Abstract
Purpose
The majority of patients with autosomal dominant optic atrophy (DOA) harbor pathogenic OPA1 mutations and certain missense mutations, mostly within the GTPase domain, have recently been shown to cause multiple mitochondrial DNA (mtDNA) deletions in skeletal muscle. This raises the possibility that the optic neuropathy could be the result of secondary mtDNA defects accumulating within retinal ganglion cells (RGCs). To explore this hypothesis, we looked for evidence of mitochondrial dysfunction in a mouse model of DOA and documented the visual and neurological progression in aging mutant mice.
Methods
Visual function was assessed with a rotating optokinetic (OKN) drum at 13 and 18 months of age and neurological phenotyping was performed using the primary SHIRPA screen at 13 months of age, comparing mutant Opa1+/− mice with wild type C57Bl/6 mice. The presence of cytochrome c oxidase (COX) deficiency and multiple mtDNA deletions was investigated in gastrocnemius muscle and eye specimens harvested from 2 and 11 month old Opa1+/+ and Opa1+/− mice.
Results
At 13 months of age, Opa1+/− mice had a statistically significant reduction in OKN responses compared to C57Bl/6 controls with both 2° and 8° gratings (P < 0.001). At 18 months of age, the difference between the two groups was significant for the 8° grating (P = 0.003) but not for the 2° grating (P = 0.082). Opa1+/− mice did not exhibit any significant neuromuscular deficits and no COX deficient areas or secondary mtDNA deletions were identified in skeletal muscle or the RGC layer. There was also no evidence of significant mtDNA depletion or proliferation in skeletal muscle from Opa1+/− mice.
Conclusions
COX deficiency and mtDNA abnormalities do not contribute to optic nerve dysfunction in pure DOA.
Keywords: deletions, dominant optic atrophy, mitochondrial diseases, mitochondrial DNA, optic nerve
Introduction
DOA (OMIM 165500) is one of the most common inherited optic neuropathies encountered in neuro-ophthalmological practice and about 60% of affected individuals will harbor a pathogenic mutation in the OPA1 gene (3q28-q29).1, 2 DOA was thought to have a relatively limited ocular phenotype, with most patients experiencing an insidious onset of central visual failure starting in early childhood, due to the focal loss of RGCs within the papillo-macular bundle.3 However, we and others have recently described DOA pedigrees where the visual loss also segregated with more severe neuromuscular deficits such as progressive external ophthalmoplegia (PEO), deafness, ataxia, myopathy and peripheral neuropathy.4-6 Interestingly, skeletal muscle biopsies were available from some of the affected family members and these showed unequivocal features of mitochondrial dysfunction, with a mosaic pattern of COX deficient muscle fibres and the presence of multiple mtDNA deletions on long-range polymerase chain reaction (PCR).4, 5 The Opa1 protein is part of the large, dynamin GTPase family of mechanoenzymes and it is located within the inner mitochondrial membrane7, providing intriguing causal links to the hitherto undescribed muscle changes in these “DOA plus” pedigrees. The underlying pathophysiology in DOA remains largely unexplained and the accumulation of secondary mtDNA abnormalities could represent an important mechanism which triggers the downstream events leading to cellular dysfunction and loss of function, especially within RGCs. Basic research in DOA has been severely restricted by the lack of affected human specimens and for this reason, we have recently developed an Opa1 mouse model to explore these fundamental molecular mechanisms in greater detail.8 To determine the role of COX deficiency and secondary mtDNA deletions in the RGC loss that characterises DOA we carried out a physiological, biochemical and molecular genetic study in our mutant Opa1 mice.
Materials and Methods
Opa1 mice
We have established a mouse model of DOA (B6;C3-Opa1Q285STOP) with a heterozygous nonsense mutation in exon 8 (c.1051C>T) of the Opa1 gene, which results in the introduction of a stop codon (Q285STOP) and a 50% reduction in the expression of the Opa1 protein.8 Breeding, maintenance and sacrifice were in compliance with the ARVO Statement for the Use of Animals in Ophthalmic and Vision Research. Homozygous mutant mice (Opa1−/−) died in utero during embryogenesis but heterozygous Opa1+/−mice faithfully replicated the human phenotype exhibiting a slowly progressive optic neuropathy and demonstrating objective reduction in visual function on psychophysical testing.
Visual assessment
Visual function was formally assessed with a rotating optokinetic (OKN) drum using high resolution 2° and low resolution 8° gratings, which correspond to 0.25 and 0.0625 cycles/degree respectively, and OKN responses were recorded using a validated protocol.9, 10 We have previously reported visual data on 6 and 12 month old Opa1+/−mice8 and in this study, further visual assessment was performed by one investigator (VJD) on an older cohort of heterozygous mutant Opa1+/− mice (N = 14 at 13 months and N = 12 at 18 months) and an age- and sex-matched group of C57Bl/6 wild type mice (N = 14 at 13 months and N = 12 at 18 months). OKN data for both wild type and mutant mice was collected on the same day, under the same experimental conditions.
