Significance
African trypanosomes are parasites that can cause African sleeping sickness in humans. Humans and some primates, but not other mammals, have a gene called APOL1 that protects against certain trypanosomes. Genetic variants in APOL1 that arose in Africa are strongly associated with kidney disease in African Americans. These kidney disease-associated variants may have risen to high frequency in Africa because they can defend humans against a particularly pathogenic trypanosome. In this paper, we show how APOL1 has evolved by analyzing the distribution of these variants in Africa and then elucidating the molecular mechanisms that enhance their trypanosome killing capacity. We also show that these antitrypanosomal APOL1 variants may have adverse consequences for the host.
Abstract
ApolipoproteinL1 (APOL1) protects humans and some primates against several African trypanosomes. APOL1 genetic variants strongly associated with kidney disease in African Americans have additional trypanolytic activity against Trypanosoma brucei rhodesiense, the cause of acute African sleeping sickness. We combined genetic, physiological, and biochemical studies to explore coevolution between the APOL1 gene and trypanosomes. We analyzed the APOL1 sequence in modern and archaic humans and baboons along with geographic distribution in present day Africa to understand how the kidney risk variants evolved. Then, we tested Old World monkey, human, and engineered APOL1 variants for their ability to kill human infective trypanosomes in vivo to identify the molecular mechanism whereby human trypanolytic APOL1 variants evade T. brucei rhodesiense virulence factor serum resistance-associated protein (SRA). For one APOL1 kidney risk variant, a two-residue deletion of amino acids 388 and 389 causes a shift in a single lysine residue that mimics the Old World monkey sequence, which augments trypanolytic activity by preventing SRA binding. A second human APOL1 kidney risk allele, with an amino acid substitution that also restores sequence alignment with Old World monkeys, protected against T. brucei rhodesiense due in part to reduced SRA binding. Both APOL1 risk variants induced tissue injury in murine livers, the site of transgenic gene expression. Our study shows that both genetic variants of human APOL1 that protect against T. brucei rhodesiense have recapitulated molecular signatures found in Old World monkeys and raises the possibility that APOL1 variants have broader innate immune activity that extends beyond trypanosomes.
Coding variants of the APOL1 gene, found only in individuals of recent African ancestry, are strongly associated with kidney disease in African Americans (1, 2). Two main coding variants associated with renal disease have been identified: the G1 allele, consisting of two amino acid substitutions (NM_003661.3 p.S342G and p.I384M) in near-perfect linkage, and the G2 allele, defined by a 2-aa deletion at the C terminus (NM_003661.3 p.delN388/Y389). Two copies of either of these renal risk variants (RRVs) cause a 7- to 10-fold increased risk of hypertension-associated end-stage renal disease (1, 3), a 10- to 17-fold increase in focal and segmental glomerulosclerosis (a primary disease of the renal microvasculature) (1, 4), and a 29-fold increase in HIV nephropathy (4). The high frequency of these RRVs in African Americans and the Yoruba (Nigeria) and their absence in Europeans and Asians suggest that positive selection increased the frequency of these APOL1 RRVs in Africa (1).
The APOL family has evolved rapidly during primate evolution (5). The APOL gene family on human chromosome 22 consists of six homologous genes in close proximity that arose through gene duplication. Although several APOL genes are widespread in the animal kingdom, APOL1 is restricted to primates. Only humans, gorillas, and baboons are known to express functional APOL1, and serum from each of these primate species is trypanolytic (6, 7). Human and gorilla sera protect against Trypanosoma brucei brucei, whereas baboon sera protect against T. b. brucei, Trypanosoma brucei rhodesiense, and potentially, Trypanosoma brucei gambiense (8, 9). Human-infective T. b. rhodesiense, which causes acute African sleeping sickness, arose from animal-infective T. b. brucei through the evolution of a virulence factor called serum resistance-associated protein (SRA) (10) that binds to (11) and prevents human APOL1-mediated lysis (12), whereas T. b. gambiense evolved different mechanisms to neutralize human APOL1 (13–15).
APOL1 is the main trypanolytic component of human trypanosome lytic factor (TLF). TLF consists of two HDL subfractions (TLF1 and TLF2) that contain apolipoprotein A-I and two unique components: APOL1 and haptoglobin-related protein (16, 17). TLF1 is a large (500 kDa), dense (1.25 g/mL) HDL complex (18, 19). T. brucei takes up this complex in a receptor-dependent process through the binding of the haptoglobin-related protein–hemoglobin (Hb) complex component of TLF by the trypanosome haptoglobin–Hb receptor (HpHbR) (20, 21). TLF2 is a 1,000-kDa complex of lipid-poor TLF1 and IgM, which is taken up independently by another mechanism (17, 20). Upon TLF binding to HpHbR, TLF1/APOL1 traffics to the lysosome, where a pH-dependent conformational switch is believed to trigger APOL1 insertion into the endosomal/lysosomal membrane (12). Here, APOL1 forms a pore permeable to monovalent ions, leading to lysosomal membrane depolarization, depolarization of the plasma membrane, osmotic swelling, and rupture (22, 23).
In humans infected with T. b. rhodesiense, the SRA virulence factor binds to and inactivates APOL1 in the endosomal and lysosomal compartments, allowing the trypanosomes to proliferate in the presence of most human APOL1 (11, 12). However, sera from subjects harboring either the G1 or G2 RRVs killed T. b. rhodesiense in an in vitro study (1). Presumably, these data indicate that APOL1 RRVs can evade SRA neutralization to maintain trypanolytic activity. Although the presence of the G2 allele did, indeed, diminish the APOL1/SRA interaction in that study, the G1 allele did not alter in vitro SRA binding (1). Given that a single copy of an APOL1 RRV allele seems sufficient to protect against acute human African trypanosomiasis (T. b. rhodesiense) in vitro, the protection conferred by APOL1 RRVs suggests a potential explanation for their rapid increase in frequency in sub-Saharan Africa (1).
In this report, we consider the evolution of APOL1 variants and their functional properties, combining genetic, physiological, and biochemical studies to understand the distant and more recent evolution of APOL1. We compare APOL1 among humans, gorillas, and Old World monkeys (OWMs) and among living and extinct human populations to define the spectrum of variation and then test the potential functional significance of that variation. The biology of these variants presented here could help illuminate the nature of APOL1/trypanosome coevolution, the molecular mechanisms providing host defense against trypanosomes, and perhaps, the biological basis of APOL1-mediated kidney injury.
Results
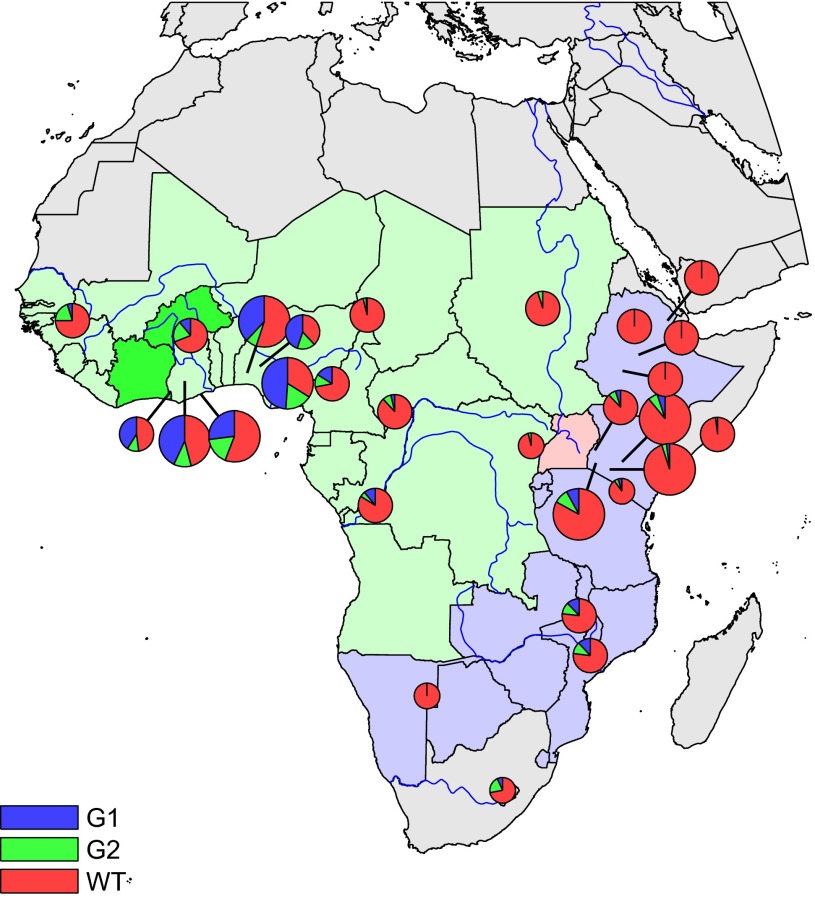
We surveyed 1,759 Africans for the presence of the G1 and G2 APOL1 RRVs. G1 consists of two coding variants in near-perfect linkage disequilibrium: a serine to glycine substitution at position 342 [p.S342G; rs73885319 (designated G1S342G in this report)] and an isoleucine to methionine substitution at position 384 [p.I384M; rs60910145 (designated G1I384M)]. G2 consists of a 2-aa deletion at positions 388 and 389 [p. delN388/Y389; rs71785313 (designated G2)]. We genotyped eight tribal groups from across sub-Saharan Africa: four groups from regions east and four groups from regions west of the Rift Valley. Frequency distributions of G1 and G2 are shown in Fig. 1 and Table 1 along with previously reported frequencies in smaller populations. High frequencies of G1 were concentrated in West Africa, whereas G2 had a more even distribution continent-wide. Given the large number of known polymorphisms that seem to be protective against malaria, we included 469 severe malaria cases and 426 nonmalarial controls from Luo in Kenya to consider severe malaria as a potential selective pressure. The frequencies of G1 and G2 did not differ between malaria cases and controls.
