Assembly of the two heme b cofactors into the respiratory chain complex III subunit cytochrome b occurs in a step-by-step process that is monitored by assembly factors and regulates additional cytochrome b synthesis through a feedback loop.
Abstract
Mitochondrial respiratory chain complexes convert chemical energy into a membrane potential by connecting electron transport with charge separation. Electron transport relies on redox cofactors that occupy strategic positions in the complexes. How these redox cofactors are assembled into the complexes is not known. Cytochrome b, a central catalytic subunit of complex III, contains two heme bs. Here, we unravel the sequence of events in the mitochondrial inner membrane by which cytochrome b is hemylated. Heme incorporation occurs in a strict sequential process that involves interactions of the newly synthesized cytochrome b with assembly factors and structural complex III subunits. These interactions are functionally connected to cofactor acquisition that triggers the progression of cytochrome b through successive assembly intermediates. Failure to hemylate cytochrome b sequesters the Cbp3–Cbp6 complex in early assembly intermediates, thereby causing a reduction in cytochrome b synthesis via a feedback loop that senses hemylation of cytochrome b.
Introduction
Oxidative phosphorylation is a key process of cellular metabolism and takes place within mitochondria. The respiratory chain complexes in the inner membrane connect the transfer of electrons from reducing equivalents to the final electron acceptor oxygen with the establishment of a membrane potential, which is in turn used for ATP synthesis. Respiratory chain complexes thus contain sequentially acting subunits equipped with prosthetic groups that allow the acceptance or donation of electrons.
In general, Fe/S clusters, FAD, and different hemes (a-, b-, and c-types) are used during electron transport. Fe/S cluster assembly and incorporation into respiratory chain complexes are partly understood (Lill, 2009), as well as certain aspects of heme attachment in c-type cytochromes (Steiner et al., 1996; Bernard et al., 2003, 2005; Kranz et al., 2009; Corvest et al., 2010). In contrast to c-type cytochromes, a- and b-type hemes of cytochrome oxidase (COX) and complex III (bc1 complex) are not covalently linked but rather noncovalently coordinated by conserved residues. The mechanism by which a- and b-type hemes are inserted is currently not known (Mick et al., 2011; Kim et al., 2012). Here, we used the central subunit of the bc1 complex, cytochrome b, as a model to understand how heme incorporation into respiratory chain subunits occurs in the mitochondrial inner membrane.
Cytochrome b contains two heme bs, one low-potential heme bL located close to the intermembrane space side of the inner membrane and one high-potential heme bH at the opposite side of the membrane (Yun et al., 1991). Together with cytochrome c1 and the Rieske Fe/S protein, cytochrome b participates in the catalytic reactions of the bc1 complex, termed the Q cycle (Mitchell, 1975; Crofts, 2004; Osyczka et al., 2005). The hemes of cytochrome b are coordinated in a four-helix bundle by four conserved histidines, two located in the second transmembrane domain (H82 and H96 in baker’s yeast) and two in the fourth transmembrane helix (H183 and H197 in baker’s yeast; Yun et al., 1991; Hunte et al., 2000).
We recently found that cytochrome b assembles through a series of four intermediates into the bc1 complex. Intermediate I, which is composed of cytochrome b, Cbp4, and the evolutionary conserved assembly factors Cbp3–Cbp6 (Tucker et al., 2013), accumulates at steady state in wild-type yeast cells and therefore likely represents a pool of cytochrome b ready to assemble (Gruschke et al., 2012). The next step in assembly is formation of intermediate II, which contains cytochrome b, Cbp4, Qcr7, and Qcr8. Incorporation of further nuclear-encoded subunits releases Cbp4 and allows the formation of the functional bc1 complex through intermediates III and IV (Gruschke et al., 2012). To date, it is not known when during these steps cytochrome b receives its heme b cofactors (Kim et al., 2012).
In this study, we designed a strategy to unravel the sequence by which cytochrome b is hemylated. By using yeast mitochondrial genetics and biochemical analyses, we reveal an obligate order in hemylation of cytochrome b (first bL and then bH) and the involvement of assembly factors that recognize the different hemylation states. The efficiency of hemylation is directly reported to the mitochondrial genetic system by a feedback loop that represses synthesis of cytochrome b when hemylation fails.
Results
A cytochrome b–Qcr7 complex retains full hemylation of cytochrome b
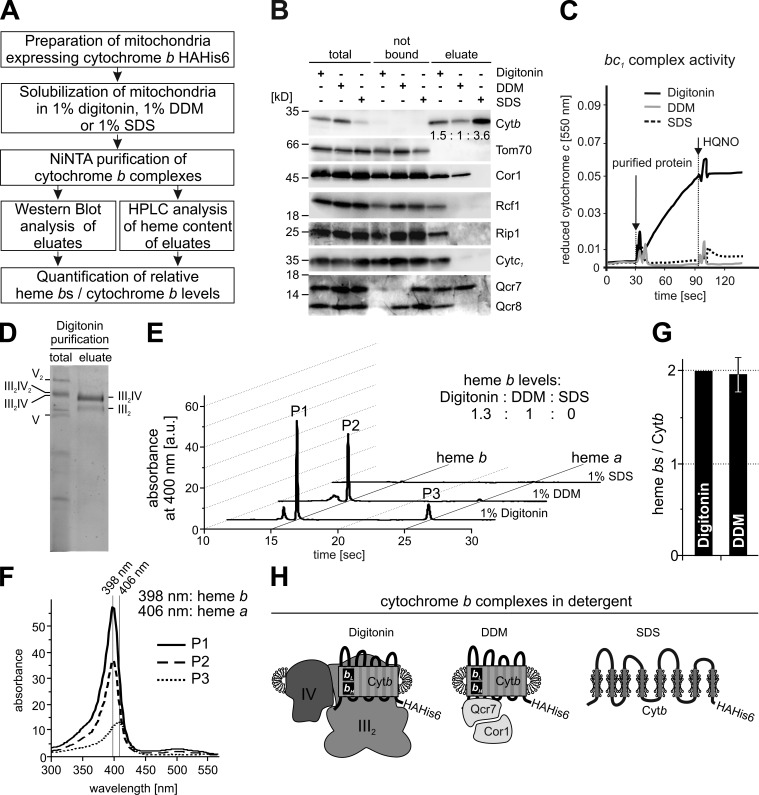
To analyze the hemylation of cytochrome b in assembly intermediates, we set up a strategy to purify cytochrome b and determine the heme b content of these preparations (Fig. 1 A). To this end, we used a yeast strain (COB-HApH) containing a mitochondrial genome coding for a C-terminally HAHis6-tagged cytochrome b. This tag enabled us to purify cytochrome b complexes from lysates prepared with three detergents that differ substantially in their stringency, namely the denaturing detergent SDS, the rather harsh but nondenaturing detergent dodecylmaltoside (DDM), or the mild detergent digitonin. When lysates were prepared with 1% DDM, we found that only a subset of the bc1 complex subunits could be co-purified with cytochrome b (Qcr7, Cor1), whereas others were removed (Cytc1, Qcr8, Rip1; Fig. 1 B). As expected, cytochrome b devoid of all other structural subunits of the bc1 complex was purified using SDS (Fig. 1 B). When cytochrome b complexes were purified from digitonin lysates, intact and enzymatically active bc1 complex (Fig. 1 C) could be purified that was mainly part of a respiratory supercomplex (Cruciat et al., 2000), as evidenced by the copurification of Rcf1 (Fig. 1 B), a supercomplex-specific COX subunit (Vukotic et al., 2012), and the migration behavior of the complexes on blue native–PAGE (Fig. 1 D).
Figure 1.
A cytochrome b–Qcr7 complex retains full hemylation of cytochrome b. (A) Schematic outline of the experiment. (B) Mitochondria were isolated from a yeast strain expressing a C-terminally HAHis6-tagged variant of cytochrome b and lysed in 1% digitonin, 1% DDM, or 1% SDS. Subsequently, cytochrome b HAHis6 was purified by affinity chromatography using NiNTA beads. Eluates were split to analyze either protein levels by Western blotting (B) with the antibodies indicated or to determine heme content by HPLC. The numbers in the Western blot of Cytb denote the relative cytochrome b levels that were densitometrically determined. (C) The cytochrome b complexes purified using the indicated detergents were tested for decyl-quinol–dependent cytochrome c reductase activity. The bc1 complex inhibitor HQNO was used to reveal specificity of the reaction. (D) The purification of cytochrome b complexes from a digitonin lysate was analyzed on blue native–PAGE stained with Coomassie. V2 and V, ATPase dimer and monomer, respectively; III2IV2 and III2IV, supercomplex composed of a bc1 dimer and either two or one COX complexes, respectively; III2, bc1 complex dimer. (E) Hemes were extracted from the identical eluates as in A and separated by HPLC. Heme absorbance was monitored at 400 nm. (F) Wavelength spectra for the individual peaks P1, P2, and P3 to confirm the identity of heme b (maximum 398 nm) and heme a (maximum 406 nm). (G) Relative heme b/cytochrome b ratios were determined from three experiments and are represented as mean values ± SEM. Wild-type levels were set to 100% (reflecting two heme bs/cytochrome b protein). (H) Schematic representation of cytochrome b complexes generated upon treatment with different detergents.
