatory BC RNAs is constitutive and specified by spatial codes that use noncanonical motif interactions (Muslimov et al., 1998, 2006, 2011). Noncanonical RNA motifs are those featuring nucleotide interactions that are distinct from Watson–Crick (WC) base pairing (Leontis and Westhof, 2003; Lescoute et al., 2005). BC RNA noncanonical targeting motifs are known as GA motifs as they are built around a core of two purine•purine base pairs of the G•A/A•G type (Muslimov et al., 2006, 2011).
BC RNAs are related to recurrent genomic elements via the retroposition mechanism (Iacoangeli and Tiedge, 2013). The dissemination of such elements, known as retroposons or retrotransposons, has been a major hallmark and driving force of mammalian phenotypic development and innovation (Brosius, 1991; Herbert, 2004; Kazazian, 2004; Cordaux and Batzer, 2009). It is estimated that the retroposition mechanism has been responsible for the generation of more than two thirds of the human genomic content (Brosius, 1999; de Koning et al., 2011). A retroposon is generated by the reverse transcription of an RNA or RNA segment followed by reinsertion into the genome. If such events occur multiple times, large numbers of retroposons can potentially be generated. Depending on the point of insertion, each newly generated retroposon can have neutral, deleterious, or beneficial cellular consequences. In the case of regulatory BC1 RNA, retroposons generated from the 5′ stem-loop domain have been dispersed in rodent genomes where they typically reside in UTRs (e.g., intergenic regions, introns, 3′ UTRs, and 5′ UTRs). Such 5′ BC1 retroposons are known as identifier (ID) elements (Kim et al., 1994). Because the 5′ BC1 domain harbors a noncanonical GA targeting motif, we reasoned that a 5′ BC1-derived ID element, inserted for instance in the 3′ UTR of a protein-coding gene, will confer dendritic targeting competence on the respective mRNA. Consistent with this notion, intron-residing ID elements have recently been found to be targeting competent if retained in cytoplasmic transcripts (Buckley et al., 2011).
Activity-dependent targeting
ID elements, retroposed from the 5′ BC1 domain, are categorized in four subtypes in rat (ID1–ID4; Fig. 1; Kim et al., 1994). We generated chimeric reporter mRNAs in which ID elements of each of the four subtypes were introduced 3′ to the coding region of α-tubulin mRNA, an mRNA that is normally restricted to neuronal somata (Bruckenstein et al., 1990; Mohr et al., 2001; Muslimov et al., 2011). While ID1 is identical to the 5′ BC1 domain, ID2 differs from the 5′ BC1 domain only inasmuch as a G·U wobble WC pair in the basal stem has been replaced with a G=C standard WC pair (Fig. 1). In either case, the noncanonical GA targeting motif is identical to the one contained in the 5′ BC1 domain, and we therefore anticipated that reporter mRNAs that harbor ID1 or ID2 elements would be constitutively delivered to dendrites. This was indeed observed in RNA transport assays (Fig. 2, A–D and I) in which we assessed RNA transport competence using our standard microinjection protocol with sympathetic neurons in primary culture (Muslimov et al., 1997, 2006, 2011).
Figure 1.
Secondary structures of the 5′ BC1 domain and ID1–ID4 elements. Rat consensus sequences (Kim et al., 1994) are shown. Secondary structure representation is based on previous evidence (Rozhdestvensky et al., 2001; Muslimov et al., 2006, 2011). Base pairs are denoted as follows: =, GC standard WC; –, AU standard WC, ·, GU wobble WC; •, noncanonical (non-WC). GA motif structures are bracketed by vertical bars. Noncanonical purine•purine base pairs (A•A, A•G, G•A, and G•G) are shown in red. GA motif WC base pairs clamping the noncanonical GA motif core are shown in blue. Differences between ID elements and the 5′ BC1 domain are indicated by arrows. The 5′ BC1 domain and ID1 are identical; 5′ BC1 nucleotide numbering is shown.
Figure 2.
Dendritic transport of ID-chimeric α-tubulin mRNAs under basal conditions. Sympathetic neurons in primary culture (nonstimulated) were microinjected with chimeric reporter mRNAs as noted in the images. Chimeric mRNAs contained the α-tubulin protein-coding sequence, an ID element, and an A98 poly(A) tail, as indicated. (A–H) Photomicrographs show subcellular localization of injected RNAs. In this figure (and in most of the following figures showing photomicrographs), dark-field images are on the left, and corresponding phase-contrast photomicrographs are depicted on the right. RNA signal appears as white silver grains in dark-field photomicrographs. Chimeric mRNAs containing ID1 or ID2 elements (A–D) were targeted to distal dendritic domains, whereas chimeric mRNAs containing ID3 or ID4 elements (E–H) were not. Number of cells analyzed: (A and B) 12 neurons, 41 dendrites; (C and D) 15 neurons, 49 dendrites; (E and F) 21 neurons, 79 dendrites; (G and H) 17 neurons, 58 dendrites. Bar, 50 µm. (I) Quantitative analysis. One-way analysis of variance (ANOVA), Dunnett’s post hoc analysis (comparison of levels of ID1-chimeric α-tubulin mRNA with levels of other ID-chimeric α-tubulin mRNA forms): comparison with ID2-chimeric α-tubulin mRNA (C and D), P > 0.5 for interval points 50 µm; comparison with ID3-chimeric α-tubulin mRNA (E and F), P < 0.001 for all interval points; comparison with ID4-chimeric α-tubulin mRNA (G and H), P < 0.001 for all interval points. Quantitative data in this and the following figures are given in the format means ± SEM; error bars indicate SEM.
ID3 and ID4, in comparison with ID1 and ID2, feature additional noncanonical content (G•G or A•G, respectively) that is directly juxtaposed with the GA targeting motif at relative position 35–41 in the apical stem-loop domain (Fig. 1). Surprisingly, and in contrast to chimeric mRNAs carrying ID1 or ID2 elements, chimeric mRNAs carrying ID3 or ID4 elements were not delivered to dendrites (Fig. 2, E–H and I). We conclude that the constitutive targeting competence of the 5′ BC1 domain is retained in ID1 and ID2, but not in ID3 or ID4, elements.
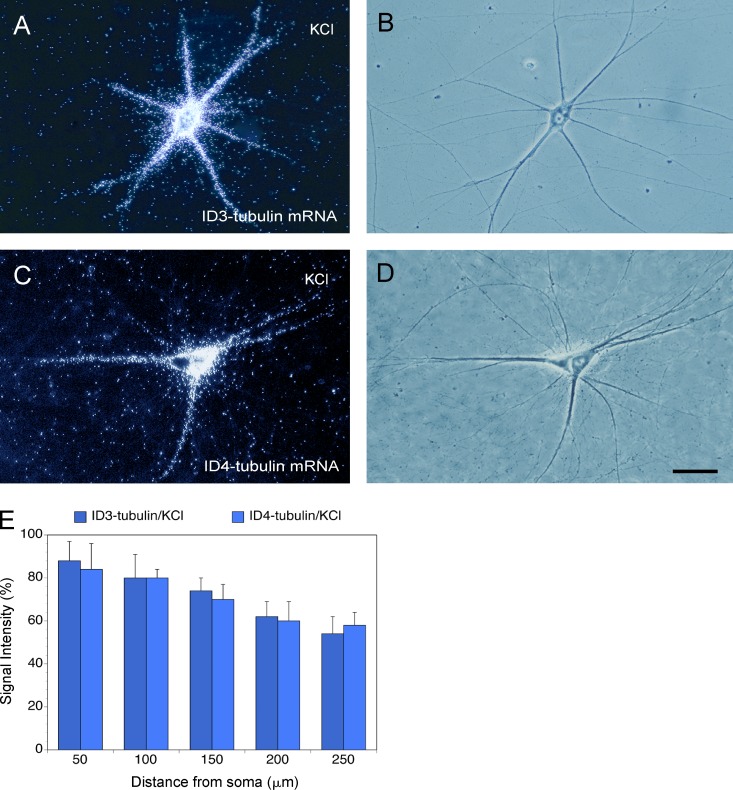
We hypothesized, as will be detailed in the following paragraphs, that conversion of the base pair at relative position 35–41 (Fig. 1) from canonical 35G=C41 (5′ BC1, ID1, and ID2) to noncanonical 35G•G41 (ID3) or 35A•G41 (ID4) will alter, but not abolish, the dendritic coding competence of the adjoining GA motif. Because the aforementioned experiments were conducted with neurons under basal steady-state (resting) conditions, we now asked whether ID3 or ID4 chimeric mRNAs are targeted to dendrites upon neuronal stimulation. To this end, we used elevated K+ to depolarize sympathetic neurons in culture, as previously described (Sun et al., 1992; Banker and Goslin, 1998; Habecker et al., 2006; see also Materials and methods). Fig. 3 shows that upon K+-induced depolarization, ID3 and ID4 α-tubulin chimeric mRNAs were indeed delivered to dendrites, and resulting somatodendritic distributions were indistinguishable from that of ID1 (5′ BC1) and ID2 α-tubulin chimeric mRNA under nonstimulated (basal) steady-state conditions. The data indicate that a noncanonical purine•purine (R•R) base pair adjoining a noncanonical GA targeting motif can render dendritic transport conditional. We refer to such motifs as R•R-linked GA targeting motifs, and we suggest that such motifs define a class of mRNAs that are conditionally targeted to dendrites.
Figure 3.
Dendritic transport of ID3- and ID4-chimeric mRNAs in stimulated neurons. (A–D) Sympathetic neurons in culture were K+ depolarized before RNA microinjection. Under these conditions, ID3 (A and B)- and ID4 (C and D)-chimeric α-tubulin mRNAs were delivered to distal dendrites. Number of cells analyzed: (A and B) 16 neurons, 55 dendrites; (C and D) 18 neurons, 60 dendrites. Bar, 50 µm. (E) Quantitative analysis. One-way ANOVA, Dunnett’s post hoc analysis (comparison of RNA levels in the basal, nonstimulated state [Fig. 2] with RNA levels after K+ depolarization): ID3-chimeric α-tubulin mRNA (A and B), P < 0.001 for all interval points; ID4-chimeric α-tubulin mRNA (C and D), P < 0.001 for all interval points. Error bars indicate SEM.
In control experiments, we used a chimeric mRNA with a reverse complementary ID4 element (ID4R), resulting in a configuration that is antisense to ID4. In ID4R elements, standard WC base pairs are retained (e.g., conversion of G=C to C=G), whereas noncanonical pairs are disrupted (e.g., G•A to C U), thus resulting in a stem loop that lacks a GA motif. No significant dendritic targeting was observed with ID4R α-tubulin mRNA in either basal or stimulated states (Fig. S1).
