DNA replication is asymmetrically regulated in the two-cell stage C. elegans embryo by the PAR-4 and PAR-1 polarity proteins, which function independently of known regulators of cell cycle timing to dampen DNA replication dynamics specifically in the posterior blastomere.
Abstract
Regulation of cell cycle duration is critical during development, yet the underlying molecular mechanisms are still poorly understood. The two-cell stage Caenorhabditis elegans embryo divides asynchronously and thus provides a powerful context in which to study regulation of cell cycle timing during development. Using genetic analysis and high-resolution imaging, we found that deoxyribonucleic acid (DNA) replication is asymmetrically regulated in the two-cell stage embryo and that the PAR-4 and PAR-1 polarity proteins dampen DNA replication dynamics specifically in the posterior blastomere, independently of regulators previously implicated in the control of cell cycle timing. Our results demonstrate that accurate control of DNA replication is crucial during C. elegans early embryonic development and further provide a novel mechanism by which PAR proteins control cell cycle progression during asynchronous cell division.
Introduction
Asymmetric division of the Caenorhabditis elegans embryo is controlled by PAR polarity proteins and results in the early partitioning of determinants that promote differential cell fates and asynchronous division of the two-cell stage blastomeres (Fig. 1 A; Kemphues and Strome, 1997; Noatynska et al., 2013). Although the mechanism regulating cell cycle asynchrony remains unclear, it was previously shown to rely on two molecular pathways downstream of PAR proteins. One pathway promotes a delay in S-phase progression in the posterior cell (the P1 blastomere) by the preferential activation of DNA replication checkpoint regulators, an event proposed to derive from a difference in blastomere size (Brauchle et al., 2003). The other pathway relies on faster mitotic entry in the anterior cell (the AB blastomere) via the preferential anterior enrichment of the mitotic regulators PLK-1 and CDC-25.1 (Budirahardja and Gönczy, 2008; Rivers et al., 2008). Embryos produced by homozygous mutants for par genes (hereafter referred to as par mutant embryos) largely fail to asymmetrically localize cell fate determinants at first mitosis and, accordingly, generate two blastomeres that aberrantly divide synchronously (Kemphues and Strome, 1997). However, in two-cell embryos mutant for the genes par-4 or par-1, the blastomeres divide synchronously despite having unequal sizes, and the anterior blastomere has wild-type cell cycle duration despite defects in PLK-1 and CDC-25.1 segregation (Morton et al., 1992; Budirahardja and Gönczy, 2008; Rivers et al., 2008). These results suggest that PAR-4 and PAR-1 control additional mechanisms to regulate asynchronous division of the early C. elegans embryo.
Figure 1.
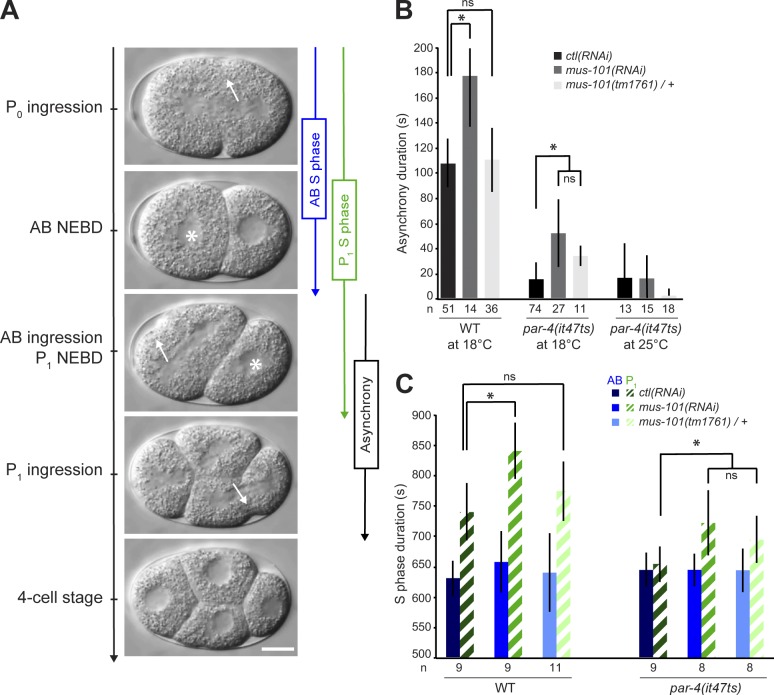
Depletion of MUS-101 lengthens the duration of asynchronous division and of P1 blastomere S phase in par-4(it47ts) mutants. (A) Time-lapse DIC images of a developing wild-type embryo. Asterisks denote nuclear envelope breakdown (NEBD), and arrows indicate sites of cortical ingression. Anterior is to the left. Bar, 10 µm. (B and C) Graphs reporting (in seconds) the measurement of asynchrony duration (B) and S-phase duration of both blastomeres (C) in the specified conditions. Empty vector was used for control (ctl) RNAi. Error bars represent standard deviations, and an asterisk denotes that the value is significantly different from the ctl(RNAi) in the same genetic background (P < 0.05, Student’s t test). The number of embryos (n) analyzed for each condition is indicated. WT, wild type.
Results and discussion
The function of PAR-4 in cell cycle regulation relies on MUS-101–dependent activity
par-4 encodes the C. elegans orthologue of the human tumor suppressor kinase LKB1 (also known as STK11) and was proposed to phosphorylate and activate PAR-1, a serine/threonine kinase of the MARK (MAP/microtubule affinity-regulating kinase) family (Guo and Kemphues, 1995; Watts et al., 2000; Lizcano et al., 2004; Griffin et al., 2011). We sought to identify regulators of asynchronous cell division by studying genetic interactors of par-4. We uncovered the gene mus-101 in an RNAi-based screen for suppressors of embryonic lethality resulting from partial loss of par-4 function (unpublished data). At the semipermissive temperature of 18°C, par-4(it47ts) mutant animals produce 13% of hatching embryos as compared with 100% for wild-type controls (Table 1). Depleting MUS-101 by RNAi in par-4(it47ts) animals resulted in 52% hatching progeny, a fourfold increase from par-4 mutant animals treated with control RNAi. Likewise, at 20°C, the viability of par-4(it47ts) embryos was enhanced by 20-fold after depletion of MUS-101 by RNAi (Table 1). Decreasing mus-101 gene dosage by half (using animals heterozygote for the presumptive null mus-101(tm1761) allele) in otherwise par-4(it47ts) mutant animals also significantly increased embryonic viability at 18°C (Table 1). The depletion of MUS-101 by RNAi in par-4(it47ts); mus-101(tm1761)/+ double mutant animals further increased viability in the progeny (Table 1). However, depleting MUS-101 at the fully restrictive temperature of 25°C could not restore viability in par-4 mutants, indicating that residual PAR-4 activity is required to allow suppression at lower temperatures. Suppression of embryonic lethality by depletion of MUS-101 was also observed in similar assays using a different mutant allele of par-4 (it57ts; unpublished data). These results indicate that mus-101 is a specific suppressor of par-4 embryonic lethality and that the activity of these two genes may be in a balanced regulation.
