The RhoGAP protein SH3BP1 mediates Sema3E-induced cell collapse through interaction with PlexinD1 and regulation of Rac1 activity.
Abstract
Extracellular signals have to be precisely interpreted intracellularly and translated into diverse cellular behaviors often mediated by cytoskeletal changes. Semaphorins are one of the largest families of guidance cues and play a critical role in many systems. However, how different cell types translate extracellular semaphorin binding into intracellular signaling remains unclear. Here we developed and performed a novel image-based genome-wide functional RNAi screen for downstream signaling molecules that convert the interaction between Semaphorin 3E (Sema3E) and PlexinD1 into cellular behaviors. One of the genes identified in this screen is a RhoGAP protein, SH3-domain binding protein 1 (SH3BP1). We demonstrate that SH3BP1 mediates Sema3E-induced cell collapse through interaction with PlexinD1 and regulation of Ras-related C3 botulinum toxin substrate 1 (Rac1) activity. The identification and characterization of SH3BP1 as a novel downstream effector of Sema3E-PlexinD1 provides an explanation for how extracellular signals are translated into cytoskeletal changes and unique cell behavior, but also lays the foundation for characterizing other genes identified from our screen to obtain a more complete picture of plexin signaling.
Introduction
The semaphorins are one of the largest families of guidance molecules, and include eight distinct classes. Some semaphorins are secreted molecules capable of long-range diffusion, whereas others are membrane-bound proteins that function as short-range guidance cues (Tran et al., 2007). Initially discovered as axon-guidance molecules, semaphorins also have much broader biological functions: they are now known to be involved in cell migration, synapse formation, and dendrite development, as well as immune and respiratory system function, vascular development, and tumor angiogenesis (Tran et al., 2007; Neufeld and Kessler, 2008). However, how different cells translate extracellular semaphorin ligand binding into intracellular signaling and cytoskeletal changes, thereby affecting diverse biological functions, is still not fully understood.
Semaphorins signal primarily through multimeric receptor complexes in which plexins (A–D), a family of large transmembrane proteins, serve as the major signaling receptor components. Secreted class 3 semaphorins generally signal through a holoreceptor composed of the ligand-binding subunit Neuropilin (Npn) and the signal transducing subunit PlexinA (Tran et al., 2007). The only known exception is the secreted Semaphorin 3E (Sema3E), which binds directly to PlexinD1 and is not dependent on Npn for binding (Gu et al., 2005). In contrast to secreted semaphorins, many membrane-bound semaphorins appear to require only plexins for signaling. So far, our limited understanding of downstream plexin signaling stems mainly from cell culture data and Drosophila melanogaster models. Studies in neurons have revealed that semaphorin binding on the cell surface triggers the depolymerization and redistribution of F-actin filaments. This reorganization causes filopodia and lamellipodia to retract, and ultimately leads to growth cone collapse. The only direct link between plexin signaling and changes in the actin cytoskeleton is the actin-binding flavoprotein monooxygenase MICAL (molecule interacting with CasL), which was identified from a Drosophila genetic screen and characterized in invertebrates (Hung et al., 2010).
In contrast to plexin signaling in invertebrate systems, our understanding of how semaphorin–plexin signaling is transduced in the vertebrate system remains elusive. Most vertebrate plexin signaling studies to date have used a candidate approach based upon plexins’ putative endogenous R-Ras GTPase-activating protein (GAP) domain (Oinuma et al., 2004a, 2006; Toyofuku et al., 2005; Ito et al., 2006; Gelfand et al., 2009). The intracellular domain of all plexins shares homology with GAPs, and in vitro studies using both cell-based experiments and purified proteins showed that this GAP activity leads to the deactivation of R-Ras, M-Ras, and Rap1 (Rohm et al., 2000; Oinuma et al., 2004a,b; Toyofuku et al., 2005; Saito et al., 2009; Uesugi et al., 2009; Wang et al., 2012). However, identifying a specific small GTPase as the effector for the individual plexin-mediated signaling has been controversial. For example, plexin-mediated deactivation of R-Ras (Oinuma et al., 2004b), activation of RhoA (Swiercz et al., 2002, 2009), and deactivation of Rap1 (Wang et al., 2012) have all been implicated as the underlying cause of axonal growth cone collapse of primary neurons. So far, whether loss of Ras-GAP activity in vivo is required for plexin-mediated biological processes has not been tested. Therefore, an unbiased approach to identify vertebrate semaphorin–plexin signaling components is necessary to fully understand how cells translate extracellular semaphorin binding to intracellular signaling and cytoskeletal changes.
Previously, we identified Sema3E and PlexinD1 as a novel ligand–receptor pair and demonstrated their in vivo requirement for heart and vascular patterning (Gu et al., 2005; Kim et al., 2011), as well as neural circuit development (Ding et al., 2012). PlexinD1 is expressed exclusively in endothelial cells (ECs) during early embryonic development, and somatic expression of Sema3E controls intersomitic vessel patterning via repulsive signaling through the PlexinD1 receptor (Van Der Zwaag et al., 2002; Gitler et al., 2004; Torres-Vázquez et al., 2004; Gu et al., 2005). Sema3E-PlexinD1 signaling is also required for the initial development of descending axon tracts in the mouse forebrain in vivo (Chauvet et al., 2007). Sema3E-PlexinD1 activity has been shown to be required for thymocyte migration in the developing immune system in an Npn1-independent manner (Choi et al., 2008). In addition, Sema3E-PlexinD1 signaling has been implicated in tumor angiogenesis and metastases (Roodink et al., 2005, 2008; Kigel et al., 2008). However, the downstream signaling components of PlexinD1 are still largely unknown. Therefore, identification of the Sema3E-PlexinD1 downstream signaling cascade is critical for understanding how it could elicit these diverse responses in different systems.
In this study, we performed an unbiased functional image-based genome-wide RNAi screen to identify downstream signaling molecules required for Sema3E-PlexinD1 signaling. This screen yielded several candidate genes that belong to different cellular processes. Here, we focused on one of the genes identified from the screen, a Rho-GAP protein, SH3-domain binding protein 1 (SH3BP1). We demonstrate that Sema3E-PlexinD1 signaling regulates cytoskeleton changes by down-regulation of Ras-related C3 botulinum toxin substrate 1 (Rac1) activity, which is mediated by SH3BP1. Moreover, SH3BP1 is colocalized with PlexinD1 at the lamellipodia, and its activity is regulated by Sema3E-PlexinD1 binding. Finally, we also demonstrated that SH3BP1 is required for the repulsive guidance effect of Sema3E-PlexinD1 on EC migration. The identification and characterization of SH3BP1 as a novel downstream effector mediating Sema3E-PlexinD1 signaling provides an explanation for how extracellular signals are translated into intracellular cytoskeletal changes.
Results
Functional identification of Sema3E-PlexinD1 downstream signalling molecules using an unbiased image-based genome-wide RNAi screen
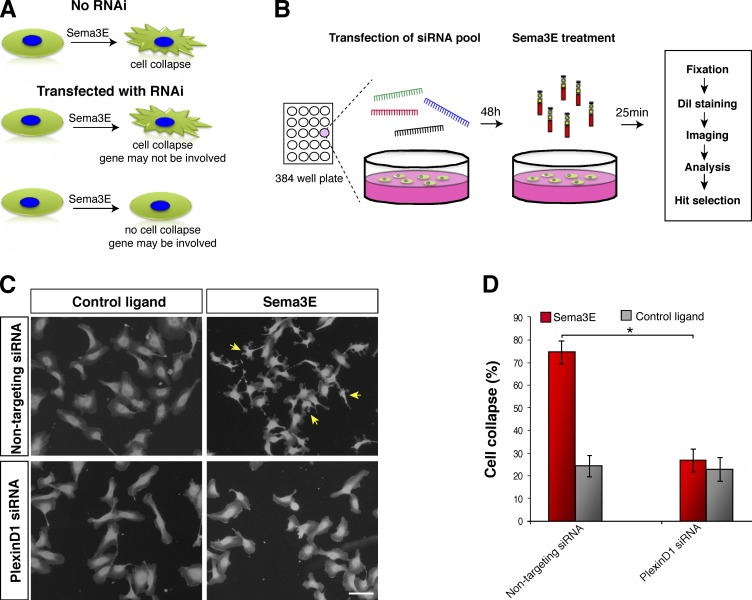
Previously, we have shown that Sema3E-PlexinD1 is required for vascular patterning in vivo (Gu et al., 2005; Kim et al., 2011). However, the intracellular signaling pathways that transduce PlexinD1 activity in ECs remain unclear. To identify proteins required for Sema3E-PlexinD1 signaling, we established an unbiased and robust cell-based assay. We first searched for a cell line that endogenously expresses PlexinD1 and shows a clear and measurable visual response to Sema3E. We and others (Casazza et al., 2010; Sakurai et al., 2010) have discovered that human umbilical vein ECs (HUVECs) endogenously express PlexinD1 (Fig. S1 A) and respond to Sema3E treatment independent of substrate context (the response is reproducible when cells are grown on glass, plastic, or collagen; unpublished data). When treated with Sema3E, HUVECs undergo rapid morphological changes that lead to a complete collapse of the cytoskeleton within 25 min at 37°C, a significant reduction in cell surface area, and the formation of distinct membrane protrusions (Fig. 1, A and C). We performed an image-based RNAi screen of the human genome using this robust Sema3E-induced HUVEC collapse assay to identify intracellular signal transduction components of Sema3E-PlexinD1 (Fig. 1 B). The hits identified from this functional screen are most likely involved in Sema3E-PlexinD1 signaling in vivo.
Figure 1.
An image-based genome-wide screen to unbiasedly identify Sema3E-PlexinD1 downstream signaling molecules. (A) Schematic illustration of the screen strategy. HUVECs endogenously express PlexinD1 and undergo cell collapse after Sema3E treatment. The RNAi screen identified genes that, when knocked down, block the Sema3E-induced cell collapse. (B) Schematic illustration of the automated screen procedure. Cells were transfected with smart pools of siRNA (four different sequence targets for each gene) for each well of the 384-well plates, and after 48 h Sema3E was added to the culture media for 25 min. Cells were fixed, stained, and imaged, and the cell collapse phenotype was quantified using the combination of CellProfiler and our custom-developed image analysis algorithm. (C) Automated imaging of a positive control well (PlexinD1 siRNA-transfected) and a negative control well (nontargeting siRNA-transfected) for each 384-well screening plate (images were generated from the DiI channel). HUVECs were transfected with nontargeting siRNA or PlexinD1 siRNA and treated with control ligand or Sema3E. Sema3E caused cell collapse resulting in decreased surface area and the appearance of protrusions (arrows). These cytoskeletal changes were completely blocked by PlexinD1 siRNA. Bar, 100 µm. (D) Quantification of cell collapse. Unlike cells treated with the control ligand, cells stimulated with Sema3E underwent cytoskeletal collapse, which was significantly blocked when PlexinD1 siRNA was used. Error bars indicate SD. *, P < 0.01. Control nontargeting siRNA did not alter the cellular response elicited by Sema3E.
