Abstract
Studies of the roles of microbial communities in the development of inflammatory bowel diseases (IBD) have reached an important milestone. A decade of genome-wide association studies and other genetic analyses have linked IBD with loci that implicate an aberrant immune response to the intestinal microbiota. More recently, profiling studies of the intestinal microbiome have associated pathogenesis of IBD with characteristic shifts in the composition of the intestinal microbiota, reinforcing the view that IBD results from altered interactions between intestinal microbes and the mucosal immune system. Enhanced technologies can increase our understanding of the interactions between the host and its resident microbiota, and their respective roles in IBD, from both a large-scale pathway view and at the metabolic level. We review important microbiome studies of patients with IBD and describe what we have learned about the mechanisms of intestinal microbiota dysfunction. We describe the recent progress in microbiome research from exploratory 16S-based studies, reporting associations of specific organisms with a disease, to more recent studies that have taken a more nuanced view, addressing the function of the microbiota by metagenomic and metabolomic methods. Finally, we propose study designs and methodologies for future investigations of the microbiome in patients with inflammatory gut and autoimmune diseases in general.
Keywords: Microbiota, Crohn’s disease, ulcerative colitis, metagenomics
Over the past decade, inflammatory bowel diseases (IBD) have emerged as one of the most studied human conditions linked to the gut microbiota.1, 2 IBD comprises both Crohn’s disease (CD) and ulcerative colitis (UC), which together affect over 3.6 million persons.3 Large scale studies of human genetics across a total of 75,000 cases and controls have revealed 163 host susceptibility loci to date.4 These loci are enriched for pathways that interact with environmental factors to modulate intestinal homeostasis.5 The incidence of the disease has been on the rise over the past few decades, further highlighting the role of environmental factors in this disease. IBD was once a very rare disorder, and only began to rise dramatically in incidence in the second half of the 20th century in North America and Europe, at times doubling every decade, and in the last two decades expanded into developing countries, although there are more cases of UC than CD in the developing world.6 In addition, several twin studies have now shown that the concordance rate for IBD between monozygotic twin pairs is significantly less than 50%, with the least concordance in CD.7 IBD is thus a multifaceted disorder in which not only germline genetics and the immune system, but also several environmental factors, play an important role.8 One such factor, the gut microbial community, is gaining increasing attention for its influence on many aspects of health in general,9 and IBD in particular (Table 1).
Table 1.
Changes in the microbiome linked to Inflammatory Bowel Disease
| Microbial composition | Decrease in alpha diversity |
| Decrease in Bacteroides and Firmicutes | |
| Increase in Gammaproteobacteria | |
| Presence of Escherichia coli, specifically AIEC | |
| Presence of Fusobacterium | |
| Decrease in Clostridia, Ruminococcaceae, Bifidobacterium, Lactobacillus | |
| Decrease in Faecalibacterium prausnitzii | |
| Microbial function | Decrease in Short Chain Fatty Acids (SCFA), butyrate |
| Decrease in butanoate and propanoate metabolism | |
| Decrease in amino acid biosynthesis | |
| Increase in auxotrophy | |
| Increase in amino acid transport | |
| Increase in sulfate transport | |
| Increased oxidative stress | |
| Increase in type II secretion system, secretion of toxins | |
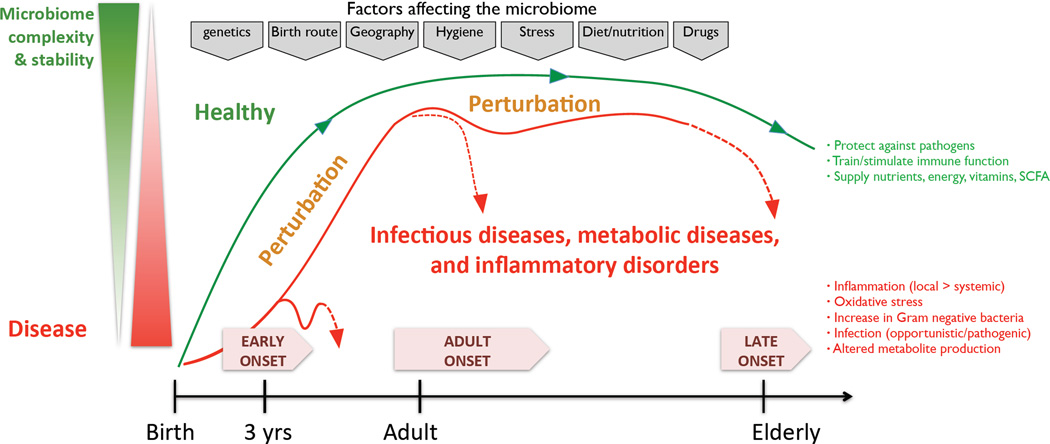
The gut microbiota, the largest reservoir of microbes in the body, coexists with its host in variable concentrations throughout the GI tract, reaching an upper level in the colon of 1011 or 1012 cells/g of luminal contents.10 This community carries out a range of useful functions for the host, including digesting substrates inaccessible to host enzymes, educating the immune system, and repressing the growth of harmful microorganisms.11 The extensive use of low-resolution surveys of the microbial community structure in the past, and renewed efforts using next-generation sequencing for a high-resolution description of composition, function, and ecology,12, 13 have improved our overall understanding of the role of the gut microbiota in health, a prerequisite for the study of disease-related dysbiosis. Several factors can intervene with microbial gut community composition, including genetics, diet, age, drug treatment, smoking, and potentially many more (Figure 1).14 The relative importance of each of these factors is still unclear, but several of them are directly or indirectly linked to disease state.
Figure 1. Factors affecting the stability and complexity of the gut microbiome in health and disease.
Key characteristics of the microbiome, including stability, resilience, and complexity, are influenced over time from infancy through adulthood and in old age. In the healthy gut, these characteristics contribute to important physiological processes such as protection against pathogens, training of the immune system, and digestion of food to supply energy and nutrients including vitamins and SCFAs. Many factors are indicated to impact the microbiome throughout microbiome development and even established assembly, including genetics, diet, medication, among others (marked in the grey boxes at the top of the figure). Some of these factors can introduce perturbations affecting the complexity and stability of the microbiome, potentially introducing microbial dysbiosis. Features of an imbalanced microbiome include, for example, an increase in Gram-negative bacteria linked to an environment of oxidative stress and inflammation, and metabolite production.
Environmental factors affect the microbiome composition
Diet
One of the most important environmental factors impacting microbial composition is dietary preference, which has been demonstrated to determine microbiome composition throughout mammalian evolution.15 Although no specific diet has been shown to directly cause, prevent, or treat IBD, it is important to take interactions between nutrients and microbiota into account when studying the role of the microbiome in disease. Thus far, only limited information on this topic has been gathered in humans, undoubtedly as a result of the challenge of setting up a large-scale controlled diet study. Wu and colleagues have shown that long-term dietary patterns affect the ratios of Bacteroides, Prevotella, and Firmicutes, and that short-term changes may not have major influences.16 In addition, Zimmer and colleagues have studied the impact of a strict vegan or vegetarian diet on the microbiota,17 and found a significant reduction in Bacteroides spp., Bifidobacterium spp., and the Enterobacteriaceae, while total bacterial load remain unaltered. Since the Enterobacteriaceae are among the taxa that are consistently found to be increased in patients with IBD (see below), it would be of value to include both short- and long-term dietary patterns in future studies of the role of the microbiome in IBD. Given the complexity of dietary effects, including such information will likely only be feasible in a large cohort study.18
Age
There is an age-related variation in the distribution of IBD phenotypes, with three distinct stages of onset. A peak age of onset is typically 15 to 30 years old, with late onset cases occurring closer to 60, and early onset less than 10 years of age. Noticeably, the latter group has seen a significant increase in incidence over the last decade.19 These stages correspond to phases in which the gut microbiota alters its diversity and stability.20 Early life is marked by a microbiome of low complexity and low stability, one that is more volatile, is affected by the birth route, and fluctuates with events such as changes in diet (switch from breastfeeding to solid foods), illness, and puberty.21 It takes until adulthood for the microbial assemblage to reach a maximal stability and complexity, with improved resilience towards perturbations.22 However, decreased stability has been observed in the elderly (60 years or older).23 Given these different characteristics of the microbiome at the three distinct stages of disease onset, a different role for the microbiome in disease initiation and progression should be considered.
