Abstract
TMEM16C belongs to the TMEM16 family, which includes the Ca2+-activated Cl– channels (CaCCs) TMEM16A and TMEM16B and a small conductance Ca2+-activated, non-selective cation channel (SCAN), TMEM16F. Here we report that in rat dorsal root ganglia (DRG) TMEM16C is expressed mainly in the IB4 positive, non-peptidergic nociceptors that also express the sodium-activated potassium (KNa) channel Slack. Together these channel proteins promote KNa channel activity and dampen neuronal excitability. DRG from TMEM16C knock out rats have reduced Slack expression, broadened action potential and increased excitability. Moreover, the TMEM16C knock out rats as well as rats with Slack knockdown via intrathecal injection of siRNA exhibit increased thermal and mechanical sensitivity. Experiments involving heterologous expression in HEK293 cells further show that TMEM16C modulates the single channel activity of Slack channels and increases its sodium sensitivity. Our study thus reveals that TMEM16C enhances KNa channel activity in DRG neurons and regulate the processing of pain messages.
Introduction
The molecular identification of TMEM16A and TMEM16B as Ca2+-activated chloride channels (CaCCs) has generated great interest in the ten member mammalian TMEM16 (transmembrane protein with unknown function) family1-3. TMEM16A-CaCC plays important roles in regulating secretion from exocrine glands and epithelial cells, smooth muscle contraction and pacemaking activity of interstitial cells of cajal (ICC) in the gut4,5. Whereas TMEM16A is expressed in dorsal root ganglia (DRG) and reported to act as a heat sensor in nociceptive neurons6, TMEM16B is responsible for CaCC in photoreceptors, olfactory sensory neurons, and hippocampal neurons. Although dispensable for olfaction, TMEM16B- CaCC regulates action potential waveform and synaptic response of hippocampal neurons7-9.
The TMEM16 family contains both anion channels and cation channels; TMEM16F generates a small-conductance Ca2+-activated non-selective cation channel (SCAN), and is linked to the bleeding disorder Scott Syndrome associated with deficient Ca2+-dependent scramblase activity required for blood coagulation10,11. Swapping a residue in the TM5 transmembrane segment between TMEM16A and TMEM16F reduces the anion selectivity of the former and the cation selectivity of the latter10, revealing that these TMEM16 family members are pore-forming subunits. It is of interest to explore the function of other members of this novel ion channel family.
As to TMEM16C, weighted gene coexpression network analysis (WGCN) of microarray data from human and chimpanzee brains positioned TMEM16C at a hub in the modules of co-expressed genes in the caudate nucleus12, and analyses of a high density genomic variant suggested an association of TMEM16C with late-onset Alzheimer's disease (LOAD)13. Moreover, a recent genetic study involving exome sequencing linked a TMEM16C mutation to human autosomal-dominant craniocervical dystonia and documented a high level of TMEM16C expression in human striatum, hippocampus and cortex14. In this study, we found that TMEM16C is mainly expressed in neuronal tissues from both the central and peripheral nervous system. We further discovered that TMEM16C is preferentially expressed in the IB4 positive, non-peptidergic nociceptors in DRG, raising the question about its role in nociception.
In mammals, pain-producing stimuli are detected by nociceptive neurons whose cell bodies are located in the DRG and extend peripheral and central processes to reach their target organs and the spinal cord, respectively. The small diameter unmyelinated “C” fibers comprise a major class of nociceptors, which are activated peripherally by noxious thermal, mechanical, and chemical stimuli15-17. The small DRG neurons can be further subdivided to the peptidergic and the non-peptidergic population and the latter binds the IB4 isolectin, and expresses G protein-coupled receptors of the Mrg family18,19. Sensory transduction in DRG neurons is achieved through the activation of specific classes of ion channels, which are the molecular sensors that may detect sensory stimuli and convert them into electrical signals15,20. Notably, pain sensation involves several members of the transient receptor potential (TRP) channel family as well as sodium channels and potassium channels15-17,21.
Here we report that TMEM16C knockout rats exhibit heightened thermal and mechanical sensitivity, which is associated with increased neuronal excitability and broadened action potential in their IB4 positive DRG neurons. Moreover, TMEM16C interacts with the sodium activated potassium channel (KNa) Slack and modulates its channel activity and sodium sensitivity. Our study indicates that TMEM16C enhances the KNa channel activity in DRG neurons and thus regulates the processing of pain messages.
Results
TMEM16C is mainly expressed in the IB4 positive nociceptors in the rat DRG
To determine the expression pattern of TMEM16C, we first performed RT-PCR and detected TMEM16C transcripts in the nervous system, including several brain regions, spinal cord and DRG, but not in the gastrointestinal tract, heart or skeletal muscle (Supplementary Fig. 1a and 2a). We then raised rabbit polyclonal antibodies against a C-terminal peptide of mouse TMEM16C (peptide sequence identical for rat TMEM16C) and then used immunocytochemistry and western blot to confirm that this antibody recognizes the mouse TMEM16C carrying an HA tag when it is heterologously expressed in HEK293 cells, (Supplementary Fig. 1b). We generated TMEM16C knockout (TMEM16C–/–) rats using the Sleeping Beauty transposon technology with the gene trap insertion site determined by 3’ and 5’ RACE. The gene trap vector with a splicing acceptor (SA) was inserted into intron 1 of rat TMEM16C genomic DNA (Supplementary Fig. 1c). This insertion creates a premature stop codon and eliminates full- length TMEM16C protein expression, as verified by western blots of neuronal tissues from the knockout and wild type rats (Supplementary Fig. 1c).
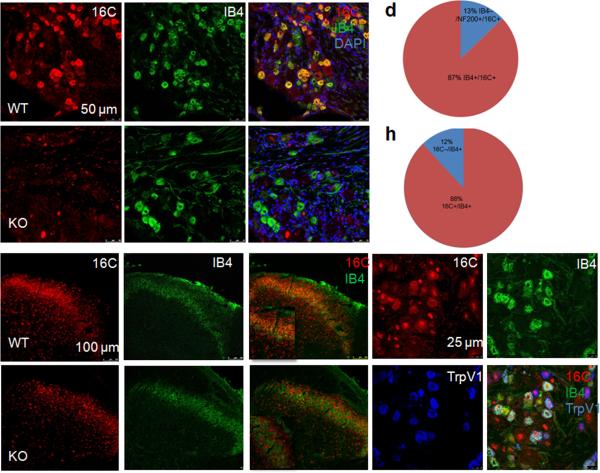
After first demonstrating robust expression of TMEM16C mRNA in the DRG (Supplementary Fig. 2a), we turned to an immunocytochemical analysis to identify the subsets of neurons in which TMEM16C is expressed. We found high levels of TMEM16C immunoreactivity mainly in the IB4-binding, non-peptidergic subpopulation of neurons (Fig. 1a–c): 87% of TMEM16C positive neurons were IB4 positive (Fig. 1d) and 88% of IB4 positive neurons were TMEM16C positive (Fig. 1h). TMEM16C immunoreactivity was also detected in some IB4 negative neurons of larger size (Fig. 1c). The latter accounted for approximately 13% of all TMEM16C positive neurons and those neurons were positive for NF200 (Fig. 1d and Supplementary Fig. 2b), which labels myelinated A delta and A beta fibers. DRGs from TMEM16C knockout rats displayed only non-specific nuclear staining (Fig. 1e), with no labeling of the IB4 positive neurons (Fig. 1e–g). TMEM16C immunoreactive and IB4-binding nerve terminals were also abundant in lamina II of the spinal cord (Fig. 1i–k), where the non-peptidergic IB4 positive afferents arborize15. TMEM16C immunoreactivity was absent in the IB4 positive nerve terminals in the spinal cord of TMEM16C knockout rats, which displayed only the non-specific nuclear staining (Fig. 1l–n). Double staining with antibodies against TrpV1 and TMEM16C revealed that around 48.9% of the TrpV1 positive neurons also express TMEM16C in the rat DRG and those neurons are all IB4 positive (Fig. 1o–r).
