Abstract
We report that dye-doped fluorescent silica nanoparticles (FSNPs) are highly efficient labels for glycans. Mono- and oligo-saccharides were conjugated to FSNPs using a general photocoupling chemistry. FSNP-labled glycans were applied to image and detect bacteria, and to study carbohydrate-lectin interactions on a lectin microarray.
A major challenge in bioanalysis, including high-throughput screening, is the means to effectively display the outcome of the ligand-receptor interactions. Fluorescence is by far the most commonly used detection method, which requires the conjugation of a fluorescent tag to the ligand to be studied.1–3 Among the fluorescent tags, organic dyes continue to be the label of choice due to the availability of a wide variety of commercial products of diverse structures, functionalities, solubility, and spectral properties. A major drawback of organic dyes, however, is their relatively poor photostability. When exposed to light, organic dyes can photobleach and lead to a decrease in fluorescence intensity. A clever solution to this problem is to entrap the fluorescent dye inside a solid matrix, for example silica nanoparticles.4 The nanoparticle protects the dye molecules from being directly exposed to the environmental oxygen, and thus greatly enhances the photostability of the entrapped dye.5 Furthermore, because a large number of dye molecules can be embedded inside a nanoparticle, high fluorescence emission can be obtained, the intensity of which exceeds the dye molecule itself or even quantum dots.6–9 In addition, silica nanoparticles are biocompatible and of low toxicity.10, 11 Uniformly sized silica nanoparticles can be readily prepared from inexpensive starting materials following simple synthetic procedures.12
Ligand labeling often requires a robust conjugation method where the labeling agent can be covalently attached to the ligand. For many biomolecules, this can be conveniently accomplished, for example, by using a commercial kit containing chemically-derivatized labeling agents. For ligands that lack functional groups or are difficult to derivatize, the task of labeling can be complex and challenging. We have developed a general coupling chemistry, based on perfluorophenyl azides (PFPAs), that can conjugate a variety of molecules regardless of their chemical structures.13–16 Upon photochemical or thermal activation, PFPA is converted to a highly reactive singlet perfluorophenylnitrene which form covalent adducts with neighboring molecules by way of CH insertion and/or C=C addition reactions. Therefore, by functionalizing FSNPs with PFPA, ligands can be conjugated to FSNPs via the photocoupling reaction of the surface PFPA. In this study, we present that PFPA-functionalized FSNPs can be used as highly efficient fluorescence labels for carbohydrate ligands. Carbohydrates are an important class of biomolecules involved in many important biorecognition processes including, for example, cell communication, immune responses, fertilization, and infections.17–20 Studies of these processes are, however, hampered by the high complexity of glycan structures and the lack of efficient bioanalytical tools.21–24 The present study seeks to address some of these challenges by developing a new method to efficiently label underivatized carbohydrates with FSNPs. We evaluate FSNP-labeled carbohydrates for their affinities with lectins. The synthesized glyco FSNPs are furthermore applied to image bacteria, and to probe carbohydrate-protein interactions on a lectin microarray.
