Significance
The abundances of predators and their prey can oscillate in time. Mathematical theory of predator–prey systems predicts that in predator–prey cycles, peaks in prey abundance precede peaks in predator abundance. However, these models do not consider how the evolution of predator and prey traits related to offense and defense will affect the ordering and timing of peaks. Here we show that predator–prey coevolution can effectively reverse the ordering of peaks in predator–prey cycles, i.e., peaks in predator abundance precede peaks in prey abundance. We present examples from three distinct systems that exhibit reversed cycles, suggesting that coevolution may be an important driver of cycles in those systems.
Keywords: eco-coevolutionary dynamics, fast–slow dynamics, population biology, community ecology
Abstract
A hallmark of Lotka–Volterra models, and other ecological models of predator–prey interactions, is that in predator–prey cycles, peaks in prey abundance precede peaks in predator abundance. Such models typically assume that species life history traits are fixed over ecologically relevant time scales. However, the coevolution of predator and prey traits has been shown to alter the community dynamics of natural systems, leading to novel dynamics including antiphase and cryptic cycles. Here, using an eco-coevolutionary model, we show that predator–prey coevolution can also drive population cycles where the opposite of canonical Lotka–Volterra oscillations occurs: predator peaks precede prey peaks. These reversed cycles arise when selection favors extreme phenotypes, predator offense is costly, and prey defense is effective against low-offense predators. We present multiple datasets from phage–cholera, mink–muskrat, and gyrfalcon–rock ptarmigan systems that exhibit reversed-peak ordering. Our results suggest that such cycles are a potential signature of predator–prey coevolution and reveal unique ways in which predator–prey coevolution can shape, and possibly reverse, community dynamics.
Population cycles, e.g., predator–prey cycles, and their ecological drivers have been of interest for the last 90 y (1–4). Classical models of predator–prey systems, developed first by Lotka (5) and Volterra (6), share a common prediction: Prey oscillations precede predator oscillations by up to a quarter of the cycle period (7). When plotted in the predator–prey phase plane, these cycles have a counterclockwise orientation (4). These cycles are driven by density-dependent interactions between the populations. When predators are scarce, prey increase in abundance. As their food source increases, predators increase in abundance. When the predators reach sufficiently high densities, the prey population is driven down to low numbers. With a scarcity of food, the predator population crashes and the cycle repeats.
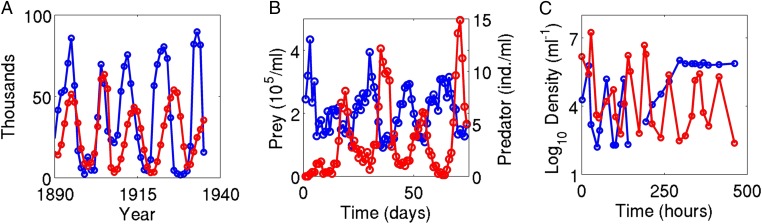
While many cycles, like the classic lynx–hare cycles (Fig. 1A) (3), exhibit the above characteristics, predator–prey cycles with different characteristics have also been observed. For example, antiphase cycles where predator oscillations lag behind prey oscillations by half of the cycle period (Fig. 1B) (8) and cryptic cycles where the predator population oscillates while the prey population remains effectively constant (Fig. 1C) (9) have been observed in experimental systems. This diversity of cycle types motivates the question, “Why do cycle characteristics differ across systems?”
Fig. 1.
Examples of different kinds of predator–prey cycles. (A) Counterclockwise lynx–hare cycles (3). (B) Antiphase rotifer–algal cycles (8). (C) Cryptic phage-bacteria cycles (9). In all time series, red and blue correspond to predator and prey, respectively. See SI Text, section C for data sources.
In Lotka–Volterra and other ecological models, predator and prey life history traits are assumed to be fixed. However, empirical studies across taxa have shown that prey (9–16) and predators (17–20) can evolve over ecological time scales. That is, changes in allele frequencies (and associated phenotypes) can occur at the same rate as changes in population densities or spatial distributions and alter the ecological processes driving the changes in population densities or distributions; this phenomenon has been termed “eco-evolutionary dynamics” (21, 22). Furthermore, predator–prey coevolution is important for driving community composition and dynamics (16, 19, 20, 23–26). This body of work suggests that the interaction between ecological and evolution processes has the potential to alter the ecological dynamics of communities.
