Abstract
Following absorption, polychlorinated biphenyls (PCBs) bind to albumin and are transported via blood into the target tissues. PCBs then accumulate in tissues and induce a variety of harmful chronic and developmental effects. The aim of the present study is to determine binding parameters, such as binding constant, quenching constant, and number of binding sites for three PCB congeners (PCB118, PCB126 and PCB153) in complex with human serum albumin (HSA). The binding parameters for the complexes of HSA-PCB118, HSA-PCB126, and HSA-PCB153 excited at 280 nm were compared with those excited at 295 nm. Quenching (static and dynamic) of HSA fluorescence was analyzed based on the Stern-Volmer method. Binding (Ka) constants were calculated according to the Scatchard method and analysis of non-linear regression was based on a two-component model with the Lavenberg–Marquardt algorithm. For all analyzed complexes, a single independent class of binding site for PCB congeners was found in HSA subdomain IIA. Tyrosine residues appear to play the most prominent role in binding of PCB126 to HSA, while tryptophan-214 played a dominant role in interactions of PCB153 with HSA. Among studied PCB congeners, PCB118 formed the most stable complexes with HSA. These results illustrate the importance of studies targeting the binding of PCBs to serum albumin as part of the strategy to understand and protect against toxicity of these environmental toxicants.
Keywords: PCB, albumin, fluorescence
1. Introduction
Polychlorinated biphenyls (PCBs) are environmental toxicants known to induce several acute and chronic effects contributing to the development of a variety of disorders and developmental changes. PCBs were produced and used for a range of industrial applications, such as insulators, engines, transformers, and other devices, which required resistance to high temperatures. PCBs were also utilized as components of paint, glue, ink, and copy papers. Although PCB production has been banned for over 30 years, disposal of these products results in PCB leaking into the environment with the subsequent pollution of soil, food, water sediments, and air. Due to their lipophilic properties and resistance to biodegradation, PCBs are bioconcentrated at each level of the food chain. Thus, both environmental and occupational sources contribute to the total body burden of PCBs in humans [1, 2, 3].
Based on their chemical structure and affinity to specific cellular receptors, PCBs are classified as coplanar or non-coplanar PCBs. PCB congeners belonging to these classes differ in the number and position of chlorines around the biphenyl rings and have distinct cellular properties. Coplanar PCBs, also called dioxin-like PCBs, are characterized by the lack of any chlorine substitution at the ortho-positions. These PCBs adopt a coplanar structure and interact with the aryl hydrocarbon receptor (AhR). Non-coplanar (or non dioxin-like) PCBs have at least two chlorine substitutions at the ortho-positions and are non-coplanar due to steric hindrance. They exert their toxic effects via mechanisms independent on the AhR binding [4, 5]. Mono-ortho PCB congeners are a separate class of PCBs, which have one chlorine substitution at the ortho-position and partially preserve both dioxin-like and non-dioxin-like properties.
To establish a structure-function relationship of PCB-mediated binding to serum albumin, representative coplanar and non-coplanar PCB congeners were used in the present study. PCB126 was employed as a coplanar PCB due to the highest toxic equivalency factor (TEF) amongst the PCBs [6]. PCB118 is a mono-ortho PCB congeners frequently found in the environment [7] and PCB153 is as a major non-coplanar PCB, which is the most prevalent PCB congener both in the environment and biological tissues [2, 4, 5]. PCBs can be absorbed via skin, inhaled, and/or taken up with contaminated food. Following absorption, they bind to serum albumin and are distributed via blood into tissues [8]. In fact, approximately 44% of PCBs present in human blood plasma were shown to be distributed in lipoprotein depleted fractions containing primarily albumin [9].