We next extracted hemes from these purifications and analyzed them by reverse-phase HPLC. As expected, the extractions from the digitonin-purified supercomplexes contained both heme b and heme a, whereas the purifications of cytochrome b using SDS completely lacked any heme signals, reflecting the fact that the hemes in cytochrome b are noncovalently coordinated to the protein (Fig. 1 E). As a result of dissociation of supercomplexes by the detergent, HPLC analysis of the DDM-purified cytochrome b revealed that it only contained heme b. Interestingly, when we compared heme b signals with the signals of cytochrome b protein, we found that the relative amounts of heme b per cytochrome b were the same for the DDM and the digitonin complexes (Fig. 1 G). This showed that although the bc1 complex was partly dissociated by DDM treatment, both hemes of cytochrome b remained bound to cytochrome b (Fig. 1 H).
Cytochrome b is semi-hemylated in assembly intermediate I but fully hemylated in assembly intermediate II
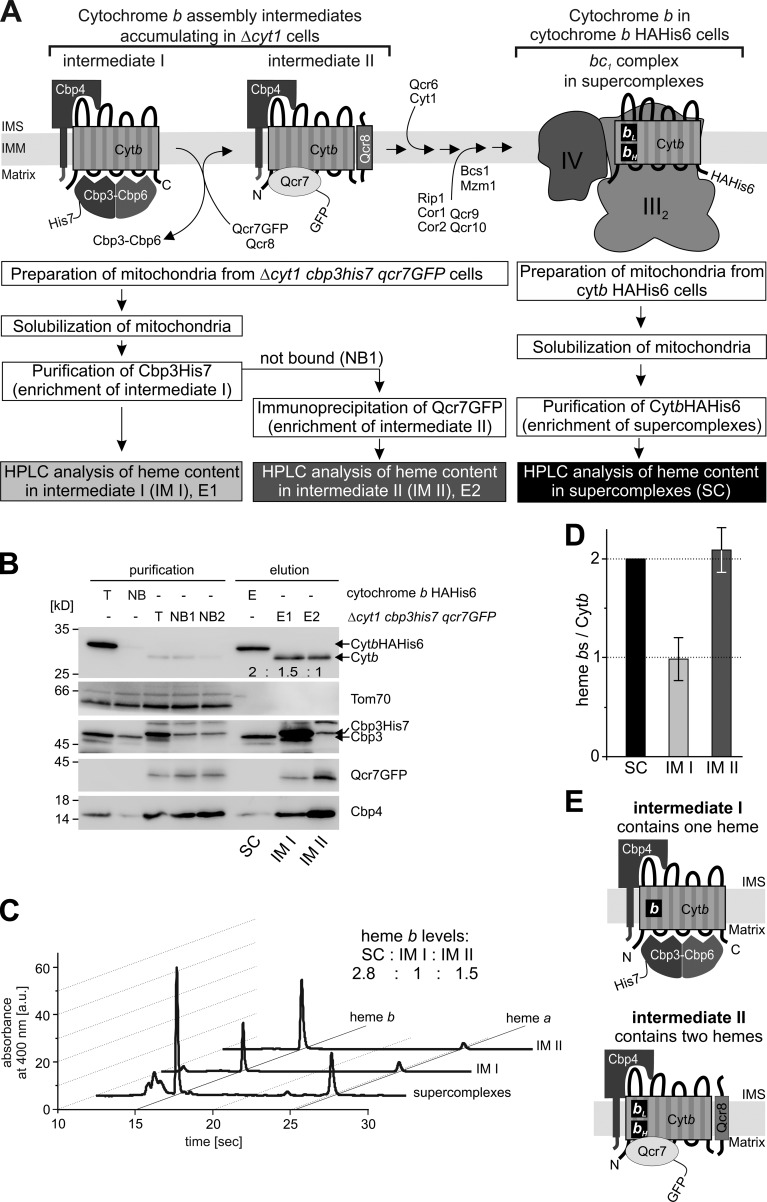
Qcr7 is the first structural subunit that interacts with newly assembling cytochrome b (Gruschke et al., 2012) and remains bound to cytochrome b in the DDM complex. The observation that cytochrome b stays fully hemylated in the DDM complexes inspired us to ask whether hemylation of cytochrome b occurs early in assembly. To test this, we sought to determine the hemylation status of early assembly intermediates of cytochrome b, namely intermediate I and intermediate II (Gruschke et al., 2012). We have previously established that only intermediate I accumulates to a detectable level in wild-type cells. To compare hemylation of cytochrome b in the two first assembly intermediates, we used a yeast mutant lacking cytochrome c1 (Δcyt1) where cytochrome b accumulates both in intermediate I and in intermediate II (Gruschke et al., 2012). To enable purification of the two intermediates from the same sample, we equipped Cbp3 with a C-terminal His7 tag and Qcr7 with a C-terminal GFP tag (Fig. 2 A). As a control, we also purified intact supercomplexes from COB-HApH cells as in Fig. 1 A. As expected, intermediate I contained cytochrome b, Cbp3–Cbp6, and Cbp4, whereas intermediate II contained cytochrome b, Cbp4, Qcr7, and Qcr8 (Fig. 2 B). As reported previously (Gruschke et al., 2012), higher quantities of Cbp4 were detected in intermediate II. Next, we extracted hemes from these preparations and analyzed the samples by HPLC (Fig. 2 C). Quantification of heme b relative to cytochrome b protein levels demonstrated that cytochrome b in assembly intermediate II is fully hemylated, just like in the bc1 complex present in supercomplexes where cytochrome b contains two heme b molecules (Fig. 2 D). In contrast, intermediate I contained only half the heme b quantities of the fully assembled bc1 complex, suggesting that only a single heme is incorporated in intermediate I (Fig. 2, D and E).
Figure 2.
Cytochrome b is semi-hemylated in assembly intermediate I but fully hemylated in assembly intermediate II. (A) Schematic representation of the experimental setup. To accumulate and specifically purify intermediate I or II from the same sample, a yeast strain with a cytochrome c1 deletion was constructed that carries both a His7-tagged Cbp3 and a GFP-tagged Qcr7. Mitochondria isolated from this stain were lysed in 1% digitonin. After intermediate I was enriched (E1) by affinity purification of Cbp3-His7, the not-bound fraction (NB1) was recovered and Qcr7-GFP was purified by immunoprecipitation using a GFP nanobody (E2). The fully assembled bc1 complex was purified from mitochondria carrying the cytochrome b HAHis6 variant (E). All eluates were split and analyzed by both Western blotting (B) with the antibodies indicated and HPLC (C) after heme extraction. The numbers in the Western blot of Cytb denote the relative cytochrome b levels that were densitometrically determined. (D) Relative heme b/cytochrome b protein ratios were calculated and the values for heme extracted from supercomplexes were set to 100% (reflecting two heme bs/cytochrome b protein). Mean values of four independent experiments ± SEM are shown. (E) Graphical summary, showing fully hemylated cytochrome b in intermediate II and a semi-hemylated cytochrome b in intermediate I. IM I, intermediate I; IM II, intermediate II; SC, supercomplexes.
Only the bL heme is inserted in intermediate I
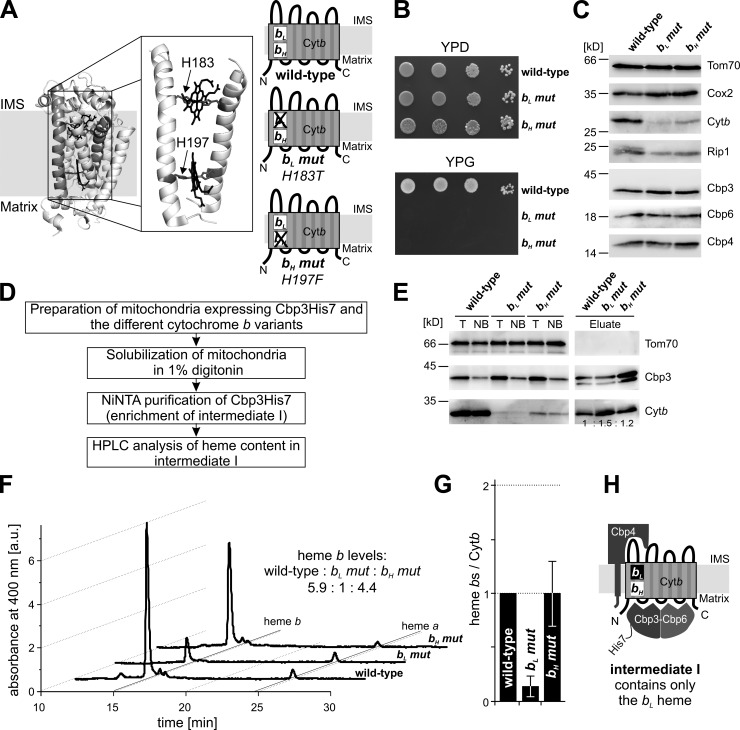
These results inspired us to test whether one specific heme is incorporated first and hence present in intermediate I. To accomplish this, we created two mitochondrial genomes expressing two different cytochrome b variants. In these mutants, one of the coordinating histidine residues of either the bL (Cytb-H183T) or bH (Cytb-H197F) site was altered, resulting in the loss of the respective heme coordination (Fig. 3 A). Because both heme cofactors are essential for the electron transport processes of the bc1 complex, the individual mutations caused a respiratory-deficient phenotype (Fig. 3 B). Consistently, cells expressing these mutated cytochrome b variants failed to accumulate normal amounts of the bc1 complex, whereas levels of the COX complex were essentially unchanged (Fig. 3 C). We then constructed yeast strains expressing these cytochrome b variants and a His7-tagged Cbp3, prepared mitochondria from these cells, and lysed them in digitonin and purified intermediate I using the His7-tag (Fig. 3, D and E). Next, we determined the heme contents of these complexes via HPLC (Fig. 3 F). Strikingly, the bH mutant exhibited identical amounts of heme bs/cytochrome b protein as intermediate I from mitochondria containing wild-type cytochrome b (Fig. 3, F and G). In contrast, intermediate I from the bL mutant almost completely lacked heme b signals (Fig. 3, F and G). These data therefore confirm that cytochrome b in intermediate I contains only one heme b and reveal that this heme is bound to the intact bL site (Fig. 3 H). Because the bL mutant contained only very little heme b, these results suggest a strict order in hemylation, namely that the bL site must be hemylated before the bH site.