Table 1.
Mutations in mus-101 suppress par-4 lethality
| Genotype | 18°C | 20°C | 25°C |
| % | % | % | |
| Wild type; ctl(RNAi)1 | 100 ± 0 (19) | 99.9 ± 0 (4) | 98.6 ± 0.4 (4) |
| Wild type; mus-101(RNAi) | 99.9 ± 0 (19) | 99.9 ± 0 (4) | 82.4 ± 2.1 (3) |
| lin-11(+) mus-101(tm1761)/lin-11(n389) mus-101(+); ctl(RNAi)2 | 100 ± 0 (4) | ND | ND |
| lin-11(+) mus-101(tm1761)/lin-11(n389) mus-101(+); mus-101(RNAi) | 100 ± 0 (4) | ND | ND |
| par-4(it47ts); ctl(RNAi) | 13.0 ± 0.8 (19) | 1.0 ± 0.6 (3) | 0 ± 0 (3) |
| par-4(it47ts); mus-101(RNAi) | 51.9 ± 0.8 (19)3 | 19.9 ± 0.8 (3)3 | 0 ± 0 (3) |
| par-4(it47ts); lin-11(+) mus-101(tm1761)/lin-11(n389) mus-101(+); ctl(RNAi) | 33.3 ± 0.9 (3)3 | ND | ND |
| par-4(it47ts); lin-11(+) mus-101(tm1761)/lin-11(n389) mus-101(+); mus-101(RNAi) | 74.5 ± 4.5 (3)3 | ND | 0 ± 0 (3) |
The values correspond to the mean percentage of hatching embryos over the total number of embryos ± SEM. The numbers in parentheses correspond to the number of triplicate assays performed. N.D., not determined.
Empty vector was used for RNAi control.
The gene lin-11 is positioned very close to the mus-101 gene on chromosome I, and heterozygote animals give rise to sterile adults (mus-101(tm1761) homozygotes), vulvaless adults (lin-11(n389) homozygotes), and viable, fertile heterozygotes.
The value is significantly different from the par-4 control at the same temperature (P < 0.05, Student’s t test).
To understand how PAR-4 and MUS-101 function together, we assessed which par-4(it47ts)–dependent defects were restored by depletion of MUS-101. The embryonic defects associated with mutations in par-4 include the mislocalization of cell fate determinants (Kemphues et al., 1988), a decrease in actomyosin dynamics during polarization and cytokinesis (Chartier et al., 2011), and a loss of cell cycle asynchrony at the two-cell stage (Morton et al., 1992). We found that depleting MUS-101 at semipermissive temperature largely failed to restore proper localization of cell fate determinants (Fig. S1 A) or actomyosin dynamics (Fig. S1, B and C) in par-4(it47ts) embryos, indicating that MUS-101 mainly acts independently of these processes. However, depletion of MUS-101 resulted in a significant increase in cell cycle asynchrony in two-cell stage par-4(it47ts) embryos when compared with control, even in conditions in which depletion of MUS-101 had no effect on cell cycle progression in wild-type embryos (such as par-4(it47ts); mus-101(tm1761)/+ double mutant animals; Fig. 1 B). This suggests that PAR-4 and MUS-101 act together to regulate cell cycle duration.
Previous studies have demonstrated that asynchrony of division at the two-cell stage is caused by a longer S-phase duration of the posterior blastomere P1 relative to that of the anterior AB blastomere (Edgar and McGhee, 1988; Brauchle et al., 2003). In par-4 mutants, S phase is accelerated in the P1-like blastomere, thus resulting in an aberrant situation in which the two blastomeres divide at the same time (Fig. 1, B and C; Rivers et al., 2008). Reduced MUS-101 levels using either heterozygous animals or mus-101(RNAi) resulted in a specific increase in S-phase duration of the posterior blastomere of par-4 mutant embryos and had no apparent effect on the anterior blastomere (Fig. 1 C). These results indicate that MUS-101 functions with PAR-4 to regulate S-phase progression in the posterior blastomere during asynchronous division of the two-cell stage embryo. The lack of observed defect in the anterior blastomere further suggests that these two proteins regulate cell cycle progression specifically in P1.