IBD genetics point to an interplay between the immune system and microbiota in IBD
A potential link between genetics and the microbiome has long been suspected. The first identified CD susceptibility gene was nucleotide-binding oligomerization domain containing 2 (NOD2),24 which stimulates an immune reaction upon recognizing muramyl dipeptide, a cell wall peptidoglycan constituent of Gram-positive and Gram-negative bacteria. NOD2 is expressed in Paneth cells, which are located predominantly in the terminal ileum at the base of intestinal crypts, and produce antimicrobial defensins.25 Therefore, it may not be surprising that mutations in NOD2 can have significant effects on the composition of the microbial milieu. Indeed, IBD patients carrying NOD2 mutations have increased numbers of mucosa-adherent bacteria2 and decreased transcription of the anti-inflammatory cytokine interleukin (IL)-10.26 IBD patients with NOD2 and autophagy related 16-like 1 (ATG16L1, an IBD susceptibility gene involved in autophagy) risk alleles have significant alterations in the structure of their gut microbiota, including decreased levels of Faecalibacterium and increases in Escherichia.27 Individuals homozygous for loss-of-function alleles for fucosyltransferase 2 (FUT2) are "nonsecretors," who do not express ABO antigen on the gastrointestinal mucosa and bodily secretions. Nonsecretors are at increased risk for CD28 and exhibit substantial alterations in the mucosa-associated microbiota.29 Host genetics may thus play a strong role in the establishment and shaping of the gut microbiota; indeed, monozygotic twins share more similar microbiomes than non-twin siblings.30 On the skin, a recent study in primary immunodeficiency patients demonstrated a bi-directional dialogue between the microbiome and the host immune system. The skin of primary immunodeficiency patients holds an altered population composition compared to immunocompetent subjects, which in turn results in increased susceptibility to infection by altering the immune response towards pathogens.31 Although currently no genome-wide studies examining the interactions between common human genetic variation and the composition of the microbial ecosystem exist, such a study could hold great value.5
An overview of gut microbiome studies in IBD
Many IBD susceptibility loci suggest an impaired response to microbes in disease, but the causality of this relationship is unclear. IBD pathogenesis may result from a dysregulation of the mucosal immune system driving a pathogenic immune response against the commensal gut flora.32 Some studies show that the gut microbiota is an essential factor in driving inflammation in IBD,1 and indeed, short-term treatment with enterically-coated antibiotics dramatically reduces intestinal inflammation33 and has been demonstrated to have some efficacy in IBD, and particularly in pouchitis.34 Specifically, rifaximin has demonstrated efficacy in recent trials in CD.35 Additionally, IBD patients show mucosal secretion of IgG antibodies36 and mucosal T cell responses against commensal microbiota.37
The dramatic improvements to DNA sequencing technology and analysis over the last decade have set the stage for investigations of the IBD microbiome. Many studies find structural imbalances, or dysbioses, that occur in IBD since the initial report,38 and a broad pattern has begun to emerge which includes a reduction in biodiversity, a decreased representation of several taxa within the Firmicutes phylum, and an increase in the Gammaproteobacteria.27, 39
Many studies consistently report a decrease in biodiversity, known as alpha-diversity or species richness in ecological terms, a measure of the total number of species in a community. There is a reduced alpha-diversity in the fecal microbiome in CD compared to healthy controls,40 which was also found in pairs of monozygotic twins discordant for CD.41 This decreased diversity has been attributed to a reduced diversity specifically within the Firmicutes phylum,42 and has also been associated with temporal instability in the dominant taxa in both UC and CD.43 There is a reduced diversity in inflamed versus non-inflamed tissues even within the same patient, and CD patients have lower overall bacterial loads at inflamed regions.44 The largest IBD-related microbiome study to date, is on new-onset Crohn’s disease in a multicenter pediatric cohort.45 This study analyzed over 1000 treatment-naïve samples, which were collected from multiple concurrent GI locations, from patients representing the variety of disease phenotypes with respect to location, severity, and behavior. In addition to a detailed characterization of the specific organisms either lost or associated with disease, this study indicates that assessing the rectal mucosa-associated microbiome offers unique potential for convenient and early diagnosis of CD.
Other non-bacterial members of the microbiota, namely the fungi, viruses, archaea, and phage may have a significant role in gastrointestinal disease;46 however, the vast majority of recent studies of the microbiota are based on 16S sequencing, thus largely ignoring these groups of organisms. For example, norovirus infection, in the context of an intact gut microflora and mutated Atg16l1, is required for the development of CD in a mouse model.47 A number of studies note a relationship between fungi and IBD48 including an overall increase of fungal diversity in UC and CD.49 The relationship between these organisms and IBD will no doubt be explored in more detail in the coming years, as microbiome studies will increasingly be performed by unbiased shotgun sequencing.
Microbes enriched in IBD may potentiate disease
Specific taxonomic shifts have been reported in IBD (Table 1). The Enterobacteriaceae are increased in relative abundance both in IBD patients and in mouse models.50 Escherichia coli, particularly adherent-invasive E. coli (AIEC) strains, have been isolated from from ileal CD (iCD) biopsies in culture-based studies,51 and are enriched in UC patients.52 This enrichment is more pronounced in mucosal samples compared to fecal samples.53 The increase in Enterobacteriaceae may indicate the preference of this clade for an inflammatory environment. In fact, treatment with mesalamine, an anti-inflammatory drug used to treat IBD, decreases intestinal inflammation and is associated with a decrease in Escherichia/Shigella.39, 54
In addition to trends seen in the lumen, a number of studies have observed a shift in microbes that are attached to the intestinal mucus layer. The small intestine has a single layer of mucus, whereas the colon has two mucus layers, a firmly attached inner mucus layer that is essentially sterile, and an outer mucus layer of variable thickness.55 The mucus layer consists of mucins, trefoil peptides, and secretory IgA.56 Though host-microbiota interactions are bidirectional, direct contact with the epithelium is limited by the mucus and the production antimicrobial factors such as defensins and RegIII-gamma.57–59 As long as the mucus layer is relatively healthy and intact, microbes will attach to the mucus and generally do not have direct access to epithelial cells. There is a greater overall density of attached bacteria on the colonic mucus layer in UC patients compared to healthy controls.2 The AIEC pathovar, in particular, is at higher abundance in mucosal biopsies from CD compared to healthy individuals, and particularly high in ileal specimens.60 AIEC invades epithelial cells and can replicate within macrophages61 and induce granuloma formation in vitro.62 In fact, E.coli has also been found at higher levels in granulomas from CD relative to other non-CD granulomas.63
A second group of adherent and invasive bacteria is the Fusobacteria. The genus Fusobacterium is a group of Gram-negative anaerobes that principally colonize the oral cavity, but can also inhabit the gut. Fusobacterium spp. have been found to be at higher abundance in the colonic mucosa of patients with UC relative to control individuals,64, 65 and human isolates of Fusobacterium varium have been shown to induce colonic mucosal erosion in mice by rectal enema.66 The invasive ability of human Fusobacterium isolates has a positive correlation with the IBD status of the host,67 suggesting that invasive Fusobacterium spp. may influence IBD pathology. Intriguingly, Fusobacterium species have recently been shown to be enriched in tumor versus noninvolved adjacent tissue in colorectal cancer68 and human Fusobacterium isolates have been demonstrated to directly promote tumorigenesis in a mouse model.69 As IBD is among the highest risk factors for the development of colorectal cancer, Fusobacterium spp. may represent a potential link between these diseases.
