Significance
The mitochondrial ribosome (mitoribosome) is a macromolecular complex that plays a central role in mitochondrial protein synthesis. Its small subunit is involved directly in the recruitment and decoding of mitochondrial mRNAs. Defects in mitochondrial translation, including mutations in components of the mitoribosome, are known to cause numerous human genetic diseases. Thus, knowledge of the molecular architecture of the mitoribosome is essential for a better understanding of those diseases and of the process of translation. To our knowledge, this article describes the first detailed cryo-EM structure of the small subunit of the mammalian mitoribosome. The study provides important clues about the evolution of this macromolecular complex and reveals unique structural features that could be important in the translation of the unusual mitochondrial mRNAs.
Keywords: mammalian mitochondrial ribosomal SSU, cryo-electron microscopy, mito-12S rRNA, mammalian MRPs
Abstract
The mammalian mitochondrial ribosomes (mitoribosomes) are responsible for synthesizing 13 membrane proteins that form essential components of the complexes involved in oxidative phosphorylation or ATP generation for the eukaryotic cell. The mammalian 55S mitoribosome contains significantly smaller rRNAs and a large mass of mitochondrial ribosomal proteins (MRPs), including large mito-specific amino acid extensions and insertions in MRPs that are homologous to bacterial ribosomal proteins and an additional 35 mito-specific MRPs. Here we present the cryo-EM structure analysis of the small (28S) subunit (SSU) of the 55S mitoribosome. We find that the mito-specific extensions in homologous MRPs generally are involved in inter-MRP contacts and in contacts with mito-specific MRPs, suggesting a stepwise evolution of the current architecture of the mitoribosome. Although most of the mito-specific MRPs and extensions of homologous MRPs are situated on the peripheral regions, they also contribute significantly to the formation of linings of the mRNA and tRNA paths, suggesting a tailor-made structural organization of the mito-SSU for the recruitment of mito-specific mRNAs, most of which do not possess a 5′ leader sequence. In addition, docking of previously published coordinates of the large (39S) subunit (LSU) into the cryo-EM map of the 55S mitoribosome reveals that mito-specific MRPs of both the SSU and LSU are involved directly in the formation of six of the 15 intersubunit bridges.
The mammalian mitochondrial genome encodes 37 genes, including two ribosomal RNAs (12S and 16S rRNAs), 22 mitochondrial tRNAs, and 13 polypeptides of the oxidative phosphorylation complexes. All the proteins required for mammalian mitochondrial translation, including mitochondrial ribosomal proteins (MRPs), are encoded in the nuclear genome, translated in the cytoplasm, and then imported into the mitochondrion. Defects in several components of the mammalian mitochondrial translational machinery are involved in a number of human genetic diseases (1–3). Thus, knowledge of the molecular architecture of the mitochondrial translational machinery is important for a better understanding these diseases. Previous low-resolution cryo-EM studies (4–6) have revealed several unique structural features of the mitoribosomes. Because mitochondria are thought to have originated through an early endosymbiotic event between an α-protobacteria and a primitive host cell (7), its ribosomes were proposed to be structurally similar to those of bacteria. However, the overall 3D structure of the mammalian mitoribosome was found to be significantly altered and porous (4) as compared with its bacterial and eukaryotic cytoplasmic counterparts (8–10).
The functional 55S mammalian mitoribosome is made up of two unequally sized subunits referred to as the small (SSU, 28S) and large (LSU, 39S) subunits. The LSU is involved in catalyzing the peptidyl-transferase reaction, and the SSU provides the platform for mRNA binding and decoding. Based on cryo-EM studies, partial molecular models of the 39S subunit have been published at 12.1-Å (6) and more recently at ∼5-Å (11) resolution. Here we present the 7-Å cryo-EM structure of the 28S mito-SSU, which is composed of a 12S rRNA molecule and 31 MRPs [numbered S2 to S39, with gaps (12)]. Fifteen of the 31 MRPs are homologous to bacterial ribosomal proteins, most of which (except for MRPs S6 and S12) have accrued mito-specific N-terminal and/or C-terminal amino acid extensions (NTE and CTE, respectively) of varying lengths (Table S1). No homologs for six bacterial ribosomal proteins—S1, S4, S8, S13, S19, and S20—are found in the mito-SSU. We have modeled the complete 12S rRNA, all of the 15 homologous MRPs, and their mito-specific extensions into the cryo-EM density. In addition, we have made an attempt to identify densities for the 16 mito-specific MRPs within the cryo-EM map. Strategic locations of some of the mito-specific MRPs and mito-specific extensions in homologous MRPs on the mito-SSU structure suggest their direct involvement in the recruitment of the unique mitochondrial mRNAs (13).