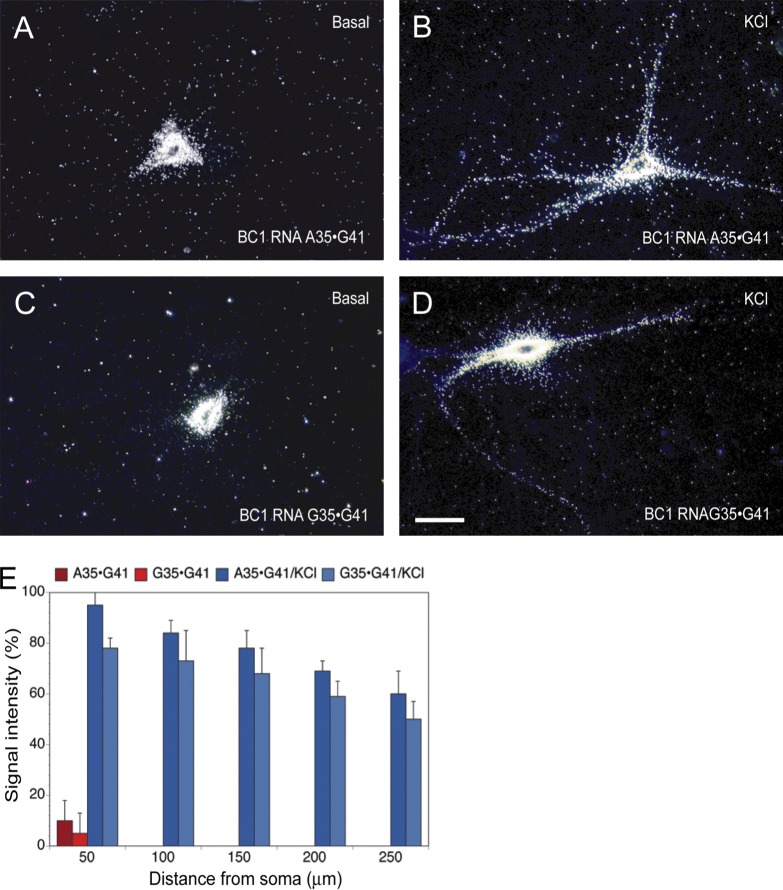
We performed a further set of experiments to test the hypothesis that noncanonical interactions at relative position 35–41 render dendritic targeting conditional. We asked whether the constitutive dendritic targeting competence of wild-type (WT) BC1 RNA can be altered to conditional, i.e., activity dependent, by introducing ID-type point mutations. We converted, in full-length BC1 RNA, WT canonical base pair G35=C41 to mutant noncanonical variant G35•G41 or A35•G41, the former corresponding to ID3 and the latter corresponding to ID4 (Kim et al., 1994). We found that G35•G41 and A35•G41 mutant BC1 RNAs both lack dendritic targeting competence under basal conditions but are delivered along the entire dendritic extent upon depolarization (Fig. 4). In contrast, conversion of wobble WC pairs to standard WC pairs in the basal stem, as indicated in Fig. 1, did not alter the constitutive targeting competence of BC1 RNA (not depicted). The data confirm that noncanonical attributes in relative position 35–41 are determinants of targeting conditionality.
Figure 4.
Mutant BC1 RNA in conditional dendritic delivery. (A–D) WT BC1 RNA (G35=C41) is constitutively targeted to dendrites of sympathetic neurons in culture (Muslimov et al., 1997, 2006, 2011). (A and C) In contrast, mutant A35•G41 BC1 RNA and mutant G35•G41 BC1 RNA were not transported under basal conditions. (B and D) Both mutant BC1 RNA forms were, however, delivered to dendrites upon K+ depolarization. Number of cells analyzed: (A) 18 cells, 57 dendrites; (B) 17 cells, 56 dendrites; (C) 16 cells, 52 dendrites; (D) 16 cells, 44 dendrites. Bar, 50 µm. (E) Quantitative analysis. One-way ANOVA, Dunnett’s post hoc analysis (comparison of levels of mutant BC1 RNAs in the basal state with levels after K+ depolarization): BC1 RNA A35•G41 (A and B), P < 0.001 for all interval points; BC1 RNA G35•G41 (C and D), P < 0.001 for all interval points. Error bars indicate SEM.
Dendritic targeting induced by receptor activation
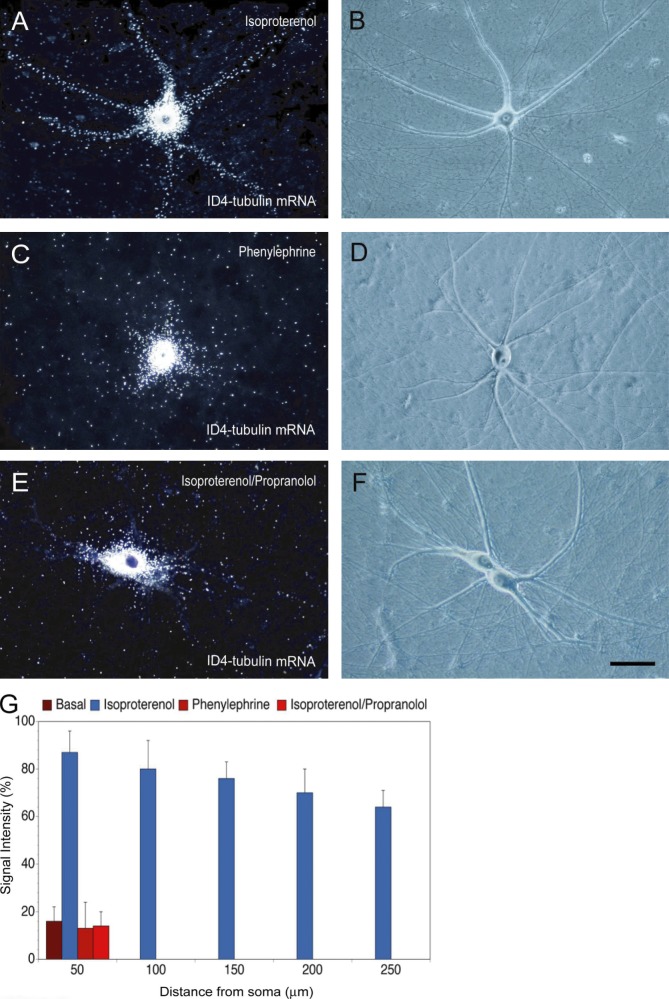
The aforementioned data on dendritic delivery after depolarization raise the question whether transport can also be induced by specific receptor activation. Noting that sympathetic neurons use adrenergic transmission (Landis, 1990), we used adrenergic agonists and antagonists to address this question. ID4 α-tubulin mRNA, which is not targeted to dendrites under basal steady-state conditions (Fig. 2), was delivered along the entire dendritic extent upon activation of β-adrenergic receptors (ARs) using the β-AR agonist isoproterenol (Fig. 5, A, B, and G). In contrast, ID4 α-tubulin mRNA remained restricted to somata after application of the α-adrenergic agonist phenylephrine (Fig. 5, C, D, and H). Induction of dendritic transport by isoproterenol was blocked by the β-adrenergic antagonist propranolol (Fig. 6, E, F, and H). Analogous results were obtained with ID3-mediated conditional dendritic transport (unpublished data). The data indicate that ID3- and ID4-mediated conditional dendritic transport is specifically induced by activation of β-ARs.
Figure 5.
Dendritic transport of ID4-chimeric mRNA after β-adrenergic activation. (A and B) After β-adrenergic activation with the agonist isoproterenol, ID4-chimeric α-tubulin mRNA was delivered to distal dendrites of sympathetic neurons. (C and D) In contrast, after α-adrenergic activation with the agonist phenylephrine, ID4-chimeric α-tubulin mRNA was not transported to dendrites to any substantial extent. (E and F) Application of the β-AR antagonist propranolol before application of agonist isoproterenol blocked the isoproterenol-induced dendritic delivery of ID4-chimeric α-tubulin mRNA. Concentrations of agonists, antagonists, and other drugs are given in Materials and methods. Number of cells analyzed: (A and B) 16 cells, 52 dendrites; (C and D) 16 cells, 57 dendrites; (E and F) 14 cells, 50 dendrites. Bar, 50 µm. (G) Quantitative analysis. One-way ANOVA, Dunnett’s post hoc analysis (comparison of RNA levels in the basal state [Fig. 2] with RNA levels after adrenergic activation/blockade): comparison with phenylephrine (A and B), P > 0.7 for interval points 50 µm; comparison with isoproterenol (C and D), P < 0.001 for all interval points; comparison with isoproterenol/propranolol (E and F), P > 0.8 for interval points 50 µm. Error bars indicate SEM.
Figure 6.
Conditional dendritic transport of CLN2 mRNA. (A and B) ID3-containing CLN2 mRNA was not targeted to dendrites of sympathetic neurons under basal conditions. (C–F) CLN2 mRNA was delivered to dendrites after β-AR activation with agonist isoproterenol (C and D) but not after α-adrenergic activation with agonist phenylephrine (E and F). Number of cells analyzed: (A and B) 19 cells, 72 dendrites; (C and D) 17 cells, 63 dendrites; (E and F) 18 cells, 66 dendrites. Bar, 50 µm. (G) Quantitative analysis. One-way ANOVA, Dunnett’s post hoc analysis (comparison of RNA levels in the basal state with RNA levels after adrenergic activation): comparison with isoproterenol (C and D), P < 0.001 for all interval points; comparison with phenylephrine (E and F), P > 0.8 for interval points 50 µm. Error bars indicate SEM.
We next asked whether inducible, activity-dependent dendritic transport can also be observed with naturally occurring mRNAs that contain ID3 or ID4 elements. As a representative of such mRNAs, we chose rat CLN2 (ceroid lipofuscinosis, neuronal 2; Jalanko and Braulke, 2009) mRNA, which contains an ID3 element in its 3′ UTR (3′ UTR nt 478–552; GenBank accession no. NM_031357). We found that this mRNA was not delivered to dendrites under basal steady-state conditions (Fig. 6, A, B, and G). However, CLN2 mRNA was transported along the dendritic extent upon K+-induced depolarization (not depicted) and upon activation of β-ARs with isoproterenol (Fig. 6, C, D, and G). In contrast, activation of α-ARs with phenylephrine did not induce dendritic delivery (Fig. 6, E, F, and G). The results further corroborate the notion that R•R-linked GA targeting motifs encode conditional dendritic transport competence as activity-dependent transport can be observed with chimeric reporter mRNAs and with naturally occurring mRNAs containing such motifs.
Conditional recognition by targeting factor hnRNP A2
We have previously shown that hnRNP A2 is a transport factor that recognizes the BC1 RNA GA targeting motif and that this interaction is necessary for the delivery of the RNA to dendrites (Muslimov et al., 2006, 2011). Now, we asked whether conditional dendritic transport, mediated by ID3/ID4-type R•R-linked GA targeting motifs, equally relies on targeting factor hnRNP A2 and whether in these cases conditional recognition of such motifs by hnRNP A2 is the basis for conditional, activity-dependent dendritic targeting.
