Significance
Current consensus indicates that modern humans originated from an ancestral African population between ∼100–200 ka. The ensuing dispersal pattern is controversial, yet has important implications for the demographic history and genetic/phenotypic structure of extant human populations. We test for the first time to our knowledge the spatiotemporal dimensions of competing out-of-Africa dispersal models, analyzing in parallel genomic and craniometric data. Our results support an initial dispersal into Asia by a southern route beginning as early as ∼130 ka and a later dispersal into northern Eurasia by ∼50 ka. Our findings indicate that African Pleistocene population structure may account for observed plesiomorphic genetic/phenotypic patterns in extant Australians and Melanesians. They point to an earlier out-of-Africa dispersal than previously hypothesized.
Keywords: modern human origins, human evolution, genome diversity, cranial diversity, SNPs
Abstract
Despite broad consensus on Africa as the main place of origin for anatomically modern humans, their dispersal pattern out of the continent continues to be intensely debated. In extant human populations, the observation of decreasing genetic and phenotypic diversity at increasing distances from sub-Saharan Africa has been interpreted as evidence for a single dispersal, accompanied by a series of founder effects. In such a scenario, modern human genetic and phenotypic variation was primarily generated through successive population bottlenecks and drift during a rapid worldwide expansion out of Africa in the Late Pleistocene. However, recent genetic studies, as well as accumulating archaeological and paleoanthropological evidence, challenge this parsimonious model. They suggest instead a “southern route” dispersal into Asia as early as the late Middle Pleistocene, followed by a separate dispersal into northern Eurasia. Here we test these competing out-of-Africa scenarios by modeling hypothetical geographical migration routes and assessing their correlation with neutral population differentiation, as measured by genetic polymorphisms and cranial shape variables of modern human populations from Africa and Asia. We show that both lines of evidence support a multiple-dispersals model in which Australo-Melanesian populations are relatively isolated descendants of an early dispersal, whereas other Asian populations are descended from, or highly admixed with, members of a subsequent migration event.
Paleontological and genetic data indicate a common ancestral population of modern humans residing in Africa between ∼100–200 ka (1–4). The timing and pattern of the modern human African diaspora continues to be strongly debated. Competing hypotheses center on either a single Late Pleistocene dispersal into Eurasia between ∼50–75 ka or multiple dispersals beginning as early as the Middle Pleistocene ∼130 ka (5–8). The observed pattern of decreasing genetic (9, 10) and cranial (11) diversity at increasing distances from sub-Saharan Africa has been interpreted as evidence for a single dispersal, characterized by a series of founder effects during global expansion. In its simplest form, a single dispersal scenario follows a series of founder events in an eastward expansion (EE) model that conforms to terrestrial routes mostly along a latitudinal axis across Asia (10, 12).
Another interpretation consistent with decreasing biological diversity from Africa is to consider multiple dispersals (MD) out of the continent. In an MD model, an initial dispersal between ∼50–100 ka occurs primarily along a coastal route through the southern Arabian Peninsula and is followed by a second dispersal through the Levant at ∼50 ka and into northern Eurasia (13, 14). This model proposes that extant, isolated populations in Asia could retain the biological signal of the initial, “southern route” dispersal. Such hypothetical, “relic” populations could include Australians, Melanesians, Papuans, Dravidian speakers of South Asia, and short-statured “Negrito” populations of Southeast Asia. A recent genetic study proposed that living Australians are direct descendants of the southern route dispersal, whereas Papuans, Melanesians, and Philippine Aeta “Negrito” populations also retain a signal of the southern route, but one that is obscured owing to admixture with members of the second dispersal (8). In this model of multiple dispersals with isolation (MDI), a southern route dispersal out of Africa commences between ∼62–75 ka and is followed by a second dispersal between ∼25–38 ka. An alternative chronology for the MDI model posits a southern route dispersal as early as the late Middle Pleistocene ∼130 ka (MDI-MP), rather than the Late Pleistocene (MDI-LP), and is based primarily on archaeological evidence in the Arabian Peninsula (6, 15).
Growing consensus on the southern route dispersal has been strengthened by the study of SNPs in hypothetical relic populations (8, 16–18). However, whether this reflects evidence of multiple dispersals from Africa continues to be debated (8, 16). A reconciling view, therefore, has been that a single dispersal from Africa might have taken place in the Late Pleistocene ∼75 ka, followed by divergence into separate migration waves outside the continent, likely in Southwest Asia (7). Like the MD and MDI models, migration into Southeast Asia is via a “beachcomber” single dispersal (BSD) route along the coast. Unlike the EE model, the BSD model implies substantial migration along a longitudinal axis in East Asia.
Because temporal and spatial dimensions are explicit in these competing out-of-Africa models, distinguishing them can be achieved by assessing the correlation of predicted spatial and temporal distances and observed neutral biological distances between modern human populations. Such a biogeographical approach accounts for the primary drivers of recent human evolution: migration, mutation, and drift. We used this test for 10 populations sampled from Africa and Asia using genetic and cranial phenotype data (Table 1). We limited our phenotype analyses to the temporal bone, because it has been shown to conserve modern human population history at higher fidelity than other parts of the cranium, from an early ontogenetic stage, and in a largely neutral manner (19, 20). For both lines of evidence we used the same quantitative evolutionary framework to assess biological distances between our sampled populations (21).