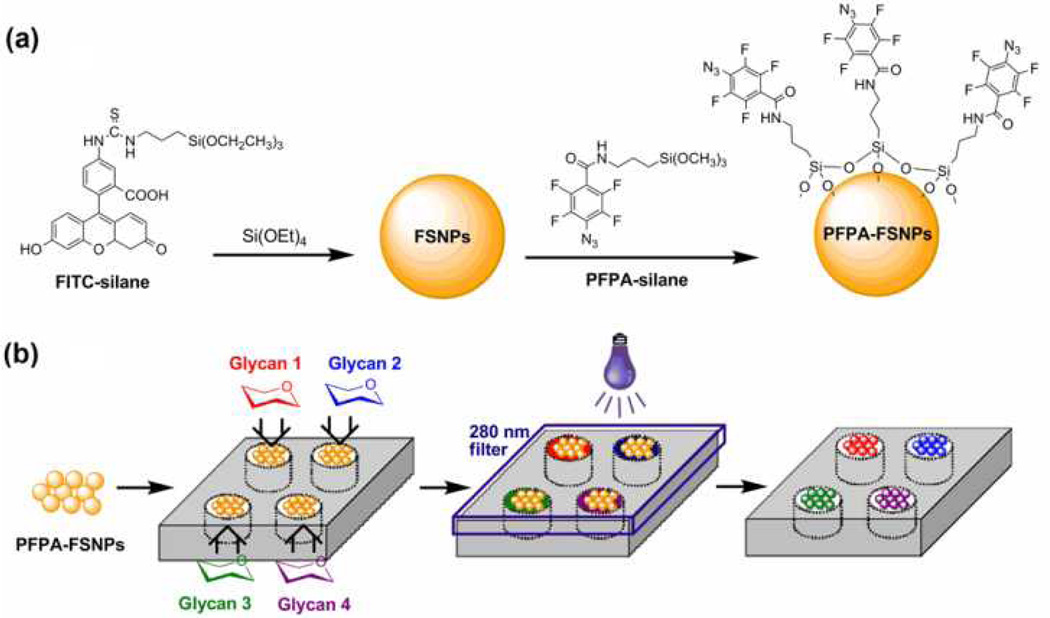
Fluorescein (FITC)-doped silica nanoparticles were synthesized using a modified Stöber method.12 Fluorescein isothiocyanate was silanized with 3-aminopropyltriethoxysilane yielded FITC-silane (Fig. 1), which was then copolymerized with tetraethyl orthosilicate (see Supplementary Information for detailed synthetic procedures).4, 7 The resulting FSNPs, ~100 nm in diameter, showed intense fluorescence even at low particle concentrations (see Fig. 1S & 2S for particle size characterization). To test whether the entrapped FITC can withstand the UV irradiation condition used for the photocoupling reaction, a solution of FSNPs was irradiated with a 450-W medium-pressure Hg lamp for 10 min. The resulting solution remained highly fluorescent, and the fluorescence intensity decreased only to a small extent (Fig. 3S, Supplementary Information). On the other hand, when a solution of FITC was irradiated under the same condition, the fluorescence intensity was reduced to about 50% of the original value (Fig. 4S, Supplementary Information). These results are consistent with the observations by others that the photostability is significantly improved when fluorescent dyes are embedded in silica nanoparticles.9, 25
Fig. 1.
(a) Synthesis of PFPA-functionalized FSNPs, and (b) high-throughput synthesis of glyco FSNPs.
Next, FSNPs were functionalized with PFPA by treating the as-prepared FSNPs with PFPA-silane (Fig. 1, See Supplementary Information for detailed procedure).26 To covalently label carbohydrates with FSNPs, our previously established procedure for coupling carbohydrates on gold nanoparticles was followed.16, 27 In the process, a solution containing the carbohydrate and FSNPs was irradiated with a medium-pressure Hg lamp for 10 min. Excess reagents were removed by dialysis, and the resulting glyco FSNPs showed excellent water solubility and high fluorescence emission intensity. The density of the immobilized carbohydrate was determined using a previously developed colorimetric assay (see Supplementary Information for detailed procedure),16 from which the coupling yields were calculated. The results showed that the photocoupling reaction was highly efficient. The coupling yield, ranging from 36% to 54% (Table 1S), increased with the size of the carbohydrate, a result that is consistent with our previous study using gold nanoparticles.16
The labeling process is well adaptable for high-throughput where the photocoupling step can be performed in parallel. A pilot study was carried out where four micro-vials containing PFPA-FSNPs and four different carbohydrates were photoactivated simultaneously (Fig. 1b). The products showed successful conjugation of each carbohydrate on the FSNPs. The number of reaction wells can be further increased by using the microarray technology to enable a rapid parallel synthesis of larger libraries of FSNP-labeled ligands.