Neurological Evaluation
The primary SHIRPA examination is a comprehensive screening technique used to define abnormal mouse phenotypes in disease models.11 It consists of 37 separate general health and neurological measures and provides quantitative information on muscle, cerebellar, sensory, neuropsychiatric and autonomic functions. (http://www.har.mrc.ac.uk/services/phenotyping/neurology/shirpa.html). Using this assessment tool, our heterozygous mutant Opa1+/− mice did not show any gross neuromuscular deficits at 6 months of age and had a normal lifespan, similar to their Opa1+/+ control littermates.8 In this study, 14 Opa1+/− mice and 14 age- and sex-matched C57Bl/6 controls were re-evaluated at 13 months of age with the primary SHIRPA protocol. This included tests of muscle tone, power and co-ordination, as well as hearing using the MRC standard click box (90dB and 18-20 Hz).
Histochemistry
The gastrocnemius muscle was harvested from the leg area and both eyes were enucleated from the following mice: 2 month old Opa1+/+ (N = 3), 2 month old Opa1+/−(N = 3), 11 month old Opa1+/+ (N = 3), and 11 month old Opa1+/− (N = 3). The muscle and eye specimens were immediately frozen in a melting isopentane bath (−150°C) and stored at −80°C. Serial sections were subsequently cut and mounted onto glass slides using a Microm™ HM560 cryostat (Thermo Fisher, Germany), at 20 μm thickness for muscle and 12 μm thickness for the eye specimens. The tissue sections were then stained using standard protocols for haematoxylin and eosin (H&E), cytochrome c oxidase (COX), succinate dehydrogenase (SDH) and dual COX-SDH.12, 13 Myofibrillar adenosine triphosphatase (ATPase) staining, with a pre-incubation pH of 4.3, was also performed on serial muscle sections.14, 15
DNA extraction
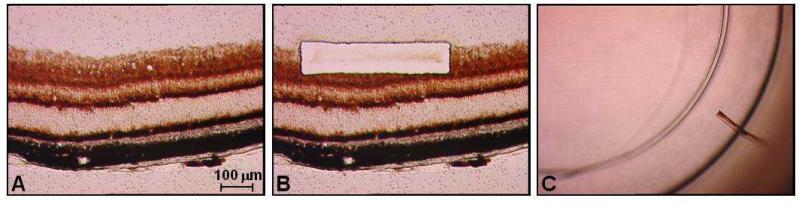
Muscle homogenate DNA was extracted using a Nucleon™ hard tissue kit (Tepnel, UK) but given the small quantities involved, an alternative method was devised to obtain an adequate yield of RGC DNA. Opa1 mouse eyes were serially sectioned onto membrane slides and stained with dual COX-SDH to localise the RGC layer and cell bodies (Figure 1A). RGC blocks (100μm × 500μm) were then cut using an LMD 6000™ laser dissecting microscope (Leica Microsystems, Germany) and collected into a 1.5ml Eppendorf™ microcentrifuge cap (Figures 1B and 1C). A total of 200 RGC blocks were pooled together for each mouse, with 10 RGC blocks dissected from 10 serial sections of both right and left eyes. The dissection block included both the RGC cell bodies and the proximal axonal segment within the retinal nerve fiber layer. DNA extraction was performed using the QIAamp™ DNA Micro Kit (Qiagen, UK) and eluted in 20μl of TE buffer.
Figure 1.
Collection of RGCs from an 11 month old Opa1+/− mouse: (A) 12μm cryostat section mounted onto a membrane slide. (B) Laser micro-dissection of a 100μm × 500μm block from the RGC layer; (C) RGC block in the microcentrifuge cap prior to cell lysis.
Long-range PCR assay
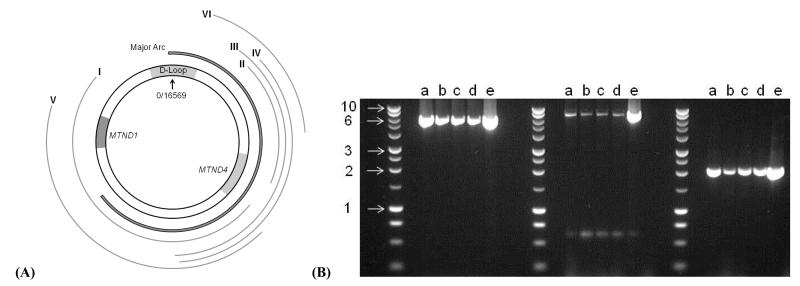
Six primer pairs, available on request, were specifically designed to amplify overlapping mtDNA regions and identify possible rearrangements, especially in the mutational hotspots along the major arc of the mitochondrial genome and involving MTND4 (Figure 2A).16 The Roche™ Expand long-range kit was used with the manufacturer’s recommended cycling procedures and the PCR products were electrophoresed in a 0.7% agarose gel at 40V for 3 hours.