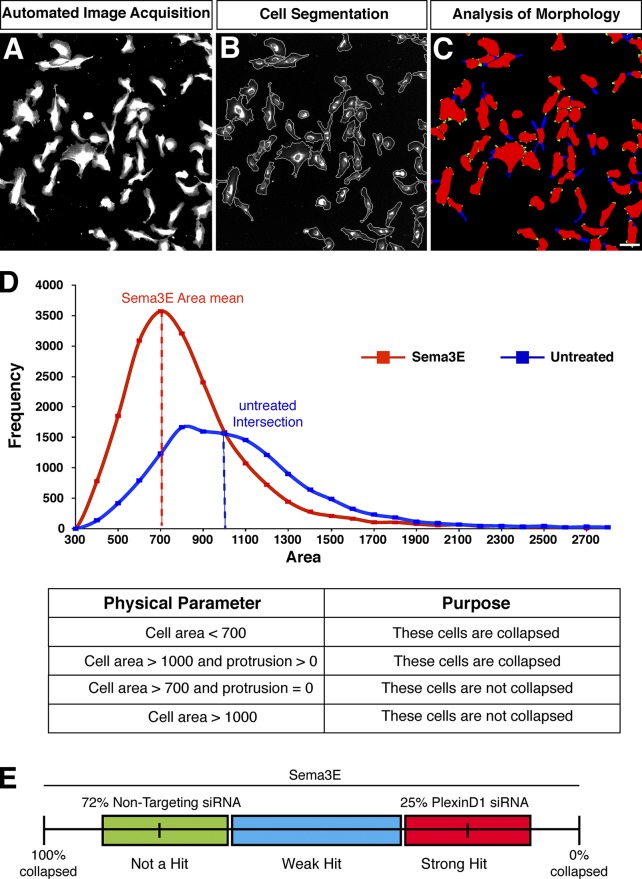
To perform a large-scale genome-wide screen using cell collapse as a readout, we then developed an automated image analysis algorithm to enable fast and unbiased identification of the collapse response at single-cell resolution. The collapse criteria were established based on images from the control conditions (Fig. 1 C). Specifically, the identification of a collapsed cell was performed based on the cell area and then the presence of protrusions on the cell surface (Fig. 2, A–D). Cells with an area <700 arbitrary units (au) were classified as collapsed, and cells with area >1,000 au were classified as noncollapsed. To systematically categorize the population of cells with an area between 700 au and 1,000 au, we looked at the presence/absence of protrusions on the cell surface, and classified any cell with at least one protrusion as collapsed. Using the algorithm that we developed based on these criteria, HUVECs transfected with a nontargeting siRNA (negative control) showed a mean of 72% collapse, whereas PlexinD1 siRNA-transfected cells (positive control) displayed 25% collapse after treatment with Sema3E (Fig. 1 D). We defined strong hits as siRNAs that reduced the percentage of cell collapse >3 SDs from the mean of the plate-matched negative controls (nontargeting siRNA-transfected wells) and <3 SDs from plate-matched positive controls (PlexinD1 siRNA-transfected wells; Fig. 2 E). Wells that were >3 SDs from both positive and negative control means were designated weak hits and were examined by manual observation. After automated imaging, the percentage of HUVECs collapsed was computed from each well and analyzed by these criteria.
Figure 2.
Automated image analysis of HUVEC morphology and Sema3E-induced cell collapse. (A) Raw images of cells acquired from a well of a screening plate by automated microscopy. DiI labeling combined with image analysis algorithms enabled automated quantification of cellular morphological changes at the screening end point. (B) Cellular membrane boundaries were overlaid on top of the raw image so that cell surface area could be automatically measured. (C) Cell protrusions were labeled in blue on top of the segmented cell masks so that the number of protrusions per cell could be counted. The green regions were the bumps on the cell surface that were too short to be classified as protrusions. Bar, 100 µm. (D) Physical parameters used to classify collapsed cells were determined based on the cell surface area distribution from the control wells (top, histogram) in combination with cell protrusion counts. Cells were classified as collapsed based on a reduction in their surface area and/or the presence of protrusions. (E) siRNAs that blocked Sema3E-induced HUVEC collapse were classified as strong hits if the percentage of cells collapsed was 3 SDs away from the plate-matched negative controls and within 3 SDs of the plate-matched positive controls (PlexinD1 siRNA; red box). siRNAs that partially blocked collapse were classified as weak hits (blue box).
HUVECs were plated in a 384-well plate (Fig. S1 B) and then transfected with 50 nM of siRNA targeting each gene in the human genome from a commercially available Dharmacon library of siRNA smart pools (four unique duplexes targeting different regions of the same mRNA) and incubated for 48 h to allow for significant protein knockdown (Fig. 1, A and B). Cells were then treated with 2 nM Sema3E or vehicle control at 37°C and 5% CO2 for 25 min. After ligand treatment, cells were fixed, stained with DiI and DAPI, and imaged. On each plate, PlexinD1 siRNA was used as a positive control to ensure that the collapse was efficiently blocked, and nontargeting siRNA was used as a negative control (Fig. S1 B). Of the total 21,121 genes screened, knockdown of 384 (∼1.8%) genes displayed significant reduction in cell collapse comparable to PlexinD1 siRNA knockdown (Fig. 1, C and D). To validate these hits, we performed a secondary screen in which four siRNA duplexes of the hits were individually tested. Of the 384 genes tested in our secondary screen, 233 (∼60.7%) were validated by at least two out of four individual duplexes. We categorized the reconfirmed genes from our screen into several groups including transcription and translation factors, kinases, and proteins known to be required for cytoskeletal regulation, cell division, endocytosis, protein trafficking, and other basic cell biological processes (Table S2).
SH3BP1 was identified from the screen as a candidate effector of Sema3E-PlexinD1 signaling, and further validated by siRNA-resistant SH3BP1 rescue and PlexinD1/SH3BP1 colocalization
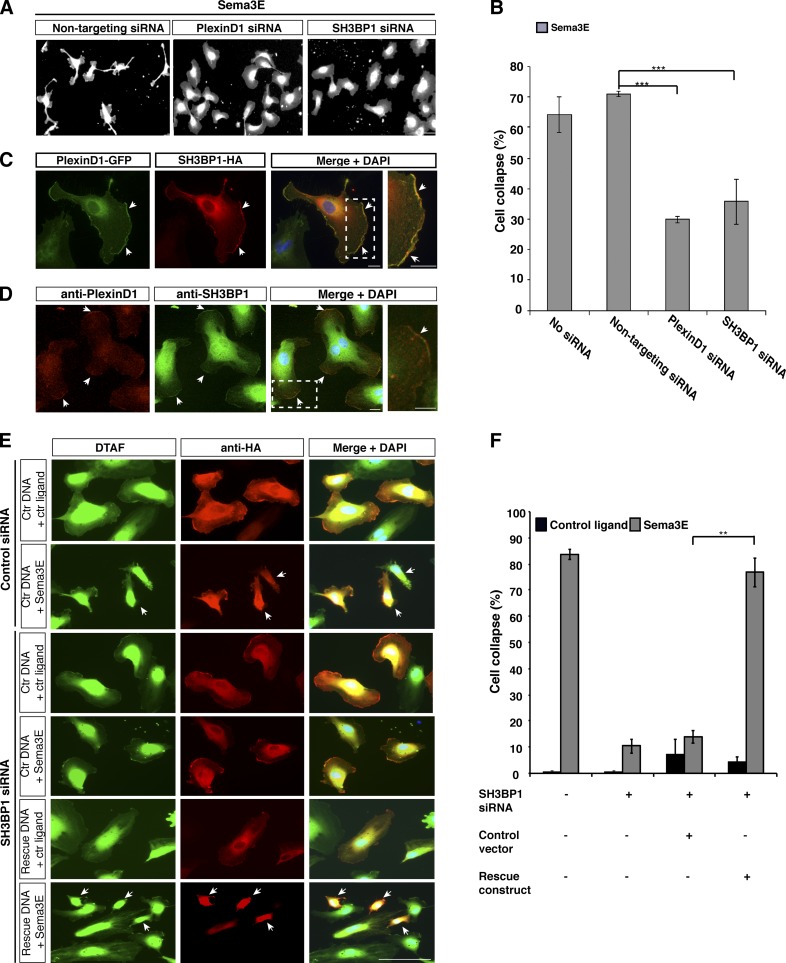
Of the 233 strong candidates identified from our secondary screen, SH3BP1, a GAP, has been further examined because of the role of GAP family proteins in regulation of small GTPases that control actin cytoskeleton dynamics. Many GAPs or regulatory proteins, such as IQGAP3, ARHGAP1, and RabGAP1, were negative hits in our screen. From the positive hits, SH3BP1 is one of the few known small GTPase regulatory proteins, down-regulation of which leads to strong blockage of Sema3E-induced cell collapse. Therefore, SH3BP1 emerged as a specific and unique candidate. SH3BP1 is expressed in HUVECs and ECs isolated from lungs and brain of embryonic day 18.5 (E18.5) mouse embryos (Fig. S2, A and B). Knockdown of SH3BP1 in HUVECs showed strong inhibition of Sema3E-induced cell collapse (Fig. 3 A). As shown from the screen of automatic imaging and collapse analysis, SH3BP1 knockdown led to a significant decrease in the percentage of the collapsed cells compared with negative control cells (∼35% vs. 70%, respectively; Fig. 3 B), a degree that is similar to that of PlexinD1 knockdown (Fig. 3, A and B).
Figure 3.
SH3BP1 was identified from the screen as a downstream component of Sema3E-PlexinD1 signaling, and further validated by siRNA-resistant SH3BP1 rescue and PlexinD1/SH3BP1 colocalization. (A) Automated image acquisition showed that SH3BP1 knockdown blocked Sema3E-induced collapse to similar extent as PlexinD1 siRNA. HUVECs treated with Sema3E underwent cell collapse that was inhibited by transfection with siRNA targeted specifically to PlexinD1 or SH3BP1. Bar, 100 µm. (B) Quantification of the Sema3E-induced cell collapse using the automated image analysis algorithm. ***, P < 0.0001. Error bars indicate SEM. (C) PlexinD1 and SH3BP1 colocalized at the leading edge of the cell (arrows). PlexinD1-GFP and SH3BP1-HA were coexpressed in HUVECs and stained with the corresponding antibodies. Bar, 10 µm. (D) Endogenous localization of PlexinD1 and SH3BP1 in HUVECs. PlexinD1 and SH3BP1 colocalized in the leading edge of the cells (arrows). The boxed regions are enlarged on the right. Bar, 10 µm. (E) Reintroducing SH3BP1 protein rescued SH3BP1 siRNA inhibition of Sema3E-induced collapse. HUVECs were transfected with SH3BP1 siRNA followed by transfection with control DNA or the siRNA-resistant SH3BP1 construct and treated with Sema3E or control ligand. Strong cell collapse was observed only in cells transfected with the rescue construct (arrows). Cell shape was visualized by DTAF labeling (green) and vector expression was detected by an HA antibody (red). Bar, 100 µm. (F) Quantification of the cell collapse demonstrated the ability of the siRNA-resistant SH3BP1 to fully rescue the Sema3E-induced collapse. **, P < 0.001. Error bars indicate SEM.
The first criterion we used to validate strong hits from our screen was to examine their protein localization, reasoning that a downstream effector should colocalize with PlexinD1 in the same subcellular compartment. To examine whether PlexinD1 and SH3BP1 colocalize, HUVECs transfected with PlexinD1-GFP and SH3BP1-HA were immunostained using GFP and HA antibodies, respectively. Interestingly, PlexinD1 protein was not uniformly expressed on the cell membrane; instead, it was highly enriched at the leading edge of the cell (Fig. 3 C). Consistent with previous reports (Parrini et al., 2011), we found a similar pattern of SH3BP1 localization (Fig. 3 C). Moreover, we investigated the localization of endogenous PlexinD1 and SH3BP1 in HUVECs using specific antibodies. Endogenous PlexinD1 and SH3BP1 localized in the same pattern as observed in overexpression experiments (Fig. 3 D and Fig. S2 C). These data further support the hypothesis that SH3BP1 functions downstream of Sema3E-PlexinD1 signaling. Interestingly, some of the strong hits from our screen, which are also small GTPase regulatory proteins, failed to pass this criterion and therefore were not pursued further.
To further validate these results, we performed rescue experiments to demonstrate that SH3BP1 siRNA knockdown was specific to the designed target (Fig. S2, D and E). Specifically, we performed rescue experiments by cotransfecting HUVECs with a plasmid encoding mouse HA-tagged SH3BP1 protein (rescue construct) containing five mismatches at the siRNA target site that rendered the gene resistant to siRNA knockdown. As shown in Fig. S2 D, Western blot analysis indicated that the messenger RNA encoded by the rescue construct was not knocked down by siRNA. In contrast, endogenous SH3BP1 was reproducibly and efficiently depleted by siRNA treatment. HUVECs were cotransfected with SH3BP1 or control siRNA with a rescue construct or control vector alone, respectively. 48 h after transfection, cells were treated with Sema3E or control alkaline phosphatase (AP) ligand, and cell collapse was analyzed. Sema3E-induced collapse was restored to normal levels when the SH3BP1-HA protein was reintroduced (Fig. 3, E and F), which demonstrates that SH3BP1 is required for Sema3E-PlexinD1–mediated HUVEC collapse. As expected, Sema3E induced strong collapse in cells transfected with control siRNA and control vector alone. Cells transfected with SH3BP1 alone or together with control vector did not show cell collapse, whereas transfection of the siRNA together with siRNA-resistant SH3BP1-HA construct resulted in cell collapse after Sema3E treatment (Fig. 3, E and F). These experiments confirmed the specificity of the siRNAs and further demonstrated that the loss of SH3BP1 function blocks Sema3E-PlexinD1 downstream signaling.