Fig. 1.
Distribution of the G1 and G2 APOL1 variants across Africa. Allele frequencies of the G1 and G2 variants are indicated as blue and green wedges, respectively. Circle size reflects the number of individuals genotyped: small, <10 individuals/20 chromosomes; medium, 10–100 individuals/20–200 chromosomes; large, >100 individuals/200 chromosomes. Countries are shaded according to the subspecies of Trypanosoma brucei that cause African sleeping sickness. Darker green, gambiense types 1 and 2; light green, gambiense type 1; pink, both rhodesiense and gambiense type 1; purple, rhodesiense.
Table 1.
Numerical data corresponding to the pie charts in Fig. 1
| Country | Population | N | Latitude | Longitude | % of G1 | % of G2 | Ref. |
| Ghana | Akan | 171 | 6.7 | −1.6 | 43 | 11 | New |
| Ghana | Ga-Adangbe | 139 | 5.6 | −0.2 | 27 | 17 | New |
| Nigeria | Ibo | 190 | 6.5 | 7.5 | 49 | 17 | New |
| Kenya | Kikuyu | 112 | −0.4 | 37 | 5 | 6 | New |
| Kenya | Luo | 895 | −0.5 | 34.7 | 8 | 9 | New |
| Kenya | Masai | 102 | −1.1 | 35.9 | 2 | 3 | New |
| Somalia | Somali | 30 | 2 | 45.4 | 0 | 2 | New |
| Nigeria | Yoruba | 113 | 7.4 | 3.9 | 38 | 7 | Hapmap |
| Kenya | Luyha | 90 | 0.6 | 34.6 | 5 | 7 | Hapmap |
| Kenya | Bantu_NE | 12 | −3 | 37 | 5 | 5 | HGDP |
| South Africa | Bantu_SA | 8 | −29.3 | 27.8 | 7 | 21 | HGDP |
| Central Africa Republic | Biaka Pygmy | 36 | 4 | 17 | 4 | 8 | HGDP |
| Senegal | Mandenka | 24 | 12 | −12 | 5 | 20 | HGDP |
| Democratic Republic of Congo | Mbuti Pygmy | 15 | 1 | 29 | 0 | 4 | HGDP |
| Namibia | San | 7 | −21 | 20 | 0 | 0 | HGDP |
| Nigeria | Yoruba | 25 | 8 | 5 | 45 | 17 | HGDP |
| Ghana | Bulsa | 22 | 10.7 | −1.3 | 11 | 20 | U.C.L. |
| Ghana | Asante | 35 | 5.8 | −2.8 | 41 | 11 | U.C.L. |
| Cameroon | Somie | 65 | 6.5 | 11.45 | 16 | 12 | U.C.L. |
| Congo | Bakongo | 55 | −4.3 | 15.28 | 11 | 5 | U.C.L. |
| Malawi | Chewa | 50 | −14 | 33.7 | 12 | 11 | U.C.L. |
| Mozambique | Sena | 51 | −17.5 | 35 | 12 | 11 | U.C.L. |
| Sudan | Kordofan | 30 | 13.1 | 30.35 | 0 | 5 | U.C.L. |
| Cameroon | Shewa | 64 | 12.5 | 14.5 | 1 | 3 | U.C.L. |
| Ethiopia | Afar | 76 | 12 | 41.5 | 0 | 0 | U.C.L. |
| Ethiopia | Amhara | 76 | 11.5 | 38.5 | 0 | 0 | U.C.L. |
| Ethiopia | Oromo | 76 | 9 | 38.7 | 0 | 0 | U.C.L. |
| Ethiopia | Maale | 76 | 7.6 | 37.2 | 0 | 0 | U.C.L. |
Country, tribe, number (N) of individuals genotyped, latitude, longitude, percent of chromosomes with G1 and G2, and reference dataset (Ref.) are shown. Datasets include HapMap (genotyped in refs. 1 and 4), Human Genome Diversity Project (HGDP; genotyped in ref. 4), and The Centre for Anthropology at University College London (U.C.L.; genotyped in ref. 2).
Previous studies in Yoruba strongly suggested positive selection for the G1 allele using long-haplotype tests (1). Long-haplotype tests are performed within one population to identify selective sweeps based on the ratio of extended haplotype homozygosity associated with the selected vs. the nonselected allele (24). Another approach to identify natural selection is testing of genetic differentiation between populations: natural selection may increase or decrease the frequency of a genetic variant depending on how it alters fitness. Here, we genotyped G1 and G2 in the Yoruba from West Africa (Nigeria) and Luhya, a tribe in Eastern Africa (Kenya) that is genetically very similar to the Yoruba across the genome [fixation index (Fst) = 0.008] (25). We compared frequency differences using a statistical test for differentiation under a neutral model of genetic drift that accounts for random variation in allele frequency (Methods) (26). Of all 1,440,616 markers from HapMap3 that we compared across the genome in the two tribes, none were as differentiated between the two tribes as G1 (P = 5.11 × 10−7) (Table 2). We concluded that genetic drift was unlikely to explain the large difference in frequency in otherwise highly genetically similar groups and that selection was responsible for this differentiation pattern. Other SNPs close to G1 showed weaker signals for selection (P = 10−4–10−5), probably caused by allelic hitchhiking, because none were as differentiated as G1.
Table 2.
SNPs with the highest population differentiation between Yoruba and Luhya
| Chromosome | rsID | NCBI36/HG18 | NCBI37/HG19 | Ref. | Alt. | YRI | LWK | P value | Gene |
| chr22 | rs73885319 | 34991852 | 36661906 | A | G | 0.38 | 0.05 | 5.11E-07 | APOL1 |
| chr3 | rs7649861 | 145653390 | 144170700 | A | G | 0.15 | 0.53 | 5.49E-07 | |
| chr7 | rs6944302 | 79942827 | 80104891 | C | T | 0.48 | 0.11 | 5.96E-07 | CD36 |
| chr5 | rs6879787 | 141817918 | 141837734 | T | G | 0.26 | 0 | 1.98E-06 | |
| chr7 | rs6967965 | 79404014 | 79566078 | T | C | 0.37 | 0.07 | 6.04E-06 | CD36 |
| chr18 | rs569176 | 53998378 | 55847380 | C | T | 0.31 | 0.04 | 8.58E-06 | |
| chr7 | rs6949761 | 79347299 | 79509363 | C | T | 0.4 | 0.09 | 9.64E-06 | CD36 |
| chr6 | rs3094159 | 29943813 | 29835834 | G | A | 0.53 | 0.85 | 1.15E-05 | HLA |
| chr6 | rs3132712 | 29949000 | 29841021 | A | G | 0.53 | 0.85 | 1.15E-05 | HLA |
| chr6 | rs3115619 | 29950858 | 29842879 | A | G | 0.53 | 0.85 | 1.15E-05 | HLA |
Frequencies of >1.4 million SNPs were tested in Yoruba (Nigeria) and Luhya (Kenya), two tribes with very high genetic similarity (Fst = 0.008). The 10 most differentiated SNPs after modeling genetic drift and sampling variance are shown here, with G1 showing the strongest statistical evidence for differentiation among all SNPs tested. Alt., alternate allele; LWK, Luhya; NCBI/HG, genomic location; Ref., reference allele; rsID, SNP ID; YRI, Yoruba.