In addressing these questions, we took into consideration that β-adrenergic downstream modes of action in neurons involve PKA-mediated stimulation of Ca2+ influx through dendritic L-type voltage-dependent calcium channels (VDCCs; Davare et al., 2001; Bloodgood and Sabatini, 2007; Hall et al., 2007; Hell, 2010). In view of these data, the possibility is raised that binding of ID3/ID4 GA targeting motifs by hnRNP A2 is Ca2+ dependent. We performed quantitative electrophoretic mobility shift assay (EMSA) analysis, as previously described (Muslimov et al., 2006, 2011), to assess this possibility.
EMSA analysis revealed that hnRNP A2 binds to ID4 GA targeting motif RNA and that this binding is dynamically modulated by Ca2+ levels (Fig. 7). Binding is robust at 500 nM Ca2+ but significantly less so at Ca2+ levels above or below this optimum, thus defining a window of effective Ca2+ concentrations. Mg2+ was found ineffective in promoting binding except at concentrations of 1 mM or above (Fig. S2). These results differ from those obtained with BC1 RNA as in the latter case, binding to hnRNP A2 is promoted by Ca2+ and Mg2+ in the millimolar concentration range (Muslimov et al., 2011). In contrast, for ID4 RNA, binding is promoted by Ca2+, but not by Mg2+, in the nanomolar concentration range. We next asked how Ca2+ levels impact the equilibrium dissociation constant (Kd) for the interaction between hnRNP A2 and the ID4 GA targeting motif. To address this question, we titrated the ID4 GA targeting motif with increasing concentrations of hnRNP A2 at three levels of Ca2+ concentrations, selected according to the results shown in Fig. 7 (A and B): 100 nM Ca2+, 500 nM Ca2+, and 2 µM Ca2+. Quantitative EMSA analysis was performed at each of these Ca2+ concentrations, and the data obtained were fitted to the Hill equation (Ryder et al., 2008; Chao et al., 2010; Muslimov et al., 2011). The following equilibrium dissociation constants were obtained: Kd = 280 nM at 100 nM Ca2+, Kd = 200 pM at 500 nM Ca2+, and Kd = 580 nM at 2 µM Ca2+. Equilibrium dissociation constants could not be established at Ca2+ concentrations <20 nM or >10 µM as no significant binding was detectable.
Figure 7.
Ca2+-dependent interaction of ID4 mRNA with hnRNP A2. In EMSA experiments using native PAGE, shifts to lower mobility reveal binding of radiolabeled RNAs to recombinant hnRNP A2. (A and B) Mobility shifts indicate that binding of ID4-chimeric α-tubulin mRNA to hnRNP A2 was dependent on Ca2+ levels. Maximal binding was observed at 500 nM Ca2+. Original data are shown on the left (A), and combined results from five experiments are shown in the diagram on the right (B). (C–H) Equilibrium binding constants were established in a series of experiments in which ID4-chimeric α-tubulin mRNA was titrated with increasing concentrations of hnRNP A2 at different concentrations of Ca2+. Original data are shown on the left (C, E, and G). The binding data were fitted to the Hill equation as previously described (Ryder et al., 2008; Chao et al., 2010; Muslimov et al., 2011) and were plotted in binding curves on the right (D, F, and H). Equilibrium dissociation constants were as follows: Kd = 280 nM at 100 nM Ca2+ (D), Kd = 200 pM at 500 nM Ca2+ (F), and Kd = 580 nM at 2 µM Ca2+ (H). Error bars indicate SEM.
The results show that binding of hnRNP A2 to the ID4 GA targeting motif is of remarkably high affinity at 500 nM Ca2+. The affinity is significantly lower (by more than three orders of magnitude) at 100 nM Ca2+ and 5 µM Ca2+, and little binding was detectable below the former or above the latter Ca2+ concentrations. We conclude that binding of hnRNP A2 to the ID4 GA motif occurs only in a narrowly demarcated Ca2+ concentration window around 500 nM.
Ca2+-dependent dendritic delivery
Because the interaction of ID4 GA motif RNA with targeting factor hnRNP A2 is Ca2+ dependent, we hypothesized that the dendritic transport of ID4 RNA would also be a function of Ca2+ concentrations. In neurons, intracellular Ca2+ levels ([Ca2+]i) are low at 50–100 nM in the resting state but can transiently rise to 500–1,000 nM upon activation (Berridge et al., 2000; Grienberger and Konnerth, 2012). Even higher transient levels may occur locally (Neher, 1998; Ross, 2012). It was thus conceivable that for ID4 GA motif RNA, induction of dendritic targeting requires transient elevation of [Ca2+]i. We performed two sets of experiments to test this possibility.
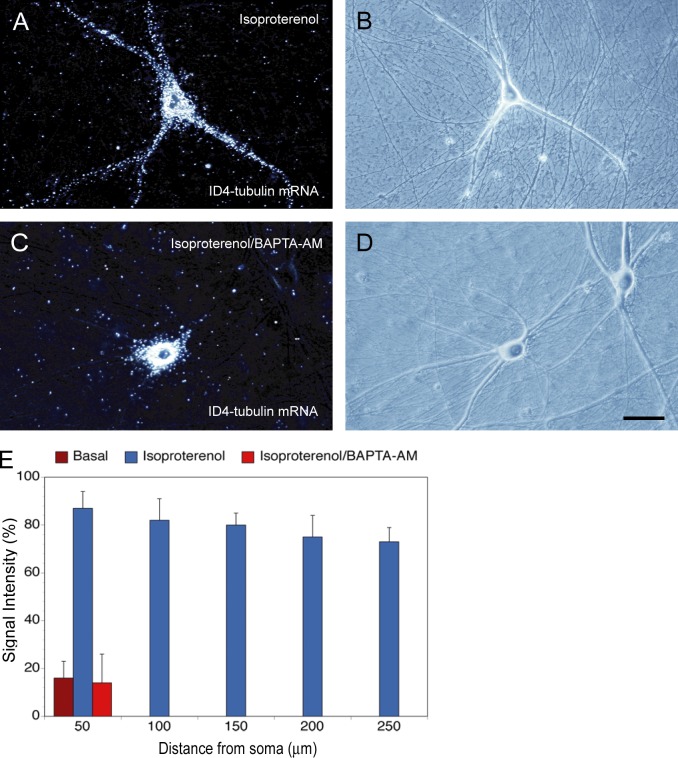
First, we induced dendritic transport of ID4 α-tubulin chimeric RNA with β-AR agonist isoproterenol, as shown in Fig. 5. In parallel experiments, isoproterenol was applied after preincubation with BAPTA-AM, a standard internal Ca2+ chelator (Bofill-Cardona et al., 2000; Paschen et al., 2003). Fig. 8 shows that isoproterenol-induced dendritic delivery of ID4 α-tubulin chimeric RNA was blocked by BAPTA-AM. Reduced ID4 GA motif targeting was also observed when BAPTA-AM was used in conjunction with neuronal depolarization (not depicted). The data suggest that intracellular Ca2+ is required for the β-adrenergic induction of ID4 GA motif conditional targeting.
Figure 8.
Ca2+-dependent dendritic transport of ID4 mRNA. (A and B) ID4-chimeric α-tubulin mRNA was delivered to dendrites of sympathetic neurons after β-adrenergic activation with isoproterenol. (C and D) In contrast, little dendritic transport was observed when the β-adrenergic agonist isoproterenol was applied after preincubation with the intracellular Ca2+ chelator BAPTA-AM. Number of cells analyzed: (A and B) 16 cells, 67 dendrites; (C and D) 15 cells, 60 dendrites. Bar, 50 µm. (E) Quantitative analysis. One-way ANOVA, Dunnett’s post hoc analysis (comparison of RNA levels in the basal state [Fig. 2] with RNA levels after β-adrenergic activation and with levels after β-adrenergic activation in the presence of BAPTA-AM): comparison with isoproterenol (A and B), P < 0.001 for all interval points; comparison with isoproterenol/BAPTA-AM (C and D), P > 0.8 for interval points 50 µm. Error bars indicate SEM.
β-Adrenergic signaling in dendrites entails influx of Ca2+ through the L-type VDCC Cav1.2 (Davare et al., 2001; Hoogland and Saggau, 2004; Bloodgood and Sabatini, 2007; Hall et al., 2007). Therefore, in a second set of experiments, we activated β-ARs in the presence of L-type VDCC blockers nimodipine/nifedipine. Fig. 9 shows that isoproterenol-induced dendritic delivery of ID4 α-tubulin chimeric RNA was significantly reduced in the presence of nimodipine (Fig. 9, C and D) or nifedipine (Fig. 9, E and F). To examine the possibility that chelation of intracellular Ca2+ by BAPTA-AM, or blockade of Ca2+ influx through L-type VDCCs, may have caused a general defect in dendritic RNA targeting, we investigated dendritic transport of PKMζ mRNA under such conditions. We have previously shown that PKMζ mRNA is constitutively targeted to dendrites and that a standard GA motif is responsible for distal dendritic delivery (Muslimov et al., 2004). We now find that dendritic targeting of PKMζ mRNA is not stimulated after β-adrenergic activation and is unaltered in the presence of BAPTA-AM or nifedipine (Fig. S3). These results suggest that β-adrenergic stimulation and Ca2+ dependence are characteristics of dendritic ID4 GA motif targeting but not of dendritic GA motif targeting in general. In summary, the aforementioned data indicate that dendritic transport of ID4 GA motif RNA is induced by β-adrenergic activation via influx of Ca2+ through L-type VDCCs.
Figure 9.
Conditional dendritic targeting dependent on influx of Ca2+ through VDCCs. (A and B) ID4-chimeric α-tubulin mRNA was delivered to dendrites of sympathetic neurons after β-adrenergic activation. (C–F) Dendritic transport was significantly reduced if β-adrenergic activation occurred in the presence of L-type VDCC blocker nimodipine (C and D) or nifedipine (E and F). Number of cells analyzed: (A and B) 16 cells, 57 dendrites; (C and D) 19 cells, 80 dendrites; (E and F) 20 cells, 78 dendrites. Bar, 50 µm. (G) Quantitative analysis. One-way ANOVA, Dunnett’s post hoc analysis (comparison of RNA levels after β-adrenergic activation with RNA levels after β-adrenergic activation in the presence of nimodipine or nifedipine): comparison with nimodipine (C and D), P < 0.001 for all interval points; comparison with nifedipine (E and F), P < 0.01 for all interval points. Error bars indicate SEM.
Activity-dependent dendritic delivery of endogenous CLN2 mRNA
The aforementioned data raise the question whether β-adrenergic activation will also induce Ca2+-dependent dendritic targeting of native (i.e., endogenous) ID3/ID4-carrying neuronal mRNAs. To address this question, we performed in situ hybridization (Muslimov et al., 1998, 2011) with probes specific for ID3-carrying rat CLN2 mRNA. These experiments were performed with both hippocampal (Muslimov et al., 1998) and sympathetic (Muslimov et al., 2011) neurons in culture.