Results and Discussion
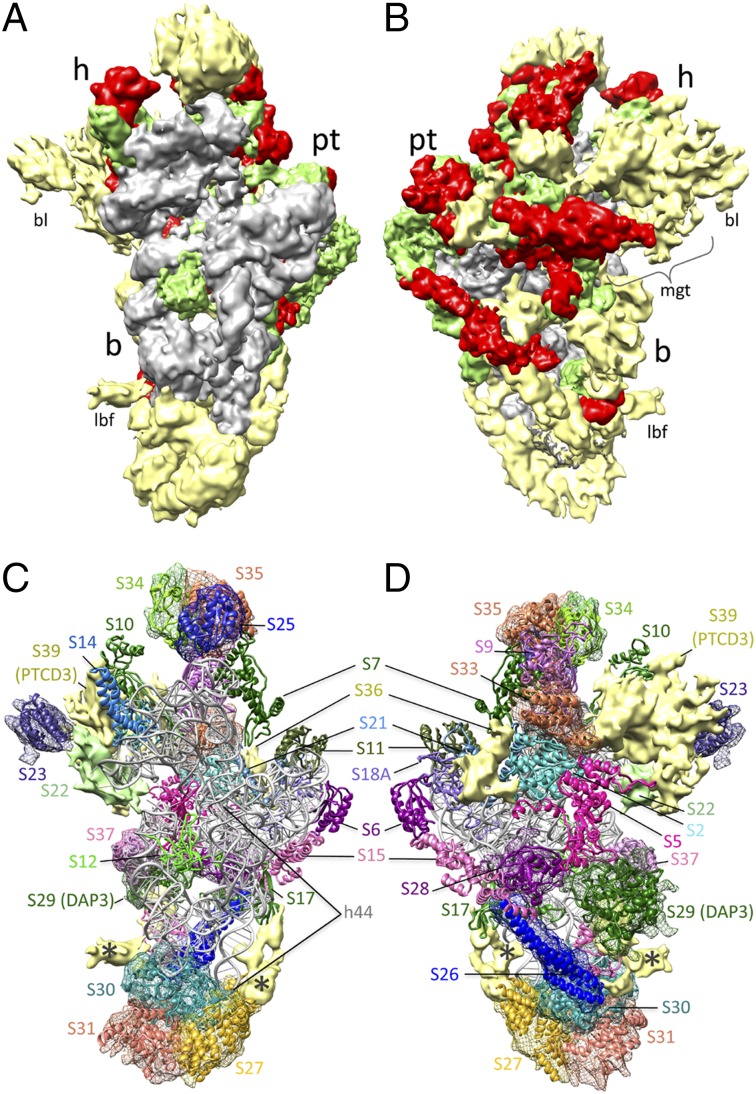
The density corresponding to the 28S SSU was well resolved in most part in our 7.0-Å resolution cryo-EM map of the 55S mitoribosome (Fig. S1), allowing modeling of the complete 12S rRNA molecule (Fig. S2) and 15 homologous MRPs and their mito-specific extensions (Fig. S1C). Although several peripheral bacterial rRNA helices are either truncated or completely lost in the mito-rRNAs (Fig. S2 and refs. 4 and 10), the tertiary structure of the core of the 12S rRNA and the overall positions of all homologous MRPs are preserved in the mito-SSU structure. However, the mito-specific extensions of the homologous MRPs are primarily oriented and exposed on the solvent side of the mito-SSU, as are the majority of the mito-specific MRPs (Fig. 1 A and B). To localize densities corresponding to the remaining 16 mito-specific MRPs (molecular mass, 11.5–77.8 kDa) (12) that do not share any significant sequence identity with the known protein structures, we took a comprehensive approach, including information available from immuno-EM, application of Segger-based multiple segmentation (14), matching the structural features of each segmented density with the ab initio model of the MRP, and establishing a correlation between the size of each segmented density and the molecular mass of the mito-specific MRPs (Methods and Materials, Table S2, and Figs. S3–S5). Using this approach, we have tentatively positioned and docked models for 12 of the 16 mito-specific MRPs within the mito-SSU map (Fig. 1 C and D and Table S3). However, further structural characterization at higher resolution would be required for more precise placement of the mito-specific MRPs. Like other ribosomal SSUs, the mito-SSU map could be divided readily into three major structural domains, namely, the body, head, and platform. The molecular architectures of these domains are described in the next three sections.
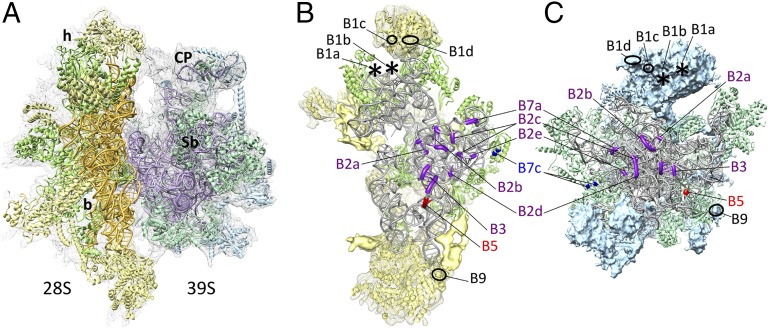
Fig. 1.
Cryo-EM structure of the mammalian mitochondrial 28S ribosomal SSU. (A and B) Segmented cryo-EM densities for 12S rRNA (gray), homologous MRPs (green), mito-specific extensions and insertions in homologous MRPs (red), and mito-specific MRPs (yellow). Landmarks: h, head; b, body; pt, platform; bl, beak lobe; lbf, lower body finger. (C and D) The 28S structure with modeled 12S rRNA (gray) and 15 homologous MRPs. Densities tentatively assigned to mito-specific MRPs (S23, S25–S31, S33–S35, and S37), along with their docked homology models, are shown as meshwork (also see Table S2 and Figs. S3–S5). Densities shown as solid yellow did not match homology models but also have been tentatively assigned, and densities marked with asterisks (*) would correspond to undocked portions of MRP S27 and S30 models. The mito-SSU is shown from the interface side in A and C and from the solvent side in B and D.
SSU Body.
As in cytoplasmic ribosomes, the 5′ major domain and a significant portion of the 3′ minor domain of the 12S rRNA (Fig. S2) provide the basic scaffold for the SSU body. Helix 44, which encompasses the mRNA decoding site and runs vertically through the main body on the subunit interface side (Fig. 1C), also is known to carry multiple genetic disease hotspots (15). (We use the bacterial numbering to refer to rRNA helices throughout, and henceforth helices are identified by a number prefixed with the letter “h.”) We find that the overall conformation of h44 is significantly altered in the mito-SSU compared with its conformation in the SSUs of cytoplasmic ribosomes (Fig. S2F). Specifically, the lower portion of h44 is oriented slightly toward the platform side of the body. However, the lowermost region of h44 is partially disordered in our map, suggesting dynamic behavior of that region in the mitoribosome.