Figure 1. Expression of TMEM16C in dorsal root ganglia and spinal cord.
a. Immunostaining for TMEM16C in wild type rat DRG. b. IB4-FITC staining in the wild type DRG. c, Overlay of a and b. (Arrow points to a double-labeled neuron and arrowhead marks one cell stained only for TMEM16C). d. Quantification of the percentage of TMEM16C positive neurons co–stained with IB4 (total of 170 TMEM16C positive neurons) and/or NF200 (total of 166 TMEM16C positive neurons). e. Absence of TMEM16C immunoreactivity in the DRG of a TMEM16C knockout rat; the bright red dots represent non-specific staining of neuronal nuclei (example pointed out by an arrow). f. IB4-FITC staining in the knockout DRG. g. Overlay of c and d.
h. Quantification of the percentage of IB4 positive neurons co-stained with TMEM16C, shows that at least 88% of IB4-positive neurons express TMEM16C (total of 125 IB4 positive neurons). i. Immunostaining of TMEM16C in the superficial dorsal horn of the spinal cord of wild type rats. j. IB4-FITC staining in the same region of spinal cord. k. Colocalization of TMEM16C in the IB4 positive nerve terminals in the spinal cord. Insert is a higher magnification image from the superficial dorsal horn. l–n. Absence of TMEM16C immunoreactivity in the IB4 positive nerve terminals in the spinal cord of TMEM16C knockout rats. Note the non-specific staining of the nuclei (bright red dots) with TMEM16C antibodies. o–r: Triple immunostaining of the DRG shows that approximately 50% of TrpV1 positive neurons express TMEM16C and those neurons are all IB4 positive. White arrow marks a neuron triple-labeled for 16C, IB4 and TrpV1; blue arrow points to a neuron stained with TrpV1 only; and yellow arrow points to a neuron stained with 16C and IB4, but not TrpV1; white arrowhead points to an example of non-specific staining of the nuclei with the TMEM16C antibody.
WT: wild type; KO: knockout. Each staining has been repeated at least 3 times.
TMEM16C knockout rats exhibit heightened thermal and mechanical sensitivity
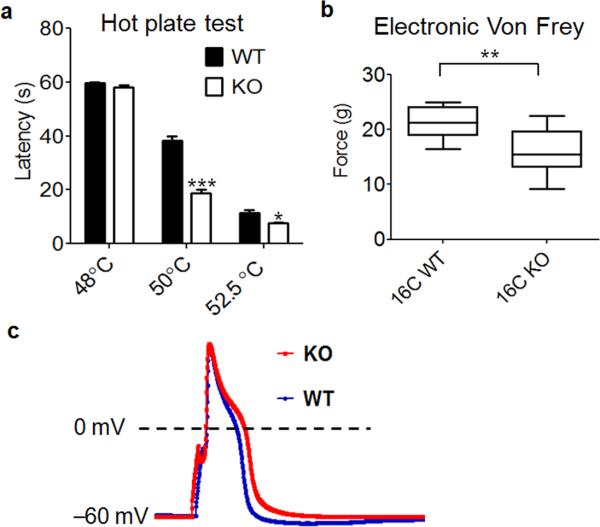
Given the concentrated expression of TMEM16C in the IB4 positive nociceptors, we next performed behavioral tests of wild type and knockout rats to assess the contribution of TMEM16C to the processing of pain messages. In the hot plate assay, which measures the latency for paw licking in response to a heated surface, TMEM16C knockout rats exhibited normal latencies at 48°C, but at temperatures greater than 50°C the mutant rats were significantly hypersensitive compared to wild type controls (Fig. 2a, two way ANOVA followed with Bonferroni test, p<0.0001, n=13 for each group). Mechanical withdrawal thresholds assayed with an electronic von Frey apparatus22 were also significantly reduced in the TMEM16C knockout rats (Fig. 2b, Student's t-test, P=0.0011, n=13 for each group).
Figure 2. TMEM16C modulates thermal and mechanical withdrawal thresholds and the action potential width of DRG neurons.
a. TMEM16C knockout rats display a significant reduction in paw withdrawal latency in the hot plate test at temperatures above 50°C (P<0.0001, two way ANOVA followed by Bonferroni test, *** P<0.001, * P<0.05, n=13). b. Compared to their wild type controls, TMEM16C knockout rats also withdraw their paws at a less intense mechanical stimulus (electronic Von Frey; Student’s t-test, ** P=0.0011, n=13). c. Representative traces of action potentials recorded in IB4 positive DRG neurons from wild type and TMEM16C knockout rats.
TMEM16C modulates action potential and excitability in the IB4 positive DRG neurons
To determine if TMEM16C influences baseline thermal and mechanical withdrawal thresholds by regulating the excitability of DRG neurons, we recorded from cultured IB4 positive DRG neurons identified by live cell labeling with IB4-FITC, and injected 2 ms currents of various amplitudes to induce action potentials. The IB4 positive neurons have broad action potentials with a hump in the falling phase23. The mean action potential width measured at 0 mV was 5.09 ± 0.33 ms for IB4 positive neurons from wild type rats (n=38); In contrast, the mean action potential width in IB4 positive neurons from TMEM16C knockout rats was significantly greater: 6.17 ± 0.38 ms (n=39) (Fig. 2c, P=0.0256, Mann Whitney test). Moreover, the voltage threshold for action potential induction was significantly reduced in the absence of TMEM16C (knockout: – 23.07 ± 0.75 mV n=25 vs wild type: –19.96 ± 0.58 mV, n=14, P=0.0007, Student's t-test). The rheobase was smaller in knockout neurons but the difference was not statistically significant (Supplementary Fig. 3a, P=0.0585, Student's t-test). The loss of TMEM16C function did not alter the resting membrane potential or the input resistance (Supplementary Fig. 3b, c, Student's t-test, P>0.05).
TMEM16C modulates a sodium-dependent potassium current in the IB4 positive DRG neurons
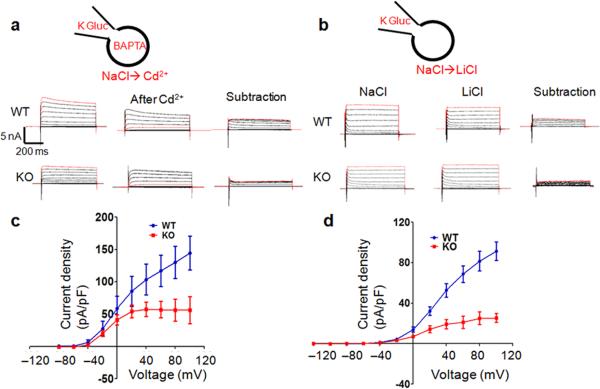
To determine how TMEM16C regulates DRG neuronal excitability, we first attempted heterologous expression of mouse TMEM16C in HEK293 cells, CHO cells and Axolotl oocytes. However, consistent with previous studies24,25, we could not detect any significant currents. Because the endogenous TMEM16C targets to nerve terminals (Fig. 1i–k), we reasoned that TMEM16C would traffic to the cell membrane of DRG neurons and therefore performed voltage clamp recordings in DRG neurons from wild type and knockout rats. Unexpectedly, we found that a sodium-activated potassium current was greatly reduced in DRG neurons lacking TMEM16C. In these recordings, we used the following solutions (in mM): external, 140 NaCl, 5 KCl, 1 MgCl2, 2 EGTA, 10 HEPES, 10 Glucose and internal, 140 K-gluconate, 10 KCl, 10 HEPES, 0.2 EGTA, 4 Mg-ATP, 0.4 Na-GTP, 5 BAPTA. After establishing voltage clamp, we applied 500 µM Cd2+ to the bath solution and obtained the Cd2+ sensitive currents by subtracting the current after Cd2+ application from the current before Cd2+ application. Cd2+, which blocks both voltage-gated calcium channels and the TTX-resistant sodium channels that contribute to the majority of sodium currents in these small DRG neurons26-32, blocked not only the inward sodium currents, but also a component of the outward currents. This outward current was significantly reduced in DRG neurons lacking TMEM16C (Fig. 3a, c, P=0.0040, paired t-test, n=7). The average current density at 100 mV is 142.8 ± 12.30 pA/pF (n=7) in control neurons and 56.33 ± 20.89 pA/pF (n=7) in DRG neurons from TMEM16C knockout rats (P=0.0040, Student's t-test). We excluded the possibility that the TMEM16C-dependent outward currents were carried by Cl– because in recordings with Cs Gluconate based internal solutions and with TEA and 4-AP in the bath to block K+ currents, there was no significant difference in the Cd2+ sensitive outward Cl– currents (due to Cl– influx) in neurons with or without TMEM16C (Supplementary Fig. 3d, paired t-test, P=0.0545). Next, we tested the possibility that the TMEM16C-dependent outward currents correspond to the sodium-activated potassium currents (KNa), previously described in DRG neurons33. Consistent with this view, replacing Na+ ions in the bath solution with Li+, which is not as potent in mediating KNa activation34, caused a reduction of the outward current in the wild type neurons and this current is greatly decreased in the knockout neurons (Fig. 3b, d. paired t-test, P=0.0233; the current at 60 mV was 68.78 ± 8.14 pA/pF in wild type, n=9, vs 24.36 ± 6.10 pA/pF in knockout, n=7, P=0.001, Student's t-test).