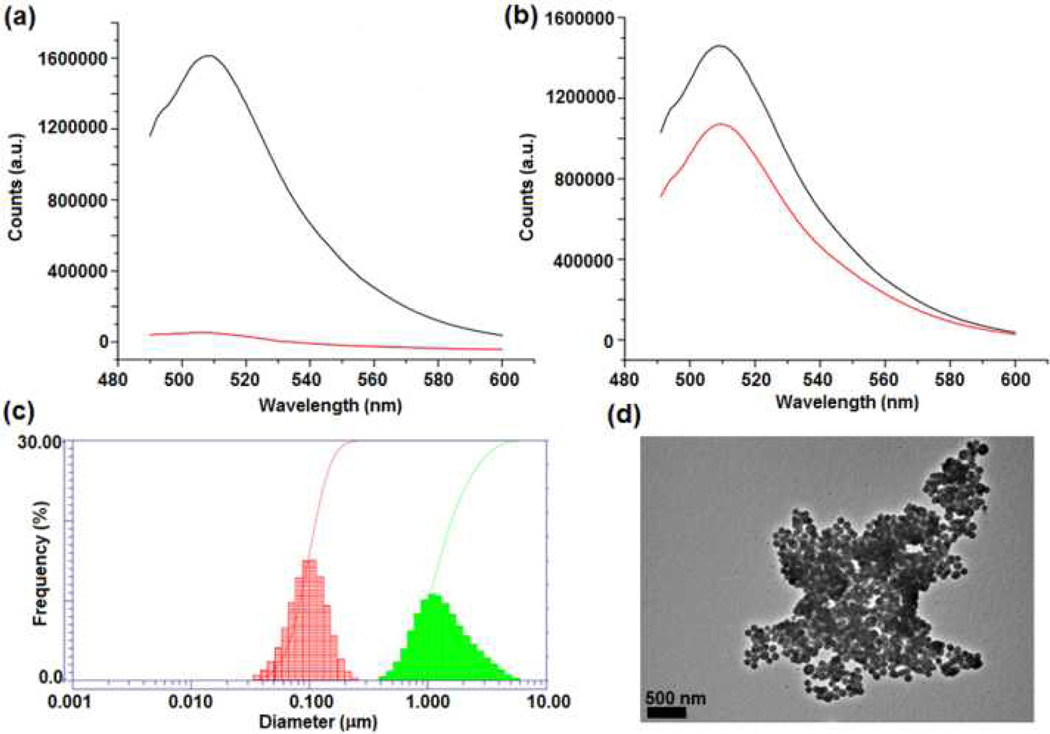
The bioactivity of FSNP-labeled carbohydrates was tested in the following studies. d-Mannose labeled with FSNP, FSNP-Man, was treated with Concanavalin A (Con A), a well-studied lectin which exhibits specific affinity to α-d-mannopyranoside, α-d-glucopyranoside, and their derivatives.28, 29 At pH>7, Con A exists as a tetramer, inducing significant nanoparticle agglomeration upon binding to Man-containing ligands.16 When FSNP-Man was incubated with Con A for 1 hour, the fluorescence intensity of the solution decreased drastically (Fig. 2a). Precipitates were observed where the nanoparticles agglomerated into clusters (Fig. 2d). In contrast, when FSNP-labeled d-galactose, FSNP-Gal, was treated with Con A following the same procedure, the fluorescence intensity decreased only slightly (Fig. 2b). The small intensity decrease is likely due to the nonspecific adsorption of Con A on the nanoparticles. The FSNP-Man-Con A aggregates were additionally examined by dynamic light scattering (DLS), which showed, on the average, a 10-time increase in the particle size in comparison to FSNP-Man (Fig. 2c).
Fig. 2.
Fluorescence spectra of (a) FSNP-Man and (b) FSNP-Gal before (black lines) and after (red lines) incubating with Con A; (c) DLS of Man-FSNPs before (red) and after (green) binding with Con A; (d) TEM image of Man-FSNPs after treating with Con A.