Dynamic changes of actin cytoskeleton during Sema3E-induced cell collapse are inhibited by PlexinD1 or SH3BP1 knockdown
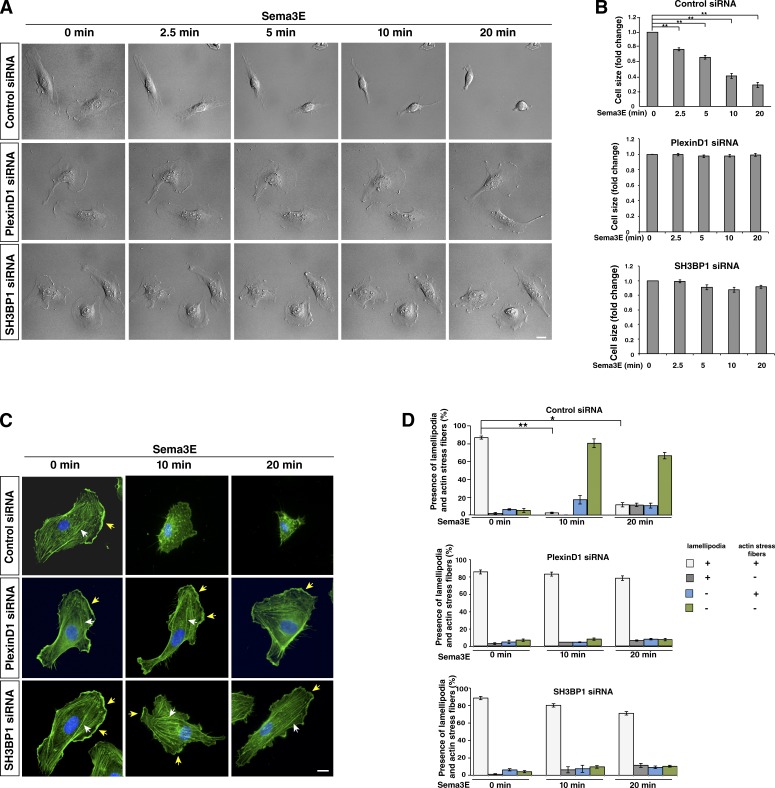
To further validate SH3BP1 as a potential downstream effector of Sema3E-PlexinD1 signaling and determine the temporal and spatial effect of SH3BP1 loss of function on Sema3E-induced cell collapse, we examined the morphological changes of the cells that undergo collapse by time-lapse microscopy. Live imaging was performed on HUVECs transfected with control, PlexinD1, or SH3BP1 siRNA and stimulated with Sema3E. Treatment of HUVECs with Sema3E caused rapid membrane retraction, which was already obvious after 2.5 min, and a complete cell collapse within 20 min (Fig. 4 A and Video 1). Knockdown of PlexinD1 or SH3BP1 by siRNA resulted in maintenance of the EC’s shape long after exposure to Sema3E (Fig. 4 A, Video 2, and Video 3). Changes in cell size were quantified for all three conditions at different time points and normalized to time zero (Fig. 4 B). These experiments demonstrated that loss of SH3BP1 function blocked the cell collapse normally induced by Sema3E-PlexinD1 signaling.
Figure 4.
Sema3E treatment causes dynamic changes in the actin cytoskeleton and cell collapse, which is inhibited by PlexinD1 or SH3BP1 knockdown. (A) Live cell imaging in control, PlexinD1, and SH3BP1 siRNA-transfected HUVECs after Sema3E treatment. Representative DIC images were taken from time-lapse videos of cells at different time points after Sema3E treatment. Control cells underwent obvious morphological changes and exhibited cell collapse in response to Sema3E treatment, whereas PlexinD1- and SH3BP1-depleted cells did not display any changes (see Videos 1, 2, and 3 for the entire video sequence). (B) Quantification of live imaging. Cell size at different time points was measure and normalized to time point 0. **, P < 0.001. Error bars indicate SEM. (C) Actin staining in control, PlexinD1, or SH3BP1 siRNA–transfected HUVECs treated with Sema3E at different time points. Cells transfected with the corresponding siRNA were treated with Sema3E, fixed, and stained with phalloidin (green). In untreated cells, well-organized actin networks with lamellipodia and F-actin stress fibers were seen. After 10 and 20 min of exposure to Sema3E, control cells lost their shape and saw a disruption of F-actin stress fibers. Sema3E treatment of PlexinD1 and SH3BP1 siRNA–transfected cells exhibited an organized network with lamellipodia (yellow arrows) and F-actin stress fibers (white arrows) at every time point. Blue, DAPI. Bar, 10 µm. (D) Quantification of changes in the presence of lamellipodia and stress actin fibers upon Sema3E treatment. In control cells, the number of cells with lamellipodia and stress fibers was significantly reduced after 10 and 20 min of Sema3E treatment. PlexinD1- and SH3BP1-transfected cells did not show significant changes in the presence of lamellipodia and actin stress fibers at 10 and 20 min from ligand introduction. n = 3. *, P < 0.01; **, P < 0.001. Error bars indicate SEM.
To understand the effect of SH3BP1 on the underlying cytoskeleton during Sema3E-induced cell collapse, we used fluorophore-conjugated phalloidin to visualize the changes in F-actin organization. HUVECs transfected with control, PlexinD1, or SH3BP1 siRNA were treated with Sema3E for different durations of time followed by fixation and staining with phalloidin. HUVECs treated with Sema3E caused noticeable actin cytoskeleton changes as early as 2.5 min and a complete actin filament collapse by 20 min, a pattern similar to the one observed in live imaging (Fig. S3). This Sema3E effect was stronger at later time points, and therefore we analyzed the presence of lamellipodia and actin stress fibers at 10 and 20 min after ligand treatment. For example, at 10 min after Sema3E treatment, control siRNA-transfected cells appeared to lose their characteristic shape as the F-actin network was disrupted and disorganized. In addition, cells displayed a perturbation in the linear arrangement of F-actin stress fibers, and a perinuclear accumulation of stained actin was detected (Fig. 4, C and D). By 20 min after ligand exposure, most of the cells still showed strong cytoskeleton collapse. In contrast, PlexinD1 and SH3BP1 siRNA-transfected cells stimulated with Sema3E retained their original size, maintained their organized network of linear F-actin stress fibers throughout the cytoplasm, and exhibited strong actin staining at the cell membrane at every time point of Sema3E treatment (Fig. 4, C and D). These experiments showed that Sema3E treatment leads to actin disorganization in ECs, which is blocked by the knockdown of PlexinD1 and SH3BP1.
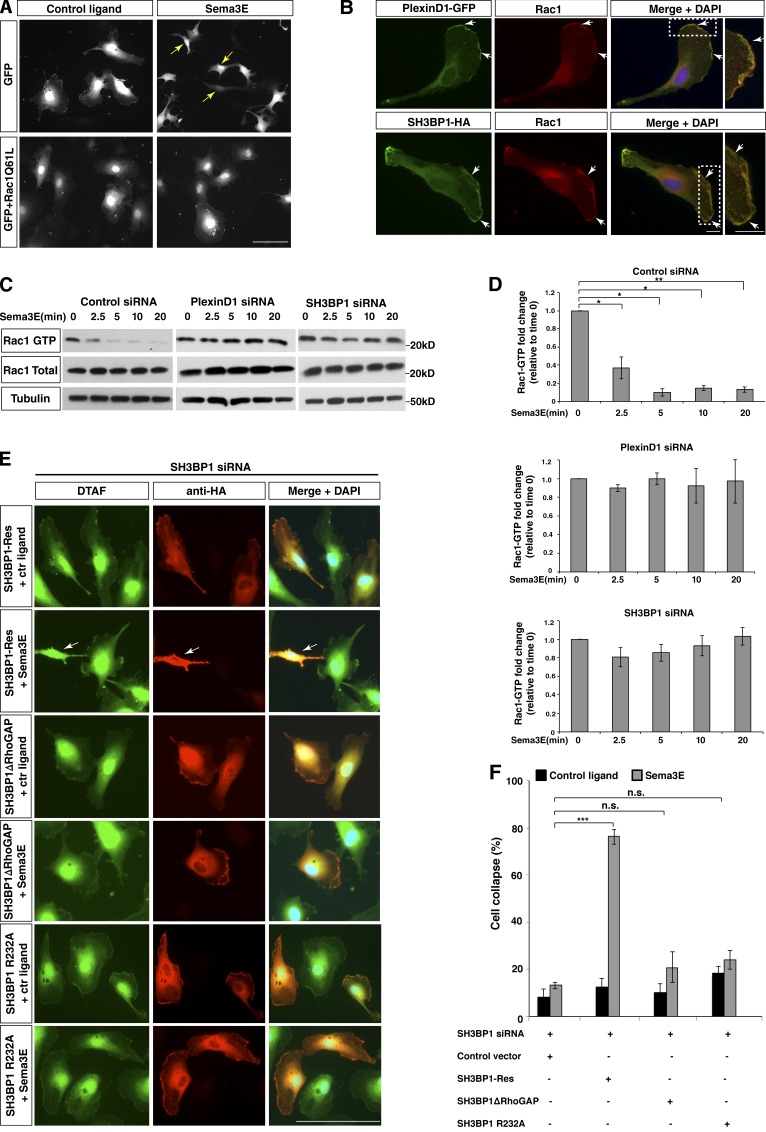
RNAi-mediated knockdown of Rac1 causes hypercollapse of ECs, and constitutively active Rac1 blocks Sema3E-induced collapse
In our RNAi screen we examined genes that suppressed cell collapse as well as those that enhanced the Sema3E-induced collapse, reasoning that the latter might reveal proteins that were down-regulated in response to Sema3E-PlexinD1 activity. One such gene was Rac1, a member of the Rho superfamily of small GTPases. We found that siRNA duplexes against Rac1 enhanced collapse in Sema3E-treated HUVECs (Fig. S4 A and Table S1). Rac1 is known to play a role in PlexinA1 and PlexinB1 signaling, where it is down-regulated after ligand treatment (Gelfand et al., 2009). Consistent with this idea, we found that overexpressing a constitutively active Rac1 mutant protein (Rac1-Q61L) in HUVECs completely blocked Sema3E-induced collapse (Fig. 5 A). These data further support the possibility that Rac1 is also involved in the Sema3E-PlexinD1 signaling cascade. Given the knowledge from previous in vitro studies that SH3BP1 is a GTPase activating protein (GAP), acting specifically as a GAP for Rac1 (Cicchetti et al., 1992, 1995; Parrini et al., 2011), it is possible that the SH3BP1–Rac1 pathway is downstream of Sema3E-PlexinD1 signaling to mediate cell collapse.
Figure 5.
Sema3E-PlexinD1 signaling down-regulates Rac1 activity, which is medicated by SH3BP1 via its GAP activity. (A) Constitutively active Rac1 blocks Sema3E-induced collapse. Cells were transfected with a constitutively active form of Rac1 (Rac1Q61L) and treated with control ligand or Sema3E. GFP construct was cotransfected to visualize cell morphology. While the addition of Sema3E to cells overexpressing only GFP led to strong cell collapse (arrows), HUVECs transfected with Rac1Q61L were unable to undergo collapse after treatment with Sema3E. Bar, 100 µm. (B) PlexinD1 and SH3BP1 colocalized with Rac1 at the lamellipodia (arrows) of HUVECs. PlexinD1-GFP and SH3BP1-HA were cotransfected in HUVECs and stained with GFP or HA antibodies combined with a Rac1 antibody. The boxed regions are enlarged on the right. Bar, 10 µm. (C) Sema3E induced a down-regulation of Rac1 activity, which was blocked by PlexinD1 or SH3BP1 siRNA. HUVECs treated with Sema3E for 0, 2.5, 5, 10, or 20 min were lysed, precipitated with the PAK Rac1-binding domain, and blotted with a Rac1 antibody. Rac1-GTP, total Rac1, and Tubulin blots are shown. (D) Quantification of a Rac1 activity assay is shown. *, P < 0.05; **, P < 0.005. Error bars indicate SEM. (E) SH3BP1 GAP activity was required for Sema3E-induced collapse. Deletion of the GAP domain of SH3BP1 as well as the GAP activity–defective mutant failed to rescue SH3BP1 siRNA inhibition of Sema3E-induced collapse. Rescue experiments with SH3BP1ΔRhoGAP, SH3BP1-R232A, and SH3BP1-Res (full-length siRNA resistant) constructs are shown. Cell shape, DTAF labeling (green); vector expression, HA antibody (red). Arrows, cell collapse. Bar, 100 µm. (F) The results from D were quantified and the percentage of cell collapse is shown. ***, P < 0.01; n ≥ 3. Error bars indicate SEM. n.s., not significant.