Table 1.
Populations, sample size, and geography
| Geographical coordinates | ||||
| Population | Genetics, n | Cranial phenotype, n | Latitude | Longitude |
| AU Australia | 12 | 20 | −33.89 | 151.24 |
| CA Central Asia | 56 | 25 | 43.29 | 68.26 |
| EA East Africa | 66 | 25 | 9.02 | 38.74 |
| JP Japan | 107 | 31 | 35.66 | 139.82 |
| ME Melanesia | 30 | 17 | −9.42 | 159.94 |
| NE Philippines Aeta/Agta “Negrito” | 16 | 23 | 14.6 | 120.98 |
| NG New Guinea | 10 | 31 | −9.48 | 147.19 |
| NI North India | 61 | 15 | 28.63 | 77.2 |
| SA South Africa | 215 | 20 | 9.02 | 38.74 |
| SI South India | 141 | 26 | 6.93 | 79.86 |
Results
We used two analytical approaches in determining the fit between interpopulation biological differentiation and the out-of-Africa dispersal models. First, we used partial Mantel tests (22, 23) to determine the correlation of population differentiation and geographical distances between populations along hypothetical dispersal routes, controlling for population divergence times in each case. Second, we considered the temporal information contained within each of the competing out-of-Africa models to validate our partial Mantel results and, in the case of the MDI model, distinguish the chronology of the southern route dispersal, commencing either in the late Middle Pleistocene (MDI-MP) or in the Late Pleistocene (MDI-LP).
In our first analysis, genetic distances, Fst, and cranial phenotypic distances, Pst, between populations were calculated using SNP data and 3D anatomical landmark data, respectively (Table 2). Geographical distances, G, were calculated as geodesic distances between populations along hypothetical dispersal routes (Fig. 1). Temporal distances between populations were calculated from the genomic data, based on levels of linkage disequilibrium, to assess when population pairs diverged in time (Table S1). Independently for Fst and Pst, we assessed their pairwise correlation with G while holding divergence time, T, constant. This approach has the effect of controlling for drift owing to the fact that populations separated at distinct points in time and space. Therefore, the partial Mantel test results indicate which out-of-Africa model best explains population differentiation when considering both spatial and temporal dimensions. The MDI model best explained both genetic (Fst) and phenotypic (Pst) differentiation (Table 3). In fact, for the phenotype dataset, only the MDI model test results were statistically significant. In the case of the genetic dataset, the control and MDI models were significant after Bonferroni correction for multiple model tests, although the MDI correlation coefficient was almost twice as large as that of the control model. A Dow–Cheverud test (22, 24) indeed differentiates MDI as a better model against the control (r = 0.588, P ≤ 0.0001).
Table 2.
Genetic and phenotypic distances
| Population | AU | CA | EA | JP | ME | NE | NG | NI | SA | SI |
| AU | 0 | 0.372231 | 0.557596 | 0.322725 | 0.365393 | 0.674131 | 0.142495 | 0.354671 | 0.35569 | 0.400061 |
| CA | 0.1099 | 0 | 0.524056 | 0.099755 | 0.571562 | 0.516308 | 0.359326 | 0.166615 | 0.448843 | 0.220931 |
| EA | 0.1907 | 0.12894 | 0 | 0.572357 | 0.636501 | 1.298547 | 0.724035 | 0.379112 | 0.333683 | 0.495417 |
| JP | 0.14366 | 0.03067 | 0.17477 | 0 | 0.55383 | 0.476002 | 0.389483 | 0.200167 | 0.450304 | 0.293722 |
| ME | 0.07893 | 0.14487 | 0.22634 | 0.17281 | 0 | 0.737244 | 0.304302 | 0.515503 | 0.497316 | 0.40173 |
| NE | 0.11885 | 0.05288 | 0.15832 | 0.06243 | 0.1588 | 0 | 0.625152 | 0.585473 | 0.723552 | 0.581859 |
| NG | 0.09229 | 0.11133 | 0.19972 | 0.13632 | 0.10713 | 0.11536 | 0 | 0.371397 | 0.419062 | 0.432981 |
| NI | 0.10533 | 0.02177 | 0.12136 | 0.07084 | 0.13882 | 0.06732 | 0.11443 | 0 | 0.328728 | 0.088056 |
| SA | 0.19096 | 0.13456 | 0.00605 | 0.17484 | 0.21913 | 0.16006 | 0.19871 | 0.12691 | 0 | 0.393329 |
| SI | 0.10408 | 0.02495 | 0.12556 | 0.07025 | 0.13477 | 0.06676 | 0.1122 | 0.00316 | 0.13087 | 0 |
Below diagonal: Fst values; above diagonal: Pst values (outlier value noted in italics; Fig. S1).