Figure 3.
The first heme is inserted into the heme bL site. (A) Crystal structure of yeast cytochrome b (Protein Data Bank accession no. 3CXH) and schematic of the three cytochrome b variants used in this study: wild-type (both heme sites intact), the bL mutant (H183T), and the bH mutant (H197F). Heme-coordinating histidine residues are highlighted in dark gray, hemes in black and mutated histidines are indicated by arrows. (B) Serial dilutions of the indicated cells were spotted on plates containing either the fermentable carbon source glucose (YPD) or the nonfermentable carbon source glycerol (YPG). (C) Isolated mitochondria of the indicated strains were lysed and their proteins separated by SDS-PAGE and analyzed by Western blotting with the antibodies indicated. (D) Strategy to purify intermediate I from cells expressing His7-tagged Cbp3 and containing either wild-type or mutated versions of cytochrome b. (E) Western blot analyses of the purifications. T, total of the starting material; NB, not-bound material. (F) HPLC analyses of the purifications. (G) Relative heme b/cytochrome b protein ratios were calculated and the mean values of three independent experiments ± SEM are shown. (H) Schematic representation of intermediate I containing one heme in the bL position.
Cbp4 interacts with the Cbp3–Cbp6-bound cytochrome b in a hemylation-dependent fashion
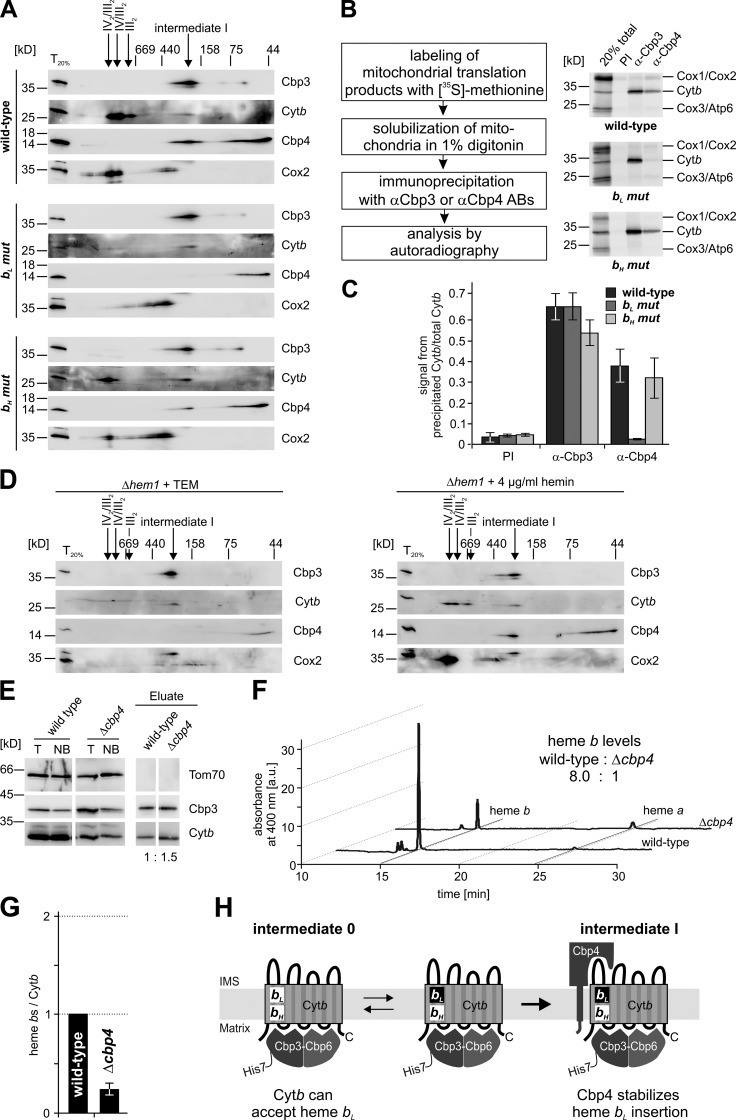
Because of this surprising result, we next analyzed the biogenesis of the two cytochrome b mutants in more detail. We first analyzed complexes formed by these two cytochrome b variants and lysed isolated mitochondria containing either variant in digitonin. Solubilized protein complexes were separated by 2D BN/SDS-PAGE followed by Western blot analysis. In wild-type mitochondria, most steady-state cytochrome b migrates at an apparent size between 600 and 750 kD, reflecting the fully assembled bc1 complex dimer alone or in supercomplexes with cytochrome oxidase (Cruciat et al., 2000). Additionally, a small fraction of cytochrome b is in association with Cbp3–Cbp6 and Cbp4 (Fig. 4 A, wild-type) in intermediate I (Gruschke et al., 2012). In the bH mutant, cytochrome b is also either localized within the 600–750 kD species or in association with Cbp3–Cbp6 and Cbp4 (Fig. 4 A, bH mut). In contrast, the bL mutant fails completely to assemble a bc1 complex. Most interestingly, the small amounts of cytochrome b detectable in this mutant are only in association with Cbp3–Cbp6, but not with Cbp4 (Fig. 4 A, bL mut).
Figure 4.
Cbp4 interacts with the Cbp3–Cbp6-bound cytochrome b in a hemylation-dependent fashion. (A) Mitochondria expressing the indicated cytochrome b variants were lysed in digitonin and analyzed by 2D BN/SDS-PAGE followed by Western blotting. (B) Left: experimental setup for coimmunoprecipitation of radiolabeled cytochrome b. Right: mitochondrial translation products were labeled with [35S]-methionine, mitochondria were lysed in 1% digitonin, and the lysates were subjected to immunoprecipitation using antibodies against Cbp3, Cbp4, or preimmune serum (PI) as a control. “20% total” corresponds to 20% of the starting material. (C) Quantification of relative cytochrome b levels that coprecipitated with the indicated antibodies. The values of three independent experiments ± SEM are shown. (D) The gene encoding the first enzyme of heme biosynthesis, HEM1, was deleted and Δhem1 cells were grown in synthetic media supplemented with either the essential products of cellular heme proteins (left; Tween 80, ergosterol, and methionine [TEM]) or with 4 µg/ml hemin (right; hemin, heme b with a chloride ligand). Mitochondria of these cells were prepared and analyzed as in A. (E) Mitochondria from strains expressing a His7-tagged Cbp3 and either containing or not containing Cbp4 were lysed in 1% digitonin and Cbp3 complexes were purified by metal affinity chromatography. The purification was analyzed by Western blotting using the indicated antibodies. T, total of the starting material; NB, not-bound material. (F) The eluates of these purifications were extracted and hemes were analyzed by HPLC. (G) Relative heme b/cytochrome b protein ratios were calculated and the mean values ± SEM of three independent purifications are shown. (H) Model of a dynamic hemylation of the bL site that is stabilized by Cbp4 binding.
To substantiate by a different approach the result that Cbp4 does not stably interact with cytochrome b when hemylation of the bL site fails, we performed coimmunoprecipitation experiments. Translation products of isolated mitochondria were labeled with [35S]-methionine for 15 min, and mitochondria were then lysed with digitonin and protein complexes precipitated with either pre-immune serum or serum against Cbp3 or Cbp4, respectively (Fig. 4 B). Immunoprecipitated proteins were separated by SDS-PAGE and detected by autoradiography. In wild-type mitochondria and the bH mutant, cytochrome b was coimmunoprecipitated with antibodies against Cbp3 and Cbp4 (Fig. 4, B and C). In contrast, cytochrome b of the bL mutant could only be coimmunoprecipitated with Cbp3, but not with Cbp4. These results demonstrate that hemylation of cytochrome b affects the formation of bc1 complex assembly intermediates. Whereas a variant of cytochrome b lacking the heme bH site forms a normal intermediate I, the bL mutant does not even reach the stage of assembly intermediate I because Cbp4 does not bind.