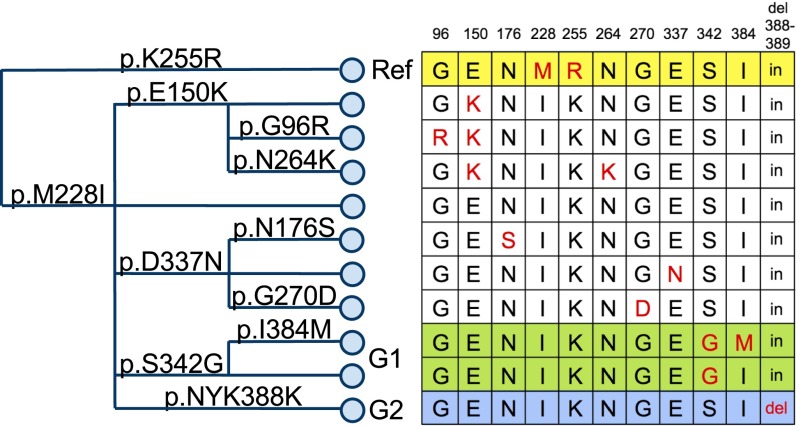
We analyzed all APOL1 variants in a haplotype analysis to assess broader African vs. non-African signatures. We focused on the 8-kbp APOL1 gene region that is between two recombination hotspots using genotype data from the 1000 Genomes Project (27) Phase I, comprising 1,092 individuals from different continents, as well as haplotype data from the Neandertal (28) and Denisova (29) genomes. Fig. 2, a hierarchical cluster tree of all unique haplotypes defined by variants in this segment with frequency ≥1% together with Neandertal and Denisova haplotypes, shows how APOL1 has diverged over time and how different APOL1 haplotypes are related to each other. Fig. 3 (with frequency data in Table S1) shows just the coding variants, which are most likely to be functional, and it includes lower frequency alleles not shown in Fig. 2. The haplotypes that we observed fall broadly into African and out-of-Africa haplotypes. One haplotype (defined by 255R and 228M) was almost exclusively found in Europeans and Asians in this dataset, whereas several haplotypes are found solely in Africans. A low frequency of East Africans with evidence of distant European admixture (Luhya) had the out-of-Africa haplotype, but the Yoruba, a West African tribe selected as most representative of pre-Bantu expansion Africans, did not. Neandertal and Denisova haplotypes, with the exception of rs132162 (and rs28391521 for Denisova), always match the out-of-Africa haplotype (p.K255R and p.I228M) at SNPs polymorphic in the human population; nine of these SNPs are derived variants specific to the out-of-Africa lineage. Notably, the hg19 human reference genome sequence at APOL1 matches this out-of-Africa haplotype shared with Neandertal and Denisova. This shared haplotype might antedate the split of these Homo lineages or may have been introduced into modern humans after the out-of-Africa expansion through interbreeding with the archaic population (introgression). The sharing of nearly all derived alleles over this 8-kbp region between the out-of-Africa lineage and Neandertal suggests that this APOL1 haplotype was reintroduced into humans after the out-of-Africa expansion by interbreeding rather than maintained as a shared haplotype in both humans and archaic humans over the ∼500,000 y since the common lineage split. The shared out-of-Africa/archaic haplotype is present in ∼22% of modern European chromosomes (which harbor 2% Neandertal DNA genome-wide). This enrichment suggests a selective event but could not be definitively ascribed to selection with currently available datasets.
Fig. 2.
APOL1 variation in modern and archaic humans. Each leaf of the polymorphism tree (Left) and the corresponding horizontal row of the haplotype table (Right) represent a different haplotype for the 8-kbp APOL1 region between rs136150 and rs9610474 observed among 1,092 samples analyzed in phase I of the 1000 Genomes Project. Numbers to the right of the polymorphism tree reflect the numbers of times that haplotype was observed (only haplotypes at a frequency of ≥1% are shown). In the polymorphism tree, the x axis measures the average distance between haplotypes in the number of base pair differences. Haplotypes observed in out-of-Africa samples are displayed in cyan, whereas all other haplotypes are displayed in red. In the haplotype table, horizontal rows represent haplotypes across the 8-kbp region; at each polymorphic locus, ancestral alleles are green, derived alleles are red, and blue indicates that the allele could not be ascertained. Each column of the haplotype table corresponds to a different biallelic marker (i.e., base pair); rs markers are color coded: nonsynonymous coding variants are in red, synonymous coding variants are in green, and noncoding variants are in black. The base pairs defining the G1 and G2 variants are boxed in blue. Introns (thin lines) and exons (wide bars) in this 8-kbp window are illustrated above the haplotype table. Importantly, the diversity observed among the out-of-Africa haplotypes is much lower than the diversity observed in the remaining haplotypes, consistent with the hypothesis that these out-of-Africa haplotypes all originated from a recent ancestor, which would be the case for introgression from archaic humans. D, deletion; I, insertion; Ref, human genome reference.
Fig. 3.
Coding variants in APOL1. The haplotype tree (Left) is constructed from coding variants in APOL1 identified at least three times in phase 1 of the 1000 Genomes Project. The tree is read from left to right, with each branch point representing an amino acid change that creates a new haplotype (i.e., set of particular variants on a single chromosome). The 11 haplotypes created by these 11 amino acid changes are illustrated in the chart (Right). In the chart, each row represents a haplotype, with the red allele marking the haplotype-defining residues. Columns show the amino acid positions where the haplotypes may differ. p.K255R defines the haplotype shared by Neanderthal and ∼22% of Europeans. This haplotype is the reference (Ref; yellow) sequence in the current draft of the human genome (hg19). Nearly all individuals with the G1 allele (green) have both p.S342G and p.I384M, but a few individuals have p.S342G without p.I384M (no individuals have yet been identified with p.I384M but without p.S342G). p.NYK388K (blue), a deletion of asparagine and tyrosine at positions 388 and 389, defines the G2 allele. Frequencies for each allele are reported in Table S1.
Because the role of APOL1 is central to the innate immunity mechanism that prevents trypanosomiasis, we sought molecular evidence that this role may have contributed to APOL1 evolution in primates and humans. We began by comparing APOL1 DNA sequences in primates known to have trypanolytic activity in their sera [humans and gorilla (Gorilla gorilla)] and cloning and sequencing APOL1 from several OWMs, including baboon (Papio), mangabey (Cercocebus), and mandrill (Mandrillus). In silico analysis predicts that all primate APOL1 proteins retain a generally conserved structural organization, defined by the positions of putative α-helices (PROFSec, prediction of secondary structure; www.predictprotein.org) and the presence of three predicted transmembrane helices, including a core hairpin of two helices and a C-terminal helix (Fig. S1, underlined) (TM-Pred, transmembrane prediction; www.expasy.org), and terminating with a predicted coiled coil/amphipathic helix. APOL1 amino acid sequences of OWMs were greater than 95% identical between species and 60% identical to gorilla and human sequences. Human and gorilla amino acid sequences were 97% identical (Fig. S1).
