Late endosomes locally regulate cell migration by transporting the p14–MP1 scaffold complex to the vicinity of focal adhesions.
Abstract
Cell migration is mediated by the dynamic remodeling of focal adhesions (FAs). Recently, an important role of endosomal signaling in regulation of cell migration was recognized. Here, we show an essential function for late endosomes carrying the p14–MP1 (LAMTOR2/3) complex in FA dynamics. p14–MP1-positive endosomes move to the cell periphery along microtubules (MTs) in a kinesin1- and Arl8b-dependent manner. There they specifically target FAs to regulate FA turnover, which is required for cell migration. Using genetically modified fibroblasts from p14-deficient mice and Arl8b-depleted cells, we demonstrate that MT plus end–directed traffic of p14–MP1-positive endosomes triggered IQGAP1 disassociation from FAs. The release of IQGAP was required for FA dynamics. Taken together, our results suggest that late endosomes contribute to the regulation of cell migration by transporting the p14–MP1 scaffold complex to the vicinity of FAs.
Introduction
Cell migration requires the coordinated activity of several modular processes, including formation and turnover of focal adhesion (FA) sites, actin dynamics, and polarized distribution of adaptor and signaling proteins. Growing evidence suggests the importance of endosomes for the local regulation of these processes (Sadowski et al., 2009; Scita and Di Fiore, 2010; Schiefermeier et al., 2011). Among the proteins suggested to use different subsets of endosomes as mobile platforms are well-known regulators of cell motility such as Rac (Palamidessi et al., 2008), Cdc42 (Osmani et al., 2010; Huang et al., 2011), Src (Tu et al., 2010), Endo 180 (Sturge et al., 2006), and PTPD1 (Carlucci et al., 2010).
The p14–MP1 (LAMTOR2/3, MAPK/ERK kinase 1 partner MP1, and its endosomal adaptor protein p14) protein complex was established as a late endosomal MAPK scaffold complex (Wunderlich et al., 2001; Kurzbauer et al., 2004). Moreover, p14–MP1 was shown to regulate mTOR signaling, organization of the late endosomal compartment, cell migration, cell spreading, and proliferation (Teis et al., 2002, 2006; Pullikuth et al., 2005; Park et al., 2009; Sancak et al., 2010). Interestingly, previous findings demonstrated that FAs in fibroblasts are specifically targeted by microtubules (MTs). Thereby, MTs deliver a so-far unidentified relaxing signal to modify FA dynamics in a kinesin-1–dependent manner (Kaverina et al., 1999; Krylyshkina et al., 2002). Recently, binding of late endosomal membranes to kinesin-1 was shown to require the Arl8b-GTP protein (Bagshaw et al., 2006; Hofmann and Munro, 2006; Rosa-Ferreira and Munro, 2011), but how Arl8b impacts on cell migration was not investigated.
Additionally, IQGAP1 was suggested to regulate cell migration in several ways. It binds directly to multiple proteins, including known cytoskeleton regulators (actin, myosin light chain-2, Rac1, Cdc42, adenomatous polyposis coli [APC], and CLIP-170 [Brown and Sacks, 2006]). IQGAP1 localizes MEK and ERK to dynamic MTs (Roy et al., 2004, 2005) and also binds components of the MAPK pathway such as B-Raf, MEK1, MEK2, ERK1, and ERK2 (Roy et al., 2004, 2005). Transfection of dominant-negative mutants or down-regulation of IQGAP1 by RNAi reduces cell motility in some cell lines (Hart et al., 1996; Mataraza et al., 2003). Recently, IQGAP1 was identified in FAs (Kuo et al., 2011; Schiller et al., 2011) and in focal complexes (FCs) of keratinocytes, where it binds to the integrin-linked kinase ILK (Wickström et al., 2010). Whether IQGAP1 interacts with FA proteins or is directly involved in regulation of FA dynamics is unknown.
Here, we report that the p14–MP1 (LAMTOR2/3) complex regulates FA dynamics and cell migration from late endosomes. Small but distinct subpopulations of the Rab7-positive late endosomes, which carry the p14–MP1 scaffold complex, move along MTs in an Arl8b-dependent manner to the cell periphery where they specifically target FAs. Using genetically modified fibroblasts from p14-deficient mice, we demonstrate that the late endosomal p14–MP1 complex is essential for FA dynamics. MT plus end–directed transport of the p14–MP1 complex regulates localization and association of IQGAP1 to mature FAs and thereby controls FA dynamics. In summary, our results suggest a new function for the p14–MP1 complex in local regulation of FAs and thus demonstrate a crucial role for specific subsets of late endosomes during cell migration.
Results
Impaired cell migration and FA remodeling in p14−/− knockout MEFs
Previously, down-regulation of p14–MP1 by RNAi was shown to inhibit migration of prostate cancer cells (Park et al., 2009). To test specifically if the knockout of the p14–MP1 complex contributes to cell migration, we performed wound-healing assays. Confluent cell layers of immortalized control p14f/− and p14−/− knockout mouse embryonic fibroblasts (MEFs; Teis et al., 2006) were scratched and wound closure was recorded by time-lapse microscopy (Fig. 1 A and Video 1). In the p14−/− knockout MEFs, MP1 no longer localizes to late endosomes and was degraded (Teis et al., 2006). The p14f/− control MEFs adopted a typical fibroblast migration behavior with a single leading edge facing the wound and closed the scratched area in approximately 10 h. In contrast, the p14−/− MEFs failed to form a clear leading edge, but instead developed multiple elongated protrusions that did not result in active migration into the scratched area (Fig. S1 A). The migration speed of control cells was 15 ± 3.5 µm/h (mean ± SD), as compared with 5 ± 0.98 µm/h for p14−/− MEFs (Fig. 1 B). The migration defect could be rescued by retroviral re-expression of a fully functional p14-GFP (11.61 ± 3.17 µm/h; Fig. 1, A and B; and Video 1; Stasyk et al., 2010). The migration speed of p14−/− MEFs in a random single-cell migration assay was also two times slower compared with their controls (p14f/− MEFs, 49.13 ± 11.36 µm/h vs. p14−/− MEFs, 23.36 ± 9.93 µm/h; Fig. S1 B).
Figure 1.
Loss of p14–MP1 endosomal signaling impairs cell migration and FA. (A) Migration of MEFs in wound-healing assay. Red lines depict edges of the wound. See also Video 1. (B) Calculation of migration speed of MEFs in wound-healing assay (µm/h, mean of cell migration speeds ± SD; **, P < 0.001, Student’s t test). (C) IF: anti-Paxillin antibody. Note formation of elongated peripheral FAs in p14−/− MEFs (red arrows) compared with FAs and FCs at the leading edge of the control p14f/− MEFs (red arrowheads). Expression of myc6-MP1 or Xpress-p14-CAAX does not restore FA pattern in p14−/− MEFs (presence of myc6-MP1 is confirmed by anti-myc antibody showing cytoplasmic distribution of MP1, presence of p14-CAAX is confirmed by anti-Xpress antibody). Reduction of FA size can be achieved by expression of p14-GFP (blue arrowheads, presence and localization of p14-GFP is confirmed by GFP image). Bar, 10 µm. (D) Quantification of average FA length and detailed analyzes of FA populations (“Length distribution”) in MEFs. Left graph: mean in percent ± SEM compared with control p14f/− MEFs (mean of FA length in control p14f/− MEFs was taken as 100%). Right graph: FAs are clustered in four specified length groups (x-axis) and are presented in percentage of all adhesions (y-axis) in each type of MEFs. See also Table S1. (E) FACS analysis of activated β1 integrins (identified with 9EG7 antibody) reveals no difference in expression in p14f/− MEFs (black line) and p14−/− MEFs (blue line; dotted lines indicate negative controls). The data shown are from a single representative experiment out of three independent experiments. For complete integrin profile, see Fig. S2.
To understand how the late endosomal p14–MP1 complex contributes to cell migration we analyzed FAs by immunostaining for Paxillin (Fig. 1 C). In control p14f/− MEFs, different populations of substrate adhesions could be distinguished, including small punctuated focal complexes at the leading edge (Fig. 1 C, red arrowheads), mature FAs of different sizes in the cell body, and trailing FAs at the cell rear. In contrast, in p14−/− MEFs, small FCs were difficult to detect, whereas elongated FAs were prominently enriched at the cell periphery (Fig. 1 C, red arrows).
On average, FA length was twofold increased in p14−/− MEFs versus p14f/− MEFs (Fig. 1 D and Table S1). Frequency distribution analysis of FA subpopulations revealed that control p14f/− MEFs contained two major groups of FAs with sizes between 0.5 and 1.0 µm (41.9% of all adhesions) or with sizes between 1.0 and 1.5 µm (30.5%). In contrast, the majority of FAs was longer than 1.5 µm in p14−/− MEFs (Fig. 1 D, length distribution graph; and Table S2). Importantly, FA length and distribution could be rescued by retroviral re-expression of p14-GFP. However, FA defects were not rescued by the expression of myc6-MP1 alone, which remains cytoplasmic due to the loss of its late endosomal binding partner p14. Expression of a CAAX-tagged p14, which redirects the p14–MP1 complex to the plasma membrane (Wunderlich et al., 2001), also failed to rescue FA length and distribution. This indicates that the late endosomal localization of the p14–MP1 complex was required for FA regulation (Fig. 1, C and D). In line with this hypothesis, we have also observed significant FA elongation in HeLa cells where MP1 was down-regulated (Fig. S1, C and D).
To test if loss of p14 affects activation of integrins, we performed FACS analyses revealing that β1, β3, α5, and αV integrins were expressed at similar levels in p14f/− MEFs and p14−/− MEFs and that β2, β4, α1, α2, and α6 integrins were not detectable (Fig. S2). Moreover, MnCl2 treatment activated β1 integrins to a similar extent in p14f/− and p14−/− MEFs (Fig. 1 E), indicating that the p14–MP1 complex neither regulated cell surface expression nor activation of integrins.
Although immunostaining for Paxillin reveals the majority of adhesion sites, another focal adhesion protein, Zyxin, was shown to incorporate into adhesions at a later maturation stage and to colocalize with Paxillin only in the larger, definitive FAs (Zaidel-Bar et al., 2003). Double immunolabeling for Paxillin and Zyxin revealed that FAs in p14−/− MEFs contained Zyxin (Fig. 2 A).
Figure 2.
Analysis of FA turnover. (A) IF of p14f/− and p14−/− MEFs with anti-Paxillin and anti-Zyxin antibodies. White arrows indicate leading edge formation areas in the control cells where no Zyxin-positive FAs are detected (magnification). In the p14 knockout MEFs the overall elongated FAs are also positive for Zyxin (magnification). (B) FRAP analysis of FAs in indicated MEFs. Image shows representative example of FAs (white arrows) before bleaching and during recovery time in p14f/− MEFs. Graph shows curves of fluorescence recovery (mean of 10 cells per group, number of measured FAs = 60) after bleaching of indicated MEFs. (C) TIRF microscopy of MEFs expressing mRuby-Paxillin. See also Videos 2 and 3. Shown are mature FAs of magnified areas of control and p14 knockout MEFs at three different timeframes (1 frame = 10 min). Merging of the three color-coded frames indicates the development of single FAs over time. Note: white FAs are stable over the shown time period (20 min). (D) Quantification of FA turnover. The left graph displays the existence of single FAs (n = 168) as dots over a time period of 60 min. All existing FAs were included. The right graph shows dynamic FA events. New growing FAs were analyzed during a time period of 80 min (P < 0.001).
