Significance
Viral infection and integration is fundamental in the generation of genetic diversity in prokaryotes. After entering an Escherichia coli cell, bacteriophage λ DNA must locate a unique site among ∼5 million possible sites on the bacterial genome, with high efficiency and within physiological times, to integrate and establish lysogeny. How does it do it? We show that λ DNA does not carry out an active search. Instead, it remains confined at its entry point where it undergoes limited diffusion, while the process of bacterial DNA replication conveys the bacterial site close to the λ DNA. This mechanism adds to the list of profligate use of host functions by λ, brought about by coevolution of host−phage processes.
Keywords: target location, establishment of lysogeny, viral transduction
Abstract
The search for specific sequences on long genomes is a key process in many biological contexts. How can specific target sequences be located with high efficiency, within physiologically relevant times? We addressed this question for viral integration, a fundamental mechanism of horizontal gene transfer driving prokaryotic evolution, using the infection of Escherichia coli bacteria with bacteriophage λ and following the establishment of a lysogenic state. Following the targeting process in individual live E. coli cells in real time revealed that λ DNA remains confined near the entry point of a cell following infection. The encounter between the 15-bp-long target sequence on the chromosome and the recombination site on the viral genome is facilitated by the directed motion of bacterial DNA generated during chromosome replication, in conjunction with constrained diffusion of phage DNA. Moving the native bacterial integration site to different locations on the genome and measuring the integration frequency in these strains reveals that the frequencies of the native site and a site symmetric to it relative to the origin are similar, whereas both are significantly higher than when the integration site is moved near the terminus, consistent with the replication-driven mechanism we propose. This novel search mechanism is yet another example of the exquisite coevolution of λ with its host.
The search for specific sequences along genomic DNA plays a key role in the location of specific sites by transcription factors (1), the repair of DNA lesions (2), and horizontal gene transfer (3). Common to these processes is a search through a very large number of possible sequences because of the long genomes involved. A fundamental question is how specific target sequences can be located with high efficiency, within physiologically relevant times. This question is crucial to understand viral transduction, one of the fundamental mechanisms of horizontal gene transfer driving the evolution of prokaryotes (3, 4). In transduction, a viral genome integrates at a unique site on a bacterial genome following infection, conferring new traits such as pathogenicity (5). A classic example of transduction is furnished by the infection of Escherichia coli cells by bacteriophage λ.
Infection of an E. coli host by the temperate bacteriophage λ begins with the binding of the phage to the E. coli maltose pore LamB (6, 7). The phage then injects its DNA into the cell, a process that lasts about 5 min (8). Infection can lead to two possible outcomes, lysis or lysogeny, which reflect alternative pathways of gene expression (9–11). In the lytic pathway, execution of a viral gene expression cascade leads to the replication of the viral DNA, resulting in cell death and lysis to release about 100 phage progeny (12). Alternatively, by establishing lysogeny, the phage shuts off the lytic cycle and locates with high efficiency (13) a unique sequence along the cellular genome where it integrates its DNA by site-specific recombination. This recombination takes place at special attachment sites called attB and attP in the bacterial and phage genomes, respectively, and requires both the phage Int and the bacterial integration host factor (IHF) proteins. Once integrated, the prophage remains in a stable, dormant state, replicating passively with the host genome.