PlexinD1, SH3BP1, and Rac1 colocalize at the lamellipodia, and Sema3E induces down-regulation of Rac1 activity
Rac1 is involved in regulating the polymerization of actin at the leading edge, and is therefore localized in the cell lamellipodia (Hall, 1998; Ridley, 2001). We then examined whether PlexinD1, SH3BP1, and Rac1 colocalize in the leading edge of ECs. Cells transfected with PlexinD1-GFP or SH3BP1-HA followed by immunostaining with GFP or HA antibodies, respectively, in combination with Rac1 antibody showed that PlexinD1 and SH3BP1 colocalize with Rac1 in the leading edge of the cell, which suggests that they may function in the same subcellular compartment (Fig. 5 B).
Because of the general role of Rac1 in regulating cytoskeletal dynamics, its effects on Sema3E-induced cell collapse could be nonspecific. To directly test whether Sema3E treatment can specifically influence Rac1 activity, we performed an activated Rac1 pull-down assay. We lysed HUVECs after 0, 2.5, 5, 10, and 20 min of Sema3E treatment and measured activated Rac1 by precipitating the lysates with beads conjugated to a GST-tagged PAK-RBD Rac1 binding domain (Rac1 binding domain of PAK protein) and blotting with Rac1 antibody. Rac1 activity was down-regulated starting from 2.5 min after Sema3E treatment, and this down-regulation was completely blocked by PlexinD1 siRNA, which demonstrates that Sema3E-PlexinD1 signaling specifically down-regulates Rac1 activity (Fig. 5, C and D). To understand how PlexinD1 down-regulates Rac1 activity, we tested whether PlexinD1 (like PlexinA and -B) can directly bind to Rac1. Interestingly, in coimmunoprecipitation (coIP) experiments we did not observe a direct interaction between Rac1 and PlexinD1 at any time point either with or without Sema3E treatment (unpublished data), which suggests that Sema3E-induced down-regulation of Rac1 activity may involve an indirect mechanism.
SH3BP1 mediates the Sema3E-induced down-regulation of Rac1 activity through its GAP domain
SH3BP1 contains three different domains with distinct functions and controls cellular processes. Besides the GAP domain, the SH3 binding motif has been shown to interact with Abl kinase (Cicchetti et al., 1992), whereas the F-BAR domain was implicated in membrane-remodeling processes and inhibits platelet-derived growth factor (PDGF)-induced membrane ruffling in fibroblasts (Cicchetti et al., 1992, 1995; Dawson et al., 2006; Heath and Insall, 2008). Therefore, together with our finding that PlexinD1, SH3BP1, and Rac1 are colocalized in the same subcellular location, it is conceivable that SH3BP1 can function to mediate the Sema3E-PlexinD1–induced cell collapse via its GAP activity (Rac1 regulation) or its BAR domain function, or as an adaptor protein through its SH3-binding motif.
To test the hypothesis that Sema3E induces Rac1 activity through the SH3BP1, we knocked down SH3BP1 using specific siRNA. Similar to PlexinD1, SH3BP1 siRNA blocked the Sema3E-induced down-regulation of Rac1 activity (Fig. 5, C and D). Therefore, SH3BP1 is upstream of Rac1, and Sema3E-PlexinD1 signaling could regulate Rac1 activity through SH3BP1.
To test the importance of SH3BP1 GAP activity for Sema3E-PlexinD1 signaling, we performed rescue experiments with an siRNA-resistant construct, SH3BP1ΔRhoGAP (lacks the GAP domain). In addition, we also generated a more refined GAP activity–defective mutant carrying a single amino acid mutation. It has been shown that the arginine 312 in the GAP domain of human SH3BP1 is critical for its GAP activity. In particular, SH3BP1-depleted cells exhibited perturbations in Rac1 localization, and the R312A SH3BP1 point mutation could not rescue this spatial defect (Parrini et al., 2011). We mutated the corresponding arginine (R312A) in the mouse SH3BP1 and generated SH3BP1-R232A mutant to check if this mutant is able to rescue the Sema3E-induced cell collapse. In SH3BP1 siRNA-transfected HUVECs, the wild-type protein rescued cell collapse after Sema3E treatment, whereas the protein lacking the RhoGAP domain or the GAP-defective R232A mutant did not (Fig. 5, E and F). These data suggest that SH3BP1 may mediate Sema3E-induced down-regulation of Rac1 activity through its RhoGAP domain.
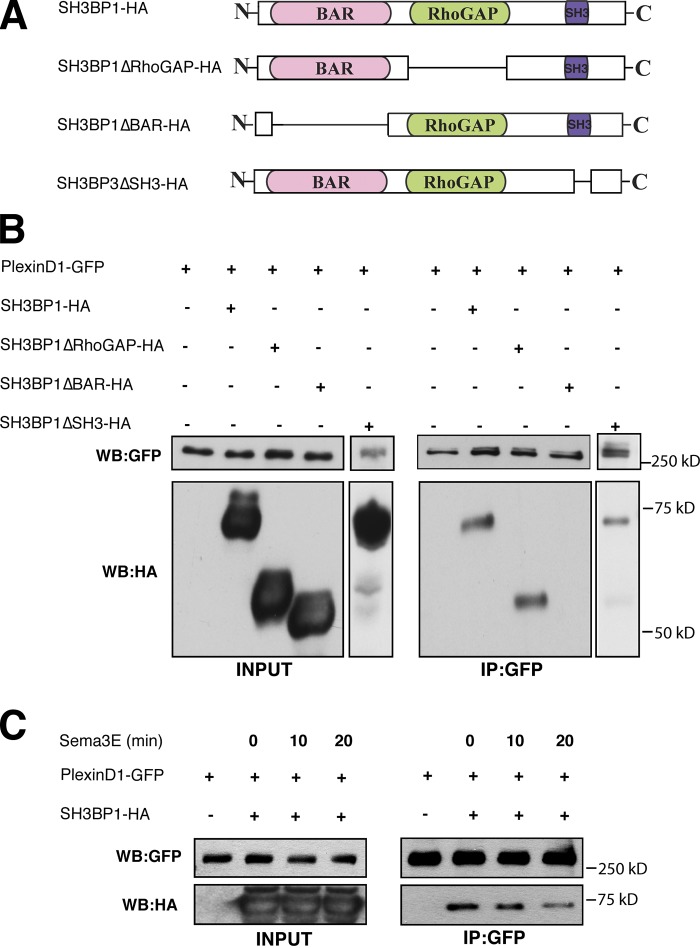
SH3BP1 forms a complex with PlexinD1 via its BAR domain
We hypothesize that Sema3E-PlexinD1 signaling regulates Rac1 by activating SH3BP1, either directly or indirectly. Because SH3BP1 and PlexinD1 are colocalized in the lamellipodia, we then tested whether they are associated in the same protein complex. To examine this, we performed coIP experiments on cells that were either transfected with PlexinD1-GFP alone (negative control) or together with full-length SH3BP1-HA construct (Fig. 6 A). Because of the low transfection efficiency of the DNA construct in HUVECs, all coIP experiments were performed in transfected HEK293T cells. Cell lysates were immunoprecipitated with GFP antibody followed by immunoblotting with HA antibody. Full-length SH3BP1 was coimmunoprecipitated with PlexinD1-GFP (Fig. 6 B), which suggests that PlexinD1 and SH3BP1 are associated in a protein complex. To identify which domain of SH3BP1 mediates this association, we performed coIP experiments with PlexinD1-GFP and either BAR or RhoGAP, or the SH3-binding motif deletion constructs SH3BP1ΔBAR-HA, SH3BP1ΔRhoGAP-HA, or SH3BP1ΔSH3, respectively (Fig. 6 A). Although RhoGAP and SH3-binding motif deletions still resulted in coIP with PlexinD1, the BAR domain deletion abolished the association between PlexinD1 and SH3BP1 (Fig. 6 B), which demonstrates that the BAR domain is required for this association. We next examined which of these three SH3BP1 domains are required for its colocalization with PlexinD1 in the lamellipodia. Consistent with the coIP result, full-length SH3BP1, SH3BP1ΔGAP, and SH3BP1ΔSH3 all colocalized with PlexinD1 in lamellipodia, but SH3BP1ΔBAR did not colocalize with PlexinD1 (Fig. S5, A and B). Together, these data demonstrate that the BAR domain is required for SH3BP1 association with PlexinD1.
Figure 6.
SH3BP1 forms a complex with PlexinD1 via its BAR domain, and Sema3E binding to PlexinD1 activates SH3BP1 by releasing it from the PlexinD1 complex. (A) Schematic illustration of the SH3BP1 constructs used in this study. The full-length SH3BP1 construct contains BAR and RhoGAP domains and SH3 motifs on the C-terminal end. In the SH3BP1ΔRhoGAP construct, the GAP domain was deleted. The SH3BP1ΔBAR construct lacked the BAR domain, and in the SH3BP1ΔSH3 construct the SH3 motif was deleted. (B) SH3BP1 is associated with PlexinD1 through the SH3BP1 BAR domain. HEK293T cells were transfected with PlexinD1-GFP and vectors expressing SH3BP1 (SH3BP1-HA or deletion constructs SH3BP1ΔBAR-HA, SH3BP1ΔRhoGAP-HA, and SH3BP1ΔSH3). Cells were lysed and immunoprecipitated with a GFP antibody and immunoblotted with an HA antibody (right). PlexinD1 and SH3BP1 expression levels in the cell lysates were determined by Western blotting with GFP and HA antibodies (left). (C) SH3BP1 dissociated from PlexinD1 upon Sema3E treatment. HEK293T cells transfected with PlexinD1-GFP and SH3BP1-HA were treated with Sema3E for 0, 10, and 20 min, then lysed and immunoprecipitated with a GFP antibody and immunoblotted with an HA antibody. The dissociation of SH3BP1 from PlexinD1 was observed 20 min after Sema3E treatment, as indicated on the right, while the protein expression levels are shown on the left.
Sema3E stimulation activates SH3BP1 in part by releasing it from PlexinD1
We next examined the effect of Sema3E treatment on the association between SH3BP1 and the PlexinD1 receptor complex. PlexinD1-GFP– and SH3BP1-HA–transfected HEK293T cells were treated with Sema3E for 0, 10, and 20 min followed by cell lysis and IP with GFP antibody. Immunoblot analysis with HA antibody showed that the interaction between PlexinD1 and SH3BP1 was decreased after 20 min of Sema3E treatment (Fig. 6 C), which suggests that Sema3E binding to PlexinD1 released SH3BP1 from the receptor complex. We note that this is consistent with our observation in HEK293T cells, where Rac1 activity is significantly down-regulated 20 min after Sema3E treatment (Fig. S4 B). Taken together, these data suggest a model in which Sema3E stimulation releases SH3BP1 from PlexinD1 and subsequently activates its GAP activity.
Releasing SH3BP1 from PlexinD1 is necessary but not sufficient for its activation
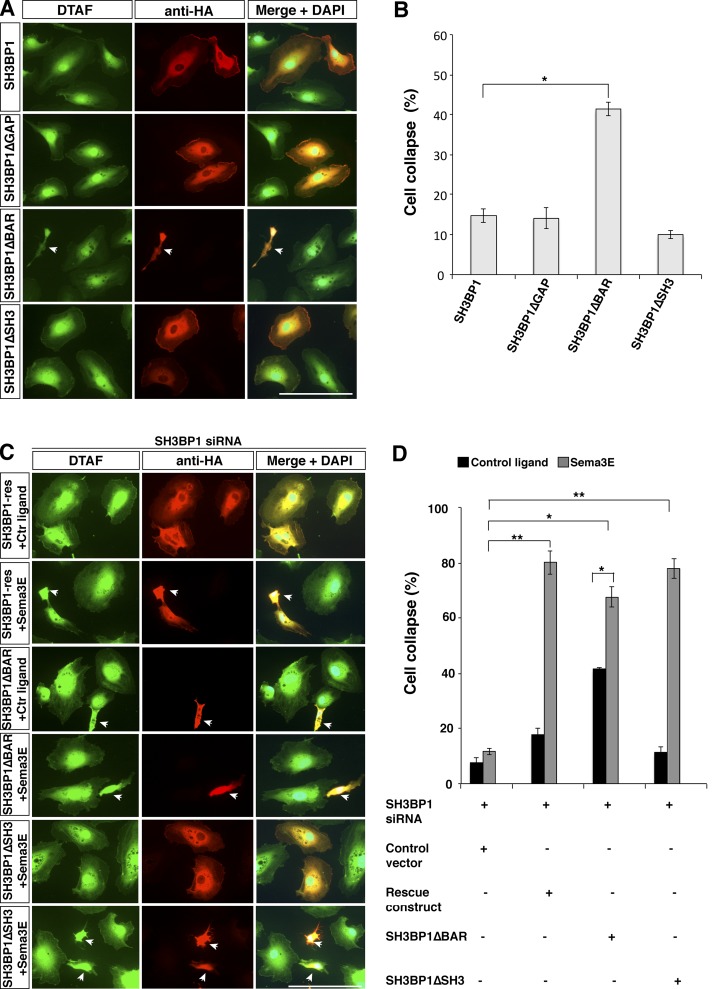
To test whether dissociating SH3BP1 from PlexinD1 is sufficient to fully activate SH3BP1 and result in cell collapse, we first analyzed if overexpression of SH3BP1ΔBAR will constitutively activate SH3BP1 to recapitulate the cell collapse induced by Sema3E ligand. SH3BP1 constructs were transfected in HUVECs and their morphology was analyzed. SH3BP1-, SH3BP1ΔGAP-, and SH3BP1ΔSH3-transfected cells showed normal cell morphology. However, ∼40% of the cells transfected with SH3BP1ΔBAR exhibited morphological changes similar to Sema3E-induced cell collapse (Fig. 7, A and B). This result indicates that although the release of SH3BP1 from PlexinD1 contributes to SH3BP1 activation, an additional signal is required to fully activate SH3BP1 function in mediating cell collapse.