To directly address if FA dynamics were altered in p14−/− MEFs, we performed FRAP of mCherry-Paxillin. Previous FRAP studies revealed that FAs differ in their dynamic protein exchange rate and density of their components, which in turn affects FA remodeling (Ballestrem et al., 2001; Wolfenson et al., 2009). We performed the FRAP experiments with MEFs that were transiently expressing mCherry-Paxillin (Laukaitis et al., 2001). FAs that were at least 1 µm in length were photobleached and FRAP was measured (Fig. 2 B). Whereas the kinetics of mCherry-Paxillin recovery did not differ in p14f/−, p14−/,− and p14−/−;p14-GFP MEFs (Thalf = 14.03 ± 1.3, 14.03 ± 1.5, and 12.05 ± 2.2 s, respectively), the extent of recovery was strongly reduced in p14−/− MEFs, reaching only ∼60% of control MEFs. The reduced overall recovery of mCherry-Paxillin revealed that Paxillin could not be efficiently exchanged, suggesting that FAs in the p14−/− MEFs contained a large proportion of immobile Paxillin molecules. Reconstitution of the late endosomal p14–MP1-signaling complex in p14−/−;p14-GFP MEFs rescued the recovery of mCherry-Paxillin (up to ∼90%; Fig. 2 B, graph).
Next, we performed total internal reflection fluorescence (TIRF) microscopy of mRuby-Paxillin–transfected MEFs and monitored FA dynamics over a period of 60–80 min (Videos 2 and 3; and Fig. 2, C and D). FA turnover was visualized by overlaying single frame shots of three different time points, summarizing a 20-min time span. The merged FA snapshots show decreased FA dynamics (white areas) in the p14−/− MEFs compared with the control cells (Fig. 2 C). Then, we manually tracked single FAs over time to analyze their stability in control and p14−/− MEFs (Fig. 2 D). We analyzed the longevity of existing (Fig. 2 D, left graph) and newly formed (Fig. 2 D, right graph) FAs. Collectively, our findings show that FA turnover is decreased in p14−/− MEFs.
MP1-carrying late endosomes target dynamic FAs
The potential contribution of local late endosomal signaling for the regulation of FAs was investigated in cells expressing GFP-MP1 (localizes to late endosomes; Wunderlich et al., 2001) and mCherry-Paxillin, which is present in FAs. Co-expression of GFP-MP1 and mCherry-Paxillin in NIH3T3 and HeLa cells revealed a partial colocalization of MP1-carrying late endosomes with FAs (Fig. 3 A). By tracking the localization of the p14–MP1 endosomes using time-lapse epifluorescence microscopy (Fig. 3 B and Video 4), we observed that a small population of late endosomes underwent directional anterograde transport and targeted FAs. Targeting events included targeting of a single FA, movement of late endosomes from one FA to another (Video 4), and targeting of a single FA repetitively (Fig. 3 B, white arrows). TIRF time-lapse microscopy and single-particle tracking revealed that the repetitive targeting of FAs by p14–MP1 endosomes occurred within less than approximately 100 nm (Fig. 3 C and Video 5). A GFP channel Z-projection of all frames recorded in Video 5, merged with mCherry-Paxillin FAs in the red channel, demonstrated an accumulation of MP1-GFP in close vicinity of FAs (Fig. 3 C).
Figure 3.
p14–MP1-carrying late endosomes target FAs. (A) IF of p14–MP1-carrying endosomes colocalizing with FAs in cells coexpressing GFP-MP1 (green) and mCherry-Paxillin (red; see white arrowheads). (B) NIH3T3 cell coexpressing GFP-MP1 (green) and mCherry-Paxillin (red). White arrows: FAs repetitively targeted by p14–MP1-carrying endosomes. See Video 4. (C) TIRF microscopy. Overlay of MP1-GFP Z-projection (green) and FAs (mCherry-Paxillin is shown in red). White arrowheads on selected time-lapse images point to individual MP1 endosomes (green) that target FAs (red). See Video 5. (D) Colocalization of MP1 with Rab7 and Transferrin. Top: images from time-lapse series of HeLa cell coexpressing GFP-MP1 (green) and mCherry-Rab7 (red). GFP-MP1 colocalizes with mCherry-Rab7 in cell center and cell periphery. Deconvolved image from the time-lapse shows colocalization of MP1 (green) and Rab7 (red) on ring-like structures of endosomes. Bottom: uptake of Transferrin (red) in HeLa cell expressing GFP-MP1 (green). (E) Quantification of the colocalization (mean ± SEM) between Rab7 and MP1 versus Transferrin and MP1. Insets in A and D are taken from white boxes in main panels.
MP1-GFP only occasionally colocalized (3–8.4%) with transferrin-positive early and recycling endosomes in transferrin uptake pulse-chase experiments (Fig. 3, D [bottom] and E). In contrast, MP1-GFP–positive late endosomes that targeted FAs (examples are shown in Fig. 3 D) colocalized with 94% of all mCherry-Rab7–positive structures, consistent with previous studies (Teis et al., 2002; Kurzbauer et al., 2004; Fig. 3, D [top] and E). These findings demonstrate that FAs were targeted by late endosomes that carry the p14–MP1 complex.
p14–MP1-carrying endosomes target mature FAs, but not FCs
Next, we asked whether all or only subpopulations of integrin adhesion sites get targeted by p14–MP1-containing endosomes. Previous experiments after siRNA depletion of MP1 in a prostate cancer line have indicated an involvement of MP1 in cell spreading (Pullikuth et al., 2005). However, we did not observe FA targeting of de novo–formed nascent FCs and FAs during cell spreading (Fig. S3 and Video 6).
FA targeting by p14–MP1 endosomes was further analyzed over a period of 2 h. The percentage of colocalization of fluorescence signals from mCherry-Paxillin (red channel) and GFP-MP1 (green channel) was calculated (Fig. 4, A and B; overall targeting in each experiment is taken as 100%; for details see Materials and methods). The overall frequency of targeting events in each frame of time-lapse sequences was 90.5 ± 1% (±SEM; n [FA] = 60; Fig. 4 B, “Targeting of FA”graph). Over 2 h, mature FAs were growing, sliding when cells moved (translocation revealed by recording changes of their XY coordinates), and some FAs were formed whereas others disappeared. The most stable FAs existed longer throughout the video sequence and were changing in XY the least. To distinguish between these particular subpopulations of FAs, we introduced the following parameters: “length of FA tracks” was used to identify sliding FAs, and “FA lifetime” to identify stationary FAs. An example of a processed video sequence and FA tracking used for calculations is shown in Fig. 4 A. Using these parameters we divided all FAs analyzed into three groups: (1) highly dynamic FAs (0–1.6-µm FA track, 0–20-min lifetime, indicated with green arrows on the graphs); (2) mature stable FAs (0–4.8 µm and 20–120 min, blue arrows); and (3) sliding FAs (4.8–11.3 µm and 0–40 min, red arrows).
Figure 4.

MP1 endosomes target mature FAs, but neither FCs nor newly formed FAs. (A) HeLa cells expressing GFP-MP1 (green) and mCherry-Paxillin (red) are used for detailed quantification of FA targeting. Top: images from the video sequence showing overlay of FAs (red) and endosomal (green) masks. Bottom: center of each FA was identified as a round sphere (purple) and used for further FA tracking. FA tracks (black arrows) during time lapse are illustrated in a line of different colors, indicating temporal changes of FA positions (as shown on the color bar below). (B) Quantification of the FA targeting by p14–MP1-carrying endosomes (mean in percentage ± SEM) and of FA targeting (per frame) during the time lapse. Green arrows indicate parameter combination of highly dynamic FAs, blue arrows of mature stable FAs, and red arrows of sliding FAs. Experiments were repeated three times, n = 10 (n = the number of cells used for quantification). (C) Schematic segmentation of FAs dissected with respect to the change of its fluorescence signal over time (left image). Right graph depictures mean calculation of fluorescence MP1–Paxillin colocalization in the FA segments. Experiments were repeated three times, n = 10 (n = the number of cells used for quantification).
All populations of FAs were targeted actively by endosomes (Fig. 4 B, “Length of FA track” and “FA lifetime” graphs). FAs with the longest FA tracks were targeted more often (38.7 ± 0.9%) compared to those with shorter FA tracks (23.3 ± 0.6%, 17 ± 0.9%, and 21 ± 1.9%; see Fig. 4 B). FAs existing in cells over 20–40 min were more often targeted (55.9 ± 6%) when compared to FAs with shorter lifetimes (0–20 min, 19.1 ± 5.1%) or very long lifetimes (40–120 min, 25 ± 2.7%). Sliding FAs were targeted most frequently (Fig. 4 B, red arrows).
Next, the colocalization of Paxillin/MP1 fluorescence signals in different parts of individual FAs was analyzed (Fig. 4 C). Each FA area was divided into segments with respect to the change of its fluorescence signal over time (schematic segmentation of FAs is shown in Fig. 4 C). “Stable” segments (unchanged over time) and “dynamic” segments (areas changing between video frames) of individual FAs were identified and fluorescence colocalization of each segment was separately calculated (Fig. 4 C, “Targeting of FA segments” graph). Interestingly, dynamic segments of FAs were targeted significantly more often when compared with stable ones (77 ± 1.4% vs. 23 ± 0.9%; Fig. 4 C).
Arl8b-dependent MT plus end–directed transport of late endosomes regulates FAs
Late endosomes are transported along MTs in a retrograde manner (Pastan and Willingham, 1981). To test whether the observed anterograde movement of p14–MP1-carrying endosomes toward FAs also utilizes MTs, we analyzed by time-lapse microscopy cells triply transfected with mCherry-Paxillin, mCherry-Tubulin, and GPF-MP1. We found that p14–MP1 endosomes were moving along MTs toward FAs (Fig. 5 A and Video 7). A representative example of an individual endosome (Fig. 5 A and Video 8) demonstrated that the endosome used a single MT to move to FAs, where it was trapped as soon as the MT stopped growing and depolymerized. Next, we treated cells with nocodazole to depolymerize MTs (Fig. 5 B). Nocodazole treatment completely abrogated the movement of p14–MP1 endosomes. A few MP1 endosomes remained at FAs, whereas the majority accumulated in the perinuclear region or displayed dispersed cytoplasmic localization (Fig. 5 B, white arrows). Thus, anterograde movement of MP1-carrying endosomes required intact MTs and can therefore be considered as plus end–directed transport.