MUS-101 suppresses par-4 cell cycle timing defects through its function in DNA replication
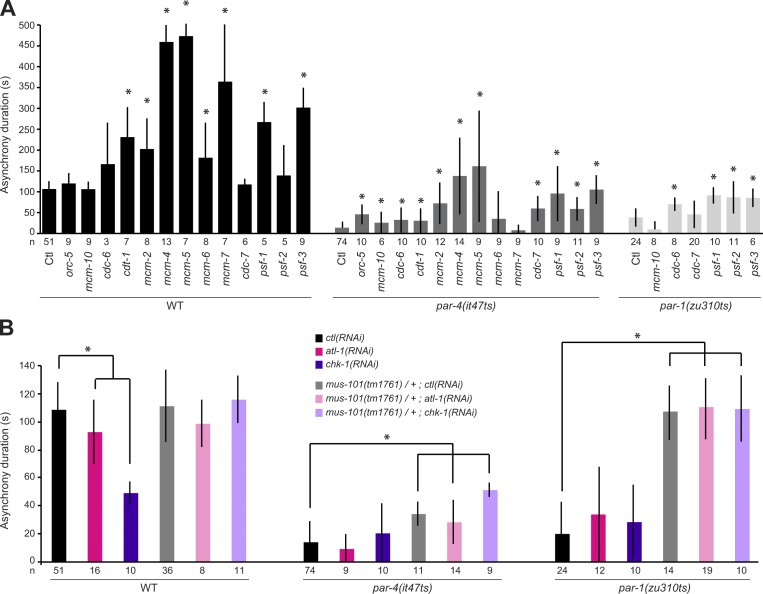
MUS-101 is the C. elegans orthologue of human TopBP1 and budding yeast Dpb11p (Holway et al., 2005). Previous studies have revealed that these orthologous proteins have two main cellular functions: they promote the initiation of DNA replication and regulate the activity of the DNA replication checkpoint (Saka and Yanagida, 1993; Araki et al., 1995; Mäkiniemi et al., 2001). To assess whether MUS-101 functions with PAR-4 through its role in DNA replication, we asked whether RNAi depletion of other regulators of DNA replication initiation restores cell cycle asynchrony in par-4 mutant embryos. In C. elegans, the initiation of DNA replication largely occurs as previously described in other organisms, and most regulators have a clear orthologue that is incorporated into the replisome following a well-defined, hierarchical order (Table S1; Sonneville et al., 2012; Gaggioli et al., 2014). Of the 13 other regulators of DNA replication initiation that were tested, RNAi depletion of 11 of them could significantly increase cell cycle asynchrony between AB-like and P1-like blastomeres in par-4 mutant embryos (Fig. 2 A). Notably, depleting some of these regulators in wild-type embryos resulted in a lengthening of cell cycle asynchrony (Fig. 2 A), suggesting, as previously reported (Brauchle et al., 2003), that the activity of the DNA replication checkpoint pathway is increased in these conditions. However, a lengthening of cell cycle asynchrony in wild-type embryos was not necessarily coupled to suppression of par-4 cell cycle timing defects. For instance, whereas MCM-7 depletion increased cell cycle asynchrony between AB and P1 blastomeres in wild-type embryos, it had no effect in par-4(it47ts) embryos (Fig. 2 A). Conversely, CDC-7 depletion had no effect in the wild type but significantly increased cell cycle asynchrony in par-4(it47ts) embryos. This suggests that hyperactivation of the DNA replication checkpoint is not the direct cause of the par-4 suppression phenotype. Accordingly, depletion of the ATL-1 or CHK-1 checkpoint regulators failed to suppress the cell cycle timing defect of par-4 mutant embryos, and depleting these regulators in par-4(it47ts); mus-101(tm1761)/+ double mutant animals did not preclude the suppression of cell cycle asynchrony defects at the embryonic two-cell stage (Fig. 2 B). Collectively, these results indicate that depleting regulators of DNA replication can suppress the cell cycle timing defects of par-4 mutant embryos and support the notion that MUS-101 modulates PAR-4 function through its role in DNA replication, independently of the DNA replication checkpoint.
Figure 2.
Depleting regulators of DNA replication initiation suppresses the asynchrony duration defect of par-4 and par-1 embryos, independently of checkpoint activity. (A and B) Graphs reporting the measurement of asynchrony duration (in seconds) after depletion of regulators of DNA replication initiation (A) or checkpoint regulators (B) in the specified genetic backgrounds. Measurements were made at 18°C for wild-type and par-4(it47ts) embryos and 22°C for par-1(zu310ts) embryos. Empty vector was used for control (Ctl) RNAi. Error bars represent standard deviations, and an asterisk denotes that the value is significantly different from the ctl(RNAi) in the same genetic background (P < 0.05, Student’s t test). The number of embryos analyzed for each condition is indicated. WT, wild type.
PAR-4 asymmetrically regulates DNA replication
The functional relationship that we uncovered between PAR-4 and MUS-101 suggests that PAR-4 may control cell cycle asynchrony at the two-cell stage by differentially regulating DNA replication in the two blastomeres. To test this hypothesis, we developed an imaging-based assay to measure the dynamics of DNA replication in the C. elegans two-cell stage embryo. The assay relies on monitoring transgenically expressed PCN-1, the C. elegans orthologue of PCNA (proliferating cell nuclear antigen; Brauchle et al., 2003), fused to GFP (see Materials and methods; Fig. 3 A). PCNA functions as a processivity factor for DNA replication and is one of the last components incorporated into the replisome upon replication origin firing (Waga and Stillman, 1998). Importantly, its accumulation in nuclear foci in other organisms was previously demonstrated to report on the active sites of DNA replication (Essers et al., 2005; Meister et al., 2007).
Figure 3.
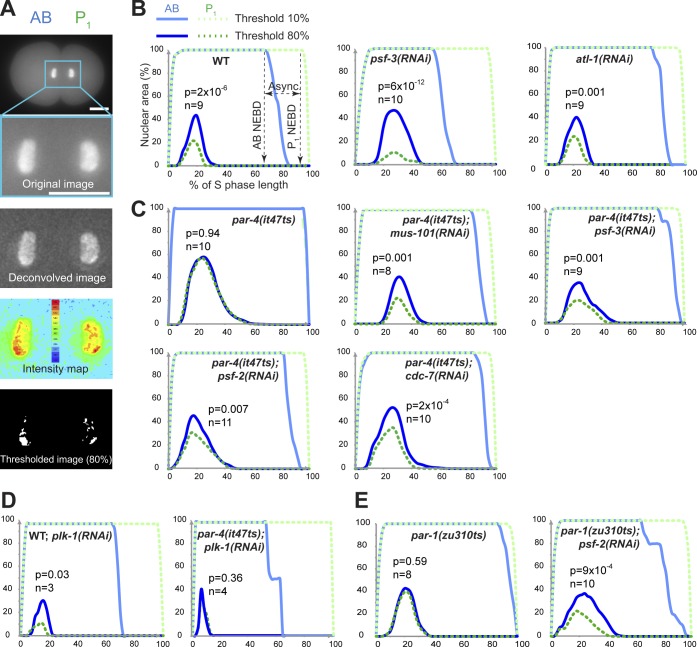
PAR-4 and PAR-1 asymmetrically regulate the initiation of DNA replication at the two-cell stage. (A) Fluorescence images of a C. elegans embryo transgenically expressing GFP::PCN-1 before (top image) and after a series of transformations that include deconvolution (second from top image), intensity measurements (second from bottom image), and threshold application (bottom image). GFP::PCN-1 nuclear distribution was measured from images after thresholds had been applied. Bars, 10 µm. (B–E) Mean percentages of AB or P1 nuclear regions occupied by GFP::PCN-1 foci after applying 10 or 80% fluorescence threshold over time during two-cell stage embryonic development. For each specified condition, analyses were made using wild-type embryos at 25°C (B), par-4(it47ts) embryos at 18°C (C and D), or par-1(zu310ts) embryos at 22°C (E). In all conditions, time is expressed in percentage of S-phase length to allow a better comparison between different embryos. For each graph, the p-value (Student’s t test) highlights the statistical difference in fluorescence distribution at the point of maximal enrichment between AB and P1 at 80% fluorescence threshold, and n corresponds to the number of embryos analyzed. WT, wild type.