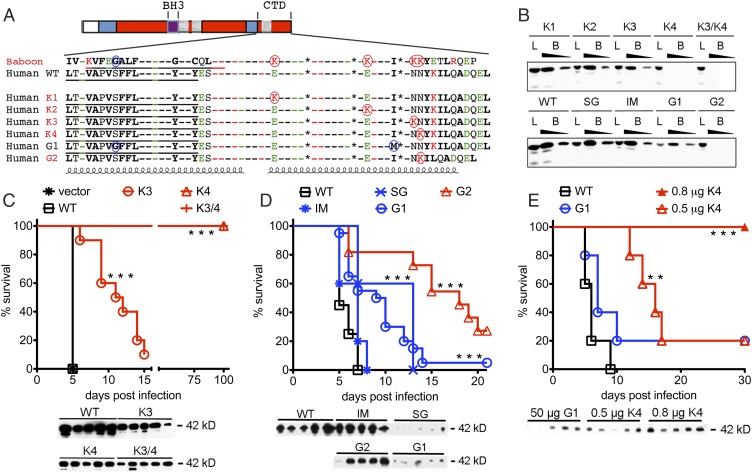
Unlike most human APOL1, baboon APOL1 can kill human-infective T. b. rhodesiense, which expresses SRA. We found that the G1 and G2 human variants partially recapitulate the molecular characteristics of trypanolytic OWM APOL1 at their C terminus. The baboon APOL1 C terminus (defined as the residues comprising the third hydrophobic helix and the C terminus) has four lysines not shared with human APOL1 (K1–K4) (Fig. 4A). The presence of tandem lysines 3 and 4 is sufficient to evade binding to, and thus neutralization by, SRA in vivo (8). All APOL1 C termini were predicted to form a coiled coil because of four heptad repeats containing nonpolar residues that generate a helix with a hydrophobic face. The G1S342G polymorphism replaces a polar for a nonpolar amino acid in the third hydrophobic helix, generating an isoform identical to baboon APOL1 at the corresponding amino acid residue. The G1I384M substitution maintains the hydrophobic face in the predicted C-terminal coiled coil domain. Similarly, the 2-aa deletion in G2 shifts the final leucine in the heptad repeat motif out of alignment while maintaining the hydrophobic face; more importantly, it places a lysine residue into register with the K4 lysine in the baboon and the OWM sequences (Fig. 4A).
Fig. 4.
C-terminal lysine residues determine APOL1/SRA binding interactions and protection against SRA-expressing trypanosomes in vivo. (A) The C-terminal domains of primate APOL1 variants. Residue identities are shown as dashes if they are identical in all sequences; otherwise, residue identities are indicated (bold, nonpolar residues; green, acidic residues; red, basic residues). Underlined hydrophobic sequences are putative membrane-spanning regions. Residues specific to G1 are circled in blue, and those lysines that are specific to baboon or G2 are circled in red. Coils denote putative α-helices; the C-terminal helix contains a heptad repeat sequence (a–g), wherein nonpolar residues occupy a and d positions. Asterisks denote the positions of the heptad-repeat leucine residues at the d position, the last of which is missing in G2 and replaced by a nonpolar alanine. Four lysine residues that are found in OWM but not human WT APOL1 are denoted K1–K4. The 2-aa deletion in the APOL1 G2 variant shifts a lysine residue into register with baboon K4. (B) Transgenic mouse sera containing comparable amounts of the indicated APOL1 variant were incubated at pH 5.0 with NHS-agarose–coupled SRA. APOL1 was detected in the unwashed suspension [load (L); diluted 1:40] and the SRA-bound fraction eluted with 0.1 M Tris [pH 8.0; bound (B); diluted 1:40 and 1:80] by immunoblotting. SG, S342G variant alone; IM, I384M variant alone. (C) Human APOL1 gene variants encoding baboon-specific lysine substitutions K3, K4, or both K3 and K4 genes were expressed in mice by HGD. (D and E) WT, SG, IM, G1, and G2 human APOL1 genes were expressed in mice by HGD. The amount of plasmid injected was 50 μg (C and D), unless otherwise indicated (E). Control mice received an empty plasmid (vector) or a plasmid containing human APOL1 (WT). On day 3 postinjection, mice were infected with 5,000 human-infective T. brucei-SRA, and their blood was monitored for parasites. When parasitemia reached 109 parasites/mL, the mouse was killed; mice that survived to 21 d never developed parasitemia. Mouse plasma protein levels of each APOL1 construct were determined by immunoblot on the day of infection using a rabbit polyclonal antibody (C; plasma diluted 1:40) or a mouse monoclonal antibody (D and E). Results in D are pooled from multiple experiments. Each experiment had at least five mice per group. Survival was significantly different from WT controls. **P < 0.01; ***P < 0.001; log-rank test.
To determine if the human APOL1 RRVs could kill SRA-expressing trypanosomes in vivo, we tested various human and primate APOL1 isoforms for trypanolytic capacity using hydrodynamic gene delivery (HGD) in Swiss–Webster mice. We included independent tests of both G1 variants together (G1dbl) and individually (G1S342G and G1I384M), because we have identified a rare African haplotype with only p.S342G (4). After injection of recombinant cDNA, the mice were screened for the production of APOL1 protein in their sera. This technique produces functional apolipoprotein in its native form that circulates bound to HDL, as it is found circulating in primate sera (8). The mice were then infected with T. b. brucei trypanosomes that express SRA, which are resistant to killing by normal human serum (T. brucei-SRA; engineered to stably produce a constant level of SRA). As previously shown, mice treated with vector or the out-of-Africa haplotype (WT) APOL1 succumbed to the infection within 7 d because of overwhelming trypanosome parasitemia in the blood. In contrast, mice producing genetically engineered human APOL1 mutants with OWM-specific K4 and K3/4 residues never developed parasitemia and were protected indefinitely from infection (Fig. 4C). Although the K3 OWM APOL1 extended median survival by 6 d compared with human WT APOL1, delaying but not preventing the onset of parasitemia, the OWM K4 lysine residue conferred full protection against T. brucei-SRA (Fig. 4C), identifying K4 as the crucial lysine for resistance to T. b. rhodesiense. Most notably, the human APOL1 G2 isoform, with a lysine in the K4 position because of the 2-aa deletion, extended the median survival by 11 d (Fig. 4D), affording significant protection against infection by SRA-expressing trypanosomes.
Although we frequently noted much lower serum levels of G1 and p.S342G APOL1 in infected mice compared with other isoforms, APOL G1dbl and G1S342G isoforms still prolonged median survival time by 7 d (Fig. 4D). Reduced protection by G1 was not caused by reduced protein levels alone, because significant protection against T. brucei-SRA was observed by K4 even after titration such that circulating levels of K4 protein were similar to G1 (Fig. 4E).
To test whether in vivo protection against T. brucei-SRA depends on reduced affinity of SRA for APOL1 variants, we used recombinant SRA N-terminal domain as bait to precipitate variant APOL1 proteins, which are integral components of HDLs, from the sera of transgenic mice. In this assay, SRA bound to APOL1 WT, K1, and K2, consistent with their susceptibility to T. brucei-SRA infection. SRA bound K3 and the APOL1 G1dbl and individual G1S342G and G1I384M variant mutations (Fig. 4B); however, K3, APOL1 G1dbl, and individual G1S342G variants were partially protective against T. brucei-SRA infection. In contrast to previously published in vitro data showing limited trypanolytic activity (1), the G1I384M variant was not trypanolytic at all in vivo. SRA did not bind to K4, K3/4, or the human APOL1 G2 variant, all of which share the lysine in the K4 position and were the most protective against infection.
To elucidate the partial protective capacity conferred by APOL1 G1 compared with other variants, we measured the binding affinity between recombinant SRA and APOL1 across a physiological range of pH. This approach mimics the environment during trafficking of TLF through the endosomes and into the lysosome of the parasite. The assay used surface plasmon resonance to measure the binding of SRA to an immobilized synthetic peptide corresponding to the C-terminal 34 residues (coiled coil domain) of human WT APOL1 and variants (Table S2 and Fig. S2). As expected, SRA binding to WT APOL1 was pH-dependent, with maximum affinity (Kd ∼ 4 nM) at pH 4.5 (equivalent to the pH of the lysosome). The affinities at pH 4.5 for SRA binding to the peptides corresponding to the G1I384M and G2 variants were reduced, with estimates of two- (G1I384M) and fivefold (G2) changes in Kd. Raising the pH decreased the binding affinity for all human peptides tested, and the first detectable binding was at pH 6.1. In contrast, no binding was detected at any of the pH values tested to a peptide corresponding to the C terminus of APOL1, consistent with the ability of baboon APOL1 to confer full protection from trypanosomes. These data confirm APOL1–SRA interactions and provide much more detail on the pH-dependent affinity of the interaction (1, 8, 30), thereby explaining some of the variability in APOL1 trypanolytic capacity.