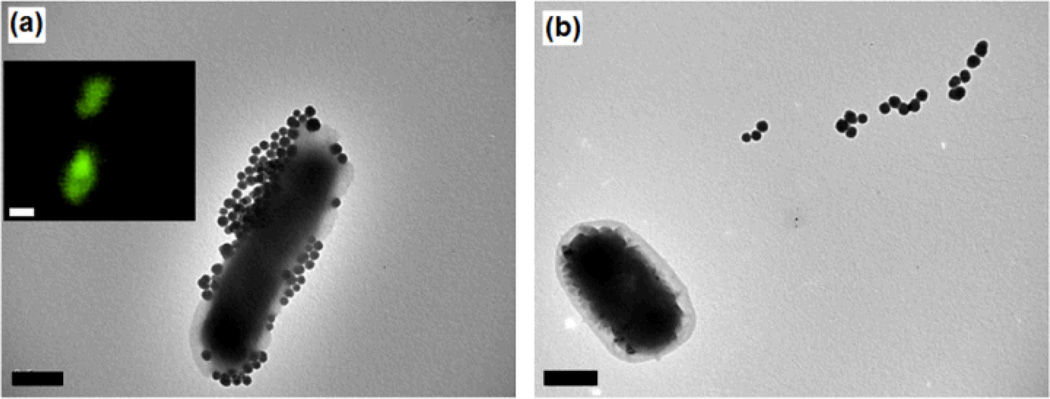
The utility of the FSNP labeling technique was next investigated by applying glyco FSNPs in imaging, and for studying glycan-lectin interactions on a lectin microarray. In the first experiment, FSNP-Man was treated with E. coli bacteria strain ORN 178. This particular strain contains a Man-specific binding domain, i.e., the FimH lectin, on type 1 pili.30, 31 The TEM image of the resulting solution displays a large number of FSNP-Man on the E. coli (Fig. 3a), which can be attributed to the multivalent interactions between Man ligands on the FSNP-Man with the FimH lectin on the bacteria.15 The strong interaction was further confirmed by fluorescence imaging where intense fluorescence was observed on the bacteria (Fig. 3a insert). In contrast, when FSNP-Man was treated with E. coli strain ORN208 that is deficient of the Man-binding FimH protein, no fluorescence was observed on the bacteria. In fact, almost no nanoparticles were attached to the bacteria surface (Fig. 3b).
Fig. 3.
TEM images after FSNP-Man was treated with E. coli strain ORN178 (a), or ORN208 (b). Insert: fluorescence image of the corresponding sample. Scale bars: 500 nm
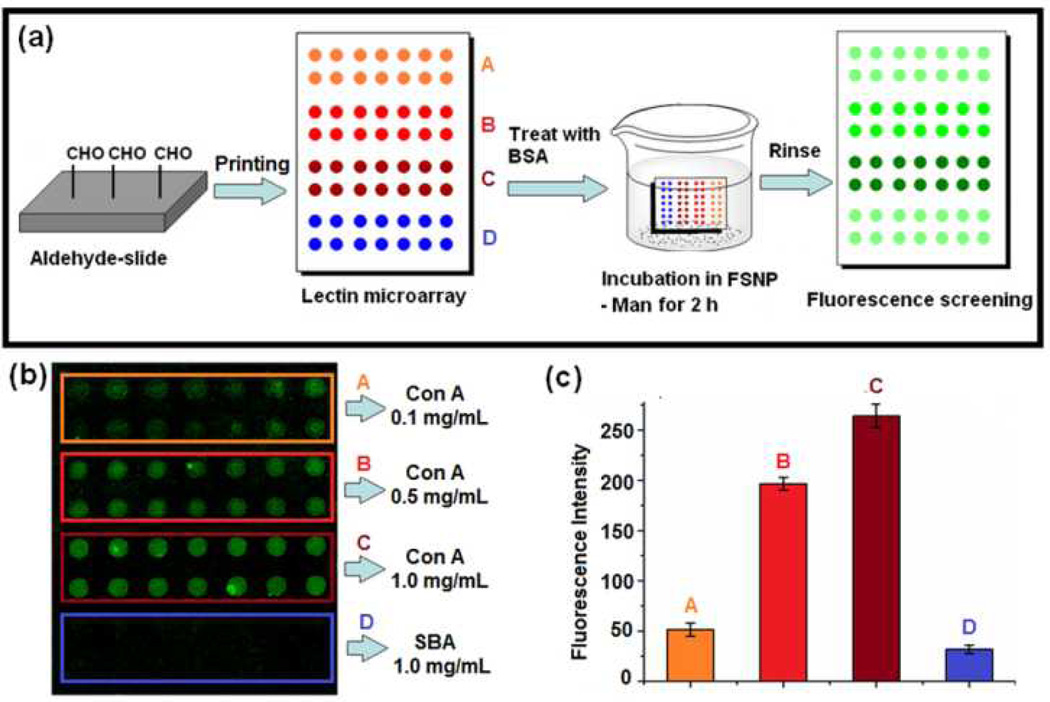
The FSNP-labeled carbohydrates were next employed to study carbohydrate-lectin interactions on a lectin microarray. The lectin microarray was fabricated on aldehyde-functionalized glass slides following the established literature procedure.32 Solutions of Con A and soybean agglutinin (SBA, a non-Man-binding lectin) in pH 7.4 phosphate-buffered saline (PBS) containing 40% glycerol were then printed on the aldehyde slides using a robotic printer.33 The lectin array slide, after blocking with bovine serum albumin (BSA), was incubated in the FSNP-Man solution for 2 hours (Fig. 4a). As anticipated, fluorescence was observed on all Con A spots (Fig. 4b). The relative fluorescence intensity on Con A were ~10 times higher than those on SBA at the printing concentration of 1 mg/mL (Fig. 4c). Even at 0.1 mg/mL, the fluorescence intensity from Con A was still noticeably higher than that from SBA, although the spot quality deteriorated at lower printing concentrations. These results clearly demonstrate that the FSNP-labeled carbohydrates are highly suited for interrogating carbohydrate-lectin interactions on microarrays. Furthermore, ligand displacement experiments can be conducted on the lectin arrays to quantitively analyze glycan-lectin interactions. Work in this aspect is underway and results will be reported in a future account.34
Fig. 4.