Figure 5.
Arl8b-dependent MT plus end–directed transport of late endosomes regulates FAs. (A) MP1 endosomes are transported along MTs. Time-lapse images of HeLa cells expressing GFP-MP1 (green), mCherry-Paxillin (red), and mCherry-tubulin (red) show colocalization of MP1 and MTs (white arrowheads). Representative individual endosome moves along MTs toward two FAs (bottom panels, white arrowheads). See also Video 7 and 8. (B) Nocodazole treatment of a cell transfected as in A results in MT depolymerization and “trapping” of few GFP-MP1 endosomes in FAs (white arrows and arrowheads). Time-lapse images of the same cell show that positions of GFP-MP1 endosomes do not change in time due to abolished MP1 transport. (C) Arl8b knockdown in MEFs. IF: anti-LAMP1 (green), anti-tubulin (red) antibodies, and Hoechst. LAMP1-positive late endosomes collapse to the perinuclear region upon Arl8b knockdown (white arrow). WB: anti-Arl8b antibody, anti-tubulin used as loading control. (D) The p14−/−;p14-GFP MEFs treated with control and Arl8b RNAi. The late p14-GFP endosomes cluster in the Arl8b RNAi-treated cells (red arrow). See also Video 9. (E) The graph on the left shows the quantification of average FA length in MEFs. Mean in percent ± SEM compared with control p14f/− MEFs treated with control RNAi (mean of FA length in control p14f/− MEFs treated with control RNAi was taken as 100%). See also Table S1. The graph on the right shows the migration speed of p14−/−;p14-GFP MEFs transfected with control (n = 26) and Arl8b siRNA (n = 66) in wound-healing assay (µm/h, mean of cell migration speeds ± SD). (F) Colocalization of Paxillin and Rab7 in MEFs. Images from time-lapse series of MEF cells coexpressing GFP-Rab7 (green) and mCherry-Paxillin (red). White arrows indicate FAs targeted by GFP-Rab7. See also Video 10.
Arl8b-GTP was shown to associate with the late endosomal membranes and to ensure binding of the late endosomes to the motor protein kinesin-1 (Bagshaw et al., 2006; Hofmann and Munro, 2006; Rosa-Ferreira and Munro, 2011). To confirm that FA targeting by p14–MP1 endosomes and its effect on FA elongation involves active kinesin-dependent transport, we depleted Arl8b protein by RNAi in MEFs (Fig. 5 C). Depletion of Arl8b had no visible effect on MT patterns and led to clustering of LAMP1-positive late endosomes in the perinuclear area (Fig. 5 C, white arrow). We performed time-lapse microscopy in p14−/− MEF;p14-GFP (Fig. 5 D and Video 9). In cells treated with control RNAi, p14-GFP endosomes moved actively throughout the cell, whereas down-regulation of Arl8b caused clustering of p14-GFP late endosomes in the perinuclear region of cells (Fig. 5 D, red arrow; and Video 9).
Next, we performed immunostaining for Paxillin and measured FA length (Fig. 5 E and Table S1). Interestingly, FA length was not further affected in p14−/− MEFs upon Arl8b knockdown (Fig. 5 E and Table S1). However, in p14f/− MEFs treated with Arl8b RNAi, length of FAs was twofold increased as compared with p14f/− MEFs treated with control RNAi (Fig. 5 E, left graph; and Table S1). Additionally, we analyzed the migration speed of Arl8b RNAi–treated cells in a wound-healing assay. In comparison to control siRNA treated cells, knockdown of Arl8b in p14−/− MEF;p14-GFP decreased the migration speed by ∼30% as compared with the control cells (Fig. 5 E, right graph). These results suggest that active transport of late endosomes requires Arl8b to regulate FAs.
Interestingly, late endosomes were transported toward FAs also in the absence of the p14–MP1 complex, as revealed by time-lapse microscopy in p14f/− MEF and p14−/− MEF cells cotransfected with mCherry-Paxillin and GPF-Rab7 (Fig. 5 F and Video 10).
IQGAP1 is a novel interacting protein of MP1
To further investigate how the p14–MP1 complex would regulate FA dynamics, we performed a two-hybrid screen using MP1 as bait in order to identify potential interaction partners. Human IQGAP1 (GenBank accession no. NM_003870) was identified as an MP1-interacting protein in 24 positive hits. To confirm this interaction we first used p14f/− MEF cells stably expressing myc6-MP1 for immunoprecipitation studies. Immunoprecipitation of endogenous IQGAP1 resulted in the co-immunoprecipitation of myc6-MP1 (Fig. 6 A) and endogenous IQGAP1 was detected in the myc6-MP1 immunoprecipitates (Fig. 6 A). We further extended these interaction studies with co-immunoprecipitation of IQGAP1 and detection of the endogenous MP1 and p18 (LAMTOR1) proteins, the latter one representing the membrane anchor of the p14–MP1 complex (Fig. 6 B). Finally, HeLa protein lysates were incubated with GST-MP1, GST-IQGAP1, or GST alone immobilized on glutathione-Sepharose.
Figure 6.

IQGAP1 and MP1 interaction. (A) Reciprocal co-immunoprecipitation of myc6-MP1 and IQGAP1. Cell extracts were prepared from myc6-MP1–expressing p14f/− MEFs. IP: with anti-myc (lane 2) and anti-IQGAP1 (lane 3) polyclonal antibodies; IgG control (lane 1). WB: anti-IQGAP1 (top) and anti-myc (bottom) antibodies, respectively. Black arrow indicates myc6-MP1 signal. (B) Immunoprecipitation of endogenous MP1 and p18. Cell extracts were prepared from HeLa cells. IP: with anti-IQGAP1 (lane 3) polyclonal antibody; cell lysates (lane 1), IgG control (lane 2). WB: anti-IQGAP1 (top), anti-p18 (middle), and anti-MP1 (bottom) antibodies, respectively. (C) Top: HeLa protein lysates were incubated with GST-MP1 or GST alone immobilized on glutathione-Sepharose beads. WB: anti-IQGAP1 antibody. Lysates not subjected to GST pull-down were processed in parallel (HeLa lysate). Bottom: HeLa protein lysates were incubated with GST-IQGAP1 or GST alone immobilized on glutathione-Sepharose beads. WB: anti-MP1 antibody. Lysates not subjected to GST pull-down were processed in parallel (HeLa lysate). Black arrow indicates MP1 signal.
In vitro–purified GST-MP1 interacted with endogenous IQGAP1 (Fig. 6 C, top) and in the reciprocal experiment in vitro–purified GST-IQGAP1 interacted with endogenous MP1 (Fig. 6 C, bottom), demonstrating that MP1 and IQGAP1 interact and hence confirming the two-hybrid interaction.
Furthermore, we confirmed the IQGAP1-MP1 interaction in situ using a Duolink proximity ligation assay (PLA). Because the available MP1 antibodies failed to detect MP1 in conventional immunofluorescence experiments, we used p14f/− MEFs stably expressing myc6-MP1, which specifically localizes to late endosomes (Fig. S4 A; Teis et al., 2006). In the PLA assay we used different sets of two primary antibodies to detect myc6-MP1 (rabbit) and IQGAP1 (mouse). The signal from each detected pair of PLA probes was visualized as an individual fluorescent dot (Fig. S4 B). IQGAP1–MP1 PLA was positive in p14f/−;myc6-MP1 MEF cells, but not in p14f/− MEFs, which we used as control cell line. This result supports our previous results and suggests that the interaction of MP1 and IQGAP1 occurs on late endosomes.
Absence of p14–MP1 or blockage of Arl8b-dependent late endosomal transport causes IQGAP1 accumulation in FAs
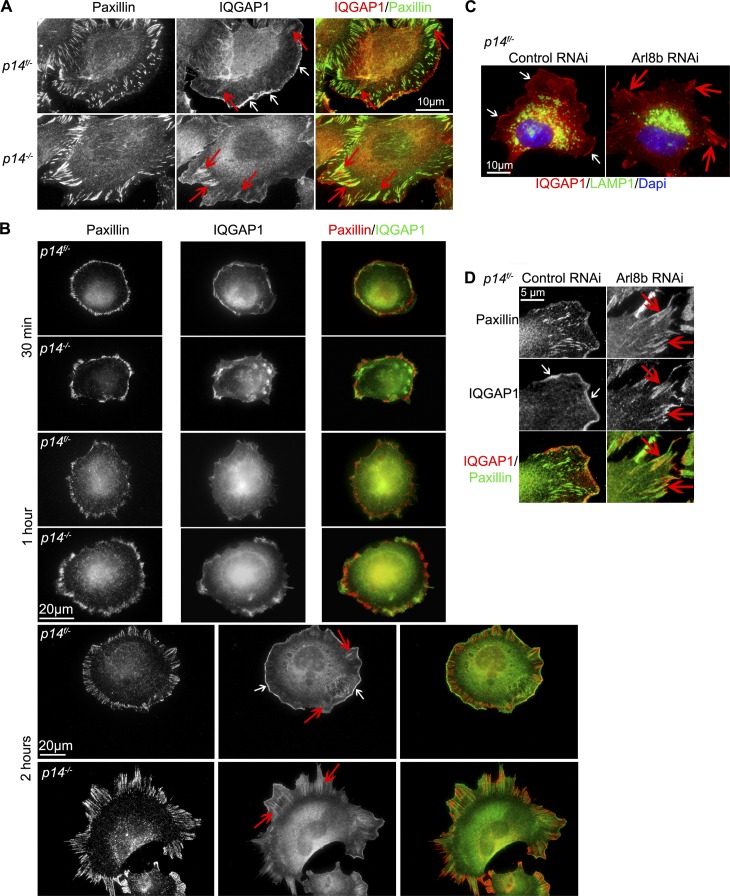
Next, we examined the subcellular localization of IQGAP1. Immunofluorescence experiments of cells migrating into a wound revealed that IQGAP1 displayed mainly cytoplasmic localization and was enriched at the leading edge of control cells (p14f/− MEFs; Fig. 7 A, white arrows) as previously described (Watanabe et al., 2004), but a small portion of IQGAP1 was also found in FAs (Fig. 7 A, red arrows). In contrast, IQGAP1 was reduced at the rim of the leading edge and strongly accumulated in FAs of p14−/− MEFs, where it colocalized with Paxillin (Fig. 7 A, red arrows). To further investigate the distribution of IQGAP1 in detail, we performed a cell-spreading assay and compared IQGAP1 localization at different time points after cell plating (Fig.7 B). At 30 min and 1 h after plating, control and p14-depleted cells looked indistinguishable in terms of morphology and IQGAP1 localization. At 2 h after plating, control as well as p14 knockout MEFs developed Paxillin-positive FAs, which in both cases showed a colocalization with IQGAP1 (red arrows). However, as described above, in p14f/− MEFs a major IQGAP1 fraction was enriched at the plasma membrane (white arrows), whereas in the p14−/− MEFs an accumulation on elongated FAs was observed (red arrows).
Figure 7.
