Abstract
Epigenetic mechanisms are proposed to underlie aberrant gene expression in systemic lupus erythematosus (SLE) that results in dysregulation of the immune system and loss of tolerance. Modifications of DNA and histones require substrates derived from diet and intermediary metabolism. DNA and histone methyltransferases depend on S-adenosylmethionine (SAM) as a methyl donor. SAM is generated from adenosine triphosphate (ATP) and methionine by methionine adenosyltransferase (MAT), a redox-sensitive enzyme in the SAM cycle. The availability of B vitamins and methionine regulate SAM generation. The DNA of SLE patients is hypomethylated, indicating dysfunction in the SAM cycle and methyltransferase activity. Acetyl-CoA, which is necessary for histone acetylation, is generated from citrate produced in mitochondria. Mitochondria are also responsible for de novo synthesis of flavin adenine dinucleotide (FAD) for histone demethylation. Mitochondrial oxidative phosphorylation is the dominant source of ATP. The depletion of ATP in lupus T cells may affect MAT activity as well as adenosine monophosphate (AMP) activated protein kinase (AMPK), which phosphorylates histones and inhibits mechanistic target of rapamycin (mTOR). In turn, mTOR can modify epigenetic pathways including methylation, demethylation, and histone phosphorylation and mediates enhanced T-cell activation in SLE. Beyond their role in metabolism, mitochondria are the main source of reactive oxygen intermediates (ROI), which activate mTOR and regulate the activity of histone and DNA modifying enzymes. In this review we will focus on the sources of metabolites required for epigenetic regulation and how the flux of the underlying metabolic pathways affects gene expression.
Keywords: Epigenetics, genetics, metabolism, SLE
Introduction
Systemic lupus erythematosus (SLE) is an autoimmune disease characterized by dysfunction of B and T cells, macrophages, dendritic cells, and production of a wide spectrum of antinuclear autoantibodies [1–5]. So far, no single, unifying hypothesis has explained the pathogenesis of SLE; rather, it appears to be caused by a complex interplay of genetic and environmental factors [6]. The genetic factors that have been associated with SLE include a large number of single nucleotide polymorphisms (SNPs) that only increase the relative risk of disease by 1.1–1.3-fold with the exception of inactivating mutations in complement genes [7,8]. Many of the lupus-linked SNPs are located in non-protein-coding DNA that regulate gene transcription [9]. Genome-wide association studies found ITGAM, STAT4, ATG5, IRF5, BLK, BANK1, IL10, IL2/IL21, ELF1, CD40, and TLR7 to be modestly associated with SLE [10,11]. There is mounting evidence that modification of DNA by direct methylation and histone modifications, which are commonly termed as epigenetic mechanisms, affect the expression of genes that regulate the function of cells within the immune system [12–16]. The pathogenic role of epigenetics was observed in SLE over two decades ago [17]. In lupus CD4+ T cells, hypomethylation of CD70, CD11a, perforin, IL4, IL6, and CD40L results in increased transcriptional activity [18]. Recently, large-scale DNA methylation analysis between SLE and control naïve CD4+ T cells identified 47 genes that are differentially methylated, including BST2, IFI44L, and STAT1 [19]. Still, much of what we know about epigenetic regulation comes from cancer biology [20,21]. The epigenetic regulation of gene transcription in autoimmune diseases, including SLE, is not as well defined and limited to a small number of genes [13].
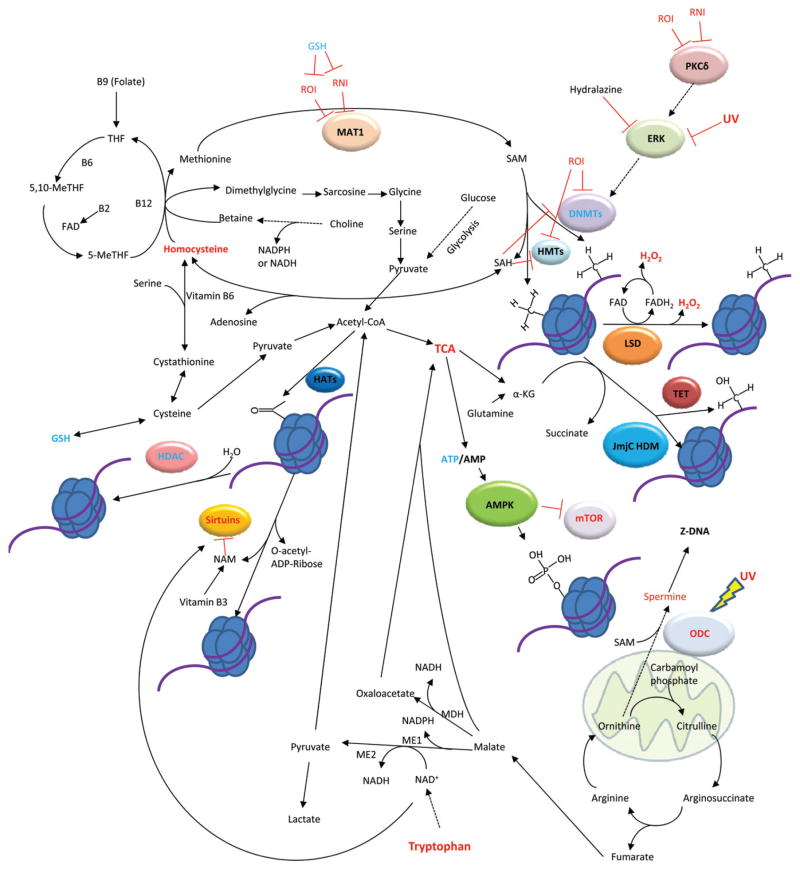
The effect of metabolism on DNA and histone modificationshas recently emerged as a major regulatory mechanism of epigenetics [20,21]. The DNA and histone modifications that encompass epigenetics rely on common metabolites such as S-adenosyl-methionine (SAM), acetyl-CoA, and nicotinamide adenine dinucleotide (NAD+) (Table 1) [21]. The de novo synthesis or regeneration of metabolites involved in epigenetic regulation are dependent on diet as well as biochemical pathways such as glycolysis, the urea cycle, the SAM cycle, and the controlled production of reactive oxygen intermediates (ROI; Figure 1) [22,23]. In particular, ROI act as signaling molecules in the immune system and play a role in SLE pathogenesis [24]. Additionally, ROI act on DNA and histone modifying enzymes to regulate transcription [25]. Mitochondria are important sources of ROI as well as other metabolites that will be discussed and serve as a focal point of dysfunction in SLE [26,27]. In this review, we aim to present the current knowledge of epigenetics with respect to metabolism in SLE while drawing heavily on what has been discovered in other fields such as yeast and cancer biology and ascribe these findings to possible mechanisms of metabolic regulation of epigenetics in SLE.
Table 1.
Metabolite requirements for DNA and histone modifications.
| Modification | Responsible enzymes | Metabolites required | Affected in SLE? | |
|---|---|---|---|---|
| DNA | ||||
| Methylation | DNA Methyltransferases (DNMT1, DNMT3A, DNMT3B) | SAM | Yes [17,36] | |
| Demethylation | TET Family Enzymes | α-ketoglutarate, O2, Fe2+ | Not described | |
| Z-DNA | N/A | Polyamines | Yes [62,63] | |
| Histones | ||||
| Methylation | Histone Methyl Transferases (HMTs) | SAM | Yes [71,72] | |
| Demethylation | LSD1/LSD2 | Flavin adenine dinculeotide | Not described | |
| Jumonji-C domain containing demethylases | α-ketoglutarate, O2, Fe2+ | Yes [72] | ||
| Acetylation | Histone Acetyl Transferases (HATs) | Acetyl-CoA | Yes [80,95,96] | |
| Deacetylation | Histone Deacetylases (HDACs) | H2O, hydrolysis reaction | Yes [95,96] | |
| Sirtuins (Sirt1, Sirt2, Sirt6, Sirt7 in nucleus) | NAD+ | Yes [95,96] | ||
| Phosphorylation | Kinases (AMPK, Akt, PKC, etc.) | ATP | Not described | |
| Ubiquitinylation | E1, E2, E3 Enzymes | Ubiquitin | Not described | |
| SUMOylation | E1, E2, E3 Enzymes | SUMO | Not described | |
| Citrullination | Peptidylargininedeiminases (PADs) | H2O, hydrolysis reaction | Not described | |
| GlcNAcylation | O-GlcNAcTransferase | O-linked N-acetylglucosamine | Yes, but no evidence for histone modification [115] | |
| Carbonylation | N/A | 4-HNE, MDA | Not described | |
Figure 1.