In hippocampal neurons, β-adrenergic activation has been shown to stimulate Ca2+ influx through dendritic L-type VDCCs (Davare et al., 2001; Hoogland and Saggau, 2004; Bloodgood and Sabatini, 2007; Hall et al., 2007). We now asked whether β-adrenergic activation can induce dendritic delivery of endogenous CLN2 mRNA in hippocampal neurons in culture. Results shown in Fig. 10 indicate that this is indeed the case. Endogenous CLN2 mRNA remained restricted to neuronal somata under basal conditions (Fig. 10, A and B) but was found distributed along dendrites, from soma to distal dendritic tips after activation of β-ARs with agonist isoproterenol (Fig. 10, C and D). β-Adrenergic induction of CLN2 mRNA transport was prevented by intracellular Ca2+ chelation with BAPTA-AM (Fig. 10, E and F), by blockade of L-type VDCCs with nifedipine (Fig. 10, G and H) and by blocking β-ARs with antagonist propranolol (not depicted). Analogous results were obtained when in situ hybridization directed at endogenous CLN2 mRNA was performed with sympathetic neurons in culture (Fig. S4). These data establish that in both hippocampal and sympathetic neurons, the dendritic delivery of a native, ID3-carrying neuronal mRNA is inducible by β-adrenergic activation and is dependent on influx of Ca2+ through L-type VDCCs.
Figure 10.
Induction of endogenous CLN2 mRNA transport by β-adrenergic activation in hippocampal neurons. In situ hybridization was performed with a probe specific for rat CLN2 mRNA. (A and B) Hybridization signal (white silver grains) indicates somatic localization of endogenous CLN2 mRNA under basal conditions. (C and D) After activation of β-ARs with agonist isoproterenol, CLN2 mRNA was found localized along the entire dendritic extent. (E–H) Preincubation of cells with intracellular Ca2+ chelator BAPTA-AM (E and F) or with L-type VDCC blocker nifedipine (G and H) prevented isoproterenol-induced dendritic delivery of CLN2 mRNA. (I and J) Little or no signal was detectable when in situ hybridization was performed with a CLN2 mRNA “sense strand” control probe. Number of cells analyzed: (A and B) 14 neurons, 71 dendrites; (C and D) 14 neurons, 72 dendrites; (E and F) 11 neurons, 53 dendrites; (G and H) 12 neurons, 60 dendrites. Bar, 50 µm. (K) Quantitative analysis. One-way ANOVA, Dunnett’s post hoc analysis (comparison of RNA levels in the basal state with RNA levels after β-adrenergic activation and after β-adrenergic activation in the presence of BAPTA-AM or nifedipine): comparison with isoproterenol (C and D), P < 0.001 for all interval points; comparison with isoproterenol/BAPTA-AM (E and F), P > 0.6 for interval points 50 µm; comparison with isoproterenol/nifedipine (G and H), P > 0.7 for interval points 50 µm. Error bars indicate SEM.
Discussion
A wealth of new structural and functional data has over the last decade resulted in the identification of various novel classes of protein-engaging RNA motifs (Steitz and Moore, 2003; Noller, 2005; Leontis et al., 2006). Leading this development have been major advances in the field of ribosome structure and function (Noller, 2005; Grandin, 2010). As a result of these and related discoveries in RNA biology, it is now recognized that, except for exclusively protein-coding regions, it is often architectural RNA motifs—rather than nucleotide sequences per se—that are subject to natural selection in evolution (Leontis et al., 2006). Such motifs often serve as recognition sites for proteins, and their structural gestalt is typically determined by noncanonical nucleotide interactions (Leontis and Westhof, 2003; Noller, 2005; Leontis et al., 2006; Grandin, 2010).
Here, we report that a newly identified type of noncanonical RNA motif, the R•R-linked GA motif, encodes conditional dendritic targeting competence. The motif, which is contained in ID3 and ID4 elements, is organized around a core of noncanonical G•A/A•G pairs. This core in turn is clamped by WC pairs, typically of the G=C type. Directly adjoining this structure is a noncanonical purine•purine pair that is critical for targeting conditionality. Recognition of this motif by the targeting factor hnRNP A2 is strongly dependent on Ca2+ levels. At 500 mM Ca2+, the motif binds to hnRNP A2 with a uniquely high affinity of Kd = 200 pM. At Ca2+ levels above or below this optimal concentration, binding is significantly lower if at all detectable. GA motifs are known to be dimorphic as they exist in an equilibrium between an extended and a kinked conformation. Divalent cations promote transition to the latter conformation, which binds cognate proteins with high affinity (Matsumura et al., 2003; Goody et al., 2004; Cojocaru et al., 2005; Lescoute et al., 2005; Rázga et al., 2006; Falb et al., 2010; Schroeder et al., 2010). Binding to a cognate protein will in turn arrest the motif in the high-affinity conformation (Turner et al., 2005). However, although such binding dynamics have been described for standard GA motifs, a narrow-window Ca2+ dependence, as described here, appears to be a feature specific to R•R-linked GA motifs. It is possible that the GA-proximal additional R•R content introduces conformational motif flexibility that is Ca2+ driven. Although a formal test of this hypothesis will require structural approaches, we propose that motif affinity to hnRNP A2 is directly Ca2+ dependent and that Ca2+ levels in the 500-nM range promote transition from a low-affinity to the high-affinity conformation.
R•R-linked GA motifs are contained in ID3 and ID4 mRNAs. Working with ID3/ID4 chimeric reporter mRNAs as well as with a naturally occurring ID3 mRNA, we found that dendritic targeting is inducible by neuronal depolarization and by receptor activation. Specifically, β-adrenergic activation (but not α-adrenergic activation) induces long-range delivery along the entire dendritic extent. β-Adrenergic transport induction is dependent on influx of Ca2+ through L-type VDCCs. β-ARs have previously been implicated in neuronal function and plasticity (Huang and Kandel, 1996; Raman et al., 1996; Moncada et al., 2011). In hippocampal neurons, β-ARs have been shown to form signaling complexes with L-type VDCCs, G proteins, adenylyl cyclase, and PKA, indicating that localized signal transduction couples receptors to channels (Davare et al., 2001; Vásquez and Lewis, 2003; Hall et al., 2007). Ca2+ influx is thus a direct consequence of β-adrenergic activation. In both sympathetic and hippocampal neurons, our data indicate that native and introduced ID3/ID4-bearing mRNAs are targeted to dendrites after β-adrenergic activation. In both cell types, β-adrenergic induction of ID3/ID4 mRNA transport is dependent on influx of Ca2+ through L-type VDCCs.
A transient rise in postsynaptic [Ca2+]i can trigger intracellular Ca2+ waves that propagate along dendrites toward the soma, and these Ca2+ transients can reach [Ca2+]i amplitudes of 1 µM or more in both somata and dendrites (Berridge et al., 2000; Grienberger and Konnerth, 2012; Ross, 2012). Any such Ca2+ transient, once it reaches a local threshold of 500 nM, will cause a switch to the high-affinity conformation of the GA targeting motif and thus enable binding of hnRNP A2. The question is then raised how a rapid rise and fall in [Ca2+]i can lead to prolonged dendritic transport (which operates on a slower time frame). We surmise that binding of hnRNP A2 to the high-affinity motif conformation will in turn fix this conformation (as has previously been reported for GA motifs; Turner et al., 2005), as a result locking the protein on the RNA and thus supporting sustained transport even after local [Ca2+]i has returned to basal resting levels.
Retroposition is a mechanism of intragenomic transfer that has been a ubiquitous and innovative force in the remodeling of eukaryotic genomes (Brosius, 1991, 2005; Herbert, 2004; Kazazian, 2004; Cordaux and Batzer, 2009). Retroposition has been responsible for the genomic dissemination of numerous 5′ BC1-derived ID elements (Kim et al., 1994). A partial list of such elements has been established, and dendritic targeting has been confirmed for selected representatives (Buckley et al., 2011). In earlier work, in contrast, ID or BC1 chimeric RNAs were not seen localized to dendrites in a conventional transgenic mouse system (Khanam et al., 2007a). However, the expression of GA motif RNAs at high levels for prolonged periods of time, as is the case in such animals, can trigger cellular anti-dsRNA mechanisms (Kaufman, 2000; Pe’ery and Mathews, 2000). Potent inducers of such mechanisms, which likely have evolved as components of antiviral defense strategies, are noncanonical GA motifs (Bevilacqua et al., 1998). Of note, the HIV-1 (human immunodeficiency virus type 1) Rev response element contains a GA motif (Jain and Belasco, 1996) that is similar to those in BC RNAs.
For a dissection of dendritic targeting competence in naturally occurring ID-containing mRNAs, we chose as a representative CLN2 mRNA that contains in its 3′ UTR an ID3 targeting element with a G•G-linked GA motif. We established that this RNA is delivered to dendrites conditionally, i.e., upon induction by depolarization or β-AR activation. CLN2 mRNA encodes TPP1 (tripeptidyl peptidase 1), a soluble lysosomal serine protease (Jalanko and Braulke, 2009). Lysosomes are located to dendrites in cultured neurons and in vivo during neurodevelopment (Roberts and Gorenstein, 1987; Nixon and Cataldo, 1995; Lee et al., 2004, 2011), suggesting that associated RNA transport is a developmental mechanism. Reduced TPP1 activity is causative of late infantile neuronal ceroid lipofuscinosis (Jalanko and Braulke, 2009; Kollmann et al., 2013), and future work will investigate whether dysregulated RNA transport contributes to disease manifestations, such as ataxia and seizures.
Because the retroposition mechanism has been shaping the phylogenetic diversification of a large number of mammalian genes, we expect ID dissemination to be only one out of several retroposition dispersals that have provided neuronal genes with constitutive and conditional targeting codes. In primates, retroposons of the Alu-J type are homologous to the 5′ BC200 domain (Tiedge et al., 1993). BC200 RNA localizes to dendrites in human brain (Tiedge et al., 1993) and is also delivered to dendrites of rat cultured neurons after microinjection (Muslimov et al., 2011). BC200 RNA and G22 RNA (a prosimian Alu-related nonprotein-coding RNA) were also found localized in dendritic fields of transgenic mouse brains (Khanam et al., 2007b). BC200 RNA interacts with hnRNP A2 via a 5′ GA motif (Muslimov et al., 2011; unpublished data). Alu-J elements are highly similar to the 5′ BC200 domain, with the critical difference of additional GA-proximal R•R content (Tiedge et al., 1993; Skryabin et al., 1998). Such an R•R-linked GA motif is contained in human CLN2 mRNA, and we found that this RNA is delivered to dendrites in a conditional manner (unpublished data). We therefore posit that the constitutive BC1-conditional ID relationship is mirrored in an analogous BC200–Alu relationship. On the other hand, and in contrast to rat CLN2 mRNA, murine CLN2 mRNA appears to be lacking an ID element or other GA motifs that may serve as dendritic targeting elements. Species-specific acquisition of RNA targeting competence has previously been described by us (Cristofanilli et al., 2004) and by others (Buckley et al., 2011). The combined evidence provides further support for the notion that the functional recruitment of mobile elements may be a determinant in the evolutionary diversification of neural systems.