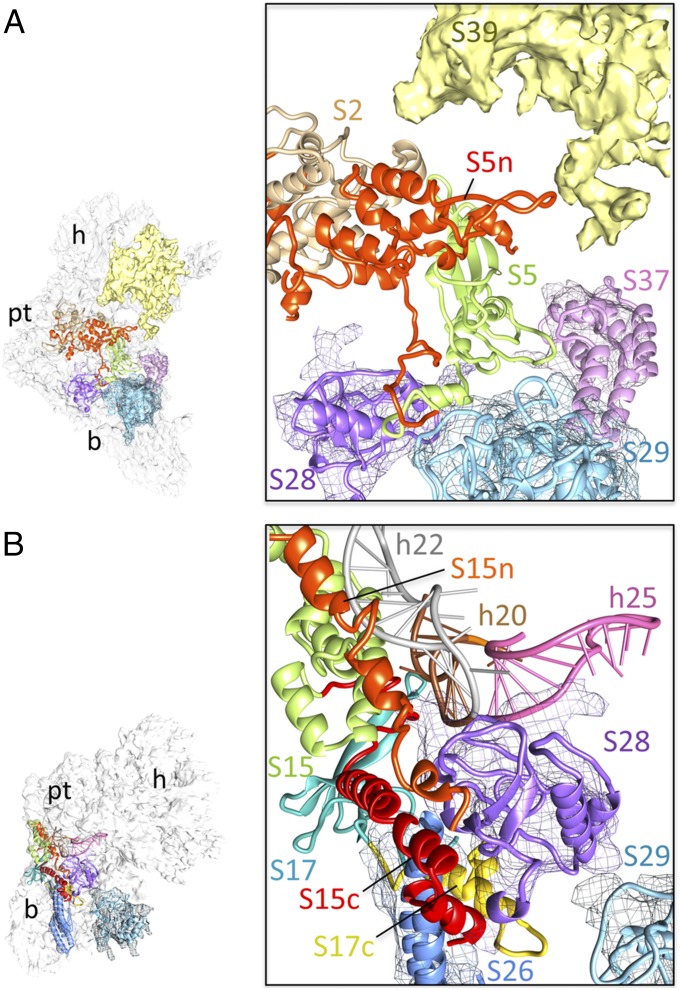
Among the conserved MRPs of the SSU body, S5 possesses large extensions at both its N and C termini (Table S1). Its NTE occupies a small portion of the structure previously described as the mRNA gate (4) and interacts with the largest of the mito-specific MRP densities, tentatively assigned to MRP S39 (or PTCD3) (12, 16), which emerges from the SSU head on the solvent side, and with the conserved domain of MRP S2 (Fig. 2A). On the solvent side, MRP S5 also interacts with densities assigned to several mito-specific MRPs, i.e., S28, S29, and S37 (Figs. 1D and 2A). Although the structure and position of MRP S12 on the mito-SSU body are highly conserved, the orientation of MRP S15, which is located at the junction of the SSU body and platform, is altered as compared with its orientation in the bacterial SSU. MRP S15 carries both an NTE and CTE that are oriented on the solvent side of the conserved domain of the protein (Fig. 2B). The CTE of MRP S15 interacts with the CTE of MRP S17, whereas both the CTE and NTE of MRP S15 and the CTE of MRP S17 interact with densities assigned to mito-specific MRPs S26 and S28. The CTEs of both MRPs S15 and S17 partially replace a small stretch of bacterial h21, which is absent in the mito-SSU (Fig. S2 A and E). Interestingly, although MRP S17 has lost almost all its rRNA-binding sites in the mito-rRNA because of deletion of the binding rRNA helices or conformational changes associated with those deletions (Fig. S6A), it retains its place in the mitoribosome. However, it is reoriented, and its interactions are redefined in the mito-SSU where it makes major interactions with partially truncated h11 (Fig. S6A).
Fig. 2.
Molecular interactions in the mito-SSU body. Environments of (A) NTE (red) of MRP S5 and (B) NTEs and CTEs of MRPs S15 (green) and S17 (sea green). Here and in subsequent figures, NTEs and CTEs of MRPs are indicated by the suffix “n” or “c”, respectively, in matching colors. “h” followed by a number indicates the 12 rRNA helix. As in Fig. 1 C and D, docked models of mito-specific MRPs are shown along with their cryo-EM densities (meshwork), and undocked densities are shown as solid masses in this and subsequent figures. Thumbnails to the left in this and subsequent figures show the overall orientations of the mito-SSU.
Except for h17 and h21, most of the bacterial rRNA helices that are known to interact with the Escherichia coli counterpart of S16 (ecS16; henceforth the prefix “ec” refers to the E. coli counterpart of an MRP) also are conserved in the mitoribosome (Fig. S2). MRP S16 has a 38-aa CTE that interacts with densities assigned to two mito-specific MRPs, S26 and S30 (Fig. 3A). A nonsense mutation in human MRP S16, from the Agr111 codon to a stop codon, leads to agenesis of the corpus callosum, dysmorphism, and fatal neonatal lactic acidosis (17). Because S16 is an assembly protein, the nonsense mutation would affect the assembly of the mito-SSU directly by disrupting interactions with mito-specific MRPs (Fig. 3B) and thereby impairing mitochondrial protein synthesis (18, 19).
Fig. 3.
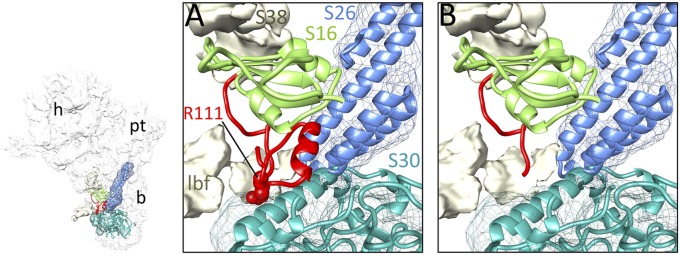
Depiction of possible effect of a nonsense mutation within the mito-specific CTE in MRP S16. (A) Close-up view of the S16 CTE region (red). (B) As in A, but the expected truncated part of the S16 CTE because the mutation of Arg111 (R111) to a stop codon has been removed computationally.