Because HGD leads to gene expression primarily in the mouse liver, we examined livers 5 d post-HGD to determine why G1 and p.S342G APOL1 levels were frequently low in sera of treated mice (Fig. 5). Whereas at 5 d postinjection, WT-, K4-, and I384M-injected mice had relatively normal liver histology or occasional areas of limited necrosis, mice transfected with G1dbl or G1S342G showed widespread severe liver necrosis, featuring dystrophic calcification and giant cell formation, as well as substantial macrophage infiltration (Fig. 5C). G2 APOL1 caused more focal, less severe liver injury than G1 variants. Removal of the signal peptide of the human variants WT, G1, and G2 reduced but did not abolish the necrosis (Fig. 5C, Sig. Del. G1 and G2) There was no histologic evidence of liver injury or inflammation caused by baboon APOL1 or saline (Fig. 5C). We were unable to quantify and compare the amount of APOL1 remaining in the livers by immunoprecipitation or ELISA, because we were technically limited due to the vast amount of liver protein/volume. We did not observe evidence of renal injury as a result of HGD or circulating APOL1 (kidneys have 4 logs less HGD gene expression than the liver) (31).
Fig. 5.
The C-terminus domain contributes to tissue pathology. (A) Sequences of APOL1 C-terminal variants and constructs screened in the livers of HGD mice. The key is as outlined in Fig. 4A. (B) Murine plasma protein levels were determined of each APOL1 construct by immunoblot 3 d post-HGD using a rabbit polyclonal antibody (plasma diluted 1:40). S, normal mouse serum; Tr, truncated APOL1 constructs; WT, full-length APOL-I. (C) The livers of HGD mice injected with saline vehicle or plasmids containing WT, baboon, K4, IM, SG, G1, G2, Tr1, Tr2, Tr2-SG, signal peptide deleted (Sig. Del.) WT, Sig. Del. G1, or Sig. Del. G2 APOL1. Livers were removed on day 5 postinjection and processed for H&E staining. Representative examples are shown of at least three mice per group. Arrowheads represent leukocyte infiltrates. Arrows represent necrosis.
To begin dissecting the domains of human APOL1 that were driving hepatocyte toxicity, we investigated the role of the C terminus by deleting just the amphipathic helix (Tr2 and Tr2-SG) or the amphipathic helix and the adjacent hydrophobic helix (Tr1) (Fig. 5A). The amphipathic helix at the C terminus of the human APOL1 variants was necessary to cause hepatocyte damage, illustrated by a marked reduction of liver injury and inflammation in its absence (Fig. 5C). Furthermore, the protein levels in sera of the truncated G1 (Tr2/p.S342G) and WT (Tr2) variant APOL1 were equivalent to WT (Fig. 5B). The Tr1 expressed much more protein in the sera than any of the other constructs, which may be due to the disruption of the hydrophobic helix at the C terminus. These data show that, in G1-expressing tissues, both the S342G substitution in the hydrophobic helix and the amphipathic helix at the C terminus are required to drive the full pathology observed.
Discussion
Trypanolytic APOL1 variants G1 and G2 show extraordinarily strong associations with kidney disease in African Americans. We examined the origins and history of APOL1 variants to explore their evolutionary role in humans and other primates. We found evidence for rapid differentiation in modern humans and functional changes that recapitulated trypanolytic features of APOL1 from OWMs. Although we found that both APOL1 renal risk variants have superior trypanolytic activity compared with WT APOL1 variants, our data suggest a complex selection process that may extend beyond African trypanosomes.
We found high rates of G1 in Nigeria and Ghana and much lower rates in East Africa, whereas the frequency of G2 was less variable across Africa. The widespread distribution of G2 and the high efficiency of G2-mediated trypanolysis in vitro and in vivo support the idea that G2 preceded G1 and evolved directly in response to the expression of SRA in T. b. rhodesiense. None of the other animal-infective trypanosomes have acquired SRA, and thus, they cannot infect humans. In contrast, human-infective T. b gambiense has evolved different mechanisms of resistance to TLF (13–15) and is resistant to G1 and G2 sera in vitro (1). The dramatic increase in frequency of G1 in West Africa, despite the presence in the gene pool of the more effective T. b. rhodesiense killer G2, hints that the two alleles were selected in response to more than one pathogen. The West African distribution of G1 may reflect the distribution of T. b. gambiense type 2, a cause of chronic sleeping sickness with active foci in the Ivory Coast and Burkina Faso (32). T .b. gambiense type 2 is an interesting candidate, because it behaves more like T. b. rhodesiense than the much more common T. b. gambiense type 1 in several respects: faster clinical course, infectivity in animal (rodent) models, variable resistance to TLF, ease of transmission by morsitans group tsetse flies, and evidence for an animal reservoir (32–34). One possibility is that G1 has activity against T .b. gambiense type 2. Alternatively, G1 may have arisen in response to entirely different pathogens. For example, the activity of WT APOL1 against Leishmania supports broad spectrum innate activity that goes beyond just African trypanosomes (35).
Our analysis of the 1000 Genome Project data showed distinct African and out-of-Africa haplotypes in this dataset. We also noted that the human APOL1 gene in the current reference genome (hg19) shared the same amino acid sequence as the recently sequenced Neandertal and Denisova APOL1 genes. The current reference human APOL1 gene was much closer to Neandertal and Denisova than to any of the other human haplotypes that we observed. Although we lack conclusive statistical support, the concordance of derived alleles in the out-of-Africa and Neandertal lineages over this 8-kbp region suggests that this APOL1 haplotype was introduced into humans after the out-of-Africa expansion by interbreeding (about 50,000 y ago) rather than by inheritance from a common ancestor (about 500,000 y ago). Genome-wide, the fraction of Neandertal DNA in modern Europeans is ∼2%. Because Neandertals had been adapting to life in Europe, Western Asia, and the Levant for hundreds of thousands of years before the arrival of modern humans, distinct properties of Neandertal APOL1 may have been adopted as beneficial by new human emigrants to the European environment. The remarkable preservation and enrichment of this small 8-kbp region of shared Neandertal DNA over more than 50 millennia could, in theory, be a chance event, but selection is a likelier explanation, and future biological studies may show that it encodes functional properties distinct from other APOL1 variants.
We also observed this Neandertal haplotype at low frequency in East Africans with evidence for European admixture (Luhya) in the 1000 Genomes Project but not in the Yoruba from West Africa, a tribe considered among the best representatives of pre-Bantu expansion Africans without significant European admixture. Recently, Ko et al. (36) found evidence of several SNPs tagging this haplotype in other Africans. Although Ko et al. (36) do not note the identity of this human haplotype (referred to as G3) with Neandertal, they do illustrate its extraordinary divergence from all other African haplotypes, arguing for its distant origins (36). Its presence in these Africans can be explained more parsimoniously by European gene flow back into Africa than coinheritance by both humans and Neandertal, and the high degree of linkage disequilibrium around G3 in Africa may be due to events other than positive selection.
Analysis of APOL1 from human, gorilla, and OWMs, including baboons, indicated that two common human-derived variants, the trypanolytic kidney disease susceptibility alleles G1 and G2, share sequence similarities with trypanolytic OWM APOL1. We engineered APOL1 variants and expressed them in mice, where they circulate bound to HDL, to test in vivo trypanolytic activity, as previous studies have determined that in vitro assays do not always faithfully reflect in vivo biology (37). Protection from trypanosome infection segregated with the OWM-like p.S342G one-half of the G1 allele, whereas p.I384M showed no protective effect in vivo. Sequential introduction of four different lysine residues into human APOL1 at their respective sites in the OWM ortholog defined K4 as the residue that confers full protection from trypanosome infection by preventing SRA binding. The G2 variant, with a similarly positioned lysine, conferred significant protection from trypanosome infection and bound SRA with reduced affinity. Taken together, the lysine substitution data (K1–K4) show that the shifted lysine position in G2 rather than the disruption of the heptad repeat of leucines in the coiled coil domain was the mechanism conferring resistance to T. brucei-SRA.