Conditional dendritic targeting can also be encoded by R•R-linked GA motifs that are not formal members of the ID or Alu retroposon families. Dendritic delivery of brain-derived neurotrophic factor mRNA and CaMKIIα mRNA is constitutive at low levels (Burgin et al., 1990; An et al., 2008) but can be induced by application of neuronal stimuli, as was shown by use of depolarization in the former and receptor activation in the latter case (Tongiorgi et al., 1997; Dictenberg et al., 2008). Both mRNAs contain GA motifs that, while sharing key attributes with ID GA motifs, feature two R•R pairs proximal to the GA core, rather than the single proximal R•R pair that is seen in ID3/ID4. In both cases, motif structure is conserved between rodent and primate mRNAs. While future work will address the significance of these GA motifs for inducible dendritic targeting, we propose that a common principle is at work in neuronal RNA transport: while noncanonical GA motifs serve as dendritic targeting elements, additional proximal R•R content renders targeting conditional.
The activity-dependent dendritic delivery of GA motif RNAs enables neurons to supply postsynaptic microdomains with requisite RNAs on demand. Work presented here indicates that a Ca2+-dependent switch to the high-affinity GA motif conformation promotes interaction with the targeting machinery and, consequently, sustained dendritic transport. How then is RNA cargo discharged upon arrival at a synaptodendritic destination? Although speculative at this time, it is tempting to hypothesize that docking of the RNA transport complex at a target site will trigger reversion to the low-affinity GA motif conformation and consequently dissolution of the RNA–protein targeting complex. We invite research to test this hypothesis.
Materials and methods
Plasmids and RNA preparation
WT BC1 RNA was transcribed from plasmid pBCX607 by T7 RNA polymerase after linearization with DraI (Muslimov et al., 2006, 2011). Analogously, the following plasmids, derived from pBCX607, were used to express mutant forms of BC1 RNA (respective mutations indicated by subscript): pBC1U5C, pBC1U67C, pBC1U5C/U67C, pBC1G35A, pBC1C41G, and pBC1G35A/C41G. ID-chimeric α-tubulin mRNAs were generated as follows. pTub-A98/TA2 was used to transcribe α-tubulin mRNA with an 98-nt poly(A) tail 3′ to the protein-coding sequence. An analogous, chimeric α-tubulin mRNA with an ID1 element (5′ BC1 domain) inserted between the protein-coding sequence and A98 was transcribed from pTID1D/T1, with an inserted ID2 element from pTID2D/T3-D, with an inserted ID3 element from pTID3D/T3, with an inserted ID4 element from pTID4D/T1-B, and with an inserted reverse complementary ID4 element from pTID4R/T2-13. These plasmids were constructed using the pBlueScript II SK+ RNA transcription kit (Agilent Technologies). They were linearized with XbaI, and transcripts were generated with SP6 RNA polymerase. CLN2 mRNA was generated from pBluescript-CLN2 (obtained from R. Crystal, Weill Cornell Medical College, New York, NY; Sondhi et al., 2005). The plasmid was linearized with SacI and transcribed with T7 RNA polymerase. For in situ hybridization, a CLN2 mRNA segment (nt 327–822) was recloned into pBluescript II SK between the SmaI and XbaI sites to give pBluescript-CLN2is. pET-9c/A2 was used to express recombinant full-length hnRNP A2 (obtained from R. Smith, University of Queensland, Brisbane, Queensland, Australia; Munro et al., 1999; Muslimov et al., 2011). The pET-9c vector (pBR322 origin and T7 promoter; EMD Millipore) was used to construct this plasmid. The hnRNPA2 expression plasmids were generated from two separate RT-PCR fragments (Mayeda et al., 1994). C- and N-terminal fragments were purified and ligated together into pET-9c digested with NdeI and BamHI. For PKMζ mRNA microinjection experiments, a segment of 1,935 nt was generated from plasmid PKMζ-(48–1,982) (Muslimov et al., 2004), representing PKMζ mRNA nt 48–1,982, inserted between the HindIII and XbaI sites in pBlueScript SK(−) (Agilent Technologies). The plasmid was linearized with XbaI and transcribed with T7. Size and integrity of all transcripts was ascertained by clear native PAGE.
Cell culture, microinjection, and in situ hybridization
Low-density primary cultures of sympathetic neurons were generated and maintained as follows (previously described in Higgins et al., 1991; Muslimov et al., 2006, 2011). Superior cervical ganglia were isolated from embryonic day 19 (E19)–E21 Sprague–Dawley rat embryos. Ganglia were dissociated by mechanical and enzymatic treatment (20 min at 37°C; TrypLE Express; Life Technologies), and neurons were plated and maintained on glass coverslips (Carolina Biological Supply) precoated with 100 mg/ml filter-sterilized poly-d-lysine (Sigma-Aldrich). Basic culture media contained a 50% (vol/vol) mixture of Ham’s F12 medium and DMEM (both from Life Technologies) supplemented with 500 µg/ml BSA (EMD Millipore), 10 µg/ml bovine insulin (Sigma-Aldrich), 20 µg/ml rat transferrin (Jackson ImmunoResearch Laboratories, Inc.), 20 µg/ml l-glutamine (Life Technologies), 5 ng/ml sodium selenite (Sigma-Aldrich), and 100 ng/ml β-NGF (Harlan Bioproducts for Science, Inc.). Neurons were grown at 35°C in an atmosphere of 5% CO2. Basement membrane extract (100 µg/ml; Matrigel; Collaborative Biomedical Products) was added on the third day in vitro to induce dendritic growth. Growth of nonneuronal cells was minimized by adding 2 µM Ara-C (cytosine arabinofuranoside; Sigma-Aldrich) on the second and fifth days after plating. Work with vertebrate animals was approved by the State University of New York Downstate Medical Center Institutional Animal Care and Use Committee.
Primary cultures of hippocampal neurons were prepared as follows (previously described by Goslin et al., 1998; Muslimov et al., 1998). Hippocampal brain tissue was prepared from E18–E19 Sprague–Dawley rat embryos. Cells were dissociated by mechanical and enzymatic treatment (20 min at 37°C; TrypLE Express) and plated on poly-l-lysine–precoated glass coverslips in MEM with 10% horse serum (Life Technologies) and 0.6% d-glucose (Sigma-Aldrich). After 4 d, cells were transferred to dishes containing monolayer cultures of astroglia. Neuronal and glial layers were facing each other without being in physical contact. Cells were then maintained in serum-free medium: MEM, 0.5 mg/ml N-2 supplement, 10 mM Hepes supplemented with 100 µg/ml rat transferrin (Jackson ImmunoResearch Laboratories, Inc.), 10 µg/ml insulin (Sigma-Aldrich), 20 nM progesterone (Sigma-Aldrich), 5 ng/ml sodium selenite (Sigma-Aldrich), 100 µM putrescine (Sigma-Aldrich), 0.1% ovalbumin (Sigma-Aldrich), and 1 mM sodium pyruvate (Sigma-Aldrich). Astroglial cell cultures were prepared from neonatal rat cerebral hemispheres dissected in a dish with Hepes-buffered, Ca2+- and Mg2+-free balanced salt solution. Cells were dissociated by mechanical and enzymatic treatment and plated onto 22-mm glass coverslips, arranged in 60-mm dishes for co-culturing with neurons. 5 µM Ara-C was added on the fifth day to reduce glial proliferation.
To depolarize neurons in culture, cells were maintained in media containing 30 mM KCl, as previously described (Sun et al., 1992; Banker and Goslin, 1998; Habecker et al., 2006) beginning 15 min before microinjection. Adrenergic agonists/antagonists were used as follows with cultured neurons: 10 µM isoproterenol (Mohney and Zigmond, 1998), 10 µM phenylephrine (Lees and Horsburgh, 1984), and 10 µM propranolol (Aguayo and Grossie, 1994). The membrane-permeable Ca2+ chelator BAPTA-AM was used at 50 µM (Bofill-Cardona et al., 2000). Ca2+ channel blockers nifedipine and nimodipine were used at 10 µM (Martínez-Pinna et al., 2002; Uhrenholt and Nedergaard, 2003). Agonists were applied 15–30 min before microinjection. Antagonists, Ca2+ chelators, and Ca2+ channel blockers were applied ≥30 min before agonists (Berchtold et al., 2007). Agonists, antagonists, Ca2+ chelators, and Ca2+ channel blockers were purchased from Sigma-Aldrich.
Microinjection of sympathetic neurons in culture was performed as follows (previously described in Muslimov et al., 1997, 2004, 2006, 2011). RNAs (35S radiolabeled at 3 × 106 cpm/µl) were microinjected, typically into cell perikarya, at volumes of a few femtoliters per pulse. Nuclear injections were performed to examine whether a “nuclear experience” was required for targeting (Wharton, 2009). No such case was observed. For each RNA, injection amounts were varied over a concentration range of at least one order of magnitude to ensure that observed dendritic targeting patterns were independent of amounts injected (Muslimov et al., 2006, 2011). We routinely co-inject 0.4% Lucifer yellow to monitor the injection process and to ascertain that injection and experimental conditions (e.g., application of agonists, antagonists, and Ca2+ chelators) are not causing alterations in cell morphology, including dendritic extent and arborization (Muslimov et al., 2006). The postinjection incubation period varied between 1 and 3 h. Although 1 h was found sufficient for GA motif RNAs to reach distal dendritic tips, a range of incubation periods was used to establish that transport velocities were constant (Muslimov et al., 2006), i.e., did not vary between different transported RNAs and were not a function of experimental paradigms, such as receptor activation or blockade. For each transcript, RNA stability was monitored preinjection by PAGE and postinjection by measuring average integrated total signal intensities per injected cell (Muslimov et al., 2006, 2011). Relative RNA stability was also ascertained by incubation with brain extract (Muslimov et al., 2011).
Neurons on coverslips were fixed, dipped in photographic emulsion (NTB-2; Kodak), and exposed at 4°C for 3–4 wk. Microscopy was performed after processing with a developer (50% strength; D-19; Kodak) and a liquid fixer (Rapid Fixer; Kodak).
Microinjection of radiolabeled transcripts is our preferred method to introduce RNAs into cells for two reasons. (1) We found microinjection superior to other delivery methods as it allows unparalleled control of amounts of RNA introduced. In combination with the high sensitivity of radiolabel detection, this approach enables us to keep the number of introduced RNA molecules lower than the number of respective endogenous RNA molecules (Muslimov et al., 2006). (2) Microinjection of radiolabeled RNAs is preferred because architectural GA motifs are intolerant of nucleotide substitutions (Goody et al., 2004), and introduced side chains (such as fluorophores) may sterically interfere with motif structure and cause altered targeting (Muslimov et al., 2011). We have developed methodology to quantify dendritic RNA transport both spatially and temporally (Muslimov et al., 2006, 2011).