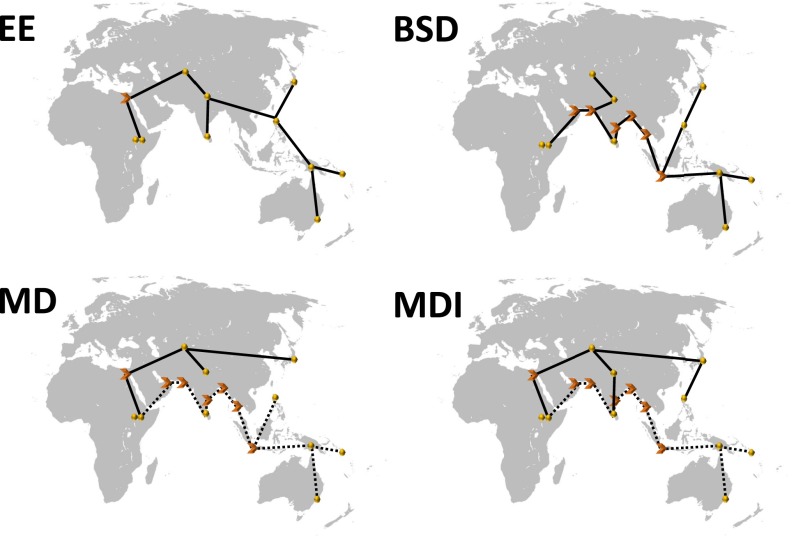
Fig. 1.
Out-of-Africa dispersal models. Spheres are approximate centroids of populations sampled (Table 1), connecting lines are dispersal routes, and arrows are geographical waypoints (Table S4). The eastward expansion (EE) model connects populations primarily along a latitudinal axis (10, 12). The beachcomber single dispersal (BSD) model connects populations primarily along a coastal route (7). The multiple dispersals model (MD) connects hypothetical relic populations along a southern route (dotted lines) and north Eurasians along a northern route (13). The multiple dispersals with isolation (MDI) model assumes that only Australo-Melanesian populations retain a strong southern route biological signal (8). For simplicity, a Holocene map outline is shown.
Table 3.
Dispersal models test
| Distances | Control | EE | BSD | MD | MDI |
| Fst | 0.405 (0.008) | 0.282 (0.085) | 0.321 (0.035) | 0.389 (0.013) | 0.782 (<0.0001) |
| Pst* | 0.138 (0.401) | 0.040 (0.822) | 0.142 (0.375) | 0.184 (0.268) | 0.464 (0.008) |
Partial Mantel test of population distances (Fst/Pst) and geodesic distances (G) for all dispersal models, while controlling for population divergence values (T). Values are Pearson correlation coefficient, r, rounded to the third digit and two-tailed probability, p, (in parentheses) after 10,000 permutations. Bold type indicates significance after Bonferroni correction (α = 0.01).
Pst correlations after removal of outlier EA-NE value (Table 2).
In our second analysis, we generated hypothetical divergence values, C, between populations, based on the chronology of dispersal for each model (Table 4). In contrast to our estimates of population divergence from the genomic data, the model chronology dates reflect estimates of modern human colonization within the geographical space of the populations we sampled. These dates contain inherent information about both time and space because they are primarily derived from archaeological, paleontological, and climatological records. Although this test is not explicit about dispersal routes, it serves to distinguish the out-of-Africa models based on the expected dates of colonization for a specific geographical region. Treating C as divergence values allowed us to exploit the relationship between population differentiation, divergence time, and effective population size, Ne (25). We calculated Ne from the genomic data (Table S2) (26) and, using C, constructed hypothetical Fst values to represent each out-of-Africa dispersal model. We then used a simple Mantel test to assess the fit between these hypothetical values and the empirical Fst and Pst values. We found that the MDI-MP model receives the best support (Table 5). Considering the Bonferroni correction for multiple model tests, results were only significant for the cranial phenotype dataset. Nevertheless, correlation and significance values were highest for the MDI-MP model in both biological datasets.
Table 4.
Dispersal models chronology
| Model | AU | CA | EA | JP | ME | NE | NG | NI | SA | SI | Ref. |
| EE | 40 | 45 | 56 | 36 | 40 | 36 | 40 | 45 | 56 | 45 | 10 |
| BSD | 55 | 25 | 75 | 40 | 55 | 40 | 55 | 40 | 75 | 45 | 7 |
| MD | 65 | 30 | 80 | 25 | 65 | 65 | 65 | 30 | 80 | 70 | 13 |
| MDI-MP | 50 | 45 | 130 | 40 | 50 | 40 | 50 | 45 | 130 | 45 | 6 |
| MDI-LP | 50 | 31.5 | 68.5 | 25 | 50 | 25 | 50 | 31.5 | 68.5 | 31.5 | 8 |
Based on approximations from the references provided, dates (approximate thousands of years before the present) are proposed times of dispersal and colonization within the geographical space of the sampled populations.
Table 5.
Dispersal models chronology test
| Distances | EE | BSD | MD | MDI-LP | MDI-MP |
| Fst | −0.146 (0.337) | 0.099 (0.524) | 0.038 (0.820) | 0.157 (0.307) | 0.335 (0.022) |
| Pst* | 0.176 (0.245) | 0.260 (0.098) | 0.237 (0.029) | 0.145 (0.409) | 0.463 (0.001) |
Simple Mantel test of empirical population distances (Fst/Pst) and hypothetical Fst distances for each dispersal model. Values are Pearson correlation coefficient, r, rounded to the third digit and two-tailed probability, p, (in parentheses) after 10,000 permutations. Bold type indicates significance after Bonferroni correction (α = 0.01).
Pst correlations after removal of outlier EA-NE value (Table 2).