Figure 7.
BAR domain deletion caused constitutive cell collapse, and Sema3E treatment enhanced the collapse. (A) Overexpression of SH3BP1ΔBAR led to changes of cell morphology in HUVECs. SH3BP1-HA, SH3BP1ΔGAP-HA, SH3BP1ΔSH3-HA, and SH3BP1ΔBAR-HA were overexpressed in HUVECs, and cells were stained with DTAF (green) and HA antibody (red). Deletion of the BAR domain caused changes in cell morphology and size compared with full-length SH3BP1 as well as GAP and SH3 deletion. Bar, 100 µm. (B) Quantification of overexpression experiments is shown. *, P < 0.01; n = 4. Error bars indicate SEM. (C) Deletion of the BAR domain of SH3BP1 partially rescued SH3BP1 siRNA inhibition of Sema3E-induced collapse. Rescue experiments with SH3BP1ΔBAR, SH3BP1ΔSH3, and SH3BP1-Res (full-length siRNA resistant) constructs are shown. Cell shape, DTAF (green); vector expression, HA antibody (red). Arrows, cell collapse. Bar, 100 µm. (D) The results from C were quantified and the percentage of cells collapsed is shown. *, P < 0.01; **, P < 0.001. n = 4. Error bars indicate SEM.
To further understand the role of Sema3E-induced dissociation of SH3BP1 from PlexinD1 and the requirement of the SH3BP1 BAR domain in Sema3E-induced cell collapse, we next performed rescue experiments with an siRNA-resistant SH3BP1ΔBAR. As expected, with control ligand treatment, SH3BP1ΔBAR exhibited ∼40% cell collapse, significantly higher than the control vector–transfected cells. Upon Sema3E treatment, the percentage of collapsed cells significantly increased with SH3BP1ΔBAR rescue (∼70%), equivalent to the level of rescue achieved by full-length SH3BP1 (Fig. 7, C and D). This suggests that the BAR domain of SH3BP1 is necessary but not sufficient for its function. It is conceivable that Sema3E treatment not only led to the release of SH3BP1 from PlexinD1, but may have triggered a “second signal” that is needed to fully activate SH3BP1 and cause cell collapse.
Because the SH3-binding motif is known to mediate protein–protein interaction, we next examined whether the SH3-binding motif of SH3BP1 plays any role in mediating the “second signal” required for Sema3E-induced cell collapse. We performed rescue experiments with siRNA-resistant SH3BP1ΔSH3, but this construct successfully rescued cell collapse (Fig. 7, C and D), which suggests that the SH3-binding motif is not required for SH3BP1 function in the context of Sema3E-PlexinD1–mediated cell collapse.
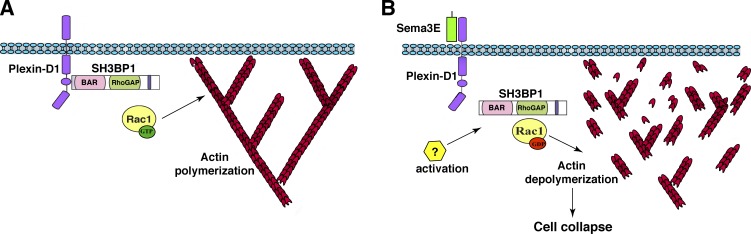
Collectively, our findings demonstrate that in the absence of Sema3E, SH3BP1 is associated with the PlexinD1 receptor complex through the SH3BP1 BAR domain. This interaction allows the active form of Rac1 to control actin polymerization (Fig. 8 A). In the presence of Sema3E ligand, SH3BP1 dissociates from the PlexinD1 receptor complex, and achieves full activation by an unknown Sema3E-induced signal followed by inactivation of Rac1 through its RhoGAP activity, which in turn triggers actin cytoskeleton depolymerization and cell collapse (Fig. 8 B).
Figure 8.
A model of how SH3BP1–Rac1 mediates Sema3E-PlexinD1 regulation of cytoskeleton stability. (A) In the absence of Sema3E, SH3BP1 is associated with the PlexinD1 complex, and the active form of Rac1 (Rac1GTP) positively regulates actin polymerization. (B) Upon Sema3E treatment, SH3BP1 dissociates from PlexinD1, becomes activated, and through its RhoGAP domain converts GTP-Rac1 to GDP-Rac1. The decreased Rac1 activity leads to actin depolymerization and cell collapse.
SH3BP1 is required for Sema3E-PlexinD1–mediated EC repulsion
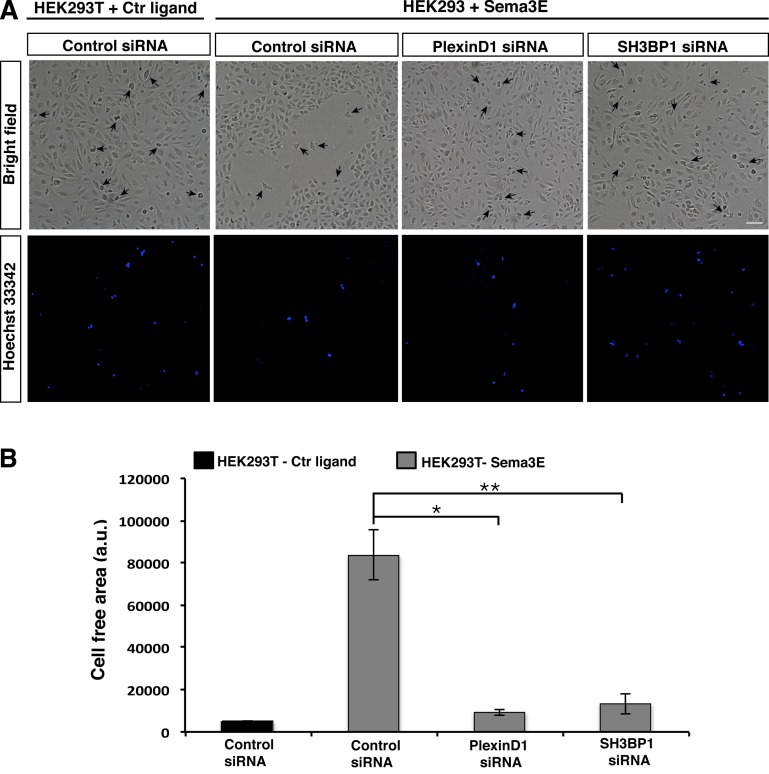
Previous in vivo and in vitro studies have demonstrated that a major function of Sema3E-PlexinD1 signaling in ECs is to serve as a repulsive signal to inhibit endothelial migration and vascular sprouting (Gu et al., 2005; Kigel et al., 2008; Casazza et al., 2010; Fukushima et al., 2011). We used the well-established “repulsion” assay to study the function of SH3BP1 in Sema3E-PlexinD1 signaling in a physiologically relevant context (Guttmann-Raviv et al., 2007; Kigel et al., 2008; Bielenberg et al., 2008). As shown previously, Sema3E-expressing HEK293T cells repelled ECs when seeded on top of a confluent monolayer of HUVECs, resulting in cell-free areas around the Sema3E-expressing cells (Fig. 9, A and B). In contrast, HEK239Ts expressing control ligand had no effect on the confluent monolayer of HUVECs. When HUVECs were transfected with PlexinD1 or SH3BP1 siRNA, Sema3E-expressing HEK293T cells no longer had any effect on the HUVECs (Fig. 9, A and B). These data demonstrate that SH3BP1 is a key effector molecule required for the repulsive guidance effect of Sema3E-PlexinD1 on EC migration.
Figure 9.
PlexinD1 and SH3BP1 play a role in Sema3E induced ECs repulsion. (A) HUVECs transfected with control, PlexinD1, and SH3BP1 siRNA were grown to a confluent monolayer. Cells were photographed 16 h later. HEK293T cells expressing control ligand or Sema3E were prelabeled with Hoechst 33342 and added on top of HUVECs. HEK293T expressing Sema3E (arrows) were surrounded by a cell-free area, whereas HEK293T expressing a control ligand did not exhibit a cell-free area. PlexinD1 and SH3BP1 siRNA-transfected cells did not repel in the presence of a Sema3E source. Bright field and Hoechst 33342 channel are shown. Bar, 200 µm. (B) Quantification of cell free area surrounding HEK293T cells. *, P < 0.01; **, P < 0.001. n = 4. Error bars indicate SEM.
Discussion
Here, we performed an image-based unbiased genome-wide RNAi screen to identify signaling molecules downstream of Sema3E-PlexinD1. We subsequently characterized one of the genes identified from our screen, a Rho-GAP protein, SH3BP1. We first validated its functional requirement for Sema3E-PleixnD1–mediated cell collapse by rescue experiments. We next demonstrated that Sema3E-PlexinD1 down-regulates Rac1 activity, and that this down-regulation is mediated by SH3BP1. Moreover, we showed that, upon Sema3E binding, the prebound SH3BP1 dissociates from the PlexinD1 complex and, through its GAP activity, converts Rac1-GTP to Rac1-GDP to initiate actin disassembly. Finally, SH3BP1 is required for Sema3E-PlexinD1–mediated EC repulsion. The identification and characterization of SH3BP1 as a novel downstream effector mediating Sema3E-PlexinD1 signaling in ECs provides insights for how extracellular signals are translated into intracellular cytoskeletal changes and unique cell behavior. Moreover, this image-based approach can be applied to other ligand receptor–mediated signaling.
Inactivation of integrin has been suggested to mediate Sema3E-induced cytoskeletal collapse (Sakurai et al., 2010). These authors showed that HUVECs or COS cells transfected with PlexinD1 collapsed in response to Sema3E only when the dishes were coated with integrin. In contrast, we performed our screening assay in HUVECs without any coating. This approach is substantiated by previous studies demonstrating that Sema3E still induces the rapid collapse of PlexinD1-transfected COS cells (Gu et al., 2005; Casazza et al., 2010) and suggests that Sema3E-PlexinD1 can initiate an intracellular signaling cascade directly linked to actin depolarization and cell collapse. Moreover, the signaling molecules and pathway we discovered from our screen represent a direct mechanism, independent of the integrin pathway.
Studies from PlexinA and -B families have implicated small GTPases as key molecules downstream of plexin signaling. However, the specific mechanisms differ between PlexinA and -B (Negishi et al., 2005; Gelfand et al., 2009; Oh and Gu, 2013a). For example, Sema4D binding to PlexinB1 reduces Rac1 activity by binding to Rac1-GTP and thereby sequestering it from its downstream effector (Kruger et al., 2005), whereas Sema3A binding to PlexinA increases Rac1 activity by activating a Rac guanine exchange factor (Rac1GEF; Toyofuku et al., 2005). Interestingly, here we found that even the same ligand–receptor pair, Sema3E-PlexinD1, uses a different signaling mechanism to regulate Rac1 GTPase activity from what was previously reported. Prior studies have suggested that, similar to the Sema4D–PlexinB1 pathway, PlexinD1 is in a conformation that enables its association with GTP-bound Rnd2 but prevents its interaction with GTP-bound Rac and R-Ras in the absence of Sema3E. Upon Sema3E binding, PlexinD1 dissociates Rnd2 and binds the active forms of both Rac and R-Ras GTPases. As a result, Rac is sequestered, thereby inactivating PAK and leading to the collapse of the actin-based cytoskeleton (Uesugi et al., 2009; Gay et al., 2011). However, here through an unbiased functional screen, we identified a RhoGAP protein, SH3BP1, as a novel key downstream signaling molecule mediating Sema3E-induced down-regulation of active Rac1GTPase and actin cytoskeleton disassembly. Although the final outcome is the same (active Rac1 down-regulation and actin-based cytoskeleton collapse), one signaling pathway functions by sequestering Rac1 and the other by activating SH3BP1 to convert active Rac1-GTP to inactive Rac1-GDP. In addition, another study has suggested that the small GTPase RhoJ might be a mediator between Sema3E-PlexinD1 and VEGF-VEGFR2 signaling through the regulation of GDP/GTP binding to RhoJ (Fukushima et al., 2011). Given the broad effect of Sema3E-PlexinD1 in diverse cell types, it is possible that the Rnd-mediated Rac1-sequesteration mechanism operates at times, while the SH3BP1 RhoGAP activity–mediated mechanism functions at other times, depending on the cell type–specific expression of these effectors.