Protective effects of microbes in IBD
Several lines of evidence suggest that specific groups of gut bacteria may have protective effects against IBD. For example, the colitis phenotype following treatment with dextran sulfate sodium is more severe in mice that are reared germ-free compared to conventionally reared mice.70 One mechanism by which the commensal microbiota may protect the host is colonization resistance, in which commensals occupy niches within the host and prevent colonization by pathogens71 and help out-compete pathogenic bacteria72 (Interestingly, the microbiota can sometimes take on the opposite role and facilitate viral infection.73) Commensal microbiota can also have direct functional effects on potential pathogens, for example in dampening virulence-related gene expression.74 In addition, the gut microbiota plays a role in shaping the mucosal immune system. Bacteroides and Clostridium species have been shown to induce the expansion of Treg cells, reducing intestinal inflammation.75 Other members of the microbiota can attenuate mucosal inflammation by regulating nuclear factor (NF)-κB activation.76
A number of bacterial species, most notably the Bifidobacterium, Lactobacillus, and Faecalibacterium genera, may protect the host from mucosal inflammation by several mechanisms, including the down-regulation of inflammatory cytokines77 or stimulation of IL-10,78 an anti-inflammatory cytokine. Faecalibacterium prausnitzii, one such proposed member of the microbiota with anti-inflammatory properties, is under-represented in IBD.79 F. prausnitzii is depleted in iCD biopsy samples concomitant with an increase in E. coli abundance,80 and low levels of mucosa-associated F. prausnitzii is associated with higher risk of recurrent CD following surgery.78 Conversely, recovery of F. prausnitzii after relapse is associated with maintenance of clinical remission of UC.81
Several constituents of the gut microbiota ferment dietary fiber, a prebiotic, to produce short-chain fatty acids (SCFAs), which include acetate, propionate, and butyrate. SCFAs are the primary energy source for colonic epithelial cells82 and have recently been shown to induce the expansion of colonic Treg cells.75 The Ruminococcaceae, particularly the butyrate-producing genus Faecalibacterium,83 is decreased in IBD, especially in iCD.38, 39, 42, 78, 80 Other SCFA-producing bacteria including Odoribacter and the Leuconostocaceae are reduced in UC, and Phascolarctobacterium and Roseburia are reduced in CD.39 Interestingly, the Ruminococcaceae consume hydrogen and produce acetate that can be utilized by Roseburia to produce butyrate,39 and it is therefore consistent that both groups together are reduced in IBD.
Functional composition of the gut microbiota in IBD
At the phylogenetic level, there is a generally high variability in the human microbiota between and within individuals over time.13 However, the functional composition (i.e. the functional potential of the gene content of the metagenome) of the gut microbiota is strikingly stable.13 Metagenomic approaches may therefore provide greater insight to the function of the gut microbiota in disease than taxonomic profiling;84, 85 indeed, one such metagenomics study of the IBD microbiome found that 12% of metabolic pathways were significantly different between IBD patients and healthy controls compared to just 2% of genus-level clades.39 Metagenomic and metaproteomic studies have confirmed a decrease in butanoate and propanoate metabolism genes in iCD39 and lower overall levels of butyrate and other SCFAs in iCD,86 consistent with the decreases in SCFA-producing Firmicutes clades seen in taxonomic profiling studies. Another metagenomic trend that has been identified in the IBD microbiome is an increase in functions characteristic of auxotrophic and pathobiont bacteria, such as a decrease in biosynthesis of amino acids, and an increase in amino acid transporter genes.39 These bacteria generally have a reduced ability to produce their own nutrients, but rather transport them from the environment as they are readily available at sites of inflammation and tissue destruction.39
A number of studies note an increase of sulfate-reducing bacteria, such as Desulfovibrio, in IBD.87 Mesalamine, a common treatment for IBD, inhibits fecal sulfide production and, intriguingly, stool samples from patients not treated with mesalamine show higher levels of sulfide.88 Genes involved in the metabolism of the sulfur-containing amino acid cysteine are increased in IBD, particularly in iCD, and there is increased sulfate transport in both UC and CD.39 Saturated fat-derived taurine conjugates to bile acids, increasing the availability of free sulfur and causing an expansion of the sulfate-reducing pathobiont Bilophila wadsworthia, driving colitis in genetically susceptible Il10−/− but not wild-type mice.89 The IBD metagenome has an increased propensity for managing oxidative stress, a hallmark of an inflammatory environment, as indicated by increased glutathione transport and riboflavin metabolism in UC.39 There is also an increase in type II secretion systems,39 which are involved in the secretion of toxins, and an increase in bacterial genes with virulence-related functions86 in patients with CD, indicative of a shift towards an inflammation-promoting microbiome.
The gut microbiota in related diseases
A number of parallels can be drawn between IBD and related metabolic diseases such as type 2 diabetes (T2D) and obesity. For example, there is an overall decrease in diversity in obesity at both the phylogenetic level (i.e. reduced number of distinct species)30 and metagenomic gene-count level (i.e. reduced number of distinct genes).90 Major shifts in clade abundances include a reduction of the Firmicutes and Clostridia in T2D91 and a significant increase in the Firmicutes-to-Bacteroidetes ratio in obesity in mice;92 however, it is less clear whether this shift also holds true in human obesity.30, 93 There is a decrease in Bifidobacterium species in obesity and T2D,93 and Bifidobacterium is reduced in children who become overweight,94 suggesting that it may act as a predictive factor. As in IBD, Faecalibacterium prausnitzii is reduced in abundance in T2D.95 In terms of gene function, there is an enrichment of genes involved in membrane transport,96 sulphate reduction, and oxidative stress resistance functions, and a decrease for functions involving cofactor and vitamin metabolism and butyrate production.97 Therefore, many of the same shifts in function of the gut microbiota -- and even specific taxa -- are seen across these diseases, suggesting the existence of generalized features that relate these diseases and the selection for an auxotrophic microbiota that can thrive in an inflammatory environment.