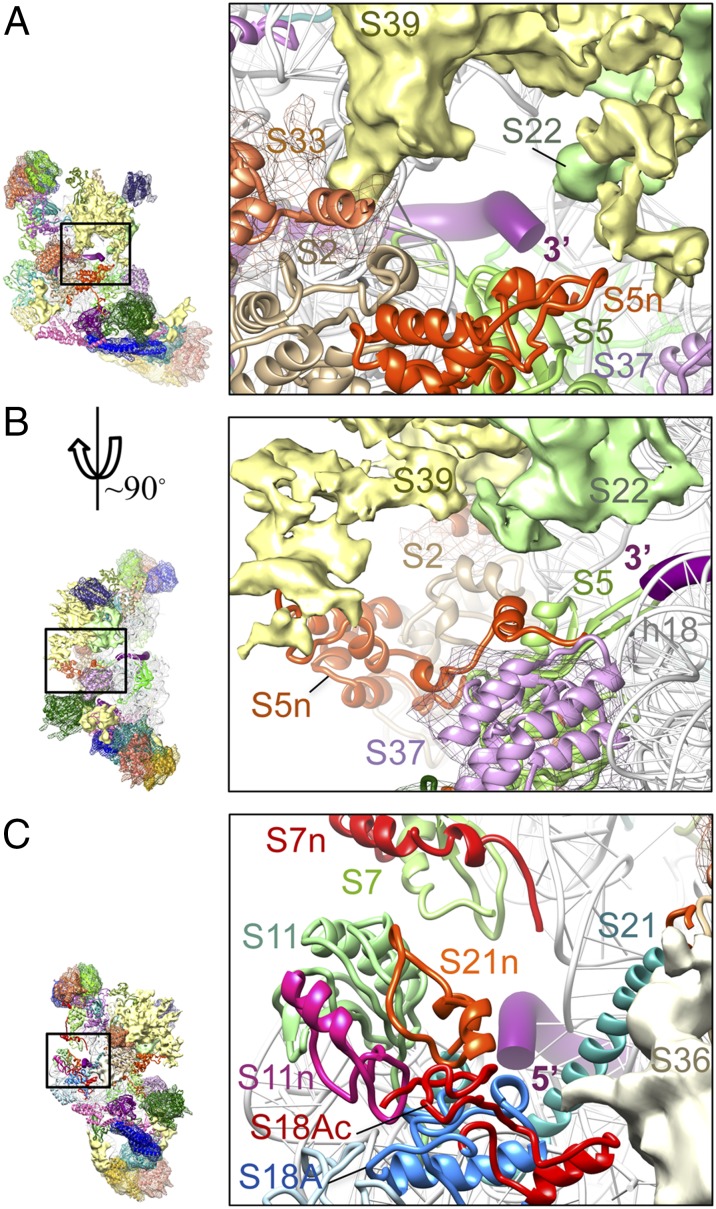
The mito-specific MRP S29 is a GTP-binding protein and has been identified as death-associated protein 3 (DAP3), which promotes apoptosis (20). The density region assigned to MRP S29 on the mito-SSU also was ascertained previously by immuno-EM (21). This mito-specific MRP interacts with MRPs S5 and with densities assigned to the mito-specific MRPs S28 and S37 (Figs. 1D and 2). The density assigned to MRP S37 is located close to the mRNA entry site on the solvent side of the SSU body, such that it interacts with h18 and MRP S5 (Figs. 1D and 2B) and partially occupies the positions of the bacterial S4 and h16 of the 16S rRNA (both absent in the mito-SSU) (Figs. 2B and 5B). The lowest portion of the SSU body carries a large mito-specific MRP mass of density (Fig. 1), previously termed the “lower body lobe” (4), which interacts with rRNA helices h15, h44, and the apical region of h6. The segmentation of this complex mass of density and motif matching suggest that this region contains three tightly interacting mito-specific MRPs, identified as S27, S30, and S31 (Fig. 1 C and D). The density corresponding to the lower body finger (4) appears to represent an unmodeled part of MRP S30 (Fig. 1C).
Fig. 5.
Molecular architectures of the mRNA entrance and exit regions and interactions in the mito-SSU platform. (A and B) The mRNA entrance is shown from two different orientations (from the solvent side in A, and from the shoulder side in B) to reveal better the features of the mRNA gate, which is composed primarily of the NTE of MRP S5 (S5n) and MRP S39. (C) The mRNA exit region. The 3′ and 5′ ends of the modeled mRNA (28) are shown as thick purple ribbons.
SSU Head.
Even though the 3′ major domain of the 12S rRNA (Fig. S2) provides its main scaffold, the head of the mito-SSU is dominated by some of the largest MRPs. MRP S2 is situated on the solvent side of the SSU and carries both NTE and CTE; it is located so that it connects components of all three major structural domains, i.e., the head, body, and platform of the SSU (Fig. 1D). ecS2 interacts with h35 and h40 of the bacterial rRNA (Fig. S6B). However, the helix-turn-helix motif that interacts with h40 is absent in MRP S2, as is h40 in the mito-12S rRNA (Figs. S2D and S6B), thus presenting an excellent example of the complementary loss of interacting RNA–protein partners in the mito-SSU during the course of evolution.
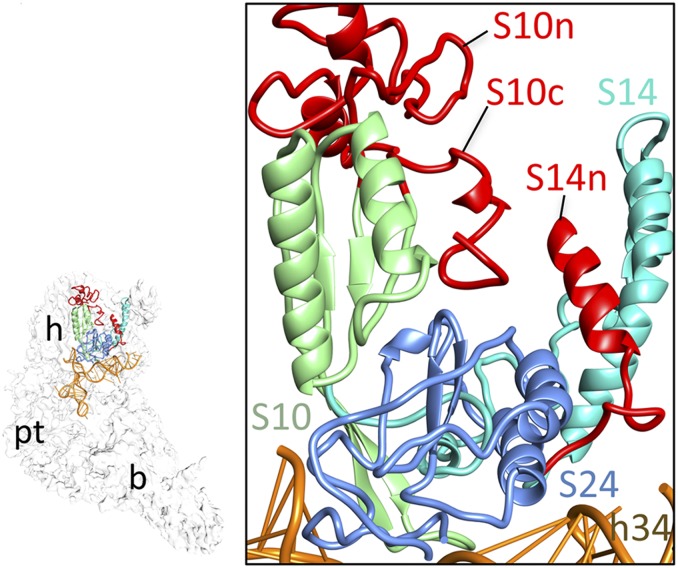
MRPs S10, S14, and S24 form a central cluster in the head of the mito-SSU (Fig. 4), above the mRNA entrance. MRP S24, as a distant ortholog of bacterial S3 (22), shows a structural similarity to the N-terminal domain of ecS3 and fits nicely into the corresponding density. Even though no MRP cryo-EM mass compensates for the lost C-terminal domain of ecS3, its interacting bacterial rRNA helices are conserved in the mito-SSU. MRP S24 is surrounded by MRPs S10, S14, and the density tentatively assigned to mito-specific MRP S39. Although the overall positions of MRPs S10, S14, and S24 are similar to those in the bacterial ribosome, the mito-specific NTE and CTE of MRP S10 and the NTE of MRP S14 are oriented on the solvent side. The CTE of MRP S10 and NTE of MRP S14 are oriented so that they both potentially might interact with MRP S24 (Fig. 4). The density assigned to MRP S39 spans the mito-SSU head, appearing on both the solvent and interface sides, covers MRP S10 and its mito-specific CTE on the solvent side (Fig. 1 C and D), and contributes in a major way to the formation of the mRNA gate that covers the entrance of the mRNA channel (4). MRP S39 is required for mitochondrial translation (16) and cross-links to mitochondrial translation initiation factor 3 (23), further supporting the strategic location of this largest of SSU MRPs close to the interface between the head and body regions.
