Significance
Chromosomal aneuploidies represent a major cause of fetal loss and birth defects. Current methods for the prenatal diagnosis of aneuploidy require invasive methods that are associated with a risk of miscarriage and other complications. Recently, noninvasive prenatal testing has been developed using cell-free fetal DNA in maternal plasma. In this study, we validated an effective method for noninvasive diagnosis of fetal aneuploidy using a semiconductor sequencer, which reduces the time and cost of sequencing. Our method is cost-effective and practical in a clinical setting with high sensitivity and specificity for the diagnosis of trisomy 13, 18, and 21 as well as sex chromosome aneuploidies.
Abstract
Massively parallel sequencing (MPS) of cell-free fetal DNA from maternal plasma has revolutionized our ability to perform noninvasive prenatal diagnosis. This approach avoids the risk of fetal loss associated with more invasive diagnostic procedures. The present study developed an effective method for noninvasive prenatal diagnosis of common chromosomal aneuploidies using a benchtop semiconductor sequencing platform (SSP), which relies on the MPS platform but offers advantages over existing noninvasive screening techniques. A total of 2,275 pregnant subjects was included in the study; of these, 515 subjects who had full karyotyping results were used in a retrospective analysis, and 1,760 subjects without karyotyping were analyzed in a prospective study. In the retrospective study, all 55 fetal trisomy 21 cases were identified using the SSP with a sensitivity and specificity of 99.94% and 99.46%, respectively. The SSP also detected 16 trisomy 18 cases with 100% sensitivity and 99.24% specificity and 3 trisomy 13 cases with 100% sensitivity and 100% specificity. Furthermore, 15 fetuses with sex chromosome aneuploidies (10 45,X, 2 47,XYY, 2 47,XXX, and 1 47,XXY) were detected. In the prospective study, nine fetuses with trisomy 21, three with trisomy 18, three with trisomy 13, and one with 45,X were detected. To our knowledge, this is the first large-scale clinical study to systematically identify chromosomal aneuploidies based on cell-free fetal DNA using the SSP and provides an effective strategy for large-scale noninvasive screening for chromosomal aneuploidies in a clinical setting.
The incidence of chromosomal abnormalities is as high as 1 in 160 live births in the United States (1) or 1 in 60 in China (2). The incidence increases with maternal age and can reach 2.5% with maternal age over 35 in China (2). Among autosomal abnormalities, Down syndrome (trisomy 21), Edward syndrome (trisomy 18), and Patau syndrome (trisomy 13) are most compatible with survival and therefore the most clinically significant. Sex chromosome aneuploidies occur in 1 in 500 male births and 1 in 850 female births in the United States (3–6) and 1 in 450 in China (2). Turner’s syndrome (45,X), Klinefelter’s syndrome (47,XXY), and 47,XYY syndrome are common sex chromosome aneuploidies that are associated with fetal loss, infertility, and language developmental delays, among other defects (7–9). Fetuses with aneuploidy account for 6–11% of all stillbirths and neonatal deaths (10). The incidence of Down syndrome increases significantly with maternal age, occurring in 25 in 100,000 births with maternal age over 35 and 30 in 100,000 births with maternal age over 40 in China. There were an estimated 27,000 babies with Down syndrome born in China in 2006, which caused an economic burden of $10,000 per capita, $48,300 per family, and a total of $2.1 billion per year (11). Diagnosis of fetal chromosomal aneuploidies is the most common indication for an invasive prenatal testing procedure such as chorionic villus sampling or amniocentesis. Currently, G-band karyotyping and molecular genetics methods including multiplex ligation-dependent probe amplification, fluorescent in situ hybridization, quantitative fluorescent PCR, and microarray-based comparative genomic hybridization have been well established for the prenatal diagnosis of common aneuploidies in clinical laboratories (12, 13). Although these testing methods are proven to be highly reliable, a major limitation remains that they depend on invasive procedures that are associated with a 0.4–0.8% chance of fetal loss (14–17). In addition, G-band karyotyping takes 7–10 d to complete, a delay that may cause significant anxiety for the family.
To overcome these limitations, methods based on the massively parallel sequencing (MPS) platform were recently developed to detect common fetal aneuploidies using noninvasive procedures (18–22). The MPS-based approach using an Illumina HiSeq platform can reliably detect common aneuploidies (trisomy 21, 18, and 13) in 7 d. A benchtop semiconductor sequencing platform (SSP) enables acquisition of ∼5 billion data points per second over a 2- to 4-h runtime with on-instrument signal processing, thus providing an alternative sequencing platform with a reduced turnaround time. Here, we report rapid noninvasive prenatal diagnosis of common aneuploidies in a large clinical sample of pregnant women in China using the SSP. In a retrospective study, we assessed the performance of the SSP for diagnosis of aneuploidy using cell-free fetal DNA in maternal plasma. We then validated the performance of the SSP in a prospective study. Our study demonstrates that the SSP can detect trisomy 13, 18, and 21 as well as sex chromosome aneuploidies with high sensitivity and specificity in a significantly shorter time frame from sample acquisition to diagnosis, thus providing an effective platform for large-scale noninvasive screening of chromosomal aneuploidies.
Results
Study Participants.