Human serum albumin (HSA) consists of 585 amino acids. It resembles an equilateral triangle with the length of the sides of ~80 Å and a depth of 30 Å [10]. The amino acid sequence contains 17 disulfide bridges, one tryptophanyl residue (Trp-214) and one thiol group (Cys-34). HSA consists of three domains, called I, II, and III. Each of these domains contains two smaller subdomains, A and B [11,12,13,14]. Albumins are in involved in several critical functions as blood components. For example, they regulate constant osmotic pressure in serum, are responsible for maintaining osmotic balance between the blood and intercellular space, and maintain fluids in vessels. Importantly, albumins bind and carry a variety of endogenous and exogenous compounds. Albumins transfer substances which do not have specific transport proteins or substances at the concentrations that exceed the capacity of specific transporters. Sudlow et al. described two sites used by albumins to bind substances: site I, where warfarin is bound, and site II utilized for benzodiazepine binding [15]. In addition, Carter et al. described six sites of structurally different ligand binding: two sites binding small heterocyclic or aromatic carboxylic acids, two sites binding long-chain fatty acids and small amino compounds, and two sites binding metal ions. It was determined that site I is localized in a hydrophobic packet in subdomain IIA, and site II is a packet in subdomain IIIA. The binding sites of fatty acids were named sites III and IV, localized in subdomains IB and IIIB [16]. Because interactions with albumins are a critical step which determines the fate and distribution of xenobiotics in living organisms, the goal of the present study was to characterize the binding of specific PCB congeners to HSA using fluorescence spectroscopy.
2. Experimental
2.1. Materials and experimental conditions
Human serum albumin fraction V (HSA, Cat. no 823022, Lot no 8234H), was purchased from MP Biomedicals (Solon, OH) and bovine albumin fraction V (BSA, Cat. no: A4503) from Sigma-Aldrich (St. Louis, MO). PCB153 (2,2’4,4’5,5’-hexachlorobiphenyl), PCB118 (2,3’,4,4’,5-pentachlorobiphenyl), PCB126 (3,3’,4,4’,5-pentachlorobiphenyl), all > 99% pure, were obtained from AccuStandard (New Haven, CT). 8-anilino-1-naphtalenesulfonic acid (ANS, Lot 70K345) was from Sigma-Aldrich.
HSA solutions were prepared in TRIS-HCl, pH 7.42 and PCBs were dissolved by mixing 1.8 mg of individual PCBs with 1 ml DMSO. The final concentration of HSA was 2×10-5 M and PCB congeners 5×10-3 M. To generate HSA-PCB complexes, individual PCB congeners were added to HSA in 3 μl aliquots. Depending on the experiment, the molar ratio of HSA to PCB was between 0.5 and 3.5. ANS solution (final concentration 100 mM) was prepared by adding 3 mg ANS to 0.1 ml of 100 mM NaOH and 0.9 ml of TRIS-HCl, pH 7.42.
2.2. Fluorescence emission spectra
Fluorescence emission spectra were recorded on Hitachi Fluorescence Spectrophotometer FL-2500 with 1 cm × 1 cm × 4 cm quartz cells 60 min after preparation of individual solutions and displayed in terms of relative fluorescence (RF). Correcting error of apparatus for wavelength (λ) was equal to ± 1 and for relative fluorescence (RF) ± 0.01. Two excitation wavelengths (λex 280 nm and λex 295 nm) were used to excite fluorophores of HSA. The highest absorbance values were ≤ 0.05; therefore, the spectra were not corrected for the inner filter effects [17].
Quenching of HSA fluorescence was analyzed based on the Stern-Volmer equation (3), which describes the ligand movement within the fluorophore microenvironment when dynamic quenching occurs [18]:
| (equation 1) |
where, RFo and RF are the fluorescence values in the absence and presence of the quencher, respectively; KSV is the dynamic quenching constant; V is the static quenching constant; and [Q] is the quencher (PCB) concentration in the binary systems of HSA-PCB.
Binding (association) constants (Ka) were calculated by using the Scatchard method modified by Hiratsuka [19]:
| (equation 2) |
where, [Lb] is the bound PCB concentration, [Lf] is free PCB concentration, [HSA] is serum albumin concentration, Ka is the binding constant, n is the number of binding sites for the independent class of PCB binding sites in albumin molecule, which corresponds to the mean number of PCB molecules bound to the independent class of PCB binding sites in albumin molecule.
The Ka values were also calculated using non-linear regression based on a two-component Scatchard model with the Lavenberg–Marquardt algorithm:
| (equation 3) |
where m = number of classes of independent binding sites, each class I having ni sites with intrinsic binding constant Kai.
2.3. Visualization of the PCB binding site on HSA molecule
Complexes of PCB153 with HSA were visualized using Molegro Virtual Docker (MVD Version 6.0.0. (June 12th, 2012)). The crystal structure of HSA was downloaded from the Protein Data Bank (1AO6). Structure of PCB153 with the lowest energy was determined using the ChemBio3D ultra.