Images of two-cell stage embryos expressing GFP::PCN-1 were acquired, and nuclear sites enriched in GFP::PCN-1 were revealed by applying various fluorescence thresholds on deconvolved images (Figs. 3 A and S2 A). With this approach, low thresholds revealed the total nuclear pool of GFP::PCN-1, whereas high thresholds highlighted the nuclear regions enriched in GFP::PCN-1, in which the number of replication forks is proportionally higher (Figs. 3 B and S2 A). The relative engagement in DNA replication between the two blastomeres at each time point was determined by measuring the percentage of nuclear area comprising GFP::PCN-1 foci in each cell, using a previously described algorithm (Maddox et al., 2006).
In control embryos, applying low fluorescence thresholds (0–20%) revealed a comparable nuclear area enriched in GFP::PCN-1 between AB and P1 blastomeres at any time point, and a decrease in fluorescence intensities was observed at nuclear envelope breakdown (Fig. 3 B). This fraction likely corresponds to both DNA-associated and nucleoplasmic pools of GFP::PCN-1 as previously reported (Brauchle et al., 2003). Applying high fluorescence thresholds (80–90%) allowed us to monitor the nuclear regions actively engaged in DNA replication and revealed significant differences between the nuclear area occupied by GFP::PCN-1 in AB and P1 blastomeres (Fig. 3 B). Specifically, GFP::PCN-1 enrichment initiated early in S phase, and maximal enrichment was reached at around the same time in AB and P1, indicating that the initial peak of DNA replication occurs concomitantly in both blastomeres. However, the AB blastomere had a significantly larger nuclear area of high GFP::PCN-1 fluorescence intensity compared with the P1 blastomere at any time point (Fig. 3 B). The ratio of high GFP::PCN-1 fluorescence intensity over distribution was equal between the two blastomeres (74 ± 10 for AB and 70 ± 9 for P1, n = 7; P = 0.51, Student’s t test), indicating that the asymmetry in fluorescence distribution does not result from a change in DNA compaction. This finding is consistent with a higher number of replication forks being active during early S phase in the AB blastomere compared with the posterior one. A comparable asymmetric distribution of fluorescence intensities was observed after performing this assay on embryos transgenically expressing GFP::DIV-1, the orthologue of the B subunit of the DNA polymerase α-primase complex (Fig. S2 B; Encalada et al., 2000; Sonneville et al., 2012).
To ascertain that this assay faithfully monitors DNA replication, we reasoned that partially depleting regulators of DNA replication should prolong the asynchrony between the two blastomeres and significantly affect the distribution of GFP::PCN-1 at high thresholds, whereas depleting replication checkpoint regulators should shorten the duration of S phase in the P1 blastomere without significantly perturbing GFP::PCN-1 distribution in early S phase. This is what we observed. Depletion of the DNA replication factor PSF-3 lengthened the asynchrony between AB and P1 blastomeres (Fig. 2 A) and significantly increased the difference in the percentage of nuclear area occupied by GFP::PCN-1 between AB and P1 at the point of maximal enrichment as compared with the control (P = 0.004, Student’s t test; Fig. 3 B). This is consistent with a preferential activation of checkpoint signaling and a defect in early DNA replication origin firing in the P1 blastomere compared with AB. Depleting the checkpoint regulator ATL-1 by RNAi caused a shortening of the time between AB and P1 nuclear envelope breakdown but no significant difference on the distribution of GFP::PCN-1 foci at high fluorescence thresholds compared with control embryos (P = 0.63, Student’s t test; Fig. 3 B). This is consistent with checkpoint signaling being largely inactivated in P1 and having no effect on the firing of early DNA replication origins in early S phase. This indicates that the assay is robust and accurately reports on the dynamics of DNA replication in vivo, independently of DNA replication checkpoint activity.
We next used this imaging assay to test whether PAR-4 plays a role in the regulation of DNA replication dynamics at the two-cell stage. In par-4(it47ts) mutant animals grown at semipermissive temperature, we observed that both blastomeres have identical nuclear distributions of GFP::PCN-1 when applying a high fluorescence intensity threshold (Fig. 3 C). This indicates that DNA replication is abnormally symmetric between the two blastomeres when PAR-4 activity is compromised. As S-phase duration is shortened in the posterior blastomere of par-4 mutant embryos (Fig. 1 C; Rivers et al., 2008), this finding is compatible with the notion that PAR-4 negatively regulates DNA replication in the P1 blastomere of wild-type embryos. Depleting the DNA replication factors MUS-101 or PSF-3 restored an asymmetry in GFP::PCN-1 nuclear distribution in par-4(it47ts) embryos (Fig. 3 C). Importantly, an asymmetry in GFP::PCN-1 nuclear distribution was also restored by depleting either PSF-2 or CDC-7 (Fig. 3 C), two DNA replication factors whose depletion can suppress par-4 defects but does not otherwise affect cell cycle timing in wild-type embryos (Fig. 2 A). As expected, GFP::PCN-1 asymmetry was not restored after depleting the replication factor CDC-7 from par-4(it47ts) embryos grown at fully restrictive temperature (Fig. S2 C), again indicating that residual PAR-4 activity is needed to allow suppression at lower temperatures. Together, these results reveal that DNA replication is asymmetric at the two-cell stage, with more active replication forks present in the nucleus of the early AB blastomere compared with P1 and that this asymmetry is controlled by PAR-4.
Previous studies demonstrated that the mitotic regulator PLK-1 promotes asynchronous division of the two-cell embryo (Budirahardja and Gönczy, 2008; Rivers et al., 2008), raising the possibility that PAR-4 acts through PLK-1 to regulate DNA replication at this stage. Partial depletion of PLK-1 was previously reported to restore embryonic viability in par-4 mutants (Fievet et al., 2013). Consistently, we found that partial depletion of PLK-1 in par-4(it47ts) mutant animals restored cell cycle asynchrony at the two-cell stage (Fig. 3 D and not depicted). However, RNAi depletion of PLK-1 in wild-type or par-4(it47ts) animals had no significant effect on GFP::PCN-1 nuclear distribution between AB and P1 blastomeres when compared with control conditions (Fig. 3 D). These results indicate that PAR-4 regulates DNA replication asymmetry at the two-cell stage independently of PLK-1 activity.