Figure 3. The Na+-activated potassium current (KNa) is significantly reduced in DRG neurons from TMEM16C knockout rats.
a. Representative traces demonstrate Cd2+ sensitive outward currents in wild type and knockout neurons, obtained by subtraction of currents before and after Cd2+ application. Recordings are made with the bath containing (mM): 140 NaCl, 5 KCl, 1 MgCl2, 2 EGTA, 10 HEPES, 10 Glucose and the internal solution in the pipette (mM): 140 K-gluconate, 10 KCl, 10 HEPES, 0.2 EGTA, 4 Mg-ATP, 0.4 Na-GTP, and 5 BAPTA.
b. Sample current traces with Li+ replacement recorded from wild type and knockout DRG neurons. Right, currents recorded with NaCl in the bath. Middle, currents recorded with LiCl. Left, KNa currents obtained by current subtraction. Recordings are made with the solution containing (mM): 140 NaCl, 5 KCl, 1 MgCl2, 2.5 CaCl2, 10 HEPES, 10 Glucose, and then the bath is switched to LiCl based solution (mM): 140 LiCl, 5 KCl, 1 MgCl2, 2.5 CaCl2, 10 HEPES, 10 Glucose. The pipette solution includes (mM): 140 K-gluconate, 10 KCl, 10 HEPES, 0.2 EGTA, 4 Mg-ATP, 0.4 Na-GTP
c. The I–V curve of the current density of the Cd2+ sensitive outward currents in the wild type and knockout DRG neurons (paired t-test, P=0.0040, n=7).
d. The I–V relationship of KNa current density in Li+ replacement experiments from wild type and knockout neurons (paired t-test, p<0.0233, n=7 and 9).
Slack expression is reduced in the IB4 positive DRG neurons that also express TMEM16C
Slack (sequence like a Ca2+-activated K+ channel; also known as Slo2.2 and KCNT1) and its close relative Slick are generally considered to underlie the sodium-activated potassium channels in neurons of both vertebrates and invertebrates35,36. Native KNa channels are heterogeneous in their single channel conductance and Na+ sensitivity, which may reflect their heterogeneity in subunit composition and local cellular environment35. To test whether TMEM16C regulates sodium-activated potassium currents in DRG neurons via an interaction with Slack or Slick, we first performed double immunocytochemical staining to examine the expression pattern of Slack and Slick in TMEM16C-expressing neurons. Indeed, Slack is expressed in the small DRG neurons that also express TMEM16C (Fig. 4a–c) and, as expected, these neurons are all IB4 positive (data no shown). In the spinal cord, TMEM16C and Slack had overlapping distributions in the same lamina that harbors the nerve terminals of the small DRG neurons (Fig. 4d–f). Of particular interest, immunocytochemical staining and western blot analysis demonstrated that immunoreactive Slack was significantly reduced in the DRG and spinal cord of TMEM16C knockout rats (Fig. 4g, h, j, P<0.0001, n=71 neurons for wild type and n=80 neurons for knockout, Student's t-test; and Fig. 4i, k, P=0.0008, n=4 rats, Student's t-test) even though the number of Slack and IB4 positive neurons did not differ between the wild type and knockout DRG.
Figure 4. Expression of Slack in the DRG and spinal cord.
a–c. Slack and TMEM16C are co-expressed in small DRG neurons. Arrow points to neurons that are doubly stained for TMEM16C and Slack; arrowhead points to a larger neuron immunostained only for TMEM16C. Staining has been repeated at 3 times.
–f. Overlapping Slack and TMEM16C nerve terminal immunoreactivity in the superficial dorsal horn. The bright green nuclear staining represents non-specific background signal (arrow) characteristic of the TMEM16C antibody. Staining has been repeated at 3 times.
g–h. Greatly reduced Slack immunostaining in the DRG of TMEM16C knockout (h) vs wild type (g) rats. i. Western blot of Slack in the spinal cord and DRG from wild type and TMEM16C knockout rats. Full-length gels/blots are shown in Supplementary Fig. 9. j. Quantification of immunostaining intensity of Slack positive neurons in the wild type and knockout DRG (P<0.0001, Student’s t test, n=71 for wild type and n=80 for knockout).
k. Plot of the normalized ratio of the density of the Slack band relative to the density of the alpha–tubulin band (P=0.0008, Student’s t–test, n=4).
TMEM16C interacts with Slack in HEK293 cells and in vivo
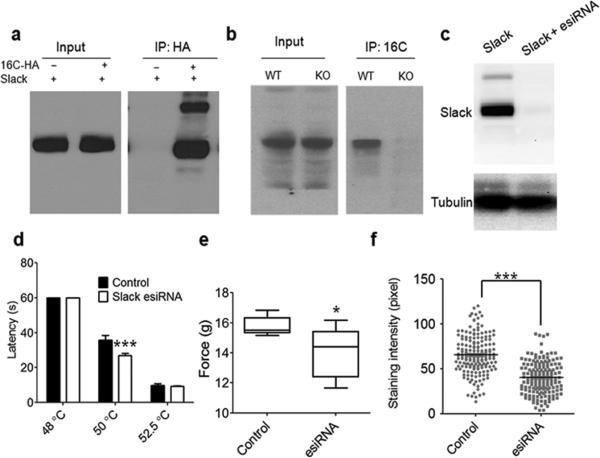
Using antibodies against TMEM16C, we found that Slack co-immunoprecipitated (co-IP) with TMEM16C in pull down assays; the specificity of the co-IP was demonstrated in control experiments that used brain tissues from TMEM16C knockout rats (Fig. 5b). To further validate the TMEM16C interaction with Slack, we transfected HEK293 cells with HA tagged TMEM16C and Slack, and demonstrated co-immunoprecipitation using antibodies against HA. The latter failed to pull down Slack from lysates of HEK293 cells transfected only with Slack (Fig. 5a). Double immunostaining with antibodies against TMEM16C and Slack revealed their co-localization in a subset of puncta in the co-transfected HEK293 cells (Supplementary Fig. 4a). The close vicinity of a fraction of TMEM16C and Slack was also demonstrated by super-resolution single molecule imaging (STORM)37 (Supplementary Fig. 4b).
Figure 5. TMEM16C interacts with Slack and knockdown of Slack in rat DRGs increases pain responsiveness.
a. Slack protein co-immunoprecipitates with TMEM16C using antibodies against HA for pull-down of associated proteins in lysates from cotransfected HEK293 cells. Note the absence of a band in the control IP from cells transfected only with Slack. Experiments were repeated 4 times. Full-length gels/blots are shown in Supplementary Fig. 9.
b. Slack protein co-immunoprecipitates with TMEM16C; antibodies against TMEM16C were used for pull-down of associated proteins in lysates from brain tissue. Note the much fainter
band of Slack in the IP from TMEM16C KO rat tissue with the same amount of input in WT and KO. Experiments were repeated 3 times. Full-length gels/blots are shown in Supplementary Fig. 9.
c. Slack esiRNA successfully knocks down Slack protein expression when co-transfected in HEK293 cells. Experiments were repeated 3 times. Full-length gels/blots are shown in Supplementary Fig. 9.
d and e. Intrathecal injection of Slack esiRNA exhibit enhanced thermal pain sensitivity at 50°C in the hot plate test, but not at 52.5°C (P=0.0006, Two Way ANOVA followed by Bonferroni's post hoc test, n=7 for control and n=8 for esiRNA) and increased mechanical sensitivity in the electronic Von Frey test (P=0.0247, Student's test, n=7 and 8).
f. Quantification of Slack immunostaining intensity in IB4 positive neurons in L4 and L5 DRGs from rats with control esiRNA and Slack esiRNA intrathecal injection (65.77 ± 1.54, n=185 vs 40.11 ± 1.35, n=171, P<0.0001, Student's t-test).