In this study, we followed in real time the search and eventual encounter between the attP site on single λ DNA molecules and the attB integration site on the genome of individual, live E. coli bacterial cells, immediately following phage infection. The results shed light on the mechanisms of search and how the encounter is achieved with high efficiency to establish integration and stable lysogeny.
Results
The attB Site Moves Toward λ DNA to Establish Lysogeny.
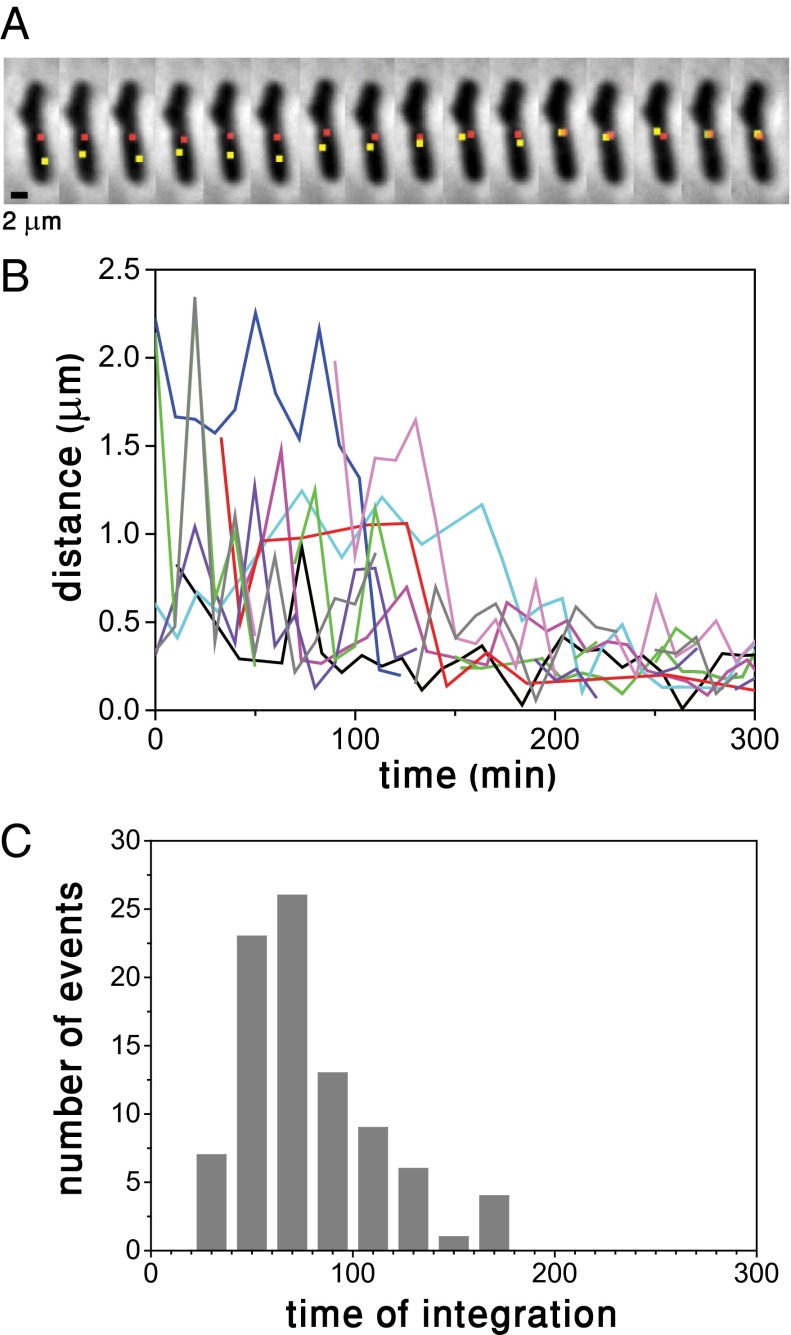
We followed the dynamics of search by labeling the bacterial and phage genomes with yellow (yGFP) and red (RFP) fluorescent protein markers, respectively, near their respective att sites, using two types of parS/ParB, which are part of bacterial partitioning systems (14–16). One type of parS sequence (P1 parS) was inserted near attP on bacteriophage λ, while another type of parS sequence (pMT1 parS) was inserted near the attB site in the bacterial genome (Materials and Methods). The two types of parS sequences are recognized by their respective ParB proteins, mCherry-P1Δ30ParB labeling the phage DNA and yGFP-pMT1Δ23ParB labeling the attB locus (Fig. S1). Control experiments measuring the integration frequency show that labeling and ParB polymerization do not affect the process under study (17) (Fig. S2). Upon establishment of lysogeny, the distance between both parS sequences is ∼12 kbp. This distance was chosen to ensure that ParB polymerization from parS sites does not interfere with phage DNA integration (17). Note that due to genome compaction, the physical separation between the two ParB foci is orders of magnitude below the optical resolution limit. We followed the fate of 241 infected cells, 36% of which followed the lysogenic pathway while the rest, 64%, followed the lytic pathway. Typical snapshots of a bacterial cell following infection are shown in Fig. 1A. The snapshots depict a single λ DNA molecule soon after its entry into the cell and subsequent RFP labeling, until successful integration at the attB site is achieved, as evidenced by the coalescence of the red and yellow loci representing the attP and attB sites. In stark contrast with our initial expectation of observing λ DNA motion as part of the search process for the integration site, the snapshots clearly show that λ DNA barely moves over the time it takes for integration to occur and that the attB site is the one to move toward a cell pole. Consistent with previous findings (18), we observe λ DNA soon after infection to be located primarily at cell poles or at midcell (65% and 35%, respectively, of 400 events total; the distribution of λ DNA along the cell axis is discussed further below). When the lytic pathway is followed, replication of λ DNA also takes place primarily at the cell poles (18) (Fig. S3). Fig. 1A also illustrates that in addition to normal cell division (Fig. S4), infection can result in growth arrest, while, in other cases, it can cause cell filamentation (47% of lysogens exhibited growth arrest, 41% normal growth and 12% filamentation). Uninfected cells continue to grow and divide normally during the experiment. Expression of the λ phage is known to alter cell physiology by inducing temporary blocks to both cell division and initiation of new rounds of replication (19). However, such effects on each cell may be dependent upon when during the cell cycle infection occurs, or other variables of infection, e.g., actual multiplicity of infection (MOI).
Fig. 1.
Search and encounter between attachment sites on the bacterial and phage genomes in the establishment of lysogeny. (A) Snapshots taken every 10 min after infection show the ParB-RFP locus (red) marking the attP site and the ParB-yGFP locus (yellow) marking the attB site on the phage DNA. ParB-RFP and the ParB-yGFP loci are separated by ∼12 kbp when lysogeny is established. (B) Distance on the focal plane between the attachment sites on the bacterial and phage DNAs as a function of time. Multiple events are shown, with each having a different color. (C) Histogram of integration times of λ phage DNA into the bacterial genome. The integration time was defined as the time the attB and attP loci established a time-independent distance of ∼0.2 μm (Fig. S5).