Figure 2.
(A) Location of the mtDNA products amplified with our long-range PCR primer pairs: I (2773-10833, 8060bp), II (11172-14446, 3274bp), III (8309-14993, 6684bp), IV (8381-14446, 6065bp), V (3106-10595, 7489bp), and VI (13053-15053, 2000bp). (B) 0.7% agarose gel showing the products generated with primer pairs IV, V and VI respectively (a-c = RGC DNA extracted from our three 11 month old Opa1+/− mice, d = RGC DNA extracted from one 11 month old Opa1+/+ mice, e = muscle homogenate DNA extracted from the same 11 month old Opa1+/+ mice, and Promega™ 1 Kb ladder with 1, 2, 3, 6 and 10 Kb bands indicated).
Quantitative PCR assay
Relative copy number was determined in homogenate muscle DNA using a well-established iQ SYBR Green protocol on the MyiQ™ real-time PCR detection system (Biorad, USA), with MTND5 as the mtDNA reference gene and GAPDH as the nuclear DNA (nDNA) reference gene.17, 18 Both assays were optimised and confirmed to be linear over an appropriate concentration range, and all measurements were done in triplicate. The mtDNA/nDNA ratio was derived from the difference in threshold cycle value (ΔCt) between MTND5 and GAPDH, using the 2−ΔCt method.
Statistical analysis
Statistical analyses were performed using SPSS™ v.15 statistical software (Chicago, Illinois). An independent sample t-test and two-way ANOVA were used to compare the OKN responses, primary SHIRPA scores and mtDNA/nDNA ratio between mutant and wild type mice, as required. The error bars in the figures provided represent the standard error of the mean.
Results
Visual function
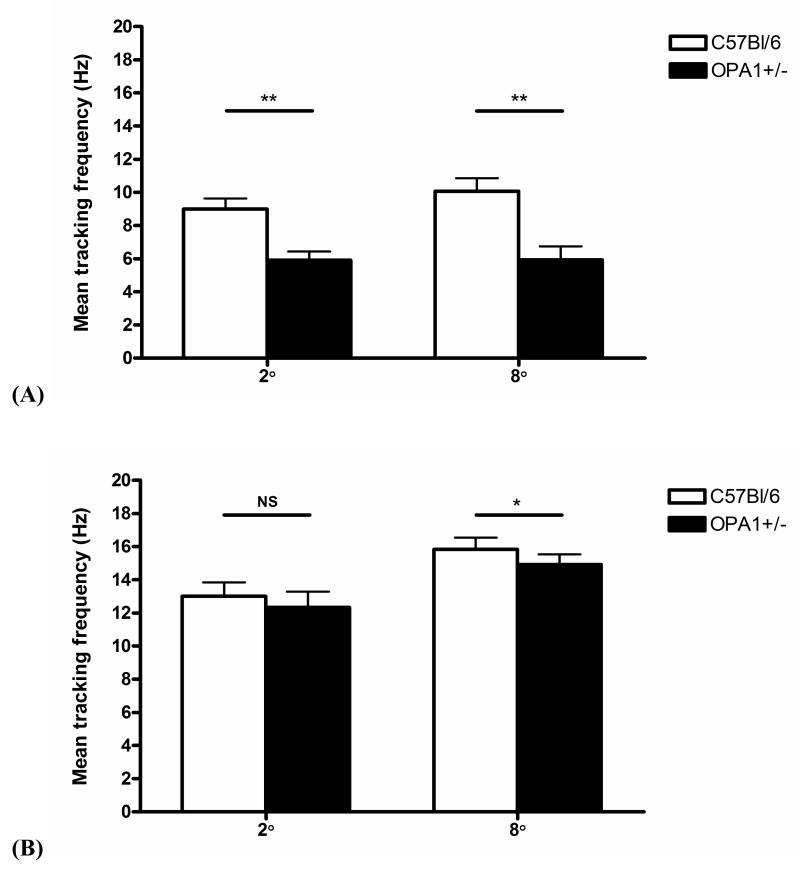
Slit lamp and dilated fundal examinations did not reveal any gross ocular pathology such as anterior segment dysgenesis, cataracts or retinal pigmentary changes in all Opa1 mice studied. In this study, 13 month old Opa1+/− mice displayed statistically significant decreased mean tracking frequency at both 2° and 8° gratings (P < 0.001) compared to wild type C57Bl/6 mice (main effect of genotype, P < 0.001; main effect of grating, P = 0.435; grating by genotype interaction, P = 0.461) (Figure 3A). At 18 months of age, the difference between mutant and control groups was significant for the 8° grating (P = 0.003) but not for the 2° grating (P = 0.082) (Figure 3B). There was also a statistically significant reduction in OKN responses between the 2° and 8° gratings at 18 months (Main effect of genotype, P = 0.325; main effect of grating, P = 0.001; grating by genotype interaction, P = 0.880). The ratio of mean tracking frequency (C57Bl/6 v/s Opa1+/−) was 1.53 for the 2° grating and 1.70 for the 8° grating at 13 months, and 1.05 for the 2° grating and 1.06 for the 8° grating at 18 months.