Fig. 4.
Molecular interactions in the mito-SSU head involving MRPs S10, S14, and S24.
MRP S7 (Figs. 1 C and D and 5C) is located on the mRNA path of the SSU head, near what would be the bacterial tRNA-exit site (E site). MRP S7 has a 43-aa NTE and a 21-aa insertion in the middle (Table S1). Its NTE forms an α-helical structure that occupies a position similar to that of protein yS28 in the yeast 40S ribosome (Fig. S6C) (24), and its mito-specific insertion interacts mainly with the density assigned to mito-specific MRP S35, right above MRP S7. MRP S9 carries the longest mito-specific NTE of ∼200 aa residues (Table S1). Its C terminus, which spans the SSU’s mRNA decoding center (25), is highly conserved. The NTE covers most of the solvent side of the homologous domain of MRP S9 and would interact with the density assigned to mito-specific MRP S35 on the upper side and with h38 and h43 on the lower side. Additional interactions of mito-specific MRPs with mito-specific extensions of MRPs S7 and S9 are described in SI Results and Discussion.
SSU Platform.
The central domain of the 12S rRNA (Fig. S2) is highly conserved, except for h26 and h21 (Fig. S2E). H21 in the bacterial SSU originates from the platform and spans the solvent side of the SSU body. MRPs on the platform, i.e., S6, S11, S18, and S21, are also conserved (Fig. 1), with MRPs S11, S18, and S21 carrying mito-specific extensions (Table S1). Three isoforms of MRP S18 (S18A, S18B, and S18C), which vary in size (143–258 aa), are present in the mammalian mitoribosome (26, 27). Both the NTE (22 aa) and CTE (60 aa) of the S18A isoform could be modeled into the density in the immediate vicinity of the conserved region of S18. The CTE of MRP S18A is located on the solvent side near the conventional mRNA-exit region (Fig. 5C) (28), and its NTE interacts primarily with h20 and h24. These observations suggest that the bovine mito-SSU under study here contained primarily the S18A isoform. MRP S11, also situated near the mRNA-exit region, carries an NTE facing the solvent side of the SSU that interacts with the CTE of MRP S18A and the NTE of MRP S21 (Fig. 5C). Bacterial S21 is known to be involved in the initiation of protein synthesis (29). The C terminus of S21 is disordered in the crystallographic structure of the E. coli ribosome (30), and S21 is absent altogether in the Thermus thermophilus ribosome (25). We have modeled the full-length MRP S21 into the cryo-EM density, which supports an α-helical structure for its C terminus (Fig. S1C). This region of MRP S21 interacts with MRP S2 and with a density that has been tentatively assigned to MRP S36 on the solvent side (Figs. 1D and 5C). The NTE of MRP S21 makes significant contacts with both conserved and mito-specific extensions of MRPs S11 and S18A in the cleft region of the platform (Fig. 5C).
Topology of the mRNA Path.
One of the unique features of the mammalian mitochondrial translation is that only three of its 11 cistrons possess 5′ UTRs, and these are only a few nucleotides in length. The remaining eight mRNAs do not have a 5′ UTR and are basically leaderless (13). In the cryo-EM structure of the mito-SSU, the mRNA entrance has a unique gate-like feature, which is composed of mito-specific MRPs (4) (“mgt” in Figs. 1B and 5 A and B) that may be involved in the recruitment of the leaderless mito-mRNAs during translation initiation. In the bacterial ribosome, proteins S3, S4, and S5 occupy the mRNA entrance (28). Of these, the proteins homologous to S4 and the C-terminal domain of S3 are absent in the mitoribosome. However, only ecS4 is replaced by a mito-specific MRP. Thus, the molecular landscape of the mRNA entrance in the mito-SSU is significantly remodeled. It is composed of MRP S5 and four other mito-specific MRPs (Fig. 5). Densities assigned to MRPs S33 and S37 also encircle the mRNA entrance from the solvent side, whereas the density tentatively assigned to MRP S22 joins the mRNA entrance from the interface side of the mito-SSU head. The NTE of MRP S5 interacts with the density assigned to mito-specific MRP S39, which apparently constitutes the majority of the mRNA gate feature emerging from the SSU head (Figs. 2A and 5 A and B).
At the mRNA-exit region, where the 5′ side of mRNA would interact with the mito-SSU, all the rRNA helices are conserved except for the anti–Shine–Dalgarno sequence, which is absent (Fig. S2F). As mentioned above, homologs of all three mRNA-exit region proteins— S11, S18, and S21—are present in the mito-SSU, and each of these possesses mito-specific NTEs and CTEs that have significantly altered the molecular landscape of that region. Intriguingly, the CTE of the S18A would be close to the 5′ end of the mRNA (Fig. 5C). Because the CTEs of the three S18 isoforms vary significantly in size (18–89 aa), these isoforms may constitute specialized mito-SSUs for the recruitment and translation of three mito-mRNAs that carry 5′-UTR segments or for the overlapping ORFs that are present in the two dicistronic mRNAs (13).
Interactions with the 39S LSU.
To understand the interaction between the two mitoribosomal subunits, we docked the components of the previously published mito-LSU models (6, 11) into the LSU portion of the 55S mitoribosome map (Fig. 6A), as described in SI Materials and Methods. We identify 14 contact points between the subunits (Fig. 6 B and C and Table S4). Locations of bridges B2a, B2b, B2c, B3, B5, and B7a are conserved between the bacterial and mitochondrial ribosomes (31); however, the interacting components of some of these bridges appear to be slightly altered (Table S4). Moreover, retention of several RNA–RNA bridges suggests strong conservation of the functional core region of the mitoribosome. An unassigned MRP density, present immediately next to the 12S rRNA helix 42 and apparently corresponds to the unmodeled C-terminal region of MRP S25 but readily segments with the mito-LSU, is involved in formation of two bridges, B1a and B1b (4). Thus, five bridges, B1a–d and B9, are formed exclusively by the mito-specific MRPs from both subunits. In summary, there are seven RNA–RNA bridges, six protein–protein bridges, and one RNA–protein bridge (Table S4). Clearly, as we previously suggested (4), there is significantly greater participation by MRPs in intersubunit communication in the mitoribosome than in the bacterial ribosome, which contains only one protein–protein bridge (31). Some of these MRPs compensate for the absence of functionally relevant bridges in the bacterial ribosome involving rRNA segments (e.g., bacterial bridge B1a involving 23S rRNA helix 38, which is absent in mito-16S rRNA and is compensated by a mito-specific MRP of the mito-LSU). The absence of the previously identified bridge B7b (4) in the present map could be attributed to dynamic nature of that bridge. Interestingly, none of the mito-specific extensions in homologous MRPs participate in the intersubunit interaction.
