Summary
The major urinary protein (MUP) family members contain a conserved β-barrel structure with a characteristic central hydrophobic pocket. They are secreted by the liver and excreted into the urine. MUPs bind via their central pockets to volatile pheromones or other lipophilic molecules, and regulate pheromone transportation in the circulation, excretion in the kidney, and release into the air from urine marks. MUPs are highly polymorphic, and the MUP profiles in urine function as individual identity signatures of the owners. The MUP signatures are detected by the main and accessory olfactory systems and trigger adaptive behavioral responses and/or developmental processes. Circulating MUPs serve as a metabolic signal to regulate glucose and lipid metabolism. Recombinant MUP1 markedly ameliorates hyperglycemia and glucose intolerance in mice with type 2 diabetes. MUP1 suppresses hepatic gluconeogenesis and promotes energy expenditure in skeletal muscle by stimulating mitochondrial biogenesis and function. MUPs are unique members of the lipocalin super-family that mediate both chemical and metabolic signaling.
Keywords: Major urinary protein, lipocalin, pheromone, chemical signaling, obesity, type 2 diabetes
I. Introduction
Animals as well as human beings have evolved a variety of communication mechanisms to exchange information. Chemical communication plays a key role in regulating both behavioral and physiological responses in the animal kingdom. Individuals generate scent substances which are excreted into the environment via sweat, urine and feces. These scent substances serve as chemical signals and are perceived by conspecifcs to trigger adaptive behavioral and physiological responses in the receivers (Brennan and Kendrick, 2006; Tirindelli et al., 2009). Most scent substances are unstable, volatile small molecules and bind to their cognate protein carries. These carries not only extend the lifetime but also regulate the release of the scent substances (Hurst, 2009; Tirindelli et al., 2009). The major urinary protein (MUP) family proteins bind to, concentrate, and stabilize many volatile scent substances (e.g. pheromones), thereby controlling both pheromone transport in circulation and pheromone release into the air from urine scent marks (Brennan and Kendrick, 2006; Hurst, 2009; Tirindelli et al., 2009). Additionally, MUPs themselves may serve as chemical signals to convey their owners’ identity information to conspecifics (Chamero et al., 2007). Recent studies reveal that the MUP family members also regulate nutrient metabolism independently of chemical signaling (Hui et al., 2009; Zhou et al., 2009). Nutrient metabolism provides energy supply to power behavioral and physiological activities. Therefore, the MUP family members appear to coordinate behavioral response and energy metabolism by serving as both chemical and metabolic signals.
II. MUP protein structure and polymorphism
MUPs belong to the lipocalin super-family (Cavaggioni and Mucignat-Caretta, 2000; Finlayson et al., 1965). The lipocalin family members have relatively low similarity in their amino acid sequences; however, their tertiary structures are extremely conserved with a characteristic β–barrel consisting of eight β–strands and an α–helix (Bocskei et al., 1992). Most lipocalin family members bind, via their central hydrophobic pockets formed by these eight β–strands, to small lipophilic molecules, including fatty acids, steroids, retinol and pheromones (Schlehuber and Skerra, 2005).
MUPs bind to pheromones via their central β-barrel cavities
The MUP family members were initially discovered in urine as a group of small proteins with molecular weights around 18 kD (Finlayson et al., 1965; Lane and Neuhaus, 1972). MUPs are mainly synthesized in the liver and secreted into the bloodstream (Shaw et al., 1983). Due to their small sizes, MUPs are efficiently filtered through the glomeruli and excreted into the urine (Kimura et al., 1991). The isoelectric points of MUPs in urine are varying from 4.6 to 5.3 (Bocskei et al., 1992; Clissold and Bishop, 1982). Each individual adult male mouse excretes approximately 8–14 different MUP isoforms in urine (Hurst, 2009).
MUP tertiary structures have been extensively studied by both X-ray crystallography and NMR spectroscopy (Bocskei et al., 1992; Darwish Marie et al., 2001; Lucke et al., 1999; Timm et al., 2001; Zidek et al., 1999). The structures of both endogenous and recombinant MUP1 protein have been characterized (Bocskei et al., 1991; Timm et al., 2001). The MUP family members contain a characteristic eight antiparallel β-sheets that are linked by seven loops to form a β-barrel (Bocskei et al., 1992; Darwish Marie et al., 2001; Lucke et al., 1999; Timm et al., 2001; Zidek et al., 1999). The first loop is a large Ω-loop that functions as a dynamic lid of the β-barrel, and the other six are typical short β-hairpin (Timm et al., 2001). The interior of the β-barrel forms a hydrophobic pocket that binds directly to hydrophobic pheromones.
The affinity of different MUP isoforms for pheromones varies. For instance, MUP4 has a 23-fold higher affinity for (±)-2-sec-butyl-4,5-dihydrothiazole than MUP1, but has a 4-fold lower affinity for 6-hydroxy-6-methyl-3-heptanone than MUP1 (Darwish Marie et al., 2001; Sharrow et al., 2002). The affinity for pheromones is determined by the amino acids of the binding pockets (Darwish Marie et al., 2001; Sharrow et al., 2002). A single-amino-acid substitution in the binding pocket can result in a dramatic change in the affinity of a MUP family member (Darwish Marie et al., 2001). The binding of pheromones to MUPs not only protects against pheromone decomposition in blood and urine but also sustains pheromone action by slowly releasing MUP-bound volatile pheromones into the air from urine marks. Therefore, MUP expression levels control pheromone levels in blood and urine (Sharrow et al., 2002).