PAR-1 regulates DNA replication
PAR-1, a conserved serine/threonine kinase that regulates cell polarity (Guo and Kemphues, 1995), was previously shown to be a downstream phosphorylation target of PAR-4/LKB1 in multiple species, including C. elegans (Lizcano et al., 2004; Narbonne et al., 2010). Accordingly, par-1 mutant embryos recapitulate many of the defects observed in par-4 mutants, including a loss of cell cycle asynchrony at the two-cell stage (Kemphues et al., 1988; Spilker et al., 2009). We therefore sought to determine whether PAR-1 also regulates DNA replication during asynchronous division of the two-cell stage C. elegans embryo. As is the case for par-4 mutants, we found that RNAi depletion of several regulators of DNA replication initiation could significantly increase cell cycle asynchrony between both blastomeres in par-1(zu310ts) mutant embryos grown at a semipermissive temperature (Fig. 2 A, light gray bars), independently of DNA replication checkpoint activity (Fig. 2 B). Furthermore, analysis of GFP::PCN-1 fluorescence distribution revealed that it is equal between the nuclei of both blastomeres in two-cell stage par-1(zu310ts) embryos, and depletion of PSF-2 by RNAi significantly restored asymmetric distribution of GFP::PCN-1 in these embryos (Fig. 3 E). These results indicate that, as is the case for PAR-4, PAR-1 regulates DNA replication dynamics during asynchronous division of the C. elegans two-cell stage embryo.
A model for the regulation of asynchronous cell division
Collectively, our results support a model in which activation of the PAR-1 kinase by PAR-4/LKB1 in the P1 blastomere of the two-cell stage embryo negatively regulates the initiation of DNA replication in early S phase. As both blastomeres have the same amount of DNA to replicate, this difference contributes to prolonged S-phase duration in P1 and asynchronous division of the two blastomeres, which was proposed to be important for proper embryonic patterning (Encalada et al., 2000; Brauchle et al., 2003). The precise targeted effector in the DNA replication initiation cascade is currently unknown. In other systems, initiation of replication was reported to occur via two distinct, consecutive steps: origin licensing and origin firing (Diffley, 2011). Performing our replication assay on wild-type embryos transgenically expressing the licensing factors ORC-2 and CDC-6 fused to GFP revealed that they are distributed symmetrically at all times (unpublished data), suggesting that PAR-4 and PAR-1 do not function at origin licensing but rather act during the step of origin firing. In yeast, the limiting amounts of many proteins controlling origin firing was previously proposed to be key in regulating early versus late origin activation and, thereby, the timing of DNA replication initiation (Mantiero et al., 2011; Tanaka et al., 2011). Although a model based on limiting amounts of critical factors was previously proposed to regulate asynchronous division of the C. elegans embryo (Brauchle et al., 2003), we consider such a mechanism unlikely for DNA replication initiation factors given that par-4 mutant blastomeres divide synchronously despite being asymmetric in size. Furthermore, we did not observe any change in cell cycle timing of the AB cell when comparing wild-type and par-4 mutant embryos (Fig. 1 C), and antibodies against PSF-3 and MUS-101, two regulators of DNA replication origin firing (Diffley, 2011), did not reveal any difference in levels between the two blastomeres in early S phase (Fig. S3). Our results therefore support a model in which PAR-4 and PAR-1 control the activity of one or more DNA replication factors specifically in the P1 cell. This provides a context in which the control of early versus late DNA replication origin firing may play a crucial biological role, that of allowing proper embryonic patterning through the regulation of cell cycle duration and asynchronous division.
Both PAR-1/MARK and PAR-4/LKB1 are conserved in other species, in which they also control cell polarity (Goldstein and Macara, 2007). Furthermore, LKB1 was shown to regulate cell growth by inhibiting the mammalian target of rapamycin pathway through direct phosphorylation and activation of AMP kinase (AMPK; Hawley et al., 2003; Woods et al., 2003). Although AMPK was shown to regulate multiple nutrient-dependent signaling events, recent studies in mammalian cells suggested that LKB1 also controls cell proliferation independently of mammalian target of rapamycin and AMPK (Gurumurthy et al., 2008, 2010; Gan et al., 2010; Nakada et al., 2010). Our finding that PAR-4/LKB1 regulates DNA replication in C. elegans further highlights the central role of this kinase in coordinating cell polarity, cell division, and cell growth.
Materials and methods
Strains and alleles
C. elegans animals were maintained according to standard procedures (Brenner, 1974) and were grown at 15°C unless otherwise stated. The wild-type strain was the N2 Bristol strain. The alleles used in this study are described in WormBase: LGI, mus-101(tm1761), lin-11(n389) (used to balance mus-101); LGIII, unc-119(ed3); LGV, par-4(it47ts and it57ts), par-1(zu310ts), zuIs45[nmy-2p::nmy-2::GFP + unc-119(+)]; and LG?, axls1462[pie-1p::GFP::pie-1 + unc-119(+)], bnIs1[pie-1p::GFP::pgl-1 + unc-119(+)], isIs17[pie-1p::GFP::pcn-1 + unc-119(+)], gtIs66[pie-1p::GFP(lap)::div-1 + unc-119(+)]. Strains were provided by the Japanese National Bioresource Project for C. elegans (Tokyo Women’s Medical University School of Medicine, Tokyo, Japan) and the Caenorhabditis Genetics Center (University of Minnesota, Minneapolis, MN). The mus-101(tm1761) allele consists in a 598-bp deletion located 794 bp away from the first ATG and that removes the fifth and sixth exons and a 5′ intron splice site. This likely results in a transcribed mRNA that is inappropriately spliced and encodes, at most, the first 194 residues of the 1,182–amino acid protein. Embryos homozygous for mus-101(tm1761) hatch and grow to become sterile adults. par-4(it47ts), par-4(it57ts), and par-1(zu310ts) are thermosensitive alleles carrying a single missense mutation in the region encoding the kinase domain and produce proteins with little or no catalytic activity at restrictive temperature (Watts et al., 2000; Spilker et al., 2009).
RNAi-mediated depletions
Protein depletion by RNAi was performed by feeding animals with individual clones from the Arhinger collection, as previously described (Kamath et al., 2001). In brief, each clone was grown up to log phase and plated overnight on nematode growth medium plates containing 50 µM carbenicillin and 1 mM IPTG. All assays were performed on embryos produced by animals grown for 48 h in the presence of a double-stranded RNA–expressing clone. Suppression of embryonic lethality was performed by counting hatching versus total progeny in at least three independent assays, as described previously (Chartier et al., 2011). Clones were systematically verified by sequencing, and empty vector (L4440) was used as a control. The efficiency of CHK-1 depletion by RNAi was confirmed by Western blotting and immunofluorescence analyses.