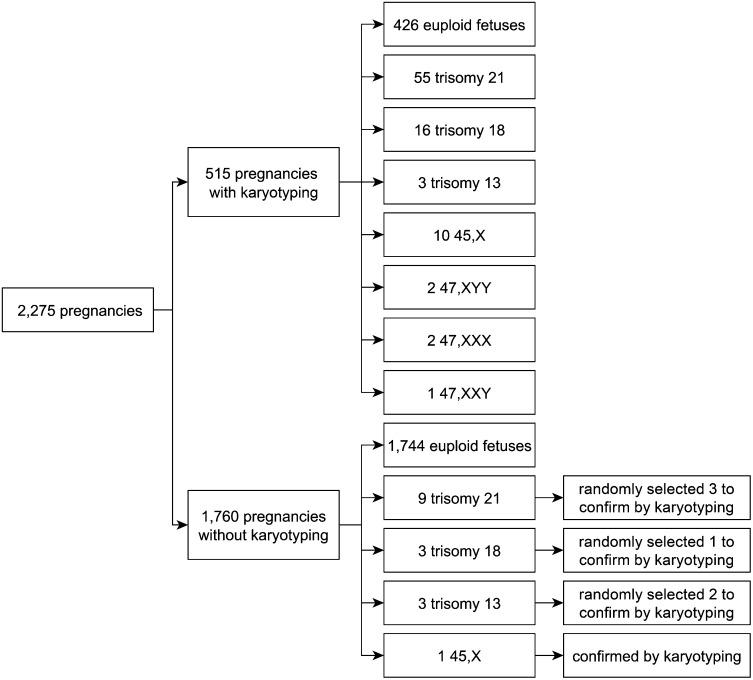
A total of 2,275 pregnant subjects included in this study was divided into two groups. Group I, which had full karyotyping results, was used for reference construction in a retrospective case-control study, and group II, which lacked karyotyping results, was used for clinical application in a prospective study. There were 515 pregnancies with karyotyping results in group I, including 55 fetuses with trisomy 21, 16 with trisomy 18, 3 with trisomy 13, and 15 with sex chromosome aneuploidies (Fig. 1). Moreover, our method detected nine fetuses with trisomy 21, three with trisomy 18, three with trisomy 13, and one with a sex chromosome aneuploidy (45,X) among the 1,760 pregnancies without karyotyping in group II (Fig. 1).
Fig. 1.
Characterization of pregnant subjects included in the retrospective and prospective studies.
Sequencing Data Collection and Analysis.
We obtained an average of 5.58 ± 1.61 million raw reads per sample. The mean length of sequencing reads was 100 bp. Although the mean size of cell-free DNA in maternal plasma is about 160 bp, and the SSP can produce ∼200-bp reads at the maximum 500 sequencing flows, we usually generated reads with a mean size of 100 bp due to use of a smaller sequencing flow number to facilitate faster turnaround time (the effect of flow number on read length is shown in Fig. S1). Only sequence reads that could be mapped to just one genome location in a reference human genome were retained by our data-filtering procedure. We termed the sequences “unique reads.” Approximately 65.5% (3.6 million) of the total reads passed the criteria and were retained as unique reads. In contrast, the Illumina HiSeq used for noninvasive prenatal diagnosis of trisomy 21, 18, and 13 generally produced ∼10 million 36-bp raw reads per sample, with only 20% (∼2 million) retained as unique reads (23–25). Shorter reads decrease the likelihood that a read can be mapped to a single, unique location (26). Next, the number of unique reads from each 20 kb bin on each chromosome was counted. To eliminate the effect of GC bias in different samples, an integrated GC correction method was applied (Materials and Methods) in which locally weighted scatterplot smoothing (LOESS) regression was used to compute the corrected number of unique reads in each 20-kb bin depending on the GC content of the genomic sequence in the corresponding bin. The corrected reads number of each chromosome was determined by summing the weighted values of all 20-kb bins of each specific chromosome in a sample. Subsequently, to overcome GC bias among different samples in a run, an intrarun normalization method was used to compute the weighted reads number of a specific chromosome among the different samples in one sequencing run. The reads ratio of each chromosome was determined by dividing the weighted reads number of a specific chromosome by the total corrected reads number of all autosomes (chromosomes 1–22). Finally, a linear model was established by plotting the reads ratio of each chromosome against GC content. To eliminate the effect of GC bias, the residual between the reads ratio based on intrarun normalization and the fitted reads ratio based on the sequencing GC content in the linear model was used to detect fetal aneuploidies using a z-score method. Trisomy 21, 18, and 13 were identified using the criteria of z score > 3.
Detection of Fetal Aneuploidy in a Retrospective Study.
Prenatal diagnosis of aneuploidy relies on identifying changes in the number of aligned sequences and thus the relative representation of the aneuploid chromosome. For our retrospective study, plasma samples were obtained from group I, in which the chromosomal status of the fetuses had been confirmed by full karyotyping (Fig. 1).