We show, in Fig. 1B, the distance on the focal plane between RFP and yGFP loci labeling attP and attB sites, respectively, in the same cells, for a number of events such as those illustrated in Fig. 1A. Each trace corresponds to an attP-attB trajectory in a single cell. Integration, indicated by the attainment of a time-independent minimal distance of ∼0.2 μm (Fig. S5), is typically established at times of 77 ± 34 min (mean ± std). A histogram of integration times is depicted in Fig. 1C. Independent measurements of integration in the bulk using PCR methods, which are sensitive and report the earliest integration events, yield a similar although shorter integration time (∼45 min) (Fig. S6), consistent with the large amount of events observed at 45 min in Fig. 1C. Note that 60 min is the cell doubling time of uninfected cells under the conditions of the present experiments. The typical behavior observed in Fig. 1B is not observed after the infection of an immune cell (Fig. S7).
λ DNA Undergoes Constrained Diffusion.
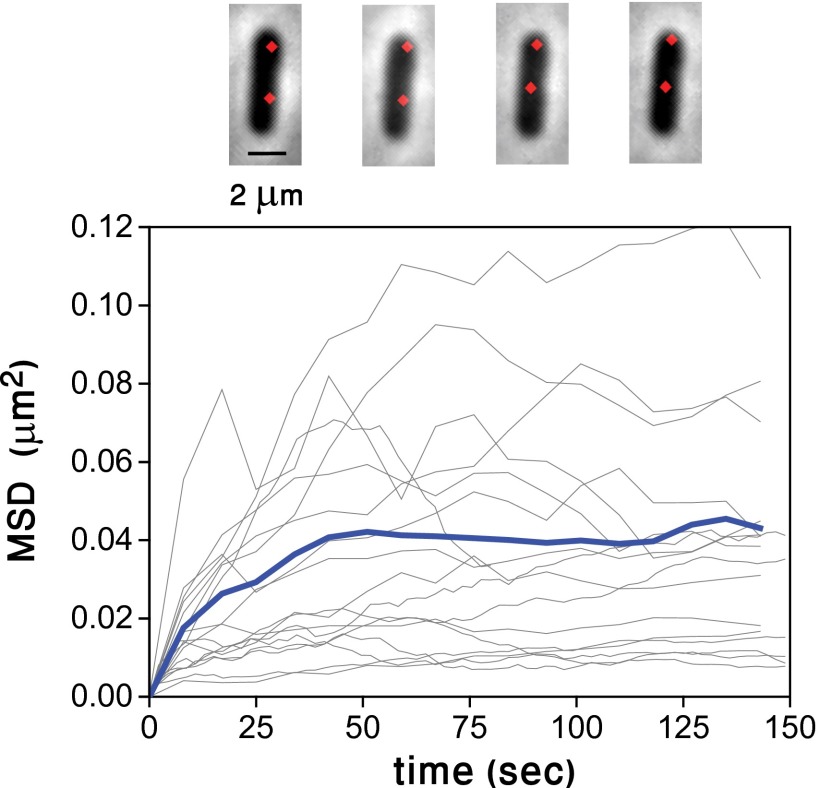
To quantify more precisely the extent to which λ DNA moves within a bacterial cell, we calculated the mean square displacement (MSD) between two individual λ DNAs within the same cell, following known procedures (20). This allows us to characterize the displacement of the molecule independently from any cellular marker such as a cell pole, which may exhibit motion due to cell growth. The MSD is a measure of the mean square change in the distance between points, and it is the canonical way to characterize quantitatively Brownian motion (20). We show the mean square displacement as a function of time for λ DNA pairs observed in different cells, as well as the average over all traces in Fig. 2. The average behavior is characteristic of constrained diffusion, with saturation reached around ∼50 s. At saturation, the mean square displacement corresponds to roughly ∼120 nm (SI Materials and Methods).
Fig. 2.
Constrained diffusion of λ DNA. Mean square displacement (MSD) between pairs of λ DNAs in doubly infected E. coli cells as a function of time. Each gray trace represents the mean square distance between a pair of λ DNAs in the same cell. The average of all traces (blue) shows constrained diffusion with a confinement radius of ∼120 nm. The snapshots, taken at 20 min intervals during one run, illustrate the small motion of both λ spots.
The Dynamical Behavior of the attB Site Is Not Affected by Infection.
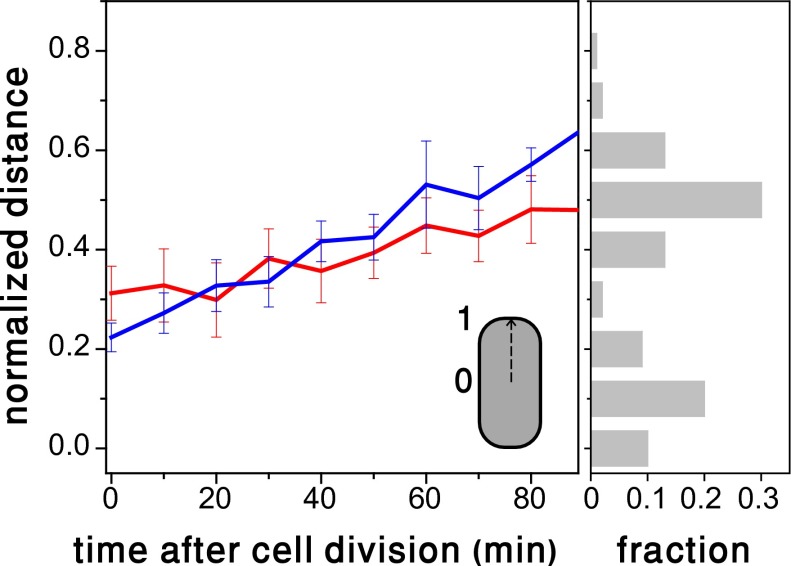
Given that λ DNA remains confined to the point of entry while the attB site moves (e.g., Fig. 1A), we asked whether this motion is driven by a phage factor to facilitate lysogeny, or by normal DNA replication. To address this, we compared the dynamical behavior of the attB site in uninfected cells with that of infected cells that eventually become lysogens. The distance between the attB marker and the cell’s center as a function of the time elapsed after the last cell division is plotted in Fig. 3. The averaged traces for infected and uninfected cells are statistically similar, and therefore we conclude that the motion of the attB site is driven by E. coli DNA segregation as it is being replicated. The histogram of λ DNAs locations along the cell’s axis on the right shows that the attB site is pushed toward regions enriched with λ DNA. Although infection may block initiation of DNA replication temporarily, replication that has already been initiated is not affected (19).