Time-lapse imaging of embryonic development
Time-lapse imaging of embryos expressing NMY-2::GFP and measurements of cortical velocity was performed as described previously (Chartier et al., 2011). Imaging of embryos was performed using a microscope (Swept Field Confocal; Nikon and Prairie Technologies) using the 45-µm pinhole setting and 1 × 1 binning on a camera (CoolSnap HQ2; Photometrics). A 100×/1.4 NA Plan Apochromat objective was used to acquire 16 confocal sections (separated by 0.5 µm) of the upper cortex exposed for 80 ms at 10-s intervals. All Swept Field Confocal acquisitions and additional components, including laser exposure setting, were controlled by Elements software (Nikon). ImageJ software (National Institutes of Health) was used to measure cortical velocity, by quantitating the mean anterior displacement of NMY-2::GFP cortical foci over time.
Differential interference contrast (DIC) imaging of developing embryos was performed as described previously (Hyenne et al., 2008). Images were acquired by a camera (AxioCam HRm; Carl Zeiss) mounted on a microscope (Axio Imager.Z1; Carl Zeiss), and the acquisition system was controlled by AxioVision software (Carl Zeiss). Plan Apochromat 63×/1.4 NA or 100×/1.4 NA objectives were used to acquire a single midsection of the embryo at 5-s intervals. ImageJ software was used to perform quantifications. Timing of nuclear envelope breakdown was defined by DIC as the first noticeable disappearance of nuclear membrane, and cytokinesis was defined by DIC as the first observable change in cortical organization after spindle assembly. As early blastomeres of C. elegans oscillate between S and M phases, the time separating cytokinesis from nuclear envelope breakdown corresponds to S phase (Edgar and McGhee, 1988; Brauchle et al., 2003).
Localization of GFP::PGL-1 was performed by imaging embryos with a microscope (DeltaVision; Applied Precision) using a 60× Plan Apochromat objective (Olympus). Images were acquired with a camera (CoolSnap HQ2), and the system was controlled by softWoRx software (Applied Precision). Localization of GFP::PIE-1 was performed by imaging embryos with the Axio Imager.Z1 microscope described in the previous paragraph. The distribution of each protein was assessed by measuring fluorescence intensities along the anteroposterior axis using ImageJ software.
Immunofluorescence
For immunofluorescence analysis, gravid hermaphrodites were cut open in 6 µl of M9 buffer with two 25-gauge needles on a 14 × 14–mm patterned slide (Cel-Line; Thermo Fisher Scientific) coated with 0.1% poly-l-lysine. A coverslip was placed on the sample, and the slide was placed for ≥5 min on a metal block cooled in dry ice. The coverslip was then removed, and the slide was placed immediately in fixative (−20°C methanol) for 20 min. The slide was rehydrated twice with PBS for 5 min and once with PBST (PBS with 0.1% Tween 20) for 5 min and then incubated in blocking buffer (PBST containing 10% goat serum) for 30 min at room temperature. Antibodies were then applied in blocking buffer and incubated overnight at 4°C (primary) or 1 h at room temperature (secondary), each followed by four washes of 5 min in PBST. The fixed specimens were mounted in 90% glycerol containing 1% N-propylgalate, and a coverslip was sealed. Embryos were visualized with a laser-scanning confocal microscope (LSM 510; Carl Zeiss) equipped with a 63×/1.4 NA Plan Apochromat objective. The mouse clone OIC1D4 anti–P granules antibody (1:500; Developmental Studies Hybridoma Bank, University of Iowa) was used. Rat polyclonal antisera recognizing amino acids 849–1,182 of the MUS-101 protein expressed in Escherichia coli as a GST fusion protein were produced by SDIX. The antisera were depleted from GST antibodies and affinity-purified using an immobilized MUS-101(849–1,182) protein fragment fused to a 6×His tag. Rabbit polyclonal antisera recognizing full-length PSF-3 expressed in E. coli as a GST fusion protein were produced by Eurogentec. Anti–PSF-3 antibodies were affinity purified using maltose binding protein–PSF-3 covalently linked to CNBr-activated Sepharose (Pharmacia), according to the manufacturer’s instructions (New England Biolabs). Anti–MUS-101 and anti–PSF-3 antibodies were diluted 1:500. Secondary antibodies were Alexa Fluor 546–coupled goat anti–mouse (1:500; Invitrogen), Cy3-coupled donkey anti–rat (1:500; Jackson ImmunoResearch Laboratories, Inc.), and Alexa Fluor 488–coupled goat anti–rabbit (1:500; Invitrogen).
Quantification of DNA replication dynamics
Embryos transgenically expressing regulators of DNA replication initiation were obtained by cutting open gravid hermaphrodites using two 25-gauge needles and mounted individually on a coverslip coated with 0.1% poly-l-lysine in 10 µl of egg buffer (Edgar, 1995). The coverslip was placed on a 3% agarose pad, and the edges were sealed with petroleum jelly. Time-lapse imaging of living embryos was performed with a camera (CoolSnap HQ2) at 1 × 1 binning mounted on a DeltaVision system and using a 60× Plan Apochromat objective. softWoRx software was used to acquire 49 sections of the nucleus, each separated by 0.25 µm, with 0.15-s exposures at 100% illumination. Acquisitions were made at 30-s intervals for the strain expressing gfp::pcn-1 and 1-min intervals for the strain expressing gfp::div-1. Images were deconvolved and stacked using softWoRx software. Using ImageJ, AB and P1 nuclei were cropped from each embryo and aligned to generate a single file containing images of all nuclei. The application of thresholds (0–95%) and measurements of fluorescence distribution (quantification of nuclear area occupied by fluorescence intensity over total nuclear area) were performed on this assembled file as described previously (Maddox et al., 2006), using a custom macro written for MetaMorph software (Molecular Devices). Results were analyzed using Excel (Microsoft). As S-phase length may slightly vary from one embryo to the next, time units were expressed as a percentage of the duration of nuclear GFP::PCN-1 accumulation in P1, and fluorescence intensity distributions were interpolated according to this time recalibration.
Online supplemental material
Fig. S1 shows that depletion of MUS-101 does not suppress the cell fate determinant localization and actomyosin contractility defects of par-4 mutants. Fig. S2 provides additional information on the PCN-1–dependent DNA replication assay. Fig. S3 shows that MUS-101 and PSF-3 are distributed symmetrically in two-cell stage C. elegans embryos. Table S1 lists DNA replication initiation homologues in C. elegans, Saccharomyces cerevisiae, Drosophila melanogaster, and Homo sapiens. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201312029/DC1.