IBD treatments affecting the microbiome
An array of antibiotics have been shown to lead to a bloom of Escherichia coli.98 Since increased Enterobacteriaceae is a distinctive feature of intestinal inflammation and oxidative stress, the relationship between microbial composition, inflammation, and antibiotic use forms an important topic for future research. In contrast, some promising data show that antibiotic therapy specifically in IBD does induce remission or prevent relapse, but this topic will require further controlled trials.99 To better understand the consequences of perturbing the gut microbiota of patients and the role of the microbiota in treatment outcome, studies that monitor the temporal response at the levels of microbial ecology and functional composition will be required.100 Thus far, several studies of healthy humans briefly exposed to some antibiotics demonstrate the substantial perturbation, and the level of resilience, of the gut microbiota.101 Repeated exposures to a single antibiotic in healthy individuals results in cumulative and persistent changes to gut microbial composition.102 Microbial homeostasis is typically disrupted by the loss of species complexity, particularly of protective microbes, thereby potentially resulting in an increased risk of infections,103 or dysbiosis.45 Another mechanism by which antibiotics lead to increased gut infections is by causing a thinning of the mucus layer, thereby weakening its barrier function.104
Instead of perturbing the existing microbiome by removing diversity through antibiotics, repopulating the gut habitat with a healthy community has gained popularity in the last few years. This will be an exciting new direction for the pharmaceutical industry, expanding the focus beyond traditional small molecules and biologics.105 The complexity and composition that will be used to repopulate the gut community will be very important. The success of probiotics in the management of IBD ranges from mixed results to considerable potential,106 and is dependent on the strains used and disease subtype targeted. In contrast, the evidence that fecal microbiota transplantation (FMT) can be highly effective in replenishing our complex microbiota has received considerable attention following a convincing clinical trial for the treatment of relapsing C. difficile infection.107 Related studies have shown that the use of a well-selected community subset rather than whole fecal communities can be sufficient for recovery.108 The high success rate reported for relapsing C. difficile infection has elevated FMT as an emerging treatment for several gastrointestinal and metabolic disorders,109 and is actively being considered for IBD.110 So far, the sparse results reported for IBD cases have been variable with regard to the success rate for inducing remission, and well-designed randomized control trials are currently still lacking.111 Although changes in the composition of the intestinal microbiota were significant, and reductions in Proteobacteria as well as an increase in Bacteroides after FMT were observed, reaching or maintaining remission has been less frequent.112 Changing protocols towards repeated FMT procedures for multiple days in a row seems to increase the chances for achieving clinical remission.113
Future directions
Studies thus far have been able to address many aspects of IBD, including genetics, immune responses, microbial dysbiosis, and microbial functional activity. However, because of the complexity of the human microbiome as a dynamically interacting system, only limited data has been produced to bridge the gap between pathogenesis in a human host, individual microbes, and alterations in microbial metabolism and function. This suggests the need for a more multifaceted approach to the microbiome in IBD (Figure 2). Since the gaps in IBD are arguably the narrowest among diseases in the microbiome field, as indicated by the progress reviewed above, it can be considered as a model for systems-level investigations of human-associated microbial communities and their interactions with the host’s immune system. Increasing our mechanistic understanding of host-microbe interactions through such a systems-level approach will provide new opportunities to develop diagnostics and treatments,114 but is certainly not without challenges.115 Recent technological advances, including improvements in sequencing and computational biology, are contributing novel methods to study human-associated microbial communities,116 with an increasing focus on functional follow-up using cultured microbes and germ-free animal models.
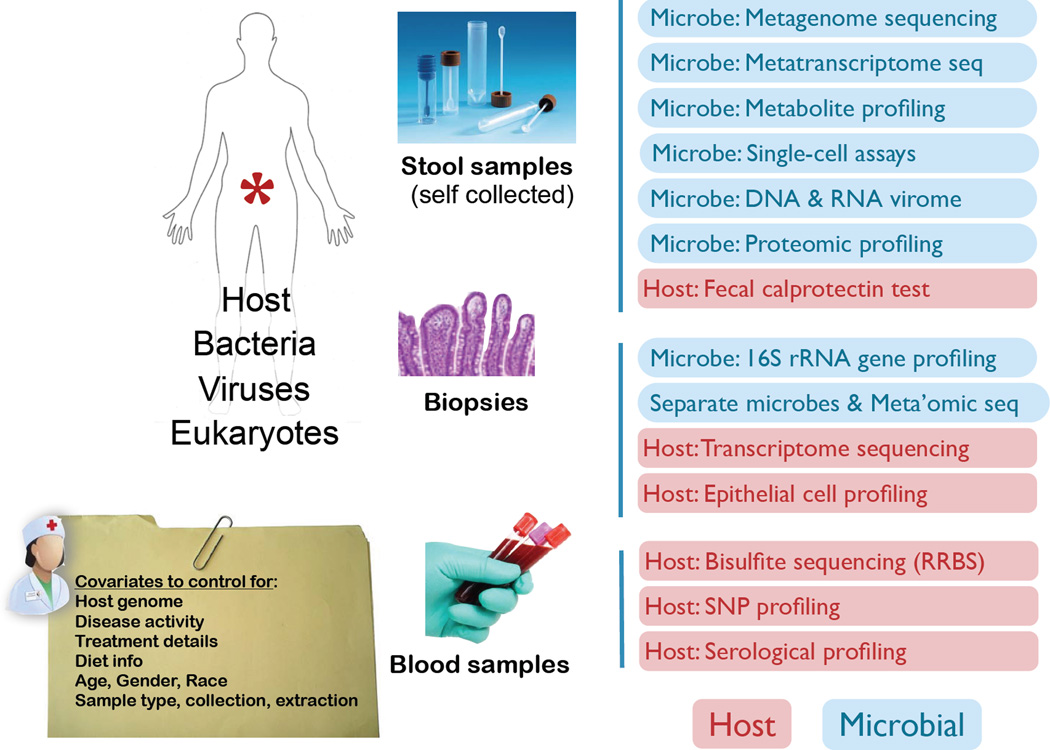
Figure 2. A multifaceted approach to study the role of the microbiome in IBD.
Future microbiome studies in the context of disease will shift towards multi-omics approaches in order to study host-microbe relations more comprehensively. Optimized sample collection, detailed clinical annotation, and sample processing will be key to expand data generation far beyond the typical marker gene and shotgun sequencing approaches. A number of assays on the host side (red) and microbial end (blue) will gain increasing attention going forward.
Essential to further explorations, more longitudinal surveys of patients before, during, and after treatment, as well as large-scale clinical trials that take into account both microbial and genetic heterogeneity, will need to be performed, and should take a multifaceted approach. One approach to increase sample size is to combine different cohort studies. However, sample type,117 collection,118 and extraction119 can introduce artifactual differences in microbial composition, which unfortunately makes it more challenging to combine datasets produced under different protocols.120 Streamlining experimental protocols across cohorts will be essential to preserve statistical power for identifying true biological effects in microbiome datasets. This will require growing efforts for standardizing the collection of patient samples and their clinical information. The discussion around the value of uniform data collection has resulted in solutions already,121 but these need further adaptation for clinical information. Pursuing a true systems biology approach will require a solution for simultaneous measurement of the host state, the microbiome, and the multi-directional signaling between them. Although solutions for sequential isolation of metabolites, RNA, DNA, and proteins from the same unique sample have been described,122 engineering a solution that can easily be deployed as a self-sampling kit for patients will require further exploration. Such challenges have been addressed with other technologies, for example, in the use of microarrays as a tool for biomarker detection in clinical applications, which led to the establishment of a consortium with a mandate to set up standards and quality measures.123 Such efforts are now also initiated in the microbiome space with both environmental and clinical relevance (see www.hmpdacc.org, http://www.mbqc.org, and http://www.earthmicrobiome.org), and could eventually allow us to combine several large well-characterized cohorts without the challenge of study-introduced biases.120
Despite promising correlations between shifts in microbial composition and disease phenotypes, to date no causative role for the microbiome has been established, and our understanding of the dynamic role of the human microbiome in IBD remains incomplete. The biological questions of interest enabled by prospective, longitudinal studies would be (1) to identify the potential role of the intestinal microbiome in triggering disease; (2) to determine if microbial composition predicts subsequent risk of activity flares; and (3) to examine whether the luminal flora predicts response to therapy. The identification of a correlative microbial pattern in humans that induces antimicrobial defense and ameliorates inflammation would have considerable promise as a novel diagnostic and therapeutic approach for the management of these complex diseases. Existing correlative genetic-microbial studies have helped to motivate this area, but cannot speak to causality, response to treatment, or risk prediction in the absence of multifaceted longitudinal measurements.
Acknowledgments
Grant support: Work was supported by grants from the Crohn’s and Colitis Foundation of America (R.J.X.) and NIH grants U54 DE023798 and R01 DK092405 (R.J.X.).
Abbreviations used in this paper
- AIEC
adherent invasive Escherichia coli
- ATG16L1
autophagy related 16-like 1
- CD
Crohn’s disease
- FMT
fecal microbiota transplantation
- FUT2
fucosyltransferase 2
- IBD
inflammatory bowel disease
- iCD
ileal Crohn’s disease
- IL
interleukin
- NF-κB
nuclear factor-κB
- NOD2
nucleotide-binding oligomerization domain containing 2
- SCFA
short-chain fatty acid
- T2D
type 2 diabetes
- UC
ulcerative colitis
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflicts of interest: The authors report no conflict of interest.