Fig. 3.
(Left) Infection does not affect the dynamics of the integration site on the E. coli chromosome. Distance between the attB site and the cell’s center as a function of the time elapsed after the last cell division, for uninfected (blue) and infected cells (red). Each plot represents an average over ten traces measured in individual bacteria, and error bars represent SEs. The distance is measured along the long cell axis in coordinates in which 0 and 1 correspond to the cell center and pole, respectively. The overlap between the two plots indicates that within experimental error, the effect of infection on the motion of the attB site is not appreciable. (Right) A histogram of λ DNA positions measured along the bacterial axis in the same coordinates.
Integration Frequency Measurements in Strains with the attB Site at Different Genomic Locations Are Consistent with a Replication-Driven Mechanism.
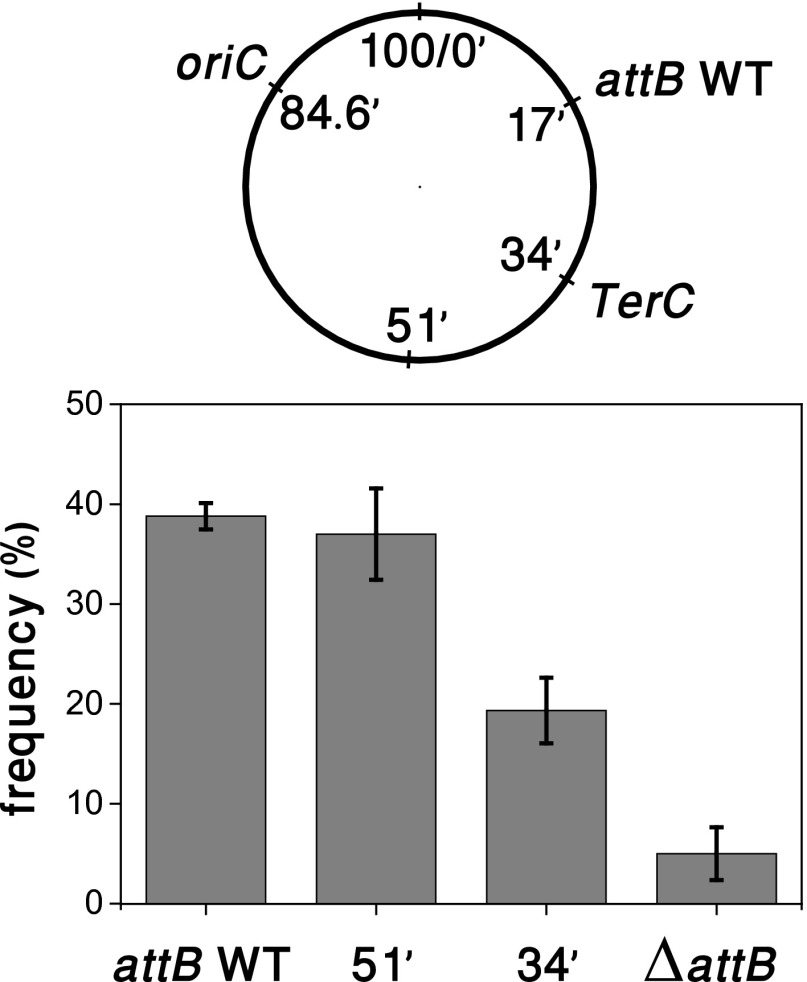
To provide further evidence that the movement of the attB site toward the phage DNA is driven by chromosome segregation, we moved the attB site at 17 min to two other genomic positions: one at 51 min in a nearly symmetrical position relative to oriC, and the other at the terminus region. Bacterial DNA at the terminus remains close to midcell throughout replication (16). The integration frequency defined as the ratio between the number of colony-forming units grown on kanamycin plates and the number of colonies on LB plates was then measured for the different strains (Fig. 4). The native attB site and its symmetrical counterpart have a similar frequency, and both DNA locations can reach the genomes of phages infecting at poles and midcell. In contrast, the frequencies of the native attB site and its symmetrical counterpart are significantly higher than when the attB site is at the terminus, because only phages infecting the midcell region may integrate when the attB site is moved to the terminus.
Fig. 4.
Integration frequency when the λ integration site is moved to other genomic positions. The frequency is defined as 100 times the ration of lysogens to the number of surviving cells. Integration frequency (Lower) was measured in strains with attB site at the native position (17’), at a nearly symmetrical position (51’) relative to oriC (84.6’), at a position in the terminus region (34’), and for a ΔattB strain, as illustrated in the diagram (Upper). Error bars denote SEs from three independent experiments. Within each experiment, the number of colonies grown on LB plates for the different strains did not vary by more than 7%, i.e., the number of survivor cells per total cell input at the start is strain independent. Proper integration in each of these sites was assayed by PCR.