3. Results and discussion
3.1. PCBs interact with HSA via Trp-214 and tyrosine residues
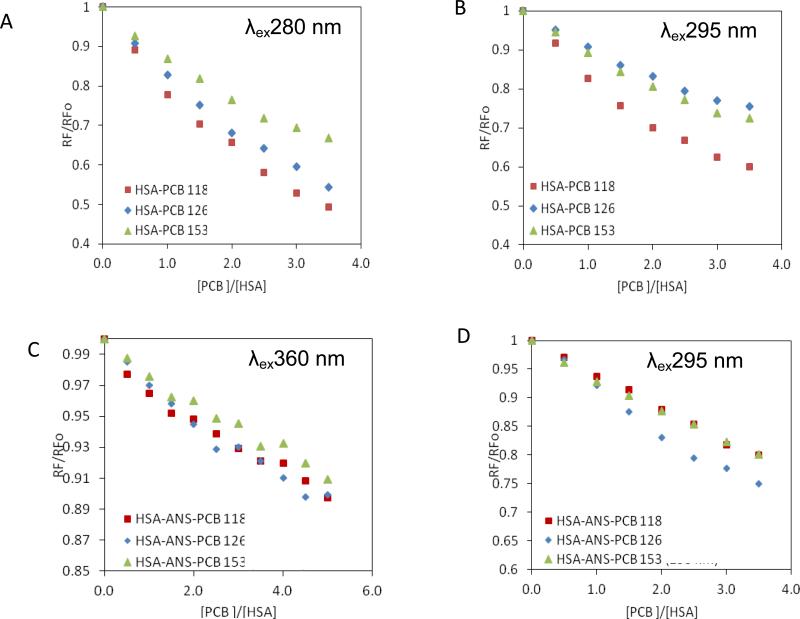
In order to evaluate the interactions of individual PCB congeners with HSA and to determine their binding sites within the tertiary structure of albumin, fluorescence emission spectra were analyzed for the complexes of HSA with PCB118, PCB126, and PCB153 (Fig. 1). Two excitation wavelengths were used, λex280 and λex295 nm. λex280 nm induces excitation of tryptophanyl (Trp) and tyrosil (Tyr) residues, while λex295 nm excites with high specificity Trp residues. Because HSA molecule contains only a single Trp residue present at the position 214, this residue (namely, Trp-214) is responsible for fluorescence emitted after excitation at λex295 nm.
Figure 1.
The quenching curves of the HSA-PCB118 (□), HSA-PCB126 (◇) and HSAPCB153 (Δ) complexes, excited at λex280 nm (A) and λex295 nm (B). The quenching curves of the HSA-PCB118 (□), HSA-PCB126 (◇) and HSA-PCB153 (Δ) complexes in the presence of ANS excited at λex360 nm (C) and λex295 (D).
Fluorescence quenching of proteins occurs when a ligand (a quencher; i.e., a PCB congener in our experimental setting) is present in a close proximity to HSA fluorophores, i.e. the Trp and Tyr residues, allowing for energy transfer between a ligand and a fluorophore [20]. Thus, fluorescence quenching observed in Fig. 1 indicates the formation of complexes between HSA and individual PCB congeners [21]. These effects were observed for the HSA-PCB complexes excited both at λex280 nm (Fig. 1A) and λex295 nm (Fig. 1B). PCB-mediated quenching of HSA fluorescence was dose-depended at the molar ratios of the individual PCB congeners to HSA from 0.5 to 3.5.
For all studied PCB congeners, quenching of HSA fluorescence was stronger at λex280 nm as compared to λex295 nm (Fig. 1A vs. Fig. 1B). For example, PCB118 at the 3.5:1 molar ratio to HSA decreased HSA fluorescence by 51% at λex280 nm as compared to 40% at λex295 nm. This effect was even more pronounced for PCB126, which (at the same molar ratio PCB to HSA) decreased HSA fluorescence by 45.7% at λex280 nm but only by 24.5% at λex295 nm. PCB153 appeared to interact with the HSA at a lower degree as compared to PCB118 or PCB126. In fact, PCB153 decreased HSA fluorescence by 33% at λex280 nm and by 27.5% at λex295 nm. The values of HSA fluorescence quenching also reflect the relative strength of PCB association with albumin. Thus, among studied PCBs, PCB118 interacted with HSA at the highest strength, followed by PCB126 and PCB153.