Schematic diagram of metabolites and pathways that control the epigenome in SLE. Metabolites or enzymes in red show elevated levels or activity, while those in blue have decreased level or activity in SLE. Artwork will only appear in color in the online journal.
DNA modifications
Methylation
DNA methylation results in the repression of gene expression. In eukaryotes, cytosine is the modified by methylation at the 5 carbon [14]. DNA methylation occurs when cytosine is adjacent to a guanosine reading in the 5′ to 3′ direction, hence the nomenclature CpG. There are clusters of CpGs in the DNA, called CpG islands, which can occur in regulatory segments of DNA. The methylation status of these islands can be controlled through metabolism and can regulate the expression of genes downstream [23].
SLE patients with active disease have hypomethylated DNA [17]. The methylation of DNA inversely correlates with lupus disease activity [28]. DNA methylation is carried out by three DNA methyltransferases (DNMT). Dnmt3a and Dnmt3b perform de novo methylation whereas Dnmt1 maintains inheritable methylation [14]. DNA methylation is dependent on SAM as a methyl group donor.SAM is generated from ATP and methionine by methionine adenosyltransferase (MAT). MAT1A knockout mice have a greater than seven-fold increase in plasma methionine while SAM and reduced glutathione (GSH) were severely reduced by 74% and 40%, respectively [29]. MAT is negatively regulated by oxidative and nitrative stress, which is reversed by the addition of GSH [30]. Thus, SAM is regulated by the cellular reduction-oxidation state.
The by-product of DNA methylation by SAM is S-adenosyl-homocysteine (SAH), which inhibits both DNMTs and histone methyltransferases (HMTs) [31]. Subsequently, SAH is hydrolyzed to adenosine and homocysteine. Adenosine generated by monocytes and regulatory T cells (Tregs) inhibits arachidonic acid release from monocytes and modifies the immune response in SLE [32]. Homocysteine can activate T cells and is elevated in children with SLE [33,34]. Homocysteine can then be re-methylated to regenerate methionine or it may be metabolized to cystathionine, a GSH precursor (Figure 1).
GSH is essential for maintaining a reducing environment and acts as a precursor of methionine. DNMTs are sensitive to ROI because of a reduced cysteine that participates directly in the methyltransferase activity [25,35]. DNMT1 expression and enzymatic activity is reduced in SLE [17,36]. The depletion of GSH and subsequent increase in oxidative stress in SLE may be driving the inhibition of DNMT1 via oxidation of catalytic cysteine residues [37]. DNMT1 and DNMT3 transcription levels and methyltransferase activity are positively regulated by protein kinase C delta (PKCδ) and ERK [38–40]. PKCδ is sensitive to oxidative and nitrosative stress and nitration of PKCδ prevents phosphorylation of the tyrosine 505 residue and subsequent activation of PKCδ [41]. Downregulated ERK activity in lupus CD4+ T cells due to less PKCδ activity results in DNA hypomethylation and more permissive transcription [39]. The importance of ERK in DNMT regulation is further supported by data showing pharmacological inhibition of ERK by hydralazine results in DNA hypomethylation [42]. Additionally, ultraviolet (UV) radiation inhibits ERK, which inhibits T cell activation [43]. Since UV induces SLE flares, T cell hypomethylation may be due to inhibited ERK signaling to DNMTs [6,17].
In addition to its role in T cell DNA methylation, PKCδ also regulates B cell proliferation in humans. PKCδ deficiency in humans results in the expansion of B cell populations and early onset SLE [44]. The methylation status of B cell DNA is important for receptor gene rearrangement and hypomethylated DNA can lead to loss of B cell tolerance [45]. The DNA methylation status was not reported in PKCδ deficient patients, but future studies may yield some information on the epigenetic regulation of B cell transcription by PKCδ. Another important regulator of B cell autoreactivity in SLE is activation-induced cytidine deaminase (AID). AID deaminates cytosine to uracil on single stranded DNA and is associated with DNA demethylation [46]. The nucleotide conversion by AID drives somatic hypermutation and immunoglobulin class switching in B cells [47]. VDJ rearrangement in B cells requires hypomethylated DNA [47]. AID associates with demethylated DNA and require histone phosphorylation, methylation, and acetylation to gain access to immunoglobulin loci [47]. The transcription of Aicda, the gene encoding AID, depends on histone 3 methylation and acetylation [47]. B cell maturation is dependent on metabolites such as glucose, tryptophan, and 5′-methylthioadenosine, a polyamine biosynthesis intermediate [48]. The flux of these metabolites in B cells may play a role in epigenetic regulation so that antibody responses are appropriate and self-tolerant.
SLE lymphocytes have hypomethylated DNA and increased oxidative stress due to the depletion of GSH [17,27,49]. This may be due to the accumulation of SAH from blockage in the SAM pathway that prevents recycling of SAH and homocysteine to methionine or cystathionine. Elevated levels of SAH inhibit methyltransferase activity [50]. SAH and homocysteine accumulation also cause demethylation in lymphocytes [51]. Increased SAH and homocysteine indicate a block in the pathways to regenerate SAM and de novo GSH synthesis, which further promotes oxidative stress. Interestingly, the depletion of GSH results in DNA hypomethylation and it is argued that methionine is diverted from the SAM cycle to regenerate GSH [52].
All of the reactions to generate GSH from methionine are reversible. Thus, it could be reasoned that GSH is a source of methionine and therefore the depletion of GSH results in less available methionine for SAM regeneration. Furthermore, MAT1A knockout mice have depleted GSH despite a dramatic increase in methionine further supporting the hypothesis that GSH is shunted towards the SAM cycle and drives methylation and not vice versa [29]. Thus, the availability of SAM and GSH coupled with less methyltransferase activity may drive T cell fate and further instigate autoimmunity.
Dietary intake of vitamins and proteins are essential for maintaining the SAM cycle. The vitamins B2, B6, B9 (folate), and B12 are all required for the re-methylation of homocysteine to methionine and are required in the diet (Figure 1) [53–56]. DNA methylation is dependent on dietary folate [14,38]. Dietary intake of methionine is likely important for maintaining SAM levels because it is an essential amino acid that cannot be generated de novo in humans. The DNA methylation pathway connects diet, the SAM cycle, and oxidative stress to T and B cell fate and autoimmunity.
Demethylation
Hypomethylated DNA in SLE patients is immunogenic and allows for increased transcription [57,58]. The hypomethylated state of SLE DNA may be a result of increased demethylation activity. The TET-family enzymes oxidize methylcytosine to generate 5-hydroxmethylcytosine, 5-formylcytosine, and 5-carboxylcytosine [59,60]. TET-family enzymes require α-ketoglutarate (α-KG), O2, and Fe2+ to demethylate DNA. α-KG is an important metabolite of the tricarboxylic acid (TCA) cycle in mitochondria and is generated from the decarboxylation of isocitrate or the deamination of glutamate. The dependence on the mitochondria for substrate makes these enzymes sensitive to cellular energetics. The transition from aerobic glycolysis to oxidative phosphorylation (OXPHOS) likely affects the availability of α-KG for demethylation and may regulate the epigenome.
DNA methylation/demethylation reactions in SLE depend on the availability of SAM, GSH, and α-KG. Within 20 minutes of T cell activation, the IL-2 promoter is demethylated [61]. The ability of the cell to regulate gene expression quickly through methylation requires coordination of metabolic flux. Thus, readily available pools of SAM, GSH, and methionine for methylation and α-KG for demethylation are essential in cells that need to quickly respond to stimuli and stress.