Fig. 6.
Location and composition of the intersubunit bridges between the two mitoribosomal subunits. (A) Structure of the 55S mitoribosome, with docked models of mitochondrial SSU (Left) and LSU (Right). The cryo-EM density is shown as semitransparent gray meshwork. Head (h) and body (b) of the mito-SSU and the central protuberance (CP) and L11 Stalk base region (Sb) of the mito-LSU are labeled. (B and C) SSU (B) and LSU (C) are shown from their interface sides to reveal intersubunit bridges. Bridges involving conserved RNA–RNA segments (purple), conserved protein–protein (blue), RNA–protein (red), and between two mito-specific MRPs (black) are highlighted. The two asterisks indicate the locations of bridges B1a and B1b, which most likely involve unmodeled segments of the same mito-specific MRP of the SSU that forms bridges B1c and B1d. rRNAs of the SSU and LSU are shown in orange and pink, respectively, in A; in B and C they are shown in gray to enhance bridges. Homologous MRPs of both subunits are shown in different shades of green. Mito-specific MRPs of the SSU and LSU are shown in yellow and blue, respectively.
In addition to illustrating the molecular topography of the significantly altered mRNA-binding path, the mito-SSU structure presented here hints at a stepwise molecular evolution of the mitoribosomal architecture starting with a conserved 12S rRNA segment and 15 homologous MRPs. In subsequent steps, NTE(s) and/or CTE(s) are added to these homologous MRPs to aid the specific functional needs of the mitoribosome, such as the recruitment of leaderless mRNAs. They also apparently fill in for some of the lost peripheral bacterial rRNA segments and partially replace missing bacterial proteins, because many of the extensions are present on the solvent side of the structure. A subsequent recruitment of mito-specific MRPs, which interact primarily with mito-specific extensions of homologous MRPs, apparently was needed to stabilize the overall architecture further. As we described earlier, these mito-specific MRPs also contribute to the formation of the mRNA entrance gate and intersubunit bridges and appear to provide an additional layer protecting the mitoribosome structure from the harsh chemical environment of the mitochondrial matrix.
Materials and Methods
Isolation and Purification of the Mitoribosome.
The 55S mitoribosome and its subunits were isolated and purified from the Bos taurus liver by following the previously reported method (32) (see SI Materials and Methods for details). For the cryo-EM studies, the 55S mitoribosome was obtained by reassociating the purified 28S and 39S subunits. The two subunits were incubated in buffer containing 20 mM Hepes-KOH (pH 7.6), 20 mM MgCl2, 40 mM KCl, and 20 mM DTT at 37 °C for 30 min. The incubation mixture then was subjected to 10–30% sucrose gradient centrifugation under the same buffer conditions at 22,000 rpm using a Beckman SW32 rotor for 16 h. The pooled reassociated 55S mitoribosome fractions were pelleted by ultracentrifugation at 42,000 rpm for 6 h in a Beckman type Ti70 rotor. The pellet was suspended in the same buffer and stored in small aliquots at −80 °C.
Cryo-EM, Image Processing, and 3D Reconstruction.
Cryo-EM grids were prepared according to standard procedures (33), by applying 4 μL of the 55S mitoribosome suspension (32 nM) onto the Quantifoil Holey carbon copper grid using Vitrobot (FEI). Data were collected on a 300-kV JEOL 3200 field emission gun electron microscope with a magnification of 59,717×. Then 832 micrographs were scanned on a Zeiss flatbed scanner (Z/I Imaging) with a step size of 7 µm, corresponding to 1.17 Å on the object scale and were sorted into 48 defocus groups. A total of 866,553 particle images were picked originally and were subjected to supervised classification (34) using our previous 55S cryo-EM map (4) and the computationally separated 39S (LSU) portion of the same map as two references and SPIDER (35). Of these images, 428,129 classified with the 55S reference (Fig. S7A). Relion-based unsupervised classification (36) estimated a conformational heterogeneity within the images classified with the 55S reference (Fig. S7B). Based on statistical information derived from the Relion classification, we removed ∼28% of the particles with low cross-correlation coefficient values with the 55S reference projections. The remaining 307,556 particle images were included in the final reconstruction. Projection-matching in conjunction with small-angle alignment (up to 0.2° angular spacing) was used for the iterative refinements using SPIDER (35). The resolution of the final contrast transfer function-corrected 3D map, estimated using the Fourier shell correlation with a cutoff value of 0.5, was 7.0 Å (Fig. S1).
Modeling of rRNA.
To model the 12S rRNA, we primarily used the structural constraints of the cryo-EM density obtained by applying a higher density threshold to the entire mito-SSU map (Fig. S2B). We used the template 16S rRNA structures (25, 30) to build the structurally homologous 12S rRNA segments as described previously (37) and a knowledge-based approach to build the single-stranded nonhomologous loop regions (see SI Materials and Methods for details). Briefly, the database of known RNA structures was searched to model the loop that was most consistent with the surrounding RNA and with the cryo-EM density corresponding to the rRNA (Fig. S2). To investigate the interaction between the loop and its surroundings, van der Waals energies (Evdw) were calculated using the standard Lennard–Jones 6–12 potential and parameters of the AMBER force field (38). The candidate loop structure that satisfies the end constraints (i.e., a small rmsd) with the minimum Evdw and the structural constraints of the cryo-EM density was selected. The final secondary structure (Fig. S2A) generally matched with the predicted secondary structure of the 12S rRNA (39), except for helices 7 and 22 and penultimate helix 44.
Modeling of MRPs.
MRPs were modeled into the cryo-EM density in multiple steps as depicted in flow diagram (Fig. S3). First, we generated the iterative threading assembly refinement (I-TASSER) (40) models for all 31 SSU MRPs. Picking the best models for the conserved regions of the homologs of the 15 bacterial MRPs and their modeling into the cryo-EM density was relatively straightforward and was performed using Situs (41) and Chimera (42) (see SI Materials and Methods for details). The extensions and insertions of homologous MRPs were modeled into densities present in the immediate vicinity of their conserved domains using Coot (43) and Chimera. Fitted 12S rRNA and homologous MRP models were combined and subjected to molecular dynamics flexible fitting (MDFF) (44).