In situ hybridization was performed with hippocampal neurons in culture (Muslimov et al., 1998) and with sympathetic neurons in culture (Muslimov et al., 2011) as previously described. 35S-labeled RNA probes directed at rat CLN2 mRNA were generated from plasmid pBluescript-CLN2is (see Plasmids and RNA preparation). To generate antisense-strand probes, the plasmid was linearized with PstI and transcribed with T3 RNA polymerase. To generate sense-strand probes, the plasmid was linearized with SacI and transcribed with T7 RNA polymerase. Probes were 35S labeled. Prehybridization and hybridization steps were performed as described with high stringency washes performed at 50°C (Muslimov et al., 1998, 2011). In brief, fixed neurons were placed in hybridization boxes containing filter paper saturated with box buffer (4× SSC and 50% formamide). For prehybridization, a small amount of hybridization buffer (10 mM DTT, 0.3 M NaCl, 20 mM Tris-HCl, pH 8.0, 0.5 mM EDTA, Denhardt’s solution, 10% dextran sulfate, and 50% formamide) was added to cover cells. Cells were prehybridized for 1 h at 42°C. The hybridization mixture contained 2 µl 35S-labeled riboprobe (300,000 cpm/µl in Tris-EDTA) and 1 µl tRNA per 100 µl prehybridization buffer. Hybridization was performed overnight at 42°C. After hybridization, slides with coverslips were rinsed twice for 10 min each at room temperature in 2× SSC containing 0.1% β-mercaptoethanol and 1 mM EDTA followed by RNase A treatment (20 µg/ml in 500 mM NaCl and 10 mM Tris-HCl, pH 8.0; Sigma-Aldrich) for 30 min at room temperature. Cells were rinsed twice with 2× SSC containing 0.1% β-mercaptoethanol and 1 mM EDTA at room temperature followed by a high stringency wash in 4 liters of 0.2× SSC containing 0.1% β-mercaptoethanol and 1 mM EDTA at 42°C for 4 h. Slides were washed twice with 0.2× SSC at room temperature. Neurons were dehydrated in graded alcohol containing 0.3 M ammonium acetate, and dried in a vacuum desiccator.
For emulsion autoradiography, dried coverslips were mounted, cell side up, on microscope slides and dipped in NTB-2 emulsion as previously described (Muslimov et al., 1998, 2011). Cells were exposed at 4°C for 7 d (microinjection) or 3–4 wk (in situ hybridization) before photographic development (D-19 developer, 50% strength; Rapid Fixer) as previously described (Muslimov et al., 1998, 2011).
Microscopy, data acquisition, and analysis
Photomicrographs (dark field and phase contrast) were taken on a microscope (Microphot-FXA; Nikon) with a camera (Digital-Sight DS-Fi1; Nikon). Photographs were taken of fixed specimens (cultured neurons mounted on coverslips) at room temperature using the following objectives: (a) Plan Fluor 10×/0.30 NA and 160×/0.17 NA; (b) PhC Plan 20×/0.50 NA and DL 160×/0.17 NA; (c) Plan 20×/0.50 NA and differential interference contrast 160×/0.17 NA; and (d) Ph3 DL Plan 40×/0.65 NA and 160×/0.17 NA. To evaluate transport of injected RNAs, silver grains were measured along dendritic shafts at 50-µm interval points, up to 250 µm (Muslimov et al., 2006, 2011). Signal intensities at the base of dendrites (0-µm points) were given a relative value of 100%. MetaMorph software (Molecular Devices) was used for image analysis. DeltaGraph 5 software (Red Rock Software) was used to design bar diagrams. Final illustrations were arranged in Illustrator (Adobe). SPSS software (IBM) was used for statistical analysis. All experiments were performed at least in quadruplicate or as noted in the figure legends.
Expression of recombinant proteins
Expression and purification of recombinant hnRNP A2 was performed as described (Munro et al., 1999; Muslimov et al., 2011). Escherichia coli BL21(DE3) was transformed with plasmid pET-9c/A2 to express hnRNP A2. Cells were induced with isopropyl-β-d-thiogalactopyranoside. Harvested by centrifugation, cells were lysed in 50 mM Tris-HCl, pH 8, 2 mM EDTA, 100 µg/ml lysozyme, and 0.1% Triton X-100. Buffer A (50 mM Tris-HCl, pH 8.5, 0.2 mM EDTA, and 5% wt/vol glycerol) was used for dialysis of the soluble fraction. Chromatography was performed on diethylaminoethyl cellulose (GE Healthcare) and, subsequently, on Sephacryl S-300 (GE Healthcare) and, for further purification, on a C4 reverse-phase HPLC column (Vydac) using a linear 10–50% acetonitrile gradient in 0.1% trifluoroacetic acid. After lyophilization, hnRNP A2 was dissolved in water. Protein identity and purity was ascertained by SDS-PAGE and by electrospray mass spectrometry on a spectrometer (Sciex 165; PerkinElmer).
Native PAGE and EMSA
Clear native PAGE and EMSA were performed as previously described (Goody et al., 2004; Muslimov et al., 2006, 2011). 4–20% Mini-PROTEAN TGX Stain-Free PAGE gels were used (Bio-Rad Laboratories). 32P-labeled RNA probes (50,000 cpm/reaction) were heated for 10 min at 70°C and allowed to cool for 5 min at room temperature. Probes were incubated for 20 min at room temperature together with proteins in binding buffer (300 mM KCl, 5 mM MgCl2, 2 mM DTT, 5% glycerol, and 20 mM Hepes, pH 7.6). The samples were electrophoresed at room temperature at 120 V and 25 mA. RNA–protein complexes were visualized by autoradiography. To quantify results, a phosphorimaging system (Storm 860) and ImageQuant software were used (GE Healthcare). Binding affinities were established by measuring the fractions of free versus bound RNA. The Hill equation was used to determine equilibrium constants (Ryder et al., 2008; Chao et al., 2010; Muslimov et al., 2011).
Online supplemental material
Fig. S1 shows control experiments in which a reverse complementary ID4 (ID4R) RNA is not delivered to dendrites under either basal or stimulation conditions. Fig. S2 provides evidence that binding of ID4 RNA to hnRNP A2 is not promoted by Mg2+ at concentrations <1 mM. Fig. S3 shows that dendritic transport of PKMζ mRNA, which depends on a 3′ UTR standard GA motif, is constitutive and remains unaltered under conditions of Ca2+ chelation or L-type VDCC blockade. Fig. S4 documents that dendritic delivery of endogenous CLN2 mRNA is induced by β-adrenergic activation in sympathetic neurons in culture and that such transport induction is prevented by chelation of intracellular Ca2+ and L-type VDCC blockade. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201310045/DC1.
Supplementary Material
Acknowledgments
We thank Dr. Ronald Crystal (Weill Cornell Medical College) for CLN2 vectors, Dr. Jeremy Weedon (State University of New York Downstate Medical Center Scientific Computing Center) for statistical consultation, and members of the Tiedge laboratory for advice and discussion.
T.H. Tang acknowledges financial support from the University of Münster Medical School where part of his work was performed. This work was supported in part by National Institutes of Health grants NS046769 and DA026110 (H. Tiedge).
The authors declare no competing financial interests.
Footnotes
Abbreviations used in this paper:
- ANOVA
- analysis of variance
- AR
- adrenergic receptor
- BC
- brain cytoplasmic
- EMSA
- electrophoretic mobility shift assay
- hnRNP
- heterogeneous nuclear RNP
- ID
- identifier
- VDCC
- voltage-dependent calcium channel
- WC
- Watson–Crick
- WT
- wild type
References
- Aguayo L.G., Grossie J. 1994. Dopamine inhibits a sustained calcium current through activation of alpha adrenergic receptors and a GTP-binding protein in adult rat sympathetic neurons. J. Pharmacol. Exp. Ther. 269:503–508 [PubMed] [Google Scholar]
- An J.J., Gharami K., Liao G.Y., Woo N.H., Lau A.G., Vanevski F., Torre E.R., Jones K.R., Feng Y., Lu B., Xu B. 2008. Distinct role of long 3′ UTR BDNF mRNA in spine morphology and synaptic plasticity in hippocampal neurons. Cell. 134:175–187 10.1016/j.cell.2008.05.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banker K., Goslin K., 1998. Culturing Nerve Cells. Vol. 2. MIT Press, Cambridge, MA: 666 pp [Google Scholar]
- Berchtold C.M., Wu Z.H., Huang T.T., Miyamoto S. 2007. Calcium-dependent regulation of NEMO nuclear export in response to genotoxic stimuli. Mol. Cell. Biol. 27:497–509 10.1128/MCB.01772-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berridge M.J., Lipp P., Bootman M.D. 2000. The versatility and universality of calcium signalling. Nat. Rev. Mol. Cell Biol. 1:11–21 10.1038/35036035 [DOI] [PubMed] [Google Scholar]
- Bevilacqua P.C., George C.X., Samuel C.E., Cech T.R. 1998. Binding of the protein kinase PKR to RNAs with secondary structure defects: role of the tandem A-G mismatch and noncontiguous helixes. Biochemistry. 37:6303–6316 10.1021/bi980113j [DOI] [PubMed] [Google Scholar]
- Bloodgood B.L., Sabatini B.L. 2007. Ca2+ signaling in dendritic spines. Curr. Opin. Neurobiol. 17:345–351 10.1016/j.conb.2007.04.003 [DOI] [PubMed] [Google Scholar]
- Bofill-Cardona E., Vartian N., Nanoff C., Freissmuth M., Boehm S. 2000. Two different signaling mechanisms involved in the excitation of rat sympathetic neurons by uridine nucleotides. Mol. Pharmacol. 57:1165–1172 [PubMed] [Google Scholar]
- Brosius J. 1991. Retroposons—seeds of evolution. Science. 251:753 10.1126/science.1990437 [DOI] [PubMed] [Google Scholar]
- Brosius J. 1999. Genomes were forged by massive bombardments with retroelements and retrosequences. Genetica. 107:209–238 10.1023/A:1004018519722 [DOI] [PubMed] [Google Scholar]
- Brosius J. 2005. Echoes from the past—are we still in an RNP world? Cytogenet. Genome Res. 110:8–24 10.1159/000084934 [DOI] [PubMed] [Google Scholar]
- Bruckenstein D.A., Lein P.J., Higgins D., Fremeau R.T., Jr 1990. Distinct spatial localization of specific mRNAs in cultured sympathetic neurons. Neuron. 5:809–819 10.1016/0896-6273(90)90340-L [DOI] [PubMed] [Google Scholar]