To investigate whether hemylation of cytochrome b is also a prerequisite for the interaction of wild-type cytochrome b with Cbp4, we tested the effect of heme depletion on the formation of intermediate I. We deleted HEM1, coding for the first gene of the heme biosynthesis pathway, and supplemented the cells either with hemin or with the essential products of cellular heme proteins (Tween 80, ergosterol, methionine [TEM]). We then isolated mitochondria from these cells and repeated the 2D BN/SDS-PAGE analyses. As in the case of the bL mutant, the bc1 complex failed to assemble when heme was depleted and Cbp4 also did not co-migrate with Cbp3–Cbp6 and cytochrome b (Fig. 4 D, left). However, in the presence of externally added heme, the bc1 complex assembled and Cbp4 was recruited into intermediate I (Fig. 4 D, right). These experiments demonstrated that cytochrome b interacts with the Cbp3–Cbp6 complex before hemylation in a novel intermediate (here termed intermediate 0) and showed that hemylation of cytochrome b is a prerequisite for the stable interaction between cytochrome b and its specific assembly factor Cbp4.
No bL hemylation of cytochrome b can be detected in the absence of Cbp4
Because Cbp4 interacts with cytochrome b in a hemylation-dependent fashion, we asked whether hemylation depends in turn on Cbp4. To determine the relative heme b amounts of cytochrome b bound to Cbp3–Cbp6, we constructed yeast strains expressing a His7-tagged Cbp3 in the absence or presence of Cbp4. When we analyzed heme b content of the purified assembly intermediates, we found that absence of Cbp4 greatly reduced the amounts of heme b that can be co-purified with cytochrome b (Fig. 4, E–G). In summary, these data demonstrate that cytochrome b receives the first heme b when bound to the Cbp3–Cbp6 complex in the novel intermediate 0 and that the interaction between this semi-hemylated cytochrome b with its assembly factor Cbp4 is important to stabilize the hemylation of the bL site (Fig. 4 H).
Failure to hemylate cytochrome b provokes reduction of cytochrome b synthesis through a translational feedback loop
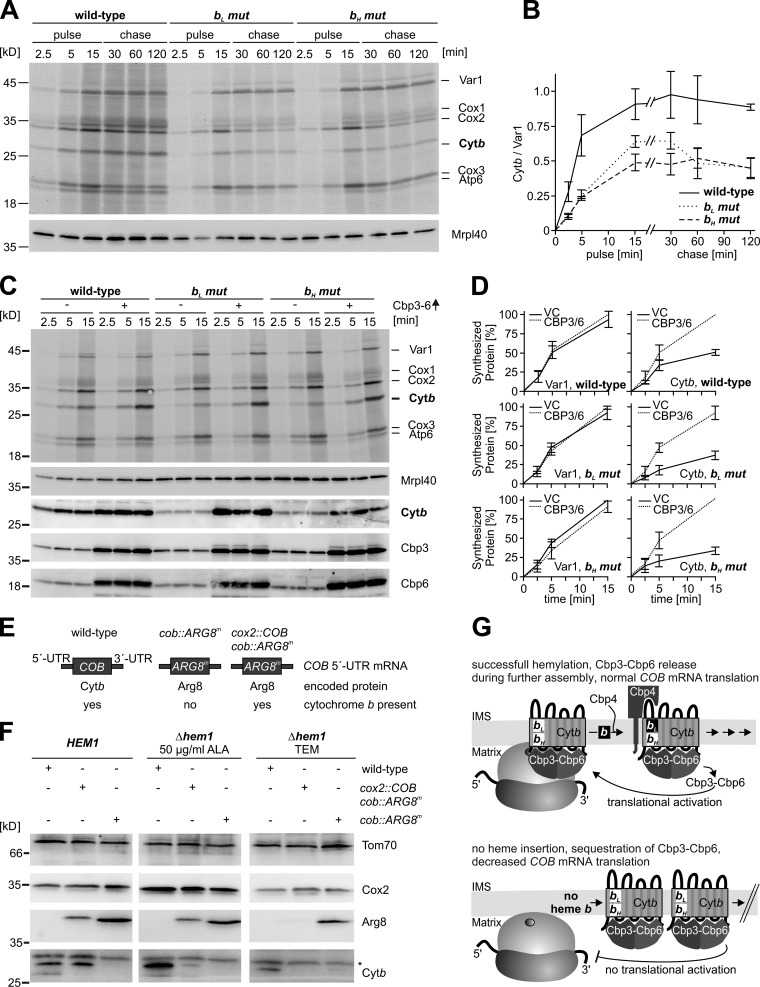
We previously demonstrated that expression of cytochrome b is subject to a translational feedback regulation (Gruschke et al., 2012). Defects at early or intermediate steps of assembly provoke an accumulation of assembly intermediates that sequesters the Cbp3–Cbp6 complex, and this in turn reduces the availability of Cbp3–Cbp6 to stimulate translation of cytochrome b (COB) mRNA. Because we found that hemylation of the bL site occurs on the Cbp3–Cbp6 complex, we asked whether hemylation efficiency might also regulate the synthesis of cytochrome b via Cbp3–Cbp6. First we tested the synthesis and stability of the bL and bH mutants of cytochrome b. We labeled mitochondrial proteins with [35S]-methionine in whole cells for up to 15 min and followed the label for 2 h. Equal amounts of proteins were extracted, separated by SDS-PAGE, and the labeled proteins were visualized by autoradiography (Fig. 5 A). In both mutants, cytochrome b was less efficiently synthesized when compared with wild-type cells. Furthermore, we observed that the newly synthesized cytochrome b was stable in all strains (Fig. 5 B).
Figure 5.
Failure to hemylate cytochrome b provokes reduction of cytochrome b synthesis through a translational feedback loop. (A) Mitochondrial translation products of the indicated strains were labeled with [35S]-methionine in whole cells. Labeling was stopped by the addition of excess unlabeled methionine. (B) The signals from three independent experiments were densitometrically quantified, normalized to the mean signal for Var1 for each strain, and are presented as mean ± SEM. (C) Cells were transformed either with empty plasmids or with plasmids overexpressing CBP3 and CBP6. Synthesis of mitochondrial translation products was analyzed in whole cells. (D) The signals of cytochrome b or Var1 from three independent experiments ± SEM were densitometrically quantified. The highest value at 15 min was set to 100%. (E) Schematic representation of mRNAs and encoded proteins in the different versions of mitochondrial genomes. (F) The indicated strains were grown for 30 h on YPGal and supplemented with either δ-aminolevulinic acid or TEM. Proteins were extracted and analyzed by Western blotting. The asterisk indicates an unspecific cross-reaction. (G) Model of the feedback loop that down-regulates synthesis of cytochrome b when hemylation fails.
Because all cytochrome b variants were similarly stable, we set out to determine whether the low efficiency of cytochrome b labeling is the result of a translational repression of COB mRNA due an inability to hemylate the heme mutants. Such regulation of translation would likely be mediated by a feedback loop involving sequestration of the Cbp3–Cbp6 complex in an assembly intermediate. To overcome a possible sequestration, we expressed simultaneously CBP3 and CBP6 from a strong promoter in either wild-type cells or the bL and bH mutant, and we again tested the synthesis of cytochrome b by short pulses. This overexpression resulted in equal rates of synthesis of all three cytochrome b variants, thus indicating that Cbp3–Cbp6 sequestration provokes the down-regulation of cytochrome b expression in the mutants (Fig. 5, C and D). Importantly, even translation of the wild-type cytochrome b could be significantly stimulated by overexpression of CBP3 and CBP6, thereby demonstrating that the amounts of Cbp3–Cbp6 are rate limiting for full synthesis of cytochrome b even under normal conditions.
Translation of COB mRNA is down-regulated in response to heme depletion
To confirm that a translational feedback mechanism indeed down-regulates cytochrome b expression when hemylation fails, we directly tested translation of COB mRNA by a different approach. To achieve this, we used a mitochondrially encoded reporter gene to assess translation of the COB mRNA in either the absence or presence of heme. The reporter we used is a recoded version of ARG8 termed ARG8m (Steele et al., 1996), a mitochondrial matrix enzyme involved in arginine biosynthesis, which replaces the authentic mitochondrially encoded COB open reading frame. By following the accumulation of Arg8, translation of COB mRNA, which depends on its 5′-untranslated regions, can be directly scored (Gruschke et al., 2011). We used two different mitochondrial genomes containing ARG8m, namely cob::ARG8m and the cox2::COB cob::ARG8m (Fig. 5 E). The cob::ARG8m genome allows us to assess the direct influence of heme depletion on translational regulation acting on the 5′-UTR of COB mRNA. Because this genome lacks a gene encoding cytochrome b, it does not allow detecting repression of COB mRNA translation through the general feedback loop that operates by the cytochrome b–dependent sequestration of Cbp3–Cbp6 in intermediate I. In contrast, cytochrome b is produced in the cox2::COB cob::ARG8m strain, but from an mRNA with the 5′-UTR of COX2 (Gruschke et al., 2012). The Arg8 in this strain can then be used to score for translational regulation of COB mRNA translation when cytochrome b is present (Fig. 5 E).