MUPs are highly polymorphic
MUP expression is sexually dimorphic in rodents, and the levels of MUPs are much higher in males than in females (Geertzen et al., 1973; Lane and Neuhaus, 1972). Androgen potently stimulates MUP expression, resulting in male-dominant expression and excretion of MUPs (Johnson et al., 1995; Kurtz and Feigelson, 1977). MUPs are synthesized mainly in the liver and excreted into the urine (Finlayson et al., 1965; Shaw et al., 1983). MUP synthesis accounts for 3.5–4% of total hepatic protein synthesis in adult male mice (Berger and Szoka, 1981). Urinary MUPs mediate chemical signaling in conspecifics (Tirindelli et al., 2009). The MUP family members are also expressed in the submaxillary, lachrymal, nasal, parotid, mammary glands, and hypothalami; however, the function of extrahepatic MUPs is unknown (Cavaggioni and Mucignat-Caretta, 2000; De Giorgio et al., 2009; Shaw et al., 1983).
The MUP family proteins are encoded by multiple paralogous genes clustered on chromosome 4 in mice and chromosome 5 in rats (Hastie et al., 1979). Rat MUPs are also called α2U-globulins (Lane and Neuhaus, 1972). The amino acid sequences of MUPs are 65% identical between mice and rats (Cavaggioni and Mucignat-Caretta, 2000). The mouse genome contains 21 MUP genes and additional 21 MUP pseudogenes (Logan et al., 2008). The MUP genes and pseudogenes have been independently evolved from a single ancestral gene (Logan et al., 2008). The MUP genes contain 6 coding exons, and the pseudogenes contain premature stop codons due to an insertion or deletion. The MUP genes and pseudogenes are classified into two groups. The first group consists of 6 MUP genes (e.g. MUP1, MUP2, MUP18, MUP24, MUP25 and MUP26) and 5 pseudogenes (Logan et al., 2008). The cDNA sequences of these six MUP genes are 82–94% identical. The second group consists of the remaining 15 MUP genes and 16 pseudogenes (Logan et al., 2008; Mudge et al., 2008). The cDNA sequences of these 15 MUP genes are >97% identical.
The MUP genes are extremely polymorphic in wild or outbred mice (Cheetham et al., 2009; Finlayson et al., 1965; Robertson et al., 2007). Each individual adult male mouse normally expresses 8–14 different MUP isoforms; therefore, the number of MUP expression patterns is extremely expanded due to MUP polymorphism (Beynon et al., 2002; Evershed et al., 1993; Hurst, 2009). Polymorphic MUP genes serve as a specific genetic marker of individual identity, and the MUP profiles in urine are recognized as an individual identity signature of the owners by conspecific receivers (Cheetham et al., 2007; Hurst et al., 2001; Sherborne et al., 2007).
III. MUP regulation of chemical communication
Pheromones are diverse, biologically active substances that are excreted to the outside by individuals. Pheromones are detected by conspecifics and trigger specific behavioral, physiological, and/or developmental responses in the receivers, including aggression, mating, territory marking, estrous cycles and pregnancy (Hurst, 2009; Tirindelli et al., 2009).
MUPs function as volatile pheromone carriers
Many pheromones are small volatile organic molecules which are unstable in aqueous environments (e.g. blood and urine) (Hurst and Beynon, 2004; Stowers and Marton, 2005). The MUP family members bound via their center hydrophobic cavities to a variety of pheromones (Bocskei et al., 1992; Peele et al., 2003; Sharrow et al., 2002). The MUP-pheromone physical interactions protect against pheromone destruction during both transportation in the bloodstream and excretion into the urine. Additionally, free volatile pheromone molecules are quickly evaporated into the air from scent urine marks. MUPs not only facilitate pheromone transportation as pheromone carries but also prolong pheromone lifetime by slowly releasing their bound pheromones into the air from scent marks (Humphries et al., 1999; Hurst et al., 1998).
MUPs act as pheromones to directly regulate behavioral and physiological responses
Interestingly, the MUP1 protein moiety is sufficient to activate sensory neurons in the vomeronasal organ (VNO) and to trigger ovulation (More, 2006). Recombinant MUP1 promotes inter-male aggression in the absence of pheromones (Chamero et al., 2007). Moreover, purified MUP1 directly stimulates Gαo-coupled V2R receptors in VNO neuron cultures (Chamero et al., 2007). Therefore, MUP proteins also act as involatile pheromones in addition to as pheromone carriers.
The MUP profiles serve as an individual identity signature
MUPs and their bound pheromones profoundly modulate the behaviors and development of conspecifics. Urine from intact but not castrated males promotes male aggression (Mugford and Nowell, 1970). Males advertize their social status to attract females via urinary pheromones (Bronson and Caroom, 1971; Jemiolo et al., 1991). MUPs accelerate female puberty and promote ovulation (More, 2006; Mucignat-Caretta et al., 1995). The urine scents of unfamiliar males block pregnancy in recently mated females (Bruce, 1959), and MUPs bind to the volatile pheromones involved in pregnancy block (Peele et al., 2003). The polymorphic patterns of MUPs serve as an individual identity signature in urine marks (Hurst and Beynon, 2004; Hurst et al., 2001). Females use the MUP signatures to recognize individual scent owners, preferentially associated with heterozygous males, and avoid inbreeding (Cheetham et al., 2007; Thom et al., 2008).