References
- 1.Sartor RB. Microbial influences in inflammatory bowel diseases. Gastroenterology. 2008;134:577–594. doi: 10.1053/j.gastro.2007.11.059. [DOI] [PubMed] [Google Scholar]
- 2.Swidsinski A, Ladhoff A, Pernthaler A, et al. Mucosal flora in inflammatory bowel disease. Gastroenterology. 2002;122:44–54. doi: 10.1053/gast.2002.30294. [DOI] [PubMed] [Google Scholar]
- 3.Loftus EV., Jr Clinical epidemiology of inflammatory bowel disease: Incidence, prevalence, and environmental influences. Gastroenterology. 2004;126:1504–1517. doi: 10.1053/j.gastro.2004.01.063. [DOI] [PubMed] [Google Scholar]
- 4.Jostins L, Ripke S, Weersma RK, et al. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature. 2012;491:119–124. doi: 10.1038/nature11582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Khor B, Gardet A, Xavier RJ. Genetics and pathogenesis of inflammatory bowel disease. Nature. 2011;474:307–317. doi: 10.1038/nature10209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Molodecky NA, Soon IS, Rabi DM, et al. Increasing incidence and prevalence of the inflammatory bowel diseases with time, based on systematic review. Gastroenterology. 2012;142:46–54. e42. doi: 10.1053/j.gastro.2011.10.001. quiz e30. [DOI] [PubMed] [Google Scholar]
- 7.Halme L, Paavola-Sakki P, Turunen U, et al. Family and twin studies in inflammatory bowel disease. World J Gastroenterol. 2006;12:3668–3672. doi: 10.3748/wjg.v12.i23.3668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pillai S. Rethinking mechanisms of autoimmune pathogenesis. J Autoimmun. 2013;45:97–103. doi: 10.1016/j.jaut.2013.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Flint HJ, Scott KP, Louis P, et al. The role of the gut microbiota in nutrition and health. Nat Rev Gastroenterol Hepatol. 2012;9:577–589. doi: 10.1038/nrgastro.2012.156. [DOI] [PubMed] [Google Scholar]
- 10.Dave M, Higgins PD, Middha S, et al. The human gut microbiome: current knowledge, challenges, and future directions. Transl Res. 2012;160:246–257. doi: 10.1016/j.trsl.2012.05.003. [DOI] [PubMed] [Google Scholar]
- 11.O'Hara AM, Shanahan F. The gut flora as a forgotten organ. EMBO Rep. 2006;7:688–693. doi: 10.1038/sj.embor.7400731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Qin J, Li R, Raes J, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Human Microbiome Project C. Structure, function and diversity of the healthy human microbiome. Nature. 2012;486:207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Blaser MJ, Falkow S. What are the consequences of the disappearing human microbiota? Nat Rev Microbiol. 2009;7:887–894. doi: 10.1038/nrmicro2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ley RE, Hamady M, Lozupone C, et al. Evolution of mammals and their gut microbes. Science. 2008;320:1647–1651. doi: 10.1126/science.1155725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu GD, Chen J, Hoffmann C, et al. Linking long-term dietary patterns with gut microbial enterotypes. Science. 2011;334:105–108. doi: 10.1126/science.1208344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zimmer J, Lange B, Frick JS, et al. A vegan or vegetarian diet substantially alters the human colonic faecal microbiota. Eur J Clin Nutr. 2012;66:53–60. doi: 10.1038/ejcn.2011.141. [DOI] [PubMed] [Google Scholar]
- 18.Moschen AR, Wieser V, Tilg H. Dietary Factors: Major Regulators of the Gut's Microbiota. Gut Liver. 2012;6:411–416. doi: 10.5009/gnl.2012.6.4.411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Martin-de-Carpi J, Rodriguez A, Ramos E, et al. Increasing incidence of pediatric inflammatory bowel disease in Spain (1996–2009): the SPIRIT Registry. Inflamm Bowel Dis. 2013;19:73–80. doi: 10.1002/ibd.22980. [DOI] [PubMed] [Google Scholar]
- 20.Spor A, Koren O, Ley R. Unravelling the effects of the environment and host genotype on the gut microbiome. Nat Rev Microbiol. 2011;9:279–290. doi: 10.1038/nrmicro2540. [DOI] [PubMed] [Google Scholar]
- 21.Dominguez-Bello MG, Blaser MJ, Ley RE, et al. Development of the human gastrointestinal microbiota and insights from high-throughput sequencing. Gastroenterology. 2011;140:1713–1719. doi: 10.1053/j.gastro.2011.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lozupone CA, Stombaugh JI, Gordon JI, et al. Diversity, stability and resilience of the human gut microbiota. Nature. 2012;489:220–230. doi: 10.1038/nature11550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Claesson MJ, Cusack S, O'Sullivan O, et al. Composition, variability, and temporal stability of the intestinal microbiota of the elderly. Proc Natl Acad Sci U S A. 2011;108(Suppl 1):4586–4591. doi: 10.1073/pnas.1000097107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ogura Y, Bonen DK, Inohara N, et al. A frameshift mutation in NOD2 associated with susceptibility to Crohn's disease. Nature. 2001;411:603–606. doi: 10.1038/35079114. [DOI] [PubMed] [Google Scholar]
- 25.Stappenbeck TS, Hooper LV, Gordon JI. Developmental regulation of intestinal angiogenesis by indigenous microbes via Paneth cells. Proc Natl Acad Sci U S A. 2002;99:15451–15455. doi: 10.1073/pnas.202604299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Philpott DJ, Girardin SE. Crohn's disease-associated Nod2 mutants reduce IL10 transcription. Nat Immunol. 2009;10:455–457. doi: 10.1038/ni0509-455. [DOI] [PubMed] [Google Scholar]
- 27.Frank DN, Robertson CE, Hamm CM, et al. Disease phenotype and genotype are associated with shifts in intestinal-associated microbiota in inflammatory bowel diseases. Inflamm Bowel Dis. 2011;17:179–184. doi: 10.1002/ibd.21339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McGovern DP, Jones MR, Taylor KD, et al. Fucosyltransferase 2 (FUT2) non-secretor status is associated with Crohn's disease. Hum Mol Genet. 2010;19:3468–3476. doi: 10.1093/hmg/ddq248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rausch P, Rehman A, Kunzel S, et al. Colonic mucosa-associated microbiota is influenced by an interaction of Crohn disease and FUT2 (Secretor) genotype. Proc Natl Acad Sci U S A. 2011;108:19030–19035. doi: 10.1073/pnas.1106408108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Turnbaugh PJ, Hamady M, Yatsunenko T, et al. A core gut microbiome in obese and lean twins. Nature. 2009;457:480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Smeekens SP, Huttenhower C, Riza A, et al. Skin Microbiome Imbalance in Patients with STAT1/STAT3 Defects Impairs Innate Host Defense Responses. J Innate Immun. 2013 doi: 10.1159/000351912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Strober W, Fuss I, Mannon P. The fundamental basis of inflammatory bowel disease. J Clin Invest. 2007;117:514–521. doi: 10.1172/JCI30587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Casellas F, Borruel N, Papo M, et al. Antiinflammatory effects of enterically coated amoxicillin-clavulanic acid in active ulcerative colitis. Inflamm Bowel Dis. 1998;4:1–5. doi: 10.1097/00054725-199802000-00001. [DOI] [PubMed] [Google Scholar]