Absence of p14–MP1 or blockage of Arl8b-dependent late endosomal transport causes IQGAP1 accumulation in FAs. (A) IF: anti-Paxillin and anti-IQGAP1 antibodies. IQGAP1 localizes to the leading edge in p14f/− (white arrows) and colocalizes with Paxillin in FAs in p14−/− MEFs (red arrows). (B) IF: anti-Paxillin and anti-IQGAP1 antibodies. Shown are different time points during spreading of p14f/− and p14−/− MEFs. White arrows indicate accumulation of IQGAP1 at the leading edge in control MEFs. Red arrows point at IQGAP1 localization at Paxillin-positive FAs in control and p14−/− MEFs. (C) IF: anti-LAMP1 (green), anti-IQGAP1 (red) antibodies, and Hoechst (blue). IQGAP1 localizes to the leading edge in p14f/− MEFs treated with control RNAi (white arrows) and localizes to FAs upon Arl8b depletion (red arrows). (D) p14f/− MEFs treated as in C. IF: anti-Paxillin and anti-IQGAP1 antibodies. Note IQGAP1 localization to the leading edge in p14f/− MEFs treated with control RNAi (white arrows) versus colocalization of IQGAP1 and Paxillin in FAs upon ARl8b depletion (red arrows).
Moreover, interruption of late endosomal traffic by the Arl8b RNAi depletion caused relocalization of IQGAP1 protein in control p14f/− MEFs similar to that observed in p14−/− MEF: IQGAP1 was reduced in the leading edge and accumulated in FAs (Fig. 7 C, white and red arrows), where it colocalized with Paxillin (Fig. 7 D, white and red arrows). Thus, knockdown of Arl8b in control p14f/− MEF cells phenocopied the loss of the p14–MP1 complex.
Down-regulation of IQGAP1 rescues FAs and cell migration in p14−/− knockout MEFs
To further investigate the interaction of IQGAP1 with p14–MP1 in regulation of FAs, we depleted IQGAP1 protein by RNAi. Reduction of IQGAP1 was efficient in p14f/− and p14−/− MEFs (immunofluorescence [IF] in Fig. 8, Western blotting [WB] in Fig. S5 A). In control p14f/− MEFs treated with IQGAP1 RNAi, FAs were somewhat elongated (22.3%) as compared with p14f/− MEFs treated with control RNAi (Fig. 8 B, left graph; and Table S1). Interestingly, the IQGAP1 knockdown in p14−/− MEFs strongly decreased the length of FAs (56.6% decrease, as compared with p14−/− MEFs treated with control RNAi).
Figure 8.
In the absence of p14–MP1 endosomal signaling, down-regulation of IQGAP1 by RNAi rescues FAs and cell migration. (A) IF: anti-Paxillin and anti-IQGAP1 antibodies. (B) Left: quantification of average FA length in MEFs transfected with control or IQGAP1 RNAi. Mean in percent ± SEM is compared with control p14f/− MEFs transfected with control RNAi (taken as 100%). Right: calculation of speed of migration in wound-healing assays. Mean in µm/h ± SEM. Importantly, only cells that had strong down-regulation of IQGAP1 (no signal on IF using anti-IQGAP1 antibodies as in A) were used for the calculation. See also Table S1.
Furthermore, we performed wound-healing assays and measured speed of cell migration (Fig. 8 B, right graph; and Fig. S5 B). Depletion of IQGAP1 had no significant effect on cell migration in control p14f/− MEFs (15 ± 0.16 µm/h, mean ± SEM in p14f/− MEFs treated with control RNAi vs. 14.7 ± 0.16 µm/h in p14f/− MEFs treated with IQGAP1 RNAi). However, knockdown of IQGAP1 in p14−/− MEFs promoted cell migration and efficiently rescued the migration defect observed in p14−/− MEFs (13.9 ± 0.20 µm/h as compared with 8.6 ± 0.10 µm/h of the respective control cells). These results suggest that in the absence of the p14–MP1 complex, IQGAP1 accumulation at FAs correlated with elongation of FAs and impaired cell migration.
Discussion
In the current work, we observed that p14–MP1 (LAMTOR2/3)-carrying late endosomes are transported from the cell center toward FAs. However, these perinuclear organelles can also undergo anterograde kinesin-dependent transport, although the biological role of the outward movement has remained so far unclear (Hollenbeck and Swanson, 1990; Feiguin et al., 1994; Nakata and Hirokawa, 1995).
Here, we demonstrated that the MT plus end–directed movement of a small but steadily p14–MP1-carrying, Rab7-positive, late endosome subpopulation was followed by directed targeting of FAs. Importantly, mature FAs (but not FCs) were actively targeted by p14–MP1-carrying endosomes, indicating that p14–MP1 is not directly involved in the recycling of FA components in newly formed adhesions. FAs can grow and translocate (slide) by treadmilling where integrins are added on the one end of FAs and removed from the other (Geiger and Bershadsky, 2001). Most of the targeting events by p14–MP1 we observed in sliding FAs, proposed to be important for cell translocation (Ballestrem et al., 2001; Rid et al., 2005). Moreover, segmentation of individual FAs into dynamic (changing) and stable segments revealed that endosomal targeting mainly took place in the dynamic areas where adhesion components are either added or removed. Thereby, the p14–MP1 complex might locally regulate FA remodeling.
Formation and disassociation of FAs is precisely controlled by diverse signaling pathways as well as by modulation of tensile forces (Broussard et al., 2008). Recent publications showed new and unexpected roles for endosomes in regulating cell migration (Sadowski et al., 2009; Scita and Di Fiore, 2010; Schiefermeier et al., 2011). Interestingly, previous findings demonstrated a specific regulation of FA disassembly by MT targeting (Kaverina et al., 1999). Blockage of the kinesin motor activity, either via microinjection of antibodies or of a kinesin-1 heavy chain construct mutated in the motor domain, induced a dramatic increase in the size and reduction in number of FAs, mimicking the effect observed after MT disruption by nocodazole (Krylyshkina et al., 2002). This would suggest the involvement of the kinesin-dependent transport of certain proteins/organelles to FAs, necessary for the release of adhesion components. Our data confirm and extend these observations because depletion of Arl8b protein that assures kinesin-1 binding to the late endosomal membrane (Bagshaw et al., 2006; Hofmann and Munro, 2006; Rosa-Ferreira and Munro, 2011) resulted in reduced cell migration and significant FA elongation in control cells.
Moreover, our data demonstrated that the absence of p14–MP1 from late endosomes caused significant FA elongation, decrease of FA turnover, and affected the ability of cells to migrate. Our FRAP experiments revealed that Paxillin molecules were not efficiently exchanged in p14−/− MEFs. We also observed that FAs in p14-depleted cells were more stable and existed longer than in control cells. Thus, elongated FAs were less dynamic and consequently led to a cell migration defect. However, not late endosomal transport per se, but the presence of the p14–MP1 complex on the late endosomes seemed to be important for the FA remodeling because neither restoration of p14–MP1 at the plasma membrane nor cytoplasmic complexes could rescue impaired FAs in p14−/−-deficient cells, and p14–MP1-depleted Rab7 endosomes still targeted FAs.
The p14–MP1 complex was previously described to scaffold MAPK signaling. MAPK plays an important role in FA formation as well as disassembly (Chen et al., 1994; Katz et al., 2007; Pullikuth and Catling, 2007). It was shown that treatment of different cell types with MEK-to-ERK inhibitors results in the formation of enlarged FAs and impaired migration of cells in response to various stimuli (Klemke et al., 1997; Webb et al., 2004), whereas MAPK-dependent phosphorylation of FA components was suggested to cause these defects (Brown et al., 1998; Parsons et al., 2000; Webb et al., 2005). However, in the MEF cells investigated here (p14−/− vs. control p14f/− MEFs), we did not identify differences in the phosphorylation of Paxillin (Ser126/Tyr118), FAK (Tyr925/576/577), Cofilin (Ser3), and IQGAP1 (unpublished data). The phospho-regulation of FA protein(s) in the absence of the p14–MP1 complex will in the future require large and comprehensive phosphoproteomics analyses.
How does the late endosomal traffic of p14–MP1 complexes regulate FAs? In our present work we have identified IQGAP1 as a novel interaction partner of MP1. We observed IQGAP1 accumulation in FAs upon loss of p14, which was also phenocopied by Arl8b RNAi depletion. Interestingly, IQGAP1 accumulated both in FAs of control and p14-depleted cells during FA maturation. Our data suggest that IQGAP1 accumulation may be the cause of impaired FA dynamics because down-regulation of IQGAP1 by RNAi could rescue FAs and the migration defect initially observed in p14−/− MEFs.
IQGAP1 was proposed to regulate the actin cytoskeleton, microtubules, and cell migration by multiple pathways including small GTPases and MAPK signaling (Hart et al., 1996; Roy et al., 2004; Brown and Sacks, 2006). Although IQGAP1 knockdown caused slight elongation of FAs and formation of protrusions in control p14f/− cells (unpublished data), in contrast with previously published observations (Fukata et al., 2002; Mataraza et al., 2003) no reduction in speed of cell migration was observed. We did not further evaluate the role of the IQGAP1 in the actin dynamics of MEFs because this effect of IQGAP1 seems to be p14–MP1 independent.
Two recent FA proteome studies identified IQGAP1 to associate with FAs (Kuo et al., 2011; Schiller et al., 2011), although its function remains unclear. IQGAP1 was found to bind to the integrin-linked kinase ILK in keratinocytes, where IQGAP1 colocalized with ILK in FCs, but not in FAs (Wickström et al., 2010). Thus, it seems that IQGAP1 functions in an ILK-dependent manner in young FCs and in a p14–MP1-dependent manner in mature FAs. Clearly the identification of downstream effectors of IQGAP1 function in FCs and FAs will help to explain how IQGAP1 functions during cell migration.
IQGAP1 was previously shown to bind myosin light chain 2 (Weissbach et al., 1998; Wu et al., 2003). Budding and fission yeast IQGAP proteins were demonstrated to anchor myosin II at the division site (Fang et al., 2010; Laporte et al., 2011). Recently, IQGAP2 was found to be antagonized by IQGAP1 in modulation of myosin-dependent mechanotransduction (Kee et al., 2012). However, the functional consequences of this interaction in mammalian cells remain to be elucidated. In fibroblasts, actomyosin contractility plays a dual role in the process of cell migration. Myosin-dependent tensile forces trigger adhesion maturation (Vicente-Manzanares et al., 2007), whereas reversing the maturation signal drives adhesion disassembly (Gupton and Waterman-Storer, 2006). Also, a balance between adhesion strength and myosin activity is required for optimal migration (Gupton and Waterman-Storer, 2006).