Because HSA fluorescence quenching at λex280 nm is caused by interaction with both Trp-214 and Tyr residues and quenching at λex295 nm is mediated only by interactions with Trp-214, these results indicate that both types of amino acid residues are involved in binding PCB congeners to the HSA molecule. The relative role of Tyr residues in binding to HSA was then quantified by subtracting the fluorescent values at λex295 nm from those at λex280 nm. Using this approach, we determined that Tyr residues appear to play the most prominent role in PCB126 binding to HSA (Δ λex280 - λex295 = 21.2%) but much lower in PCB118 or PCB153 binding (Δ = 11% and 5.66%, respectively). These differences correspond well to the differences in structural properties of individual PCB congeners. PCB126 is a coplanar PCB without ortho substitutes; PCB118 has one chlorine substitute in the ortho position, while PCB153 has two ortho chlorine substitutes. Thus, it appears that coplanar structure of PCBs favors interactions with Tyr residues, while ortho-substitutes target binding of PCBs almost exclusively to Trp-214.
3.2. Subdomain IIIA is not involved in HSA-PCB interactions
Subdomain IIIA of the HSA molecule is characterized by hydrophobic properties and contains several Tyr residues. Therefore, the interactions of PCB congeners with this subdomain were evaluated in the next series of experiments by employing ANS (Fig. 1C,D), which binds with high specificity to subdomain IIIA when used at the 1:1 molar ratio to HSA. ANS has low fluorescent properties in water solutions; however, it emits strong fluorescence in non-polar solutes or after binding with proteins.
When fluorescence of the HSA-ANS complex was induced at λex360 nm, adding increasing concentrations of individual PCB congeners did not affect the fluorescence values (Fig 1C), indicating that PCBs do not compete with ANS for its binding site. This finding is important because it indicates that PCB congeners evaluated in the present study do not have affinity to the subdomains IIIA. Thus, binding to Tyr residues detected in experiments shown on Fig 1A,B occurs only in subdomain IIA.
Fluorescence of the ANS-HSA complex induced at λex295 nm results from energy transfer from Trp-214 to ANS. When λex295 nm was employed, increasing concentrations of PCBs diminished HSA fluorescence by ~20-25% (Fig. 1D), indicating that PCBs can compete with ANS for Trp-214 binding site. These data further confirm the role of Trp-214 in interactions of PCBs with the HSA molecule.
3.3. Dynamic and static quenching of HSA fluorescence by individual PCB congeners
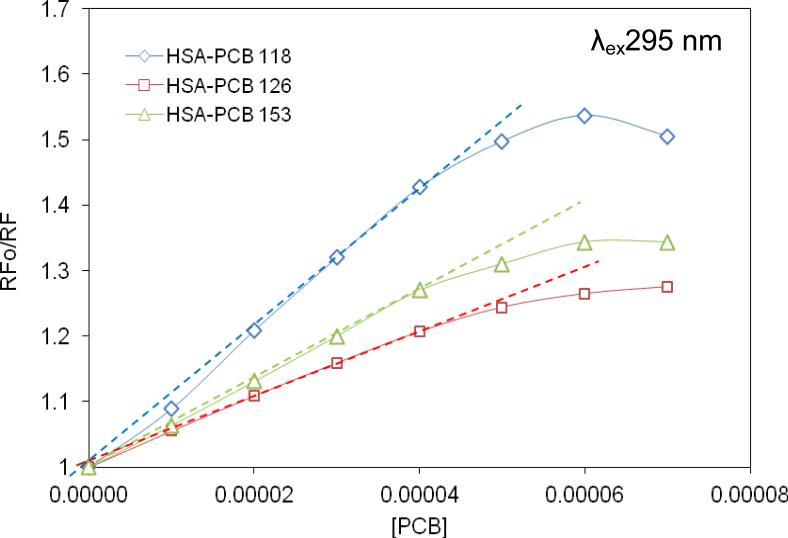
We next analyzed PCB-mediated fluorescence quenching of HSA based on the Stern-Volmer equation model. A linear Stern-Volmer curve indicates dynamic or static quenching, while deviations from linearity point to coexistence of both the dynamic and passive quenching [19]. The Stern-Volmer curves were linear for all studied PCB congeners when used at the molar ratio of PCB-HSA ≤ 2:1 (Fig. 2). Because these analyses were performed at λex295 nm, the results indicate that interference with the Trp-214 residue is involved in PCB- mediated dynamic quenching of HSA fluorescence. At higher molar ratios to HSA, PCBs appear to saturate this binding site, resulting in decreased ability to quench HSA fluorescence, a phenomenon which is reflected by negative deviations.