- 34.Sartor RB. Therapeutic manipulation of the enteric microflora in inflammatory bowel diseases: antibiotics, probiotics, and prebiotics. Gastroenterology. 2004;126:1620–1633. doi: 10.1053/j.gastro.2004.03.024. [DOI] [PubMed] [Google Scholar]
- 35.Rietdijk ST, D'Haens GR. Recent developments in the treatment of inflammatory bowel disease. J Dig Dis. 2013;14:282–287. doi: 10.1111/1751-2980.12048. [DOI] [PubMed] [Google Scholar]
- 36.Macpherson A, Khoo UY, Forgacs I, et al. Mucosal antibodies in inflammatory bowel disease are directed against intestinal bacteria. Gut. 1996;38:365–375. doi: 10.1136/gut.38.3.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pirzer U, Schonhaar A, Fleischer B, et al. Reactivity of infiltrating T lymphocytes with microbial antigens in Crohn's disease. Lancet. 1991;338:1238–1239. doi: 10.1016/0140-6736(91)92104-a. [DOI] [PubMed] [Google Scholar]
- 38.Frank DN, St Amand AL, Feldman RA, et al. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc Natl Acad Sci U S A. 2007;104:13780–13785. doi: 10.1073/pnas.0706625104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Morgan XC, Tickle TL, Sokol H, et al. Dysfunction of the intestinal microbiome in inflammatory bowel disease and treatment. Genome Biol. 2012;13:R79. doi: 10.1186/gb-2012-13-9-r79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Manichanh C, Rigottier-Gois L, Bonnaud E, et al. Reduced diversity of faecal microbiota in Crohn's disease revealed by a metagenomic approach. Gut. 2006;55:205–211. doi: 10.1136/gut.2005.073817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dicksved J, Halfvarson J, Rosenquist M, et al. Molecular analysis of the gut microbiota of identical twins with Crohn's disease. ISME J. 2008;2:716–727. doi: 10.1038/ismej.2008.37. [DOI] [PubMed] [Google Scholar]
- 42.Kang S, Denman SE, Morrison M, et al. Dysbiosis of fecal microbiota in Crohn's disease patients as revealed by a custom phylogenetic microarray. Inflamm Bowel Dis. 2010;16:2034–2042. doi: 10.1002/ibd.21319. [DOI] [PubMed] [Google Scholar]
- 43.Martinez C, Antolin M, Santos J, et al. Unstable composition of the fecal microbiota in ulcerative colitis during clinical remission. Am J Gastroenterol. 2008;103:643–648. doi: 10.1111/j.1572-0241.2007.01592.x. [DOI] [PubMed] [Google Scholar]
- 44.Sepehri S, Kotlowski R, Bernstein CN, et al. Microbial diversity of inflamed and noninflamed gut biopsy tissues in inflammatory bowel disease. Inflamm Bowel Dis. 2007;13:675–683. doi: 10.1002/ibd.20101. [DOI] [PubMed] [Google Scholar]
- 45.Gevers D, Kugathasan S, Denson LA, et al. The treatment-naïve microbiome in new-onset Crohn’s disease. Cell Host Microbe. doi: 10.1016/j.chom.2014.02.005. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hunter P. The secret garden's gardeners. Research increasingly appreciates the crucial role of gut viruses for human health and disease. EMBO Rep. 2013;14:683–685. doi: 10.1038/embor.2013.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cadwell K, Patel KK, Maloney NS, et al. Virus-plus-susceptibility gene interaction determines Crohn's disease gene Atg16L1 phenotypes in intestine. Cell. 2010;141:1135–1145. doi: 10.1016/j.cell.2010.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Trojanowska D, Zwolinska-Wcislo M, Tokarczyk M, et al. The role of Candida in inflammatory bowel disease. Estimation of transmission of C. albicans fungi in gastrointestinal tract based on genetic affinity between strains. Med Sci Monit. 2010;16:CR451–CR457. [PubMed] [Google Scholar]
- 49.Ott SJ, Kuhbacher T, Musfeldt M, et al. Fungi and inflammatory bowel diseases: Alterations of composition and diversity. Scand J Gastroenterol. 2008;43:831–841. doi: 10.1080/00365520801935434. [DOI] [PubMed] [Google Scholar]
- 50.Lupp C, Robertson ML, Wickham ME, et al. Host-mediated inflammation disrupts the intestinal microbiota and promotes the overgrowth of Enterobacteriaceae. Cell Host Microbe. 2007;2:119–129. doi: 10.1016/j.chom.2007.06.010. [DOI] [PubMed] [Google Scholar]
- 51.Darfeuille-Michaud A, Boudeau J, Bulois P, et al. High prevalence of adherent-invasive Escherichia coli associated with ileal mucosa in Crohn's disease. Gastroenterology. 2004;127:412–421. doi: 10.1053/j.gastro.2004.04.061. [DOI] [PubMed] [Google Scholar]
- 52.Sokol H, Lepage P, Seksik P, et al. Temperature gradient gel electrophoresis of fecal 16S rRNA reveals active Escherichia coli in the microbiota of patients with ulcerative colitis. J Clin Microbiol. 2006;44:3172–3177. doi: 10.1128/JCM.02600-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chassaing B, Darfeuille-Michaud A. The commensal microbiota and enteropathogens in the pathogenesis of inflammatory bowel diseases. Gastroenterology. 2011;140:1720–1728. doi: 10.1053/j.gastro.2011.01.054. [DOI] [PubMed] [Google Scholar]
- 54.Benjamin JL, Hedin CR, Koutsoumpas A, et al. Smokers with active Crohn's disease have a clinically relevant dysbiosis of the gastrointestinal microbiota. Inflamm Bowel Dis. 2012;18:1092–1100. doi: 10.1002/ibd.21864. [DOI] [PubMed] [Google Scholar]
- 55.Johansson ME, Larsson JM, Hansson GC. The two mucus layers of colon are organized by the MUC2 mucin, whereas the outer layer is a legislator of host-microbial interactions. Proc Natl Acad Sci U S A. 2011;108(Suppl 1):4659–4665. doi: 10.1073/pnas.1006451107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cario E. Microbiota and innate immunity in intestinal inflammation and neoplasia. Curr Opin Gastroenterol. 2013;29:85–91. doi: 10.1097/MOG.0b013e32835a670e. [DOI] [PubMed] [Google Scholar]
- 57.Shanahan F. The colonic microbiota in health and disease. Curr Opin Gastroenterol. 2013;29:49–54. doi: 10.1097/MOG.0b013e32835a3493. [DOI] [PubMed] [Google Scholar]
- 58.Vaishnava S, Yamamoto M, Severson KM, et al. The antibacterial lectin RegIIIgamma promotes the spatial segregation of microbiota and host in the intestine. Science. 2011;334:255–258. doi: 10.1126/science.1209791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hooper LV, Littman DR, Macpherson AJ. Interactions between the microbiota and the immune system. Science. 2012;336:1268–1273. doi: 10.1126/science.1223490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Martinez-Medina M, Aldeguer X, Lopez-Siles M, et al. Molecular diversity of Escherichia coli in the human gut: new ecological evidence supporting the role of adherent-invasive E. coli (AIEC) in Crohn's disease. Inflamm Bowel Dis. 2009;15:872–882. doi: 10.1002/ibd.20860. [DOI] [PubMed] [Google Scholar]
- 61.Glasser AL, Boudeau J, Barnich N, et al. Adherent invasive Escherichia coli strains from patients with Crohn's disease survive and replicate within macrophages without inducing host cell death. Infect Immun. 2001;69:5529–5537. doi: 10.1128/IAI.69.9.5529-5537.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Meconi S, Vercellone A, Levillain F, et al. Adherent-invasive Escherichia coli isolated from Crohn's disease patients induce granulomas in vitro. Cell Microbiol. 2007;9:1252–1261. doi: 10.1111/j.1462-5822.2006.00868.x. [DOI] [PubMed] [Google Scholar]