In humans, APOL1 circulates in blood in the 5-μg/mL range and does not differ between WT and trypanolytic variants (38). Based on both in vitro and in vivo data, we conclude that the protective capacity of APOL1 variants against T. b. rhodesiense is baboon > K4 > G2 > G1 > WT and that it is governed, in part, by their affinity for binding to SRA at a physiological pH range (pH 6.1–4.5) found within endosomes and lysosomes of the parasite.
We found widespread and severe hepatic necrosis in mice expressing G1dbl and G1S342G which circulated at variable levels in sera. The p.I384M variant circulated at high levels, conferred no protection, and caused little or no harm to the liver. This finding is consistent with genetic data showing that S342G seems equivalent to G1 with respect to kidney disease susceptibility (4). The S342G occurs within a putative transmembrane domain of APOL1. Glycine facilitates helix packing and protein stability in hydrophobic transmembrane environments (39) and may stabilize the membrane association of this variant. G2 also caused some liver necrosis, albeit more focal and less severe than G1 or G1S342G, despite G2 circulating at levels similar to the less toxic K4 and WT variants. When the signal peptide was deleted, we still observe necrosis caused by G1 and G2, suggesting that cell damage is unrelated to the APOL1 export process. These findings show the propensity of the human but not baboon trypanolytic variants to induce cell death and tissue injury. We did not observe glomerular injury consistent with focal and segmental glomerulosclerosis on light microscopy from circulating APOL1 variants, although kidney injury may take more time to manifest.
Primates and trypanosomes have coevolved over many millennia, and pieces of this history are written in the genomes of both families. SRA and trypanolytic APOL1 variants are adaptations that resulted in important selective advantages. Both adaptations also seem to confer disadvantages. SRA is an inducible parasite gene, and its expression disappears when the parasite grows in a T. brucei-permissive experimental host, suggesting that its selective advantage in primates could carry a reproductive cost (40). The human APOL1 variants seem to confer an advantage against trypanosomes that express SRA but also predispose the host to kidney failure when allelic frequencies rise to permit homozygosity, at least in the modern environment. Our study defines several important evolutionary events in this story at the population and molecular levels. However, questions remain. For example, it is surprising that gorillas retain an APOL1 gene, whereas chimpanzees and bonobos have lost it (Ensembl.org). Phylogenetically, it is surprising, because humans, chimpanzees, and bonobos are closer evolutionary relatives, meaning that chimpanzees and bonobos have lost APOL1 along their own evolutionary lineage. Furthermore, gorillas and chimpanzees are sympatric over some of their range, and both inhabit rainforest environments, suggesting that they may be exposed to similar pathogens. Similarly, the trypanolytic factors of other African monkeys remain to be investigated. We expect the continued study of trypanosome epidemiology, APOL1 molecular biology, and evolution in both humans and nonhuman primates to add additional chapters to this evolutionary story.
Methods
Genotyping.
Individuals were genotyped for the G1 and G2 variants using Sequenom iPlex; rs73885319 was used as a proxy for G1 (r2 between rs73885319 and rs60910145 is nearly one). The East African populations genotyped in this study are described in the work by Ayodo et al. (26). The West African populations genotyped in this study have been previously described (41, 42), and the samples were originally obtained from participants in a genetic epidemiology study of type 2 diabetes in West Africa (the Africa America Diabetes Mellitus Study) (43). Informed consent was obtained from all participants.
Population Differentiation.
To test for statistically significant differentiation of allele frequency in between two populations, we used all 113 Yoruba and 120 Luhya founders from Hapmap3 (44) genotyped on a panel of 1,440,616 SNPs as well as APOL1 coding variants rs73885319 and rs71785313. We assume that the difference in frequencies for a given polymorphism has a mean of zero and variance of cp(1 − p), where p is the ancestral frequency and c = 2 × Fst (26). The small sample size in the two analyzed populations requires modeling sampling noise, which has variance of p(1 − p)(1/N1 + N2), where N1 and N2 are the total counts for the alleles for the two populations. Therefore, to test for differentiation of frequency at a given allele, we model the difference as a normal random variable with a mean of zero and variance of p(1 − p)(c + 1/N1 + 1/N2), and we compute for each allele a χ2 statistic with 1 degree of freedom.
APOL1 Haplotype Structure.
Phased genotype data for the 1000 Genomes Project were downloaded at ftp://ftp-trace.ncbi.nih.gov/1000genomes/ftp/release/20110521/ (National Center for Biotechnology Information), consisting of phased and inferred data from 1,092 individuals with 2,184 haplotypes. Data from the Neandertal and Denisova were downloaded from the University of California, Santa Cruz browser (ftp://hgdownload.cse.ucsc.edu/gbdb/hg18/neandertal/ and ftp://hgdownload.cse.ucsc.edu/gbdb/hg18/denisova/), and genotype was computed using the Genome Analysis Toolkit (44, 45). The haplotype that we analyzed was bounded by variants rs136150 and rs9610474, a short region of about 8 kbp within which we assume limited ancestral recombination.
Cloning of Primate APOL1 Genes.
Mandrill DNA and baboon (Papio papio) liver were obtained from the Texas Biomedical Research Institute; these biological materials are funded by the National Center for Research Resources (p51 RR013986) and currently supported by the Office of Research Infrastructure Programs/OD P51 OD011133. Mangabey blood was obtained from Yerkes National Primate Research Center of Emory University, and baboon (Papio anubis) kidney was a gift from Afzal A. Siddiqui (Texas Tech University Health Sciences Center, Lubbock, TX). RNA and DNA were extracted from 0.2 g liver or kidney using a Brinkman homogenizer and 3 mL Tri Reagent (MRC) according to the manufacturer’s instructions. Whole-blood RNA was immediately stabilized using PAXgene RNA stabilization tubes and purified using the PAXgene blood RNA validation kit according to the manufacturer’s instructions. Baboon APOL1 cDNAs were reverse-transcribed from whole RNA using MuLV reverse transcriptase (Roche) for 1 h at 42 °C using the baboon APOL1 3′ UTR specific primer bLR1: 5′-GGTGGTTGCCCTGCCCTGTGG. Mangabey APOL1 cDNA was reverse-transcribed using a second 3′ UTR specific primer bLRA: 5′-TGGCCCGTCCCAGGCATATCTCCCCTGG. RT-PCR on baboon cDNA was performed using nested primer pairs bLF1: 5′-GGAGGAGGCCCCGCAGTGAC and bLR1: 5′-GGTGGTTGCCCTGCCCTGTGG followed by bLF2: 5′-CTCGAGGCCACCATGGAGGGAGCTGCTTTGCTGAGAC and bLR2: 5′-GTCACGGTTCTTGGCTCAGAGTCTCATAC. Sequences at the 3′ end of the coding sequence were confirmed by a second PCR on baboon cDNA using primer pair Sp5: 5′-GCCTTGACACAGTGGAGGAG and bLR1. In the case of mangabey blood-derived cDNA, heminested primer pairs Sp5 and bLRA followed by sp5 and bLR1 were used. PCR on mandrill DNA was performed with the heminested primer pairs P1: 5′-GTGGCTACTGCTGAACTGCCCAGG and bLR1 followed by P2: 5′-CTGGAAGAGTTTCCTCGG and bLR1. All PCRs were done with Pfu-Ultra High-Fidelity DNA Polymerase (Stratagene). PCR products were either directly sequenced or first subcloned into pCR4-TOPO (Invitrogen). All APOL1 isoforms and mutants were generated using the Quick-Change Site-Directed Mutagenesis Kit (Stratagene) and verified by sequencing. All sequences have been deposited in the National Center for Biotechnology Information (accession nos. KC197809, KC197810, KC197811, and KC197812).
In Vivo Mouse Studies and Serum Analysis.