Z-DNA
Transcriptionally active DNA is found in three different conformations: A, B, and Z-DNA. Z-DNA has a left handed helix that turns in the opposite direction of A and B-DNA. The Z-DNA structure prevents histone inhibition of transcription due to the low affinity of Z-DNA for histones [15]. Furthermore, anti-dsDNA antibodies in SLE have a greater affinity for Z-DNA than B-DNA indicating that Z-DNA is common in SLE patients and immunogenic [62]. Z-DNA occurs transiently in cells, but can be stabilized in the presence of polyamines such as spermine [15]. Polyamines are increased in the sera of children with SLE [63]. Hydralazine induced lupus also stabilizes Z-DNA indicating that its inhibition of the ERK pathway plays a role in Z-DNA stabilization or it affects polyamine synthesis [64].
Ornithine, generated in the urea cycle, is a precursor in spermine synthesis. SAM is also necessary for the synthesis of spermine. Ornithine decarboxylase converts ornithine into putrescine through decarboxylation to initiate spermine synthesis. Ornithine decarboxylase is activated by UV radiation [65]. In SLE patients, this means that UV radiation could contribute to the stabilization of Z-DNA and promote transcription by increasing spermine levels. Spermine levels in SLE have not been investigated thus far, but coupled with demethylation this may lead to increased transcription in CD4+ T cells.
It has been proposed that spermine synthase and spermidine/spermine acetyl transferase can act as a futile cycle that wastes SAM in SLE cells [15]. In wasting SAM, cells could draw on GSH to replenish SAM, effectively depleting GSH. Spermine synthase and spermidine/spermine acetyl transferaseare located on the X chromosome, and demethylation of the inactivated X chromosome in females would allow for increased gene dosage in SLE [15]. Furthermore, polyamines induce histone acetylation which promotes transcription [66,67]. The sensitivity of SAM generation to oxidative stress and ornithine decarboxylase to UV make spermine and other polyamines possible biomarkers for disease activity in SLE.
Histone modifications
Methylation
Much like DNA methylation, histone methylation is also dependent on SAM as a methyl donor and HMTs are inhibited by SAH (Figure 1) [31]. A distinction of histone methylation from DNA methylation is that the methylation of histones does not act as a simple on/off switch for transcription. Instead, the methylation code of histones is determined by both which lysine is methylated as well as the number methyl groups attached to the lysines [68]. When lysine 79 of H3is mono or dimethylated, transcription is activated [15,69]. Conversely, trimethylation of H3K79 represses transcription [69]. Much like DNMTs, HMTs maintain reduced cysteine residues at that are involved in the catalysis of histone methylation [70]. These cysteines are targets of ROI and implicate oxidative stress and GSH depletion in histone methylation dysfunction [25]. Instances of increased histone methylation have been reported in SLE T cells indicating HMT activity may be increased [71,72]. The complexity of the histone methylation code does not lend itself to straightforward regulation by metabolism and more studies are required to determine how SAM, GSH, and SAH availability affects histone methylation in SLE.
Demethylation
Metabolically, there are 2 classes of histone demethylases. Lysine-specific demethylases (LSD) are flavin adenine dinucleotide (FAD) dependent and the JumonjiC (JmjC) domain containing histone demethylases which have the same substrates as TET-family demethylases [21]. FAD is formed from ATP and riboflavin (B2) in mitochondria [20]. The demethylation of histones by LSD enzymes generates reduced FAD (FADH2). To recycle FADH2, the cell uses O2 to oxidize FADH2 into FAD which results in the production of H2O2 [21]. In addition, each demethylation reaction catalyzed by LSD1 generates one molecule of H2O2 and can result in the oxidation of nearby DNA bases [73]. Thus, demethylation of histones ultimately causes oxidative stress through the production of H2O2. Consequently, LSD1 negatively regulates the transcription of many genes involved in energetic metabolism such as PGC-1α, PDK4, and ATGL [74].
Diminished JmjC demethylase 3 activity results in increased trimethylation of H3K27 and is implicated in increased T and B cell activation in SLE [72]. The JmjC demethylases have the same requirements as the TET family DNA demethylases, and thus the same metabolic disturbances that affect α-KG concentration and TET function also affect JmjC demethylases. Succinate is a product of TET and JmjC demethylation, which acts through feedback inhibition to block further DNA and histone demethylation [75]. Furthermore, both JmjC and TET demethylases are negatively regulated by ROI [76].
The oxidative environment of lupus T cells may inhibit the function of these demethylases. The mammalian target of rapamycin (mTOR) may also regulate the JmjC histone demethylases through its control of hypoxia inducible factor 1 (Hif1) transcription. HIF1 acts as a transcription factor to upregulate the transcription of some of the JmjC demethylases [77]. mTOR is sensitive to amino acid and glucose levels and positively regulates Hif1 expression [78]. Conversely, the EgIN prolyl hydxroylases negatively regulate Hif1 [20]. The EgIN prolyl hydroxylases are induced by α-KG and inhibited by succinate and fumarate, all products of the TCA cycle [20]. The dependence of demethylase enzymes on mitochondrial substrates makes understanding mitochondrial dysfunction in SLE that much more essential [27].
Acetylation
The positive charge of lysines allows for tight binding of histones to negatively charged DNA, which inhibits transcription by hindering access of transcription enzymes to DNA. Acetylation by histone acetylases (HATs) eliminates the charge differences between histones and DNA allowing transcription factors to access DNA [79]. The expression of the HAT p300 is protective against lupus and its loss results in a SLE phenotype in mice [80]. HATs utilize acetyl-CoA for acetylation reactions. Acetyl-CoA can be generated in mitochondria from pyruvate or in the cytosol during fatty acid synthesis. The citrate that is used for fatty acid synthesis is generated by the TCA cycle from glucose or α-KG in the forward and reverse cycle directions, respectively.
The export of citrate from mitochondria, typically in a well-fed state, results in the generation of cytosolic acetyl-CoA by ATP-citrate lyase. Cytosolic acetyl-CoA can then be used to acetylate histones in the nucleus. The concentration of acetyl-CoA is essential for acetylation because its abundance and depletion induce acetylation and hypoacetylation, respectively, in yeast [81,82]. The dependence on the TCA cycle for both acetyl-CoA and α-KG link acetylation and demethylation processes of the cell.
The ERK pathway may also affect acetylation indirectly. ERK positively regulates pyruvate dehydrogenase (PDH), the enzyme responsible for converting pyruvate to acetyl-CoA [83]. Due to low PKCδ and ERK activity in SLE, one might expect less flux of glucose into the TCA due to less PDH activity. TCA cycle activity is actually increased in chronically activated lymphocytes so the interrelationship between ERK and glycolysis/TCA activity may not be as clear cut [84]. ERK also disrupts the TSC1/TSC2 complex which prevents TSC2 from inhibiting mTOR [85]. Low ERK in SLE would then allow for mTOR inhibition in CD4+ T cells, which is not the case, indicating that mTOR activation is independent of ERK in lupus CD4+ T cells. Rather, mTOR is elevated in lupus CD4+ T cells and causes dysfunction through increased recycling of CD4 and TCRζ via HRES/Rab4 and HIF1 as mentioned above [78,86].
Deacetylation
Histone deacetylases (HDACs) and NAD+ dependent sirtuins act on histones to cleave acetyl moieties. HDACs hydrolyze acetyl groups and thus are not dependent on metabolite cofactors to be active. Cell metabolism does affect HDAC activity through ROI and RNS signaling. The activities of HDAC1, HDAC2, and HDAC3 are reduced due to nitration and HDAC2 is degraded under nitrative and oxidative stress [87,88]. Sirtuins 1, 2, 6, and 7 are localized to the nucleus. Sirtuins require NAD+ as a cofactor and their activity is dependent on the NAD+/NADH ratio [89]. Sirtuins are sensitive to the oxidative state of the cell because NAD+ and NADH are used in reduction-oxidation reactions [89]. NAD+ is an important substrate in glycolysis and it dominates over its reduced form, NADH [90]. NAD+ can be generated de novo from tryptophan, an essential amino acid further implicating diet in epigenetic regulation [91]. Interestingly, tryptophan is depleted in SLE and its catabolite, kynurenine, is increased which may have an effect on de novo NAD+ synthesis [92,93]. Niacin and aspartate are also NAD+ precursors making NAD+ central to many metabolic pathways that affect epigenetics.