We disrupted HEM1 in these strains and grew them either under heme-depleted conditions (TEM) or under conditions where heme could be synthesized by adding δ-aminolevulinic acid to the growth media to bypass the HEM1 deletion. Western blotting revealed that Arg8 accumulated normally in the cob::ARG8m strains regardless of whether or not the cells were able to synthesize heme (Fig. 5 F), thereby demonstrating that translation of COB mRNA is not directly influenced by heme levels. In contrast, accumulation of Arg8 of the cox2::COB cob::ARG8m strain was strongly suppressed by heme depletion (Fig. 5 F), in line with the notion that unhemylated cytochrome b sequestered Cbp3–Cbp6 in early assembly intermediates upon heme depletion. Taken together, these data establish that a failure to hemylate cytochrome b provokes a reduction of cytochrome b synthesis through the general feedback loop that regulates COB mRNA translation (Fig. 5 G).
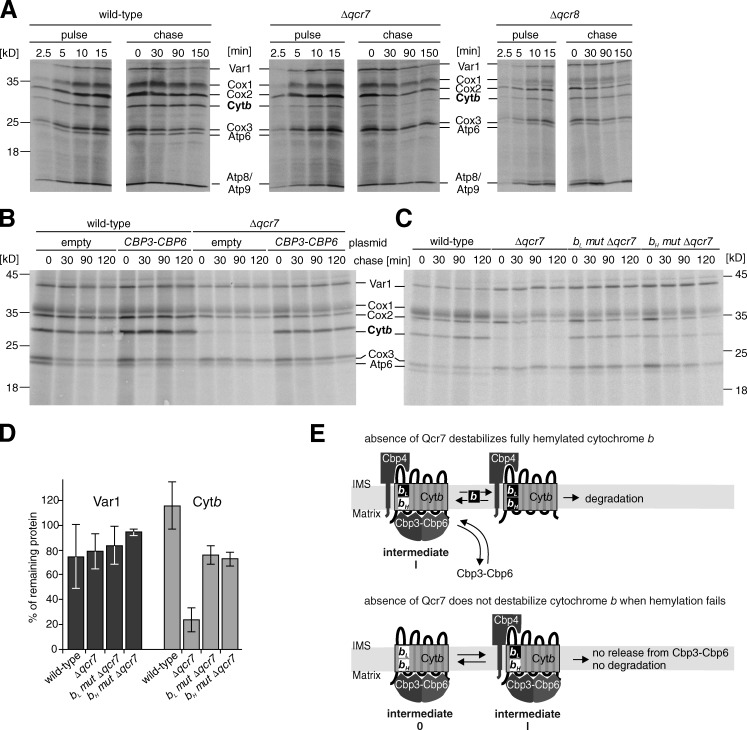
Absence of Qcr7 destabilizes newly synthesized cytochrome b in a hemylation-dependent fashion
We have observed that cytochrome b changes from a semi-hemylated form to a fully hemylated form when it transits from intermediate I to intermediate II (Fig. 2). During the course of this transformation, the interaction with the Cbp3–Cbp6 complex is lost and interactions with the two first structural subunits, namely Qcr7 and Qcr8, are established. To obtain insights into how this transition occurs, we analyzed the fate of newly synthesized cytochrome b in the absence of either Qcr7 or Qcr8. Although the newly synthesized cytochrome b was stable in both wild-type and Δqcr8 cells, the absence of Qcr7 greatly destabilized cytochrome b (Fig. 6 A). This destabilization in the absence of Qcr7 is indistinguishable from that observed in Δcbp3 mutants (Gruschke et al., 2011, 2012). This result is puzzling because newly synthesized cytochrome b is specifically stabilized by binding to Cbp3–Cbp6 during the first steps in its biogenesis, whereas Qcr7 interacts with cytochrome b first after its release from Cbp3–Cbp6 (Gruschke et al., 2012). Because no cytochrome b complex lacking Cbp3–Cbp6 can be observed in Δqcr7 cells (Gruschke et al., 2012), we postulated that the transit generates an unstable form of cytochrome b that loses its interaction with Cbp3–Cbp6 and that needs to interact with Qcr7 for stabilization. In the absence of Qcr7, the transiting cytochrome b interacts with the stabilizing Cbp3–Cbp6 complex, if it is available, and if not, the cytochrome b is proteolytically degraded.
Figure 6.
Absence of Qcr7 provokes destabilization of newly synthesized cytochrome b, which can be reversed by impairment of hemylation. (A) Mitochondrial translation products of the indicated strains were labeled with [35S]-methionine. After 15 min, labeling was stopped by the addition of unlabeled methionine and the cells were incubated for 150 min. (B) The indicated strains were transformed with either empty plasmids or plasmids overexpressing CBP3 and CBP6 and the stability of mitochondrial translation products was analyzed after pulse-labeling with [35S]-methionine. (C) Stability of mitochondrial translation products of the indicated cells were analyzed after 15 min of labeling. (D) The signals of cytochrome b or Var1 that remained after the 120-min chase were quantified. Mean values from three independent experiments ± SEM are shown. (E) Model of the fates of cytochrome b in the absence of Qcr7.
If this scenario is correct, then increased levels of Cbp3–Cbp6 should stabilize newly synthesized cytochrome b in the absence of Qcr7. To test this hypothesis, we simultaneously overexpressed CBP3 and CBP6 in either wild-type or Δqcr7 cells and analyzed the stability of cytochrome b for up to 2 h. Although cytochrome b was very unstable in the absence of Qcr7, a higher level of the Cbp3–Cbp6 complex substantially stabilized the protein (Fig. 6 B).
We next sought to identify the trigger that induces the transit from intermediate I to intermediate II. Because we have observed that cytochrome b is fully hemylated in intermediate II, we asked whether hemylation of the bH site is the trigger that releases cytochrome b from Cbp3–Cbp6. Because cytochrome b is unstable in the absence of Qcr7, we were not able to purify the transit intermediate. Instead, we used the instability of cytochrome b in the absence of Qcr7 as readout to test for the release of cytochrome b from the Cbp3–Cbp6 complex. To ascertain whether the inability to hemylate cytochrome b would impair release from Cbp3–Cbp6 and thus stabilize cytochrome b in the absence of Qcr7, we disrupted QCR7 in cells containing the bL and bH mutations and followed the stability of newly synthesized cytochrome b. As predicted, the radioactively labeled cytochrome b variants were well detectable and substantially stable in the absence of Qcr7 (Fig. 6, C and D), thus indicating that hemylation of cytochrome b dictates release of cytochrome b from Cbp3–Cbp6. This release must be accompanied by a subsequent binding to Qcr7 to stabilize the protein and its bound cofactors. At this stage in intermediate II, cytochrome b is fully hemylated and ready to assemble further into a functional bc1 complex.
Discussion
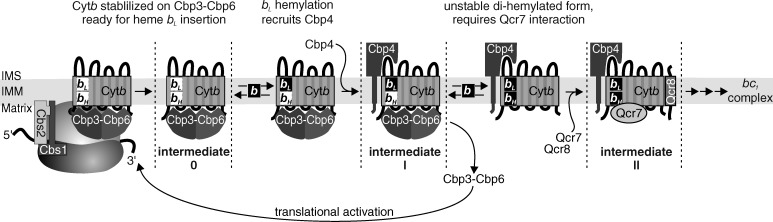
Respiratory chain complexes contain noncovalently bound a- and b-type hemes that are essential for electron transport in cytochrome oxidase and the bc1 complex. A yet completely unresolved aspect is how these hemes are incorporated during assembly (Mick et al., 2011; Kim et al., 2012; Smith et al., 2012). Based on the data presented here, we suggest the following model for heme b incorporation into mitochondrial cytochrome b (Fig. 7): cytochrome b is synthesized by ribosomes that have the Cbp3–Cbp6 complex bound in close proximity to the tunnel exit (Gruschke et al., 2011). Fully synthesized cytochrome b directly interacts with Cbp3–Cbp6 and this intermediate 0 is released from the ribosome. Cytochrome b in this complex is in a conformation that allows the incorporation of heme bL. Upon insertion of heme bL, Cbp4 binds to cytochrome b to stabilize the cofactor. The resulting cytochrome b–Cbp3–Cbp6–Cbp4 complex represents the previously described intermediate I (Gruschke et al., 2012). The trigger to transfer cytochrome b from intermediate I to intermediate II is the incorporation of the bH heme. This releases the Cbp3–Cbp6 complex and makes cytochrome b proteolytically unstable. To stabilize the protein and its bound hemes, cytochrome b must interact with Qcr7. In this fully hemylated form in intermediate II, cytochrome b is ready to be further assembled into a functional bc1 complex.