Figure 3.
Performance on the optokinetic visual screening test expressed as the mean frequency of tracking a moving 2° and 8° grating for 2 minutes at (A) 13 months: Opa1+/− mice (N = 14) and C57Bl/6 controls (N = 14) and, (B) 18 months: Opa1+/− mice (N = 12) and C57B6 controls (N = 12). ** P < 0.001, * P = 0.003, and NS = non significant at P = 0.082.
Neurological phenotype
The primary SHIRPA neurological examination did not reveal any functional neurological deficits in the Opa1+/− mice at 13 months of age. No gait abnormalities or tremor were present at resting level and there was no statistically significant difference in limb tone, grip strength and muscle coordination between mutant and wild type mice. Compared to wild type C57Bl/6 controls, Opa1+/− mice had increased transfer arousal and locomotor activity scores, indicating that they were possibly more anxious and fearful in novel environments (P < 0.05). All the Opa1+/− mice tested had normal hearing test results with the MRC standard click box.
Histology and mtDNA analysis
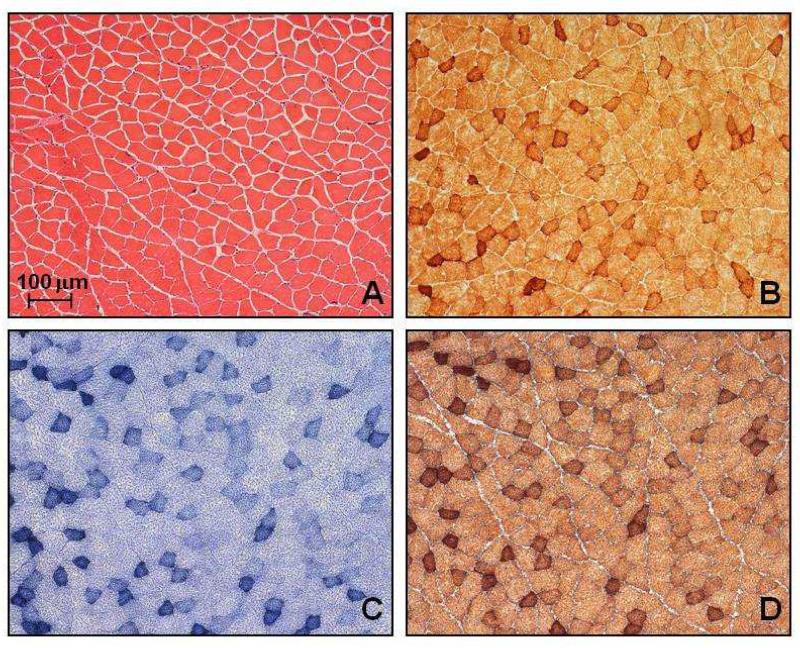
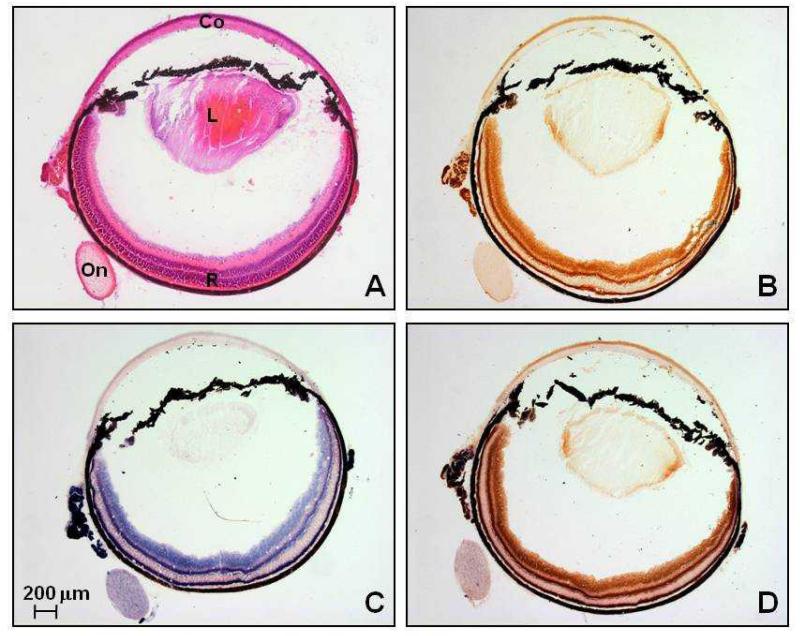
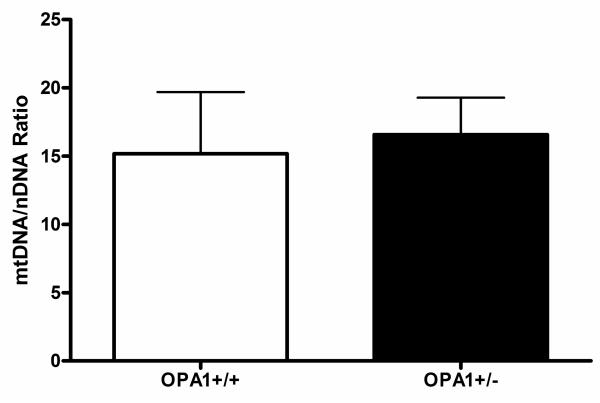
In the 12 Opa1 mice that were studied, H&E and myofibrillar ATPase staining showed normal muscle fibre morphology, with no inflammatory or neuropathic changes such as muscle fibre necrosis, regeneration, or fibre type grouping. There were no ragged red fibres (RRFs) on SDH staining and no COX deficient fibres were identified, with an average sampling of 3793 muscle fibres for each mouse (Standard deviation (SD) = 1049, range = 1690-5302) (Figure 4). Similarly, we found no evidence of COX deficient regions within the RGC layer of both the 2 month and 11 month old Opa1+/+ and Opa1+/− mice (Figure 5). The long-range PCR assay using all 6 primer pairs only amplified full mtDNA fragments with no evidence of smaller amplicons in both control and mutant groups (Figure 2B). There was no statistically significant difference in mtDNA/nNDA ratio in muscle from Opa1+/− (Mean = 16.58, SD = 2.71, N = 6) and Opa1+/+ mice (Mean = 15.17, SD = 4.51, N = 6, P = 0.795) (Figure 6).
Figure 4.
Serial 20 μm thick limb muscle sections from an 11 month old Opa1+/− mouse stained with (A) H&E, (B) COX, (C) SDH, and (D) COX-SDH. The darker staining muscle fibres are type I and reflect their higher mitochondrial oxidative capacity. A similar fibre type distribution was observed in all Opa1 mice studied, irrespective of age or mutational status.
Figure 5.
Serial 12 μm thick eye sections from an 11 month old Opa1+/− mouse stained with (A) H&E, (B) COX, (C) SDH, and (D) COX-SDH (Co: cornea, L: lens, On: optic nerve, R: retina).
Figure 6.
Comparison of mitochondrial copy number in homogenate muscle DNA extracted from OPA1+/+ and OPA1+/− mice.
Discussion