- Buckley P.T., Lee M.T., Sul J.Y., Miyashiro K.Y., Bell T.J., Fisher S.A., Kim J., Eberwine J. 2011. Cytoplasmic intron sequence-retaining transcripts can be dendritically targeted via ID element retrotransposons. Neuron. 69:877–884 10.1016/j.neuron.2011.02.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgin K.E., Waxham M.N., Rickling S., Westgate S.A., Mobley W.C., Kelly P.T. 1990. In situ hybridization histochemistry of Ca2+/calmodulin-dependent protein kinase in developing rat brain. J. Neurosci. 10:1788–1798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao X., Yeo G., Muotri A.R., Kuwabara T., Gage F.H. 2006. Noncoding RNAs in the mammalian central nervous system. Annu. Rev. Neurosci. 29:77–103 10.1146/annurev.neuro.29.051605.112839 [DOI] [PubMed] [Google Scholar]
- Chao J.A., Patskovsky Y., Patel V., Levy M., Almo S.C., Singer R.H. 2010. ZBP1 recognition of beta-actin zipcode induces RNA looping. Genes Dev. 24:148–158 10.1101/gad.1862910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cojocaru V., Nottrott S., Klement R., Jovin T.M. 2005. The snRNP 15.5K protein folds its cognate K-turn RNA: a combined theoretical and biochemical study. RNA. 11:197–209 10.1261/rna.7149605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordaux R., Batzer M.A. 2009. The impact of retrotransposons on human genome evolution. Nat. Rev. Genet. 10:691–703 10.1038/nrg2640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cristofanilli M., Thanos S., Brosius J., Kindler S., Tiedge H. 2004. Neuronal MAP2 mRNA: species-dependent differential dendritic targeting competence. J. Mol. Biol. 341:927–934 10.1016/j.jmb.2004.06.045 [DOI] [PubMed] [Google Scholar]
- Darnell J.C. 2011. Defects in translational regulation contributing to human cognitive and behavioral disease. Curr. Opin. Genet. Dev. 21:465–473 10.1016/j.gde.2011.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davare M.A., Avdonin V., Hall D.D., Peden E.M., Burette A., Weinberg R.J., Horne M.C., Hoshi T., Hell J.W. 2001. A β2 adrenergic receptor signaling complex assembled with the Ca2+ channel Cav1.2. Science. 293:98–101 10.1126/science.293.5527.98 [DOI] [PubMed] [Google Scholar]
- de Koning A.P., Gu W., Castoe T.A., Batzer M.A., Pollock D.D. 2011. Repetitive elements may comprise over two-thirds of the human genome. PLoS Genet. 7:e1002384 10.1371/journal.pgen.1002384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dictenberg J.B., Swanger S.A., Antar L.N., Singer R.H., Bassell G.J. 2008. A direct role for FMRP in activity-dependent dendritic mRNA transport links filopodial-spine morphogenesis to fragile X syndrome. Dev. Cell. 14:926–939 10.1016/j.devcel.2008.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle M., Kiebler M.A. 2011. Mechanisms of dendritic mRNA transport and its role in synaptic tagging. EMBO J. 30:3540–3552 10.1038/emboj.2011.278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberwine J., Belt B., Kacharmina J.E., Miyashiro K. 2002. Analysis of subcellularly localized mRNAs using in situ hybridization, mRNA amplification, and expression profiling. Neurochem. Res. 27:1065–1077 10.1023/A:1020956805307 [DOI] [PubMed] [Google Scholar]
- Falb M., Amata I., Gabel F., Simon B., Carlomagno T. 2010. Structure of the K-turn U4 RNA: a combined NMR and SANS study. Nucleic Acids Res. 38:6274–6285 10.1093/nar/gkq380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goody T.A., Melcher S.E., Norman D.G., Lilley D.M. 2004. The kink-turn motif in RNA is dimorphic, and metal ion-dependent. RNA. 10:254–264 10.1261/rna.5176604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goslin K., Banker G., Asmussen H. 1998. Rat hippocampal neurons in low density cultures. Culturing Nerve Cells. Banker K., Goslin K., MIT Press, Cambridge, MA: 339–370 [Google Scholar]
- Grandin K., editor 2010. The Nobel prizes 2009. Les Prix Nobel Stockholm: Nobel Foundation [Google Scholar]
- Grienberger C., Konnerth A. 2012. Imaging calcium in neurons. Neuron. 73:862–885 10.1016/j.neuron.2012.02.011 [DOI] [PubMed] [Google Scholar]
- Habecker B.A., Willison B.D., Shi X., Woodward W.R. 2006. Chronic depolarization stimulates norepinephrine transporter expression via catecholamines. J. Neurochem. 97:1044–1051 10.1111/j.1471-4159.2006.03792.x [DOI] [PubMed] [Google Scholar]
- Hall D.D., Davare M.A., Shi M., Allen M.L., Weisenhaus M., McKnight G.S., Hell J.W. 2007. Critical role of cAMP-dependent protein kinase anchoring to the L-type calcium channel Cav1.2 via A-kinase anchor protein 150 in neurons. Biochemistry. 46:1635–1646 10.1021/bi062217x [DOI] [PubMed] [Google Scholar]
- Hell J.W. 2010. β-adrenergic regulation of the L-type Ca2+ channel Ca(V)1.2 by PKA rekindles excitement. Sci. Signal. 3:pe33 10.1126/scisignal.3141pe33 [DOI] [PubMed] [Google Scholar]
- Herbert A. 2004. The four Rs of RNA-directed evolution. Nat. Genet. 36:19–25 10.1038/ng1275 [DOI] [PubMed] [Google Scholar]
- Higgins D., Lein P.J., Osterhout D.J., Johnson M.I. 1991. Tissue culture of mammalian autonomic neurons. Culturing Nerve Cells. Banker G., Goslin K., MIT Press, Cambridge, MA: 177–205 [Google Scholar]
- Hoogland T.M., Saggau P. 2004. Facilitation of L-type Ca2+ channels in dendritic spines by activation of β2 adrenergic receptors. J. Neurosci. 24:8416–8427 10.1523/JNEUROSCI.1677-04.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y.Y., Kandel E.R. 1996. Modulation of both the early and the late phase of mossy fiber LTP by the activation of β-adrenergic receptors. Neuron. 16:611–617 10.1016/S0896-6273(00)80080-X [DOI] [PubMed] [Google Scholar]
- Iacoangeli A., Tiedge H. 2013. Translational control at the synapse: role of RNA regulators. Trends Biochem. Sci. 38:47–55 10.1016/j.tibs.2012.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain C., Belasco J.G. 1996. A structural model for the HIV-1 Rev-RRE complex deduced from altered-specificity rev variants isolated by a rapid genetic strategy. Cell. 87:115–125 10.1016/S0092-8674(00)81328-8 [DOI] [PubMed] [Google Scholar]
- Jalanko A., Braulke T. 2009. Neuronal ceroid lipofuscinoses. Biochim. Biophys. Acta. 1793:697–709 10.1016/j.bbamcr.2008.11.004 [DOI] [PubMed] [Google Scholar]
- Jin P., Duan R., Qurashi A., Qin Y., Tian D., Rosser T.C., Liu H., Feng Y., Warren S.T. 2007. Pur α binds to rCGG repeats and modulates repeat-mediated neurodegeneration in a Drosophila model of fragile X tremor/ataxia syndrome. Neuron. 55:556–564 10.1016/j.neuron.2007.07.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Job C., Eberwine J. 2001. Localization and translation of mRNA in dendrites and axons. Nat. Rev. Neurosci. 2:889–898 10.1038/35104069 [DOI] [PubMed] [Google Scholar]
- Kaufman R.J. 2000. The double-stranded RNA-activated protein kinase (PKR). Translational Control of Gene Expression. Second edition Sonenberg N., Hershey J.W.B., Mathews M.B., Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY: 503–527 [Google Scholar]
- Kazazian H.H., Jr 2004. Mobile elements: drivers of genome evolution. Science. 303:1626–1632 10.1126/science.1089670 [DOI] [PubMed] [Google Scholar]
- Khanam T., Raabe C.A., Kiefmann M., Handel S., Skryabin B.V., Brosius J. 2007a. Can ID repetitive elements serve as cis-acting dendritic targeting elements? An in vivo study. PLoS ONE. 2:e961 10.1371/journal.pone.0000961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khanam T., Rozhdestvensky T.S., Bundman M., Galiveti C.R., Handel S., Sukonina V., Jordan U., Brosius J., Skryabin B.V. 2007b. Two primate-specific small non-protein-coding RNAs in transgenic mice: neuronal expression, subcellular localization and binding partners. Nucleic Acids Res. 35:529–539 10.1093/nar/gkl1082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Martignetti J.A., Shen M.R., Brosius J., Deininger P. 1994. Rodent BC1 RNA gene as a master gene for ID element amplification. Proc. Natl. Acad. Sci. USA. 91:3607–3611 10.1073/pnas.91.9.3607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Krichevsky A., Grad Y., Hayes G.D., Kosik K.S., Church G.M., Ruvkun G. 2004. Identification of many microRNAs that copurify with polyribosomes in mammalian neurons. Proc. Natl. Acad. Sci. USA. 101:360–365 10.1073/pnas.2333854100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kindler S., Wang H., Richter D., Tiedge H. 2005. RNA transport and local control of translation. Annu. Rev. Cell Dev. Biol. 21:223–245 10.1146/annurev.cellbio.21.122303.120653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kollmann K., Uusi-Rauva K., Scifo E., Tyynelä J., Jalanko A., Braulke T. 2013. Cell biology and function of neuronal ceroid lipofuscinosis-related proteins. Biochim. Biophys. Acta. 1832:1866–1881 10.1016/j.bbadis.2013.01.019 [DOI] [PubMed] [Google Scholar]
- Kosik K.S., Krichevsky A.M. 2005. The elegance of the microRNAs: a neuronal perspective. Neuron. 47:779–782 10.1016/j.neuron.2005.08.019 [DOI] [PubMed] [Google Scholar]
- Landis S.C. 1990. Target regulation of neurotransmitter phenotype. Trends Neurosci. 13:344–350 10.1016/0166-2236(90)90147-3 [DOI] [PubMed] [Google Scholar]
- Lee S.H., Simonetta A., Sheng M. 2004. Subunit rules governing the sorting of internalized AMPA receptors in hippocampal neurons. Neuron. 43:221–236 10.1016/j.neuron.2004.06.015 [DOI] [PubMed] [Google Scholar]
- Lee S., Sato Y., Nixon R.A. 2011. Lysosomal proteolysis inhibition selectively disrupts axonal transport of degradative organelles and causes an Alzheimer’s-like axonal dystrophy. J. Neurosci. 31:7817–7830 10.1523/JNEUROSCI.6412-10.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lees G.J., Horsburgh R.J. 1984. Retrograde transport of dopamine beta-hydroxylase antibodies in sympathetic neurons: effects of drugs modifying noradrenergic transmission. Brain Res. 301:281–286 10.1016/0006-8993(84)91097-7 [DOI] [PubMed] [Google Scholar]
- Leontis N.B., Westhof E. 2003. Analysis of RNA motifs. Curr. Opin. Struct. Biol. 13:300–308 10.1016/S0959-440X(03)00076-9 [DOI] [PubMed] [Google Scholar]
- Leontis N.B., Lescoute A., Westhof E. 2006. The building blocks and motifs of RNA architecture. Curr. Opin. Struct. Biol. 16:279–287 10.1016/j.sbi.2006.05.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lescoute A., Leontis N.B., Massire C., Westhof E. 2005. Recurrent structural RNA motifs, Isostericity Matrices and sequence alignments. Nucleic Acids Res. 33:2395–2409 10.1093/nar/gki535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Pinna J., Lamas J.A., Gallego R. 2002. Calcium current components in intact and dissociated adult mouse sympathetic neurons. Brain Res. 951:227–236 10.1016/S0006-8993(02)03165-7 [DOI] [PubMed] [Google Scholar]