Figure 2.
The non-modified Stern-Volmer curves for HSA-PCB118 (◇), HSA-PCB126 (□), and HSA-PCB153 (Δ) complexes. HSA concentration was 2 × 10-5 M, λex295 nm.
The Stern-Volmer curves illustrated in Fig. 2 are characterized by different slopes, with the lowest inclination for the complexes of HSA with PCB126. These results are consistent with the more prominent role of the Tyr residues in PCB126 association with HSA as compared to other PCB congeners
Based on the exponential equation (equation 1) and non-linear regression curves, we next determined the dynamic (KSV) and static (V) quenching constants for the HSA-PCB118, HSA-PCB126, and HSA-PCB153 complexes (Table 1). The obtained values were similar for all studied PCB congeners and were in the range of 103 [M-1]. The only exception was ~2 fold higher KSV for the HSA-PCB153 complex at λex280 nm as compared to other PCB congeners. These results are consistent with those illustrated in Fig. 1 further supporting the finding that the Tyr residues play less important role in binding of PCB153 to HSA as compared to coplanar PCB126.
Table 1.
Dynamic (KSV) and static (V) quenching constants [M−1] determined for HSA-PCB118, HSA-PCB126, and HSA-PCB153 complexes at λex 295 and λex 280 nm.
| λ ex | Complex | KSV [M−1] | V [M−1] |
|---|---|---|---|
| 295 nm | HSA-PCB118 | 13.43 ×103 | 2.11×103 |
| HSA-PCB126 | 11.66 ×103 | 2.93×103 | |
| HSA-PCB153 | 12.33×103 | 5.36×103 | |
| 280 nm | HSA-PCB118 | 6.01×103 | 5.54×103 |
| HSA-PCB126 | 6.37×103 | 3.70×103 | |
| HSA-PCB153 | 11.78×103 | 4.15×103 | |
3.4. Affinity assessment of PCB-HSA interactions
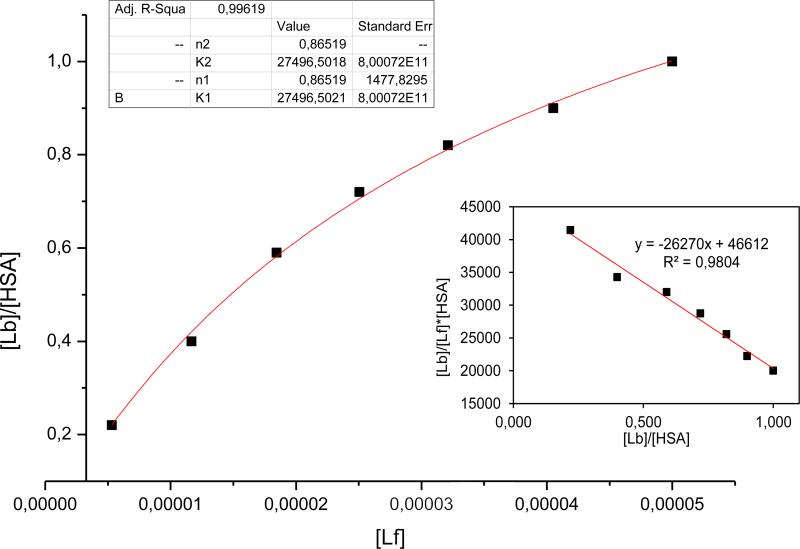
Analysis of the Scatchard equation (equation 2) was used for the assessment of the affinity constant Ka of the binding of individual PCB congeners to HSA. The Scatchard curve prepared for the HSA-PCB153 complex at λex280 nm (Fig. 3, insert) indicated a linear relationship, which is typical for a ligand binding to one class of the binding sites. Similar results were obtained for the complexes of HSA with PCB118 or PCB126 (data not shown). This relationship was then confirmed by assessing the isotherms based on the analysis of non-linear regression (equation 3) (Fig. 3).