We previously reported a significant reduction in OKN responses in our heterozygous mutant Opa1+/− mice aged 12 month old compared to their littermate controls. In this report, we extended our observations to a more aged colony and further confirm that the pathogenic variant in exon 8 (c.1051C>T) of the Opa1 gene leads to optic nerve dysfunction in our heterozygous mutant mice. The optic neuropathy is relatively mild as none of the Opa1+/− mice completely failed to track the gratings, consistent with the better visual prognosis in DOA compared to other inherited optic neuropathies such as Leber hereditary optic neuropathy (LHON, OMIM 535000).3 There was a statistically significant reduction in mean tracking frequency between Opa1+/− mice and C57Bl/6 controls for both 2° and 8° gratings at 13 months, but the difference was only significant for the 8° grating at 18 months. The most likely explanation is a loss of visual acuity occurring with age for the C57Bl/6 mice, which reduced the difference between mutant and wild type mice at the higher resolution 2° grating but not at the lower resolution 8° grating. The OKN ratios at 13 and 18 months also suggest that visual acuity does not deteriorate further with advancing age in our Opa1+/− mice, which is similar to the natural history of DOA in humans, where < 50% of affected patients experience further, albeit gradual, deterioration in their visual function on long-term follow-up.19-21
A comprehensive histochemical analysis of muscle and eye specimens failed to demonstrate any COX deficient areas and no mtDNA deletions were identified using our long-range PCR assay. Quantitative analysis on homogenate muscle DNA also showed no evidence of significant mitochondrial depletion or proliferation in the Opa1+/− mice compared to Opa1+/+ controls. If the pathogenic Opa1 mutation induced these secondary mitochondrial changes, we would have expected these to be apparent in the 11 month old Opa1+/− mice, an age group where visual dysfunction had already developed. In addition, we previously showed with electron microscopy that significant morphological changes were clearly visible by 9 months of age within the optic nerve, with abnormally swollen and distorted RGC axons, and irregular areas of demyelination and myelin aggregates along the nerve fibre bundles.8 A limitation of our experimental procedures is the inability to visualize individual RGC cell bodies and axons due to their small size and it is possible that isolated COX deficient RGCs were present that have been missed. However, long-range PCR is a very sensitive molecular technique because of its preferential amplification of smaller mtDNA species and yet, enriching for RGCs by laser micro-dissection did not reveal any mtDNA deletions.22 Another possible argument is that COX deficient RGCs were present in the pre-clinical stages but these were then lost coincident with the onset of optic nerve dysfunction. Although we cannot absolutely exclude this scenario, it is somewhat unlikely given the consistent observations of viable COX-deficient neurons in brain biopsies from patients with neurodegenerative disorders such as Parkinson’s disease, where COX-deficient neurons accumulate both with increasing age and progression of the disease process.23, 24 Furthermore, previous retinal histological analysis of our Opa1+/− mice did not show any significant reduction in total RGC count.8