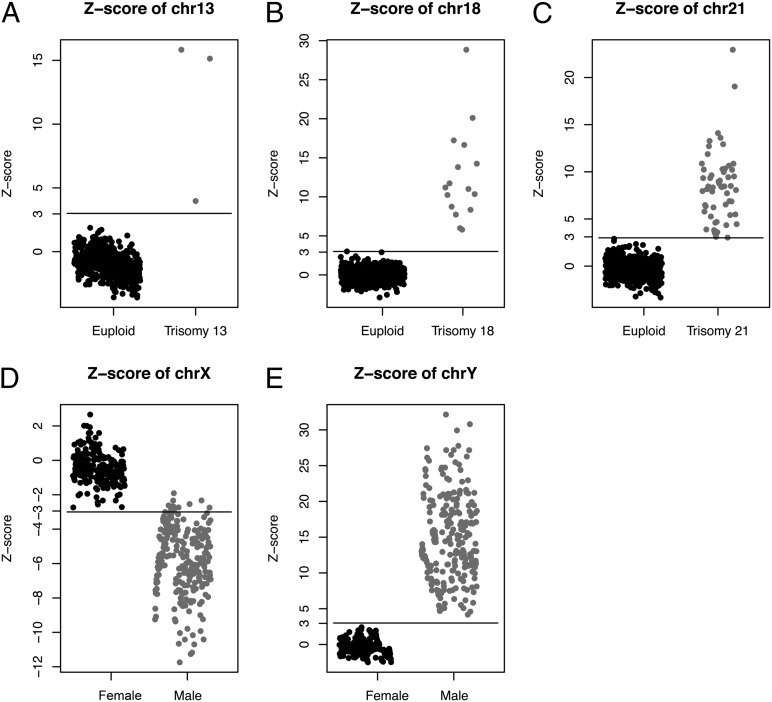
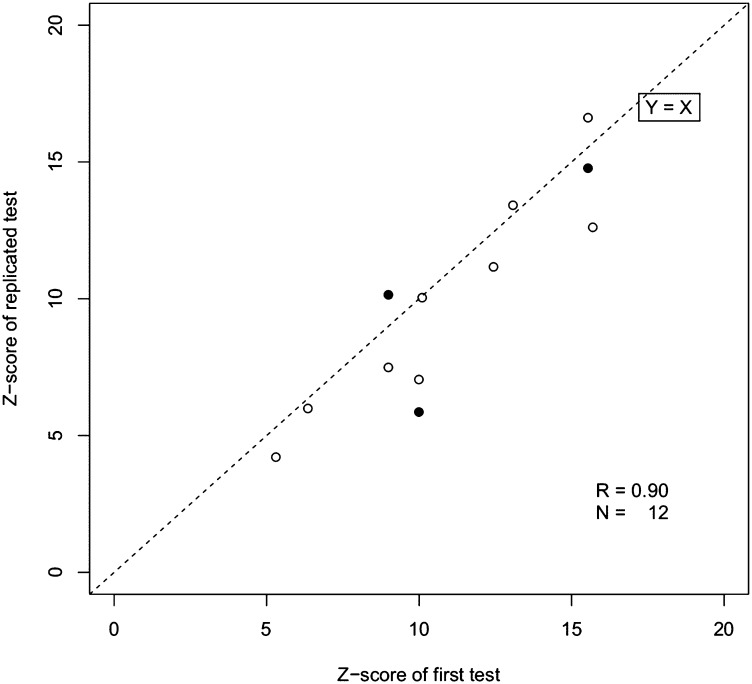
To objectively quantify the degree of overrepresentation in the sequence tags of an aneuploid chromosome, we used data from pregnancies with euploid fetuses as a reference population to calculate the mean and SD of the number of usable reads per chromosome. Using these reference values, we calculated the z scores for each affected chromosome among the pregnancies with aneuploid fetuses (Fig. 2). Table 1 summarizes the diagnostic performance of this strategy for detecting trisomy 21, 18, and 13. We then applied the cross-validation method to evaluate the sensitivity and specificity of our approach. Using a z score of 3 as the diagnostic cutoff point, we detected 55 trisomy 21 fetuses with 99.94% sensitivity and 99.46% specificity, 16 trisomy 18 fetuses with 100% sensitivity and 99.24% specificity, and 3 trisomy 13 fetuses with 100% sensitivity and 100% specificity. To evaluate the reproducibility of our method, we randomly reprocessed 12 of the samples with aneuploidy. Similar z scores were obtained when replicate experiments were run on each sample (Fig. 3). This demonstrated the stability of the SSP for detection of trisomy 21, 18, and 13.
Fig. 2.
Z scores obtained for each sample in group I (n = 515) and the cutoffs for detection of fetal aneuploidy. (A–C) Z scores for chromosome 13 (A), chromosome 18 (B), and chromosome 21 (C). (D and E) Z scores for male and female fetuses for chromosome X (D) and chromosome Y (E).
Table 1.
Diagnostic performance of the SSP for identifying trisomy 21, trisomy 18, and trisomy 13
| Type | Sensitivity, % | Specificity, % |
| Trisomy 21 | 99.94 | 99.46 |
| Trisomy 18 | 100 | 99.24 |
| Trisomy 13 | 100 | 100 |
Fig. 3.
Comparison of z scores obtained from technical repeat experiments. Results from nine samples involving a fetus with trisomy are shown (open circles), including three samples that were tested a third time (solid circles).
Detection of Fetal Aneuploidy in a Prospective Study.
For our prospective study, plasma samples were obtained from 1,760 pregnancies in group II. Using the reference values established from group I, we detected nine fetuses with trisomy 21, three with trisomy 18, three with trisomy 13, and one with 45,X. We randomly selected six positive trisomy samples and the 45,X sample to perform full karyotyping, in which we confirmed all of the diagnoses (Fig. 1). This validation experiment demonstrated that noninvasive prenatal diagnosis by the SSP was reliable.
Sex Chromosome Aneuploidy Detection.