As a cofactor in sirtuin driven deacetylation, NAD+ is cleaved into nicotinamide (NAM) and O-acetyl-ADP-ribose [21]. NAM acts to inhibit sirtuins and thus the product of deacetylation acts to inhibit further sirtuin activity [94]. NAM can also be recycled to regenerate NAD+. NADH is also an inhibitor of sirtuins [89]. Glycolysis and redox are thus equally important for acetylation reactions as deacetylation reactions.
Hypoacetylation of histone H3 correlates directly with SLE disease activity [95]. In MRL/lpr mice there is increased expression of Sirt1 and decreased expression of HDAC2 and HDAC7 in CD4+ T cells [95,96]. Despite the downregulation of HDAC proteins, HDAC inhibitors increase histone acetylation and improve disease in mouse lupus models [97]. The overexpression of Sirt1 drives histone hypoacetylation and may deprive glycolysis of NAD+ or drive increased degradation of tryptophan to replenish NAD+.
Acetylation in SLE is very dynamic and is different depending on cell type [13]. Monocyte H4 histones are hyperacetylated in SLE [98]. Interestingly, it was found that hyperacetylation of monocytes histones leads to increased production of the pro-inflammatory cytokine TNF-α [99]. Lupus monocytes have impaired glycolysis and thus less carbon is being shunted into the TCA cycle and lactate production [100]. The low glycolytic activity of these cells may provide less NAD+ for deacetylation because there is not likely an increase in acetyl-CoA to drive acetylation. In contrast to lupus monocytes, lupus CD4+ T cells have general hypoacetylation [95]. One would expect the abundance of acetyl-CoA from increased TCA cycle activity in chronically activated lymphocytes to increase acetylation [84]. Consequently, the hypoacetylation may then be driven by sirtuins rather than HATs in lupus CD4+ cells.
Phosphorylation
Histone phosphorylation is another method of transcriptional regulation. ATP is not considered a limiting factor for kinases that act on histones [21]. There are kinases that act as metabolic sensors which then phosphorylate histones based on metabolite levels. A metabolic sensor that has the capacity to phosphorylate histones is AKT [101]. AKT is phosphorylated by mTORC2, which is sensitive to amino acid levels in the cell [102]. Thus, the cellular concentration of amino acids can drive histone phosphorylation. AMP activated protein kinase (AMPK) is sensitive to low ATP/AMP ratios and is activated under these conditions. During energy stress, AMPK translocates to the nucleus and phosphorylates histones to activate transcription of stress related genes [103].
In SLE, ATP is depleted and thus a low ATP/AMP activates AMPK [104,105]. There are currently no studies found upon cross-referencing SLE and AMPK, but there is a great opportunity to explore the regulatory effects of AMPK on the epigenome of SLE T cells. Furthermore, AMPK negatively regulates mTOR, which is activated in lupus T cells [106]. Many other kinases phosphorylate histones but metabolic control of their activities is not as well described [101].
Lipid peroxidation/carbonylation
The oxidative environment in SLE can generate reactive lipid species. Peroxidation of lipids generates malondialdehyde (MDA) and 4-hydroxynonenal (4-HNE). MDA reacts with adenosine and guanosine in DNA and forms adducts which can inhibit RNA polymerase II (RNAP II) activity [107,108]. RNAP II is responsible for the synthesis of mRNA during transcription. Thus, MDA inhibition of transcription could result in aberrant gene expressionin SLE. 4-HNE reacts with HDACs 1-3 to carbonylate cysteine residues [109]. 4-HNE repression of HDACs permits Gadd45 transcription, which is overexpressed in SLE T cells [13,110]. Gadd45 promotes demethylation and gene transcription of CD11a, and CD70 in autoreactive cells [110,111]. Gadd45 is also activated by UV radiation which causes lupus flares and induces oxidative stress [6,12,110]. Gadd45 connects oxidative stress, lipid peroxidation, histone acetylation, UV damage, and demethylation to lupus flares.
GlcNAcylation/ubiquitination
A recently discovered histone modification is O-linked N-acetylglucosamine (GlcNAc)[112]. GlcNAc is formed by amination of glucose. The TET-family DNA demethylase enzymes TET2 and TET3 regulate GlcNAcylation of histones by interacting with O-linked GlcNAc transferase (OGT) [113,114]. The TET2/3-OGT interaction with chromatin tends to induce transcription as well as histone ubiquitination [112]. With respect to SLE, it is not currently known how histone glycosylation may affect pathogenesis, but a link may exist between TET driven demethylation and glycosylation. Although histone glycosylation has not been described in SLE, the addition of GlcNAc to the transcription factor Elf1 is defective in SLE [115]. Elf1 is responsible for the transcription of TCRζ in T cells. The loss of glycosylation prevents Elf1 from translocating to the nucleus and inducing TCRζ transcription [115]. As stated above, the activation of the mTOR results in increased degradation of TCRζ through increased recycling by HRES/Rab4 endocytic trafficking [106]. Thus, the downregulation of TCRζ in SLE is regulated by two different metabolic pathways. GlcNAc is relevant in SLE pathogenesis and further studies are needed to determine how histone glycosylation may affect SLE activity.
Lymphocyte activation and metabolism
Acutely activated lymphocytes block OXPHOS and instead favor glycolysis as the major source of ATP [116]. In blocking OXPHOS, there is increased lactate production in acutely activated lymphocytes which diverts carbon from the TCA. This would effectively prevent the generation of α-KG for demethylation enzymes. Thus, acute activation of lymphocytes could affect the methylation status of histones and DNA. Upregulation of glycolysis by acutely activated lymphocytes also acts to protect the cells from ROI [117]. In contrast, chronically activated lymphocytes, such as lupus T cells, generate ATP predominantly from OXPHOS [84]. Chronically activated CD4+ and CD8+ T cells from NZB/W lupus mice showed less lactate production than their BALB/c controls indicating that these lymphocytes are not utilizing glycolysis for their chief source of ATP [84]. The shift from acute to chronic activation in lupus T cells likely alters ROI production as well as the availability of SAM, GSH, and α-KG. The transition from acute to chronic activation is suitable for metabolome studies and epigenetic changes that occur as a result.
Conclusion
The role of metabolism in epigenetic regulation of SLE is still greatly underexplored. With recent advances in the ability to detect a large number of metabolites via mass spectrometry, the metabolome of SLE monocytes, T cells, and B cells is well within reach [118]. Identifying the relative differences between healthy controls and SLE patients with flares will provide a metabolic profile of SLE. Future studies will also need to focus on the subcellular localization of metabolites. The mitochondria generates many of the metabolites necessary for epigenetic regulation but the impermeability of the inner mitochondrial membrane makes the availability of these metabolites dependent on transmembrane transport [119].
The nuclear membrane, on the other hand, allows passive transport for molecules up to 60 kilodaltons which permits small metabolites to easily enter the nucleus [120]. Key metabolites will act as nodes that connect pathways involved in epigenetic regulation of SLE and serve as biomarkers of disease activity. When building the metabolome of SLE, we can then identify pathways that may be sensitive to changes in metabolite availability due to diet, transport, or enzymatic turnover.
Acknowledgments
This work was supported in part by grant AI072648 from the National Institutes of Health, the Alliance for Lupus Research, and the Central New York Community Foundation.
Footnotes
Declaration of interest
The authors alone are responsible for the content and writing of the paper.