We also found that TMEM16C and Slack co-expression in HEK293 cells slightly increased their trafficking to the cell membrane, as revealed by surface biotinylation (Supplementary Fig. 5, Student's t-test, p=0.0228 and p=0.0130, n=3).
siRNA knockdown of Slack in DRG increased the pain sensitivity
Endoribonuclease-prepared siRNAs (esiRNA), which can robustly knock down gene expression in diverse cells38, are a mixture of siRNA oligos produced by cleavage of long double-stranded RNA (dsRNA) with an endoribonuclease, such as Escherichia coli RNase III. To determine if Slack in DRG neurons indeed contributes to the processing of pain messages, we generated esiRNA for Slack and confirmed its efficacy in HEK293 cells. Cotransfection of esiRNA with Slack pcDNA3 almost completely knocked down Slack expression in HEK293 cells (n=3, Fig. 5c). To knock down Slack in DRG in vivo, we made daily intrathecal injection of each rat with 2 µg of esiRNA mixed with i–Fect in vivo transfection reagents (Neuromics, MN) for 3 consecutive days and tested the rats on the 4th day, as previously reported6,39. We found that, compared to control rats (n=7) injected with nonspecific esiRNA, Slack knockdown (n=8) significantly reduced the 50°C hot plate latencies (Fig. 5d, P=0.0006, Two way ANOVA followed by post hoc Bonferroni test). Slack knockdown also reduced mechanical thresholds in the electronic Von Frey test (P=0.0247, Student's t-test, n=7 and 8, Fig. 5e). We then confirmed the knockdown of Slack proteins in L4 and L5 DRG neurons by comparing Slack immunoreactivity in IB4 positive neurons (Fig. 5f, P<0.0001, Student's t-test, n=184 neurons for control and n=171 neurons for Slack esiRNA; Supplementary Fig. 6).
TMEM16C modulates the single channel activity of Slack in HEK293 cells
Having identified the interaction between TMEM16C and Slack, we then asked whether TMEM16C co-expression with Slack alters Slack channel properties. Slack generates sodium-activated potassium channels, whose single channel conductance strongly depends on the intracellular and extracellular solution compositions, particularly the K+ concentration. For example, when expressed in oocytes, with 80 mM K+o/ K+i (80 mM Na+, 160 mM Cl–), the single channel conductance is around 88 pS, while with 160 mM K+o/ K+i(0 mM Na+, 100 mM Cl–), the single channel conductance is around 165 pS36. In another report, the single channel conductance of Slack expressed in CHO cells varied in the range of 40–65 pS with symmetric K+ and 25–65 pS with physiological solutions, likely reflecting multiple subconductance states40.
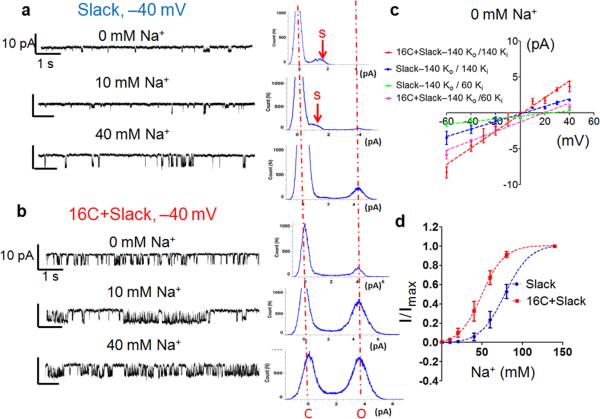
To determine if TMEM16C affects the biophysical properties of Slack at the single channel level, we recorded from excised inside-out patches with the pipette solution containing (mM): 140 KCl, 1 EGTA, 2 MgCl2, 10 HEPES and with the cytoplasmic side of the patch exposed to solutions with Na+ concentrations ranging from 0 mM to 80 mM. In patches from untransfected HEK293 cells or cells transfected with TMEM16C only, we could not detect single channel activities (>20 cells), regardless of the intracellular sodium concentration. In patches from HEK293 cells expressing Slack alone, very few channel openings could be detected with 0 mM Na+ and the single channel current amplitude was around 1.8 pA at –40 mV (Fig. 6a, upper trace and histogram on the right). With 10 mM Na+, the single channel activity was slightly increased and, in addition to the single channel current of around 1.8 pA, we observed a second single channel current of around 4 pA (Fig. 6a, middle trace and histogram). We reasoned that the 1.8 pA single channel current likely reflects subconductance states while the 4 pA single channel current corresponds to the full conductance. Indeed, with 60 mM Na+, the 4 pA current was much more prominent (Fig. 6a, bottom trace and histogram). When TMEM16C and Slack were co-expressed, we found that the channels were more active at 0 mM Na+ than with Slack expressed alone and the major peak in the all point histogram corresponds to the full conductance (Fig. 6b, upper trace and histogram). The single channel activity is further enhanced by an increase of the intracellular sodium concentration (Fig. 6b, the middle trace and histogram, 10 mM Na+; the bottom trace and histogram, 40 mM Na+). Similar results were obtained for 12 cells.
Figure 6. TMEM16C modulates Slack channel activity in HEK293 cells.
a .The left panel shows representative traces of inside-out excised patch recordings at –40 mV from HEK293 cells expressing Slack in the solution containing 0 mM Na+ (with 140 mM [K+]o / 60 mM [K+]i, 0 mM [Na+], 80 mM [Choline]), 10 mM Na (140 mM [K+]o / 60 mM [K+]i, 10 mM [Na]i, 70 mM [Choline]i) and 40 mM Na+ (140 mM [K+o/ 60 mM [K]i, 40 mM [Choline]i); The right panel shows the all point histograms. The subconductance is indicated by arrows. Similar results were obtained in 12 cells with Slack expression.
b. Representative traces and all point histogram from excised patch recordings at –40 mV from HEK293 cells expressingTMEM16C and Slack in solutions containing 0 mM, 10 mM or 40 mM Na+. The two dashed red lines in the all point histogram indicate the closed (C) and full open state (O). Note the full conductance peak at 0 mM and 10 mM Na+. Similar results were obtained in 12 cells.
c. I–V curve of KNa single channels in inside-out patches excised from cells with or without TMEM16C co-expression, exposed to 0 mM Na+ solution and at two different sets of intracellular solutions: 140 mM [K+] and 140 mM [K+ o]with Erev 0 mV and 140 mM [K+]
Slack is reportedly insensitive to low concentrations of TEA and thus TEA is generally included to eliminate TEA sensitive endogenous potassium channels33,35. By comparing the traces before and after TEA application (Supplementary Fig. 7), we confirmed that 1 mM TEA did not significantly alter the Slack single channel activity. With 1 mM TEA in the intracellular solution, TMEM16C co-expression with Slack also promoted its full conductance at low sodium concentrations (Supplementary Fig. 8, see the example traces recorded at –40 mV and the corresponding all point histograms). Similar results were obtained for 6 cells.
Since we used intracellular solutions with different potassium concentrations in the two sets of experiments described above, we also confirmed that the single channel conductance of Slack is strongly dependent on the K+ concentration. With 0 mM intracellular Na+ and 140 mM [K+]o /140 mM [K+]i, the single channel conductance in the cells expressing TMEM16C and Slack is 109.5 ± 8.2 pS (n=6), while in the recordings with 140 mM [K+] / 60 mM [K+]i], the single channel conductance is 79.3 ± 3.2 pS (n=10). For Slack expressing cells, only the subconductance was observed at 0 mM Na+ and the conductance was 47.34 ± 4.7 pS (n=9) with 140 mM [K+]o /140 mM [K+] and 19.8 ±1.33 pS (n=6) with 140 mM [K+]o / 60 mM [K+]i (Fig. 6c).
We determined the Na+ dose-response curve by plotting the average currents at different Na+ concentrations (from 0 mM to 140 mM) in excised patched with multiple channels. We found that the response curve was shifted to the left by TMEM16C co-expression (Fig. 6d, Slack: EC50: 75.74 ± 2.04 mM vs 16C+Slack: EC50: 45.00 ± 6.46 mM, P=0.0011, Student’s t-test, n=6).