Small GTPases and their regulators (GEFs and GAPs) comprise a large portion of the genome, and regulation of their activities is a major mechanism underlying a vast range of biological activities. Emerging evidence demonstrates that specific members of the small GTPase superfamily mediate specific aspects of biological functions. For example, α2-chimaerin, a Rho GAP, has been shown to be essential for ephrin-mediated corticospinal axon guidance in rodents (Beg et al., 2007; Iwasato et al., 2007; Wegmeyer et al., 2007). A recent study also showed that the RacGAP β2-Chimaerin selectively mediates axonal pruning in the hippocampus (Riccomagno et al., 2012). Investigating the specific in vivo requirement of SH3BP1 in semaphorin-mediated neural and vascular development will be a future focus.
SH3BP1 is a relatively lesser known GAP protein. It was originally identified from an SH3 domain screening (Cicchetti and Baltimore, 1995). More recently, SH3BP1 has been shown to partner with an exocyst complex (Parrini et al., 2011) and mediate epithelial junction formation by regulating Cdc42 activity (Elbediwy et al., 2012). Our results are the first study linking SH3BP1 to an external stimulus (Sema3E) through its interaction with the PlexinD1 receptor. The interaction of SH3BP with PlexinD1 through its BAR domain and the down-regulation of Rac1 through its GAP activity explain how Sema3E-PlexinD1 signals are transmitted to ultimately produce intracellular cytoskeleton changes. Whether SH3BP1’s ability to regulate Cdc42, exocytosis, and junction formation plays any role in Sema3E-PlexinD1 signaling will be investigated in the future.
We also identified several other candidate genes in our RNAi screen that might potentially be involved in Sema3E-PlexinD1 signaling, and it will be interesting to examine how these genes might intersect with SH3BP1 signaling. For example, our data so far suggest that dissociation of SH3BP1 from PlexinD1 contributes to the activation of its GAP activity, another Sema3E-dependent signal that is also needed to fully activate SH3BP1. Such a signal could be one of the strong hits from our screen. It will be interesting to conduct similar screens at different time points using actin morphology as a readout to identify downstream molecules that act at different phases of the collapse. This unbiased functional screen will allow us to not only visualize a more complete picture of plexin signaling but also assemble the entire pathway to reveal the logical progression from signal integration, cytoskeleton regulation, and cellular behavior. We will also test whether the signaling pathways we have discovered are generally used by other plexins. It will also be important to examine whether the diverse functions of plexins in different cell types and processes are mediated by the same or distinct set of signaling mechanisms. In the long term, we hope to understand the overall properties of a signaling network in neurons and ECs and to predict how this network will behave in response to new stimuli, or how it can be modified or rebuilt to give a desired effect. Finally, given the important and diverse biological functions of semaphorins and plexins, a complete understanding of the signaling mechanisms resulting from these ligand receptor pairs will ultimately provide potential therapeutic targets for diseases.
Materials and methods
Cell cultures
HEK293T cells were cultured in DMEM supplemented with 10% (vol/vol) FBS and 1% (vol/vol) penicillin/streptomycin. HUVECs were cultured in EGM-2 medium supplemented with a SingleQuots kit (Lonza). All cells were cultured in 5% CO2 at 37°C.
AP-tagged ligand production
HEK293T cells were transfected with AP-Sema3E or AP expression constructs using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. Media was changed after 4 h. Cells were cultured for an additional 48 h in DMEM + 2% FBS. After 48 h, the media was collected and filtered to remove cell debris, and AP activity was measured. The AP-Sema3E (delineated throughout manuscript as Sema3E) and AP (control ligand) were frozen at −80°C until use.
Plasmids and RNAi
A full-length human PlexinD1 (available from GenBank under accession no. NM_015103) was used to generate GFP-tagged constructs that were cloned into a pBK-CMV vector. For SH3BP1 HA fusion protein constructs, the sequences encoding the full-length mouse SH3BP1 (accession no. BC004598) were cloned into pCAG vector to produce a fusion protein. To generate the HA-tagged deletion constructs SH3BP1ΔBAR (amino acids deleted: 2–174), SH3BP1ΔGAP (amino acids deleted: 210–356), or SH3BP1ΔSH3 (amino acids deleted: 528–538), fusion protein constructs of the sequences without the indicated domains were cloned into a pCAG vector. To generate a mutant allele defective in GAP activity, arginine 232 was replaced by an alanine residue using the mutagenesis kit (QuikChange; Agilent Technologies). The pGEX-KG-PAK-RBD vector encoding a GST-fused Rac-binding domain (RBD, amino acids 72–152) of mouse PAK-A was provided by K.-L. Guan (University of California San Diego, La Jolla, CA), pcDNA3 mammalian expression vector encoding human Rac1-Q61L was provided by M. Kirschner (Harvard Medical School, Boston, MA), and pAPtag-5-Sema3E vector encoding mouse Sema3E cloned in aAPtag-5 vector was obtained from F. Mann (Aix-Marseille Université, Centre National de la Recherche Scientifique, Marseille, France). Empty pAPtag-5 vector (GenHunter) was used as a control. Sequences targeted in the siRNA screen were: PlexinD1, 5′-GCAAGGAUUCGCCAACCAA-3′; and SH3BP1, 5′-GAUGACAGCCACCCACUUC-3′.
Preparation of PAK GST fusion protein and Rac activity assay
The p21-activated protein kinase (PAK1) was expressed in bacteria as a GST fusion protein and coupled to agarose beads. The bacteria culture was grown until the OD at 600 nm reached 0.6–0.8. Transcription was induced by the addition of 0.1 mM IPTG and bacteria were grown for 3 h at 30°C. The culture was centrifuged for 10 min at 2,300 g at 4°C. The pellet was washed once with cold PBS followed by centrifugation, resuspension in cold bacterial lysis buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 5 mM MgCl2, 1 mM EDTA, 1 mM DTT, 1 mM PMSF, and 1 µg/ml aprotinin), incubation on ice for 10 min, and sonication three times for 30 s. The lysate was cleared by centrifugation (15 min, 20,000 g, 4°C). Sepharose 4B beads (GE Healthcare) were added to the supernatant and incubated for 2 h at 4°C on a rotator. Beads were washed five times with ice-cold bacterial lysis buffer, resuspended in washing buffer with 10% vol/vol glycerol (1:1 slurry), and stored at −80°C.
HUVECs treated with 2 nM Sema3E at 0, 10, and 20 min were washed rapidly with ice-cold PBS, then lysed in cell lysis buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 30 mM MgCl2, 1% NP-40, 1 mM DTT, protease inhibitor, and 2 mM sodium orthovanadate). The lysate was incubated on ice for 15 min for efficient extraction, then vortexed and clarified by centrifugation for 10 min at 12,000 rpm and 4°C. Protein concentration was determined via the Bradford assay. After sample normalization, one-twentieth of the original sample was kept for immunoblotting to determine the total Rac1 content in the lysate. 10 µl of purified GST-PAK-RBD beads was added to the rest of each sample and incubated for 1 h at 4°C while rotating. The samples were centrifuged at 400 g for 2 min, the supernatant was aspirated, and the pellet was washed three times in cell lysis buffer. Protein was eluted by the addition of 2× SDS-PAGE sample buffer and boiling for 5 min.
Rescue experiments
Reverse transfection with Dharmafect (Thermo Fisher Scientific) was used for delivering siRNAs into HUVECs. After 24 h, cells were transfected with the SH3BP1 and control (pCAG-GFP-HA) constructs using Lipofectamine 2000 (Invitrogen). 24 h after the DNA transfection, cells were treated with 2 nM Sema3E, fixed in 4% PFA, washed in 1× PBS, and stained with 5-([4,6-Dichlorotriazin-2-yl]amino)fluorescein hydrochloride (DTAF) in 1× PBT (1× PBS + 0.1% Triton X-100) for 1 h. Then cells were washed three times in PBT, blocked in 3% BSA, and incubated with mouse anti-HA (Covance) primary antibody overnight at 4°C. Cells were washed three times for 5 min in 1× PBS and incubated with Alexa Fluor 567 anti–mouse secondary antibodies for 1 h at room temperature. Coverslips were mounted on slides using ProLong Gold antifade reagent with DAPI (Life Technologies). Cells collapse was analyzed manually. A minimum of 50 cells were quantified for each condition.
Primary mouse EC isolation
Primary mouse ECs were isolated from mouse embryonic lungs and brain at E18.5 using a previously described protocol (Deng et al., 2013), with some modification. In brief, lungs and brain were dissected from embryos, minced finely with scissors, and then digested in 1 mg/ml Collagenase/Dispase (Roche) for 45 min at 37°C on a rotator. The digested tissue suspension was transferred to a syringe with a 14-g cannula attached, and clumps were triturated into a single cell suspension. Cells were centrifuged at 400 g for 5 min, then the pellet was resuspended in 0.1% BSA/PBS and incubated with anti–mouse CD31 (BD) antibody–coated Dynabeads (Invitrogen) at room temperature for 10 min with rotation. Using a magnetic separator, the bead-bound cells were washed with 0.1% BSA/PBS and recovered, then cultured in EGM-2 medium supplemented with a SingleQuots kit. The cells were then used for gene expression analysis by RT-PCR.
RNA isolation, cDNA synthesis, and RT-PCR
Total RNA was isolated using TRIzol (Invitrogen) according to the manufacturer’s protocol. Afterward, 1 µg of RNA from each sample was reverse transcribed into cDNA using the SuperScript III Reverse transcription kit (Invitrogen). RT-PCR was performed with taq polymerase (QIAGEN) with the listed primers: GAPDH Fw, 5′-GTCTACATGTTCCAGTATGACTCCACTCAC-3′; GAPDH Rev, 5′-CAATCTTGAGTGAGTTGTCATATTTCTCGT-3′; SH3BP1 Fw, 5′-GCCCTCCATGATGTTTGC-3′; and SH3BP1 Rev, 5′-ACAGCCTCAGGGCCTTCT-3′.
EC repulsion assay
HUVECs transfected with control, PlexinD1, or SH3BP1 siRNA were grown to confluence. HEK293T transfected with AP-Sema3E or AP were stained with NucBlue Live ReadyProbes Reagent (Invitrogen), then plated on top of the HUVECs and incubated for 16 h. The cells were then washed with PBS and fixed with 4% PFA for 15 min at room temperature, and washed in PBS. 10 randomly selected areas were imaged in PBS for each sample. Cell repulsion was quantified by calculating the cell free area using ImageJ software (National Institutes of Health). A minimum of 35 cell-free areas were measured and quantified for each condition.
CoIP
HEK293T cells were transfected with the indicated constructs using Lipofectamine 2000 (Invitrogen) and grown in DMEM + 10% fetal bovine serum + 1% penicillin/streptomycin. 48 h after transfection, cells were washed rapidly with ice-cold PBS. Cells were lysed in lysis buffer (50 mM Tris/HCl, pH 7.5, 150 mM NaCl, 1% Triton X-100, 2 mM EDTA, and 2 mM DTT) containing complete proteinase inhibitors (Roche). After 30 min of rotation at 4°C and subsequent centrifugation, protein was quantified and 20 µg of protein was frozen down as input controls. 0.5 µg of rabbit anti-GFP antibody (Invitrogen) was added to 500 µg of protein and rotated at 4°C for 1 h. Then, 30 µl of Protein A/G beads (Thermo Fisher Scientific) were added to the protein and rotated overnight at 4°C. Beads were washed three times with lysis buffer and two times with wash buffer (lysis buffer with 300 mM NaCl). Protein was eluted by the addition of 2× SDS-PAGE sample buffer and boiling for 10 min.