References
- 1.Kyttaris VC, Tsokos GC. T lymphocytes in systemic lupus erythematosus: an update. Curr Opin Rheumatol. 2004;16:548–552. doi: 10.1097/01.bor.0000132646.55056.e0. [DOI] [PubMed] [Google Scholar]
- 2.Anolik J, Sanz I. B cells in human and murine systemic lupus erythematosus. Curr Opin Rheumatol. 2004;16:505–512. doi: 10.1097/01.bor.0000133660.52599.f6. [DOI] [PubMed] [Google Scholar]
- 3.Nagy G, Koncz A, Perl A. T- and B-cell abnormalities in systemic lupus erythematosus. Crit Rev Immunol. 2005;25:123–140. doi: 10.1615/critrevimmunol.v25.i2.30. [DOI] [PubMed] [Google Scholar]
- 4.Pascual V, Banchereau J, Palucka AK. The central role of dendritic cells and interferon-alpha in SLE. Curr Opin Rheumatol. 2003;15:548–556. doi: 10.1097/00002281-200309000-00005. [DOI] [PubMed] [Google Scholar]
- 5.Crispin JC, Alcocer-Varela J. The role myeloid dendritic cells play in the pathogenesis of systemic lupus erythematosus. Autoimmun Rev. 2007;6:450–456. doi: 10.1016/j.autrev.2007.01.014. [DOI] [PubMed] [Google Scholar]
- 6.Rao T, Richardson B. Environmentally induced autoimmune diseases: potential mechanisms. Environ Health Perspect. 1999;107:737–742. doi: 10.1289/ehp.99107s5737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Manderson AP, Botto M, Walport MJ. The role of complement in the development of systemic lupus erythematosus. Annu Rev Immunol. 2004;22:431–456. doi: 10.1146/annurev.immunol.22.012703.104549. [DOI] [PubMed] [Google Scholar]
- 8.Gateva V, Sandling JK, Hom G, et al. A large-scale replication study identifies TNIP1, PRDM1, JAZF1, UHRF1BP1 and IL10 as risk loci for systemic lupus erythematosus. Nat Genet. 2009;41:1228–1233. doi: 10.1038/ng.468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nagy G, Ward J, Mosser DD, et al. Regulation of CD4 expression via recycling by HRES-1/RAB4 controls susceptibility to HIV infection. J Biol Chem. 2006;281:34574–34591. doi: 10.1074/jbc.M606301200. [DOI] [PubMed] [Google Scholar]
- 10.Vaughn SE, Kottyan LC, Munroe ME, Harley JB. Genetic susceptibility to lupus: the biological basis of genetic risk found in B cell signaling pathways. J Leukoc Biol. 2012;92:577–591. doi: 10.1189/jlb.0212095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Flesher DL, Sun X, Behrens TW, et al. Recent advances in the genetics of systemic lupus erythematosus. Expert Rev Clin Immunol. 2010;6:461–479. doi: 10.1586/eci.10.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Richardson B. DNA methylation and autoimmune disease. Clin Immunol. 2003;109:72–79. doi: 10.1016/s1521-6616(03)00206-7. [DOI] [PubMed] [Google Scholar]
- 13.Patel DR, Richardson BC. Epigenetic mechanisms in lupus. Curr Opin Rheumatol. 2010;22:478–482. doi: 10.1097/BOR.0b013e32833ae915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Richardson B. Primer: epigenetics of autoimmunity. Nat Clin Pract Rheumatol. 2007;3:521–527. doi: 10.1038/ncprheum0573. [DOI] [PubMed] [Google Scholar]
- 15.Brooks WH, Le Dantec C, Pers JO, et al. Epigenetics and autoimmunity. J Autoimmun. 2010;34:J207–J219. doi: 10.1016/j.jaut.2009.12.006. [DOI] [PubMed] [Google Scholar]
- 16.Zouali M. Epigenetics in lupus. Ann N Y Acad Sci. 2011;1217:154–165. doi: 10.1111/j.1749-6632.2010.05831.x. [DOI] [PubMed] [Google Scholar]
- 17.Richardson B, Scheinbart L, Strahler J, et al. Evidence for impaired T cell DNA methylation in systemic lupus erythematosus and rheumatoid arthritis. Arthritis Rheum. 1990;33:1665–1673. doi: 10.1002/art.1780331109. [DOI] [PubMed] [Google Scholar]
- 18.Zhang Y, Zhao M, Sawalha AH, et al. Impaired DNA methylation and its mechanisms in CD4(+)T cells of systemic lupus erythematosus. J Autoimmun. 2013;41:92–99. doi: 10.1016/j.jaut.2013.01.005. [DOI] [PubMed] [Google Scholar]
- 19.Coit P, Jeffries M, Altorok N, et al. Genome-wide DNA methylation study suggests epigenetic accessibility and transcriptional poising of interferon-regulated genes in naive CD4+ T cells from lupus patients. J Autoimmun. 2013;43:78–84. doi: 10.1016/j.jaut.2013.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kaelin WG, Jr, McKnight SL. Influence of metabolism on epigenetics and disease. Cell. 2013;153:56–69. doi: 10.1016/j.cell.2013.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lu C, Thompson CB. Metabolic regulation of epigenetics. Cell Metab. 2012;16:9–17. doi: 10.1016/j.cmet.2012.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ray PD, Huang BW, Tsuji Y. Reactive oxygen species (ROS) homeostasis and redox regulation in cellular signaling. Cell Signal. 2012;24:981–990. doi: 10.1016/j.cellsig.2012.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ulrey CL, Liu L, Andrews LG, Tollefsbol TO. The impact of metabolism on DNA methylation. Hum Mol Genet. 2005;14:R139–R147. doi: 10.1093/hmg/ddi100. [DOI] [PubMed] [Google Scholar]
- 24.Perl A. Oxidative stress and endosome recycling are complementary mechanisms reorganizing the T-cell receptor signaling complex in SLE. Clin Immunol. 2012;142:219–222. doi: 10.1016/j.clim.2011.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Goswami SK. Cellular redox, epigenetics and diseases. Subcell Biochem. 2012;61:527–542. doi: 10.1007/978-94-007-4525-4_23. [DOI] [PubMed] [Google Scholar]
- 26.Adam-Vizi V. Production of reactive oxygen species in brain mitochondria: contribution by electron transport chain and non-electron transport chain sources. Antioxid Redox Signal. 2005;7:1140–1149. doi: 10.1089/ars.2005.7.1140. [DOI] [PubMed] [Google Scholar]
- 27.Perl A, Hanczko R, Doherty E. Assessment of mitochondrial dysfunction in lymphocytes of patients with systemic lupus erythematosus. Methods Mol Biol. 2012;900:61–89. doi: 10.1007/978-1-60761-720-4_4. [DOI] [PubMed] [Google Scholar]
- 28.Corvetta A, Della Bitta R, Luchetti MM, Pomponio G. 5-methylcytosine content of DNA in blood, synovial mononuclear cells and synovial tissue from patients affected by autoimmune rheumatic diseases. J Chromatogr. 1991;566:481–491. doi: 10.1016/0378-4347(91)80265-e. [DOI] [PubMed] [Google Scholar]
- 29.Lu SC, Alvarez L, Huang ZZ, et al. Methionine adenosyltransferase 1A knockout mice are predisposed to liver injury and exhibit increased expression of genes involved in proliferation. Proc Natl Acad Sci USA. 2001;98:5560–5565. doi: 10.1073/pnas.091016398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Avila MA, Corrales FJ, Ruiz F, et al. Specific interaction of methionine adenosyltransferase with free radicals. Biofactors. 1998;8:27–32. doi: 10.1002/biof.5520080106. [DOI] [PubMed] [Google Scholar]
- 31.Selhub J, Miller JW. The pathogenesis of homocysteinemia: interruption of the coordinate regulation by S-adenosylmethionine of the remethylation and transsulfuration of homocysteine. Am J Clin Nutr. 1992;55:131–138. doi: 10.1093/ajcn/55.1.131. [DOI] [PubMed] [Google Scholar]