Discussion
In order for bacteriophage λ DNA to integrate and establish stable lysogeny after infecting an E. coli cell, a unique 15-bp-long integration site must be located among ∼5 million possible sites on the bacterial chromosome. In this work, we aimed at elucidating the strategy used by λ phage to find the integration site with high efficiencies and within a physiologically relevant time. We have visualized the dynamics of the attB and attP sites in live E. coli cells during infection. Contrary to our initial expectations, it is not λ DNA that carries out a search throughout the cellular interior, either by diffusion or by exploiting native mechanisms to move directionally as many viruses infecting eukaryotic cells do (21, 22), nor by deploying facilitated transport mechanisms as transcription factors do in their search for their cognate sites (1, 23). Instead, λ DNA remains confined near the point of entry into an infected cell, and takes advantage of the directed motion provided by the segregation of the nascent nucleoid driven by bacterial DNA replication machinery (15, 24), to bring the attB site of the bacterium close to the phage attP site. Once in close proximity, confined diffusion of both attachment sites facilitates their final encounter.
What constrains λ DNA to remain near the point of entry into the cell? Most likely, λ DNA becomes anchored to a site on the cellular membrane through which the λ DNA entered the cell. Extant experimental evidence suggests association of the phage DNA to the membrane is mediated by early gene N expression (25–27). Despite λ DNA’s circularization and its likely compaction upon entry into the cell (28), both of which reduce the effective physical size of the molecule within the cell, reduced diffusion by crowding is unlikely to hinder λ DNA movement significantly (29).
To address whether the genomic location of the attB site affects the integration frequency, we moved the attB site to two different locations on the E. coli chromosome. The chromosome in E. coli is organized with the origin of replication (oriC site) and the terminus region at midcell, and the two chromosome halves located at separate cell halves (16, 30). Chromosomal replication is initiated at the oriC site and proceeds in both directions (31). In slow-growing E. coli cells, replication and chromosome segregation are continuous and simultaneous (32). The replicated oriC sites are segregated and captured approximately at each quarter-cell position, which will become the new cell centers after division, and other loci lie roughly in the position defined by their order of replication (32). The chromosomes in the nascent daughter cells are arranged in a left-right-left-right organization (31), indicating that one of the replicated attB sites will move toward the pole, whereas the other will cross the midcell plane. Because both the poles and the midcell region are enriched with λ DNA (Fig. 3), attB segregation increases the chance of encounter between the attB and attP sites. The order in the organization and segregation of the E. coli chromosome, together with the required movement of the attB site toward regions enriched with λ DNA, suggests that the genomic location of the attB site plays an important role in determining the likelihood of the encounter between the attB and the attP sites. We tested this hypothesis by deleting the wild-type attB site and moving it to two different locations: the terminus region at 34 min and the symmetric location to the wild-type relative to oriC on the left replichore at 51 min. The integration frequency was reduced significantly when the attB site was moved to the terminus region because this region does not move to the poles during DNA replication, and the strain with attB at the symmetric location was similar to the wild-type, consistent with the DNA segregation being symmetric to the left and right replichores. These results are consistent with a search driven by DNA segregation during replication.
We believe that this mechanism can be generalized to other temperate bacteriophages infecting different types of bacteria, because many of its key features are preserved. It has been shown that different bacteriophages enter cells preferentially through the poles (18). Evidence has also been provided that once intracellular, the phage DNA of different bacteriophages remains attached to the bacterial membrane (33). Furthermore, many temperate phages integrate at specific sites on the bacterial chromosome [e.g., ϕ80 infecting E. coli (34), ϕ105 infecting Bacillus subtilis (35), and P22, which infects Salmonella typhimurium (36)]. Although different types of bacteria exhibit differences in chromosome organization and segregation, chromosome segregation, either due to entropic (24) or other forces (37), will drive global movement of chromosomal loci within the bacteria, and we anticipate that this movement will assist in the encounter between the bacterial and phage DNA attachment sites. These findings add to the list of profligate use of host functions by λ, such as LamB porins for phage docking and injection (6), GroEL for folding of capsid proteins (38), and IHF for integration into the bacterial chromosome (39), brought about by coevolution of host−phage interactions (40).
Materials and Methods
Bacterial Strains and Phage Construction.
A general recombineering protocol (41) was used to create the recombinant derivatives of phage λ and MG1655, containing the parS sites used in this paper. To create the MG1655 strains carrying the pMT parS sites on the chromosome, the λ Red recombination functions were supplied by pSIM18. Following isolation of recombinants, pSIM18 was removed by growing at 42 °C. The P1 parS (near attP on bacteriophage λ) and pMT parS (near attB in the bacterial genome) recombinants were selected by a linked kanR or cmR cassette, respectively, screened by PCR, and confirmed by sequence analysis. These parS sites serve as specific and unique binding sites for the two different fluorescently tagged ParB proteins, P1 ParB and pMT ParB, expressed from the plasmid (see SI Materials and Methods). The sequences of P1 parS-kan and pMT parS-cat are shown in SI Materials and Methods (Sequences S1 and S2) and were PCR amplified from a bacterial strain generously supplied by the Stuart Austin laboratory.