Figure 3.
The binding isotherm for the HSA-PCB153 complex excited at λex 280 nm. The insert reflects the Scatchard plot for the same complex. Abbreviations: Lf, ligand free; Lb, ligand bound.
The affinity constant for the individual HSA-PCB complexes are provided in Table 2. The results indicate that PCB118, PCB126, and PCB153 can form weak bonds with HSA as determined by the Ka values in the range of 104. Although the exact character of these associations remains unknown, one can suggest the involvement of the π-π interactions between the biphenyl rings of PCBs with aromatic amino acids of the HSA molecule. Such interactions are characteristic not only for PCBs but several other xenobiotics, including prescribed drugs. While interactions between PCBs and HSA allow for PCBs distribution via the bloodstream, their relatively weakness facilitates separation of PCBs from HSA, followed by tissue and cellular uptake. The differences in binding to HSA may influence the differences in tissue distribution and toxicity of individual PCB congeners. Based on the Scatchard equation, we also determined the number of PCB molecules binding to the independent class of binding sites present on the HSA molecule. These values were assessed between 1.31-1.89 at λex280 nm and below 1 at λex295 nm.
Table 2.
Binding constants Ka [M−1] and mean number of PCB molecules bound to the independent class of HSA binding sites (n) determined for HSA-PCB complexes using the Scatchard method and non-linear regression model, λex 295 and λex 280 nm.
| λ ex | Complex | Ka (Scatchard) [M−1] | n | Ka (non-linear regression) [M−1] | n |
|---|---|---|---|---|---|
| 295 nm | HSA-PCB118 | 2.150×104 | 1.89 | 2.182×104 | 0.70 |
| HSA-PCB126 | 1.744 ×104 | 1.33 | 1.998×104 | 0.99 | |
| HSA-PCB153 | 1.511×104 | 1.47 | 1.652×104 | 0.88 | |