How can we then account for this apparent disparity between our Opa1 mouse model and the recent reports of COX deficiency and multiple mtDNA deletions in “DOA plus” pedigrees? We recently investigated a cohort of 21 patients with multi-systemic neuromuscular disorders, COX deficient muscle fibres and multiple mtDNA deletions.25 Initial mutational screen for genes previously described in patients with multiple mtDNA deletions was negative: POLG1, POLG2, SLC25A4 and PEO1, but subsequent OPA1 gene sequencing identified pathogenic variants in three probands who also had visual failure i.e. 14% of our study cohort. Although the underlying mechanisms are unknown, this very interesting finding provides additional robust evidence that some OPA1 mutations do lead to the formation and clonal expansion of mtDNA deletions.
All of the causative OPA1 mutations in these “DOA plus” families have so far been missense mutations with most, but not all of them, within the catalytic GTPase site of the protein.4-6 Although functional studies are lacking, it has been speculated that the more severe phenotype is the consequence of the mutant Opa1 protein exerting a dominant negative effect, and the more severe cellular dysfunction becomes apparent not only within RGCs, but also in other “at-risk mitochondrial” tissues such as extra-ocular muscles, muscle and brain.26 If this hypothesis is substantiated, it is perhaps not surprising that we failed to detect these abnormalities in our Opa1 mutant mice where the pathology is limited to the optic nerve i.e. “pure DOA”, with no evidence to suggest additional neurological deficits such as muscle weakness, ataxia or deafness, using the well-validated primary SHIRPA screen.11 The heterozygous exon 8 mutation (c.1051C>T) in our Opa1 mice is also truncative (Q285STOP) and overall results in a 50% reduction in the expression of the Opa1 protein, thereby representing a haploinsufficiency, and not a dominant negative, disease model. We previously showed the following percentage decrease in Opa1 protein levels in a panel of post-mitotic tissues harvested from Opa1+/− mice: retina (55%), brain (50%), skeletal muscle (80%), heart (55%), liver (80%), kidney (35%) and spleen (65%).8
Although our results only relate to this specific mouse model, one could speculate that secondary mtDNA abnormalities do not contribute to optic nerve dysfunction among patients with truncative OPA1 mutations and a “pure DOA” phenotype. It is perhaps also premature to conclude that COX deficiency and multiple mtDNA deletions are a sine qua non for a more severe disease progression in DOA. Spinazzi and colleagues have recently described a large, multi-generational pedigree where the optic atrophy segregated with mild myopathy and an axonal sensory-motor peripheral neuropathy.27 In contrast to previous “DOA plus” pedigrees, COX deficient muscle fibres and multiple mtDNA deletions were absent and a novel OPA1 deletion in the GTPase domain (c.1410_1443+4del38) was identified which reduced the level of protein expression by half i.e. led to haploinsufficiency. Functional studies performed on fibroblast and myoblast cultures showed no demonstrable respiratory chain defects, in contrast to the common c.2708_2711delTTAG OPA1 deletion which has been shown to inhibit mitochondrial oxidative phosphorylation in both in vitro fibroblast28 and in vivo muscle assays.29 However, the c.1410_1443+4del38 mutation led to marked fragmentation of the mitochondrial network, an effect also seen with other OPA1 mutations3 and in fibroblasts derived from our mouse model8, highlighting the additional important pro-fusion function of the Opa1 protein.