The PCR product of P1 parS‐kan was made with chimeric primers RG3 and RG4 (Table S1) that contain homologies at their 5′ ends to insert the cassette near attP on λ. To do this, strain LT447, MG1655(λcI857 ind−), carrying a single copy of λ in the prophage state was grown to midlog, and Red functions were induced from the intact prophage at 42 °C to carry out the recombination step (41). Following electroporation, cells were left to grow at 42 °C to allow continued λ gene expression and lytic growth. A phage lysate containing recombinants was harvested and used to lysogenize MG1655 at 32 °C. Recombinant λ lysogens were selected as kanR colonies. A kanR single-copy lysogen was used to generate a high-titer, pure λcI857 P1 parS‐kanR lysate. The P1 parS site was confirmed by sequence analysis.
A deletion of the native attB site in MG1655 was made using oligonucleotide RG21 (Table S1) following standard recombineering protocols. Colonies were screened for recombinants by mismatch amplification mutation assay PCR (41), and the identified recombinants were purified and confirmed by sequence analysis. This MG1655 derivative carrying ΔattB was used to insert the wild-type attB with the cat-pMT parS cassette at two different locations on the genome by recombineering as described below.
A PCR product of the pMT parS site linked to cmR was made using chimeric primers RG11 and RG12 (within the bioD gene) or RG17 and RG18 (within the bioA gene) that carried homologies at their 5′ ends to target insertion of the pMT parS site near attB at 17 min (Table S1). The PCR product was recombined into MG1655 using standard protocols, selecting CmR. We then PCR-amplified the entire attB–cat–pMT parS cassette at bioA using chimeric primers with 5′ homologies to target the cassette to the chromosome locations at 34 or 51 min. The primer sequences are given in Table S1; RG25 and RG26 were used to make the PCR product targeting the 34-min region, and RG29 and 30 were used for the 51-min region product. The MG1655 ΔattB mutant was the recipient strain so that the newly inserted intact attB site associated with the pMT parS site becomes the only cellular site for λ integration. Each parS site was verified by sequence analysis. Note that the homology segments of the primers (Table S1) can be used by the reader to determine the exact location of each attB-P1 parS insertion within the genome sequence (see Sequences S1 and S2 in SI Materials and Methods).
Microscopy.
Images were taken with a modified Zeiss Axiovert 135TV microscope. Phase contrast and fluorescence images were collected using a Neoplan 100× 1.3 phase contrast objective (ZEISS) with temperature controlled to 32 °C and an iXon EMCCD (Andor Technology). The objective lens is placed on a piezo mount that is controlled by a Mipos100 piezo controller (Piezosystem Jena). The piezo is used for z-scanning in a contrast detection autofocus algorithm, before image acquisition. All filters were purchased from Semrock. For RFP and yGFP images, filters FF01-503/572-25 for excitation, FF444/520/590-Di01 as dichroic were used. The yGFP and RFP emissions were split using a FF562-Di02 dichroic mirror, and are then imaged on half of the camera CCD chip with the RFP and yGFP emission filters FF01-628/32 and FF01-535/22, respectively. Pictures were taken with 2-s integration. The microscope is equipped with a XY motorized stage (PI), which enables automatic data acquisition and taking multiple fields of view in every experimental run. Phase contrast and fluorescent images were taken at 10-min intervals.
Experimental Growth Conditions.
Cells from the bacterial strain with parS in the bioD region were propagated in LBMM medium (LB supplemented with 0.2% maltose and 10 mM MgSO4) supplemented with 50 μg/mL ampicillin at 32 °C overnight from a single-colony inoculum. The cultures were diluted 1:100 into AB minimal media supplemented with 10 mM MgSO4, 0.2% maltose, 0.4% glycerol, 0.05% casamino acids, 1 μg/mL thiamine, 1 μg/mL uracil, and 100 μM isopropyl β-D-1-thiogalactopyranoside and grown at 32 °C to OD600 of 0.6. Cells were then mixed with λ phage particles at a MOI of ∼3 for 5 min in room temperature. Cells were then deposited on a low-melting agarose pad (1.5%), prepared with AB medium with supplements, and covered with glass slides before mounting on the microscope.
Measurement of Integration Frequency.
Cells from the bacterial strain with parS in the bioA region (WT) and the strains with the attB site at different locations were propagated in LBMM medium supplemented with 50 μg/mL ampicillin at 37 °C overnight from a single-colony inoculum. Cells were then diluted 1:200 into 8 mL TB medium (10 g/L Bacto-tryptone, 5 g/L NaCl, pH 7.2) supplemented with 0.2% maltose and 10 mM MgSO4 and grown for 5 h. Cells were then centrifuged and resuspended in 4 mL, TMG buffer [10 mM Tris (PH 7.4), 10 mM MgSO4, 100 μg/mL gelatin] for 30 min at 30 °C. Cells were diluted 1:5 in TM buffer [10 mM Tris (PH 7.4), 10 mM MgSO4], the OD of all strains was set to OD = 0.5 and then mixed with phage lysate at MOI = 3 to a 100-μL final volume at 30 °C for 1 h. Four milliliters of LB were then added, and the cells were incubated for 1 h at 30 °C before plating. The cells were serially diluted to give single colonies and plated on LB and LB supplemented with 30 μg/mL Kanamycin plates overnight at 30 °C. For calculation of integration frequency, the number of lysogens was measured as the number of colonies growing on Kanamycin plates; the number of cells surviving infection was measured as the number of colonies growing on LB plates. The integration frequency is defined as 100 times the ratio between the number of lysogens and the number of cells surviving infection, as described in Rutkai et al. (42). Insertion of the λ phage into the attB site was verified using colony PCR with primers RG-15 and AT-FWR that amplify the DNA fragment from the phage to the kan insertion cassette. For primer sequences, see Table S1.