- 63.Ryan P, Kelly RG, Lee G, et al. Bacterial DNA within granulomas of patients with Crohn's disease--detection by laser capture microdissection and PCR. Am J Gastroenterol. 2004;99:1539–1543. doi: 10.1111/j.1572-0241.2004.40103.x. [DOI] [PubMed] [Google Scholar]
- 64.Ohkusa T, Sato N, Ogihara T, et al. Fusobacterium varium localized in the colonic mucosa of patients with ulcerative colitis stimulates species-specific antibody. J Gastroenterol Hepatol. 2002;17:849–853. doi: 10.1046/j.1440-1746.2002.02834.x. [DOI] [PubMed] [Google Scholar]
- 65.Ohkusa T, Yoshida T, Sato N, et al. Commensal bacteria can enter colonic epithelial cells and induce proinflammatory cytokine secretion: a possible pathogenic mechanism of ulcerative colitis. J Med Microbiol. 2009;58:535–545. doi: 10.1099/jmm.0.005801-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ohkusa T, Okayasu I, Ogihara T, et al. Induction of experimental ulcerative colitis by Fusobacterium varium isolated from colonic mucosa of patients with ulcerative colitis. Gut. 2003;52:79–83. doi: 10.1136/gut.52.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Strauss J, Kaplan GG, Beck PL, et al. Invasive potential of gut mucosa-derived Fusobacterium nucleatum positively correlates with IBD status of the host. Inflamm Bowel Dis. 2011;17:1971–1978. doi: 10.1002/ibd.21606. [DOI] [PubMed] [Google Scholar]
- 68.Kostic AD, Gevers D, Pedamallu CS, et al. Genomic analysis identifies association of Fusobacterium with colorectal carcinoma. Genome Res. 2012;22:292–298. doi: 10.1101/gr.126573.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rubinstein MR, Wang X, Liu W, et al. Fusobacterium nucleatum promotes colorectal carcinogenesis by modulating E-cadherin/beta-catenin signaling via its FadA adhesin. Cell Host Microbe. 2013;14:195–206. doi: 10.1016/j.chom.2013.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kitajima S, Morimoto M, Sagara E, et al. Dextran sodium sulfate-induced colitis in germ-free IQI/Jic mice. Exp Anim. 2001;50:387–395. doi: 10.1538/expanim.50.387. [DOI] [PubMed] [Google Scholar]
- 71.Callaway TR, Edrington TS, Anderson RC, et al. Probiotics, prebiotics and competitive exclusion for prophylaxis against bacterial disease. Anim Health Res Rev. 2008;9:217–225. doi: 10.1017/S1466252308001540. [DOI] [PubMed] [Google Scholar]
- 72.Kamada N, Chen G, Nunez G. A complex microworld in the gut: Harnessing pathogen-commensal relations. Nat Med. 2012;18:1190–1191. doi: 10.1038/nm.2900. [DOI] [PubMed] [Google Scholar]
- 73.Kane M, Case LK, Kopaskie K, et al. Successful transmission of a retrovirus depends on the commensal microbiota. Science. 2011;334:245–249. doi: 10.1126/science.1210718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Medellin-Pena MJ, Wang H, Johnson R, et al. Probiotics affect virulence-related gene expression in Escherichia coli O157:H7. Appl Environ Microbiol. 2007;73:4259–4267. doi: 10.1128/AEM.00159-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Atarashi K, Tanoue T, Oshima K, et al. Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature. 2013;500:232–236. doi: 10.1038/nature12331. [DOI] [PubMed] [Google Scholar]
- 76.Kelly D, Campbell JI, King TP, et al. Commensal anaerobic gut bacteria attenuate inflammation by regulating nuclear-cytoplasmic shuttling of PPAR-gamma and RelA. Nat Immunol. 2004;5:104–112. doi: 10.1038/ni1018. [DOI] [PubMed] [Google Scholar]
- 77.Llopis M, Antolin M, Carol M, et al. Lactobacillus casei downregulates commensals' inflammatory signals in Crohn's disease mucosa. Inflamm Bowel Dis. 2009;15:275–283. doi: 10.1002/ibd.20736. [DOI] [PubMed] [Google Scholar]
- 78.Sokol H, Pigneur B, Watterlot L, et al. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc Natl Acad Sci U S A. 2008;105:16731–16736. doi: 10.1073/pnas.0804812105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sokol H, Seksik P, Furet JP, et al. Low counts of Faecalibacterium prausnitzii in colitis microbiota. Inflamm Bowel Dis. 2009;15:1183–1189. doi: 10.1002/ibd.20903. [DOI] [PubMed] [Google Scholar]
- 80.Willing B, Halfvarson J, Dicksved J, et al. Twin studies reveal specific imbalances in the mucosa-associated microbiota of patients with ileal Crohn's disease. Inflamm Bowel Dis. 2009;15:653–660. doi: 10.1002/ibd.20783. [DOI] [PubMed] [Google Scholar]
- 81.Varela E, Manichanh C, Gallart M, et al. Colonisation by Faecalibacterium prausnitzii and maintenance of clinical remission in patients with ulcerative colitis. Aliment Pharmacol Ther. 2013;38:151–161. doi: 10.1111/apt.12365. [DOI] [PubMed] [Google Scholar]
- 82.Ahmad MS, Krishnan S, Ramakrishna BS, et al. Butyrate and glucose metabolism by colonocytes in experimental colitis in mice. Gut. 2000;46:493–499. doi: 10.1136/gut.46.4.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Duncan SH, Hold GL, Barcenilla A, et al. Roseburia intestinalis sp. nov., a novel saccharolytic, butyrate-producing bacterium from human faeces. Int J Syst Evol Microbiol. 2002;52:1615–1620. doi: 10.1099/00207713-52-5-1615. [DOI] [PubMed] [Google Scholar]
- 84.Meyer F, Trimble WL, Chang EB, et al. Functional predictions from inference and observation in sequence-based inflammatory bowel disease research. Genome Biol. 2012;13:169. doi: 10.1186/gb-2012-13-9-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Presley LL, Ye J, Li X, et al. Host-microbe relationships in inflammatory bowel disease detected by bacterial and metaproteomic analysis of the mucosal-luminal interface. Inflamm Bowel Dis. 2012;18:409–417. doi: 10.1002/ibd.21793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Erickson AR, Cantarel BL, Lamendella R, et al. Integrated metagenomics/metaproteomics reveals human host-microbiota signatures of Crohn's disease. PLoS One. 2012;7:e49138. doi: 10.1371/journal.pone.0049138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Rowan F, Docherty NG, Murphy M, et al. Desulfovibrio bacterial species are increased in ulcerative colitis. Dis Colon Rectum. 2010;53:1530–1536. doi: 10.1007/DCR.0b013e3181f1e620. [DOI] [PubMed] [Google Scholar]
- 88.Pitcher MC, Beatty ER, Cummings JH. The contribution of sulphate reducing bacteria and 5-aminosalicylic acid to faecal sulphide in patients with ulcerative colitis. Gut. 2000;46:64–72. doi: 10.1136/gut.46.1.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Devkota S, Wang Y, Musch MW, et al. Dietary-fat-induced taurocholic acid promotes pathobiont expansion and colitis in Il10−/− mice. Nature. 2012;487:104–108. doi: 10.1038/nature11225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Le Chatelier E, Nielsen T, Qin J, et al. Richness of human gut microbiome correlates with metabolic markers. Nature. 2013;500:541–546. doi: 10.1038/nature12506. [DOI] [PubMed] [Google Scholar]
- 91.Larsen N, Vogensen FK, van den Berg FW, et al. Gut microbiota in human adults with type 2 diabetes differs from non-diabetic adults. PLoS One. 2010;5:e9085. doi: 10.1371/journal.pone.0009085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Turnbaugh PJ, Ley RE, Mahowald MA, et al. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027–1031. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- 93.Schwiertz A, Taras D, Schafer K, et al. Microbiota and SCFA in lean and overweight healthy subjects. Obesity (Silver Spring) 2010;18:190–195. doi: 10.1038/oby.2009.167. [DOI] [PubMed] [Google Scholar]