Pheromones are believed to stimulate sensory neurons in VNO when animals make nasal contact with scent marks (Breer et al., 2006; Halpern and Martinez-Marcos, 2003; Meredith, 1994). Each pheromone activates a specific subset of sensory neurons that convey unique signals to the brain (Dulac and Torello, 2003). VNO neurons project to the accessory olfactory bulb, and the second-order neurons in the accessory olfactory bulb project to amygdala that innervates hypothalamic neurons either directly or indirectly (Tirindelli et al., 2009). In contrast, the airborne volatile odorants are believed to stimulate sensory neurons in the main olfactory epithelium (MOE) which project to the main olfactory bulb (Tirindelli et al., 2009). The second-order neurons in the main olfactory bulb project to higher centers in the brain, including the piriform cortex and the cortical amygdale (Tirindelli et al., 2009). However, recent studies suggest that both the vomeronasal and the main olfactory systems are involved in pheromone detection (Hurst, 2009).
IV. MUP regulation of nutrient metabolism
Behavioral and developmental responses are powered by energy derived from nutrient metabolism. It is not surprising that many factors simultaneously regulate both behaviors and metabolism. Glucose and fatty acids are the primary fuel substrates to power cellular activity that underlies behavioral and developmental responses. Animals have evolved a sophisticated neuroendocrine system that maintains glucose and lipid homeostasis. For instance, a rise in blood glucose derived from ingested food stimulates pancreatic β cells to secrete insulin. Insulin in return reduces blood glucose levels by stimulating glucose uptake into skeletal muscle and adipose tissue as well as by suppressing glucose production from the liver (Saltiel and Kahn, 2001). In contrast, a fall in blood glucose during fasting stimulates the secretion of counterregulatory hormones (e.g. glucagon and catecholamines) which increase blood glucose levels by stimulating liver glucose production (Jiang and Zhang, 2003). Therefore, blood glucose homeostasis is maintained mainly by a balance between insulin and counterregulatory hormones. Impaired ability of insulin to decrease blood glucose (insulin resistance) is the primary risk factor for the development of type 2 diabetes. Insulin sensitivity is regulated by multiple humoral factors, including MUP1.
MUP1 is involved in nutrient sensing
Recent studies show that the expression and secretion of MUP1 are regulated by nutrient signals. Fasting markedly reduced MUP1 expression in the liver, which is reversed by refeeding (Hui et al., 2009). The liver plays a key role in nutrient sensing and metabolism. In agreement with this observation, caloric restriction (chronic malnutrition) also dramatically reduces MUP1 expression in mouse livers (Dhahbi et al., 2004; Miller et al., 2002). The expression of other MUP family members, including MUP4 and MUP5, is also suppressed in calorie-restricted mice (Dhahbi et al., 2004). Interestingly, MUP1 deficiency is associated with obesity and type 2 diabetes. Two groups reported independently that hepatic MUP1 expression and circulating MUP1 levels are markedly reduced in mice with either genetic (leptin receptor-deficient db/db) or dietary fat-induced obesity (Hui et al., 2009; Zhou et al., 2009). Interestingly, MUP1 is also expressed in several extrahepatic tissues, and MUP1 expression is similarly reduced in both adipose tissues and the hypothalamus in response to nutrient deprivation (De Giorgio et al., 2009; van Schothorst et al., 2006). Adipocytes and hypothalamic neurons are also key players in nutrient sensing. These observations suggest that MUP1 and/or the other MUP family members are likely involved in the nutrient sensing process, and defects in MUP-mediated nutrient sensing might contribute to the development of metabolic diseases, including type 2 diabetes.
MUP1 regulates nutrient metabolism in multiple tissues
There are multiple lines of evidence supporting an important role of MUP1 in glucose metabolism. In mice with either genetic (db/db) or dietary-induced type 2 diabetes, liver-specific overexperssion of MUP1 markedly reduces hyperglycemia and glucose intolerance (Zhou et al., 2009). Similarly, chronic administration of purified recombinant MUP1 proteins also ameliorates hyperglycemia and improves glucose intolerance in db/db mice (Hui et al., 2009). The MUP1 therapy also improves systemic insulin sensitivity in diabetic mice as expected (Hui et al., 2009; Zhou et al., 2009). Interestingly, rosiglitazone (a potent PPARβ agonist) and resveratrol (a natural product abundant in grape skins), two chemically distinct compounds that decrease hyperglycemia and glucose intolerance in diabetic mice, also stimulate MUP1 expression in the liver (Baur et al., 2006; Hui et al., 2009).