Figure 7.
Model for the stepwise hemylation of cytochrome b. Cytochrome b is synthesized by mitochondrial ribosomes that have the Cbp3–Cbp6 complex bound for efficient translation of the cytochrome b encoding mRNA. Newly synthesized cytochrome b interacts with Cbp3–Cbp6 in an unhemylated form in intermediate 0. Upon hemylation of the bL site, the assembly factor Cbp4 is recruited to stabilize this heme incorporation. Hemylation of the bH site provokes release of Cbp3–Cbp6 and the fully hemylated cytochrome b must interact with Qcr7 for stabilization. Cytochrome b in intermediate II is fully hemylated and ready for further assembly. Hemylation efficiency modulates synthesis of cytochrome b by a feedback loop involving Cbp3–Cbp6 sequestration in assembly intermediates 0 or I. When hemylation proceeds normally, Cbp3–Cbp6 is released for further stimulation of cytochrome b synthesis by mitochondrial ribosomes.
Both hemes of cytochrome b are coordinated by conserved histidines in a four-helix bundle. Heme incorporation likely changes the conformation of the four transmembrane segments and their soluble loops. During this study we obtained evidence that at least two structural changes might occur during the maturation of cytochrome b. First, hemylation of the bL site is necessary for the interaction of Cbp4 with the cytochrome b–Cbp3–Cbp6 complex, suggesting that hemylation may induce a structural rearrangement in cytochrome b that allows binding of Cbp4. In this scenario, Cbp4 might bind only after hemylation. Alternatively, Cbp4 could be part of a mechanism by which the bL heme is delivered, and the subsequent interaction between Cbp4 and cytochrome b could then stabilize the hemylation. The observed increase in Cbp4 levels in intermediate II in comparison to intermediate I could indicate that full hemylation of cytochrome b promotes tighter binding of the assembly factor. Importantly, Cbp4 is necessary for the formation of a stable, semi-hemylated intermediate containing the bL heme, an event that is a prerequisite for bH insertion. The next transition in the assembly line, namely the release of Cbp3–Cbp6 from cytochrome b, is also accompanied by a hemylation event. Interestingly, it is the binding of heme b to the bH site which is close to the matrix side of the inner membrane and therefore close to the site where Cbp3–Cbp6 is binding to cytochrome b. Hence, it is possible that a conformational change induced by heme bH insertion decreases the affinity of the Cbp3–Cbp6 complex toward cytochrome b, thereby triggering release of the fully hemylated protein and its subsequent interaction with Qcr7. The interaction of Qcr7 with the fully hemylated cytochrome b is rather tight as it also withstands harsh lysis in 1% DDM. Because full hemylation of cytochrome b is maintained in these DDM complexes, it is possible that Qcr7 serves as a lid to lock the acquired hemes in place. Clearly, structural insights into these assembly intermediates are necessary to address directly these possible conformational changes in the future.
The obligate order of hemylation, i.e., first the bL and then the bH site, as reported here for the mitochondrial cytochrome b appears to be conserved. Both chloroplast cytochrome b6 as well as cytochrome b of bacteria were reported to follow the same principle, although the involvement of assembly factors in these systems has not yet been described (Yun et al., 1991; Kuras et al., 1997; Dreher et al., 2008). Because Cbp3 proteins are found in all systems having cytochrome b–type redox proteins, it is tempting to speculate that at least the function of this protein during cytochrome b biogenesis is conserved.
A special aspect in the biogenesis of mitochondrial bc1 complexes is that they are assembled from subunits derived from two different genetic systems. To allow for an efficient assembly process, both systems must be coordinated to supply equal quantities of subunits. In yeast mitochondria, specific feedback loops have been identified that adjust mitochondrial translation of the key respiratory chain subunits to a level that can successfully assemble into complexes (Fox, 2012; Fontanesi, 2013). We recently demonstrated that Cbp3–Cbp6 coordinates cytochrome b translation with bc1 complex assembly (Gruschke et al., 2012). Cbp3–Cbp6 thus has a dual function, acting as an assembly factor as well as a translational activator necessary for efficient translation of the COB mRNA. This feedback control regulating COB translation is achieved by sequestration of Cbp3–Cbp6 in intermediate I when further assembly is blocked, thereby ensuring that Cbp3–Cbp6 is not available to activate new rounds of COB translation. Successful hemylation of cytochrome b releases Cbp3–Cbp6 from intermediate I, thereby stimulating COB mRNA translation. When hemylation fails, Cbp3–Cbp6 is sequestered and synthesis of cytochrome b is down-regulated by the general feedback loop. This mechanism is different to that of COX1 translation, where a specific mechanism connects heme biosynthesis with Cox1 synthesis through the action of Mss51 that in itself is a heme b–binding protein (Soto et al., 2012). Although these feedback loops have now been well described, a detailed understanding is missing of exactly how and through which molecular mechanisms the protein synthesis machinery in mitochondria is regulated. This will be an exciting area for future research.
Materials and methods
Yeast strains and growth media
All strains used in this study were isogenic to the wild-type strain W303-1B (Mat-α ade2-1 his3-11 leu2-3,112 trp1-1 ura3-1) with an intronless wild-type mitochondrial genome. The parental strain carrying engineered mitochondrial genomes had a nuclear arg8::HIS3 mutation. In the mitochondrial genome cob::ARG8m, the coding sequence of COB is replaced by a recoded version of ARG8 (termed ARG8m), leaving the 5′- and 3′-untranslated regions of the COB mRNA intact (Gruschke et al., 2011). Into this mitochondrial genome, a novel gene was inserted into a silent region of the DNA that consists of the promoter and 5′-untranslated portion of COX2, followed by the coding sequence of COB, which was again flanked by the 3′-untranslated portion and the terminator of COX2 to generate the cox2:COB cob::ARG8m mitochondrial genome (Gruschke et al., 2012). Yeast cultures were grown at 30°C in YP (1% yeast extract and 2% peptone) medium supplemented with 2% dextrose, 2% galactose, or 2% glycerol. To deplete heme, Δhem1 cells were grown in medium with TEM (0.5% Tween 80, 12 µg/ml ergosterol, and 55 µg/ml methionine); to allow heme protein biogenesis in Δhem1 cells, medium was supplemented with either 50 µg/ml δ-aminolevulinic acid or 4 µg/ml hemin (heme b with a chloride ligand), respectively.
Construction of cytochrome b heme mutant strains
The mutant Cytb-H183T was obtained by random mutagenesis as described previously (Meunier et al., 1993). The mutant Cytb-H197F was constructed by site-directed mutagenesis: The plasmid pBM5 carrying the wild-type intronless sequence of the COB gene was constructed by blunt-end cloning of a PCR product of COB into the pCRscript vector (Agilent Technologies). Mutagenesis was performed using the QuikChange site-directed mutagenesis kit (Agilent Technologies). After verification of the sequence, the plasmid carrying the mutated gene was used for biolistic transformation. Mitochondrial transformation by microprojectile bombardment and the identification of the mitochondrial transformants were performed as described previously (Meunier, 2001; Hill et al., 2003; Fisher et al., 2004). The mutated genes were then introduced into a rho+ mitochondrial genome via recombination, by mating the mitochondrial transformants (or synthetic rho−) with CKWT (Mat a leu1 kar1-1), carrying the wild-type intronless mitochondrial genome and the nuclear mutation kar1-1, which is required for cytoduction (Conde and Fink, 1976). Recombinant rho+ colonies with mutated COB were identified by crossing with tester strains. The mutated mitochondrial genome of these strains was then transferred into W303-1B/rho0. The resulting strain was used for analysis.
Construction of the COB-HApH strain
The COB-HApH mitochondrial genome that encodes a version of cytochrome b equipped with an HA and polyhistidine tag was constructed by A. Tzagoloff (Columbia University, New York, NY). To this end, a plasmid carrying the cytochrome b ORF plus an HA tag first was constructed. This was accomplished by amplifying 300 bp of the 5′-untranslated region plus the coding sequence and, separately, the sequence coding for the HA tag plus 300 bp of the 3′-untranslated region using the primer pairs 5′-GGCGGATCCGATATCATTAATATTAATATAATCGTC-3′/5′-GGCCTGCAGTTAAGCGTAGTCTGGGACGTCGTATGGGTATTTATTAACTCTACCGATATAG-3′ and 5′-GGCCTGCAGATTAAATTAATACATAGATATAATAT-3′/5′-GGGTCTAGAGATTCTATAATAATTATGCTTTATG-3′. DNA from the respiratory competent haploid strain MR6 (Rak et al., 2007) served as the template. Amplified DNA were digested with BamHI–PstI and PstI–XbaI, respectively, and ligated to the BamHI and XbaI sites of pJM2 (Steele et al., 1996). This plasmid (pCOB/ST6) was used for a PCR to generate the plasmid that carried the sequence COB-HApH using the primers 5′-GGCGGATCCGATATCATTAATATTAATATAATCGTC-3′/5′-GGCCTGCAGTTAGTGGTGATGATGGTGGTGAGCGTAGTCTGGGACGTCGTA-3′. The PCR product was ligated into pJM2 (Steele et al., 1996). Biolistic transformation (Bonnefoy and Fox, 2007) was used to introduce this plasmid, pCOB/ST7, into the kar1-1 strain αDFS160ρ0. Correct transformants could rescue the cox2 mutation of M9-94/A3 (Tzagoloff et al., 1975). One transformant (αDFS160/COB/ST7) was crossed to the respiratory-deficient mutant MRSI0ΔCOB that carries a cob::ARG8m mitochondrial genome (Gruschke et al., 2011). Cytoductants in which the coding sequence of ARG8m had been replaced by that of COB-HApH were identified by arginine auxotrophy and their ability to grow on media requiring respiratory growth.