- 94.Kalliomaki M, Collado MC, Salminen S, et al. Early differences in fecal microbiota composition in children may predict overweight. Am J Clin Nutr. 2008;87:534–538. doi: 10.1093/ajcn/87.3.534. [DOI] [PubMed] [Google Scholar]
- 95.Furet JP, Kong LC, Tap J, et al. Differential adaptation of human gut microbiota to bariatric surgery-induced weight loss: links with metabolic and low-grade inflammation markers. Diabetes. 2010;59:3049–3057. doi: 10.2337/db10-0253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Greenblum S, Turnbaugh PJ, Borenstein E. Metagenomic systems biology of the human gut microbiome reveals topological shifts associated with obesity and inflammatory bowel disease. Proc Natl Acad Sci U S A. 2012;109:594–599. doi: 10.1073/pnas.1116053109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Qin J, Li Y, Cai Z, et al. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature. 2012;490:55–60. doi: 10.1038/nature11450. [DOI] [PubMed] [Google Scholar]
- 98.Looft T, Allen HK. Collateral effects of antibiotics on mammalian gut microbiomes. Gut Microbes. 2012;3:463–467. doi: 10.4161/gmic.21288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Khan KJ, Ullman TA, Ford AC, et al. Antibiotic therapy in inflammatory bowel disease: a systematic review and meta-analysis. Am J Gastroenterol. 2011;106:661–673. doi: 10.1038/ajg.2011.72. [DOI] [PubMed] [Google Scholar]
- 100.Lemon KP, Armitage GC, Relman DA, et al. Microbiota-targeted therapies: an ecological perspective. Sci Transl Med. 2012;4:137rv5. doi: 10.1126/scitranslmed.3004183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Relman DA. The human microbiome: ecosystem resilience and health. Nutr Rev. 2012;70(Suppl 1):S2–S9. doi: 10.1111/j.1753-4887.2012.00489.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Dethlefsen L, Relman DA. Incomplete recovery and individualized responses of the human distal gut microbiota to repeated antibiotic perturbation. Proc Natl Acad Sci U S A. 2011;108(Suppl 1):4554–4561. doi: 10.1073/pnas.1000087107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Khosravi A, Mazmanian SK. Disruption of the gut microbiome as a risk factor for microbial infections. Curr Opin Microbiol. 2013;16:221–227. doi: 10.1016/j.mib.2013.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Wlodarska M, Willing B, Keeney KM, et al. Antibiotic treatment alters the colonic mucus layer and predisposes the host to exacerbated Citrobacter rodentium-induced colitis. Infect Immun. 2011;79:1536–1545. doi: 10.1128/IAI.01104-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Fischbach MA, Bluestone JA, Lim WA. Cell-based therapeutics: the next pillar of medicine. Sci Transl Med. 2013;5:179ps7. doi: 10.1126/scitranslmed.3005568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Whelan K, Quigley EM. Probiotics in the management of irritable bowel syndrome and inflammatory bowel disease. Curr Opin Gastroenterol. 2013;29:184–189. doi: 10.1097/MOG.0b013e32835d7bba. [DOI] [PubMed] [Google Scholar]
- 107.van Nood E, Vrieze A, Nieuwdorp M, et al. Duodenal infusion of donor feces for recurrent Clostridium difficile. N Engl J Med. 2013;368:407–415. doi: 10.1056/NEJMoa1205037. [DOI] [PubMed] [Google Scholar]
- 108.Shahinas D, Silverman M, Sittler T, et al. Toward an understanding of changes in diversity associated with fecal microbiome transplantation based on 16S rRNA gene deep sequencing. MBio. 2012;3 doi: 10.1128/mBio.00338-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Smits LP, Bouter KE, de Vos WM, et al. Therapeutic Potential of Fecal Microbiota Transplantation. Gastroenterology. 2013 doi: 10.1053/j.gastro.2013.08.058. [DOI] [PubMed] [Google Scholar]
- 110.Damman CJ, Miller SI, Surawicz CM, et al. The microbiome and inflammatory bowel disease: is there a therapeutic role for fecal microbiota transplantation? Am J Gastroenterol. 2012;107:1452–1459. doi: 10.1038/ajg.2012.93. [DOI] [PubMed] [Google Scholar]
- 111.Anderson JL, Edney RJ, Whelan K. Systematic review: faecal microbiota transplantation in the management of inflammatory bowel disease. Aliment Pharmacol Ther. 2012;36:503–516. doi: 10.1111/j.1365-2036.2012.05220.x. [DOI] [PubMed] [Google Scholar]
- 112.Kump PK, Grochenig HP, Lackner S, et al. Alteration of intestinal dysbiosis by fecal microbiota transplantation does not induce remission in patients with chronic active ulcerative colitis. Inflamm Bowel Dis. 2013;19:2155–2165. doi: 10.1097/MIB.0b013e31829ea325. [DOI] [PubMed] [Google Scholar]
- 113.Kunde S, Pham A, Bonczyk S, et al. Safety, tolerability, and clinical response after fecal transplantation in children and young adults with ulcerative colitis. J Pediatr Gastroenterol Nutr. 2013;56:597–601. doi: 10.1097/MPG.0b013e318292fa0d. [DOI] [PubMed] [Google Scholar]
- 114.Olle B. Medicines from microbiota. Nat Biotechnol. 2013;31:309–315. doi: 10.1038/nbt.2548. [DOI] [PubMed] [Google Scholar]
- 115.Brown J, de Vos WM, DiStefano PS, et al. Translating the human microbiome. Nat Biotechnol. 2013;31:304–308. doi: 10.1038/nbt.2543. [DOI] [PubMed] [Google Scholar]
- 116.Gevers D, Pop M, Schloss PD, et al. Bioinformatics for the Human Microbiome Project. PLoS Comput Biol. 2012;8:e1002779. doi: 10.1371/journal.pcbi.1002779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Stearns JC, Lynch MD, Senadheera DB, et al. Bacterial biogeography of the human digestive tract. Sci Rep. 2011;1:170. doi: 10.1038/srep00170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Rubin BE, Gibbons SM, Kennedy S, et al. Investigating the impact of storage conditions on microbial community composition in soil samples. PLoS One. 2013;8:e70460. doi: 10.1371/journal.pone.0070460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Momozawa Y, Deffontaine V, Louis E, et al. Characterization of bacteria in biopsies of colon and stools by high throughput sequencing of the V2 region of bacterial 16S rRNA gene in human. PLoS One. 2011;6:e16952. doi: 10.1371/journal.pone.0016952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Lozupone CA, Stombaugh J, Gonzalez A, et al. Meta-analyses of studies of the human microbiota. Genome Res. 2013;23:1704–1714. doi: 10.1101/gr.151803.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Yilmaz P, Kottmann R, Field D, et al. Minimum information about a marker gene sequence (MIMARKS) and minimum information about any (x) sequence (MIxS) specifications. Nat Biotechnol. 2011;29:415–420. doi: 10.1038/nbt.1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Roume H, Heintz-Buschart A, Muller EE, et al. Sequential isolation of metabolites, RNA, DNA, and proteins from the same unique sample. Methods Enzymol. 2013;531:219–236. doi: 10.1016/B978-0-12-407863-5.00011-3. [DOI] [PubMed] [Google Scholar]
- 123.Tillinghast GW. Microarrays in the clinic. Nat Biotechnol. 2010;28:810–812. doi: 10.1038/nbt0810-810. [DOI] [PubMed] [Google Scholar]