HGD, immunoblotting, and parasite infections were performed exactly as described previously using the same expression vector (8). Briefly, Swiss–Webster mice were injected with 50 μg plasmid pRG977 containing various APOL1 gene variants. APOL1 protein was detected in mouse plasma by immunoblotting with either rabbit anti-human APOL1 polyclonal IgG (1:10,000; Protein Tech) or mouse monoclonal antibody SF13.11 (1:10,000), which was a gift of Stephen Hajduk (University of Georgia, Athens, GA). Secondary antibodies were anti-rabbit IgG-HRP or anti-mouse IgG-HRP (1:10,000; Promega). In some experiments, anti-mouse TrueBlot-HRP (1:1,000; eBioscience) was used to eliminate background binding to mouse plasma IgGs. After 3 d, mice were infected with 5,000 human serum-resistant T. brucei strain 427-SRA, which constitutively expresses the SRA gene (46). Infected mice were monitored for blood parasitemia and survival. All animal experiments were approved by the Institutional Animal Care and Use Committee at New York University Medical Center and Hunter College, which both have currently approved Animal Welfare Assurance Agreements with the National Institues of Health Office for Protection from Research Risks.
SRA Binding Assays.
Recombinant N-terminal His-tagged SRA was produced in Escherichia coli as described previously (8). The His-tag was cleaved off with tobacco etch virus protease. After dialysis into PBS (pH 7.4), 5 μg SRA was coupled to 50 μL NHS-activated agarose beads (Pierce) for 30 min at room temperature. The reaction was quenched by the addition of 0.1 M Tris, and the beads were washed according to the manufacturer’s instructions, except that a higher pH was used (0.1 M acetate, pH 4.6) to avoid SRA denaturation. Blood was obtained from HGD mice either 2 or 3 d after plasmid injection (protein levels peaked at 2 d) and allowed to clot for at least 30 min at room temperature. Serum was obtained by centrifugation at 11,000 × g and stored at −80 °C. Immunoblotting, followed by densitometry (Image J), was used to estimate serum levels of primate APOL1 variants. Serum volumes were adjusted so that similar amounts of APOL1 were added to SRA beads, and the serum volume was made up to 20 μL with normal mouse serum. A complete protease inhibitor mixture was added (Pierce). Immediately before binding, serum was titrated to pH 5.0 with a precalibrated volume of 1 M acetic acid, and SRA beads were equilibrated in binding buffer (0.1 M acetate, pH 5.0, 0.15 M NaCl). Serum was incubated with SRA beads for 2 h at 4 °C with rotation. Beads were washed three times in binding buffer and then eluted for 10 min at 25 °C in 0.1 M Tris (pH 8). Beads with serum (load) and eluted APOL1 (bound) were diluted 1:40 and 1:80, and proteins were separated by SDS/PAGE before transfer to PVDF for immunoblotting.
Surface Plasmon Resonance Analysis.
Measurements were performed on a Biacore T100 instrument with a constant flow rate of 30 μL/min using 20 mM citrate-phosphate buffers at different pH values (obtained by mixing 100 mM citric acid with 200 mM disodium hydrogen phosphate, allowing single-buffer systems to be used to obtain pH values from 4.0 to 7.6) in 150 mM NaCl. A streptavidin chip was coupled with biotinylated APOL1 peptides diluted in 20 mM citrate-phosphate (pH 5.8) and 150 mM NaCl to a final concentration of 10 μg/mL, giving a total response of 2,000 resonance units. Flow cell 1 was left blank.
SRA was equilibrated using PD10 columns (Amersham Biosciences) into citrate-phosphate (pH 5.6) and 150 mM NaCl and concentrated to 100 μM. All peptide-coupled channels were equilibrated with appropriate buffer before injection of SRA. To perform the pH titration experiments, SRA was diluted to a final concentration of 3 μM into the appropriate citrate-phosphate buffer. The chip was primed into the same buffer before protein injection. To investigate the binding to different peptides, the same procedure was used with a series of twofold dilutions from a maximum of 0.8 μM injected over the chip. The flow cells were regenerated with a 30-s injection of 20 mM citrate-phosphate (pH 7.6) and 150 mM NaCl. The level of specific binding was obtained from a subtraction of the response from channel 2 from that of channel 1. Sensorgrams were fitted to a 1:1 interaction model using the BIAevaluation software to allow determination of ka, kd, and Kd.
Histology.
Livers were fixed by immersion in 4% (g/vol) paraformaldehyde for 2 h at 25 °C and at least 16 h at 4 °C. After washing with phosphate buffered saline, livers were immersed in 70% ethanol, embedded, sectioned, and stained with H&E using standard techniques [New York University (NYU) Langone Medical Center Histopathology Core NYU Cancer Institute Center Support Grant, National Institutes of Health/National Cancer Institute 5 P30CA16087-31].
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health (NIH)/National Institute of Allergy and Infectious Diseases (NIAID) Grant R01AI041233 (to J.R.), National Science Foundation Bread Award IOS-1249166 (to J.R.), NIH/NIAID Training Grant T32 AI007180 (to R.T.), NIH/National Institute of Diabetes and Digestive and Kidney Diseases Grant (NIDDK) DK76868 (to D.J.F.), the Doris Duke Charitable Foundation (D.J.F.), Satellite Healthcare Foundation Coplon Award (to D.J.F.), NIH/National Institue on Minority Health and Health Disparities Grant R01MD007092 (to M.R.P.), Wellcome Trust Project Grants 085256/Z/08/Z (to M.C.) and 087692/Z/08/Z (to M.K.H.), NIH Intramural Research Program of the Center for Research on Genomics and Global Health Grant Z01HG200362 (to C.R.), and the NIH/NIDDK and National Cancer Institute Intramural Research Programs (C.A.W. and J.K.). D.J.F. is a recipient of a Doris Duke Clinical Scientist Development Award.
Footnotes
Conflict of interest statement: G.G., M.R.P., and D.J.F. have filed for patents related to APOL1 and kidney disease.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1400699111/-/DCSupplemental.
References
- 1.Genovese G, et al. Association of trypanolytic ApoL1 variants with kidney disease in African Americans. Science. 2010;329(5993):841–845. doi: 10.1126/science.1193032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tzur S, et al. Missense mutations in the APOL1 gene are highly associated with end stage kidney disease risk previously attributed to the MYH9 gene. Hum Genet. 2010;128(3):345–350. doi: 10.1007/s00439-010-0861-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Friedman DJ, Kozlitina J, Genovese G, Jog P, Pollak MR. Population-based risk assessment of APOL1 on renal disease. J Am Soc Nephrol. 2011;22(11):2098–2105. doi: 10.1681/ASN.2011050519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kopp JB, et al. APOL1 genetic variants in focal segmental glomerulosclerosis and HIV-associated nephropathy. J Am Soc Nephrol. 2011;22(11):2129–2137. doi: 10.1681/ASN.2011040388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Smith EE, Malik HS. The apolipoprotein L family of programmed cell death and immunity genes rapidly evolved in primates at discrete sites of host-pathogen interactions. Genome Res. 2009;19(5):850–858. doi: 10.1101/gr.085647.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lugli EB, Pouliot M, Portela MdelP, Loomis MR, Raper J. Characterization of primate trypanosome lytic factors. Mol Biochem Parasitol. 2004;138(1):9–20. doi: 10.1016/j.molbiopara.2004.07.004. [DOI] [PubMed] [Google Scholar]
- 7.Seed JR, Sechelski JB, Loomis MR. A survey for a trypanocidal factor in primate sera. J Protozool. 1990;37(5):393–400. doi: 10.1111/j.1550-7408.1990.tb01163.x. [DOI] [PubMed] [Google Scholar]
- 8.Thomson R, Molina-Portela P, Mott H, Carrington M, Raper J. Hydrodynamic gene delivery of baboon trypanosome lytic factor eliminates both animal and human-infective African trypanosomes. Proc Natl Acad Sci USA. 2009;106(46):19509–19514. doi: 10.1073/pnas.0905669106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kageruka P, et al. Infectivity of Trypanosoma (Trypanozoon) brucei gambiense for baboons (Papio hamadryas, Papio papio) Ann Soc Belg Med Trop. 1991;71(1):39–46. [PubMed] [Google Scholar]
- 10.Xong HV, et al. A VSG expression site-associated gene confers resistance to human serum in Trypanosoma rhodesiense. Cell. 1998;95(6):839–846. doi: 10.1016/s0092-8674(00)81706-7. [DOI] [PubMed] [Google Scholar]