Discussion
The test of current competing out-of-Africa models shows unambiguous support for a multiple dispersals model in which Australians, Papuans, and Melanesians remain relatively isolated after an early dispersal from Africa via a southern route. Although some degree of Holocene admixture between our sampled Indian and Australian populations has been previously proposed (17), our results are generally consistent with the view that extant Australians are descended from a relatively isolated lineage that first occupied that continent ∼50 ka (8). They differ from previous findings in that our dispersal chronology test suggests an initial African dispersal closer to the Middle–Late Pleistocene boundary. This is consistent with archaeological evidence for modern human occupation in the southern Arabian Peninsula at ∼125 ka (6, 15). This date is in intriguingly closer correspondence with the genetic divergence estimates for our sampled populations, with a calendar date of divergence between Melanesians and South Africans at ∼116 ka, for example (Table S1). No modern human fossils have been discovered in the southern Arabian Peninsula, but lithic artifacts show affinities with African assemblages, including those discovered alongside the fossil remains at Herto, Ethiopia, dated between ∼154–160 ka (2, 27). Importantly, the geological age of these specimens falls within the recent estimates for the common ancestor of all modern human populations (3, 4). This implies that an initial dispersal occurred not long after modern human origins in Africa, rather than much later, as an EE or BSD model would predict. The environmental and geographical viability for the MDI-MP model has been confirmed with a recent synthesis of available Middle–Late Pleistocene climate proxy data for Africa (28). Likewise, spatially explicit simulations developed from climate and microsatellite genetic data are in agreement with a southern route dispersal and earlier dates of Eurasian occupation than previously hypothesized (29). Moreover, it has been proposed that severe East African droughts occurring between 135–75 ka may have prompted human population fragmentation and bottlenecks (30), also possibly resulting in dispersals out of the continent. The modern human fossil series of Qafzeh and Skhul from the Levant, dated between ∼90–120 ka, could therefore correspond to this initial dispersal. Although often considered to represent a short-lived extension of African ecosystems rather than evidence of a long-range dispersal into Eurasia (31), in comparative craniometric studies the Levantine series and other early modern humans from Africa have consistently closer affinity to recent Australians than to other modern human populations (2, 32–34).
Presently, clear evidence of modern human occupation eastward of the Arabian Peninsula during the early Late Pleistocene is lacking. Occupation of Australia is documented by the human paleontological record at ∼50 ka and in continental Southeast Asia at a maximum date of ∼63 ka (8, 35). Specimens before this time period are fragmentary and taxonomically ambiguous but have, in some cases, been claimed to represent anatomically modern humans (6, 7, 35–37). The MDI-MP model tested here suggests that whereas Southeast Asia may have been populated by modern humans, replacement of these descendants from subsequent migrants may obscure a southern route biological signal in extant populations of that region (6). Our dataset conforms to this hypothesis in that neither the genetic nor the cranial phenotype dataset from our sampled populations separate the Indo-European and Dravidian speakers from India, as might be expected if the latter where relic descendants of the southern route dispersal (Supporting Information, The “Negrito” Hypothesis). Instead, both Indian samples exhibit closer genetic and phenotypic affinity to the hypothetical second dispersal descendants (the Japanese, Aeta/Agta, and Central Asian populations). Sampling of other isolated, relic populations will serve to further support this hypothesis (8, 18).
Although the models tested do not explicitly account for archaic admixture, the continued validation of the southern route dispersal and support for the MDI-MP model have important implications for understanding the degree, timing, and location of such events. Presently, the favored explanation for genetic resemblance between Neanderthals and non-African modern human populations is a hypothetical admixture event in the Middle East (38). Likewise, shared polymorphisms between Denisovans and certain relic descendants of a southern route dispersal are explained by admixture in Southeast Asia (16). Identifying the presence of Neanderthal and Denisovan occupation along the southern route geographical space and within the Late Pleistocene temporal boundary is therefore crucial. The paleontological and archaeological records thus far remain elusive. An important consideration, therefore, is the persistence of population substructure in Africa (18, 38–41), which has been inferred from the human paleontological record (33, 34) and is concordant with climate fluctuations in the continent (28, 30).
Population substructure implies that differential lineage assortment could be pronounced if populations in Africa remained spatially and temporally separated, affecting the subsequent diversity that is exported outside of the continent, as in an MDI-MP scenario. Genetically, polymorphisms within a parental population are randomly distributed into daughter lineages during speciation. In the recent human lineage, modern humans, Neanderthals, and Denisovans can be considered the daughter lineages of a common parental ancestor. Therefore, expression of shared genetic polymorphisms with Neanderthals and Denisovans in certain extant populations would be the consequence of biogeographical contingency and drift instead of, or in addition to, admixture with other hominins (18, 39–41). In a similar vein, expression of a plesiomorphic skeletal phenotype in extant and extinct populations has been interpreted as evidence for admixture with, or “assimilation” of, other hominin populations (42). Instead, population substructure implies that such expression reflects the retention of traits inherited from the parental population and could be more prominent in descendants of the southern route dispersal, who are chronologically closer to the parental ancestor. These findings do not imply that dispersing modern people from Africa did not interbreed with other hominin populations but suggest that, at present, other hypotheses also seem to be compatible with the biological evidence.