MUP1 treatment enhances insulin signaling in the skeletal muscle but not livers of diabetic mice, suggesting that skeletal muscle is a physiological target of MUP1 (Hui et al., 2009). Moreover, recombinant MUP1 directly suppresses glucose production in primary hepatocyte cultures independently of insulin (Zhou et al., 2009). Additionally, liver-specific overexpresion of MUP1 markedly decreases triglyceride levels in the livers of db/db mice (Zhou et al., 2009). Therefore, MUP1 may also regulate hepatic glucose and lipid metabolism in an autocrine and/or paracrine fashion. Interestingly, MUP1 expression is also regulated by nutrients in adipose tissue and the hypothalamus, suggesting that MUP1 may regulate the metabolic activity of these two tissues in a similar autocrine and/or paracrine manner (De Giorgio et al., 2009; van Schothorst et al., 2006).
MUP1 regulates metabolism by multiple mechanisms
MUP1 reduces blood glucose levels at least in part by suppressing the hepatic gluconeogenic program (Zhou et al., 2009). In both animals and primary hepatocyte cultures, recombinant MUP1 markedly inhibits the expression of phosphoenolpyruvate carboxykinase (PEPCK) and glucose-6-phosphatase (G6Pase), two rate-limiting enzymes for gluconeogenesis (Zhou et al., 2009). Insulin is well known to suppress hepatic gluconeogenesis; however, MUP1 suppresses hepatic glucose production independently of insulin, suggesting that MUP1 regulates the hepatic gluconeogenic program by a novel mechanism (Zhou et al., 2009). Hepatic gluconeogenesis is abnormally elevated in type 2 diabetes, thus significantly contributing to hyperglycemia and glucose intolerance (Ali and Drucker, 2009; Jiang and Zhang, 2003). Interestingly, type 2 diabetes is associated with a marked reduction in MUP1expression, suggesting that reduced expression of hepatic MUP1 contributes to abnormally-elevated hepatic gluconeogenesis (Hui et al., 2009; Zhou et al., 2009).
Chronic MUP1 treatment also decreases the levels of plasma lipids in db/db mice (Hui et al., 2009; Zhou et al., 2009). Moreover, liver-specific overexpression of MUP1 results in a marked reduction in hepatic lipid levels, presumably due to suppression of lipogenic genes in the liver, including the stearoyl-CoA desaturase-1, fatty acid synthase, carbohydrate response element binding protein, and peroxisome proliferator-activated receptor-β (PPARβ) genes (Zhou et al., 2009). Chronic administration of purified recombinant MUP1 also decreases lipid levels in the skeletal muscles of db/db mice (Hui et al., 2009). Together, these observations suggest that MUP1 regulates both glucose and lipid metabolism in multiple tissues.
MUP1 improves insulin sensitivity in skeletal muscle at least in part by increasing energy expenditure (Hui et al., 2009). Chronic administration of purified MUP1 proteins increases energy expenditure, body temperature and ambulatory locomotion in db/db mice (Hui et al., 2009). MUP1 increases not only mitochondrial biogenesis but also the capacity of mitochondrial oxidative phosphorylation (Hui et al., 2009). Interestingly, MUP1 promotes mitochondrial biogenesis and function specifically in the skeletal muscle but not other tissues (e.g. adipose tissues and livers) of db/db mice (Hui et al., 2009). An increase in mitochondrial content and function is likely to result in an increase in fatty acid β-oxidation and a decrease in lipid levels in skeletal muscles, thereby ameliorating lipotoxicity and insulin resistance in MUP1-treated mice with type 2 diabetes.
Recombinant MUP1 inhibits the hepatic gluconeogenic program directly in primary hepatocyte cultures, suggesting that MUP1 regulates metabolic function in the liver by activating its own cognate receptors (Zhou et al., 2009). Additionally, in animals, circulatory MUP1 binds to, concentrates, and slowly releases various lipophilic molecules (Cavaggioni and Mucignat-Caretta, 2000; Sharrow et al., 2002). These lipophilic molecules may be bioactive and regulate nutrient metabolism; therefore, MUP1 may also regulate metabolism indirectly by controlling the stability, concentrations, and/or activity of these bioactive lipophilic molecules.