In summary, we found that TMEM16C facilitates Slack channel activity by promoting its full single channel conductance at very low sodium concentrations, even in the absence of sodium, and by increasing its sodium sensitivity.
Discussions
Sodium-activated potassium (KNa) currents have been identified in cardiac cells, DRG neurons, medium spiny neurons of the striatum and tufted-mitral cells of the olfactory bulb35. Because of the lack of specific pharmacological blockers and mutant animal models, little is known about the composition of native KNa channels35,36. It has also been difficult to reconcile the diverse physiological functions of KNa channels with the high Na+ concentrations required for their activation in heterologous expression systems, given that the EC50 for Na+ activation of Slack channels is around 80 mM41. The ability of TMEM16C to interact with Slack so as to modulate its channel properties, thereby enhancing the sodium sensitivity and increasing the single channel activity at low sodium concentrations, may enable the KNa channels to be activated under normal physiological conditions and thus regulate neuronal excitability and synaptic transmission35,42.
In IB4 positive DRG neurons, the TTX-resistant sodium channels account for a substantial fraction (80–90%) of the inward current during the rising phase of the action potential43-45. The two major TTX-resistant sodium channels NaV1.8 and NaV1.9 display much slower inactivation compared to TTX-sensitive sodium channels, and thus remain active well into the repolarization phase of the action potential10. This prolonged sodium current may be optimal for activating the KNa channels in DRG neurons. This in turn will shape the action potential waveform and modulate neuronal excitability, as indicated by the action potential broadening and the decrease in threshold in neurons lacking TMEM16C (Fig. 2). The great reduction of KNa currents in DRG neurons from TMEM16C KO rats is likely due to a reduction of Slack protein as well as a decrease in sodium sensitivity. It will be interesting to determine how exactly TMEM16C and Slack interact to regulate KNa channel activity in DRG neurons. Besides regulating the KNa channels, it remains possible that TMEM16C may form channels or regulate other ion channels in other neuronal types.
At the behavioral level, TMEM16C KO rats show enhanced sensitivity to heat in the hot plate test and greater mechanosensitivity in the Von Frey test. This multimodal increase in pain sensitivity likely reflects the contribution of KNa currents in the IB4 subpopulation of nociceptors and possibly a small portion of the A type myelinated low threshold mechanoreceptors. Since TMEM16C is also expressed in the CNS, future studies using conditional knockout rodents, to selectively delete TMEM16C from nociceptors, will be necessary to determine how selective is the primary afferent contribution. Finally, given that recent genetic studies have linked TMEM16C to human autosomal-dominant craniocervical dystonia and documented a high level of TMEM16C expression in human striatum, hippocampus and cortex14, whether TMEM16C modulates KNa currents and physiological functions of neurons in these brain regions is one of the interesting open questions for future investigation.
Material and Methods
TMEM16C knockout rats
Genetic modification of Rattus norvegicus gene Tmem16C (a.k.a. ANO3) was carried out by DNA transposon insertional mutagenesis as described previously (Mamm Genome (2007) 18:338–346). The DNA transposon–modified allele is designated Brinp3Tn(sb– T2/Bart3)2.189Mcwi. The mutant strain symbol for the genetically modified rat is Tmem16cTn(sb–T2/Bart3)2.307Mcwi. The DNA transposon insertion is located within intron 1 of the Tmem16c gene in the Fischer (F344) rat strain. The genomic DNA flanking the insertion is the following:
TATACTACATACTATTTTCCAAAGATAATTTTTTTAATGTGTGTCTGTGT GGTGTGTGTGTCTATGTATATCTCTGTGTGTTTATGTGTCTCTATGTGTC
TATGTGTCTGTGTGTGTGTCTCTGTGTGTATTCATGTGTGTATTTTTGTC TCTTTGTCTTTGAAGA
Additional information about this rat model can be found at the Transposagen
Biopharmaceuticals, Inc website (www.transposagen.com).
PCR genotyping was performed on tail DNA extracted from the offspring.
All animal procedures were approved by the UCSF Institutional Animal Care and Use Committee and were performed according to the guidelines provided. Animals were housed in group of 5 rats/cage in a controlled environment, on a 12-h light/dark cycle, with free access to food and water.
Immunocytochemistry, western blot, surface biotinylation and immunoprecipitation
TMEM16C polyclonal antibodies were raised against the C-terminal peptide of mouse TMEM16C (EHLQQQRRKSGQPIHHE, peptide sequence identical to rat TMEM16C). Tissues were dissected from TMEM16C KO rats and wild type siblings and fresh frozen on a dry ice/ethanol mixture. Tissues from TMEM16C knockout rats and wild type controls were embedded together in OCT (Tissue-Tek); 20 µm cryostat sections were mounted on the same slide, air dried at room temperature (RT) for 10 min, fixed with cold 4% paraformaldehyde (PFA) or Methanol at –20°C, and then immunostained with the rabbit TMEM16C polyclonal antibody (1:500). For double staining, we used FITC-conjugated Griffonia simplicifolia isolectin B4 (IB4; L2895, Sigma, St, Louis), guinea pig anti-TrpV1 (generated by David Julius lab, 1:2000,), mouse anti-NF200 (N0142, clone N52, 1:5000, Sigma) or mouse anti-Slack (1:1000, Neuromab) together with the rabbit anti-TMEM16C antibody. Secondary antibodies conjugated with Alexa Fluor 568, 488 or 633 (1:1000; Invitrogen) were used as well as DAPI for nuclear staining. Images were taken with a Leica SP5 confocal microscope and processed with ImageJ (NIH).
For Western blot, cells or tissues were homogenized in RIPA buffer and the lysate was separated with 4–12% Bis-Tris precast gel and transferred to a PVDF membrane. TMEM16C polyclonal antibodies (1:1000), mouse anti-tubulin (T9026, Sigma, 1:2000) and mouse anti-Slack antibody (1:1000, Neuromab) were diluted in blocking buffer and incubated for 2 hrs at RT. The signal was detected with ECL plus kit and exposed with X-ray film or captured by Versadoc MP imaging system (Bio-Rad).
For immunoprecipitation, the tissue or cultured cells were homogenized in RIPA buffer with a proteinase inhibitor cocktail, sonicated briefly and then spun at 14000 rpm for 15 mins. The supernatant were collected and 2 µg of polyclonal TMEM16C antibodies with 50 µl Protein A agarose beads (Sigma) or 40 µl Anti-HA Affinity Matrix from rat IgG1 (#11815016001, Roche) were added for the immunoprecipitation. After overnight incubation, the beads were spun down and washed 4 times with RIPA buffer. The binding proteins were eluted with LDS sample buffer (Invitrogen) with 100 mM DTT.
For surface biotinylation, transfected HEK293 cells were washed 3 times with cold PBS and then incubated with 1mg/ml Sulfo-NHS-SS-Biotin (Thermo Scientific) for 20 min on ice. The cells were subsequently washed 2 times with 100 mM glycine/PBS and 2 times with PBS, and then lysed in RIPA buffer with proteinase inhibitors. Samples were incubated with 100 µl of streptavidin beads and rotated overnight at 4 °C. The samples were then washed 4 times with RIPA buffer and the beads were resuspended in LDS sample buffer with 100 mM DTT and incubated at 50 °C for 30 mins.
Slack esiRNA production and intrathecal injection
Slack esiRNA was prepared as previously reported38. Briefly, a target region of rat Slack mRNA was chosen from the RiDDLE database (http://cluster-12.mpi-cbg.de/cgi-bin/riddle/search) and PCR products flanked with T7 promoter sequences were amplified from Slack-pcDNA3 plasmids. dsRNA was generated by in vitro transcription and annealing. The resulting long dsRNA was enzymatically digested with ShortCut RNase III (NEB) into a pool of overlapping esiRNAs (around 22 nt), followed by a clean-up with the Qiagen RNeasy Kit. The esiRNA knockdown efficacy was first tested in HEK293 cells by co-transfection of 0.5 µg Slack pcDNA3 plasmids with 200 ng of esiRNA.