- 11.Vanhamme L, et al. Apolipoprotein L-I is the trypanosome lytic factor of human serum. Nature. 2003;422(6927):83–87. doi: 10.1038/nature01461. [DOI] [PubMed] [Google Scholar]
- 12.Stephens NA, Hajduk SL. Endosomal localization of the serum resistance-associated protein in African trypanosomes confers human infectivity. Eukaryot Cell. 2011;10(8):1023–1033. doi: 10.1128/EC.05112-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Uzureau P, et al. Mechanism of Trypanosoma brucei gambiense resistance to human serum. Nature. 2013;501(7467):430–434. doi: 10.1038/nature12516. [DOI] [PubMed] [Google Scholar]
- 14.Capewell P, et al. The TgsGP gene is essential for resistance to human serum in Trypanosoma brucei gambiense. PLoS Pathog. 2013;9(10):e1003686. doi: 10.1371/journal.ppat.1003686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.DeJesus E, Kieft R, Albright B, Stephens NA, Hajduk SL. A single amino acid substitution in the group 1 Trypanosoma brucei gambiense haptoglobin-hemoglobin receptor abolishes TLF-1 binding. PLoS Pathog. 2013;9(4):e1003317. doi: 10.1371/journal.ppat.1003317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Raper J, Fung R, Ghiso J, Nussenzweig V, Tomlinson S. Characterization of a novel trypanosome lytic factor from human serum. Infect Immun. 1999;67(4):1910–1916. doi: 10.1128/iai.67.4.1910-1916.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bullard W, et al. Haptoglobin-hemoglobin receptor independent killing of African trypanosomes by human serum and trypanosome lytic factors. Virulence. 2012;3(1):72–76. doi: 10.4161/viru.3.1.18295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Davidson WS, et al. Proteomic analysis of defined HDL subpopulations reveals particle-specific protein clusters: Relevance to antioxidative function. Arterioscler Thromb Vasc Biol. 2009;29(6):870–876. doi: 10.1161/ATVBAHA.109.186031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shiflett AM, Bishop JR, Pahwa A, Hajduk SL. Human high density lipoproteins are platforms for the assembly of multi-component innate immune complexes. J Biol Chem. 2005;280(38):32578–32585. doi: 10.1074/jbc.M503510200. [DOI] [PubMed] [Google Scholar]
- 20.Vanhollebeke B, et al. A haptoglobin-hemoglobin receptor conveys innate immunity to Trypanosoma brucei in humans. Science. 2008;320(5876):677–681. doi: 10.1126/science.1156296. [DOI] [PubMed] [Google Scholar]
- 21.Drain J, Bishop JR, Hajduk SL. Haptoglobin-related protein mediates trypanosome lytic factor binding to trypanosomes. J Biol Chem. 2001;276(32):30254–30260. doi: 10.1074/jbc.M010198200. [DOI] [PubMed] [Google Scholar]
- 22.Molina-Portela MdelP, Lugli EB, Recio-Pinto E, Raper J. Trypanosome lytic factor, a subclass of high-density lipoprotein, forms cation-selective pores in membranes. Mol Biochem Parasitol. 2005;144(2):218–226. doi: 10.1016/j.molbiopara.2005.08.018. [DOI] [PubMed] [Google Scholar]
- 23.Pérez-Morga D, et al. Apolipoprotein L-I promotes trypanosome lysis by forming pores in lysosomal membranes. Science. 2005;309(5733):469–472. doi: 10.1126/science.1114566. [DOI] [PubMed] [Google Scholar]
- 24.Sabeti PC, et al. Detecting recent positive selection in the human genome from haplotype structure. Nature. 2002;419(6909):832–837. doi: 10.1038/nature01140. [DOI] [PubMed] [Google Scholar]
- 25.Altshuler DM, et al. International HapMap 3 Consortium Integrating common and rare genetic variation in diverse human populations. Nature. 2010;467(7311):52–58. doi: 10.1038/nature09298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ayodo G, et al. Combining evidence of natural selection with association analysis increases power to detect malaria-resistance variants. Am J Hum Genet. 2007;81(2):234–242. doi: 10.1086/519221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Abecasis GR, et al. 1000 Genomes Project Consortium A map of human genome variation from population-scale sequencing. Nature. 2010;467(7319):1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Green RE, et al. A draft sequence of the Neandertal genome. Science. 2010;328(5979):710–722. doi: 10.1126/science.1188021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Meyer M, et al. A high-coverage genome sequence from an archaic Denisovan individual. Science. 2012;338(6104):222–226. doi: 10.1126/science.1224344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lecordier L, et al. C-terminal mutants of apolipoprotein L-I efficiently kill both Trypanosoma brucei brucei and Trypanosoma brucei rhodesiense. PLoS Pathog. 2009;5(12):e1000685. doi: 10.1371/journal.ppat.1000685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu F, Song Y, Liu D. Hydrodynamics-based transfection in animals by systemic administration of plasmid DNA. Gene Ther. 1999;6(7):1258–1266. doi: 10.1038/sj.gt.3300947. [DOI] [PubMed] [Google Scholar]
- 32.Gibson W. The origins of the trypanosome genome strains Trypanosoma brucei brucei TREU 927, T. b. gambiense DAL 972, T. vivax Y486 and T. congolense IL3000. Parasit Vectors. 2012;5:71. doi: 10.1186/1756-3305-5-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Truc P, Formenty P, Diallo PB, Komoin-Oka C, Lauginie F. Confirmation of two distinct classes of zymodemes of Trypanosoma brucei infecting man and wild mammals in Côte d’Ivoire: Suspected difference in pathogenicity. Ann Trop Med Parasitol. 1997;91(8):951–956. doi: 10.1080/00034989760356. [DOI] [PubMed] [Google Scholar]
- 34.Capewell P, et al. Differences between Trypanosoma brucei gambiense groups 1 and 2 in their resistance to killing by trypanolytic factor 1. PLoS Negl Trop Dis. 2011;5(9):e1287. doi: 10.1371/journal.pntd.0001287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Samanovic M, Molina-Portela MP, Chessler AD, Burleigh BA, Raper J. Trypanosome lytic factor, an antimicrobial high-density lipoprotein, ameliorates Leishmania infection. PLoS Pathog. 2009;5(1):e1000276. doi: 10.1371/journal.ppat.1000276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ko WY, et al. Identifying Darwinian selection acting on different human APOL1 variants among diverse African populations. Am J Hum Genet. 2013;93(1):54–66. doi: 10.1016/j.ajhg.2013.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Molina-Portela MP, Samanovic M, Raper J. Distinct roles of apolipoprotein components within the trypanosome lytic factor complex revealed in a novel transgenic mouse model. J Exp Med. 2008;205(8):1721–1728. doi: 10.1084/jem.20071463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bruggeman LA, et al. Plasma Apolipoprotein L1 Levels Do Not Correlate with CKD. J Am Soc Nephrol. 2014;25(3):634–644. doi: 10.1681/ASN.2013070700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Javadpour MM, Eilers M, Groesbeek M, Smith SO. Helix packing in polytopic membrane proteins: Role of glycine in transmembrane helix association. Biophys J. 1999;77(3):1609–1618. doi: 10.1016/S0006-3495(99)77009-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.De Greef C, Hamers R. The serum resistance-associated (SRA) gene of Trypanosoma brucei rhodesiense encodes a variant surface glycoprotein-like protein. Mol Biochem Parasitol. 1994;68(2):277–284. doi: 10.1016/0166-6851(94)90172-4. [DOI] [PubMed] [Google Scholar]
- 41.Bentley AR, et al. Variation in APOL1 Contributes to Ancestry-Level Differences in HDLc-Kidney Function Association. Int J Nephrol. 2012;2012:748984. doi: 10.1155/2012/748984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Adeyemo AA, Chen G, Chen Y, Rotimi C. Genetic structure in four West African population groups. BMC Genet. 2005;6:38. doi: 10.1186/1471-2156-6-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rotimi CN, et al. In search of susceptibility genes for type 2 diabetes in West Africa: The design and results of the first phase of the AADM study. Ann Epidemiol. 2001;11(1):51–58. doi: 10.1016/s1047-2797(00)00180-0. [DOI] [PubMed] [Google Scholar]
- 44.McKenna A, et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20(9):1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.DePristo MA, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43(5):491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang J, Böhme U, Cross GA. Structural features affecting variant surface glycoprotein expression in Trypanosoma brucei. Mol Biochem Parasitol. 2003;128(2):135–145. doi: 10.1016/s0166-6851(03)00055-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.