V. Conclusions and future directions
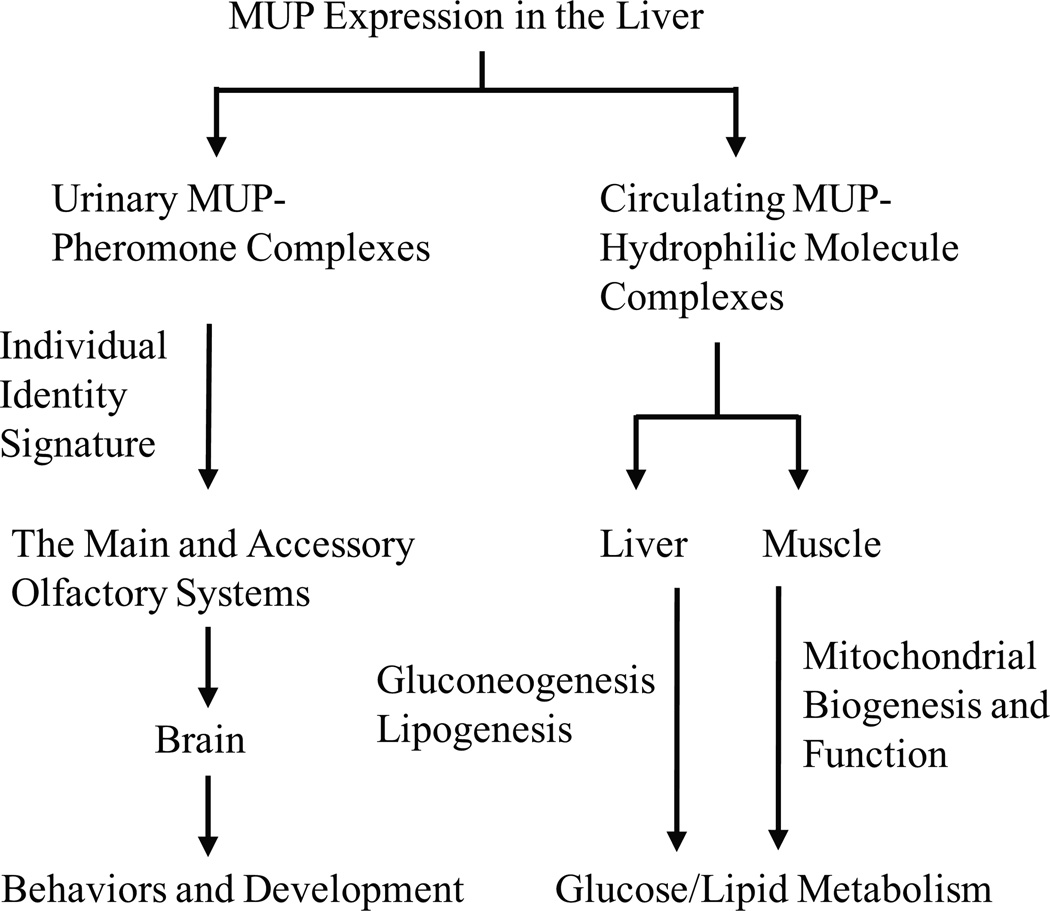
MUPs belong to the lipocalin super-family whose tertiary structure contains a conserved β-barrel with a characteristic central hydrophobic pocket. The MUP family members are expressed mainly by the liver and secreted into the bloodstream (Fig. 1). Various pheromones and other small lipophilic molecules bind to the central pockets of MUPs and are transported through the circulation. MUPs are excreted into the urine in the kidney, and urinary MUPs prolong pheromone lifetime by slowing the release of MUP-bound pheromones into the air from urine scent marks. MUPs are highly polymorphic, and the urinary MUP profiles are recognized as an individual identity signature of the scent owners by conspecifics. MUPs and MUP-bound pheromones are detected by both the main and the accessory olfactory systems. These two systems act coordinately to convey the information about the individual identity of signalers to the brain of conspecific receivers and to trigger behavioral responses and/or developmental processes. However, it remains completely unclear how the MUP detection system and the central nervous system extract the individual identify information encoded in the MUP profiles. Interestingly, circulating MUPs may play an important role in regulating nutrient metabolism. MUPs, particularly MUP1, suppress the hepatic gluconeogenic and lipogenic programs. MUP1 also promotes mitochondrial biogenesis and oxidative phosphorylation in skeletal muscles, thus increasing energy expenditure and insulin sensitivity. However, it is unclear whether MUP1 regulates metabolism directly through its own cognate receptors or indirectly by controlling the stability, the release, and/or the activity of MUP-bound small molecules. It also remains unclear whether hypothalamic and adipose MUP1, whose expression is regulated by nutrients, regulates metabolism. Additionally, the therapeutic potential of MUP1 in treating type 2 diabetes and metabolic disorders remains to be determined.
Figure 1. A model of MUP action.
The MUP family members are expressed mainly by the liver and secreted into the bloodstream. MUPs bind to various volatile pheromones or other lipophilic small molecules, and regulate the transportation and bioactivity of these small molecules. MUPs and MUP-bound pheromones are excreted into the urine and detected by the main and accessory olfactory systems of conspecifics. MUPs are highly polymorphic, and the MUP profiles in urine are recognized as an identity signature of the owners by receivers. Additionally, circulating MUPs and MUP-bound bioactive molecules also regulate metabolism by suppressing the hepatic gluconeogenic and/or lipogenic programs as well as by promoting mitochondrial biogenesis and function and insulin sensitivity in skeletal muscles.
Acknowledgements
This study was supported by RO1 DK 065122 and RO1 DK073601 from NIH.