We followed a previously reported protocol for intrathecal esiRNA injection 39. 2 µg of esiRNA mixed with 10 µl of i-Fect transfection reagent (1:5 w/v, 12 µl total, see instructions from Neuromics) were injected into the L4–L5 lumbar region of 20 days old rats. The esiRNAs were injected at 24-hour intervals for 3 consecutive days and we performed behavioral tests 1 day after the last esiRNA injection. The esiRNA for GFP served as a control. Gene knockdown in DRG neurons was confirmed by immunofluorescence for Slack. The L4 and L5 DRGs from control and slack esiRNA treated rats were dissected, freshly frozen on dry ice, and then embedded in the same OCT block to ensure identical sectioning and staining conditions. The cryostat sections were doubly stained with IB4 and Slack and images were acquired with a Leica SP5 confocal microscope with identical setting for control and slack esiRNA samples. The staining intensities were quantified with LAS AF lite from Leica, with background staining subtracted. Average intensity of Slack staining in each IB4 positive neuron was obtained and plotted with Prism.
Reverse-transcriptase PCR
Tissue from adult rats was harvested and freshly frozen with dry ice. Total RNA was extracted, isolated and prepared with ToTALLY RNA™ Kit (Ambion). cDNA was synthesized with an Invitrogen SuperScript® III kit. The following primers were used for rat TMEM16C: Set1: Forward: TCCACCACTCAGGCTCCATTC, Reverse: CAGTTGACTGAAACAGCGAGG; Set2: Forward: CAAGTATGCAGAGAGGCTGAAC, Reverse: GCTCTGGAAACGCTGACTTG; beta-actin: Forward: GATGGTGGGAATGGGTCAGAAG, Reverse: CAGTAACAGTCCGCCTAGAAGC.
Plasmids and transfection
Mouse TMEM16C was cloned into the mammalian expression vector prk5 so as to include an N-terminal hemagglutinin (HA) tag and a C-terminal GFP tag. Mouse Slack was cloned into a pcDNA3.1 vector. Cultured HEK293 cells were transfected with Lipofectamine 2000 (Invitrogen, CA) with equal quantities of TMEM16C or EGFP N1 vector and Slack plasmids. Cells were cultured for 1–2 days before recording.
DRG neuron culture
DRGs from TMEM16C knockout rats and age matched wild type rats (postnatal 12 to 25 days) were dissected and digested with collagenase/Dispase (1 mg/ml, Roche) for 15 min, then with 0.25% Trypsin for 15 min at 37 °C. Next the digested DRGs were triturated with a fire-polished Pasteur pipette until the cell mixture turned cloudy. The cells were then plated on poly-L-lysine/laminin coated coverslips. The culture medium was MEM Eagles media with Earle's BSS supplemented with glutamine, penicillin-streptomycin, MEM vitamin solution and 10% horse serum.
Hot plate test and electronic Von Frey
Age matched (5-6 week old) WT and TMEM16C KO rats were placed on a hot plate at 48 °C, 50 °C or 52.5 °C. We measured the latency for the rat to lick its paws. We used a cut off (maximum response) time of 60 s to prevent injury. Mechanical sensitivity was assessed with an electronic version of the von Frey test (Dynamic Plantar Aesthesiometer). The filament was applied perpendicular to the plantar surface of the hind paw and the force and time when the rat lifted its paw were recorded. All experiments were performed from 9 am to 2 pm of the experimental day. The investigator was blinded to the genotype of the animal groups during the behavioral tests.
Whole cell recordings and data analysis
DRG neurons were incubated with FITC (Fluorescein) conjugated griffonia isolectin B4 (IB4, Sigma) for 10 min before recording. The IB4 stained neurons were easily recognized under epifluorescence illumination. After formation of a tight seal (>1 GΩ), we obtained a whole cell patch (< 13 MΩ) configuration by rupturing the cell membrane within the patch electrode. Whole cell capacitance and series resistance were adjusted with series resistance compensation >80%. Recordings were amplified with MultiClamp 700B (Axon Instruments), and data were analyzed with Clampfit 10 and plotted with Microsoft Excel and Prism.
All internal and external solutions were at pH 7.2-7.4 and ~300 mOsm. All experiments were performed at room temperature.
For action potential recordings, we cultured DRG neurons for 5–7 days and then recorded under current clamp. We injected a small hyperpolarizing current to set the starting potential close to –60 mV. The bath solution contained (mM): 140 NaCl, 5 KCl, 1 MgCl2, 2.5 CaCl2, 10 HEPES, 10 glucose and the pipette was filled with normal internal solution: 125 K-Gluconate, 15 KCl, 10 HEPES, 3.0 MgATP, 0.3 NaGTP, 3.0 sodium phosphocreatine, 0.2 EGTA. To induce action potential firing, we injected 2 ms current pulses with progressively greater amplitude (Δ=25 pA) until a single action potential was elicited.
For voltage clamp experiments, to ensure sufficient voltage clamp, we cultured DRG neurons within 30 hours after dissociation, before extensive processes formed. To record sodium-activated potassium currents, we used the following solutions (mM): external, 140 NaCl, 5 KCl, 1 MgCl2, 2.0 EGTA, 10 HEPES, 10 Glucose and internal, 140 K-gluconate, 10 KCl, 10 HEPES, 0.2 EGTA, 4.0 Mg-ATP, 0.4 Na-GTP, 5.0 BAPTA. The cells were held at –70 mV and voltage gated currents were evoked by 500 ms voltage steps that ranged between −100 mV and +100 mV, in 10 mV increments. We then added 500 µM CdCl2 to the bath and repeated this protocol to again induce currents. The current-voltage (I–V) relationship for the total outward current was determined at intervals of 250 ms–300 ms after initiation of voltage step pulses. Total outward currents are shown before and after the addition of Cd2+. The Cd2+ sensitive currents were obtained by subtraction of the current after Cd2+ application from the current before Cd2+ application.
In experiments that substituted LiCl for NaCl, stable whole cell voltage clamp recordings were established in Ringer's buffer (mM): 140 NaCl, 5.0 KCl, 1.0 MgCl2, 2.5 CaCl2, 10 HEPES, 10 Glucose, and then the bath was changed to LiCl based Ringer's (mM) (140 LiCl, 5.0 KCl, 1.0 MgCl2, 2.5 CaCl2, 10 HEPES, 10 Glucose). The pipette solution included (mM): 140 K-gluconate, 10 KCl, 10 HEPES, 0.2 EGTA, 4.0 Mg-ATP, 0.4 Na-GTP. 500 ms voltage steps ranging from −100 mV to +100 mV in 10 mV increments were applied and then the Na+ dependent currents were obtained by current subtraction.
To detect the Cd2+ sensitive chloride currents, the following solution was used (in mM): external, 120 NaCl, 20 TEACl, 5 KCl, 1 MgCl2, 2.5 CaCl2, 10 HEPES, 10 Glucose, 5 4-AP; internal (mM): 10 HEPES, 0.2 EGTA, 125 Cs-gluconate, 10 CsCl, 5 TEACl. 500 µM CdCl2 was added to the bath and then the protocol was repeated to induce currents. The Cd2+ sensitive Cl– currents were obtained by subtraction of the current after Cd2+ application from the current before Cd2+ application. All data are presented as MEAN ± S.E. M with the appropriate statistical analysis, as stated individually in the figures.
Single channel recordings and data analysis
Transfected cells with GFP expression were visualized with a Nikon inverted fluorescence microscope. Data were acquired using an Axopatch 200B patch clamp amplifier and pClamp9 software (Molecular Devices). All experiments were performed at room temperature. For inside-out excised patch recordings, cells were initially bathed in a solution containing (in mM): 140 NaCl, 1.0 CaCl2, 5.0 KCl, 2.0 MgCl2, 10 Glucose, and 10 HEPES, pH 7.2. The pipette solution contained (in mM): 140 KCl, 2.0 MgCl2, 1.0 EGTA, and 10 HEPES, pH 7.2. After gigaohm seal formation, the cells were perfused with 0 mM Na+ solution, which contains (in mM): 60 KCl, 2.0 MgCl2, 1.0 EGTA, 80 Choline Cl and 10 HEPES, pH 7.2. The inside out excised patches were then pulled in the 0 mM Na+ solution and the single channel activity at different holding potentials were recorded, while the cytoplasmic side of the membrane was perfused with different concentrations of Na+. The 10 mM Na+ solution contained (in mM): 60 KCl, 10 NaCl, 2.0 MgCl2, 1.0 EGTA, 10 HEPES and 70 Choline Cl, pH 7.2 and the 40 mM Na+ solution contained (in mM): 60 KCl, 40 NaCl, 2.0 MgCl2, 1.0 EGTA, 10 HEPES and 40 Choline Cl, pH 7.2.