Supplementary Material
Acknowledgments
We are grateful to the Japanese National Bioresource Project for C. elegans and the Caenorhabditis Genetics Center for strains and reagents. We also thank Alain Verreault, Paul Maddox, Daniel Durocher, Jonas Dorn, and members of the Maddox and Labbé laboratories for helpful discussions and comments on the manuscript, Paulina Salazar, Marianne Desrosiers, Corentin Monfort, and Julien Burger for technical assistance, and Christian Charbonneau of the Institute of Research in Immunology and Cancer (IRIC) Bio-imaging Facility for technical help with image acquisition.
C. Descoteaux held studentships from the Mach–Gaensslen Foundation of Canada, awarded through the COPSÉ (Comité d’organisation du programme des stages d’été), and from the Faculté de médecine de l’Université de Montréal. N.T. Chartier held a fellowship from the Fonds de Recherche du Québec–Santé (FRQS). L. Pintard is supported by the City of Paris and by the French National Research Agency (under grant no. ANR-2012-BSV2-0001-01). This work was funded by a grant from the Canadian Institutes of Health Research (no. 230043) to J.-C. Labbé, who holds the Canada Research Chair in Cell Division and Differentiation. IRIC is supported in part by the Canadian Center of Excellence in Commercialization and Research, the Canada Foundation for Innovation, and the FRQS.
The authors declare no competing financial interests.
Footnotes
Abbreviations used in this paper:
- AMPK
- AMP kinase
- DIC
- differential interference contrast
References
- Araki H., Leem S.H., Phongdara A., Sugino A. 1995. Dpb11, which interacts with DNA polymerase II(epsilon) in Saccharomyces cerevisiae, has a dual role in S-phase progression and at a cell cycle checkpoint. Proc. Natl. Acad. Sci. USA. 92:11791–11795 10.1073/pnas.92.25.11791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brauchle M., Baumer K., Gönczy P. 2003. Differential activation of the DNA replication checkpoint contributes to asynchrony of cell division in C. elegans embryos. Curr. Biol. 13:819–827 10.1016/S0960-9822(03)00295-1 [DOI] [PubMed] [Google Scholar]
- Brenner S. 1974. The genetics of Caenorhabditis elegans. Genetics. 77:71–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budirahardja Y., Gönczy P. 2008. PLK-1 asymmetry contributes to asynchronous cell division of C. elegans embryos. Development. 135:1303–1313 10.1242/dev.019075 [DOI] [PubMed] [Google Scholar]
- Chartier N.T., Salazar Ospina D.P., Benkemoun L., Mayer M., Grill S.W., Maddox A.S., Labbé J.C. 2011. PAR-4/LKB1 mobilizes nonmuscle myosin through anillin to regulate C. elegans embryonic polarization and cytokinesis. Curr. Biol. 21:259–269 10.1016/j.cub.2011.01.010 [DOI] [PubMed] [Google Scholar]
- Diffley J.F. 2011. Quality control in the initiation of eukaryotic DNA replication. Philos. Trans. R. Soc. Lond. B Biol. Sci. 366:3545–3553 10.1098/rstb.2011.0073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar L.G. 1995. Blastomere culture and analysis. Methods Cell Biol. 48:303–321 10.1016/S0091-679X(08)61393-X [DOI] [PubMed] [Google Scholar]
- Edgar L.G., McGhee J.D. 1988. DNA synthesis and the control of embryonic gene expression in C. elegans. Cell. 53:589–599 10.1016/0092-8674(88)90575-2 [DOI] [PubMed] [Google Scholar]
- Encalada S.E., Martin P.R., Phillips J.B., Lyczak R., Hamill D.R., Swan K.A., Bowerman B. 2000. DNA replication defects delay cell division and disrupt cell polarity in early Caenorhabditis elegans embryos. Dev. Biol. 228:225–238 10.1006/dbio.2000.9965 [DOI] [PubMed] [Google Scholar]
- Essers J., Theil A.F., Baldeyron C., van Cappellen W.A., Houtsmuller A.B., Kanaar R., Vermeulen W. 2005. Nuclear dynamics of PCNA in DNA replication and repair. Mol. Cell. Biol. 25:9350–9359 10.1128/MCB.25.21.9350-9359.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fievet B.T., Rodriguez J., Naganathan S., Lee C., Zeiser E., Ishidate T., Shirayama M., Grill S., Ahringer J. 2013. Systematic genetic interaction screens uncover cell polarity regulators and functional redundancy. Nat. Cell Biol. 15:103–112 10.1038/ncb2639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaggioli V., Zeiser E., Rivers D., Bradshaw C.R., Ahringer J., Zegerman P. 2014. CDK phosphorylation of SLD-2 is required for replication initiation and germline development in C. elegans. J. Cell Biol. 204:507–522 10.1083/jcb.201310083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan B., Hu J., Jiang S., Liu Y., Sahin E., Zhuang L., Fletcher-Sananikone E., Colla S., Wang Y.A., Chin L., Depinho R.A. 2010. Lkb1 regulates quiescence and metabolic homeostasis of haematopoietic stem cells. Nature. 468:701–704 10.1038/nature09595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein B., Macara I.G. 2007. The PAR proteins: fundamental players in animal cell polarization. Dev. Cell. 13:609–622 10.1016/j.devcel.2007.10.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin E.E., Odde D.J., Seydoux G. 2011. Regulation of the MEX-5 gradient by a spatially segregated kinase/phosphatase cycle. Cell. 146:955–968 10.1016/j.cell.2011.08.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo S., Kemphues K.J. 1995. par-1, a gene required for establishing polarity in C. elegans embryos, encodes a putative Ser/Thr kinase that is asymmetrically distributed. Cell. 81:611–620 10.1016/0092-8674(95)90082-9 [DOI] [PubMed] [Google Scholar]