Conclusions
Considering two independent biological datasets, using a common quantitative evolutionary framework, and using a biogeographical approach, we have tested the primary hypotheses for the modern human out-of-Africa event. Our results are unambiguous in their support of multiple dispersals into Eurasia, with Australians, Papuans, and Melanesians retaining the signal of a southern route dispersal that commenced closer to the temporal boundary of the Middle–Late Pleistocene. Furthermore, these results suggest that models of ancient admixture events with other hominin populations should enclose the South Asian, southern route geographical space and a Late Pleistocene time frame—areas that have been largely understudied and where neither Neanderthal nor Denisovan occupation has been confirmed by the fossil record. This study suggests that ancient population substructure, in addition or as an alternative to hominin interbreeding, may contribute to the observed pattern of resemblance between certain modern human populations and other hominins, ultimately generating the structure of extant modern human genetic and phenotypic diversity. Continued field work, alongside rapid advances in modern and ancient genome sequencing, will allow for greater resolution in modeling the spatial and temporal dimensions of modern human origins and dispersals.
Materials and Methods
Genetic Data.
We combined SNP data from published datasets for n = 714 individuals and grouped the samples into 10 ethnolinguistically and geographically related populations using the Greenberg language classification (Table 1 and Table S3). Using the PLINK 1.07 software (43), we selected only the autosomal SNPs with genotyping success rate >98% and minor allele frequency >0.01. To optimize strand alignment, we also removed from the merged genotype data file the alleles carrying ambiguities in strand flipping, namely, A/T and C/G polymorphisms. Following these quality control procedures, 3,345 SNPs were available for subsequent analysis. For measures of biological distances, we estimated the Weir–Cockerham Fst (25) values between pairs of populations (Table 2). Each value represents the average of the pairwise Fst calculated for each SNP, over all SNPs in the dataset. Fst does not by itself provide information on the mechanisms involved in generating the differentiation between populations, namely, parameters of demography, time, and space. Because our objective was to test spatial dispersal scenarios, a measure informative of both demography and time is required to assess the relationship between biological distance and geographical distance. Under neutrality, genetic differences between populations accumulate because of genetic drift, and so their extent, represented by Fst, is inversely proportional to Ne and directly proportional to the time, T, elapsed since their separation. Therefore, to estimate T from genetic differences between populations, independent estimates of Ne are needed. Levels of linkage disequilibrium (LD) also depend on Ne and on the recombination rate between the SNPs considered (44). However, LD between SNPs separated by large distances along the chromosome reflects relatively recent Ne, whereas LD over short recombination distances depends on relatively ancient Ne (26). Thus, we estimated LD independently in each population considering the number of polymorphic markers available for that population, which depended on the sequencing platform in which the data were originally typed (Table S3). For example, ∼54,000 SNPs were used for the Aeta/Agta population and ∼600,000 SNPs were used for the Australian sample. We assigned to each SNP a genetic map position based on the HapMap2 (release 22) recombination data. For each pair of SNPs separated by less than 0.25 cM, we quantified LD as r2LD (45). All of the observed r2LD values were then binned into 50 recombination distance classes, from 0.005 to 0.25 cM, with incremental upper boundaries of 0.005 cM. Pairs of SNPs separated by less than 0.005 cM were not considered because at such short distances gene conversion may mimic the effects of recombination (44). We also adjusted the r2LD value for the sample size using (44). We estimated Ne for each population in each recombination distance class as , corresponding to the effective population size generations ago, where c is the distance between loci, expressed in Morgans (26, 46, 47). Finally, the long-term Ne for each population was calculated as the harmonic mean of Ne over all recombination distance classes up to 0.25 cM. At this point, based on the independently estimated values of Ne (Table S2), we calculated the separation time between populations as (25), expressed in generations (Table S1). All procedures were performed with the NeON and 4P software packages developed by the Barbujani laboratory and available online at (www.unife.it/dipartimento/biologia-evoluzione/ricerca/evoluzione-e-genetica/software). Because our objective was to test competing dispersal models, we did not include parameters of migration or admixture events in these calculations.
Cranial Phenotype Data.
We matched the sampled genetic populations with n = 233 modern human (Holocene) crania (Table 1), balancing population samples by sex to the extent possible. Crania, housed at the Musée de l’Homme, National Museum of Natural History in Paris, were selected on the basis of adult ontogeny and the absence of bone pathology. Congruence with the genetic populations was assessed first by ethnolinguistic affiliation and second by geographical provenance (Table S3). Following ref. 19, a total of 13 anatomical landmarks were collected by H.R.-C. for the right-side temporal bone of each specimen (Dataset S1). Landmarks were collected in the form of 3D coordinate data using a MicroScribe G2X desktop digitizer. Landmark measurement error was tested by digitizing a specimen 10 times across the span of a week and ranged from 0.25 to 1.157 mm, or 0.3–1.35%. All specimens were subjected to generalized Procrustes analysis, which superimposes the specimens following a least-squares procedure that rotates and translates the specimen landmark configurations and scales them to unit centroid size (19, 20, 33, 34). Because the number of variables (a total of 39 Procrustes shape variables per specimen) exceeded the number of specimens per population, we performed a principal component analysis (PCA) using the MorphoJ 1.05 software (48) and used PC scores to arrive at pairwise Pst. By convention, seven degrees of freedom are lost following Procrustes superimposition in three dimensions, accounting for scaling and for translation and rotation along each axis; therefore, a total of 32 PCs were used for arriving at Pst. We included in the calculation of Pst the parameters of Ne (Table S2) derived from the genetic data, as well as the cranial trait heritability value h2 = 0.23 ascertained for the basicranium in a modern human population (49). Pst calculations were made in the RMET 5.0 software and corrected for sampling bias (50).
