Estimating false conviction rate among death sentences
When criminal courts convict an innocent defendant, the mistake is often unobserved and rarely identified subsequently. As a result, some researchers have held that the rate of false convictions is not merely unknown but unknowable. Samuel Gross et al. (pp. 7230–7235) focused on death sentences, a small subset of criminal convictions that produces a far higher exoneration rate than any other context. Noting that most death-sentenced defendants are resentenced to life in prison and do not remain on death row, the authors analyzed data on exonerations among defendants between 1973, when the modern form of the death penalty was established in the United States, and 2004 from the Department of Justice and the Death Penalty Information Center, a nonprofit that tracks exonerations among capital defendants. The authors found that the high exoneration rate for death-sentenced defendants applies only while the defendants remain under the threat of execution. Using survival analysis, a technique in epidemiology, the authors estimate that if all death-sentenced defendants were to remain under a threat of execution indefinitely at least 4.1% would be exonerated. The finding is a conservative estimate of the proportion of false convictions among death sentences in the United States, according to the authors. — T.J.
Uncovering elicitors of silent bacterial gene clusters
Bacterial genomes house an immense reservoir of biosynthetic gene clusters that can be used to produce beneficial drugs. Although bacterial natural products account for most antibiotics in clinical use today, efforts to find new drugs have largely been thwarted because the majority of these gene clusters appear to be inactive with no discernible triggering mechanism. Mohammad Seyedsayamdost (pp. 7266–7271) outlines a strategy for rapidly identifying small molecules that activate silent gene clusters. The method, the author reports, uses reporter gene fusions to monitor the activity of a silent cluster of interest while high-throughput screening sifts through small molecule libraries to provide candidate activators. Demonstrating the approach, the author reports a prominent activation mechanism for two cryptic gene clusters in the pathogen Burkholderia thailandensis and identifies a previously unreported metabolite. Furthermore, the study reveals that almost all activating molecules were antibiotics, suggesting a key role for these drugs in modulating silent biosynthetic pathways. In addition to providing access to cryptic gene clusters in bacteria, further applications of this method might engender new antibiotics, thus facilitating efforts to combat the rise of multidrug-resistant bacteria, according to the author. — T.J.
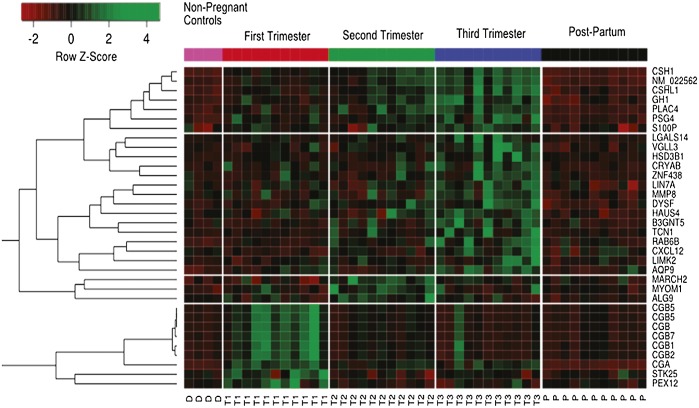
Noninvasive analysis of human gene expression
Heatmap of time-varying genes from microarray analysis.
In 2013, an estimated half million women underwent noninvasive prenatal genetic testing based on cell-free, fetal DNA circulating in maternal plasma. Such tests indicate gene content but do not reveal gene expression status, crucial to understanding maternal and fetal health. Winston Koh et al. (pp. 7361–7366) used high-throughput microarrays and next-generation sequencing to characterize the circulating RNA complement of 4 healthy adults—two men and two nonpregnant women—and 11 pregnant women who provided plasma samples during each trimester of pregnancy and after delivery. The authors report that RNA transcript levels helped monitor temporal and tissue-specific fetal gene expression across pregnancy; the number of expressed genes rose as pregnancy progressed and plummeted after delivery. The temporal pattern of appearance of RNA transcripts of a hormone used to diagnose pregnancy and monitor gestational health reflected the hormone’s documented temporal expression pattern, suggesting that RNA might serve as an alternative biomarker. In addition, the authors measured the RNA transcript levels of a protein precursor of beta-amyloid, implicated in Alzheimer’s disease (AD), in the blood plasma of 6 AD patients and 10 healthy adults, and found increased levels in the patients’ plasma, suggesting that plasma RNA might serve as a potential AD biomarker, the diagnostic sensitivity and specificity of which remain to be determined. According to the authors, cell-free RNA in maternal plasma might aid the future diagnosis of pregnancy-related complications. — P.N.
Semiconductor sequencing might help improve fetal genetic testing

Cell-free fetal DNA sequencing can help detect aneuploidies. Image courtesy of National Human Genome Research Institute (www.genome.gov).
Recent methods based on massively parallel sequencing have boosted rapid, noninvasive prenatal genetic testing for chromosomal copy number abnormalities called aneuploidies through the analysis of cell-free, circulating fetal DNA in maternal plasma. Can Liao et al. (pp. 7415–7420) developed a semiconductor-based benchtop sequencing platform to obtain 5 billion data points per second in 2–4 hours, thus increasing the efficiency of rapid, portable, and cost-effective prenatal diagnosis for fetal aneuploidies. The authors validated the sequencing platform by testing its diagnostic sensitivity and specificity on 515 pregnant women at two hospitals in China for whom aneuploidy data was previously obtained through karyotyping. The authors report that the platform identified all 55 cases of Down syndrome, 16 cases of Edward syndrome, and three cases of Patau syndrome—all marked by extra chromosome copies—with specificity and sensitivity between 99% and 100%. Further, the method helped identify 15 fetuses with aneuploid sex chromosomes. Next, the authors applied the platform to 1,760 pregnant women without karyotype data, and detected 15 cases of Down, Edward, or Patau syndromes, and one case of X chromosome aneuploidy. According to the authors, the platform might pave the way toward reducing the time and cost of sensitive and specific prenatal genetic diagnosis in clinics. — P.N.
Brain size, diet, and cognitive evolution

The pictured chimpanzee and members of 36 other animal species were tested for their abilities to exert self-control in two experimental tasks.
Several hypotheses propose to explain the forces that shape cognitive evolution, but few have been rigorously tested. Evan MacLean et al. (pp. E2140–E2148) integrated experimental and phylogenetic approaches to discern which forces exert the greatest influence on cognitive evolution. The authors measured the cognitive abilities of 567 animals representing 36 different species by testing their abilities to exert self-control in two experimental tasks. The authors looked for correlations between the species’ cognitive abilities and a range of ecological, social, and brain measures to investigate four hypotheses about cognitive evolution. Two hypotheses propose brain volume changes as a mechanism underlying cognitive evolution: one proposes that brain size predicts cognitive ability, whereas the other proposes that brain size relative to body size is the most important factor for cognitive ability. The authors also tested two hypotheses regarding the selective pressures that may have stimulated cognitive evolution: social complexity and dietary complexity. According to the study, species with greater absolute, but not relative, brain volume exhibited greater self-control than species with smaller brain volume, as did primate species with greater dietary complexity than other primates. According to the authors, the findings suggest that feeding ecology and brain size may influence cognitive evolution. — J.P.J.