Our data show that most of the FA targeting by p14–MP1 takes place in mature sliding FAs, where the balance of myosin-dependent contractile forces plays an important role (Vicente-Manzanares et al., 2007). It is tempting to speculate that p14–MP1 interaction with IQGAP1 plays a role in regulation of contractility. Furthermore, both MP1 and IQGAP1 were shown to interact with PAK1 (Noritake et al., 2005; Pullikuth et al., 2005). PAK1 plays a complex role in regulation of cell migration; interaction partners of PAK1 include Rac, MAPK, and myosins (Manser et al., 1994; Frost et al., 1998; Sanders et al., 1999; Delorme-Walker et al., 2011). Future studies should evaluate the exact relations between MP1, IQGAP1, PAK1, and myosin at local FA levels.
Based on our data we propose the following model. In migrating cells, IQGAP1 is localized to the plasma membrane and to FCs. Upon maturation of FCs into FAs and further dynamic FA turnover, FAs are targeted by p14–MP1 late endosomes. Consequently, IQGAP1 is removed from FAs and recycles back to the plasma membrane. This removal of IQGAP1 no longer occurs in the absence of p14 and IQGAP1 begins to accumulate at FAs. It is likely that interaction partners of IQGAP1 will also begin to accumulate and thereby contribute to the loss of FA dynamics. Thus, loss of p14 results in a phenotype where new FCs are not efficiently formed, mature FAs are elongated and less dynamic, and cell migration is impaired. These results led us to conclude that the removal of IQGAP1 excess could be essential for the regulation of FA dynamics during cell migration and requires local p14–MP1 late endosomal targeting of FAs.
Materials and methods
Tissue culture, transfections, and RNAi
HeLa, NIH3T3, and MEF cells were grown in high glucose DMEM supplemented with 10 mM Hepes, pH 7.3, 100 IU/ml penicillin, 100 µg/ml streptomycin, and 10% FC at 37°C, in 5% CO2 and 95% humidity. Media and reagents for tissue culture were purchased from Gibco. Cells were transfected with Lipofectamine 2000 (Invitrogen) following the manufacturer’s instructions. RNA interference of IQGAP1 and Arl8b were performed using Thermo Fisher Scientific’s SMARTpool siRNAs against the corresponding targets and Saint-Red (Synvolux Therapeutics) as a transfection reagent. As a control, siCONTROL Nontargeting siRNA Pool was used (Thermo Fisher Scientific).
Antibodies and reagents
The monoclonal mouse anti–α-tubulin antibody, DAPI, and Hoechst (Bisbenzimide H33258) were obtained from Sigma-Aldrich. The rabbit anti-Arl8b polyclonal antibody was bought from ProteinTech. The rabbit polyclonal antibody against IQGAP1 was obtained from Santa Cruz Biotechnology, Inc., and the mouse monoclonal anti-IQGAP1 was purchased from BD. The mouse monoclonal anti-Paxillin antibody was bought from EMD Millipore, and the rat anti-LAMP1 (CD107a) antibody was purchased from Merck. The rabbit polyclonal anti-p18 antibody was obtained from Atlas Antibodies. For the rabbit polyclonal anti-p14 antibody the serum was raised against the GST fusion protein of p14 and for the rabbit polyclonal anti-MP1 antibody the serum was raised against the peptide Kp532 (CVSDRDGVPVIKVANDSAPEHALR) as described previously (Teis et al., 2002). For the HeLa coimmunoprecipitation the rabbit monoclonal MP1 antibody from Epitomics was used. Rabbit polyclonal anti-Myc antibody was from Gramsch Laboratories. The rabbit polyclonal anti-Zyxin antibody was obtained from ProteinTech. The Alexa Fluor 568 phalloidin–, Alexa Fluor 488–, and Alexa Fluor 568–conjugated anti–mouse and anti–rabbit secondary antibodies and mouse monoclonal anti-Xpress antibody was purchased from Molecular Probes. Nocodazole was bought from EMD Millipore and applied on cells (10 µM) for 1 h. Western blotting was performed as published previously (Fialka et al., 1997).
Plasmid DNA constructions
mCherry-Paxillin-C3 was generated by replacing the YFP sequence of the pYFP-Paxillin-C3 expression vector (vector backbone: pYFP-C3) provided by B. Geiger (The Weizmann Institute of Science, Rehovot, Israel; Zaidel-Bar et al., 2007) and introducing mCherry cDNA, provided by R.Y. Tsien (University of California, San Diego, La Jolla, CA; Shaner et al., 2004). The YFP sequence from pYFP-Paxillin-C3 was removed upon digestion with HINDIII and AgeI enzymes following the manufacturer’s instructions (Fermentas). PCR products introducing HindIII sites in mCherry fragment sequence were amplified and were further subcloned into pYFP-Paxillin-C3 using the Rapid DNA Ligation kit (Roche). For the mRuby-Paxillin construct, Paxillin cDNA was PCR amplified and ligated into a pENTR4-mRuby2 vector using EcoRV and XbaI restriction sites N terminally and in frame of the mRuby2 fluorescent protein and sequence verified. pENTR4-mRuby2 was generated by ligating mRuby2 cDNA into the multiple cloning site of a pENTR4 vector (Invitrogen) via NcoI and HindIII restriction sites. mRuby2-tagged Paxillin was subsequently cloned out of the pENTR4 vector backbone in the retroviral expression vector pQCXIN-DEST using Gateway multicloning technology (Invitrogen). GFP-Rab7 (vector backbone: pEGFP-C1) was provided by C. Bucci (Department of Clinical and Experimental Medicine, Federicoll, Napoli, Italy; Bucci et al., 2000). Cloning of mCherry-Rab7 PCR was performed using GFP-Rab7 as a template. Insertion of HindIII and SalI sites on the ends of Rab7 cDNA allowed for the digestion and subsequent ligation into pmCherry-C1. MP1-GFP (vector backbone: pEGP-C1) and p14-CAAX were described previously (Wunderlich et al., 2001). In brief: CAAX-tagged p14 was constructed by placing a linker sequence encoding the last 21 amino acids of human K-ras at the C terminus of the p14 cDNA, replacing the STOP codon. Further, p14-CAAX was cloned in frame with the His6/Xpress tag into pEF4/HisC vector (Invitrogen).
IF, time-lapse microscopy, TIRF, and PLA
IF was performed as described previously (Teis et al., 2002). In brief, cells were fixed with pre-warmed 4% paraformaldehyde in cytoskeleton buffer (10 mM Pipes, pH 6.8, 150 mM NaCl, 5 mM EGTA, 5 mM glucose, 5 mM MgCl2, 5 mM Na4P2O7, 2 mM Na3OV4, 50 mM NaF, and 20 mM β-glycerophosphate) for 20 min at room temperature, after washing in CB and 2 min permeabilization in 0.1% Triton X-100 in CB. Washed cells were blocked for 1 h in a blocking solution (1% BSA, 5% horse serum, and 15% goat serum [both from Gibco] in PBS), incubated for 1 h with first antibody, washed in PBS, incubated for 30 min with secondary antibody, washed in PBS, and mounted in Mowiol (EMD Millipore). Images were taken at room temperature with either an inverted microscope (Axiovert 200; Carl Zeiss) with a 0.4 NA/32× air objective or a confocal microscope (AxioImager M1.2; Carl Zeiss) with a 1.4 NA/63× oil objective equipped with a CCD camera (CoolSNAP HQ2; Photometrics), and acquisition was controlled by the AxioVision release 4.5 SP1 software (Carl Zeiss). For time-lapse microscopy, cells were kept in phenol red–free DMEM medium (Sigma-Aldrich), supplemented with 10% FC (Gibco) Time-lapse movies were taken on the Axiovert 200 microscope equipped with a 0.3 NA/10× and 0.4 NA/32× air objective and a 1.4 NA/63× and 1.4 NA/100× oil objective. All figures were prepared in Photoshop, videos in ImageJ (National Institutes of Health) and Imaris (Bitplane). Image shown in Fig. 3 D was deconvolved using Imaris, as stated in the Fig. 3 legend. Scripts used in Imaris software are described in the Supplemental material. Images were taken at 37°C in a CO2 incubation chamber.
TIRF microscopy.
Images were captured using an inverted microscope (Axiovert 200M; Carl Zeiss) with a 1.6 NA/100× oil objective and a CCD camera (CoolSNAP HQ; Photometrics). Acquisition was controlled by MetaMorph Software (Molecular Devices). TIRF images were collected at room temperature. For TIRF movies concerning FA turnover, cells stably expressing mRuby-paxilin were plated in Leibovitz’s L-15 medium (Invitrogen) without phenol red on 35-mm Petri dishes with a glass bottom (ibidi). Images were taken with an iMIC microscope (FEI Munich GmbH) equipped with a 1.49 NA/60× oil objectives and a CMOS camera (Orca Flash 4.0; Hamamatsu Photonics) at 37°C. Acquisition was controlled by Life Acquisition software (FEI Munich GmbH). For the random single-cell migration assay, cell were plated with full media in a 24-well plate and covered with a CellSedure lid (Chip-Man Technologies, Ltd.). Time-lapse imaging was done using a Cell-IQ v.2 system (Chip-Man Technologies, Ltd.) equipped with a 0.13 NA/4× air objective (Nikon). Acquisition was controlled by live-cell imaging analysis system software (Cell-IQ Imagen v.2.9.5.c; Chip-Man Technologies, Ltd.). Cells were kept at 37°C and 5% CO2 during imaging. To analyze single-cell migration in the wound-healing assay and random cell migration assay, the “Manual Tracking” plugin (ImageJ v1.48o) was used to track single cells. The speed parameter was calculated by analyzing the acquired data with the Chemotaxis and Migration Tool 1.01 plugin (ibidi).
Detection of protein–protein interactions by PLA (Duolink) was performed in situ according to the manufacturer’s suggestions (Olink Bioscience). In brief, cells were cultured on coverslips, fixed for 20 min in cold (−20°C) methanol, and blocked and incubated in a primary antibody as described above for cell staining, followed by a 2-h incubation in rabbit and goat PLA PLUS and PLA MINUS probes and a 15-min hybridization, 15-min ligation, 90-min amplification, and 60-min detection according to the manufacturer’s protocol. Nuclei were counterstained with DAPI (blue). Rabbit anti-myc (to detect MP1) was coupled with mouse anti-IQGAP1. To maximize signal strength, samples were incubated in primary antibody overnight (4°C). All steps after primary antibody incubation were performed at 37°C.
FRAP
FRAP data of mCherry-Paxillin–expressing MEFs were acquired with a confocal microscope (model SP5; Leica) equipped with a CO2 incubation chamber using a 1.2 NA/63× water objective and LAS AF acquisition software (Leica). mCherry fluorescence was acquired as follows: excitation, 561-nm laser; emission, 575–732 nm. Several 12-µm2 FA areas (1.5 µm × 8 µm) were photobleached (bleach pulse with the 561-nm DPSS laser and the 488-nm argon laser lines) in each cell. FRAP was monitored for 40 frames at 0.7 s/frame and additional 15 frames at 10 s/frame. FRAP data for the FRAP regions, a background region, and the “whole-cell region” (reference region) were exported from the LAS AF software and further analyzed with MATLAB. In brief, FRAP data were processed according to the double-normalization method, correcting for both background and bleaching in order to enable comparisons between different cells. Double-normalized FRAP data were averaged and fitted to a single exponential recovery curve to calculate recovery half-life and immobile fraction.