In a different set of experiments, we used the same pipette internal solution as above. The cytoplasmic side of the membrane was perfused with different concentration of Na+ solutions and 1 mM TEA to block backgound endogenous potassium channels33. 0 mM Na+ solution contains (in mM): 140 KCl, 2.0 MgCl2, 1.0 EGTA, 1 TEA and 10 HEPES, pH 7.2. The 20 mM Na+ solution contained (in mM): 80 KCl, 20 NaCl, 2.0 MgCl2, 1.0 EGTA, 1 TEA, 10 HEPES and 40 Choline Cl, pH 7.2 and the 40 mM Na+ solution contained (in mM): 80 KCl, 40 NaCl, 2.0 MgCl2, 1.0 EGTA, 1 TEA, 10 HEPES and 20 Choline Cl, pH 7.2.
The major unitary current level was measured as previously reported40,46,47. All points histograms were constructed with pClamp 10.0 (Molecular Devices, Union City, CA) from 5–10 seconds of recording traces digitally filtered at 0.5 kHz (bin size 0.01 pA) and Gaussian fit was used to determine mean amplitude. Because Slack channels exhibit multiple subconductances and the weight of subconductance is sodium concentration dependent, we used the average current amplitude at each sodium concentrations for sodium sensitivity analysis.
Statistics
Average data are expressed as MEAN ± SEM. Two-way ANOVA tests followed by Bonferroni's post hoc test or two tailed Student's t-test (detailed in specific figures) were applied for statistical analysis using GraphPad Prism and normality distribution were tested by Shapiro-Wilk test and F-test for equal variance. When normality assumption was not met, non-parametric Mann Whitney test was used. P value of < 0.05 was considered to be significant.
No statistical methods were used to predetermine sample sizes, but our sample size are similar to those reported in previous publications6,8-10. For the behavioral studies, the data were collected with the investigator blind to the rat genotype and there was no blinding in the other experiments.
For behavioral experiments, animals from different genotypes were randomly picked for testing. For electrophysiogical experiments, the IB4-FITC labeled DRG neurons or transfected HEK cells were randomly picked for patch clamp recordings.
STORM
Slack and TMEM16C were co-transfected in ND7/23 cells and then cultured in medium containing DMEM-H21/10%FBS/2mM L-Glutamine. The transfected cells were fixed and stained with antibodies against TMEM16C and Slack. The cells were washed and then incubated in donkey anti-mouse antibody dual labeled with Alexa Fluor 405 and Alexa Fluor 647 and donkey anti-rabbit antibody dual-labeled with Cy3 and Alexa Fluor 647 (unlabeled secondary antibody from Jackson ImmunoResearch). The coverslips were mounted in buffer containing PBS, 1 M mercaptoethylamine, pH 8.5, 50% glucose in MilliQ water and oxygen scavenging solution (10 mg of glucose oxidase, 25 μl of catalase in 100 μl of PBS) in the ratio of 80:10:10:1. The STORM setup has been described previously48. Two-color imaging was achieved using different wavelengths of light to activate the dye pairs. The crosstalk between the two channels was subtracted according to previous descriptions49.
Supplementary Material
Acknowledgements
We thank Marena Tynan La Fontaine and Denan Wang for the assistance with the rodent genotyping and husbandry. We thank Taihao Jin, Huanghe Yang, Wei Zhang, Wooping Ge, Tongfei Wang, Christian Peters, Shaohua Xiao, Tong Cheng, James Berg and Zhonghui Guan for technical help and discussions. We thank Mr. Jack Crawford at Transposagen for administrative assistance.
This study is supported by grants from the NIH to L.Y.J. and A.I.B. and grants from the NIH and the Commonwealth of Kentucky to E. O. Y.N.J. and L.Y.J. are Howard Hughes Medical Institute Investigators.
Footnotes
Contributions
F.H. carried out the immunocytochemistry, molecular biology and electrophysiological studies and analyzed the data; F.H. and X.D.W. carried out behavioral studies and analyzed the data. T.W and B.H. performed the STORM imaging and data analysis. E.O. generated the knockout rats. F.H., L.Y.J. and A.I.B wrote the manuscript. L.Y.J. and Y.N.J. supervised the studies.
References
- 1.Yang YD, et al. TMEM16A confers receptor-activated calcium-dependent chloride conductance. Nature. 2008;455:1210–1215. doi: 10.1038/nature07313. [DOI] [PubMed] [Google Scholar]
- 2.Schroeder BC, Cheng T, Jan YN, Jan LY. Expression cloning of TMEM16A as a calcium-activated chloride channel subunit. Cell. 2008;134:1019–1029. doi: 10.1016/j.cell.2008.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Caputo A, et al. TMEM16A, a membrane protein associated with calcium-dependent chloride channel activity. Science. 2008;322:590–594. doi: 10.1126/science.1163518. [DOI] [PubMed] [Google Scholar]
- 4.Huang F, et al. Calcium-activated chloride channel TMEM16A modulates mucin secretion and airway smooth muscle contraction. Proc Natl Acad Sci U S A. 2012;109:16354–16359. doi: 10.1073/pnas.1214596109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huang F, et al. Studies on expression and function of the TMEM16A calcium-activated chloride channel. Proc Natl Acad Sci U S A. 2009;106:21413–21418. doi: 10.1073/pnas.0911935106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cho H, et al. The calcium-activated chloride channel anoctamin 1 acts as a heat sensor in nociceptive neurons. Nat Neurosci. 2012;15:1015–1021. doi: 10.1038/nn.3111. [DOI] [PubMed] [Google Scholar]
- 7.Stohr H, et al. TMEM16B, a novel protein with calcium-dependent chloride channel activity, associates with a presynaptic protein complex in photoreceptor terminals. J Neurosci. 2009;29:6809–6818. doi: 10.1523/JNEUROSCI.5546-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huang WC, et al. Calcium-activated chloride channels (CaCCs) regulate action potential and synaptic response in hippocampal neurons. Neuron. 2012;74:179–192. doi: 10.1016/j.neuron.2012.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Billig GM, Pal B, Fidzinski P, Jentsch TJ. Ca2+-activated Cl- currents are dispensable for olfaction. Nat Neurosci. 2011;14:763–769. doi: 10.1038/nn.2821. [DOI] [PubMed] [Google Scholar]
- 10.Yang H, et al. TMEM16F forms a Ca2+-activated cation channel required for lipid scrambling in platelets during blood coagulation. Cell. 2012;151:111–122. doi: 10.1016/j.cell.2012.07.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Suzuki J, Umeda M, Sims PJ, Nagata S. Calcium-dependent phospholipid scrambling by TMEM16F. Nature. 2010;468:834–838. doi: 10.1038/nature09583. [DOI] [PubMed] [Google Scholar]
- 12.Oldham MC, Horvath S, Geschwind DH. Conservation and evolution of gene coexpression networks in human and chimpanzee brains. Proc Natl Acad Sci U S A. 2006;103:17973–17978. doi: 10.1073/pnas.0605938103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Briones N, Dinu V. Data mining of high density genomic variant data for prediction of Alzheimer's disease risk. BMC Med Genet. 2012;13:7. doi: 10.1186/1471-2350-13-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Charlesworth G, et al. Mutations in ANO3 Cause Dominant Craniocervical Dystonia: Ion Channel Implicated in Pathogenesis. Am J Hum Genet. 2012;91:1041–1050. doi: 10.1016/j.ajhg.2012.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Basbaum AI, Bautista DM, Scherrer G, Julius D. Cellular and molecular mechanisms of pain. Cell. 2009;139:267–284. doi: 10.1016/j.cell.2009.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dhaka A, et al. TRPM8 is required for cold sensation in mice. Neuron. 2007;54:371–378. doi: 10.1016/j.neuron.2007.02.024. [DOI] [PubMed] [Google Scholar]