Segmentation and Placement of Mito-Specific MRPs.
Densities for 16 mito-specific MRPs are distributed mostly at the map’s peripheral regions, where the resolution of the map is somewhat lower (7–9 Å) than within the core of the molecule. Therefore, to localize these MRPs, we took the following approach (Fig. S3; also see SI Materials and Methods for details). We separated the remaining unmodeled cryo-EM density as an ensemble mass and subjected it to Segger-based segmentation (14 and Fig. S4). To validate the segmentation results for the mito-specific MRPs, we also applied Segger tools to the complete protein density, including the homologous MRPs. Results were highly reproducible and consistent for the homologous MRPs (Fig. S4 C and F). Next, we determined the voxel volume and surface area for each of the segmented densities and initially assigned the density to a mito-specific MRP based on its relative correlation with MRP’s molecular mass (Table S2 and Fig. S4G). The final assignments were made after comparing the shape of the each segmented density with that of an I-TASSER homology model (Fig. S5). Independent assignment of the density for MRP S29 using this method also matched the immuno-EM localization of this protein on the mito-SSU (21). After the docking, we individually applied MDFF (44) for 100 ps for each mito-specific MRP; if there was an indication of melting of secondary structures, that model was not taken into account.
Supplementary Material
Acknowledgments
We thank the staff of the New York Structural Biology Center for help with cryo-EM data collection on their 300 kV FEG JEOL electron microscope; Kae Yokoyama for help with manual screening of particle images; Nilesh Banavali for helpful discussions on MRP modeling; and Wadsworth Center computing facilities for all image processing and MDFF simulations. This work was supported by National Institutes of Health Grants R01 GM061576 (to R.K.A.) and R01 GM072686 (to K.Y.S.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The cryo-EM map of the 28S subunit of the mammalian mitochondrial ribosome has been deposited in the EM Database, http://emdep.rutgers.edu (accession no. EMD-5941). The modeled and fitted coordinates of the 12 S rRNA and 15 homologous MRPs with their mito-specific insertions/extensions have been deposited in the Protein Data Bank, www.rcsb.org (accession no. 3J6V).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1401657111/-/DCSupplemental.
References
- 1.O’Brien TW, O’Brien BJ, Norman RA. Nuclear MRP genes and mitochondrial disease. Gene. 2005;354:147–151. doi: 10.1016/j.gene.2005.03.026. [DOI] [PubMed] [Google Scholar]
- 2.Pearce S, Nezich CL, Spinazzola A. Mitochondrial diseases: Translation matters. Mol Cell Neurosci. 2013;55:1–12. doi: 10.1016/j.mcn.2012.08.013. [DOI] [PubMed] [Google Scholar]
- 3.Watanabe K. Unique features of animal mitochondrial translation systems. The non-universal genetic code, unusual features of the translational apparatus and their relevance to human mitochondrial diseases. Proc Jpn Acad, Ser B, Phys Biol Sci. 2010;86(1):11–39. doi: 10.2183/pjab.86.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sharma MR, et al. Structure of the mammalian mitochondrial ribosome reveals an expanded functional role for its component proteins. Cell. 2003;115(1):97–108. doi: 10.1016/s0092-8674(03)00762-1. [DOI] [PubMed] [Google Scholar]
- 5.Sharma MR, Booth TM, Simpson L, Maslov DA, Agrawal RK. Structure of a mitochondrial ribosome with minimal RNA. Proc Natl Acad Sci USA. 2009;106(24):9637–9642. doi: 10.1073/pnas.0901631106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mears JA, et al. A structural model for the large subunit of the mammalian mitochondrial ribosome. J Mol Biol. 2006;358(1):193–212. doi: 10.1016/j.jmb.2006.01.094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gray MW, Burger G, Lang BF. The origin and early evolution of mitochondria. Genome Biol. 2001;2(6):Reviews1018.1–1018.5. doi: 10.1186/gb-2001-2-6-reviews1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Agrawal RK, Sharma MR, Yassin AS, Lahiri I, Spremulli L. In: Structure and Function of Organellar Ribosomes as Revealed by Cryo-EM. Ribosomes: Structure, Function, and Dynamics. Rodnina MV, Wintermeyer W, Green R, editors. New York: Springer; 2011. pp. 83–96. [Google Scholar]
- 9.Agrawal RK, Sharma MR. Structural aspects of mitochondrial translational apparatus. Curr Opin Struct Biol. 2012;22(6):797–803. doi: 10.1016/j.sbi.2012.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sharma MR, Kaushal PS, Gupta M, Banavali NK, Agrawal RK. Insights into Structural Basis of Mammalian Mitochondrial Translation. In: Duchêne A-M, editor. Translation in Mitochondria and Other Organelles. Berlin: Springer; 2013. pp. 1–28. [Google Scholar]
- 11.Greber BJ, et al. Architecture of the large subunit of the mammalian mitochondrial ribosome. Nature. 2014;505(7484):515–519. doi: 10.1038/nature12890. [DOI] [PubMed] [Google Scholar]
- 12.Koc EC, et al. Identification and characterization of CHCHD1, AURKAIP1, and CRIF1 as new members of the mammalian mitochondrial ribosome. Front Physiol. 2013;4:183. doi: 10.3389/fphys.2013.00183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Temperley RJ, Wydro M, Lightowlers RN, Chrzanowska-Lightowlers ZM. Human mitochondrial mRNAs—like members of all families, similar but different. Biochim Biophys Acta. 2010;1797(6-7):1081–1085. doi: 10.1016/j.bbabio.2010.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pintilie GD, Zhang J, Goddard TD, Chiu W, Gossard DC. Quantitative analysis of cryo-EM density map segmentation by watershed and scale-space filtering, and fitting of structures by alignment to regions. J Struct Biol. 2010;170(3):427–438. doi: 10.1016/j.jsb.2010.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rötig A. Human diseases with impaired mitochondrial protein synthesis. Biochim Biophys Acta. 2011;1807(9):1198–1205. doi: 10.1016/j.bbabio.2011.06.010. [DOI] [PubMed] [Google Scholar]