- 32.Sipka S. Adenosine inhibits the release of arachidonic acid in activated human peripheral mononuclear cells. A proposed model for physiologic and pathologic regulation in systemic lupus erythematosus. Scientific World J. 2011;11:972–980. doi: 10.1100/tsw.2011.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dawson H, Collins G, Pyle R, et al. The immunoregulatory effects of homocysteine and its intermediates on T-lymphocyte function. Mech Ageing Dev. 2004;125:107–110. doi: 10.1016/j.mad.2003.11.013. [DOI] [PubMed] [Google Scholar]
- 34.do Prado R, D’Almeida VM, Guerra-Shinohara E, et al. Increased concentration of plasma homocysteine in children with systemic lupus erythematosus. Clin Exp Rheumatol. 2006;24:594–598. [PubMed] [Google Scholar]
- 35.Svedruzic ZM, Reich NO. DNA cytosine C5 methyltransferase Dnmt1: catalysis-dependent release of allosteric inhibition. Biochemistry. 2005;44:9472–9485. doi: 10.1021/bi050295z. [DOI] [PubMed] [Google Scholar]
- 36.Deng C, Kaplan MJ, Yang J, et al. Decreased ras-mitogen-activated protein kinase signaling may cause DNA hypomethylation in T lymphocytes from lupus patients. Arthritis Rheum. 2001;44:397–407. doi: 10.1002/1529-0131(200102)44:2<397::AID-ANR59>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 37.Perl A. Pathogenic mechanisms in systemic lupus erythematosus. Autoimmunity. 2010;43:1–6. doi: 10.3109/08916930903374741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Deng C, Yang J, Scott J, et al. Role of the ras-MAPK signaling pathway in the DNA methyltransferase response to DNA hypomethylation. Biol Chem. 1998;379:1113–1120. doi: 10.1515/bchm.1998.379.8-9.1113. [DOI] [PubMed] [Google Scholar]
- 39.Gorelik G, Fang JY, Wu A, et al. Impaired T cell protein kinase C delta activation decreases ERK pathway signaling in idiopathic and hydralazine-induced lupus. J Immunol. 2007;179:5553–5563. doi: 10.4049/jimmunol.179.8.5553. [DOI] [PubMed] [Google Scholar]
- 40.Januchowski R, Wudarski M, Chwalinska-Sadowska H, Jagodzinski PP. Prevalence of ZAP-70, LAT, SLP-76, and DNA methyltransferase 1 expression in CD4+ T cells of patients with systemic lupus erythematosus. Clin Rheumatol. 2008;27:21–27. doi: 10.1007/s10067-007-0644-8. [DOI] [PubMed] [Google Scholar]
- 41.Gorelik GJ, Yarlagadda S, Richardson BC. Protein kinase cdelta oxidation contributes to ERK inactivation in lupus T cells. Arthritis Rheum. 2012;64:2964–2974. doi: 10.1002/art.34503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cornacchia E, Golbus J, Maybaum J, et al. Hydralazine and procainamide inhibit T cell DNA methylation and induce autoreactivity. J Immunol. 1988;140:2197–2200. [PubMed] [Google Scholar]
- 43.Li-Weber M, Treiber MK, Giaisi M, et al. Ultraviolet irradiation suppresses T cell activation via blocking TCR-mediated ERK and NF-kappa B signaling pathways. J Immunol. 2005;175:2132–2143. doi: 10.4049/jimmunol.175.4.2132. [DOI] [PubMed] [Google Scholar]
- 44.Belot A, Kasher PR, Trotter EW, et al. Protein kinase C delta deficiency causes mendelian systemic lupus erythematosus with B-cell defective apoptosis and hyperproliferation. Arthritis Rheum. 2013;65:2161–2171. doi: 10.1002/art.38008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mazari L, Ouarzane M, Zouali M. Subversion of B lymphocyte tolerance by hydralazine, a potential mechanism for drug-induced lupus. Proc Natl Acad Sci USA. 2007;104:6317–6322. doi: 10.1073/pnas.0610434104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Franchini DM, Schmitz KM, Petersen-Mahrt SK. 5-methylcytosine DNA demethylation: more than losing a methyl group. Annu Rev Genet. 2012;46:419–441. doi: 10.1146/annurev-genet-110711-155451. [DOI] [PubMed] [Google Scholar]
- 47.Zan H, Casali P. Regulation of aicda expression and AID activity. Autoimmunity. 2013;46:83–101. doi: 10.3109/08916934.2012.749244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Garcia-Manteiga JM, Mari S, Godejohann M, et al. Metabolomics of B to plasma cell differentiation. J Proteome Res. 2011;10:4165–4176. doi: 10.1021/pr200328f. [DOI] [PubMed] [Google Scholar]
- 49.Wu T, Xie C, Han J, et al. Metabolic disturbances associated with systemic lupus erythematosus. PLoS One. 2012;7:e37210. doi: 10.1371/journal.pone.0037210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hoffman DR, Marion DW, Cornatzer WE, Duerre JA. S-adenosylmethionine and S-adenosylhomocystein metabolism in isolated rat liver. effects of L-methionine, L-homocystein, and adenosine. J Biol Chem. 1980;255:10822–10827. [PubMed] [Google Scholar]
- 51.Yi P, Melnyk S, Pogribna M, et al. Increase in plasma homocysteine associated with parallel increases in plasma S-adenosylhomocysteine and lymphocyte DNA hypomethylation. J Biol Chem. 2000;275:29318–29323. doi: 10.1074/jbc.M002725200. [DOI] [PubMed] [Google Scholar]
- 52.Lertratanangkoon K, Wu CJ, Savaraj N, Thomas ML. Alterations of DNA methylation by glutathione depletion. Cancer Lett. 1997;120:149–156. doi: 10.1016/s0304-3835(97)00300-5. [DOI] [PubMed] [Google Scholar]
- 53.Poirier LA, Wise CK, Delongchamp RR, Sinha R. Blood determinations of S-adenosylmethionine, S-adenosylhomocysteine, and homocysteine: correlations with diet. Cancer Epidemiol Biomarkers Prev. 2001;10:649–655. [PubMed] [Google Scholar]
- 54.Kim YI. Nutritional epigenetics: impact of folate deficiency on DNA methylation and colon cancer susceptibility. J Nutr. 2005;135:2703–2709. doi: 10.1093/jn/135.11.2703. [DOI] [PubMed] [Google Scholar]
- 55.Feil R, Fraga MF. Epigenetics and the environment: emerging patterns and implications. Nat Rev Genet. 2012;13:97–109. doi: 10.1038/nrg3142. [DOI] [PubMed] [Google Scholar]
- 56.Kimura M, Umegaki K, Higuchi M, et al. Methylenetetrahydrofolate reductase C677T polymorphism, folic acid and riboflavin are important determinants of genome stability in cultured human lymphocytes. J Nutr. 2004;134:48–56. doi: 10.1093/jn/134.1.48. [DOI] [PubMed] [Google Scholar]
- 57.Huck S, Zouali M. DNA methylation: a potential pathway to abnormal autoreactive lupus B cells. Clin Immunol Immunopathol. 1996;80:1–8. doi: 10.1006/clin.1996.0087. [DOI] [PubMed] [Google Scholar]
- 58.Huck S, Deveaud E, Namane A, Zouali M. Abnormal DNA methylation and deoxycytosine-deoxyguanine content in nucleosomes from lymphocytes undergoing apoptosis. FASEB J. 1999;13:1415–1422. doi: 10.1096/fasebj.13.11.1415. [DOI] [PubMed] [Google Scholar]
- 59.He YF, Li BZ, Li Z, et al. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science. 2011;333:1303–1307. doi: 10.1126/science.1210944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ito S, Shen L, Dai Q, et al. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science. 2011;333:1300–1303. doi: 10.1126/science.1210597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bruniquel D, Schwartz RH. Selective, stable demethylation of the interleukin-2 gene enhances transcription by an active process. Nat Immunol. 2003;4:235–240. doi: 10.1038/ni887. [DOI] [PubMed] [Google Scholar]
- 62.Krishna P, Fritzler MJ, Van de Sande JH. Interactions of anti-DNA antibodies with Z-DNA. Clin Exp Immunol. 1993;92:51–57. doi: 10.1111/j.1365-2249.1993.tb05947.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Puri H, Campbell RA, Puri-Harner V, et al. Serum-free polyamines in children with systemic lupus erythematosus. Vol. 2. New York: Raven Press; 1978. pp. 359–367. [Google Scholar]