Because males have one copy of the Y chromosome and one fewer copy of the X chromosome than females, we hypothesized that there is underrepresentation of chromosome X and the presence of chromosome Y in pregnancies with a male fetus compared with those with a female fetus (Fig. 2). We derived a model () to define the relationship between the z scores of chromosome X and Y. From this model, sex chromosome aneuploidies (45,X, 47,XYY, 47,XXX, 47,XXY) could be detected. As expected, 45,X fetuses had a z score < −3 for the X chromosome and a z score between −3 and 3 for the Y chromosome, 47,XXX fetuses had a z score > 3 for the X chromosome and a z score < 3 for the Y chromosome, and the 47,XYY fetus had a high z score (32.88) for the Y chromosome and a low z score (−11.96) for the X chromosome (Table 2).
Table 2.
Criteria used to detect sex chromosome aneuploidy
| Criterion | Predicted sex chromosomes |
| < 3, < −3 | XO |
| < 3, < 3 | XX |
| < 3 and > 3, | XXX |
| > 3, < 3 and > | XXY |
| > 3, < −3 and > | XYY |
| > 3 and | XY |
GC Correction as a Quality Control Measure to Obtain More Robust Data.
GC bias introduced during PCR in library preparation and cluster generation can influence the accuracy of the data, as reported for other high-throughput platforms (27). We therefore investigated the relationship between the GC content of the chromosomes and the number of reads. Fig. S2 illustrates GC bias on the SSP. The number of sequence tags within every 20-kb nonoverlapping window was summed to obtain the distribution of sequence tag densities for each chromosome. Regions of high (>50%) or low (<30%) GC content had below average tag densities and greater variability in the number of sequence tags. Therefore, we applied an integrated GC correction method to eliminate GC bias in the raw data. We then calculated the coefficient of variation (CV) for measuring the genomic representation of each autosome among reference samples to evaluate the effect of GC correction. As expected, the CVs of chromosomes 21, 18, and 13 decreased after LOESS correction and intrarun normalization (Fig. S3). The average CVs for measuring the genomic representations of chromosomes 21, 18, and 13 without GC correction were 1.097%, 0.773%, and 1.738%, respectively. After LOESS correction and intrarun normalization, the CVs of chromosomes 21, 18, and 13 were significantly reduced to 0.625%, 0.494%, and 0.480%, respectively. When we analyzed the relationship of the unique reads ratio and sequencing GC content, a significant linear model was established (Fig. S4). Each chromosome has a different GC content and consequent variable GC bias; therefore, the slopes of GC content (GC content of unique reads derived by each sequencing run) and ratio (unique read ratio of each chromosome) varied among different chromosomes. Finally, to eliminate the GC bias, the residual (ε) between the real reads ratio and fitted predicted reads ratio was used as a statistical value for z score testing. Thus, GC correction was an effective way to ensure robust data quality.
Quantification of Variability in the Number of Sequence Tags.
The accuracy of fetal aneuploidy detection by the SSP was limited by variability in the relative read coverage. This variability was quantified using the SD in the number of unique reads that were counted. The depth of sequencing was a major factor in determining the accuracy of aneuploidy detection. We randomly selected 30 plasma samples with euploid fetuses to examine the relationship between the number of usable reads and the SD of the relative z score (Fig. S5). For each chromosome, the SD of the relative z score among the 30 samples had a significantly negative correlation with the number of unique reads. We calculated that more than 3.5 million usable reads was sufficient to obtain a robust and reliable z score for each chromosome.
The Relationship Between Data Variability and Read Length.
The sequencing read length is an important factor that could alter the proportion of aligned reads. Unlike the Illumina platform, the SSP produces different read lengths on one chip. Read length depends on the number of sequencing flows on the SSP. Thus, we ran sequencing samples at the maximum 400 flows to generate the raw data. The mean length of reads was about 120 bp. With a reduced number of flows, the average read length shortened (Fig. S1A). Moreover, if the flow number was reduced to 160, the ratio of usable reads decreased significantly, due to the stricter Burrows–Wheeler Aligner (BWA) mapping criteria with short reads (Fig. S1B).
Discussion
MPS has proved useful in the noninvasive prenatal diagnosis of trisomy 21, trisomy 18, and trisomy 13 based on cell-free fetal DNA in maternal plasma. Here, in a large sample cohort from China, we demonstrated that the SSP can be used in noninvasive prenatal diagnosis with high specificity and sensitivity and may offer several advantages. It is shown to be practical and reliable for the large-scale prenatal diagnosis of fetal aneuploidy. Theoretically, it is possible to identify the presence of aneuploidy in any chromosome, but recent studies have found that a high SD in particular chromosomes affects the precision of aneuploidy determination using the cutoffs of z score > 3 for trisomy and z score < −3 for monosomy.
In our study, we show that, compared with euploid fetuses, differences in the amount of fetal DNA in maternal plasma from chromosomes 21, 18, and 13 contributed by fetuses with trisomy 21, 18, and 13 can be unambiguously detected. Furthermore, differences in the amounts of X chromosome and Y chromosome DNA sequences in maternal plasma contributed by male fetuses compared with female fetuses can be observed robustly and used to diagnose sex chromosome aneuploidies. We first obtained maternal plasma samples from 515 pregnancies that had full karyotyping and used them in a retrospective study to validate our protocol. We detected trisomy 21 with 99.94% sensitivity and 99.46% specificity, which is notable given the importance of diagnosing Down syndrome. Then we used maternal plasma samples from 1,760 pregnancies without karyotyping in a prospective study. In both studies, our method accurately detected trisomy 21, trisomy 18, trisomy 13, and sex chromosome aneuploidy. However, because there is a lack of karyotype results for the prospective study, there is uncertainty about the true sensitivity and specificity of our diagnostic.