Geographical Data.
Geodesic distances, G, were calculated using the PASSaGE 2 software (51), which assumes a spherical terrestrial shape and a radius of 6,379.336847 km. Latitude and longitude coordinates (Table 1) are an approximate centroid for each population, although we placed both African samples at Addis Ababa, Ethiopia to avoid assumptions about internal migrations within the continent. Our control model was calculated using the pairwise geodesic distances between populations, without consideration for geographical barriers (24). Waypoints were used in the other models to represent the complex geography of hypothetical dispersal routes (Fig. 1 and Table S4). The EE model connects populations by geographical proximity and primarily along a latitudinal axis (10, 12), with Cairo as a waypoint into Eurasia. The BSD model follows the migration pattern proposed in ref. 7, following a migration into Eurasia via the southern Arabian Peninsula. A broad MD model represents the hypothesis that Dravidian-speaking Indians, Philippine Aeta/Agta “Negritos,” Papuans, Melanesians, and Australians are relic southern route descendants, whereas Indo-European–speaking Indians, Central Asians, and Japanese are descendants of the second dispersal along a northern inland migration route through the Levant (13, 14). The MDI model assumes the same geographical dispersal scenario as the MD model but considers only Australians, Papuans, and Melanesians as southern route dispersal descendants (8) (Supporting Information, The “Negrito” Hypothesis, Figs. S1–S4, and Table S5).
Chronological Data and Hypothetical Fst.
Per dispersal model, hypothetical divergence values, C, between populations were determined by averaging the estimated dates of expected colonization in their indigenous region (Table 4). For example, the hypothetical divergence between Australians and Central Asians was at 42.5 ka under the EE model or 47.5 ka under the MDI-MP model. We treated these as T to take advantage of the known relationship between population differentiation and Ne. Using the Ne values derived from the genomic data, we then calculated hypothetical Fst values as .
Mantel Tests.
When distance matrices (as opposed to paired observations) are considered, the significance of their association can only be evaluated by comparison with an empirical null distribution (i.e., by Mantel tests). Simple Mantel tests were used to explore the correlation of the Fst and Pst values, because they are expected to be proportional under neutrality and thus display a linear correlation (21). The phenotypic distance between the Aeta/Agta (NE) and East African (EA) populations was a clear outlier in our dataset, greater than expected in an otherwise linear relationship between Fst and Pst values (Table 2 and Fig. S1). This demonstrates that these populations are the most phenotypically differentiated when considering the apportionment of variance between populations, and proportionally greater than their genetic differentiation. Given the statistical framework of our study (i.e., Pearson product-moment correlations), we removed this outlier from subsequent analyses. Simple Mantel tests were also used in our second analysis to assess the correlation between the hypothetical Fst values and the Fst or Pst values empirically derived from our datasets.
Partial Mantel correlations, estimated from the residuals of a previous correlation, allow one to keep constant the effects of a third matrix over the matrices being compared (23). The partial Mantel test (Table 3) assessed the correlation of the pairwise biological population differentiation values (Fst or Pst) against the dispersal models (G), while controlling for population divergence values (T). To assess whether one model could be favored over another when more than one competing model was correlated significantly after Bonferroni correction for multiple model tests, we conducted a Dow–Cheverud test (22, 24). In all cases, we ran 10,000 permutations to assess correlation significance. The population differentiation matrix (Fst or Pst) was permuted before the regression with T. This method is preferred over permuting the rows and columns of the residual matrices (23). Calculations were made in the PASSaGE 2 software (51).
Supplementary Material
Acknowledgments
We thank Aurélie Fort, Alain Froment, Liliana Huet, Véronique Laborde, and Philippe Mennecier for discussion and access to the cranial collections; Katarzyna Bryc, Carlos Bustamante, Irina Pugach, and Mark Stoneking for access to, and assistance with, their genetic datasets; and two anonymous reviewers for their constructive comments and suggestions. This work was supported by the Senckenberg Gesellshaft für Naturforschung and the European Commission (Erasmus Mundus International Master in Quaternary and Prehistory; European Research Council Advanced Grant Agreement 295733, “LanGeLin” Project).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
See Commentary on page 7170.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1323666111/-/DCSupplemental.