Two-hybrid screen
A bait fragment was generated by insertion of mouse MP1(1–124) into pB29 (LexA, N-terminal fusion). As prey library, the human placenta RP4 from Hybrigenics was used. The screen was performed using 20 mM of 3-amino-1,2,4-triazole. Overall, 327 primary clones were processed, leading to the identification of 56 interactions. From the obtained clones, 120 corresponded to antisense sequences that were not considered as relevant or included in the results. Among the interacting partners of MP1 we found, as expected, five clones corresponded to p14. Based on the number of hits and on results from previous two-hybrid screens (particularly important for the identification of false positives), Hybrigenics has derived a global prediction score for the confidence of each interaction. A very high confidence score was asserted for six interacting proteins, among them human IQGAP1 (GenBank accession no. NM_003870). IQGAP1 was found in 24 of the 327 clones processed.
Generation of stable cell lines
p14f/− and p14−/− MEFs were generated as described previously (Teis et al., 2006). In brief, the single mouse p14 gene (NCBI Gene no. 83409) is located on chromosome 3. Exons 1–4 of p14 were flanked by loxP sites to create a conditional allele in HM1-ES cells, which were injected into C57BL/6 blastocysts. All mice were maintained on a mixed genetic background of C57BL/6 and 129/Sv. To generate floxed p14 alleles, p14targ/targ mice were crossed with hACTB::Flpe mice. The p14-null allele, p14−/+, was generated by crossing p14targ/targ with MORE mice. p14f/− MEFs were derived from day 13.5 embryos, E1A immortalized, and infected with an adenovirus-expressing Cre to yield p14−/− MEFs. The p14-GFP MEFs were generated as described previously (Obexer et al., 2007; Stasyk et al., 2010). In short, pEGFP-p14 was expressed in p14−/− MEFs using retroviruses. The pEGFPp14 fusion protein was constructed by inserting p14 in a pEGFP-C1 vector (Takara Bio Inc.) and subcloned in a pLib-MCS2-iresPURO vector.
p14f/− and p14−/− MEFs stably expressing myc6-MP1 were generated as follows. PCR products introducing BamHI and SalI sites in mouse Myc6-MP1wt were amplified, and upon digestion with the same enzymes the products were subcloned into the retroviral transfer vector pLIB-MCS2-iresPURO.
The inducible MP1 knockdown HeLa cells were produced as follows. The construct (V2THS_98045 = RHS4696-99636658) with mature sense 5′-CACAGAAATGGTTCAGTCT-3′ in pTRIPZ was from Thermo Fisher Scientific.
Lentiviral supernatant was prepared by introducing the vector together with the Trans-Lentiviral Packaging Mix (Thermo Fisher Scientific) and Arrest-In transfection reagent (Thermo Fisher Scientific) into the 293LTV packaging cell line (LTV-100; Cell Biolabs, Inc.). Three days after transfection, the culture supernatant was filtered through a polyethersulfone filter (514-0075l; VWR International), and polybrene was added to 4 µg/ml and used to infect HeLa cells (Sigma-Aldrich). The cells were then selected 3 days after transduction with 1 µg/ml puromycin for 2 weeks.
Flow cytometry (FACS)
Flow cytometry was performed as described previously (Czuchra et al., 2006). In brief, 500,000 MEFs were used per staining. Cells were washed in 1% BSA/PBS and incubated with first antibody for 10 min at room temperature after a 30–40-min incubation on ice. First antibody was diluted either in FACS-Tris solution (24 mM Tris-HCl, pH 7.4, 137 mM NaCl, and 2.7 mM KCl) or in FACS-Tris + 5 mM MnCl2. Washing was performed in 1% BSA/PBS. Then cells were incubated with secondary antibody for 15 min on ice and washed in 1% BSA/PBS. Propidium iodide was added before measurements. The following antibodies were used for the assay: anti-α1 (AbD Serotec), -α5, -α6, -αV, -β1, -β2, -β3, -β4, and -β7 integrins (all obtained from BD). Biotinylated (anti-β3, -α5, and -αv integrins) antibodies were detected with streptavidin-Cy5 (Jackson ImmunoResearch Laboratories, Inc.). The 9EG7 antibody was from BD. Anti–rat FITC and anti–hamster FITC were from BD. To assess autofluorescence and nonspecific staining, cells were stained with IgM isotype-FITC, IgG2A isotype-FITC, or streptavidin-Cy5 (all obtained from BD).
Preparation of fusion proteins
As a control we used the pGEX-6P3 GST expression vector from GE Healthcare. The wild-type mouse MP1 sequence flanked by BamHI and EcoRI sites was amplified by PCR. The obtained product was then subcloned into pCR-blunt using the Topo-kit. The intermediate construct was digested with BamHI and EcoRI and the obtained insert ligated into the corresponding sites of pGEX4T1 (GE Healthcare). Wild-type human IQGAP1 cDNA was subcloned into the pGEX-2T vector. This construct was provided by D. Sacks (Department of Laboratory Medicine, National Institutes of Health, Bethesda, MD; Li and Sacks, 2003). GST fusion proteins were expressed in Escherichia coli and isolated with glutathione-Sepharose as described previously (Ho et al., 1999). In brief, GST and GST-MP1 were induced with 1 mM IPTG for 3 h at 37°C. GST-IQGAP1 was induced with 0.1 mM IPTG for 2 h at 16°C. Bacterial cultures were lysed by sonication in 50 mM Tris-HCl, pH 8.0, 2 mM DTT, 150 mM NaCl, 1 mg/ml lysosime, 10 µg/ml DNaseI, and protease inhibitors.
Pull-down experiments
The obtained E. coli lysates expressing GST, GSTMP1, or GSTIQGAP1 were bound to glutathione-Sepharose beads at 4°C. The beads were then washed with GST lysis buffer in preparation for the pull-down experiment. In parallel, HeLa cells were lysed in 50 mM Tris-HCl, pH 7.5, 250 mM NaCl, 1% NP-40, 50 mM NaF, 1 mM Na3VO4, 0.5 mM PMSF, and protease inhibitors and precleared with glutathione-Sepharose beads. The precleared HeLa lysates were incubated with immobilized GST, GST-IQGAP1, or GST-MP1 on a rotator at 4°C for at least 3 h. After washing in 50 mM Tris-HCl, pH 7.5, 250 mM NaCl, 1% NP-40, and 50 mM NaF, beads were resuspended in sample buffer and boiled.
Calculation of focal adhesion targeting
Image analysis was performed with ImageJ and Imaris software using custom-designed scripts (for details see Supplemental material). First, Paxillin-positive FAs and MP1-positive endosomes were thresholded and masked using ImageJ. Next, individual endosomes and FAs were identified on the basis of their fluorescence intensities using a local maxima thresholding approach essentially as described previously (Teis et al., 2006) and their geometric center was used for further analysis. In short, images have been preprocessed with an unsharp mask filter (sigma of 3 pixels and weight of 0.8 pixels) in order to define the borders of the organelles more clearly and suppress background noise. Images were consequently manually thresholded based on intensity levels. Image histograms were used as indication for separation of background and signal and transferred to binary masks. The image sequences were next transferred to Imaris for further processing, tracking, and determination of their geometric centers (see Supplemental material). FA targeting was evaluated by quantification of overlaying positions of individual endosomes onto the FA mask (in percentage of overlay of two fluorescent signals array) per frame of the video sequence. Temporal changes of FA positions (FA tracking) were calculated using Imaris and were shown using pseudocolored lines. Overall targeting was calculated over the whole sequence. To calculate which part of the individual FA is targeted, a method modified from di Penta et al. (2009) was used. FA signal was dissected in respect to the time domain. Dynamic and stable segments were dissected by simple arithmetical analysis along the time course of the adhesion signals (for example, subtracting the previous image frame from the current image frame identified newly added parts of the adhesion sites). The other parts were identified respectively. Analysis was completed in 10 cells from 10 independent time-lapse experiments. The analysis was realized using the ImageJ package and the JACoP plug-in (Bolte and Cordelières, 2006; http://rsb.info.nih.gov/ij/).
Coimmunoprecipitation
MEFs were lysed by sonication in 50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 1% Triton X-100, 0.5% sodium deoxycholate, 10 mM EDTA, and protease inhibitor cocktail. HeLa cells were lysed in 50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 1% NP-40, 5 mM EDTA, and protease inhibitors. Subsequent steps were similar for both cell types. In brief, lysates containing 3 mg of total protein per sample were used for immunoprecipitation. After 20 min of preclearing with Ultralink G beads (Thermo Fisher Scientific), the supernatant was subjected to immunoprecipitation using the respective antibody for 30 min on ice. Ultralink G beads were added after incubation with rotation for 1 h at 4°C. Beads were washed three times in the corresponding lysis buffer. The immune complex was eluted by 5 min of incubation at 95°C in sample buffer and separated by SDS-PAGE (Fialka et al., 1997).
Transferrin uptake experiments
MEFs were incubated on ice for 1 h with 50 ng/ml Alexa Fluor 594–transferrin (Invitrogen) and allowed to uptake for 0–30 min at 37°C. Cells were fixed in 4% paraformaldehyde, mounted in Mowiol, and analyzed by microscope (AxioImager M1.2; Carl Zeiss).
Statistics
Length of FAs was measured in ImageJ. Values were transferred into SPSS and Excel programs and analyzed by Student’s t tests. Results are expressed as means ± SD and SEM in absolute numbers (µm) and percentages (%). SEM was calculated as follows: standard deviation was divided by the square root of the sample size (amount of calculated FAs in a given sample). F = 334,396 and P < 0.001 were considered significant for these studies.
Online supplemental material
Fig. S1 shows quantification of single leading edge and protrusions, quantification of migration speed, anti-MP1 WB, and effect of MP1 knockdown on FA length in HeLa cells. Fig. S2 shows integrin expression profile in p14f/− and p14−/− MEFs. Fig. S3 illustrates absence of endosomal targeting to FAs during cell spreading. Fig. S4 shows interaction of IQGAP1 and MP1 in PLA assay. Fig. S5 shows anti-IQGAP1 WB (A) and migration data from time-lapse microcopy of MEFs treated with IQGAP1 RNAi. Video 1 shows the time-lapse images depicted in Fig. 1 A. Video 2 shows the time-lapse images depicted in Fig. 2 C. Video 3 shows the time-lapse images depicted in Fig. 2 C. Video 4 shows the time-lapse images depicted in Fig. 3 B. Video 5 shows the time-lapse images depicted in Fig. 3 C. Video 6 shows the time-lapse images depicted in Fig. S3. Videos 7 and 8 show the time-lapse images depicted in Fig. 5 A. Video 9 shows the time-lapse images depicted in Fig. 5 D. Video 10 shows the time-lapse images depicted in Fig. 5 F. Tables S1 and S2 present statistics of the FA length quantification. The supplemental text describes Imaris and ImageJ scripts used for the FA-targeting calculations shown in Fig. 4, B and C. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201310043/DC1. Additional data are available in the JCB DataViewer at http://dx.doi.org/10.1083/jcb.201310043.dv.