References List
- Ali S, Drucker DJ. Benefits and limitations of reducing glucagon action for the treatment of type 2 diabetes. Am J Physiol Endocrinol Metab. 2009;296:E415–E421. doi: 10.1152/ajpendo.90887.2008. [DOI] [PubMed] [Google Scholar]
- Baur JA, Pearson KJ, Price NL, Jamieson HA, Lerin C, Kalra A, Prabhu VV, Allard JS, Lopez-Lluch G, Lewis K, Pistell PJ, Poosala S, Becker KG, Boss O, Gwinn D, Wang M, Ramaswamy S, Fishbein KW, Spencer RG, Lakatta EG, Le Couteur D, Shaw RJ, Navas P, Puigserver P, Ingram DK, de Cabo R, Sinclair DA. Resveratrol improves health and survival of mice on a high-calorie diet. Nature. 2006;444:337–342. doi: 10.1038/nature05354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger FG, Szoka P. Biosynthesis of the major urinary proteins in mouse liver: a biochemical genetic study. Biochem Genet. 1981;19:1261–1273. doi: 10.1007/BF00484578. [DOI] [PubMed] [Google Scholar]
- Beynon RJ, Veggerby C, Payne CE, Robertson DH, Gaskell SJ, Humphries RE, Hurst JL. Polymorphism in major urinary proteins: molecular heterogeneity in a wild mouse population. J Chem Ecol. 2002;28:1429–1446. doi: 10.1023/a:1016252703836. [DOI] [PubMed] [Google Scholar]
- Bocskei Z, Findlay JB, North AC, Phillips SE, Somers WS, Wright CE, Lionetti C, Tirindelli R, Cavaggioni A. Crystallization of and preliminary X-ray data for the mouse major urinary protein and rat alpha-2u globulin. J Mol Biol. 1991;218:699–701. doi: 10.1016/0022-2836(91)90258-8. [DOI] [PubMed] [Google Scholar]
- Bocskei Z, Groom CR, Flower DR, Wright CE, Phillips SE, Cavaggioni A, Findlay JB, North AC. Pheromone binding to two rodent urinary proteins revealed by X-ray crystallography. Nature. 1992;360:186–188. doi: 10.1038/360186a0. [DOI] [PubMed] [Google Scholar]
- Breer H, Fleischer J, Strotmann J. The sense of smell: multiple olfactory subsystems. Cell Mol Life Sci. 2006;63:1465–1475. doi: 10.1007/s00018-006-6108-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan PA, Kendrick KM. Mammalian social odours: attraction and individual recognition. Philos Trans R Soc Lond B Biol Sci. 2006;361:2061–2078. doi: 10.1098/rstb.2006.1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bronson FH, Caroom D. Preputial gland of the male mouse; attractant function. J Reprod Fertil. 1971;25:279–282. doi: 10.1530/jrf.0.0250279. [DOI] [PubMed] [Google Scholar]
- Bruce HM. An exteroceptive block to pregnancy in the mouse. Nature. 1959;184:105. doi: 10.1038/184105a0. [DOI] [PubMed] [Google Scholar]
- Cavaggioni A, Mucignat-Caretta C. Major urinary proteins, alpha(2U)-globulins and aphrodisin. Biochim Biophys Acta. 2000;1482:218–228. doi: 10.1016/s0167-4838(00)00149-7. [DOI] [PubMed] [Google Scholar]
- Chamero P, Marton TF, Logan DW, Flanagan K, Cruz JR, Saghatelian A, Cravatt BF, Stowers L. Identification of protein pheromones that promote aggressive behaviour. Nature. 2007;450:899–902. doi: 10.1038/nature05997. [DOI] [PubMed] [Google Scholar]
- Cheetham SA, Smith AL, Armstrong SD, Beynon RJ, Hurst JL. Limited variation in the major urinary proteins of laboratory mice. Physiol Behav. 2009;96:253–261. doi: 10.1016/j.physbeh.2008.10.005. [DOI] [PubMed] [Google Scholar]
- Cheetham SA, Thom MD, Jury F, Ollier WE, Beynon RJ, Hurst JL. The genetic basis of individual-recognition signals in the mouse. Curr Biol. 2007;17:1771–1777. doi: 10.1016/j.cub.2007.10.007. [DOI] [PubMed] [Google Scholar]
- Clissold PM, Bishop JO. Variation in mouse major urinary protein (MUP) genes and the MUP gene products within and between inbred lines. Gene. 1982;18:211–220. doi: 10.1016/0378-1119(82)90158-5. [DOI] [PubMed] [Google Scholar]
- Darwish Marie A, Veggerby C, Robertson DH, Gaskell SJ, Hubbard SJ, Martinsen L, Hurst JL, Beynon RJ. Effect of polymorphisms on ligand binding by mouse major urinary proteins. Protein Sci. 2001;10:411–417. doi: 10.1110/ps.31701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Giorgio MR, Yoshioka M, St-Amand J. Feeding induced changes in the hypothalamic transcriptome. Clin Chim Acta. 2009;406:103–107. doi: 10.1016/j.cca.2009.06.004. [DOI] [PubMed] [Google Scholar]
- Dhahbi JM, Kim HJ, Mote PL, Beaver RJ, Spindler SR. Temporal linkage between the phenotypic and genomic responses to caloric restriction. Proc Natl Acad Sci U S A. 2004;101:5524–5529. doi: 10.1073/pnas.0305300101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dulac C, Torello AT. Molecular detection of pheromone signals in mammals: from genes to behaviour. Nat Rev Neurosci. 2003;4:551–562. doi: 10.1038/nrn1140. [DOI] [PubMed] [Google Scholar]