Western blotting
Protein samples were loaded on 8% or 12% polyacrylamide gels and run until the appropriate protein separation was achieved. Samples were transferred onto polyvinylidene difluoride (PVDF) membrane and blocked for 1 h in 5% nonfat milk in TBST (Tris-buffered saline + 0.1% Tween 20). The membranes were then incubated overnight (see next paragraph) with the following primary antibodies at 4°C: rabbit anti-GFP (Invitrogen), mouse anti-HA (Covance), mouse anti-Rac1 (BD), rabbit polyclonal antiserum raised against PlexinD1 peptide (CELVEPKKSHRQSHRKK; anti-PlexinD1 was a gift from Y. Yoshida, Cincinnati Children’s Hospital Medical Center, Cincinnati, OH; Oh and Gu, 2013b), goat anti-SH3BP1 (Everest Biotech Ltd.; Parrini et al., 2011; Elbediwy et al., 2012), rabbit VE-Cadherin (Abcam), and mouse anti-Tubulin (Sigma-Aldrich). The intensity of individual bands was quantified using ImageJ.
Immunohistochemistry
HUVECs were grown on coverslips and then transfected with PlexinD1-GFP and/or SH3BP1-HA. After 48 h, cells were fixed in 4% PFA for 15 min, washed twice for 5 min in PBS, and permeabilized for 10 min in 1× PBT (1× PBS + 0.1% Triton X-100). The samples were then incubated for 1 h in 10% donkey serum in PBT. After blocking, the samples were incubated with primary antibodies: rabbit anti-GFP (Invitrogen), mouse or rabbit anti-HA (Covance; Abcam), mouse anti-Rac1 (EMD Millipore), goat or rabbit anti-PlexinD1 (R&D Systems; a gift from Y. Yoshida), and goat anti-SH3BP1 (Everest Biotech Ltd.) overnight at 4°C. Cells were washed three times for 5 min in 1× PBS and incubated with Alexa Fluor 488 anti–rabbit and Alexa Fluor 567 anti–mouse secondary antibodies for 1 h at room temperature. Coverslips were mounted on slides using ProLong Gold antifade reagent with DAPI (Life Technologies). Cells used for colocalization studies were counted manually.
Actin staining
HUVECs were grown on coverslips and then transfected with control, PlexinD1, or SH3BP1 siRNA. 48 h after transfection, cells were treated with Sema3E, rapidly washed with PBS, fixed in 4% PFA for 15 min, washed twice for 5 min in PBS, and permeabilized for 10 min in 1× PBT (1× PBS + 0.1% Triton X-100). Cells were incubated with phalloidin Alexa Fluor 488 (Invitrogen) for 30 min at room temperature then washed five times for 5 min in PBS and mounted on slides. The presence of lamellipodia and actin stress fibers was counted manually in a minimum of 50 cells.
Live imaging
HUVEC were grown on glass-bottom culture dishes (MatTek Corporation) overnight. DIC images were acquired at intervals of 10 s with a microscope (VivaView). First baseline images were acquired for 5–10 min, then cells were treated with AP-Sema3E ligand at a concentration of 2 nM, and images were acquired for the next 20 min. Cell size changes were analyzed in ImageJ. 7–12 cells were quantified for each condition.
Microscopy
All epifluorescent images of fixed specimens were acquired with a fluorescence microscope (Eclipse 80i; Nikon) using a 20×/0.75 NA or 60×/1.4 NA oil immersion objective lens fitted with a digital camera (DS-2; Nikon) and NIS-Elements BR 3.0 software (Nikon). Images were adjusted for brightness and contrast using ImageJ. Images from endothelial repulsion assays were acquired with an inverted florescent microscope (Eclipse TE2000-S; Nikon) using a 4×/0.2 NA objective lens, a digital camera (DS-5M; Nikon), and NIS-Elements BR 3.0 software (Nikon). Images were adjusted for brightness and contrast using ImageJ. Time-lapse videos were recorded at 37°C using an incubator live cell imaging microscope (VivaView FL LCV-110; Olympus) with a UPlan-SApochromat 40×/0.95 NA, WD 0.18 mm objective lens (Olympus) and a camera (Orca R2 CCD; Hamamatsu Photonics). Videos were acquired with MetaMorph for Olympus VivaView FL LCV-110 software, and separated images were adjusted for brightness and contrast using ImageJ.
Screen protocol
An RNAi library composed of 21,121 smart pools of four siRNA oligos, each targeting the whole human genome (siARRAY siRNA Library, human genome, G-005000-05; Thermo Fisher Scientific) was used. For the secondary screen, individual siRNA oligos from the pool in the primary screen were assayed. Reverse transfection with Dharmafect (Thermo Fisher Scientific) was used for delivering siRNAs into HUVECs in triplicate in a 384-well plate format.
Reverse transfection
A 384-well assay plate was loaded with 2 µl of 1 µM siRNA using the Velocity11 Bravo robot inside a biological safety cabinet (Bioprotect II; Baker). Each well of the 384-well plates contained a smart pool of four siRNA oligos targeting a single gene or a control siRNA. Then 6 µl of Opti-MEM media and Dharmafect No. 1 transfection reagent mixed at a ratio of 1:300 were added using an automated solution dispenser (WellMate; Matrix). Samples were spun down to ensure proper mixing and incubated at room temperature for 20 min to allow lipid siRNA complexes to form. Finally, HUVECs were plated at a density of ∼280 cells/well in 32 µl of media to bring the total reaction volume to 40 µl per well and the siRNA concentration to 50 nM. The final concentration of 50 nM is optimized to provide effective mRNA knockdown while avoiding off-target effects. Assay plates were run so that each library plate was treated with Sema3E in duplicate.
Ligand treatment and Dil staining
48 h after transfection, cells in the screening plates were treated with either Sema3E or control ligand. Plates were removed from the incubator and solution was aspirated using a microplate washer (ELx405; BioTek) until 25 µl remained. Using the automated solution dispenser (Wellmate; Matrix), 25 µl of 4 nM Sema3E or control ligand was added for a final concentration of 2 nM. The plates were then returned to the incubator for 25 min. After ligand treatment, cells were fixed using 4% PFA and stained with 0.003% DiI in 50% ethanol for 10 min and DAPI (0.5 µg/ml) for 5 min. All fixation and staining steps were done with high-throughput automation accomplished using the Wellmate and the ELx405 microplate washer.
Imaging and image analysis
After immunostaining, plates were imaged using a high-content screening system (ImageXpress Micro; Molecular Devices) using a 10× 0.3 NA Plan Fluor objective lens and suitable filters (DAPI and Texas red for DiI). Four sites were imaged from each well. For image analysis, the images were preprocessed using CellProfiler. During the preprocessing, nuclei and whole cells are segmented using the DAPI and DiI channels, respectively. The segmented cellular masks were then passed onto our custom-developed image analysis algorithm in MATLAB, which detects the distinct membrane protrusions present on the cell surface of collapsed cells. The custom algorithm then combines the information of membrane protrusions with cellular area to classify individual cells into collapsed and intact populations. We also took into account the neighborhood environment of each cell by eliminating all the cells that are completely surrounded by their neighboring cells from the analysis. The percentage of cells that are classified as collapsed is computed for each well, and is used as the metric to identify the potential hits in the screen. The total cell numbers counted for each condition are as follows. Fig. 1 C: nontargeting siRNA control ligand treatment, 20,624; nontargeting siRNA Sema3E treatment, 25,365; PlexinD1 siRNA control ligand treatment, 2,684; and PlexinD1 siRNA Sema3E treatment, 1,887. Fig. 3 B: no siRNA, 18,850; nontargeting siRNA, 9,996; PlexinD1 siRNA, 7,197; and SH3BP1, 833. The Z′ score is 0.32. The large-scale image analysis was performed on the Orchestra high-performance computing cluster at the Harvard Medical School.
Statistical analysis
The SEM or SD of the mean was calculated from the mean of at least three independent experiments. Bars in the graph of the screen data represent SD. In the rest of the graphs, SEM is provided. Respective n values are provided in the figure legends. The indicated P-values were obtained with the two-tailed Student’s t test.
Online supplemental material
Fig. S1 shows that endogenous PlexinD1 expression in HUVECs could be down-regulated by specific PlexinD1 siRNA and used as a positive control in the screen. Fig. S2 shows the expression of SH3BP1 and the specificity of SH3BP1 siRNA. Fig. S3 shows Sema3E-regulated cytoskeleton changes. Fig. S4 shows that Rac1 is regulated by Sema3E. Fig. S5 shows that SH3BP1ΔBAR did not colocalize with PlexinD1 in the lamellipodia. Video 1 shows that control siRNA–transfected cells underwent morphological changes and exhibited cell collapse in response to Sema3E treatment. Video 2 shows that PlexinD1 siRNA-transfected cells did not display any morphological changes after Sema3E treatment. Video 3 shows that SH3BP1 siRNA-transfected cells did not display any morphological changes after Sema3E treatment. Table S1 shows exemplar genes demonstrating Sema3E-induced hypercollapse after gene down-regulation. Table S2 shows a list of genes identified in the secondary screen. A custom MATLAB algorithm is provided as raw code in a TXT file. This program processes the images after they have been segmented by CellProfiler, analyzes the protrusions of each cell by shrinking the cells and comparing with the original one, measures object properties, and merges sections that belong to the same cell back together. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201309004/DC1.
Supplementary Material
Acknowledgments
We thank members of the Gu laboratory and Dr. Purushothama Rao Tata for helpful comments on the manuscript; Lauren Byrnes and Maya Stawnychy for technical support; The Institute of Chemistry and Cell Biology (ICCB) Screening core facility at Harvard Medical School; the Neurobiology Imaging Facility in the Neurobiology Department of Harvard Medical School for consultation and instrument availability that supported this work (this facility is supported in part by the Neural Imaging Center as part of an NINDS P30 Core Center grant #NS072030). We greatly thank Dr. Kun-Liang Guan, Dr. Marc Kirschner, and Dr. Fanny Mann for providing the pGEX-KG-PAK-RBD, pcDNA3-Rac1-Q61L, and pAPtag-5-Sema3E constructs and Dr. Yutaka Yoshida for providing the PlexinD1 antibody.
This work was supported by the Alice and Joseph Brooks Fund Postdoctoral Fellowship (A. Tata), the Goldenson postdoctoral fellowship (A. Ben-Zvi), and the following grants to C. Gu: Sloan research fellowship, Armenise junior faculty award, the Genise Goldenson fund, and National Institutes of Health grant R01NS064583.
The authors declare no competing financial interests.