Because the z score of aneuploid chromosomes positively correlated with fetal DNA concentration (Materials and Methods), maternal plasma with a low fetal DNA concentration may be assigned a lower z-score cutoff to avoid the incidence of false negatives. In a clinical diagnosis setting, to decrease the risk of false negatives and false positives, we assumed new criteria to identify aneuploidies. A negative result was defined as |z score| < 2, whereas a positive result was defined as |z score| > 4. We found 2 in 1,700 cases in which the z score was between 2 and 3, which led to an ambiguous diagnosis of chromosome number. Repeat sampling confirmed our hypothesis that these maternal plasma samples had a low fetal DNA concentration. Other methods such as karyotyping of fetal cells are required in these instances.
We have also demonstrated that the stability of z scores for chromosomes was related to GC content bias during sequencing. Recent studies have shown the existence of substantial GC bias in MPS platforms such as Illumina/Solexa and ABI/SOLiD, and this limits the sensitivity for detecting trisomy or monosomy. Thus, GC correction was essential to improve the performance of fetal aneuploidy diagnosis. Here, we demonstrate an integrated method to compensate for GC bias in SSP data that is appropriate for the noninvasive detection of fetal aneuploidy from cell-free DNA in maternal plasma. Our method of removing GC bias in sequencing data reveals that the difference in representation among chromosomes within a sample can be normalized. Although our diagnostic performance was of sufficient quality for detection of common fetal aneuploidies, future studies will aim to further reduce the CV so that aneuploidy in any chromosome can be detected.
One of the major factors that limit the sensitivity of diagnosis is the sequencing depth. The amount of sequencing reads per sample could be increased, ensuring that measurements can be made precisely enough to detect quantitative differences in other chromosomes. Consequently, fetal aneuploidy should be detectable not only for the common chromosomal aneuploidies such as trisomy 21, 18, and 13, but for other chromosomal aneuploidies as well. As the fetal DNA fraction varies in the maternal plasma of different individuals and at different stages of pregnancy, it is important to determine the minimum fetal DNA concentration and corresponding sequencing depth required. For instance, by extending our analysis of Fig. S5, we predicted that trisomy or monosomy could be detected by a sequencing throughput of 3.5 million reads. Theoretically, higher sequencing depth could achieve more accurate results, but it would also increase the cost significantly. Therefore, a balance of sequencing cost and diagnostic sensitivity and specificity should be considered to achieve the goal of cost-effective large-scale population screening.
We tested different sequencing read lengths generated by either 400, 260, or 160 flows (see Materials and Methods for definition of flows) on the SSP to determine optimal reads that are long enough to be aligned to the reference genome balanced by a short enough sequencing time to be practical for a diagnostic test. Although longer reads may contribute to a slightly higher alignment rate, the trade-off is a longer sequencing and analysis time. We preferred a 260 flows parameter for optimized performance, due to reads produced by a 160 flows parameter having a significantly lower mapping ratio.
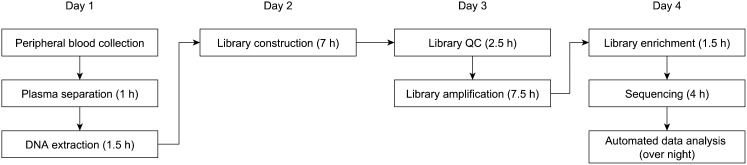
Unlike G-band karyotyping, which is the gold standard for diagnosing fetal aneuploidy in clinical practice, MPS can be performed at the ninth gestational week and uses noninvasive technology. Cell-free fetal DNA sequencing with GC bias statistical correction can accurately identify fetal trisomy 21, trisomy 18, and trisomy 13 with a high detection and low false-positive rate. A minimal 100 ng of genomic DNA extracted from 600 μL of maternal plasma is sufficient to produce reliable diagnostic detection of common fetal aneuploidies using the SSP with a turnaround time of 4 d (Fig. 4).
Fig. 4.
Workflow for the noninvasive prenatal diagnosis of trisomy 21, 18, and 13 using the SSP.
At present, the SSP has a throughput of 13–15 samples per run, and each sample can produce 6–8 million sequencing reads. We expect the cost of sequencing to decrease rapidly and throughput to increase significantly in the near future, which in turn will make this method more robust and affordable.
In summary, we demonstrate a rapid and robust methodology to detect fetal aneuploidies based on the SSP and conducted a large-scale clinical study to systematically identify autosomal and sex chromosomal aneuploidies using cell-free fetal DNA. The SSP is small and portable; therefore, it can be deployed and placed in clinical diagnostic laboratories and is expected to play an increasingly significant role in prenatal diagnosis.
Materials and Methods
Subject Recruitment.
This study was approved by the Institutional Review Boards of Guangzhou Women and Children’s Medical Center, Guangdong Women and Children Hospital, and Guangzhou DaAn Clinical Laboratory Center (Guangzhou, China). Informed consent was obtained from all participants. A total of 2,275 pregnant subjects was recruited. In the retrospective analysis, 515 pregnant subjects with karyotyping results were recruited from Guangzhou Women and Children’s Medical Center and Guangdong Women and Children Hospital. Karyotyping analysis was indicated in these subjects because of increased risk for aneuploidy. Risk factors included advanced maternal age (>30 y old), history of previous miscarriage, positive serum marker screening, or abnormal fetal ultrasound results. Maternal blood was collected before serum marker screening and karyotyping. Samples from pregnancies with known aneuploidies as well as samples from euploid pregnancies were selected to establish our computational diagnostic method. Analysis was performed with blinding to karyotyping results, and subsequently validated using karyotyping results. The remaining 1,760 subjects in the prospective study were randomly selected from Guangzhou DaAn Clinical Laboratory Center. Subjects were excluded from the study if they had undergone in vitro fertilization, blood transfusion within the past year, or immunotherapy within the past 4 wk.