- 17.Davis JB, et al. Vanilloid receptor-1 is essential for inflammatory thermal hyperalgesia. Nature. 2000;405:183–187. doi: 10.1038/35012076. [DOI] [PubMed] [Google Scholar]
- 18.Snider WD, McMahon SB. Tackling pain at the source: new ideas about nociceptors. Neuron. 1998;20:629–632. doi: 10.1016/s0896-6273(00)81003-x. [DOI] [PubMed] [Google Scholar]
- 19.Dong X, Han S, Zylka MJ, Simon MI, Anderson DJ. A diverse family of GPCRs expressed in specific subsets of nociceptive sensory neurons. Cell. 2001;106:619–632. doi: 10.1016/s0092-8674(01)00483-4. [DOI] [PubMed] [Google Scholar]
- 20.Julius D, Basbaum AI. Molecular mechanisms of nociception. Nature. 2001;413:203–210. doi: 10.1038/35093019. [DOI] [PubMed] [Google Scholar]
- 21.Noel J, et al. The mechano-activated K+ channels TRAAK and TREK-1 control both warm and cold perception. Embo J. 2009;28:1308–1318. doi: 10.1038/emboj.2009.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vivancos GG, et al. An electronic pressure-meter nociception paw test for rats. Braz J Med Biol Res. 2004;37:391–399. doi: 10.1590/s0100-879x2004000300017. [DOI] [PubMed] [Google Scholar]
- 23.Stucky CL, Lewin GR. Isolectin B(4)-positive and -negative nociceptors are functionally distinct. J Neurosci. 1999;19:6497–6505. doi: 10.1523/JNEUROSCI.19-15-06497.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tian Y, Schreiber R, Kunzelmann K. Anoctamins are a family of Ca2+ activated Cl- channels. J Cell Sci. 2012 doi: 10.1242/jcs.109553. [DOI] [PubMed] [Google Scholar]
- 25.Duran C, Qu Z, Osunkoya AO, Cui Y, Hartzell HC. ANOs 3-7 in the anoctamin/Tmem16 Cl- channel family are intracellular proteins. Am J Physiol Cell Physiol. 2012;302:C482–493. doi: 10.1152/ajpcell.00140.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Leffler A, Herzog RI, Dib-Hajj SD, Waxman SG, Cummins TR. Pharmacological properties of neuronal TTX-resistant sodium channels and the role of a critical serine pore residue. Pflugers Arch. 2005;451:454–463. doi: 10.1007/s00424-005-1463-x. [DOI] [PubMed] [Google Scholar]
- 27.Kuo CC, Lin TJ, Hsieh CP. Effect of Na(+) flow on Cd(2+) block of tetrodotoxin- resistant Na(+) channels. J Gen Physiol. 2002;120:159–172. doi: 10.1085/jgp.20018536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rush AM, Cummins TR, Waxman SG. Multiple sodium channels and their roles in electrogenesis within dorsal root ganglion neurons. J Physiol. 2007;579:1–14. doi: 10.1113/jphysiol.2006.121483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huang Y, Quayle JM, Worley JF, Standen NB, Nelson MT. External cadmium and internal calcium block of single calcium channels in smooth muscle cells from rabbit mesenteric artery. Biophys J. 1989;56:1023–1028. doi: 10.1016/S0006-3495(89)82747-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ryglewski S, Pflueger HJ, Duch C. Expanding the neuron's calcium signaling repertoire: intracellular calcium release via voltage-induced PLC and IP3R activation. PLoS Biol. 2007;5:e66. doi: 10.1371/journal.pbio.0050066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Catterall WA, Perez-Reyes E, Snutch TP, Striessnig J. International Union of Pharmacology. XLVIII. Nomenclature and structure-function relationships of voltage-gated calcium channels. Pharmacol Rev. 2005;57:411–425. doi: 10.1124/pr.57.4.5. [DOI] [PubMed] [Google Scholar]
- 32.Blair NT, Bean BP. Roles of tetrodotoxin (TTX)-sensitive Na+ current, TTX- resistant Na+ current, and Ca2+ current in the action potentials of nociceptive sensory neurons. J Neurosci. 2002;22:10277–10290. doi: 10.1523/JNEUROSCI.22-23-10277.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tamsett TJ, Picchione KE, Bhattacharjee A. NAD+ activates KNa channels in dorsal root ganglion neurons. J Neurosci. 2009;29:5127–5134. doi: 10.1523/JNEUROSCI.0859-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nanou E, et al. Na+-mediated coupling between AMPA receptors and KNa channels shapes synaptic transmission. Proc Natl Acad Sci U S A. 2008;105:20941–20946. doi: 10.1073/pnas.0806403106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bhattacharjee A, Kaczmarek LK. For K+ channels, Na+ is the new Ca2+. Trends Neurosci. 2005;28:422–428. doi: 10.1016/j.tins.2005.06.003. [DOI] [PubMed] [Google Scholar]
- 36.Yuan A, et al. The sodium-activated potassium channel is encoded by a member of the Slo gene family. Neuron. 2003;37:765–773. doi: 10.1016/s0896-6273(03)00096-5. [DOI] [PubMed] [Google Scholar]
- 37.Kamiyama D, Huang B. Development in the STORM. Dev Cell. 2012;23:1103–1110. doi: 10.1016/j.devcel.2012.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Heninger AK, Buchholz F. Production of Endoribonuclease-Prepared Short Interfering RNAs (esiRNAs) for Specific and Effective Gene Silencing in Mammalian Cells. CSH Protoc. 2007;2007 doi: 10.1101/pdb.prot4824. pdb prot4824. [DOI] [PubMed] [Google Scholar]
- 39.Luo MC, et al. An efficient intrathecal delivery of small interfering RNA to the spinal cord and peripheral neurons. Mol Pain. 2005;1:29. doi: 10.1186/1744-8069-1-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Joiner WJ, et al. Formation of intermediate-conductance calcium-activated potassium channels by interaction of Slack and Slo subunits. Nat Neurosci. 1998;1:462–469. doi: 10.1038/2176. [DOI] [PubMed] [Google Scholar]
- 41.Bhattacharjee A, et al. Slick (Slo2.1), a rapidly-gating sodium-activated potassium channel inhibited by ATP. J Neurosci. 2003;23:11681–11691. doi: 10.1523/JNEUROSCI.23-37-11681.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Budelli G, et al. Na+-activated K+ channels express a large delayed outward current in neurons during normal physiology. Nat Neurosci. 2009;12:745–750. doi: 10.1038/nn.2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Renganathan M, Cummins TR, Waxman SG. Contribution of Na(v)1.8 sodium channels to action potential electrogenesis in DRG neurons. J Neurophysiol. 2001;86:629–640. doi: 10.1152/jn.2001.86.2.629. [DOI] [PubMed] [Google Scholar]
- 44.Fang X, et al. The presence and role of the tetrodotoxin-resistant sodium channel Na(v)1.9 (NaN) in nociceptive primary afferent neurons. J Neurosci. 2002;22:7425–7433. doi: 10.1523/JNEUROSCI.22-17-07425.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fang X, McMullan S, Lawson SN, Djouhri L. Electrophysiological differences between nociceptive and non-nociceptive dorsal root ganglion neurones in the rat in vivo. J Physiol. 2005;565:927–943. doi: 10.1113/jphysiol.2005.086199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Brown MR, et al. Amino-termini isoforms of the Slack K+ channel, regulated by alternative promoters, differentially modulate rhythmic firing and adaptation. J Physiol. 2008;586:5161–5179. doi: 10.1113/jphysiol.2008.160861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yang B, Desai R, Kaczmarek LK. Slack and Slick K(Na) channels regulate the accuracy of timing of auditory neurons. J Neurosci. 2007;27:2617–2627. doi: 10.1523/JNEUROSCI.5308-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Beaudoin GM, 3rd, et al. Afadin, a Ras/Rap effector that controls cadherin function, promotes spine and excitatory synapse density in the hippocampus. J Neurosci. 2012;32:99–110. doi: 10.1523/JNEUROSCI.4565-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dani A, Huang B, Bergan J, Dulac C, Zhuang X. Superresolution imaging of chemical synapses in the brain. Neuron. 2010;68:843–856. doi: 10.1016/j.neuron.2010.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.