- 16.Davies SM, et al. Pentatricopeptide repeat domain protein 3 associates with the mitochondrial small ribosomal subunit and regulates translation. FEBS Lett. 2009;583(12):1853–1858. doi: 10.1016/j.febslet.2009.04.048. [DOI] [PubMed] [Google Scholar]
- 17.Miller C, et al. Defective mitochondrial translation caused by a ribosomal protein (MRPS16) mutation. Ann Neurol. 2004;56(5):734–738. doi: 10.1002/ana.20282. [DOI] [PubMed] [Google Scholar]
- 18.Emdadul Haque M, Grasso D, Miller C, Spremulli LL, Saada A. The effect of mutated mitochondrial ribosomal proteins S16 and S22 on the assembly of the small and large ribosomal subunits in human mitochondria. Mitochondrion. 2008;8(3):254–261. doi: 10.1016/j.mito.2008.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Koc EC, Haque ME, Spremulli LL. Current views of the structure of the mammalian mitochondrial ribosome. Isr J Chem. 2010;50:45–59. [Google Scholar]
- 20.Cavdar Koc E, et al. A new face on apoptosis: Death-associated protein 3 and PDCD9 are mitochondrial ribosomal proteins. FEBS Lett. 2001;492(1-2):166–170. doi: 10.1016/s0014-5793(01)02250-5. [DOI] [PubMed] [Google Scholar]
- 21.Sharma MR, et al. Three-dimensional localization of the GTP-binding protein within the mammalian mitochondrial ribosome. Mitochondrion. 2011;11(4):657. [Google Scholar]
- 22.Smits P, Smeitink JA, van den Heuvel LP, Huynen MA, Ettema TJ. Reconstructing the evolution of the mitochondrial ribosomal proteome. Nucleic Acids Res. 2007;35(14):4686–4703. doi: 10.1093/nar/gkm441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Haque ME, Koc H, Cimen H, Koc EC, Spremulli LL. Contacts between mammalian mitochondrial translational initiation factor 3 and ribosomal proteins in the small subunit. Biochim Biophys Acta. 2011;1814(12):1779–1784. doi: 10.1016/j.bbapap.2011.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ben-Shem A, et al. The structure of the eukaryotic ribosome at 3.0 Å resolution. Science. 2011;334(6062):1524–1529. doi: 10.1126/science.1212642. [DOI] [PubMed] [Google Scholar]
- 25.Wimberly BT, et al. Structure of the 30S ribosomal subunit. Nature. 2000;407(6802):327–339. doi: 10.1038/35030006. [DOI] [PubMed] [Google Scholar]
- 26.Cavdar Koc E, Burkhart W, Blackburn K, Moseley A, Spremulli LL. The small subunit of the mammalian mitochondrial ribosome. Identification of the full complement of ribosomal proteins present. J Biol Chem. 2001;276(22):19363–19374. doi: 10.1074/jbc.M100727200. [DOI] [PubMed] [Google Scholar]
- 27.Suzuki T, et al. Proteomic analysis of the mammalian mitochondrial ribosome. Identification of protein components in the 28 S small subunit. J Biol Chem. 2001;276(35):33181–33195. doi: 10.1074/jbc.M103236200. [DOI] [PubMed] [Google Scholar]
- 28.Yusupova GZ, Yusupov MM, Cate JH, Noller HF. The path of messenger RNA through the ribosome. Cell. 2001;106(2):233–241. doi: 10.1016/s0092-8674(01)00435-4. [DOI] [PubMed] [Google Scholar]
- 29.Van Duin J, Wijnands R. The function of ribosomal protein S21 in protein synthesis. Eur J Biochem. 1981;118(3):615–619. doi: 10.1111/j.1432-1033.1981.tb05563.x. [DOI] [PubMed] [Google Scholar]
- 30.Schuwirth BS, et al. Structures of the bacterial ribosome at 3.5 A resolution. Science. 2005;310(5749):827–834. doi: 10.1126/science.1117230. [DOI] [PubMed] [Google Scholar]
- 31.Yusupov MM, et al. Crystal structure of the ribosome at 5.5 A resolution. Science. 2001;292(5518):883–896. doi: 10.1126/science.1060089. [DOI] [PubMed] [Google Scholar]
- 32.Spremulli LL. Large-scale isolation of mitochondrial ribosomes from mammalian tissues. Methods Mol Biol. 2007;372:265–275. doi: 10.1007/978-1-59745-365-3_19. [DOI] [PubMed] [Google Scholar]
- 33.Grassucci RA, Taylor DJ, Frank J. Preparation of macromolecular complexes for cryo-electron microscopy. Nat Protoc. 2007;2(12):3239–3246. doi: 10.1038/nprot.2007.452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Valle M, et al. Cryo-EM reveals an active role for aminoacyl-tRNA in the accommodation process. EMBO J. 2002;21(13):3557–3567. doi: 10.1093/emboj/cdf326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shaikh TR, et al. SPIDER image processing for single-particle reconstruction of biological macromolecules from electron micrographs. Nat Protoc. 2008;3(12):1941–1974. doi: 10.1038/nprot.2008.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Scheres SH. RELION: Implementation of a Bayesian approach to cryo-EM structure determination. J Struct Biol. 2012;180(3):519–530. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tung CS, Joseph S, Sanbonmatsu KY. All-atom homology model of the Escherichia coli 30S ribosomal subunit. Nat Struct Biol. 2002;9(10):750–755. doi: 10.1038/nsb841. [DOI] [PubMed] [Google Scholar]
- 38.Weiner SJ, et al. A new force field for molecular mechanical simulation of nucleic acids and proteins. J Am Chem Soc. 1984;106(3):765–784. [Google Scholar]
- 39.Cannone JJ, et al. The comparative RNA web (CRW) site: An online database of comparative sequence and structure information for ribosomal, intron, and other RNAs. BMC Bioinformatics. 2002;3:2. doi: 10.1186/1471-2105-3-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Roy A, Kucukural A, Zhang Y. I-TASSER: A unified platform for automated protein structure and function prediction. Nat Protoc. 2010;5(4):725–738. doi: 10.1038/nprot.2010.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wriggers W. Using Situs for the integration of multi-resolution structures. Biophys Rev. 2010;2(1):21–27. doi: 10.1007/s12551-009-0026-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pettersen EF, et al. UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem. 2004;25(13):1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 43.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 4):486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Trabuco LG, Villa E, Schreiner E, Harrison CB, Schulten K. Molecular dynamics flexible fitting: A practical guide to combine cryo-electron microscopy and X-ray crystallography. Methods. 2009;49(2):174–180. doi: 10.1016/j.ymeth.2009.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.