- Evershed RP, Robertson DH, Beynon RJ, Green BN. Application of electrospray ionization mass spectrometry with maximum-entropy analysis to allelic 'fingerprinting' of major urinary proteins. Rapid Commun Mass Spectrom. 1993;7:882–886. doi: 10.1002/rcm.1290071005. [DOI] [PubMed] [Google Scholar]
- Finlayson JS, Asofsky R, Potter M, Runner CC. Major urinary protein complex of normal mice: origin. Science. 1965;149:981–982. doi: 10.1126/science.149.3687.981. [DOI] [PubMed] [Google Scholar]
- Geertzen HG, Ouderaa FJv, Kassenaar AA. Isolation and metabolism of male sex-dependent urinary protein from rats. Acta Endocrinol (Copenh) 1973;72:197–208. doi: 10.1530/acta.0.0720197. [DOI] [PubMed] [Google Scholar]
- Halpern M, Martinez-Marcos A. Structure and function of the vomeronasal system: an update. Prog Neurobiol. 2003;70:245–318. doi: 10.1016/s0301-0082(03)00103-5. [DOI] [PubMed] [Google Scholar]
- Hastie ND, Held WA, Toole JJ. Multiple genes coding for the androgen-regulated major urinary proteins of the mouse. Cell. 1979;17:449–457. doi: 10.1016/0092-8674(79)90171-5. [DOI] [PubMed] [Google Scholar]
- Hui X, Zhu W, Wang Y, Lam KS, Zhang J, Wu D, Kraegen EW, Li Y, Xu A. Major urinary protein-1 increases energy expenditure and improves glucose intolerance through enhancing mitochondrial function in skeletal muscle of diabetic mice. J Biol Chem. 2009;284:14050–14057. doi: 10.1074/jbc.M109.001107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphries RE, Robertson DH, Beynon RJ, Hurst JL. Unravelling the chemical basis of competitive scent marking in house mice. Anim Behav. 1999;58:1177–1190. doi: 10.1006/anbe.1999.1252. [DOI] [PubMed] [Google Scholar]
- Hurst JL. Female recognition and assessment of males through scent. Behav Brain Res. 2009;200:295–303. doi: 10.1016/j.bbr.2008.12.020. [DOI] [PubMed] [Google Scholar]
- Hurst JL, Beynon RJ. Scent wars: the chemobiology of competitive signalling in mice. Bioessays. 2004;26:1288–1298. doi: 10.1002/bies.20147. [DOI] [PubMed] [Google Scholar]
- Hurst JL, Payne CE, Nevison CM, Marie AD, Humphries RE, Robertson DH, Cavaggioni A, Beynon RJ. Individual recognition in mice mediated by major urinary proteins. Nature. 2001;414:631–634. doi: 10.1038/414631a. [DOI] [PubMed] [Google Scholar]
- Hurst JL, Robertson DHL, Tolladay U, Beynon RJ. Proteins in urine scent marks of male house mice extend the longevity of olfactory signals. Anim Behav. 1998;55:1289–1297. doi: 10.1006/anbe.1997.0650. [DOI] [PubMed] [Google Scholar]
- Jemiolo B, Xie TM, Novotny M. Socio-sexual olfactory preference in female mice: attractiveness of synthetic chemosignals. Physiol Behav. 1991;50:1119–1122. doi: 10.1016/0031-9384(91)90570-e. [DOI] [PubMed] [Google Scholar]
- Jiang G, Zhang BB. Glucagon and regulation of glucose metabolism. Am J Physiol Endocrinol Metab. . 2003;284:E671–E678. doi: 10.1152/ajpendo.00492.2002. [DOI] [PubMed] [Google Scholar]
- Johnson D, al-Shawi R, Bishop JO. Sexual dimorphism and growth hormone induction of murine pheromone-binding proteins. J Mol Endocrinol. 1995;14:21–34. doi: 10.1677/jme.0.0140021. [DOI] [PubMed] [Google Scholar]
- Kimura H, Odani S, Nishi S, Sato H, Arakawa M, Ono T. Primary structure and cellular distribution of two fatty acid-binding proteins in adult rat kidneys. J Biol Chem. 1991;266:5963–5972. [PubMed] [Google Scholar]
- Kurtz DT, Feigelson P. Multihormonal induction of hepatic alpha2u-globulin mRNA as measured by hybridization to complementary DNA. Proc Natl Acad Sci U S A. 1977;74:4791–4795. doi: 10.1073/pnas.74.11.4791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane SE, Neuhaus OW. Multiple forms of 2 u, a sex-dependent urinary protein of the adult male rat. Biochim Biophys Acta. 1972;263:433–440. doi: 10.1016/0005-2795(72)90095-5. [DOI] [PubMed] [Google Scholar]
- Logan DW, Marton TF, Stowers L. Species specificity in major urinary proteins by parallel evolution. PLoS One. 2008;3:e3280. doi: 10.1371/journal.pone.0003280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucke C, Franzoni L, Abbate F, Lohr F, Ferrari E, Sorbi RT, Ruterjans H, Spisni A. Solution structure of a recombinant mouse major urinary protein. Eur J Biochem. 1999;266:1210–1218. doi: 10.1046/j.1432-1327.1999.00984.x. [DOI] [PubMed] [Google Scholar]
- Meredith M. Chronic recording of vomeronasal pump activation in awake behaving hamsters. Physiol Behav. 1994;56:345–354. doi: 10.1016/0031-9384(94)90205-4. [DOI] [PubMed] [Google Scholar]