- Matsumura S., Ikawa Y., Inoue T. 2003. Biochemical characterization of the kink-turn RNA motif. Nucleic Acids Res. 31:5544–5551 10.1093/nar/gkg760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayeda A., Munroe S.H., Cáceres J.F., Krainer A.R. 1994. Function of conserved domains of hnRNP A1 and other hnRNP A/B proteins. EMBO J. 13:5483–5495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyashiro K.Y., Bell T.J., Sul J.Y., Eberwine J. 2009. Subcellular neuropharmacology: the importance of intracellular targeting. Trends Pharmacol. Sci. 30:203–211 10.1016/j.tips.2009.01.005 [DOI] [PubMed] [Google Scholar]
- Mohney R.P., Zigmond R.E. 1998. Vasoactive intestinal peptide enhances its own expression in sympathetic neurons after injury. J. Neurosci. 18:5285–5293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohr E., Prakash N., Vieluf K., Fuhrmann C., Buck F., Richter D. 2001. Vasopressin mRNA localization in nerve cells: characterization of cis-acting elements and trans-acting factors. Proc. Natl. Acad. Sci. USA. 98:7072–7079 10.1073/pnas.111146598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moncada D., Ballarini F., Martinez M.C., Frey J.U., Viola H. 2011. Identification of transmitter systems and learning tag molecules involved in behavioral tagging during memory formation. Proc. Natl. Acad. Sci. USA. 108:12931–12936 10.1073/pnas.1104495108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muddashetty R.S., Nalavadi V.C., Gross C., Yao X., Xing L., Laur O., Warren S.T., Bassell G.J. 2011. Reversible inhibition of PSD-95 mRNA translation by miR-125a, FMRP phosphorylation, and mGluR signaling. Mol. Cell. 42:673–688 10.1016/j.molcel.2011.05.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munro T.P., Magee R.J., Kidd G.J., Carson J.H., Barbarese E., Smith L.M., Smith R. 1999. Mutational analysis of a heterogeneous nuclear ribonucleoprotein A2 response element for RNA trafficking. J. Biol. Chem. 274:34389–34395 10.1074/jbc.274.48.34389 [DOI] [PubMed] [Google Scholar]
- Mus E., Hof P.R., Tiedge H. 2007. Dendritic BC200 RNA in aging and in Alzheimer’s disease. Proc. Natl. Acad. Sci. USA. 104:10679–10684 10.1073/pnas.0701532104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muslimov I.A., Santi E., Homel P., Perini S., Higgins D., Tiedge H. 1997. RNA transport in dendrites: a cis-acting targeting element is contained within neuronal BC1 RNA. J. Neurosci. 17:4722–4733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muslimov I.A., Banker G., Brosius J., Tiedge H. 1998. Activity-dependent regulation of dendritic BC1 RNA in hippocampal neurons in culture. J. Cell Biol. 141:1601–1611 10.1083/jcb.141.7.1601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muslimov I.A., Nimmrich V., Hernandez A.I., Tcherepanov A., Sacktor T.C., Tiedge H. 2004. Dendritic transport and localization of protein kinase Mzeta mRNA: implications for molecular memory consolidation. J. Biol. Chem. 279:52613–52622 10.1074/jbc.M409240200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muslimov I.A., Iacoangeli A., Brosius J., Tiedge H. 2006. Spatial codes in dendritic BC1 RNA. J. Cell Biol. 175:427–439 10.1083/jcb.200607008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muslimov I.A., Patel M.V., Rose A., Tiedge H. 2011. Spatial code recognition in neuronal RNA targeting: role of RNA–hnRNP A2 interactions. J. Cell Biol. 194:441–457 10.1083/jcb.201010027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neher E. 1998. Vesicle pools and Ca2+ microdomains: new tools for understanding their roles in neurotransmitter release. Neuron. 20:389–399 10.1016/S0896-6273(00)80983-6 [DOI] [PubMed] [Google Scholar]
- Nixon R.A., Cataldo A.M. 1995. The endosomal-lysosomal system of neurons: new roles. Trends Neurosci. 18:489–496 10.1016/0166-2236(95)92772-I [DOI] [PubMed] [Google Scholar]
- Noller H.F. 2005. RNA structure: reading the ribosome. Science. 309:1508–1514 10.1126/science.1111771 [DOI] [PubMed] [Google Scholar]
- Oostra B.A., Willemsen R. 2009. FMR1: a gene with three faces. Biochim. Biophys. Acta. 1790:467–477 10.1016/j.bbagen.2009.02.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paschen W., Hotop S., Aufenberg C. 2003. Loading neurons with BAPTA-AM activates xbp1 processing indicative of induction of endoplasmic reticulum stress. Cell Calcium. 33:83–89 10.1016/S0143-4160(02)00195-1 [DOI] [PubMed] [Google Scholar]
- Pe’ery T., Mathews M.B. 2000. Viral translation strategies and host defense mechanisms. Translational Control of Gene Expression. Second edition Sonenberg N., Hershey J.W.B., Mathews M.B., Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY: 400–424 [Google Scholar]
- Qureshi I.A., Mehler M.F. 2012. Emerging roles of non-coding RNAs in brain evolution, development, plasticity and disease. Nat. Rev. Neurosci. 13:528–541 10.1038/nrn3234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raman I.M., Tong G., Jahr C.E. 1996. β-adrenergic regulation of synaptic NMDA receptors by cAMP-dependent protein kinase. Neuron. 16:415–421 10.1016/S0896-6273(00)80059-8 [DOI] [PubMed] [Google Scholar]
- Rázga F., Zacharias M., Réblová K., Koca J., Sponer J. 2006. RNA kink-turns as molecular elbows: hydration, cation binding, and large-scale dynamics. Structure. 14:825–835 10.1016/j.str.2006.02.012 [DOI] [PubMed] [Google Scholar]
- Roberts V.J., Gorenstein C. 1987. Examination of the transient distribution of lysosomes in neurons of developing rat brains. Dev. Neurosci. 9:255–264 10.1159/000111628 [DOI] [PubMed] [Google Scholar]
- Ross W.N. 2012. Understanding calcium waves and sparks in central neurons. Nat. Rev. Neurosci. 13:157–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozhdestvensky T.S., Kopylov A.M., Brosius J., Hüttenhofer A. 2001. Neuronal BC1 RNA structure: evolutionary conversion of a tRNA(Ala) domain into an extended stem-loop structure. RNA. 7:722–730 10.1017/S1355838201002485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryder S.P., Recht M.I., Williamson J.R. 2008. Quantitative analysis of protein-RNA interactions by gel mobility shift. Methods Mol. Biol. 488:99–115 10.1007/978-1-60327-475-3_7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schratt G.M., Tuebing F., Nigh E.A., Kane C.G., Sabatini M.E., Kiebler M., Greenberg M.E. 2006. A brain-specific microRNA regulates dendritic spine development. Nature. 439:283–289 10.1038/nature04367 [DOI] [PubMed] [Google Scholar]
- Schroeder K.T., McPhee S.A., Ouellet J., Lilley D.M. 2010. A structural database for k-turn motifs in RNA. RNA. 16:1463–1468 10.1261/rna.2207910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skryabin B.V., Kremerskothen J., Vassilacopoulou D., Disotell T.R., Kapitonov V.V., Jurka J., Brosius J. 1998. The BC200 RNA gene and its neural expression are conserved in Anthropoidea (primates). J. Mol. Evol. 47:677–685 10.1007/PL00006426 [DOI] [PubMed] [Google Scholar]
- Smith R. 2004. Moving molecules: mRNA trafficking in mammalian oligodendrocytes and neurons. Neuroscientist. 10:495–500 10.1177/1073858404266759 [DOI] [PubMed] [Google Scholar]
- Sondhi D., Peterson D.A., Giannaris E.L., Sanders C.T., Mendez B.S., De B., Rostkowski A.B., Blanchard B., Bjugstad K., Sladek J.R., Jr, et al. 2005. AAV2-mediated CLN2 gene transfer to rodent and non-human primate brain results in long-term TPP-I expression compatible with therapy for LINCL. Gene Ther. 12:1618–1632 10.1038/sj.gt.3302549 [DOI] [PubMed] [Google Scholar]
- Steitz T.A., Moore P.B. 2003. RNA, the first macromolecular catalyst: the ribosome is a ribozyme. Trends Biochem. Sci. 28:411–418 10.1016/S0968-0004(03)00169-5 [DOI] [PubMed] [Google Scholar]
- Sun Y., Rao M.S., Landis S.C., Zigmond R.E. 1992. Depolarization increases vasoactive intestinal peptide- and substance P-like immunoreactivities in cultured neonatal and adult sympathetic neurons. J. Neurosci. 12:3717–3728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson M.S., Orr H.T. 2007. Fragile X tremor/ataxia syndrome: blame the messenger! Neuron. 55:535–537 10.1016/j.neuron.2007.07.032 [DOI] [PubMed] [Google Scholar]
- Tiedge H., Chen W., Brosius J. 1993. Primary structure, neural-specific expression, and dendritic location of human BC200 RNA. J. Neurosci. 13:2382–2390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tongiorgi E., Righi M., Cattaneo A. 1997. Activity-dependent dendritic targeting of BDNF and TrkB mRNAs in hippocampal neurons. J. Neurosci. 17:9492–9505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tongiorgi E., Armellin M., Giulianini P.G., Bregola G., Zucchini S., Paradiso B., Steward O., Cattaneo A., Simonato M. 2004. Brain-derived neurotrophic factor mRNA and protein are targeted to discrete dendritic laminas by events that trigger epileptogenesis. J. Neurosci. 24:6842–6852 10.1523/JNEUROSCI.5471-03.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner B., Melcher S.E., Wilson T.J., Norman D.G., Lilley D.M. 2005. Induced fit of RNA on binding the L7Ae protein to the kink-turn motif. RNA. 11:1192–1200 10.1261/rna.2680605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhrenholt T.R., Nedergaard O.A. 2003. Calcium channels involved in noradrenaline release from sympathetic neurones in rabbit carotid artery. Pharmacol. Toxicol. 92:226–233 10.1034/j.1600-0773.2003.920505.x [DOI] [PubMed] [Google Scholar]
- Vásquez C., Lewis D.L. 2003. The β2-adrenergic receptor specifically sequesters Gs but signals through both Gs and Gi/o in rat sympathetic neurons. Neuroscience. 118:603–610 10.1016/S0306-4522(03)00024-1 [DOI] [PubMed] [Google Scholar]
- Wells D.G., Fallon J.R. 2000. Dendritic mRNA translation: deciphering the uncoded. Nat. Neurosci. 3:1062–1064 10.1038/80560 [DOI] [PubMed] [Google Scholar]
- Wharton R.P. 2009. A splicer that represses (translation). Genes Dev. 23:133–137 10.1101/gad.1768509 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.