(a) Preparation of lectin microarray, incubation with FSNP-Man, and fluorescence imaging. (b) Fluorescence image and (c) fluorescence intensities of lectin microarray after treating with FSNP-Man. Each data point was the average of the 7 spots on the microarray.
In conclusion, a simple and general method was developed to label carbohydrates with dye-doped silica nanoparticles. The strategy applies to underivatized carbohydrate structures, thus avoiding complex synthesis and purification steps that are often involved in the chemical derivatization of these ligands. The labeling is highly efficient, and the resulting FSNP-labeled carbohydrates retained their binding affinity and selectivity towards lectins. The utility of this labeling technique was successfully demonstrated where FSNP-labeled carbohydrates were applied in bacteria detection and imaging, and in probing glycan-lectin interactions on microarray. These results illustrate that, although the labeling chemistry, i.e., the CH insertion reaction of PFPA, yields a random orientation of the attached ligands, the labeled ligands retain their binding selectivities nonetheless. Further, the labeling reaction may allow the exposure of all epitopes on the ligands, and thus a biased epitope selection can be avoided. The technique developed can in principle be readily applied to other biologically significant molecules including pharmaceuticals and metabolites. The advantage of this method resides in its generality and simplicity, where the labeling process employs a single labeling agent, PFPA-FSNP, and a uniform coupling chemistry. Ligands are labeled in their native forms without undergoing prior chemical derivatization. These features, combined with the straightforward preparation of dye-doped silica nanoparticles and the low material cost, may open up a myriad of opportunities for this technique in bioanalysis and diagnostic applications.
Supplementary Material
Acknowledgments
This work was supported by the National Institutes of General Medical Science (NIGMS) under NIH Award Numbers 1R01GM080295 and 2R15GM066279.
Footnotes
Electronic Supplementary Information (ESI) available: Experimental details, nanoparticle characterization, photo-stability studies, ligand density determination and coupling yields. See DOI: 10.1039/b000000x/
References
- 1.Pepperkok R, Ellenberg J. Nat. Rev. Mol. Cell Biol. 2006;7:690–696. doi: 10.1038/nrm1979. [DOI] [PubMed] [Google Scholar]
- 2.Krutzik PO, Nolan GP. Nat. Methods. 2006;3:361–368. doi: 10.1038/nmeth872. [DOI] [PubMed] [Google Scholar]
- 3.Kuno A, Uchiyama N, Koseki-Kuno S, Ebe Y, Takashima S, Yamada M, Hirabayashi J. Nat. Methods. 2005;2:851–856. doi: 10.1038/nmeth803. [DOI] [PubMed] [Google Scholar]
- 4.Van Blaaderen A, Vrij A. Langmuir. 1992;8:2921–2931. [Google Scholar]
- 5.Yao G, Wang L, Wu YR, Smith J, Xu JS, Zhao WJ, Lee EJ, Tan WH. Anal. Bioanal. Chem. 2006;385:518–524. doi: 10.1007/s00216-006-0452-z. [DOI] [PubMed] [Google Scholar]
- 6.Ow H, Larson DR, Srivastava M, Baird BA, Webb WW, Wiesner U. Nano Lett. 2005;5:113–117. doi: 10.1021/nl0482478. [DOI] [PubMed] [Google Scholar]
- 7.Larson DR, Ow H, Vishwasrao HD, Heikal AA, Wiesner U, Webb WW. Chem. Mater. 2008;20:2677–2684. [Google Scholar]