| 280 nm | HSA-PCB118 | 4.730 ×104 | 1.52 | 4.922×104 | 0.72 |
| HSA-PCB126 | 6.291 ×104 | 1.31 | 7.180×104 | 0.79 | |
| HSA-PCB153 | 2.627 ×104 | 1.77 | 2.749×104 | 0.86 | |
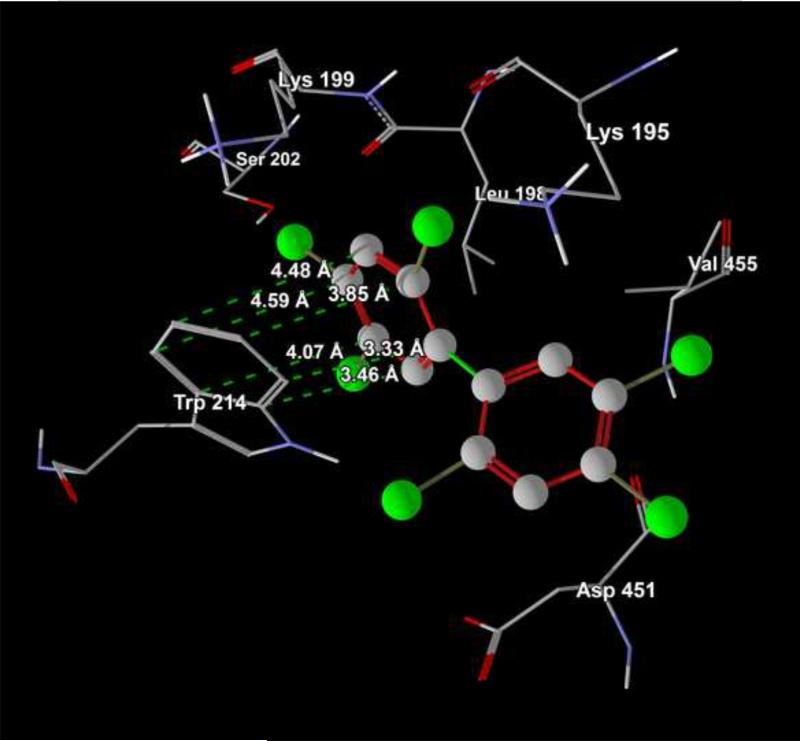
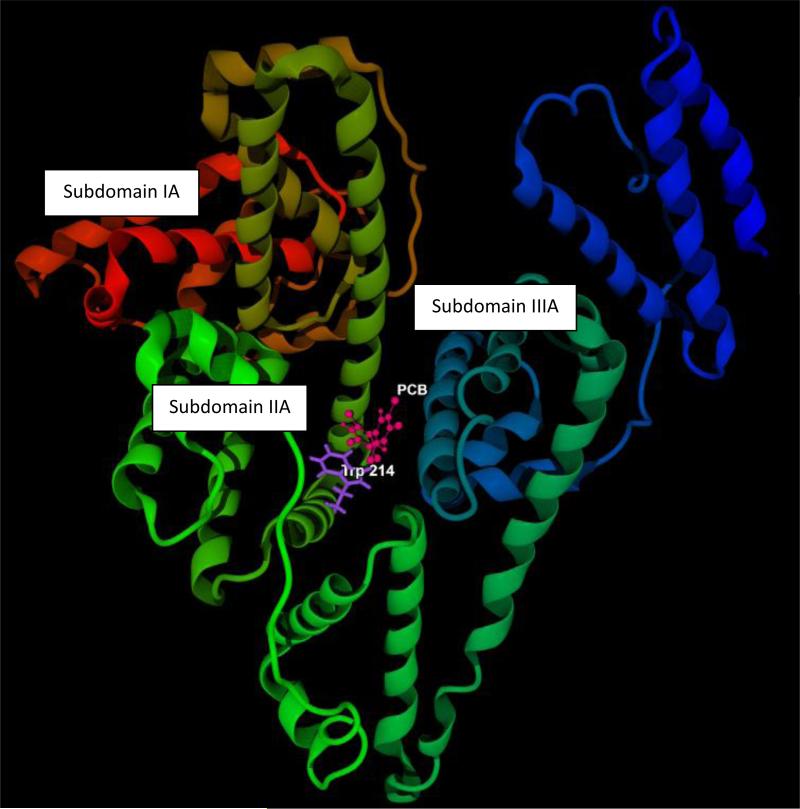
3.5. Modeling of PCB153 interactions with HSA
Computer modeling using was employed to verify the major findings of the present study. PCB153 was used as a model compounds in these analyses. Fig 4A confirms that binding site of PCB153 in HSA molecule is located in subdomain IIA, close (the distance is from 3.33 to 4.59 A) to Trp-214. Fig 4B shows that PCB153 does not interact with HSA via H-bond(s), favoring weak hydrophobic and/or steric interactions with amino acids located in subdomain IIA, e.g. Trp-214.
Figure 4.
Computer visualization of the PCB153 binding site on the HSA molecule (A), Visualization of PCB153 interactions with Trp-214 on the HSA molecule (B).
4. Conclusions
Due to their hydrophobic properties, PCBs interact with HSA, which serves as a carrier to distribute PCBs into tissues via the bloodstream. Within the HSA molecule, PCBs bind preferentially to Trp-214 and the Tyr residues located in the IIA subdomain. Interactions with Trp-214 were most prominent for ortho-substituted PCB153, while binding to the Tyr residues appeared to play the most important role for coplanar PCB126. These findings based on fluorescence spectroscopy analyses were confirmed using computer modeling for binding of PCB153 with HSA. Interactions within the PCB-HSA complexes are weak, suggesting that PCBs can easily separate from HSA and be taken up by cells, where they can induce cellular and systemic toxicity. Overall, these results illustrate the importance of studies targeting the binding of individual PCB congeners to HSA as part of the strategy to protect against toxicity of these environmental toxicants.