Cell-Free DNA Preparation and Sequencing.
Five to 10 mL of peripheral venous blood was collected from each participating pregnant woman in EDTA-containing tubes or Streck blood collection tubes. The blood samples were first centrifuged at 1,600 × g for 10 min at 4 °C to separate the plasma from peripheral blood cells. The plasma portion was carefully transferred to a polypropylene tube and subjected to centrifugation at 16,000 × g for 10 min at 4 °C to pellet the remaining cells (28). Cell-free DNA from 600 μL of maternal plasma was extracted using the QIAamp DSP DNA Blood Mini Kit (Qiagen) following the blood and body fluid protocol. For the SSP, DNA from maternal plasma was used for library construction according to the Ion Plus Fragment Library Kit (Life Technologies), and semiconductor sequencing was performed using an Ion Proton sequencer at 400 flows according to the manufacturer’s instructions (Life Technologies). The procedure for the SSP can be roughly divided into nine steps: sample collection, plasma separation (1 h), DNA extraction (1.5 h), library construction (7 h), library quality control (2.5 h), library amplification (7.5 h; emulsion PCR using Ion OneTouch 2 Instrument, which enables automated delivery of templated Ion Sphere particles), library enrichment (1.5 h; isolation of template-positive Ion Sphere particles that can be loaded directly onto the Ion semiconductor chip for sequencing), sequencing (4 h), and automated data analysis (8 h overnight) (Fig. 4).
The SSP determines the read sequences not base by base, but by measuring the number of consecutive A, T, G, or C in a sequence (29). Sequencing occurs by flowing one dNTP (base) at a time over the template. dNTPs are flown over the sequencing plate in a determined order. When the nucleotide in the flow is complementary to the template base, the nucleotide is incorporated into the nascent strand by the bound polymerase. This increases the length of the sequencing reads by one base (or more, if a homopolymer stretch is directly downstream).
Data Analysis.
Raw reads with different lengths obtained from the Ion Torrent Suite Software were trimmed from the 3′ end by sequencing quality value of >15 and filtered by read length (<50 bp). The retained reads were aligned to the human genomic reference sequences (hg19) using the BWA (30). Reads that were unmapped or had multiple primary alignment records were filtered by FLAG field in the alignment file, using an in-house Perl script. Duplicate reads were identified by Picard (http://picard.sourceforge.net/) with the parameters “java -jar MarkDuplicates.jar M=picard_duplication_metrics REMOVE_DUPLICATES=true ASSUME_SORTED=true 1” and removed by an in-house Perl script. The remaining reads were considered unique reads for further analysis. To eliminate the effect of GC bias, we applied an integrated method for GC correction using a three-step process: LOESS regression (31), intrarun normalization (32), and linear model regression (33). Briefly, LOESS regression was used to smooth the sequencing bias produced by variable GC content on different chromosomes. All chromosomes were first divided into segments with a bin size of 20 kb. The number of unique reads and GC content (rounded to 0.1%) in each bin were determined. Bins including reference sequences with undeterminable bases and bins without any reads were filtered. Then, using LOESS regression, the fit predicted value (URloess) of each bin was obtained by the number of unique reads (UR) in each bin against the GC content (GCbin) of the corresponding bin according to the equation: . The LOESS-corrected reads number (URcorrected) was calculated using the following equation:
where e(UR) was the expected value for unique reads (UR) of each bin, which was set to the overall average unique reads number in each bin. After LOESS correction, the corrected unique reads number for each chromosome (CR) was added using the LOESS-corrected reads number (URcorrected) in each bin for the corresponding chromosome. In the second step, intrarun normalization was applied to normalize the deviation between samples in one sequencing run. For each sequencing run, the corrected unique reads (CRi,j) on chromosome j in sample i were obtained. Because of the differences among samples, the normalized corrected unique reads () on chromosome j were computed using the equation: . Then the reads ratio (RRi,j) on chromosome j of sample i was calculated as follows: . Because there were GC content disturbances of reads between different sequencing runs and a correlation between the reads ratio (RRi,j) and GC content (Fig. S4), linear regression was used. The linear model was established according to the equation , where GCi was the GC content in sample i, and β was the coefficient factor between the reads ratio (RRi,j) and GC content. The statistical significance was calculated using the linear regression model. The fitting predicted reads ratio () was calculated as . Finally, the residual (ε) obtained by the equation could be fitted to a normal distribution and used to test for chromosomal aneuploidies.
Statistical Analysis.
We derived a z score for each of the chromosomes 21, 18, and 13 in a test sample by subtracting the mean chromosome ratio in a reference set of euploid control pregnancies from the chromosome ratio in a test case and dividing by the SD of the chromosome ratio in the reference set according to the following equation:
where, for a given chromosome, is the residuals of the chromosome to autosomes in the test sample, is the average value of the residuals in reference samples, and is the SD of the residuals in reference samples. In previous studies, a cutoff value of z score > 3 was used to determine whether the ratio of chromosome 21, 18, or 13 was increased and hence fetal trisomy 21, 18, or 13 was present. A z score > 3 represented a chromosome ratio greater than that of the 99.9th percentile of the reference sample set for a one-tailed distribution (23).