Supplementary Material
Acknowledgments
The authors are indebted to B. Geiger, R.Y. Tsien, J.V. Small, D. Sacks, G. Geley, and C. Bucci for providing expression constructs; and to M. Baccarini for providing cells as outlined in the manuscript. We also thank the Core Facility Biooptics, Innsbruck Medical University, for technical assistance in image acquisition and analysis.
This work was supported by the Austrian Science Funds (SFB021 to L.A. Huber and R. Fässler and P19486-B12 to M.W. Hess). This work was also supported by the COMET Center Oncotyrol (to L.A. Huber) and by the Federal Ministry for Transport Innovation and Technology (BMVIT) and HFSP-CDA (to D. Teis).
The authors declare no competing financial interests.
Footnotes
Abbreviations used in this paper:
- FA
- focal adhesion
- FC
- focal complex
- IF
- immunofluorescence
- MEF
- mouse embryonic fibroblast
- MT
- microtubule
- PLA
- proximity ligation assay
- TIRF
- total internal reflection fluorescence
- WB
- Western blotting
References
- Bagshaw R.D., Callahan J.W., Mahuran D.J. 2006. The Arf-family protein, Arl8b, is involved in the spatial distribution of lysosomes. Biochem. Biophys. Res. Commun. 344:1186–1191 10.1016/j.bbrc.2006.03.221 [DOI] [PubMed] [Google Scholar]
- Ballestrem C., Hinz B., Imhof B.A., Wehrle-Haller B. 2001. Marching at the front and dragging behind: differential alphaVbeta3-integrin turnover regulates focal adhesion behavior. J. Cell Biol. 155:1319–1332 10.1083/jcb.200107107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolte S., Cordelières F.P. 2006. A guided tour into subcellular colocalization analysis in light microscopy. J. Microsc. 224:213–232 10.1111/j.1365-2818.2006.01706.x [DOI] [PubMed] [Google Scholar]
- Broussard J.A., Webb D.J., Kaverina I. 2008. Asymmetric focal adhesion disassembly in motile cells. Curr. Opin. Cell Biol. 20:85–90 10.1016/j.ceb.2007.10.009 [DOI] [PubMed] [Google Scholar]
- Brown M.C., Perrotta J.A., Turner C.E. 1998. Serine and threonine phosphorylation of the paxillin LIM domains regulates paxillin focal adhesion localization and cell adhesion to fibronectin. Mol. Biol. Cell. 9:1803–1816 10.1091/mbc.9.7.1803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown M.D., Sacks D.B. 2006. IQGAP1 in cellular signaling: bridging the GAP. Trends Cell Biol. 16:242–249 10.1016/j.tcb.2006.03.002 [DOI] [PubMed] [Google Scholar]
- Bucci C., Thomsen P., Nicoziani P., McCarthy J., van Deurs B. 2000. Rab7: a key to lysosome biogenesis. Mol. Biol. Cell. 11:467–480 10.1091/mbc.11.2.467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlucci A., Porpora M., Garbi C., Galgani M., Santoriello M., Mascolo M., di Lorenzo D., Altieri V., Quarto M., Terracciano L., et al. 2010. PTPD1 supports receptor stability and mitogenic signaling in bladder cancer cells. J. Biol. Chem. 285:39260–39270 10.1074/jbc.M110.174706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q., Kinch M.S., Lin T.H., Burridge K., Juliano R.L. 1994. Integrin-mediated cell adhesion activates mitogen-activated protein kinases. J. Biol. Chem. 269:26602–26605 [PubMed] [Google Scholar]
- Czuchra A., Meyer H., Legate K.R., Brakebusch C., Fässler R. 2006. Genetic analysis of beta1 integrin “activation motifs” in mice. J. Cell Biol. 174:889–899 10.1083/jcb.200604060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delorme-Walker V.D., Peterson J.R., Chernoff J., Waterman C.M., Danuser G., DerMardirossian C., Bokoch G.M. 2011. Pak1 regulates focal adhesion strength, myosin IIA distribution, and actin dynamics to optimize cell migration. J. Cell Biol. 193:1289–1303 10.1083/jcb.201010059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- di Penta A., Mercaldo V., Florenzano F., Munck S., Ciotti M.T., Zalfa F., Mercanti D., Molinari M., Bagni C., Achsel T. 2009. Dendritic LSm1/CBP80-mRNPs mark the early steps of transport commitment and translational control. J. Cell Biol. 184:423–435 10.1083/jcb.200807033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang X., Luo J., Nishihama R., Wloka C., Dravis C., Travaglia M., Iwase M., Vallen E.A., Bi E. 2010. Biphasic targeting and cleavage furrow ingression directed by the tail of a myosin II. J. Cell Biol. 191:1333–1350 10.1083/jcb.201005134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feiguin F., Ferreira A., Kosik K.S., Caceres A. 1994. Kinesin-mediated organelle translocation revealed by specific cellular manipulations. J. Cell Biol. 127:1021–1039 10.1083/jcb.127.4.1021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fialka I., Pasquali C., Lottspeich F., Ahorn H., Huber L.A. 1997. Subcellular fractionation of polarized epithelial cells and identification of organelle-specific proteins by two-dimensional gel electrophoresis. Electrophoresis. 18:2582–2590 10.1002/elps.1150181414 [DOI] [PubMed] [Google Scholar]
- Frost J.A., Khokhlatchev A., Stippec S., White M.A., Cobb M.H. 1998. Differential effects of PAK1-activating mutations reveal activity-dependent and -independent effects on cytoskeletal regulation. J. Biol. Chem. 273:28191–28198 10.1074/jbc.273.43.28191 [DOI] [PubMed] [Google Scholar]
- Fukata M., Watanabe T., Noritake J., Nakagawa M., Yamaga M., Kuroda S., Matsuura Y., Iwamatsu A., Perez F., Kaibuchi K. 2002. Rac1 and Cdc42 capture microtubules through IQGAP1 and CLIP-170. Cell. 109:873–885 10.1016/S0092-8674(02)00800-0 [DOI] [PubMed] [Google Scholar]
- Geiger B., Bershadsky A. 2001. Assembly and mechanosensory function of focal contacts. Curr. Opin. Cell Biol. 13:584–592 10.1016/S0955-0674(00)00255-6 [DOI] [PubMed] [Google Scholar]
- Gupton S.L., Waterman-Storer C.M. 2006. Spatiotemporal feedback between actomyosin and focal-adhesion systems optimizes rapid cell migration. Cell. 125:1361–1374 10.1016/j.cell.2006.05.029 [DOI] [PubMed] [Google Scholar]
- Hart M.J., Callow M.G., Souza B., Polakis P. 1996. IQGAP1, a calmodulin-binding protein with a rasGAP-related domain, is a potential effector for cdc42Hs. EMBO J. 15:2997–3005 [PMC free article] [PubMed] [Google Scholar]
- Ho Y.D., Joyal J.L., Li Z., Sacks D.B. 1999. IQGAP1 integrates Ca2+/calmodulin and Cdc42 signaling. J. Biol. Chem. 274:464–470 10.1074/jbc.274.1.464 [DOI] [PubMed] [Google Scholar]
- Hofmann I., Munro S. 2006. An N-terminally acetylated Arf-like GTPase is localised to lysosomes and affects their motility. J. Cell Sci. 119:1494–1503 10.1242/jcs.02958 [DOI] [PubMed] [Google Scholar]
- Hollenbeck P.J., Swanson J.A. 1990. Radial extension of macrophage tubular lysosomes supported by kinesin. Nature. 346:864–866 10.1038/346864a0 [DOI] [PubMed] [Google Scholar]
- Huang M., Satchell L., Duhadaway J.B., Prendergast G.C., Laury-Kleintop L.D. 2011. RhoB links PDGF signaling to cell migration by coordinating activation and localization of Cdc42 and Rac. J. Cell. Biochem. 112:1572–1584 10.1002/jcb.23069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz M., Amit I., Yarden Y. 2007. Regulation of MAPKs by growth factors and receptor tyrosine kinases. Biochim. Biophys. Acta. 1773:1161–1176 10.1016/j.bbamcr.2007.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaverina I., Krylyshkina O., Small J.V. 1999. Microtubule targeting of substrate contacts promotes their relaxation and dissociation. J. Cell Biol. 146:1033–1044 10.1083/jcb.146.5.1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kee Y.S., Ren Y., Dorfman D., Iijima M., Firtel R., Iglesias P.A., Robinson D.N. 2012. A mechanosensory system governs myosin II accumulation in dividing cells. Mol. Biol. Cell. 23:1510–1523 10.1091/mbc.E11-07-0601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klemke R.L., Cai S., Giannini A.L., Gallagher P.J., de Lanerolle P., Cheresh D.A. 1997. Regulation of cell motility by mitogen-activated protein kinase. J. Cell Biol. 137:481–492 10.1083/jcb.137.2.481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krylyshkina O., Kaverina I., Kranewitter W., Steffen W., Alonso M.C., Cross R.A., Small J.V. 2002. Modulation of substrate adhesion dynamics via microtubule targeting requires kinesin-1. J. Cell Biol. 156:349–359 10.1083/jcb.200105051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo J.C., Han X., Hsiao C.T., Yates J.R., III, Waterman C.M. 2011. Analysis of the myosin-II-responsive focal adhesion proteome reveals a role for β-Pix in negative regulation of focal adhesion maturation. Nat. Cell Biol. 13:383–393 10.1038/ncb2216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurzbauer R., Teis D., de Araujo M.E., Maurer-Stroh S., Eisenhaber F., Bourenkov G.P., Bartunik H.D., Hekman M., Rapp U.R., Huber L.A., Clausen T. 2004. Crystal structure of the p14/MP1 scaffolding complex: how a twin couple attaches mitogen-activated protein kinase signaling to late endosomes. Proc. Natl. Acad. Sci. USA. 101:10984–10989 10.1073/pnas.0403435101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laporte D., Coffman V.C., Lee I.J., Wu J.Q. 2011. Assembly and architecture of precursor nodes during fission yeast cytokinesis. J. Cell Biol. 192:1005–1021 10.1083/jcb.201008171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laukaitis C.M., Webb D.J., Donais K., Horwitz A.F. 2001. Differential dynamics of alpha 5 integrin, paxillin, and alpha-actinin during formation and disassembly of adhesions in migrating cells. J. Cell Biol. 153:1427–1440 10.1083/jcb.153.7.1427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Sacks D.B. 2003. Elucidation of the interaction of calmodulin with the IQ motifs of IQGAP1. J. Biol. Chem. 278:4347–4352 10.1074/jbc.M208579200 [DOI] [PubMed] [Google Scholar]