Acknowledgements
This work was supported by the grant from Medical University of Silesia: KNW-1-001/K/3/0 and from the National Institutes of Health P42 ES 07380 and R01 CA133257.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Longnecker MP, Rogan WJ, Lucier G. The human health effects of DDT (dichlorodiphenyltrichloroethane) and PCBS (polychlorinated biphenyls) and an overview of organochlorines in public health. Ann. Rev. Public Health. 1997;18:211. doi: 10.1146/annurev.publhealth.18.1.211. [DOI] [PubMed] [Google Scholar]
- 2.Crinnion WJ. Polychlorinated biphenyls: persistent pollutants with immunological, neurological, and endocrinological consequences. Altern. Med. Rev. 2011;16:5. [PubMed] [Google Scholar]
- 3.Robertson LW, Ludewig G. Polychlorinated Biphenyl (PCB) carcinogenicity with special emphasis on airborne PCBs. Gefahrst. Reinhalt. Luft. 2011;71:25. [PMC free article] [PubMed] [Google Scholar]
- 4.Kimbrough RD. Polychlorinated biphenyls (PCBs) and human health: an update. Crit. Rev. Toxicol. 1995;25:133. doi: 10.3109/10408449509021611. [DOI] [PubMed] [Google Scholar]
- 5.Hansen LG. The ortho side of PCBs: Occurrence and disposition. Kluwer Academic Publishers; Boston: 1999. [Google Scholar]
- 6.Bhavsar SP, Reiner EJ, Hayton A, Fletcher R, MacPherson K. Converting Toxic Equivalents (TEQ) of dioxins and dioxin-like compounds in fish from one Toxic Equivalency Factor (TEF) scheme to another. Environ Int. 2008;34:915–921. doi: 10.1016/j.envint.2008.02.001. [DOI] [PubMed] [Google Scholar]
- 7.McFarland VA, Clarke JU. Environmental occurrence, abundance, and potential toxicity of polychlorinated biphenyl congeners: considerations for a congener-specific analysis. Environ Health Perspect. 1989;81:225–239. doi: 10.1289/ehp.8981225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lotti M. Pharmacokinetics and blood levels of polychlorinated biphenyls. Toxicol. Rev. 2003;22:203. doi: 10.2165/00139709-200322040-00003. [DOI] [PubMed] [Google Scholar]
- 9.Norén K, Weistrand C, Karpe F. Distribution of PCB congeners, DDE, hexachlorobenzene, and methylsulfonyl metabolites of PCB and DDE among various fractions of human blood plasma. Arch. Environ. Contam. Toxicol. 1999;37:408. doi: 10.1007/s002449900532. [DOI] [PubMed] [Google Scholar]
- 10.Carter DC, Ho JX. Structure of serum albumin. Adv. Protein Chem. 1994;45:153–203. doi: 10.1016/s0065-3233(08)60640-3. [DOI] [PubMed] [Google Scholar]
- 11.Brown JR. Structural origins of albumin. Feder. Proc. 1976;35:2141. [PubMed] [Google Scholar]
- 12.Trivedi V, Saxena J. Interaction of bromocresol green with different serum albumins studied by fluorescence quenching. Biochem. Mol. Biol. Int. 1997;43:1. doi: 10.1080/15216549700203751. [DOI] [PubMed] [Google Scholar]
- 13.Carter DC, He XM. Structure of serum albumin. Science. 1990;249:302. doi: 10.1126/science.2374930. [DOI] [PubMed] [Google Scholar]
- 14.Curry S, Mandelkow H. Crystal structure of human serum albumin complexed with fatty acids reveals an asymmetric distribution of binding sites. Nat. Struct. Biol. 1998;9:827. doi: 10.1038/1869. [DOI] [PubMed] [Google Scholar]
- 15.Sudlow G, Birkett D, Wade DN. The characterization of two specific drug binding sites on human serum albumin. Mol. Pharmacol. 1975;11:824. [PubMed] [Google Scholar]
- 16.Peters T. Serum albumin, Jr. Adv. Protein Chem. 1985;37:161. doi: 10.1016/s0065-3233(08)60065-0. [DOI] [PubMed] [Google Scholar]
- 17.Steinem RF, Weinryb I. Fluorescence instrumentation and Methodology. Inc. Maryland: 1970. p. 39. [Google Scholar]
- 18.Eftink MR, Ghiron CA. Exposure of tryptophanyl residues in proteins. Quantitative determination by fluorescence quenching studies. Biochem. 1976;15:672. doi: 10.1021/bi00648a035. [DOI] [PubMed] [Google Scholar]
- 19.Hiratsuka T. Conformational changes in the 23-kilodalton NH2-terminal peptide segment of myosin ATPase associated with ATP hydrolysis. J. Biol. Chem. 1990;265:18786. [PubMed] [Google Scholar]
- 20.Valeur B, Berberan-Santos M. Molecular Fluorescence Principles and Applications. 2nd edition Willey/VCH; Germany: 2012. [Google Scholar]
- 21.Lakowicz JR. Principles of Fluorescence Spectroscopy. 3rd edition USA: 2006. [Google Scholar]