We also adapted a method to identify fetuses with sex chromosome aneuploidies. First, z scores for the X and Y chromosomes were generated as described for the autosomes. Then, a least-squares method was applied to establish the relationship between the X and Y chromosomes of a female fetus based on the formula represents the z score for the X chromosome and r represents the coefficient between the X chromosome and Y chromosome. Because human males have one fewer X chromosome than human females, the ratio of read counts for chromosome X in male fetuses should indicate monosomy, in contrast to that of female fetuses. Meanwhile, males have one Y chromosome, but females do not have a Y chromosome; thus, the unique reads number of chromosome Y in male fetuses should be much higher than that in female fetuses. Table 2 shows the criteria used to determine sex chromosome status. represents the fitted z score of chromosome X according to the model
To evaluate the sensitivity and specificity of our approach, we performed cross-validation on pregnancies with full karyotyping. Briefly, we randomly divided the pregnancies into two groups. One was the training set (90% of the samples), and the other was the validation set (10% of the samples). Then, using the training set as a reference, we calculated the z score for each chromosome to be used in the validation set. Sensitivity and specificity were obtained by concordance of the z-score method and karyotyping results. After repeating this method 1,000 times, the final sensitivity and specificity were generated.
The Relationship Between z score and DNA Concentration in Plasma.
For a fetus with trisomy 21, the z score could be calculated by the traditional formula , where R is the reads ratio of the test sample for chromosome 21, Rreference is the mean reads ratio of the reference set for chromosome 21, and SD is the SD of the reads ratio of the reference set for chromosome 21. R is also calculated by the following:
where r is the ratio of normal chromosome 21, and Fc is the DNA concentration in maternal plasma.
In general, r and SD are constant. The z score of the abnormal chromosome correlated positively with plasma DNA concentration.
Supplementary Material
Acknowledgments
This work was supported in part by National Natural Science Foundation of China (Grant 81100435), National Science Foundation for Young Scholars of China (Grant 81000255), National Natural Science Foundation of Guangdong (Grant 915101800 5000001), Key Project of Guangzhou Health Bureau (Grant 201102A212026), Major Project of Guangzhou Science and Technology and Information Bureau (Grant 201300000086), and 973 program (2014CB964900 and 2013CB967500), and Burroughs Welcome Fund.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1321997111/-/DCSupplemental.
References
- 1.Driscoll DA, Gross S. Clinical practice. Prenatal screening for aneuploidy. N Engl J Med. 2009;360(24):2556–2562. doi: 10.1056/NEJMcp0900134. [DOI] [PubMed] [Google Scholar]
- 2.Zhang YP, et al. [Karyotype analysis of amniotic fluid cells and comparison of chromosomal abnormality rate during second trimester] Zhonghua Fu Chan Ke Za Zhi. 2011;46(9):644–648. [PubMed] [Google Scholar]
- 3.Rives N, Siméon N, Milazzo JP, Barthélémy C, Macé B. Meiotic segregation of sex chromosomes in mosaic and non-mosaic XYY males: Case reports and review of the literature. Int J Androl. 2003;26(4):242–249. doi: 10.1046/j.1365-2605.2003.00421.x. [DOI] [PubMed] [Google Scholar]
- 4.Park JH, et al. Effects of sex chromosome aneuploidy on male sexual behavior. Genes Brain Behav. 2008;7(6):609–617. doi: 10.1111/j.1601-183X.2008.00397.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tartaglia NR, Howell S, Sutherland A, Wilson R, Wilson L. A review of trisomy X (47,XXX) Orphanet J Rare Dis. 2010;5(1):8. doi: 10.1186/1750-1172-5-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sybert VP, McCauley E. Turner’s syndrome. N Engl J Med. 2004;351(12):1227–1238. doi: 10.1056/NEJMra030360. [DOI] [PubMed] [Google Scholar]
- 7.Bouchlariotou S, et al. Turner’s syndrome and pregnancy: Has the 45,X/47,XXX mosaicism a different prognosis? Own clinical experience and literature review. J Matern Fetal Neonatal Med. 2011;24(5):668–672. doi: 10.3109/14767058.2010.520769. [DOI] [PubMed] [Google Scholar]
- 8.Leggett V, Jacobs P, Nation K, Scerif G, Bishop DV. Neurocognitive outcomes of individuals with a sex chromosome trisomy: XXX, XYY, or XXY: A systematic review. Dev Med Child Neurol. 2010;52(2):119–129. doi: 10.1111/j.1469-8749.2009.03545.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hofherr SE, Wiktor AE, Kipp BR, Dawson DB, Van Dyke DL. Clinical diagnostic testing for the cytogenetic and molecular causes of male infertility: The Mayo Clinic experience. J Assist Reprod Genet. 2011;28(11):1091–1098. doi: 10.1007/s10815-011-9633-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Korteweg FJ, et al. Cytogenetic analysis after evaluation of 750 fetal deaths: Proposal for diagnostic workup. Obstet Gynecol. 2008;111(4):865–874. doi: 10.1097/AOG.0b013e31816a4ee3. [DOI] [PubMed] [Google Scholar]