Supplementary Material
Acknowledgments
We thank S. Austin for comments and plasmids, L. Thomason for the gift of lambda strain cI857 bor::kanR, and O. Kobiler, and J. Sawitzke for a careful reading of the manuscript. This work was supported in part by the Yeda-Sela Center of the Weizmann Institute of Science (J.S.), and by the Intramural Research Program of the National Institutes of Health, National Cancer Institute, Center for Cancer Research (D.L.C.).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1324066111/-/DCSupplemental.
References
- 1.Halford SE, Marko JF. How do site-specific DNA-binding proteins find their targets? Nucleic Acids Res. 2004;32(10):3040–3052. doi: 10.1093/nar/gkh624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barzel A, Kupiec M. Finding a match: How do homologous sequences get together for recombination? Nat Rev Genet. 2008;9(1):27–37. doi: 10.1038/nrg2224. [DOI] [PubMed] [Google Scholar]
- 3.Ochman H, Lawrence JG, Groisman EA. Lateral gene transfer and the nature of bacterial innovation. Nature. 2000;405(6784):299–304. doi: 10.1038/35012500. [DOI] [PubMed] [Google Scholar]
- 4.Kenzaka T, Tani K, Nasu M. High-frequency phage-mediated gene transfer in freshwater environments determined at single-cell level. ISME J. 2010;4(5):648–659. doi: 10.1038/ismej.2009.145. [DOI] [PubMed] [Google Scholar]
- 5.Brüssow H, Canchaya C, Hardt WD. Phages and the evolution of bacterial pathogens: From genomic rearrangements to lysogenic conversion. Microbiol Mol Biol Rev. 2004;68(3):560–602. doi: 10.1128/MMBR.68.3.560-602.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chatterjee S, Rothenberg E. Interaction of bacteriophage l with its E. coli receptor, LamB. Viruses. 2012;4(11):3162–3178. doi: 10.3390/v4113162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rothenberg E, et al. Single-virus tracking reveals a spatial receptor-dependent search mechanism. Biophys J. 2011;100(12):2875–2882. doi: 10.1016/j.bpj.2011.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Van Valen D, et al. A single-molecule Hershey-Chase experiment. Curr Biol. 2012;22(14):1339–1343. doi: 10.1016/j.cub.2012.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Oppenheim AB, Kobiler O, Stavans J, Court DL, Adhya S. Switches in bacteriophage lambda development. Annu Rev Genet. 2005;39:409–429. doi: 10.1146/annurev.genet.39.073003.113656. [DOI] [PubMed] [Google Scholar]
- 10.Ptashne M. A Genetic Switch. 3rd Ed. Cold Spring Harbor, NY: Cold Spring Harbor Lab Press; 2004. [Google Scholar]
- 11.St-Pierre F, Endy D. Determination of cell fate selection during phage lambda infection. Proc Natl Acad Sci USA. 2008;105(52):20705–20710. doi: 10.1073/pnas.0808831105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ellis EL, Delbrück M. The growth of bacteriophage. J Gen Physiol. 1939;22(3):365–384. doi: 10.1085/jgp.22.3.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zeng L, et al. Decision making at a subcellular level determines the outcome of bacteriophage infection. Cell. 2010;141(4):682–691. doi: 10.1016/j.cell.2010.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Youngren B, Radnedge L, Hu P, Garcia E, Austin S. A plasmid partition system of the P1-P7par family from the pMT1 virulence plasmid of Yersinia pestis. J Bacteriol. 2000;182(14):3924–3928. doi: 10.1128/jb.182.14.3924-3928.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Youngren B, Nielsen HJ, Jun S, Austin S. The multifork Escherichia coli chromosome is a self-duplicating and self-segregating thermodynamic ring polymer. Genes Dev. 2014;28(1):71–84. doi: 10.1101/gad.231050.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nielsen HJ, Ottesen JR, Youngren B, Austin SJ, Hansen FG. The Escherichia coli chromosome is organized with the left and right chromosome arms in separate cell halves. Mol Microbiol. 2006;62(2):331–338. doi: 10.1111/j.1365-2958.2006.05346.x. [DOI] [PubMed] [Google Scholar]
- 17.Rodionov O, Lobocka M, Yarmolinsky M. Silencing of genes flanking the P1 plasmid centromere. Science. 1999;283(5401):546–549. doi: 10.1126/science.283.5401.546. [DOI] [PubMed] [Google Scholar]
- 18.Edgar R, et al. Bacteriophage infection is targeted to cellular poles. Mol Microbiol. 2008;68(5):1107–1116. doi: 10.1111/j.1365-2958.2008.06205.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sergueev K, Court D, Reaves L, Austin S. E.coli cell-cycle regulation by bacteriophage lambda. J Mol Biol. 2002;324(2):297–307. doi: 10.1016/s0022-2836(02)01037-9. [DOI] [PubMed] [Google Scholar]
- 20.Marshall WF, et al. Interphase chromosomes undergo constrained diffusional motion in living cells. Curr Biol. 1997;7(12):930–939. doi: 10.1016/s0960-9822(06)00412-x. [DOI] [PubMed] [Google Scholar]