- 64.Thomas TJ, Seibold JR, Adams LE, Hess EV. Hydralazine induces Z-DNA conformation in a polynucleotide and elicits anti(Z-DNA) antibodies in treated patients. Biochem J. 1993;294:419–425. doi: 10.1042/bj2940419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ahmad N, Gilliam AC, Katiyar SK, et al. A definitive role of ornithine decarboxylase in photocarcinogenesis. Am J Pathol. 2001;159:885–892. doi: 10.1016/S0002-9440(10)61764-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hobbs CA, Gilmour SK. High levels of intracellular polyamines promote histone acetyltransferase activity resulting in chromatin hyperacetylation. J Cell Biochem. 2000;77:345–360. [PubMed] [Google Scholar]
- 67.Hobbs CA, Paul BA, Gilmour SK. Deregulation of polyamine biosynthesis alters intrinsic histone acetyltransferase and deacetylase activities in murine skin and tumors. Cancer Res. 2002;62:67–74. [PubMed] [Google Scholar]
- 68.Upadhyay AK, Cheng X. Dynamics of histone lysine methylation: structures of methyl writers and erasers. Prog Drug Res. 2011;67:107–124. doi: 10.1007/978-3-7643-8989-5_6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Barski A, Cuddapah S, Cui K, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 70.Zhang X, Yang Z, Khan SI, et al. Structural basis for the product specificity of histone lysine methyltransferases. Mol Cell. 2003;12:177–185. doi: 10.1016/s1097-2765(03)00224-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rauen T, Grammatikos AP, Hedrich CM, et al. cAMP-responsive element modulator alpha (CREMalpha) contributes to decreased notch-1 expression in T cells from patients with active systemic lupus erythematosus (SLE) J Biol Chem. 2012;287:42525–42532. doi: 10.1074/jbc.M112.425371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhang Q, Long H, Liao J, et al. Inhibited expression of hematopoietic progenitor kinase 1 associated with loss of jumonji domain containing 3 promoter binding contributes to autoimmunity in systemic lupus erythematosus. J Autoimmun. 2011;37:180–189. doi: 10.1016/j.jaut.2011.09.006. [DOI] [PubMed] [Google Scholar]
- 73.Perillo B, Ombra MN, Bertoni A, et al. DNA oxidation as triggered by H3K9me2 demethylation drives estrogen-induced gene expression. Science. 2008;319:202–206. doi: 10.1126/science.1147674. [DOI] [PubMed] [Google Scholar]
- 74.Hino S, Sakamoto A, Nagaoka K, et al. FAD-dependent lysine-specific demethylase-1 regulates cellular energy expenditure. Nat Commun. 2012;3:758. doi: 10.1038/ncomms1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Letouze E, Martinelli C, Loriot C, et al. SDH mutations establish a hypermethylator phenotype in paraganglioma. Cancer Cell. 2013;23:739–752. doi: 10.1016/j.ccr.2013.04.018. [DOI] [PubMed] [Google Scholar]
- 76.Chervona Y, Costa M. The control of histone methylation and gene expression by oxidative stress, hypoxia, and metals. Free Radic Biol Med. 2012;53:1041–1047. doi: 10.1016/j.freeradbiomed.2012.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mimura I, Nangaku M, Kanki Y, et al. Dynamic change of chromatin conformation in response to hypoxia enhances the expression of GLUT3 (SLC2A3) by cooperative interaction of hypoxia-inducible factor 1 and KDM3A. Mol Cell Biol. 2012;32:3018–3032. doi: 10.1128/MCB.06643-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hudson CC, Liu M, Chiang GG, et al. Regulation of hypoxia-inducible factor 1alpha expression and function by the mammalian target of rapamycin. Mol Cell Biol. 2002;22:7004–7014. doi: 10.1128/MCB.22.20.7004-7014.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lee DY, Hayes JJ, Pruss D, Wolffe AP. A positive role for histone acetylation in transcription factor access to nucleosomal DNA. Cell. 1993;72:73–84. doi: 10.1016/0092-8674(93)90051-q. [DOI] [PubMed] [Google Scholar]
- 80.Forster N, Gallinat S, Jablonska J, et al. P300 protein acetyltransferase activity suppresses systemic lupus erythematosus-like autoimmune disease in mice. J Immunol. 2007;178:6941–6948. doi: 10.4049/jimmunol.178.11.6941. [DOI] [PubMed] [Google Scholar]
- 81.Cai L, Sutter BM, Li B, Tu BP. Acetyl-CoA induces cell growth and proliferation by promoting the acetylation of histones at growth genes. Mol Cell. 2011;42:426–437. doi: 10.1016/j.molcel.2011.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Takahashi H, McCaffery JM, Irizarry RA, Boeke JD. Nucleocytosolic acetyl-coenzyme a synthetase is required for histone acetylation and global transcription. Mol Cell. 2006;23:207–217. doi: 10.1016/j.molcel.2006.05.040. [DOI] [PubMed] [Google Scholar]
- 83.Grassian AR, Metallo CM, Coloff JL, et al. Erk regulation of pyruvate dehydrogenase flux through PDK4 modulates cell proliferation. Genes Dev. 2011;25:1716–1733. doi: 10.1101/gad.16771811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wahl DR, Petersen B, Warner R, et al. Characterization of the metabolic phenotype of chronically activated lymphocytes. Lupus. 2010;19:1492–1501. doi: 10.1177/0961203310373109. [DOI] [PubMed] [Google Scholar]
- 85.Ma L, Chen Z, Erdjument-Bromage H, et al. Phosphorylation and functional inactivation of TSC2 by erk implications for tuberous sclerosis and cancer pathogenesis. Cell. 2005;121:179–193. doi: 10.1016/j.cell.2005.02.031. [DOI] [PubMed] [Google Scholar]
- 86.Perl A. Systems biology of lupus: mapping the impact of genomic and environmental factors on gene expression signatures, cellular signaling, metabolic pathways, hormonal and cytokine imbalance, and selecting targets for treatment. Autoimmunity. 2010;43:32–47. doi: 10.3109/08916930903374774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Osoata GO, Yamamura S, Ito M, et al. Nitration of distinct tyrosine residues causes inactivation of histone deacetylase 2. Biochem Biophys Res Commun. 2009;384:366–371. doi: 10.1016/j.bbrc.2009.04.128. [DOI] [PubMed] [Google Scholar]
- 88.Yang SR, Chida AS, Bauter MR, et al. Cigarette smoke induces proinflammatory cytokine release by activation of NF-kappaB and posttranslational modifications of histone deacetylase in macrophages. Am J Physiol Lung Cell Mol Physiol. 2006;291:L46–L57. doi: 10.1152/ajplung.00241.2005. [DOI] [PubMed] [Google Scholar]
- 89.Reilly CM, Regna N, Mishra N. HDAC inhibition in lupus models. Mol Med. 2011;17:417–425. doi: 10.2119/molmed.2011.00055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Veech RL, Eggleston LV, Krebs HA. The redox state of free nicotinamide-adenine dinucleotide phosphate in the cytoplasm of rat liver. Biochem J. 1969;115:609–619. doi: 10.1042/bj1150609a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lin SJ, Guarente L. Nicotinamide adenine dinucleotide, a metabolic regulator of transcription, longevity and disease. Curr Opin Cell Biol. 2003;15:241–246. doi: 10.1016/s0955-0674(03)00006-1. [DOI] [PubMed] [Google Scholar]