References
- 1.McDougall I, Brown FH, Fleagle JG. Stratigraphic placement and age of modern humans from Kibish, Ethiopia. Nature. 2005;433(7027):733–736. doi: 10.1038/nature03258. [DOI] [PubMed] [Google Scholar]
- 2.White TD, et al. Pleistocene Homo sapiens from Middle Awash, Ethiopia. Nature. 2003;423(6941):742–747. doi: 10.1038/nature01669. [DOI] [PubMed] [Google Scholar]
- 3.Fu Q, et al. A revised timescale for human evolution based on ancient mitochondrial genomes. Curr Biol. 2013;23(7):553–559. doi: 10.1016/j.cub.2013.02.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Poznik GD, et al. Sequencing Y chromosomes resolves discrepancy in time to common ancestor of males versus females. Science. 2013;341(6145):562–565. doi: 10.1126/science.1237619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mellars P, Gori KC, Carr M, Soares PA, Richards MB. Genetic and archaeological perspectives on the initial modern human colonization of southern Asia. Proc Natl Acad Sci USA. 2013;110(26):10699–10704. doi: 10.1073/pnas.1306043110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Petraglia MD, Haslam M, Fuller DQ, Boivin N, Clarkson C. Out of Africa: New hypotheses and evidence for the dispersal of Homo sapiens along the Indian Ocean rim. Ann Hum Biol. 2010;37(3):288–311. doi: 10.3109/03014461003639249. [DOI] [PubMed] [Google Scholar]
- 7.Oppenheimer S. Out-of-Africa, the peopling of continents and islands: Tracing uniparental gene trees across the map. Philos Trans R Soc Lond B Biol Sci. 2012;367(1590):770–784. doi: 10.1098/rstb.2011.0306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rasmussen M, et al. An Aboriginal Australian genome reveals separate human dispersals into Asia. Science. 2011;334(6052):94–98. doi: 10.1126/science.1211177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ramachandran S, et al. Support from the relationship of genetic and geographic distance in human populations for a serial founder effect originating in Africa. Proc Natl Acad Sci USA. 2005;102(44):15942–15947. doi: 10.1073/pnas.0507611102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu H, Prugnolle F, Manica A, Balloux F. A geographically explicit genetic model of worldwide human-settlement history. Am J Hum Genet. 2006;79(2):230–237. doi: 10.1086/505436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Manica A, Amos W, Balloux F, Hanihara T. The effect of ancient population bottlenecks on human phenotypic variation. Nature. 2007;448(7151):346–348. doi: 10.1038/nature05951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ramachandran S, Rosenberg NA. A test of the influence of continental axes of orientation on patterns of human gene flow. Am J Phys Anthropol. 2011;146(4):515–529. doi: 10.1002/ajpa.21533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mirazón Lahr M, Foley R. Multiple dispersals and modern human origins. Evol Anthropol. 1994;3(2):48–60. [Google Scholar]
- 14.Mirazón Lahr M. The Evolution of Modern Human Diversity: A Study of Cranial Variation. Cambridge, UK: Cambridge Univ Press; 1996. [Google Scholar]
- 15.Armitage SJ, et al. The southern route “out of Africa”: Evidence for an early expansion of modern humans into Arabia. Science. 2011;331(6016):453–456. doi: 10.1126/science.1199113. [DOI] [PubMed] [Google Scholar]
- 16.Reich D, et al. Denisova admixture and the first modern human dispersals into Southeast Asia and Oceania. Am J Hum Genet. 2011;89(4):516–528. doi: 10.1016/j.ajhg.2011.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pugach I, Delfin F, Gunnarsdóttir E, Kayser M, Stoneking M. Genome-wide data substantiate Holocene gene flow from India to Australia. Proc Natl Acad Sci USA. 2013;110(5):1803–1808. doi: 10.1073/pnas.1211927110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ghirotto S, Penso-Dolfin L, Barbujani G. Genomic evidence for an African expansion of anatomically modern humans by a Southern route. Hum Biol. 2011;83(4):477–489. doi: 10.3378/027.083.0403. [DOI] [PubMed] [Google Scholar]
- 19.Harvati K, Weaver TD. Human cranial anatomy and the differential preservation of population history and climate signatures. Anat Rec A Discov Mol Cell Evol Biol. 2006;288(12):1225–1233. doi: 10.1002/ar.a.20395. [DOI] [PubMed] [Google Scholar]
- 20.Smith HF, Ritzman T, Otárola-Castillo E, Terhune CE. A 3-D geometric morphometric study of intraspecific variation in the ontogeny of the temporal bone in modern Homo sapiens. J Hum Evol. 2013;65(5):479–489. doi: 10.1016/j.jhevol.2013.01.017. [DOI] [PubMed] [Google Scholar]
- 21.Roseman CC, Weaver TD. Molecules versus morphology? Not for the human cranium. Bioessays. 2007;29(12):1185–1188. doi: 10.1002/bies.20678. [DOI] [PubMed] [Google Scholar]
- 22.Pinhasi R, von Cramon-Taubadel N. Craniometric data supports demic diffusion model for the spread of agriculture into Europe. PLoS ONE. 2009;4(8):e6747. doi: 10.1371/journal.pone.0006747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Legendre P. Comparison of permutation methods for the partial correlation and partial mantel tests. J Statist Comput Simulation. 2000;67(1):37–73. [Google Scholar]