- Manser E., Leung T., Salihuddin H., Zhao Z.S., Lim L. 1994. A brain serine/threonine protein kinase activated by Cdc42 and Rac1. Nature. 367:40–46 10.1038/367040a0 [DOI] [PubMed] [Google Scholar]
- Mataraza J.M., Briggs M.W., Li Z., Entwistle A., Ridley A.J., Sacks D.B. 2003. IQGAP1 promotes cell motility and invasion. J. Biol. Chem. 278:41237–41245 10.1074/jbc.M304838200 [DOI] [PubMed] [Google Scholar]
- Nakata T., Hirokawa N. 1995. Point mutation of adenosine triphosphate-binding motif generated rigor kinesin that selectively blocks anterograde lysosome membrane transport. J. Cell Biol. 131:1039–1053 10.1083/jcb.131.4.1039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noritake J., Watanabe T., Sato K., Wang S., Kaibuchi K. 2005. IQGAP1: a key regulator of adhesion and migration. J. Cell Sci. 118:2085–2092 10.1242/jcs.02379 [DOI] [PubMed] [Google Scholar]
- Obexer P., Geiger K., Ambros P.F., Meister B., Ausserlechner M.J. 2007. FKHRL1-mediated expression of Noxa and Bim induces apoptosis via the mitochondria in neuroblastoma cells. Cell Death Differ. 14:534–547 10.1038/sj.cdd.4402017 [DOI] [PubMed] [Google Scholar]
- Osmani N., Peglion F., Chavrier P., Etienne-Manneville S. 2010. Cdc42 localization and cell polarity depend on membrane traffic. J. Cell Biol. 191:1261–1269 10.1083/jcb.201003091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palamidessi A., Frittoli E., Garré M., Faretta M., Mione M., Testa I., Diaspro A., Lanzetti L., Scita G., Di Fiore P.P. 2008. Endocytic trafficking of Rac is required for the spatial restriction of signaling in cell migration. Cell. 134:135–147 10.1016/j.cell.2008.05.034 [DOI] [PubMed] [Google Scholar]
- Park E.R., Pullikuth A.K., Bailey E.M., Mercante D.E., Catling A.D. 2009. Differential requirement for MEK Partner 1 in DU145 prostate cancer cell migration. Cell Commun. Signal. 7:26 10.1186/1478-811X-7-26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons J.T., Martin K.H., Slack J.K., Taylor J.M., Weed S.A. 2000. Focal adhesion kinase: a regulator of focal adhesion dynamics and cell movement. Oncogene. 19:5606–5613 10.1038/sj.onc.1203877 [DOI] [PubMed] [Google Scholar]
- Pastan I.H., Willingham M.C. 1981. Journey to the center of the cell: role of the receptosome. Science. 214:504–509 10.1126/science.6170111 [DOI] [PubMed] [Google Scholar]
- Pullikuth A.K., Catling A.D. 2007. Scaffold mediated regulation of MAPK signaling and cytoskeletal dynamics: a perspective. Cell. Signal. 19:1621–1632 10.1016/j.cellsig.2007.04.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pullikuth A., McKinnon E., Schaeffer H.J., Catling A.D. 2005. The MEK1 scaffolding protein MP1 regulates cell spreading by integrating PAK1 and Rho signals. Mol. Cell. Biol. 25:5119–5133 10.1128/MCB.25.12.5119-5133.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rid R., Schiefermeier N., Grigoriev I., Small J.V., Kaverina I. 2005. The last but not the least: the origin and significance of trailing adhesions in fibroblastic cells. Cell Motil. Cytoskeleton. 61:161–171 10.1002/cm.20076 [DOI] [PubMed] [Google Scholar]
- Rosa-Ferreira C., Munro S. 2011. Arl8 and SKIP act together to link lysosomes to kinesin-1. Dev. Cell. 21:1171–1178 10.1016/j.devcel.2011.10.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy M., Li Z., Sacks D.B. 2004. IQGAP1 binds ERK2 and modulates its activity. J. Biol. Chem. 279:17329–17337 10.1074/jbc.M308405200 [DOI] [PubMed] [Google Scholar]
- Roy M., Li Z., Sacks D.B. 2005. IQGAP1 is a scaffold for mitogen-activated protein kinase signaling. Mol. Cell. Biol. 25:7940–7952 10.1128/MCB.25.18.7940-7952.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadowski L., Pilecka I., Miaczynska M. 2009. Signaling from endosomes: location makes a difference. Exp. Cell Res. 315:1601–1609 10.1016/j.yexcr.2008.09.021 [DOI] [PubMed] [Google Scholar]
- Sancak Y., Bar-Peled L., Zoncu R., Markhard A.L., Nada S., Sabatini D.M. 2010. Ragulator-Rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Cell. 141:290–303 10.1016/j.cell.2010.02.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders L.C., Matsumura F., Bokoch G.M., de Lanerolle P. 1999. Inhibition of myosin light chain kinase by p21-activated kinase. Science. 283:2083–2085 10.1126/science.283.5410.2083 [DOI] [PubMed] [Google Scholar]
- Schiefermeier N., Teis D., Huber L.A. 2011. Endosomal signaling and cell migration. Curr. Opin. Cell Biol. 23:615–620 10.1016/j.ceb.2011.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiller H.B., Friedel C.C., Boulegue C., Fässler R. 2011. Quantitative proteomics of the integrin adhesome show a myosin II-dependent recruitment of LIM domain proteins. EMBO Rep. 12:259–266 10.1038/embor.2011.5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scita G., Di Fiore P.P. 2010. The endocytic matrix. Nature. 463:464–473 10.1038/nature08910 [DOI] [PubMed] [Google Scholar]
- Shaner N.C., Campbell R.E., Steinbach P.A., Giepmans B.N., Palmer A.E., Tsien R.Y. 2004. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat. Biotechnol. 22:1567–1572 10.1038/nbt1037 [DOI] [PubMed] [Google Scholar]
- Stasyk T., Holzmann J., Stumberger S., Ebner H.L., Hess M.W., Bonn G.K., Mechtler K., Huber L.A. 2010. Proteomic analysis of endosomes from genetically modified p14/MP1 mouse embryonic fibroblasts. Proteomics. 10:4117–4127 10.1002/pmic.201000258 [DOI] [PubMed] [Google Scholar]
- Sturge J., Wienke D., Isacke C.M. 2006. Endosomes generate localized Rho-ROCK-MLC2-based contractile signals via Endo180 to promote adhesion disassembly. J. Cell Biol. 175:337–347 10.1083/jcb.200602125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teis D., Wunderlich W., Huber L.A. 2002. Localization of the MP1-MAPK scaffold complex to endosomes is mediated by p14 and required for signal transduction. Dev. Cell. 3:803–814 10.1016/S1534-5807(02)00364-7 [DOI] [PubMed] [Google Scholar]
- Teis D., Taub N., Kurzbauer R., Hilber D., de Araujo M.E., Erlacher M., Offterdinger M., Villunger A., Geley S., Bohn G., et al. 2006. p14-MP1-MEK1 signaling regulates endosomal traffic and cellular proliferation during tissue homeostasis. J. Cell Biol. 175:861–868 10.1083/jcb.200607025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu C., Ortega-Cava C.F., Winograd P., Stanton M.J., Reddi A.L., Dodge I., Arya R., Dimri M., Clubb R.J., Naramura M., et al. 2010. Endosomal-sorting complexes required for transport (ESCRT) pathway-dependent endosomal traffic regulates the localization of active Src at focal adhesions. Proc. Natl. Acad. Sci. USA. 107:16107–16112 10.1073/pnas.1009471107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicente-Manzanares M., Zareno J., Whitmore L., Choi C.K., Horwitz A.F. 2007. Regulation of protrusion, adhesion dynamics, and polarity by myosins IIA and IIB in migrating cells. J. Cell Biol. 176:573–580 10.1083/jcb.200612043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe T., Wang S., Noritake J., Sato K., Fukata M., Takefuji M., Nakagawa M., Izumi N., Akiyama T., Kaibuchi K. 2004. Interaction with IQGAP1 links APC to Rac1, Cdc42, and actin filaments during cell polarization and migration. Dev. Cell. 7:871–883 10.1016/j.devcel.2004.10.017 [DOI] [PubMed] [Google Scholar]
- Webb D.J., Donais K., Whitmore L.A., Thomas S.M., Turner C.E., Parsons J.T., Horwitz A.F. 2004. FAK-Src signalling through paxillin, ERK and MLCK regulates adhesion disassembly. Nat. Cell Biol. 6:154–161 10.1038/ncb1094 [DOI] [PubMed] [Google Scholar]
- Webb D.J., Schroeder M.J., Brame C.J., Whitmore L., Shabanowitz J., Hunt D.F., Horwitz A.R. 2005. Paxillin phosphorylation sites mapped by mass spectrometry. J. Cell Sci. 118:4925–4929 10.1242/jcs.02563 [DOI] [PubMed] [Google Scholar]
- Weissbach L., Bernards A., Herion D.W. 1998. Binding of myosin essential light chain to the cytoskeleton-associated protein IQGAP1. Biochem. Biophys. Res. Commun. 251:269–276 10.1006/bbrc.1998.9371 [DOI] [PubMed] [Google Scholar]
- Wickström S.A., Lange A., Hess M.W., Polleux J., Spatz J.P., Krüger M., Pfaller K., Lambacher A., Bloch W., Mann M., et al. 2010. Integrin-linked kinase controls microtubule dynamics required for plasma membrane targeting of caveolae. Dev. Cell. 19:574–588 10.1016/j.devcel.2010.09.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfenson H., Lubelski A., Regev T., Klafter J., Henis Y.I., Geiger B. 2009. A role for the juxtamembrane cytoplasm in the molecular dynamics of focal adhesions. PLoS ONE. 4:e4304 10.1371/journal.pone.0004304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J.Q., Kuhn J.R., Kovar D.R., Pollard T.D. 2003. Spatial and temporal pathway for assembly and constriction of the contractile ring in fission yeast cytokinesis. Dev. Cell. 5:723–734 10.1016/S1534-5807(03)00324-1 [DOI] [PubMed] [Google Scholar]
- Wunderlich W., Fialka I., Teis D., Alpi A., Pfeifer A., Parton R.G., Lottspeich F., Huber L.A. 2001. A novel 14-kilodalton protein interacts with the mitogen-activated protein kinase scaffold mp1 on a late endosomal/lysosomal compartment. J. Cell Biol. 152:765–776 10.1083/jcb.152.4.765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaidel-Bar R., Ballestrem C., Kam Z., Geiger B. 2003. Early molecular events in the assembly of matrix adhesions at the leading edge of migrating cells. J. Cell Sci. 116:4605–4613 10.1242/jcs.00792 [DOI] [PubMed] [Google Scholar]
- Zaidel-Bar R., Milo R., Kam Z., Geiger B. 2007. A paxillin tyrosine phosphorylation switch regulates the assembly and form of cell-matrix adhesions. J. Cell Sci. 120:137–148 10.1242/jcs.03314 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.