- 8.Zhao XJ, Bagwe RP, Tan WH. Adv. Mater. 2004;16:173–176. [Google Scholar]
- 9.Wang L, Wang KM, Santra S, Zhao XJ, Hilliard LR, Smith JE, Wu JR, Tan WH. Anal. Chem. 2006;78:646–654. [Google Scholar]
- 10.De M, Ghosh PS, Rotello VM. Adv. Mater. 2008;20:4225–4241. [Google Scholar]
- 11.Rosi NL, Mirkin CA. Chem. Rev. 2005;105:1547–1562. doi: 10.1021/cr030067f. [DOI] [PubMed] [Google Scholar]
- 12.Stöber W, Fink A. J. Colloid Interface Sci. 1968;26:62–69. [Google Scholar]
- 13.Yan M. Polym. News. 2002;27:6–12. [Google Scholar]
- 14.Yan M. Chem. Eur. J. 2007;13:4138–4144. doi: 10.1002/chem.200700317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu LH, Dietsch H, Schurtenberger P, Yan M. Bioconjugate Chem. 2009;20:1349–1355. doi: 10.1021/bc900110x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang X, Ramström O, Yan M. J. Mater. Chem. 2009;19:8944–8949. doi: 10.1039/B917900C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Koeller KM, Wong CH. Nat. Biotechnol. 2000;18:835–841. doi: 10.1038/78435. [DOI] [PubMed] [Google Scholar]
- 18.Liu F-T, Rabinovich GA. Nat. Rev. Cancer. 2005;5:29–41. doi: 10.1038/nrc1527. [DOI] [PubMed] [Google Scholar]
- 19.Blow N. Nature. 2009;457:617–620. doi: 10.1038/457617a. [DOI] [PubMed] [Google Scholar]
- 20.Ohtsubo K, Marth JD. Cell. 2006;126:855–867. doi: 10.1016/j.cell.2006.08.019. [DOI] [PubMed] [Google Scholar]
- 21.Dwek RA. Chem. Rev. 1996;96:683–720. doi: 10.1021/cr940283b. [DOI] [PubMed] [Google Scholar]
- 22.Varki A. Glycobiology. 1993;3:97–130. doi: 10.1093/glycob/3.2.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sears P, Wong CH. Proc. Natl. Acad. Sci. U. S. A. 1996;93:12086–12093. doi: 10.1073/pnas.93.22.12086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Feizi T, Chai W. Nat. Rev. Mol. Cell Biol. 2004;5:582–588. doi: 10.1038/nrm1428. [DOI] [PubMed] [Google Scholar]
- 25.Tapec R, Zhao XJJ, Tan WH. J. Nanosci. Nanotechnol. 2002;2:405–409. doi: 10.1166/jnn.2002.114. [DOI] [PubMed] [Google Scholar]
- 26.Gann JP, Yan M. Langmuir. 2008;24:5319–5323. doi: 10.1021/la7029592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang X, Ramström O, Yan M. Anal. Chem. 2010;82:9082–9089. doi: 10.1021/ac102114z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bittiger H, Schnebli HP. Concanavalin A as a Tool. London: John Wiley and Sons; 1976. [Google Scholar]
- 29.Lis H, Sharon N. Chem. Rev. 1998;98:637–674. doi: 10.1021/cr940413g. [DOI] [PubMed] [Google Scholar]
- 30.Harris SL, Spears PA, Havell EA, Hamrick TS, Horton JR, Orndorff PE. J. Bacteriol. 2001;183:4099–4102. doi: 10.1128/JB.183.13.4099-4102.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Krogfelt KA, Bergmans H, Klemm P. Infect. Immun. . 1990;58:1995–1998. doi: 10.1128/iai.58.6.1995-1998.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.MacBeath G, Schreiber SL. Science. 2000;289:1760–1763. doi: 10.1126/science.289.5485.1760. [DOI] [PubMed] [Google Scholar]
- 33.Lian W, Litherland SA, Badrane H, Tan WH, Wu DH, Baker HV, Gulig PA, Lim DV, Jin SG. Anal. Biochem. 2004;334:135–144. doi: 10.1016/j.ab.2004.08.005. [DOI] [PubMed] [Google Scholar]
- 34.Wang X, Ramström O, Yan M. to be submitted. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.