- Gurumurthy S., Hezel A.F., Sahin E., Berger J.H., Bosenberg M.W., Bardeesy N. 2008. LKB1 deficiency sensitizes mice to carcinogen-induced tumorigenesis. Cancer Res. 68:55–63 10.1158/0008-5472.CAN-07-3225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurumurthy S., Xie S.Z., Alagesan B., Kim J., Yusuf R.Z., Saez B., Tzatsos A., Ozsolak F., Milos P., Ferrari F., et al. 2010. The Lkb1 metabolic sensor maintains haematopoietic stem cell survival. Nature. 468:659–663 10.1038/nature09572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawley S.A., Boudeau J., Reid J.L., Mustard K.J., Udd L., Mäkelä T.P., Alessi D.R., Hardie D.G. 2003. Complexes between the LKB1 tumor suppressor, STRAD alpha/beta and MO25 alpha/beta are upstream kinases in the AMP-activated protein kinase cascade. J. Biol. 2:28 10.1186/1475-4924-2-28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holway A.H., Hung C., Michael W.M. 2005. Systematic, RNA-interference-mediated identification of mus-101 modifier genes in Caenorhabditis elegans. Genetics. 169:1451–1460 10.1534/genetics.104.036137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyenne V., Desrosiers M., Labbé J.C. 2008. C. elegans Brat homologs regulate PAR protein-dependent polarity and asymmetric cell division. Dev. Biol. 321:368–378 10.1016/j.ydbio.2008.06.037 [DOI] [PubMed] [Google Scholar]
- Kamath R.S., Martinez-Campos M., Zipperlen P., Fraser A.G., Ahringer J. 2001. Effectiveness of specific RNA-mediated interference through ingested double-stranded RNA in Caenorhabditis elegans. Genome Biol. 2:H0002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemphues K., Strome S. 1997. Fertilization and establishment of polarity in the embryo. In C. elegans II Second edition Riddle D.L., Blumenthal T., Meyer B.J., Priess J.R., Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY: 335–359 [PubMed] [Google Scholar]
- Kemphues K.J., Priess J.R., Morton D.G., Cheng N.S. 1988. Identification of genes required for cytoplasmic localization in early C. elegans embryos. Cell. 52:311–320 10.1016/S0092-8674(88)80024-2 [DOI] [PubMed] [Google Scholar]
- Lizcano J.M., Göransson O., Toth R., Deak M., Morrice N.A., Boudeau J., Hawley S.A., Udd L., Mäkelä T.P., Hardie D.G., Alessi D.R. 2004. LKB1 is a master kinase that activates 13 kinases of the AMPK subfamily, including MARK/PAR-1. EMBO J. 23:833–843 10.1038/sj.emboj.7600110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddox P.S., Portier N., Desai A., Oegema K. 2006. Molecular analysis of mitotic chromosome condensation using a quantitative time-resolved fluorescence microscopy assay. Proc. Natl. Acad. Sci. USA. 103:15097–15102 10.1073/pnas.0606993103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mäkiniemi M., Hillukkala T., Tuusa J., Reini K., Vaara M., Huang D., Pospiech H., Majuri I., Westerling T., Mäkelä T.P., Syväoja J.E. 2001. BRCT domain-containing protein TopBP1 functions in DNA replication and damage response. J. Biol. Chem. 276:30399–30406 10.1074/jbc.M102245200 [DOI] [PubMed] [Google Scholar]
- Mantiero D., Mackenzie A., Donaldson A., Zegerman P. 2011. Limiting replication initiation factors execute the temporal programme of origin firing in budding yeast. EMBO J. 30:4805–4814 10.1038/emboj.2011.404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meister P., Taddei A., Ponti A., Baldacci G., Gasser S.M. 2007. Replication foci dynamics: replication patterns are modulated by S-phase checkpoint kinases in fission yeast. EMBO J. 26:1315–1326 10.1038/sj.emboj.7601538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morton D.G., Roos J.M., Kemphues K.J. 1992. par-4, a gene required for cytoplasmic localization and determination of specific cell types in Caenorhabditis elegans embryogenesis. Genetics. 130:771–790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakada D., Saunders T.L., Morrison S.J. 2010. Lkb1 regulates cell cycle and energy metabolism in haematopoietic stem cells. Nature. 468:653–658 10.1038/nature09571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narbonne P., Hyenne V., Li S., Labbé J.C., Roy R. 2010. Differential requirements for STRAD in LKB1-dependent functions in C. elegans. Development. 137:661–670 10.1242/dev.042044 [DOI] [PubMed] [Google Scholar]
- Noatynska A., Tavernier N., Gotta M., Pintard L. 2013. Coordinating cell polarity and cell cycle progression: what can we learn from flies and worms? Open Biol. 3:130083 10.1098/rsob.130083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivers D.M., Moreno S., Abraham M., Ahringer J. 2008. PAR proteins direct asymmetry of the cell cycle regulators Polo-like kinase and Cdc25. J. Cell Biol. 180:877–885 10.1083/jcb.200710018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saka Y., Yanagida M. 1993. Fission yeast cut5+, required for S phase onset and M phase restraint, is identical to the radiation-damage repair gene rad4+. Cell. 74:383–393 10.1016/0092-8674(93)90428-S [DOI] [PubMed] [Google Scholar]
- Sonneville R., Querenet M., Craig A., Gartner A., Blow J.J. 2012. The dynamics of replication licensing in live Caenorhabditis elegans embryos. J. Cell Biol. 196:233–246 10.1083/jcb.201110080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spilker A.C., Rabilotta A., Zbinden C., Labbé J.C., Gotta M. 2009. MAP kinase signaling antagonizes PAR-1 function during polarization of the early Caenorhabditis elegans embryo. Genetics. 183:965–977 10.1534/genetics.109.106716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka S., Nakato R., Katou Y., Shirahige K., Araki H. 2011. Origin association of Sld3, Sld7, and Cdc45 proteins is a key step for determination of origin-firing timing. Curr. Biol. 21:2055–2063 10.1016/j.cub.2011.11.038 [DOI] [PubMed] [Google Scholar]
- Waga S., Stillman B. 1998. The DNA replication fork in eukaryotic cells. Annu. Rev. Biochem. 67:721–751 10.1146/annurev.biochem.67.1.721 [DOI] [PubMed] [Google Scholar]
- Watts J.L., Morton D.G., Bestman J., Kemphues K.J. 2000. The C. elegans par-4 gene encodes a putative serine-threonine kinase required for establishing embryonic asymmetry. Development. 127:1467–1475 [DOI] [PubMed] [Google Scholar]
- Woods A., Johnstone S.R., Dickerson K., Leiper F.C., Fryer L.G., Neumann D., Schlattner U., Wallimann T., Carlson M., Carling D. 2003. LKB1 is the upstream kinase in the AMP-activated protein kinase cascade. Curr. Biol. 13:2004–2008 10.1016/j.cub.2003.10.031 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.