- 92.Widner B, Sepp N, Kowald E, et al. Degradation of tryptophan in patients with systemic lupus erythematosus. Adv Exp Med Biol. 1999;467:571–577. doi: 10.1007/978-1-4615-4709-9_71. [DOI] [PubMed] [Google Scholar]
- 93.Widner B, Sepp N, Kowald E, et al. Enhanced tryptophan degradation in systemic lupus erythematosus. Immunobiology. 2000;201:621–630. doi: 10.1016/S0171-2985(00)80079-0. [DOI] [PubMed] [Google Scholar]
- 94.Yamamori T, DeRicco J, Naqvi A, et al. SIRT1 deacetylates APE1 and regulates cellular base excision repair. Nucleic Acids Res. 2010;38:832–845. doi: 10.1093/nar/gkp1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Hu N, Qiu X, Luo Y, et al. Abnormal histone modification patterns in lupus CD4+ T cells. J Rheumatol. 2008;35:804–810. [PubMed] [Google Scholar]
- 96.Hu N, Long H, Zhao M, et al. Aberrant expression pattern of histone acetylation modifiers and mitigation of lupus by SIRT1-siRNA in MRL/lpr mice. Scand J Rheumatol. 2009;38:464–471. doi: 10.3109/03009740902895750. [DOI] [PubMed] [Google Scholar]
- 97.Mishra N, Reilly CM, Brown DR, et al. Histone deacetylase inhibitors modulate renal disease in the MRL-lpr/lpr mouse. J Clin Invest. 2003;111:539–552. doi: 10.1172/JCI16153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Zhang Z, Song L, Maurer K, et al. Global H4 acetylation analysis by ChIP-chip in systemic lupus erythematosus monocytes. Genes Immun. 2010;11:124–133. doi: 10.1038/gene.2009.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Lee JY, Kim NA, Sanford A, Sullivan KE. Histone acetylation and chromatin conformation are regulated separately at the TNF-alpha promoter in monocytes and macrophages. J Leukoc Biol. 2003;73:862–871. doi: 10.1189/jlb.1202618. [DOI] [PubMed] [Google Scholar]
- 100.Phillips R, Lomnitzer R, Wadee AA, Rabson AR. Defective monocyte function in patients with systemic lupus erythematosus. Clin Immunol Immunopathol. 1985;34:69–76. doi: 10.1016/0090-1229(85)90008-x. [DOI] [PubMed] [Google Scholar]
- 101.Banerjee T, Chakravarti D. A peek into the complex realm of histone phosphorylation. Mol Cell Biol. 2011;31:4858–4873. doi: 10.1128/MCB.05631-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Tato I, Bartrons R, Ventura F, Rosa JL. Amino acids activate mammalian target of rapamycin complex 2 (mTORC2) via PI3K/akt signaling. J Biol Chem. 2011;286:6128–6142. doi: 10.1074/jbc.M110.166991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Bungard D, Fuerth BJ, Zeng PY, et al. Signaling kinase AMPK activates stress-promoted transcription via histone H2B phosphorylation. Science. 2010;329:1201–1205. doi: 10.1126/science.1191241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Gergely P, Jr, Grossman C, Niland B, et al. Mitochondrial hyperpolarization and ATP depletion in patients with systemic lupus erythematosus. Arthritis Rheum. 2002;46:175–190. doi: 10.1002/1529-0131(200201)46:1<175::AID-ART10015>3.0.CO;2-H. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Griffey RH, Brown MS, Bankhurst AD, et al. Depletion of high-energy phosphates in the central nervous system of patients with systemic lupus erythematosus, as determined by phosphorus-31 nuclear magnetic resonance spectroscopy. Arthritis Rheum. 1990;33:827–833. doi: 10.1002/art.1780330609. [DOI] [PubMed] [Google Scholar]
- 106.Fernandez DR, Telarico T, Bonilla E, et al. Activation of mammalian target of rapamycin controls the loss of TCRzeta in lupus T cells through HRES-1/Rab4-regulated lysosomal degradation. J Immunol. 2009;182:2063–2073. doi: 10.4049/jimmunol.0803600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Cline SD, Riggins JN, Tornaletti S, et al. Malondialdehyde adducts in DNA arrest transcription by T7 RNA polymerase and mammalian RNA polymerase II. Proc Natl Acad Sci USA. 2004;101:7275–7280. doi: 10.1073/pnas.0402252101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Marnett LJ. Lipid peroxidation-DNA damage by malondialdehyde. Mutat Res. 1999;424:83–95. doi: 10.1016/s0027-5107(99)00010-x. [DOI] [PubMed] [Google Scholar]
- 109.Doyle K, Fitzpatrick FA. Redox signaling, alkylation (carbonylation) of conserved cysteines inactivates class I histone deacetylases 1, 2, and 3 and antagonizes their transcriptional repressor function. J Biol Chem. 2010;285:17417–17424. doi: 10.1074/jbc.M109.089250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Li Y, Zhao M, Yin H, et al. Overexpression of the growth arrest and DNA damage-induced 45alpha gene contributes to autoimmunity by promoting DNA demethylation in lupus T cells. Arthritis Rheum. 2010;62:1438–1447. doi: 10.1002/art.27363. [DOI] [PubMed] [Google Scholar]
- 111.Barreto G, Schafer A, Marhold J, et al. Gadd45a promotes epigenetic gene activation by repair-mediated DNA demethylation. Nature. 2007;445:671–675. doi: 10.1038/nature05515. [DOI] [PubMed] [Google Scholar]
- 112.Fujiki R, Hashiba W, Sekine H, et al. GlcNAcylation of histone H2B facilitates its monoubiquitination. Nature. 2011;480:557–560. doi: 10.1038/nature10656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Deplus R, Delatte B, Schwinn MK, et al. TET2 and TET3 regulate GlcNAcylation and H3K4 methylation through OGT and SET1/COMPASS. EMBO J. 2013;32:645–655. doi: 10.1038/emboj.2012.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Chen Q, Chen Y, Bian C, et al. TET2 promotes histone O-GlcNAcylation during gene transcription. Nature. 2013;493:561–564. doi: 10.1038/nature11742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Tsokos GC, Nambiar MP, Juang YT. Activation of the ets transcription factor elf-1 requires phosphorylation and glycosylation: defective expression of activated elf-1 is involved in the decreased TCR zeta chain gene expression in patients with systemic lupus erythematosus. Ann N Y Acad Sci. 2003;987:240–245. doi: 10.1111/j.1749-6632.2003.tb06054.x. [DOI] [PubMed] [Google Scholar]
- 116.Guppy M, Greiner E, Brand K. The role of the crabtree effect and an endogenous fuel in the energy metabolism of resting and proliferating thymocytes. Eur J Biochem. 1993;212:95–99. doi: 10.1111/j.1432-1033.1993.tb17637.x. [DOI] [PubMed] [Google Scholar]
- 117.Brand KA, Hermfisse U. Aerobic glycolysis by proliferating cells: a protective strategy against reactive oxygen species. FASEB J. 1997;11:388–395. doi: 10.1096/fasebj.11.5.9141507. [DOI] [PubMed] [Google Scholar]
- 118.Yuan M, Breitkopf SB, Yang X, Asara JM. A positive/negative ion-switching, targeted mass spectrometry-based metabolomics platform for bodily fluids, cells, and fresh and fixed tissue. Nat Protoc. 2012;7:872–881. doi: 10.1038/nprot.2012.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Lemasters JJ. Modulation of mitochondrial membrane permeability in pathogenesis, autophagy and control of metabolism. J Gastroenterol Hepatol. 2007;2:S31–S37. doi: 10.1111/j.1440-1746.2006.04643.x. [DOI] [PubMed] [Google Scholar]
- 120.Allen TD, Cronshaw JM, Bagley S, et al. The nuclear pore complex: mediator of translocation between nucleus and cytoplasm. J Cell Sci. 2000;113:1651–1659. doi: 10.1242/jcs.113.10.1651. [DOI] [PubMed] [Google Scholar]