- 11.Bin W, Yingyao C, Qi S. The economic burden of Down's syndrome in China. Chin Health Econ. 2006;25(3):24–26. [Google Scholar]
- 12.Williams ES, Cornforth MN, Goodwin EH, Bailey SM. CO-FISH, COD-FISH, ReD-FISH, SKY-FISH. Methods Mol Biol. 2011;735(1):113–124. doi: 10.1007/978-1-61779-092-8_11. [DOI] [PubMed] [Google Scholar]
- 13.Leung TY, et al. Identification of submicroscopic chromosomal aberrations in fetuses with increased nuchal translucency and apparently normal karyotype. Ultrasound Obstet Gynecol. 2011;38(3):314–319. doi: 10.1002/uog.8988. [DOI] [PubMed] [Google Scholar]
- 14.Vermeesch JR, et al. Guidelines for molecular karyotyping in constitutional genetic diagnosis. Eur J Hum Genet. 2007;15(11):1105–1114. doi: 10.1038/sj.ejhg.5201896. [DOI] [PubMed] [Google Scholar]
- 15.American College of Obstetricians and Gynecologists ACOG Practice Bulletin No. 88, December 2007. Invasive prenatal testing for aneuploidy. Obstet Gynecol. 2007;110(6):1459–1467. doi: 10.1097/01.AOG.0000291570.63450.44. [DOI] [PubMed] [Google Scholar]
- 16.Nanal R, Kyle P, Soothill PW. A classification of pregnancy losses after invasive prenatal diagnostic procedures: An approach to allow comparison of units with a different case mix. Prenat Diagn. 2003;23(6):488–492. doi: 10.1002/pd.623. [DOI] [PubMed] [Google Scholar]
- 17.Tabor A, Alfirevic Z. Update on procedure-related risks for prenatal diagnosis techniques. Fetal Diagn Ther. 2010;27(1):1–7. doi: 10.1159/000271995. [DOI] [PubMed] [Google Scholar]
- 18.Norton ME, et al. Non-Invasive Chromosomal Evaluation (NICE) Study: Results of a multicenter prospective cohort study for detection of fetal trisomy 21 and trisomy 18. Am J Obstet Gynecol. 2012;207(2):e1–e8. doi: 10.1016/j.ajog.2012.05.021. [DOI] [PubMed] [Google Scholar]
- 19.Nicolaides KH, Syngelaki A, Ashoor G, Birdir C, Touzet G. Noninvasive prenatal testing for fetal trisomies in a routinely screened first-trimester population. Am J Obstet Gynecol. 2012;207(5):e1–e6. doi: 10.1016/j.ajog.2012.08.033. [DOI] [PubMed] [Google Scholar]
- 20.Palomaki GE, et al. DNA sequencing of maternal plasma to detect Down syndrome: An international clinical validation study. Genet Med. 2011;13(11):913–920. doi: 10.1097/GIM.0b013e3182368a0e. [DOI] [PubMed] [Google Scholar]
- 21.Bianchi DW, et al. Genome-wide fetal aneuploidy detection by maternal plasma DNA sequencing. Obstet Gynecol. 2012;119(5):890–901. doi: 10.1097/AOG.0b013e31824fb482. [DOI] [PubMed] [Google Scholar]
- 22.Bianchi DW, et al. DNA sequencing versus standard prenatal aneuploidy screening. N Engl J Med. 2014;370(9):799–808. doi: 10.1056/NEJMoa1311037. [DOI] [PubMed] [Google Scholar]
- 23.Chiu RW, et al. Noninvasive prenatal diagnosis of fetal chromosomal aneuploidy by massively parallel genomic sequencing of DNA in maternal plasma. Proc Natl Acad Sci USA. 2008;105(51):20458–20463. doi: 10.1073/pnas.0810641105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fan HC, Blumenfeld YJ, Chitkara U, Hudgins L, Quake SR. Noninvasive diagnosis of fetal aneuploidy by shotgun sequencing DNA from maternal blood. Proc Natl Acad Sci USA. 2008;105(42):16266–16271. doi: 10.1073/pnas.0808319105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dan S, et al. Clinical application of massively parallel sequencing-based prenatal noninvasive fetal trisomy test for trisomies 21 and 18 in 11,105 pregnancies with mixed risk factors. Prenat Diagn. 2012;32(13):1225–1232. doi: 10.1002/pd.4002. [DOI] [PubMed] [Google Scholar]
- 26.Koboldt DC, Ding L, Mardis ER, Wilson RK. Challenges of sequencing human genomes. Brief Bioinform. 2010;11(5):484–498. doi: 10.1093/bib/bbq016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fan HC, Quake SR. Sensitivity of noninvasive prenatal detection of fetal aneuploidy from maternal plasma using shotgun sequencing is limited only by counting statistics. PLoS One. 2010;5(5):e10439. doi: 10.1371/journal.pone.0010439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chiu RW, et al. Effects of blood-processing protocols on fetal and total DNA quantification in maternal plasma. Clin Chem. 2001;47(9):1607–1613. [PubMed] [Google Scholar]
- 29.Rothberg JM, et al. An integrated semiconductor device enabling non-optical genome sequencing. Nature. 2011;475(7356):348–352. doi: 10.1038/nature10242. [DOI] [PubMed] [Google Scholar]
- 30.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Alkan C, et al. Personalized copy number and segmental duplication maps using next-generation sequencing. Nat Genet. 2009;41(10):1061–1067. doi: 10.1038/ng.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jiang F, et al. Noninvasive Fetal Trisomy (NIFTY) test: An advanced noninvasive prenatal diagnosis methodology for fetal autosomal and sex chromosomal aneuploidies. BMC Med Genomics. 2012;5(1):57. doi: 10.1186/1755-8794-5-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen EZ, et al. Noninvasive prenatal diagnosis of fetal trisomy 18 and trisomy 13 by maternal plasma DNA sequencing. PLoS One. 2011;6(7):e21791. doi: 10.1371/journal.pone.0021791. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.