Opa1 is proving to be a multi-faceted protein with several crucial molecular roles relating to mtDNA replication, oxidative phosphorylation, maintenance of the mitochondrial network and apoptosis. The challenge ahead will be to unravel how OPA1 mutations actually lead to loss of function, not only in RGCs, but also in other post-mitotic tissues to explain the expanding clinical spectrum being documented in DOA families. In this respect another Opa1 mouse model with more severe neurological deficits in addition to the optic neuropathy would provide invaluable insights into these molecular mechanisms and how they interact to cause disease. Understanding these key pathophysiological steps will hopefully lead to the development of effective neuroprotective strategies with clinical benefits to patients with DOA.
Acknowledgements
PYWM is an MRC Clinical Research Fellow and PFC is a Wellcome Trust Senior Fellow in Clinical Science. This work was funded by a grant from the Medical Research Council (MRC, UK) to MV.
Footnotes
Conflict of Interest: None declared
References
- 1.Newman NJ, Biousse V. Hereditary optic neuropathies. Eye. 2004;18:1144–1160. doi: 10.1038/sj.eye.6701591. [DOI] [PubMed] [Google Scholar]
- 2.Ferre M, Amati-Bonneau P, Tourmen Y, Malthiery Y, Reynier P. eOPA1: An online database for OPA1 mutations. Hum Mutat. 2005;25:423–428. doi: 10.1002/humu.20161. [DOI] [PubMed] [Google Scholar]
- 3.Yu-Wai-Man P, Griffiths PG, Hudson G, Chinnery PF. Inherited mitochondrial optic neuropathies. J Med Genet. 2009;46 doi: 10.1136/jmg.2007.054270. doi:10.1136/jmg.2007.054270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hudson G, Amati-Bonneau P, Blakely EL, et al. Mutation of OPA1 causes dominant optic atrophy with external ophthalmoplegia, ataxia, deafness and multiple mitochondrial DNA deletions: a novel disorder of mtDNA maintenance. Brain. 2008;131:329–337. doi: 10.1093/brain/awm272. [DOI] [PubMed] [Google Scholar]
- 5.Amati-Bonneau P, Valentino ML, Reynier P, et al. OPA1 mutations induce mitochondrial DNA instability and optic atrophy plus phenotypes. Brain. 2008;131:338–351. doi: 10.1093/brain/awm298. [DOI] [PubMed] [Google Scholar]
- 6.Ferraris S, Clark S, Garelli E, et al. Progressive external ophthalmoplegia and vision and hearing loss in a patient with mutations in POLG2 and OPA1. Arch Neurol. 2008;65:125–131. doi: 10.1001/archneurol.2007.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Davies V, Votruba M. Focus on molecules: the OPA1 protein. Experimental Eye Research. 2006;83:1003–1004. doi: 10.1016/j.exer.2005.11.021. [DOI] [PubMed] [Google Scholar]
- 8.Davies VJ, Hollins AJ, Piechota MJ, et al. Opa1 deficiency in a mouse model of autosomal dominant optic atrophy impairs mitochondrial morphology, optic nerve structure and visual function. Hum Mol Genet. 2007;16:1307–1318. doi: 10.1093/hmg/ddm079. [DOI] [PubMed] [Google Scholar]
- 9.Thaung C, Arnold K, Jackson IJ, Coffey PJ. Presence of visual head tracking differentiates normal sighted from retinal degenerate mice. Neurosci Lett. 2002;325:21–24. doi: 10.1016/s0304-3940(02)00223-9. [DOI] [PubMed] [Google Scholar]
- 10.Hart AW, McKie L, Morgan JE, et al. Genotype-phenotype correlation of mouse Pde6b mutations. Invest Ophthalmol Vis Sci. 2005;46:3443–3450. doi: 10.1167/iovs.05-0254. [DOI] [PubMed] [Google Scholar]
- 11.Rogers DC, Fisher EMC, Brown SDM, Peters J, Hunter AJ, Martin JE. Behavioral and functional analysis of mouse phenotype: SHIRPA, a proposed protocol for comprehensive phenotype assessment. Mamm Genome. 1997;8:711–713. doi: 10.1007/s003359900551. [DOI] [PubMed] [Google Scholar]
- 12.Johnson MA, Barron MJ. Muscle biopsy analysis. In: Lane RJM, editor. Handbook of muscle disease. Marcel Dekker; New York, USA: 1996. pp. 61–79. [Google Scholar]