Analysis of the interaction of cytochrome b with Cbp3 and Cbp4 by coimmunoprecipitation
Translation products of isolated mitochondria from wild-type cells or cytochrome b heme mutants (1 mg each) were labeled with [35S]-methionine as described previously (Hell et al., 2001). After the labeling was stopped by the addition of 10 mM unlabeled methionine and 80 µM puromycin, mitochondria were re-isolated and lysed for 30 min in 1 ml buffer containing 1% digitonin, 150 mM KCl, 20 mM Hepes/KOH, pH 7.4, 1 mM PMSF, 1× Complete protease inhibitor mix, and 10% glycerol. Cbp3 or Cbp4 was purified from these lysates by immunoprecipitation using serum against Cbp3, Cbp4, or preimmune serum (as a negative control) and protein A–Sepharose beads (Invitrogen). Beads were washed three times with lysis buffer containing 0.1% digitonin. Bound proteins were eluted with sample buffer. Samples were separated via SDS-PAGE and analyzed by autoradiography.
Analysis of bc1 complex activity
The activity of purified cytochrome bc1 complexes was measured at room temperature in 50 mM KCl, 2.5 mM MgCl2, 40 µM cytochrome c from bovine heart (Sigma-Aldrich), 20 µM KCN, 20 mM Hepes/KOH, pH 7.4, and 40 µM reduced decyl-quinol (Trumpower and Edwards, 1979). The reaction was started by adding the purified protein and reduction of cytochrome c was monitored at 550 nm. The activity of the bc1 complex was inhibited by the addition of 10 µM HQNO.
Heme extraction and HPLC analysis of hemes in cytochrome b
Complexes containing His-tagged cytochrome b or Cbp3, or a GFP-tagged Qcr7 were purified from digitonin, DDM, or SDS (all at 1%) mitochondrial lysates using Ni-NTA purification or immunoprecipitation using a GFP nanobody (ChromoTek). Hemes were extracted from the bound material of the washed beads with 0.14% HCl in acetone (Del Arenal et al., 1997; Weinstein and Beale, 1983) and clarified by centrifugation. The total heme composition of the supernatant was analyzed by reverse-phase HPLC. Hemes were separated and analyzed on an HPLC system (Agilent Technologies or Shimadzu Scientific Instruments) using a modified procedure (Lübben and Morand, 1994; Del Arenal et al., 1997; Brown et al., 2002). The solvents 0.1% TFA/H2O (buffer A) and 0.1% TFA/CH3-CN (buffer B) were degassed by ultrasonic treatment. The hemes were loaded at 0.5 ml/min onto a 150-mm YMC ODS-A column (5 µm, 300 Å) in 25% buffer B and resolved using a 1%/min gradient from 55% to 75% buffer B and detected at 400 nm. The isolated hemes were analyzed by UV/Vis spectroscopy.
Miscellaneous
Mitochondria were isolated from yeast spheroplasts broken with a Teflon potter, followed by differential centrifugation (Gruschke et al., 2011). For 2D BN/SDS-PAGE analyses, digitonin complexes were first separated on blue native–PAGE, then full lanes were excised and mounted on denaturing SDS-PAGE to resolve individual proteins according to their size (Gruschke et al., 2012). Mitochondrial translation products were labeled with [35S]-methionine either in isolated mitochondria or in whole cells where cytosolic protein synthesis was inhibited by cycloheximide (Prestele et al., 2009). Signals from autoradiography or Western blotting were quantified with ImageJ software (National Institutes of Health). All antibodies used were raised in rabbits with the following antigens: recombinantly expressed Arg8 (a gift from T. Fox, Cornell University, Ithaca, NY); a peptide representing amino acids 143–159 of yeast Rcf1 (a gift from P. Rehling, University of Göttingen, Göttingen, Germany); recombinantly expressed soluble domain of yeast Tom70 (a gift from D. Rapaport, Univeristy of Tübingen, Tübingen, Germany); cytochrome c1, Rip1, Cor1, and Cox2 purified from yeast mitochondria (a gift from W. Neupert, Max Planck Institute for Biochemistry, Munich, Germany); and recombinantly expressed mature Cbp3, MBP-Cbp6, Cbp4(61–171), MBP-Qcr7, MBP-Qcr8, and a peptide representing amino acids 204–226 of yeast cytochrome b (Gruschke et al., 2011, 2012).
Acknowledgments
We especially thank Alex Tzagoloff for sharing the COB-HApH strain and for many helpful suggestions. We thank Tom Fox, Peter Rehling, Doron Rapaport, and Walter Neupert for antibodies; Christoph von Ballmoos (Stockholm University, Sweden) for help with the bc1 complex activity assay; and John Wright (Michigan State University, East Lansing, MI) for helping with heme quantification.
This work was supported by the Swedish research council, the Center for Biomembrane Research at Stockholm University, the Carl Tryggers foundation, and the Knut and Alice Wallenberg Foundation (to M. Ott); and the National Institutes of Health (GM101386) to E.L. Hegg. The Wenner-Gren Foundation (Stockholm, Sweden) supported M. Hildenbeutel through a post-doctoral stipend and E.L. Hegg’s stay in Stockholm.
The authors declare no competing financial interests.
Footnotes
Abbreviations used in this paper:
- COB
- cytochrome b (gene or mRNA name)
- COX
- cytochrome oxidase
- DDM
- dodecyl maltoside
- TEM
- Tween 80, ergosterol, methionine
References
- Bernard D.G., Gabilly S.T., Dujardin G., Merchant S., Hamel P.P. 2003. Overlapping specificities of the mitochondrial cytochrome c and c1 heme lyases. J. Biol. Chem. 278:49732–49742 10.1074/jbc.M308881200 [DOI] [PubMed] [Google Scholar]
- Bernard D.G., Quevillon-Cheruel S., Merchant S., Guiard B., Hamel P.P. 2005. Cyc2p, a membrane-bound flavoprotein involved in the maturation of mitochondrial c-type cytochromes. J. Biol. Chem. 280:39852–39859 10.1074/jbc.M508574200 [DOI] [PubMed] [Google Scholar]
- Bonnefoy N., Fox T.D. 2007. Directed alteration of Saccharomyces cerevisiae mitochondrial DNA by biolistic transformation and homologous recombination. Methods Mol. Biol. 372:153–166 10.1007/978-1-59745-365-3_11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown K.R., Allan B.M., Do P., Hegg E.L. 2002. Identification of novel hemes generated by heme A synthase: evidence for two successive monooxygenase reactions. Biochemistry. 41:10906–10913 10.1021/bi0203536 [DOI] [PubMed] [Google Scholar]
- Conde J., Fink G.R. 1976. A mutant of Saccharomyces cerevisiae defective for nuclear fusion. Proc. Natl. Acad. Sci. USA. 73:3651–3655 10.1073/pnas.73.10.3651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corvest V., Murrey D.A., Bernard D.G., Knaff D.B., Guiard B., Hamel P.P. 2010. c-type cytochrome assembly in Saccharomyces cerevisiae: a key residue for apocytochrome c1/lyase interaction. Genetics. 186:561–571 10.1534/genetics.110.120022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crofts A.R. 2004. The cytochrome bc1 complex: function in the context of structure. Annu. Rev. Physiol. 66:689–733 10.1146/annurev.physiol.66.032102.150251 [DOI] [PubMed] [Google Scholar]
- Cruciat C.M., Brunner S., Baumann F., Neupert W., Stuart R.A. 2000. The cytochrome bc1 and cytochrome c oxidase complexes associate to form a single supracomplex in yeast mitochondria. J. Biol. Chem. 275:18093–18098 10.1074/jbc.M001901200 [DOI] [PubMed] [Google Scholar]
- Del Arenal I.P., Contreras M.L., Svlateorova B.B., Rangel P., Lledías F., Dávila J.R., Escamilla J.E. 1997. Haem O and a putative cytochrome bo in a mutant of Bacillus cereus impaired in the synthesis of haem A. Arch. Microbiol. 167:24–31 10.1007/s002030050412 [DOI] [PubMed] [Google Scholar]
- Dreher C., Prodöhl A., Hielscher R., Hellwig P., Schneider D. 2008. Multiple step assembly of the transmembrane cytochrome b6. J. Mol. Biol. 382:1057–1065 10.1016/j.jmb.2008.07.025 [DOI] [PubMed] [Google Scholar]