- 21.McDonald D, et al. Visualization of the intracellular behavior of HIV in living cells. J Cell Biol. 2002;159(3):441–452. doi: 10.1083/jcb.200203150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Seisenberger G, et al. Real-time single-molecule imaging of the infection pathway of an adeno-associated virus. Science. 2001;294(5548):1929–1932. doi: 10.1126/science.1064103. [DOI] [PubMed] [Google Scholar]
- 23.Elf J, Li GW, Xie XS. Probing transcription factor dynamics at the single-molecule level in a living cell. Science. 2007;316(5828):1191–1194. doi: 10.1126/science.1141967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jun S, Wright A. Entropy as the driver of chromosome segregation. Nat Rev Microbiol. 2010;8(8):600–607. doi: 10.1038/nrmicro2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hallick L, Boyce RP, Echols H. Membrane association by bacteriophage lambda-DNA: Possible direct role of regulator gene N. Nature. 1969;223(5212):1239–1242. doi: 10.1038/2231239a0. [DOI] [PubMed] [Google Scholar]
- 26.Kolber AR, Sly WS. Association of lambda bacteriophage DNA with a rapidly sedimenting Escherichia coli component. Virology. 1971;46(3):638–654. doi: 10.1016/0042-6822(71)90067-5. [DOI] [PubMed] [Google Scholar]
- 27.Siegel PJ, Schaechter M. The role of the host cell membrane in the replication and morphogenesis of bacteriophages. Annu Rev Microbiol. 1973;27:261–282. doi: 10.1146/annurev.mi.27.100173.001401. [DOI] [PubMed] [Google Scholar]
- 28.Murphy LD, Zimmerman SB. Condensation and cohesion of lambda DNA in cell extracts and other media: Implications for the structure and function of DNA in prokaryotes. Biophys Chem. 1995;57(1):71–92. doi: 10.1016/0301-4622(95)00047-2. [DOI] [PubMed] [Google Scholar]
- 29.Derman AI, Lim-Fong G, Pogliano J. Intracellular mobility of plasmid DNA is limited by the ParA family of partitioning systems. Mol Microbiol. 2008;67(5):935–946. doi: 10.1111/j.1365-2958.2007.06066.x. [DOI] [PubMed] [Google Scholar]
- 30.Wang X, Liu X, Possoz C, Sherratt DJ. The two Escherichia coli chromosome arms locate to separate cell halves. Genes Dev. 2006;20(13):1727–1731. doi: 10.1101/gad.388406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang X, Montero Llopis P, Rudner DZ. Organization and segregation of bacterial chromosomes. Nat Rev Genet. 2013;14(3):191–203. doi: 10.1038/nrg3375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nielsen HJ, Li Y, Youngren B, Hansen FG, Austin S. Progressive segregation of the Escherichia coli chromosome. Mol Microbiol. 2006;61(2):383–393. doi: 10.1111/j.1365-2958.2006.05245.x. [DOI] [PubMed] [Google Scholar]
- 33.Earhart CF. The association of host and phage DNA with the membrane of Escherichia coli. Virology. 1970;42(2):429–436. [PubMed] [Google Scholar]
- 34.Rybchin VN. Genetics of bacteriophage phi 80—A review. Gene. 1984;27(1):3–11. doi: 10.1016/0378-1119(84)90233-6. [DOI] [PubMed] [Google Scholar]
- 35.Ellis DM, Dean DH. Location of the Bacillus subtilis temperate bacteriophage phi 105 attP attachment site. J Virol. 1986;58(1):223–224. doi: 10.1128/jvi.58.1.223-224.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Susskind MM, Botstein D. Molecular genetics of bacteriophage P22. Microbiol Rev. 1978;42(2):385–413. doi: 10.1128/mr.42.2.385-413.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Di Ventura B, et al. Chromosome segregation by the Escherichia coli Min system. Mol Syst Biol. 2013;9(1):686. doi: 10.1038/msb.2013.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Georgopoulos CP, Hendrix RW, Casjens SR, Kaiser AD. Host participation in bacteriophage lambda head assembly. J Mol Biol. 1973;76(1):45–60. doi: 10.1016/0022-2836(73)90080-6. [DOI] [PubMed] [Google Scholar]
- 39.Kikuchi Y, Nash HA. The bacteriophage lambda int gene product. A filter assay for genetic recombination, purification of int, and specific binding to DNA. J Biol Chem. 1978;253(20):7149–7157. [PubMed] [Google Scholar]
- 40.Meyer JR, et al. Repeatability and contingency in the evolution of a key innovation in phage lambda. Science. 2012;335(6067):428–432. doi: 10.1126/science.1214449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thomason L, et al. Current Protocols in Molecular Biology, ed Ausubel FJ. New York: Wiley; 2007. Recombineering: Genetic engineering in bacteria using homologous recombination; pp. 1–16. [DOI] [PubMed] [Google Scholar]
- 42.Rutkai E, György A, Dorgai L, Weisberg RA. Role of secondary attachment sites in changing the specificity of site-specific recombination. J Bacteriol. 2006;188(9):3409–3411. doi: 10.1128/JB.188.9.3409-3411.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.