- 13.Taylor RW, Schaefer AM, Barron MJ, McFarland R, Turnbull DM. The diagnosis of mitochondrial muscle disease. Neuromusc Disord. 2004;14:237–245. doi: 10.1016/j.nmd.2003.12.004. [DOI] [PubMed] [Google Scholar]
- 14.Carpenter S, Karpati G. Methods of tissue removal and preparation. In: Carpenter S, Karpati G, editors. Pathology of skeletal muscle. Oxford University Press; New York, USA: 2001. pp. 8–27. [Google Scholar]
- 15.Carpenter S, Karpati G. Major pathological reactions and their consequences for skeletal muscle cells. In: Carpenter S, Karpati G, editors. Pathology of skeletal muscle. Oxford University Press; New York, USA: 2001. pp. 63–130. [Google Scholar]
- 16.Krishnan KJ, Reeve AK, Samuels DC, et al. What causes mitochondrial DNA deletions in human cells? Nat Genet. 2008;40:275–279. doi: 10.1038/ng.f.94. [DOI] [PubMed] [Google Scholar]
- 17.Cree LM, Patel SK, Pyle A, et al. Age-related decline in mitochondrial DNA copy number in isolated human pancreatic islets. Diabetologia. 2008;51:1440–1443. doi: 10.1007/s00125-008-1054-4. [DOI] [PubMed] [Google Scholar]
- 18.Cree LM, Samuels DC, Lopes S, et al. A reduction of mitochondrial DNA molecules during embryogenesis explains the rapid segregation of genotypes. Nat Genet. 2008;40:249–254. doi: 10.1038/ng.2007.63. [DOI] [PubMed] [Google Scholar]
- 19.Votruba M, Moore AT, Bhattacharya SS. Clinical features, molecular genetics, and pathophysiology of dominant optic atrophy. J Med Genet. 1998;35:793–800. doi: 10.1136/jmg.35.10.793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Puomila A, Huoponen K, Mantyjarvi M, et al. Dominant optic atrophy: correlation between clinical and molecular genetic studies. Acta Ophthalmol Scand. 2005;83:337–346. doi: 10.1111/j.1600-0420.2005.00448.x. [DOI] [PubMed] [Google Scholar]
- 21.Cohn AC, Toomes C, Hewitt AW, et al. The Natural History of OPA1-related Autosomal Dominant Optic Atrophy. Br J Ophthalmol. 2008;92:1333–1336. doi: 10.1136/bjo.2007.134726. [DOI] [PubMed] [Google Scholar]
- 22.Lightowlers RH, Jacobs HT, Kajander OA. Mitochondrial DNA - all things bad? Trends Genet. 1999;15:91–93. doi: 10.1016/s0168-9525(98)01684-9. [DOI] [PubMed] [Google Scholar]
- 23.Bender A, Krishnan KJ, Morris CM, et al. High levels of mitochondrial DNA deletions in substantia nigra neurons in aging and Parkinson disease. Nat Genet. 2006;38:515–517. doi: 10.1038/ng1769. [DOI] [PubMed] [Google Scholar]
- 24.Kraytsberg Y, Kudryavtseva E, McKee AC, Geula C, Kowall NW, Khrapko K. Mitochondrial DNA deletions are abundant and cause functional impairment in aged human substantia nigra neurons. Nat Genet. 2006;38:518–520. doi: 10.1038/ng1778. [DOI] [PubMed] [Google Scholar]
- 25.Stewart JD, Hudson G, Yu-Wai-Man P, et al. Opa1 in Multiple Mitochondrial DNA Deletion Disorders. Neurology. 2008;71:1829–U1159. doi: 10.1212/01.wnl.0000335931.54095.0a. [DOI] [PubMed] [Google Scholar]
- 26.Zeviani M. OPA1 mutations and mitochondrial DNA damage: keeping the magic circle in shape. Brain. 2008;131:314–317. doi: 10.1093/brain/awm339. [DOI] [PubMed] [Google Scholar]
- 27.Spinazzi M, Cazzola S, Bortolozzi M, et al. A novel deletion in the GTPase domain of OPA1 causes defects in mitochondrial morphology and distribution, but not in function. Hum Mol Genet. 2008;17:3291–3302. doi: 10.1093/hmg/ddn225. [DOI] [PubMed] [Google Scholar]
- 28.Zanna C, Ghelli A, Porcelli AM, et al. OPA1 mutations associated with dominant optic atrophy impair oxidative phosphorylation and mitochondrial fusion. Brain. 2008;131:352–367. doi: 10.1093/brain/awm335. [DOI] [PubMed] [Google Scholar]
- 29.Lodi R, Tonon C, Valentino ML, et al. Deficit of in vivo mitochondrial ATP production in OPA1-related dominant optic atrophy. Ann Neurol. 2004;56:719–723. doi: 10.1002/ana.20278. [DOI] [PubMed] [Google Scholar]