- Miller RA, Chang Y, Galecki AT, Al-Regaiey K, Kopchick JJ, Bartke A. Gene expression patterns in calorically restricted mice: partial overlap with long-lived mutant mice. Mol Endocrinol. 2002;16:2657–2666. doi: 10.1210/me.2002-0142. [DOI] [PubMed] [Google Scholar]
- More L. Mouse major urinary proteins trigger ovulation via the vomeronasal organ. Chem Senses. 2006;31:393–401. doi: 10.1093/chemse/bjj043. [DOI] [PubMed] [Google Scholar]
- Mucignat-Caretta C, Caretta A, Cavaggioni A. Acceleration of puberty onset in female mice by male urinary proteins. J Physiol. 1995;486(Pt 2):517–522. doi: 10.1113/jphysiol.1995.sp020830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mudge JM, Armstrong SD, McLaren K, Beynon RJ, Hurst JL, Nicholson C, Robertson DH, Wilming LG, Harrow JL. Dynamic instability of the major urinary protein gene family revealed by genomic and phenotypic comparisons between C57 and 129 strain mice. Genome Biol. 2008;9:R91. doi: 10.1186/gb-2008-9-5-r91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mugford RA, Nowell NW. Pheromones and their effect on aggression in mice. Nature. 1970;226:967–968. doi: 10.1038/226967a0. [DOI] [PubMed] [Google Scholar]
- Peele P, Salazar I, Mimmack M, Keverne EB, Brennan PA. Low molecular weight constituents of male mouse urine mediate the pregnancy block effect and convey information about the identity of the mating male. Eur J Neurosci. 2003;18:622–628. doi: 10.1046/j.1460-9568.2003.02790.x. [DOI] [PubMed] [Google Scholar]
- Robertson DH, Hurst JL, Searle JB, Gunduz I, Beynon RJ. Characterization and comparison of major urinary proteins from the house mouse, Mus musculus domesticus, and the aboriginal mouse, Mus macedonicus. J Chem Ecol. 2007;33:613–630. doi: 10.1007/s10886-006-9247-0. [DOI] [PubMed] [Google Scholar]
- Saltiel AR, Kahn CR. Insulin signalling and the regulation of glucose and lipid metabolism. Nature. 2001;414:799–806. doi: 10.1038/414799a. [DOI] [PubMed] [Google Scholar]
- Schlehuber S, Skerra A. Lipocalins in drug discovery: from natural ligand-binding proteins to "anticalins". Drug Discov Today. 2005;10:23–33. doi: 10.1016/S1359-6446(04)03294-5. [DOI] [PubMed] [Google Scholar]
- Sharrow SD, Vaughn JL, Zidek L, Novotny MV, Stone MJ. Pheromone binding by polymorphic mouse major urinary proteins. Protein Sci. 2002;11:2247–2256. doi: 10.1110/ps.0204202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw PH, Held WA, Hastie ND. The gene family for major urinary proteins: expression in several secretory tissues of the mouse. Cell. 1983;32:755–761. doi: 10.1016/0092-8674(83)90061-2. [DOI] [PubMed] [Google Scholar]
- Sherborne AL, Thom MD, Paterson S, Jury F, Ollier WE, Stockley P, Beynon RJ, Hurst JL. The genetic basis of inbreeding avoidance in house mice. Curr Biol. 2007;17:2061–2066. doi: 10.1016/j.cub.2007.10.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stowers L, Marton TF. What is a pheromone? Mammalian pheromones reconsidered. Neuron. 2005;46:699–702. doi: 10.1016/j.neuron.2005.04.032. [DOI] [PubMed] [Google Scholar]
- Thom MD, Stockley P, Jury F, Ollier WE, Beynon RJ, Hurst JL. The direct assessment of genetic heterozygosity through scent in the mouse. Curr Biol. 2008;18:619–623. doi: 10.1016/j.cub.2008.03.056. [DOI] [PubMed] [Google Scholar]
- Timm DE, Baker LJ, Mueller H, Zidek L, Novotny MV. Structural basis of pheromone binding to mouse major urinary protein (MUP-I) Protein Sci. 2001;10:997–1004. doi: 10.1110/ps.52201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirindelli R, Dibattista M, Pifferi S, Menini A. From pheromones to behavior. Physiol Rev. 2009;89:921–956. doi: 10.1152/physrev.00037.2008. [DOI] [PubMed] [Google Scholar]
- van Schothorst EM, Keijer J, Pennings JL, Opperhuizen A, van den Brom CE, Kohl T, Franssen-van Hal NL, Hoebee B. Adipose gene expression response of lean and obese mice to short-term dietary restriction. Obesity (Silver Spring) 2006;14:974–979. doi: 10.1038/oby.2006.111. [DOI] [PubMed] [Google Scholar]
- Zhou Y, Jiang L, Rui L. Identification of MUP1 as a regulator for glucose and lipid metabolism in mice. J Biol Chem. 2009;284:11152–11159. doi: 10.1074/jbc.M900754200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zidek L, Stone MJ, Lato SM, Pagel MD, Miao Z, Ellington AD, Novotny MV. NMR mapping of the recombinant mouse major urinary protein I binding site occupied by the pheromone 2-sec-butyl-4,5-dihydrothiazole. Biochemistry. 1999;38:9850–9861. doi: 10.1021/bi990497t. [DOI] [PubMed] [Google Scholar]