Footnotes
Abbreviations used in this paper:
- AP
- alkaline phosphatase
- au
- arbitrary unit
- E
- embryonic day
- EC
- endothelial cell
- GAP
- GTPase-activating protein
- HUVEC
- human umbilical vein EC
- Rac1
- Ras-related C3 botulinum toxin substrate 1
- Sema3E
- Semaphorin 3E
- SH3BP1
- SH3-domain binding protein 1
References
- Beg A.A., Sommer J.E., Martin J.H., Scheiffele P. 2007. α2-Chimaerin Is an Essential EphA4 Effector in the Assembly of Neuronal Locomotor Circuits. Neuron. 55:768–778 10.1016/j.neuron.2007.07.036 [DOI] [PubMed] [Google Scholar]
- Bielenberg D.R., Shimizu A., Klagsbrun M. 2008. Semaphorin-induced cytoskeletal collapse and repulsion of endothelial cells. Methods Enzymol. 443:299–314 10.1016/S0076-6879(08)02015-6 [DOI] [PubMed] [Google Scholar]
- Casazza A., Finisguerra V., Capparuccia L., Camperi A., Swiercz J.M., Rizzolio S., Rolny C., Christensen C., Bertotti A., Sarotto I., et al. 2010. Sema3E-Plexin D1 signaling drives human cancer cell invasiveness and metastatic spreading in mice. J. Clin. Invest. 120:2684–2698 (published erratum appears in J. Clin. Invest. 2011. 121:2945) 10.1172/JCI42118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chauvet S., Cohen S., Yoshida Y., Fekrane L., Livet J., Gayet O., Segu L., Buhot M.-C., Jessell T.M., Henderson C.E., Mann F. 2007. Gating of Sema3E/PlexinD1 signaling by neuropilin-1 switches axonal repulsion to attraction during brain development. Neuron. 56:807–822 10.1016/j.neuron.2007.10.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi Y.I., Duke-Cohan J.S., Ahmed W.B., Handley M.A., Mann F., Epstein J.A., Clayton L.K., Reinherz E.L. 2008. PlexinD1 glycoprotein controls migration of positively selected thymocytes into the medulla. Immunity. 29:888–898 10.1016/j.immuni.2008.10.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cicchetti P., Baltimore D. 1995. Identification of 3BP-1 in cDNA expression library by SH3 domain screening. Methods Enzymol. 256:140–148 10.1016/0076-6879(95)56019-X [DOI] [PubMed] [Google Scholar]
- Cicchetti P., Mayer B.J., Thiel G., Baltimore D. 1992. Identification of a protein that binds to the SH3 region of Abl and is similar to Bcr and GAP-rho. Science. 257:803–806 10.1126/science.1379745 [DOI] [PubMed] [Google Scholar]
- Cicchetti P., Ridley A.J., Zheng Y., Cerione R.A., Baltimore D. 1995. 3BP-1, an SH3 domain binding protein, has GAP activity for Rac and inhibits growth factor-induced membrane ruffling in fibroblasts. EMBO J. 14:3127–3135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson J.C., Legg J.A., Machesky L.M. 2006. Bar domain proteins: a role in tubulation, scission and actin assembly in clathrin-mediated endocytosis. Trends Cell Biol. 16:493–498 10.1016/j.tcb.2006.08.004 [DOI] [PubMed] [Google Scholar]
- Deng Y., Larrivée B., Zhuang Z.W., Atri D., Moraes F., Prahst C., Eichmann A., Simons M. 2013. Endothelial RAF1/ERK activation regulates arterial morphogenesis. Blood. 121:3988–3996: S1–S9 10.1182/blood-2012-12-474601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding J.B., Oh W.-J., Sabatini B.L., Gu C. 2012. Semaphorin 3E-Plexin-D1 signaling controls pathway-specific synapse formation in the striatum. Nat. Neurosci. 15:215–223 10.1038/nn.3003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbediwy A., Zihni C., Terry S.J., Clark P., Matter K., Balda M.S. 2012. Epithelial junction formation requires confinement of Cdc42 activity by a novel SH3BP1 complex. J. Cell Biol. 198:677–693 10.1083/jcb.201202094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukushima Y., Okada M., Kataoka H., Hirashima M., Yoshida Y., Mann F., Gomi F., Nishida K., Nishikawa S.-I., Uemura A. 2011. Sema3E-PlexinD1 signaling selectively suppresses disoriented angiogenesis in ischemic retinopathy in mice. J. Clin. Invest. 121:1974–1985 10.1172/JCI44900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gay C.M., Zygmunt T., Torres-Vázquez J. 2011. Diverse functions for the semaphorin receptor PlexinD1 in development and disease. Dev. Biol. 349:1–19 10.1016/j.ydbio.2010.09.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelfand M.V., Hong S., Gu C. 2009. Guidance from above: common cues direct distinct signaling outcomes in vascular and neural patterning. Trends Cell Biol. 19:99–110 10.1016/j.tcb.2009.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gitler A.D., Lu M.M., Epstein J.A. 2004. PlexinD1 and semaphorin signaling are required in endothelial cells for cardiovascular development. Dev. Cell. 7:107–116 10.1016/j.devcel.2004.06.002 [DOI] [PubMed] [Google Scholar]
- Gu C., Yoshida Y., Livet J., Reimert D.V., Mann F., Merte J., Henderson C.E., Jessell T.M., Kolodkin A.L., Ginty D.D. 2005. Semaphorin 3E and plexin-D1 control vascular pattern independently of neuropilins. Science. 307:265–268 10.1126/science.1105416 [DOI] [PubMed] [Google Scholar]
- Guttmann-Raviv N., Shraga-Heled N., Varshavsky A., Guimaraes-Sternberg C., Kessler O., Neufeld G. 2007. Semaphorin-3A and semaphorin-3F work together to repel endothelial cells and to inhibit their survival by induction of apoptosis. J. Biol. Chem. 282:26294–26305 10.1074/jbc.M609711200 [DOI] [PubMed] [Google Scholar]
- Hall A. 1998. Rho GTPases and the actin cytoskeleton. Science. 279:509–514 10.1126/science.279.5350.509 [DOI] [PubMed] [Google Scholar]
- Heath R.J.W., Insall R.H. 2008. F-BAR domains: multifunctional regulators of membrane curvature. J. Cell Sci. 121:1951–1954 10.1242/jcs.023895 [DOI] [PubMed] [Google Scholar]
- Hung R.-J., Yazdani U., Yoon J., Wu H., Yang T., Gupta N., Huang Z., van Berkel W.J.H., Terman J.R. 2010. Mical links semaphorins to F-actin disassembly. Nature. 463:823–827 10.1038/nature08724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito Y., Oinuma I., Katoh H., Kaibuchi K., Negishi M. 2006. Sema4D/plexin-B1 activates GSK-3β through R-Ras GAP activity, inducing growth cone collapse. EMBO Rep. 7:704–709 10.1038/sj.embor.7400737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasato T., Katoh H., Nishimaru H., Ishikawa Y., Inoue H., Saito Y.M., Ando R., Iwama M., Takahashi R., Negishi M., Itohara S. 2007. Rac-GAP α-Chimerin Regulates Motor-Circuit Formation as a Key Mediator of EphrinB3/EphA4 Forward Signaling. Cell. 130:742–753 10.1016/j.cell.2007.07.022 [DOI] [PubMed] [Google Scholar]
- Kigel B., Varshavsky A., Kessler O., Neufeld G. 2008. Successful inhibition of tumor development by specific class-3 semaphorins is associated with expression of appropriate semaphorin receptors by tumor cells. PLoS ONE. 3:e3287 10.1371/journal.pone.0003287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Oh W.-J., Gaiano N., Yoshida Y., Gu C. 2011. Semaphorin 3E-Plexin-D1 signaling regulates VEGF function in developmental angiogenesis via a feedback mechanism. Genes Dev. 25:1399–1411 10.1101/gad.2042011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruger R.P., Aurandt J., Guan K.-L. 2005. Semaphorins command cells to move. Nat. Rev. Mol. Cell Biol. 6:789–800 10.1038/nrm1740 [DOI] [PubMed] [Google Scholar]
- Negishi M., Oinuma I., Katoh H. 2005. Plexins: axon guidance and signal transduction. Cell. Mol. Life Sci. 62:1363–1371 10.1007/s00018-005-5018-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neufeld G., Kessler O. 2008. The semaphorins: versatile regulators of tumour progression and tumour angiogenesis. Nat. Rev. Cancer. 8:632–645 10.1038/nrc2404 [DOI] [PubMed] [Google Scholar]
- Oh W.-J., Gu C. 2013a. The role and mechanism-of-action of Sema3E and Plexin-D1 in vascular and neural development. Semin. Cell Dev. Biol. 24:156–162 10.1016/j.semcdb.2012.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh W.-J., Gu C. 2013b. Establishment of neurovascular congruency in the mouse whisker system by an independent patterning mechanism. Neuron. 80:458–469 10.1016/j.neuron.2013.09.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oinuma I., Ishikawa Y., Katoh H., Negishi M. 2004a. The Semaphorin 4D receptor Plexin-B1 is a GTPase activating protein for R-Ras. Science. 305:862–865 10.1126/science.1097545 [DOI] [PubMed] [Google Scholar]
- Oinuma I., Katoh H., Negishi M. 2004b. Molecular dissection of the semaphorin 4D receptor plexin-B1-stimulated R-Ras GTPase-activating protein activity and neurite remodeling in hippocampal neurons. J. Neurosci. 24:11473–11480 10.1523/JNEUROSCI.3257-04.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oinuma I., Katoh H., Negishi M. 2006. Semaphorin 4D/Plexin-B1-mediated R-Ras GAP activity inhibits cell migration by regulating β1 integrin activity. J. Cell Biol. 173:601–613 10.1083/jcb.200508204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parrini M.C., Sadou-Dubourgnoux A., Aoki K., Kunida K., Biondini M., Hatzoglou A., Poullet P., Formstecher E., Yeaman C., Matsuda M., et al. 2011. SH3BP1, an exocyst-associated RhoGAP, inactivates Rac1 at the front to drive cell motility. Mol. Cell. 42:650–661 10.1016/j.molcel.2011.03.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riccomagno M.M., Hurtado A., Wang H., Macopson J.G.J., Griner E.M., Betz A., Brose N., Kazanietz M.G., Kolodkin A.L. 2012. The RacGAP β2-Chimaerin selectively mediates axonal pruning in the hippocampus. Cell. 149:1594–1606 10.1016/j.cell.2012.05.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridley A.J. 2001. Rho GTPases and cell migration. J. Cell Sci. 114:2713–2722 [DOI] [PubMed] [Google Scholar]
- Rohm B., Rahim B., Kleiber B., Hovatta I., Püschel A.W. 2000. The semaphorin 3A receptor may directly regulate the activity of small GTPases. FEBS Lett. 486:68–72 10.1016/S0014-5793(00)02240-7 [DOI] [PubMed] [Google Scholar]
- Roodink I., Raats J., van der Zwaag B., Verrijp K., Kusters B., van Bokhoven H., Linkels M., de Waal R.M.W., Leenders W.P.J. 2005. Plexin D1 expression is induced on tumor vasculature and tumor cells: a novel target for diagnosis and therapy? Cancer Res. 65:8317–8323 10.1158/0008-5472.CAN-04-4366 [DOI] [PubMed] [Google Scholar]
- Roodink I., Kats G., van Kempen L., Grunberg M., Maass C., Verrijp K., Raats J., Leenders W. 2008. Semaphorin 3E expression correlates inversely with Plexin D1 during tumor progression. Am. J. Pathol. 173:1873–1881 10.2353/ajpath.2008.080136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito Y., Oinuma I., Fujimoto S., Negishi M. 2009. Plexin-B1 is a GTPase activating protein for M-Ras, remodelling dendrite morphology. EMBO Rep. 10:614–621 10.1038/embor.2009.63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakurai A., Gavard J., Annas-Linhares Y., Basile J.R., Amornphimoltham P., Palmby T.R., Yagi H., Zhang F., Randazzo P.A., Li X., et al. 2010. Semaphorin 3E initiates antiangiogenic signaling through plexin D1 by regulating Arf6 and R-Ras. Mol. Cell. Biol. 30:3086–3098 10.1128/MCB.01652-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swiercz J.M., Kuner R., Behrens J., Offermanns S. 2002. Plexin-B1 directly interacts with PDZ-RhoGEF/LARG to regulate RhoA and growth cone morphology. Neuron. 35:51–63 10.1016/S0896-6273(02)00750-X [DOI] [PubMed] [Google Scholar]
- Swiercz J.M., Worzfeld T., Offermanns S. 2009. Semaphorin 4D Signaling Requires the Recruitment of Phospholipase Cγ into the Plexin-B1 Receptor Complex. Mol. Cell. Biol. 29:6321–6334 10.1128/MCB.00103-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres-Vázquez J., Gitler A.D., Fraser S.D., Berk J.D., Pham Van N., Fishman M.C., Childs S., Epstein J.A., Weinstein B.M. 2004. Semaphorin-plexin signaling guides patterning of the developing vasculature. Dev. Cell. 7:117–123 10.1016/j.devcel.2004.06.008 [DOI] [PubMed] [Google Scholar]
- Toyofuku T., Yoshida J., Sugimoto T., Zhang H., Kumanogoh A., Hori M., Kikutani H. 2005. FARP2 triggers signals for Sema3A-mediated axonal repulsion. Nat. Neurosci. 8:1712–1719 10.1038/nn1596 [DOI] [PubMed] [Google Scholar]
- Tran T.S., Kolodkin A.L., Bharadwaj R. 2007. Semaphorin regulation of cellular morphology. Annu. Rev. Cell Dev. Biol. 23:263–292 10.1146/annurev.cellbio.22.010605.093554 [DOI] [PubMed] [Google Scholar]
- Uesugi K., Oinuma I., Katoh H., Negishi M. 2009. Different requirement for Rnd GTPases of R-Ras GAP activity of Plexin-C1 and Plexin-D1. J. Biol. Chem. 284:6743–6751 10.1074/jbc.M805213200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Der Zwaag B., Hellemons A.J.C.G.M., Leenders W.P.J., Burbach J.P.H., Brunner H.G., Padberg G.W., Van Bokhoven H. 2002. PLEXIN-D1, a novel plexin family member, is expressed in vascular endothelium and the central nervous system during mouse embryogenesis. Dev. Dyn. 225:336–343 10.1002/dvdy.10159 [DOI] [PubMed] [Google Scholar]
- Wang Y., He H., Srivastava N., Vikarunnessa S., Chen Y.B., Jiang J., Cowan C.W., Zhang X. 2012. Plexins are GTPase-activating proteins for Rap and are activated by induced dimerization. Sci. Signal. 5:ra6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegmeyer H., Egea J., Rabe N., Gezelius H., Filosa A., Enjin A., Varoqueaux F., Deininger K., Schnütgen F., Brose N., et al. 2007. EphA4-dependent axon guidance is mediated by the RacGAP alpha2-chimaerin. Neuron. 55:756–767 10.1016/j.neuron.2007.07.038 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.