- Fisher N., Castleden C.K., Bourges I., Brasseur G., Dujardin G., Meunier B. 2004. Human disease-related mutations in cytochrome b studied in yeast. J. Biol. Chem. 279:12951–12958 10.1074/jbc.M313866200 [DOI] [PubMed] [Google Scholar]
- Fontanesi F. 2013. Mechanisms of mitochondrial translational regulation. IUBMB Life. 65:397–408 10.1002/iub.1156 [DOI] [PubMed] [Google Scholar]
- Fox T.D. 2012. Mitochondrial protein synthesis, import, and assembly. Genetics. 192:1203–1234 10.1534/genetics.112.141267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruschke S., Kehrein K., Römpler K., Gröne K., Israel L., Imhof A., Herrmann J.M., Ott M. 2011. Cbp3–Cbp6 interacts with the yeast mitochondrial ribosomal tunnel exit and promotes cytochrome b synthesis and assembly. J. Cell Biol. 193:1101–1114 10.1083/jcb.201103132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruschke S., Römpler K., Hildenbeutel M., Kehrein K., Kühl I., Bonnefoy N., Ott M. 2012. The Cbp3–Cbp6 complex coordinates cytochrome b synthesis with bc(1) complex assembly in yeast mitochondria. J. Cell Biol. 199:137–150 10.1083/jcb.201206040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hell K., Neupert W., Stuart R.A. 2001. Oxa1p acts as a general membrane insertion machinery for proteins encoded by mitochondrial DNA. EMBO J. 20:1281–1288 10.1093/emboj/20.6.1281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill P., Kessl J., Fisher N., Meshnick S., Trumpower B.L., Meunier B. 2003. Recapitulation in Saccharomyces cerevisiae of cytochrome b mutations conferring resistance to atovaquone in Pneumocystis jiroveci. Antimicrob. Agents Chemother. 47:2725–2731 10.1128/AAC.47.9.2725-2731.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunte C., Koepke J., Lange C., Rossmanith T., Michel H. 2000. Structure at 2.3 A resolution of the cytochrome bc(1) complex from the yeast Saccharomyces cerevisiae co-crystallized with an antibody Fv fragment. Structure. 8:669–684 10.1016/S0969-2126(00)00152-0 [DOI] [PubMed] [Google Scholar]
- Kim H.J., Khalimonchuk O., Smith P.M., Winge D.R. 2012. Structure, function, and assembly of heme centers in mitochondrial respiratory complexes. Biochim. Biophys. Acta. 1823:1604–1616 10.1016/j.bbamcr.2012.04.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kranz R.G., Richard-Fogal C., Taylor J.S., Frawley E.R. 2009. Cytochrome c biogenesis: mechanisms for covalent modifications and trafficking of heme and for heme-iron redox control. Microbiol. Mol. Biol. Rev. 73:510–528 10.1128/MMBR.00001-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuras R., de Vitry C., Choquet Y., Girard-Bascou J., Culler D., Büschlen S., Merchant S., Wollman F.A. 1997. Molecular genetic identification of a pathway for heme binding to cytochrome b6. J. Biol. Chem. 272:32427–32435 10.1074/jbc.272.51.32427 [DOI] [PubMed] [Google Scholar]
- Lill R. 2009. Function and biogenesis of iron-sulphur proteins. Nature. 460:831–838 10.1038/nature08301 [DOI] [PubMed] [Google Scholar]
- Lübben M., Morand K. 1994. Novel prenylated hemes as cofactors of cytochrome oxidases. Archaea have modified hemes A and O. J. Biol. Chem. 269:21473–21479 [PubMed] [Google Scholar]
- Meunier B. 2001. Site-directed mutations in the mitochondrially encoded subunits I and III of yeast cytochrome oxidase. Biochem. J. 354:407–412 10.1042/0264-6021:3540407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meunier B., Lemarre P., Colson A.M. 1993. Genetic screening in Saccharomyces cerevisiae for large numbers of mitochondrial point mutations which affect structure and function of catalytic subunits of cytochrome-c oxidase. Eur. J. Biochem. 213:129–135 10.1111/j.1432-1033.1993.tb17742.x [DOI] [PubMed] [Google Scholar]
- Mick D.U., Fox T.D., Rehling P. 2011. Inventory control: cytochrome c oxidase assembly regulates mitochondrial translation. Nat. Rev. Mol. Cell Biol. 12:14–20 10.1038/nrm3029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell P. 1975. Protonmotive redox mechanism of the cytochrome b-c1 complex in the respiratory chain: protonmotive ubiquinone cycle. FEBS Lett. 56:1–6 10.1016/0014-5793(75)80098-6 [DOI] [PubMed] [Google Scholar]
- Osyczka A., Moser C.C., Dutton P.L. 2005. Fixing the Q cycle. Trends Biochem. Sci. 30:176–182 10.1016/j.tibs.2005.02.001 [DOI] [PubMed] [Google Scholar]
- Prestele M., Vogel F., Reichert A.S., Herrmann J.M., Ott M. 2009. Mrpl36 is important for generation of assembly competent proteins during mitochondrial translation. Mol. Biol. Cell. 20:2615–2625 10.1091/mbc.E08-12-1162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rak M., Tetaud E., Duvezin-Caubet S., Ezkurdia N., Bietenhader M., Rytka J., di Rago J.P. 2007. A yeast model of the neurogenic ataxia retinitis pigmentosa (NARP) T8993G mutation in the mitochondrial ATP synthase-6 gene. J. Biol. Chem. 282:34039–34047 10.1074/jbc.M703053200 [DOI] [PubMed] [Google Scholar]
- Smith P.M., Fox J.L., Winge D.R. 2012. Biogenesis of the cytochrome bc(1) complex and role of assembly factors. Biochim. Biophys. Acta. 1817:276–286 10.1016/j.bbabio.2011.11.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soto I.C., Fontanesi F., Myers R.S., Hamel P., Barrientos A. 2012. A heme-sensing mechanism in the translational regulation of mitochondrial cytochrome c oxidase biogenesis. Cell Metab. 16:801–813 10.1016/j.cmet.2012.10.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steele D.F., Butler C.A., Fox T.D. 1996. Expression of a recoded nuclear gene inserted into yeast mitochondrial DNA is limited by mRNA-specific translational activation. Proc. Natl. Acad. Sci. USA. 93:5253–5257 10.1073/pnas.93.11.5253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiner H., Kispal G., Zollner A., Haid A., Neupert W., Lill R. 1996. Heme binding to a conserved Cys-Pro-Val motif is crucial for the catalytic function of mitochondrial heme lyases. J. Biol. Chem. 271:32605–32611 10.1074/jbc.271.51.32605 [DOI] [PubMed] [Google Scholar]
- Trumpower B.L., Edwards C.A. 1979. Purification of a reconstitutively active iron-sulfur protein (oxidation factor) from succinate. cytochrome c reductase complex of bovine heart mitochondria. J. Biol. Chem. 254:8697–8706 [PubMed] [Google Scholar]
- Tucker E.J., Wanschers B.F., Szklarczyk R., Mountford H.S., Wijeyeratne X.W., van den Brand M.A., Leenders A.M., Rodenburg R.J., Reljić B., Compton A.G., et al. 2013. Mutations in the UQCC1-interacting protein, UQCC2, cause human complex III deficiency associated with perturbed cytochrome b protein expression. PLoS Genet. 9:e1004034 10.1371/journal.pgen.1004034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzagoloff A., Akai A., Needleman R.B. 1975. Assembly of the mitochondrial membrane system. Characterization of nuclear mutants of Saccharomyces cerevisiae with defects in mitochondrial ATPase and respiratory enzymes. J. Biol. Chem. 250:8228–8235 [PubMed] [Google Scholar]
- Vukotic M., Oeljeklaus S., Wiese S., Vögtle F.N., Meisinger C., Meyer H.E., Zieseniss A., Katschinski D.M., Jans D.C., Jakobs S., et al. 2012. Rcf1 mediates cytochrome oxidase assembly and respirasome formation, revealing heterogeneity of the enzyme complex. Cell Metab. 15:336–347 10.1016/j.cmet.2012.01.016 [DOI] [PubMed] [Google Scholar]
- Weinstein J.D., Beale S.I. 1983. Separate physiological roles and subcellular compartments for two tetrapyrrole biosynthetic pathways in Euglena gracilis. J. Biol. Chem. 258:6799–6807 [PubMed] [Google Scholar]
- Yun C.H., Crofts A.R., Gennis R.B. 1991. Assignment of the histidine axial ligands to the cytochrome bH and cytochrome bL components of the bc1 complex from Rhodobacter sphaeroides by site-directed mutagenesis. Biochemistry. 30:6747–6754 10.1021/bi00241a017 [DOI] [PubMed] [Google Scholar]