- 24.Hubbe M, Neves WA, Harvati K. Testing evolutionary and dispersion scenarios for the settlement of the new world. PLoS ONE. 2010;5(6):e11105. doi: 10.1371/journal.pone.0011105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Holsinger KE, Weir BS. Genetics in geographically structured populations: defining, estimating and interpreting F(ST) Nat Rev Genet. 2009;10(9):639–650. doi: 10.1038/nrg2611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hayes BJ, Visscher PM, McPartlan HC, Goddard ME. Novel multilocus measure of linkage disequilibrium to estimate past effective population size. Genome Res. 2003;13(4):635–643. doi: 10.1101/gr.387103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Clark JD, et al. Stratigraphic, chronological and behavioural contexts of Pleistocene Homo sapiens from Middle Awash, Ethiopia. Nature. 2003;423(6941):747–752. doi: 10.1038/nature01670. [DOI] [PubMed] [Google Scholar]
- 28.Blome MW, Cohen AS, Tryon CA, Brooks AS, Russell J. The environmental context for the origins of modern human diversity: A synthesis of regional variability in African climate 150,000-30,000 years ago. J Hum Evol. 2012;62(5):563–592. doi: 10.1016/j.jhevol.2012.01.011. [DOI] [PubMed] [Google Scholar]
- 29.Eriksson A, et al. Late Pleistocene climate change and the global expansion of anatomically modern humans. Proc Natl Acad Sci USA. 2012;109(40):16089–16094. doi: 10.1073/pnas.1209494109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Scholz CA, et al. East African megadroughts between 135 and 75 thousand years ago and bearing on early-modern human origins. Proc Natl Acad Sci USA. 2007;104(42):16416–16421. doi: 10.1073/pnas.0703874104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Klein RG. Archeology and the evolution of human behavior. Evol Anthropol. 2000;9(1):17–36. [Google Scholar]
- 32.Grine FE, et al. Late Pleistocene human skull from Hofmeyr, South Africa, and modern human origins. Science. 2007;315(5809):226–229. doi: 10.1126/science.1136294. [DOI] [PubMed] [Google Scholar]
- 33.Gunz P, et al. Early modern human diversity suggests subdivided population structure and a complex out-of-Africa scenario. Proc Natl Acad Sci USA. 2009;106(15):6094–6098. doi: 10.1073/pnas.0808160106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Harvati K, et al. The Later Stone Age calvaria from Iwo Eleru, Nigeria: Morphology and chronology. PLoS ONE. 2011;6(9):e24024. doi: 10.1371/journal.pone.0024024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Demeter F, et al. Anatomically modern human in Southeast Asia (Laos) by 46 ka. Proc Natl Acad Sci USA. 2012;109(36):14375–14380. doi: 10.1073/pnas.1208104109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu W, et al. Human remains from Zhirendong, South China, and modern human emergence in East Asia. Proc Natl Acad Sci USA. 2010;107(45):19201–19206. doi: 10.1073/pnas.1014386107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mijares AS, et al. New evidence for a 67,000-year-old human presence at Callao Cave, Luzon, Philippines. J Hum Evol. 2010;59(1):123–132. doi: 10.1016/j.jhevol.2010.04.008. [DOI] [PubMed] [Google Scholar]
- 38.Green RE, et al. A draft sequence of the Neandertal genome. Science. 2010;328(5979):710–722. doi: 10.1126/science.1188021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ghirotto S, Tassi F, Benazzo A, Barbujani G. No evidence of Neandertal admixture in the mitochondrial genomes of early European modern humans and contemporary Europeans. Am J Phys Anthropol. 2011;146(2):242–252. doi: 10.1002/ajpa.21569. [DOI] [PubMed] [Google Scholar]
- 40.Eriksson A, Manica A. Effect of ancient population structure on the degree of polymorphism shared between modern human populations and ancient hominins. Proc Natl Acad Sci USA. 2012;109(35):13956–13960. doi: 10.1073/pnas.1200567109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lowery RK, et al. Neanderthal and Denisova genetic affinities with contemporary humans: Introgression versus common ancestral polymorphisms. Gene. 2013;530(1):83–94. doi: 10.1016/j.gene.2013.06.005. [DOI] [PubMed] [Google Scholar]
- 42.Smith FH, Falsetti AB, Donnelly SM. Modern human origins. Am J Phys Anthropol. 1989;32(S10):35–68. [Google Scholar]
- 43.Purcell S, et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81(3):559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tenesa A, et al. Recent human effective population size estimated from linkage disequilibrium. Genome Res. 2007;17(4):520–526. doi: 10.1101/gr.6023607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hill WG, Robertson A. Linkage disequilibrium in finite populations. Theor Appl Genet. 1968;38(6):226–231. doi: 10.1007/BF01245622. [DOI] [PubMed] [Google Scholar]
- 46.McVean GAT. A genealogical interpretation of linkage disequilibrium. Genetics. 2002;162(2):987–991. doi: 10.1093/genetics/162.2.987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sved JA. Linkage disequilibrium and homozygosity of chromosome segments in finite populations. Theor Popul Biol. 1971;2(2):125–141. doi: 10.1016/0040-5809(71)90011-6. [DOI] [PubMed] [Google Scholar]
- 48.Klingenberg CP. MorphoJ: an integrated software package for geometric morphometrics. Mol Ecol Resour. 2011;11(2):353–357. doi: 10.1111/j.1755-0998.2010.02924.x. [DOI] [PubMed] [Google Scholar]
- 49.Martínez-Abadías N, et al. Heritability of human cranial dimensions: Comparing the evolvability of different cranial regions. J Anat. 2009;214(1):19–35. doi: 10.1111/j.1469-7580.2008.01015.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Relethford JH, Crawford MH, Blangero J. Genetic drift and gene flow in post-famine Ireland. Hum Biol. 1997;69(4):443–465. [PubMed] [Google Scholar]
- 51.Rosenberg MS, Anderson CD. PASSaGE: Pattern Analysis, Spatial Statistics and Geographic Exegesis. Version 2. Methods Ecol Evol. 2011;2(3):229–232. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.