Experimental (8, 9, 13, 14) and theoretical studies (13, 27, 28) have shown that prey or predator evolution alone can alter the characteristics of predator–prey cycles and drive antiphase (Fig. 1B) and cryptic (Fig. 1C) cycles. Additional theoretical work has shown that predator–prey coevolution can also drive antiphase and cryptic cycles (29). Thus, evolution in one or both species is one mechanism through which antiphase or cryptic predator–prey cycles can arise. However, it is unclear if coevolution can drive additional kinds of cycles with characteristics different from those in Fig. 1.
The main contribution of this study is to show that predator–prey coevolution can drive unique cycles where peaks in predator abundance precede peaks in prey abundance, the opposite of what is predicted by classical ecological models. We refer to these reversed cycles as “clockwise cycles.” The theoretical and empirical finding of clockwise cycles represents an example of how evolution over ecological time scales can alter community-level dynamics.
Models
Discrete Trait Clonal Predator–Prey Model.
We first consider a particular mechanism of eco-coevolutionary dynamics: a clonal predator–prey system where individuals can only have particular discrete trait values. The prey population is composed of low- and high-vulnerability prey clones with trait values and , respectively. The predator population is composed of low- and high-offense predator clones with trait values and , respectively. The variables , , , and denote the densities of the respective clones.
The dynamics of the clonal types can be written as
| [1] |
where represents the growth rates of prey clones, is the predation rate of prey by predator , is the growth rate of predator due to consumption of prey , and is the death rate of predator . We work with the general functional forms in system 1 to gain insight into how coevolution shapes community dynamics across models. However, in simulations we use functional forms that are standard in ecological models. For example, in our simulations prey exhibit logistic growth and predation rates are Type I or II functional responses (30).
In the clonal model, evolution occurs via temporal fluctuations in the frequencies of the clonal types. Key to our approach is the assumption that clones differ in their life history traits and that there are costs for low vulnerability and high offense. Trade-offs between species traits have been observed in predators (31, 32) and prey (13, 33, 34). In the clonal model, low-vulnerability prey clones are consumed at a lower rate than high-vulnerability clones at the cost of a lower growth rate (i.e., a lower intraspecific competitive ability). High-offense predator clones have higher predation rates that come at the cost of a higher mortality rate. See SI Text, section A for additional details.
Continuous Trait Eco-coevolutionary Model.
While some populations comprise genetically distinct clonal subpopulations, phenotypes may also vary continuously. We extend prior theoretical work on an eco-evolutionary predator–prey system with one evolving species (28) to a general eco-coevolutionary system where predators and prey coevolve and selection favors extreme phenotypes. In the continuous trait model, x and y denote the densities of the total prey and total predator populations, respectively, and α and β denote the mean trait values of those populations, respectively. Larger prey trait values correspond to higher intraspecific competitive ability, which comes at the cost of increased vulnerability to predation. Larger predator trait values correspond to increased offense, which comes at the cost of increased mortality.
The continuous trait model follows from the quantitative genetics approach derived in Lande (35) and Abrams et al. (36). The continuous trait model can be written as
| [2] |
where F is the prey growth rate in the absence of predation, G is the predation rate, H is the composition of the predation rate and the predator to prey conversion, and D is the predator death rate. As in the clonal model, our approach uses general functional forms, however in numerical simulations we use the functional forms that are standard in ecological models. The terms and represent the genetic variances of the traits. The remaining terms in the trait equations represent the individual fitness gradients for the traits, where the derivatives are taken with respect to an individual’s phenotype ( or ); see SI Text, section B for details. In system 2, evolution drives the mean trait values in the direction of increasing fitness.
The functions and also bound the mean trait values between their lowest ( and ) and highest ( and ) allowable values. In numerical simulations we use and . However, our qualitative results (e.g., cycle orientation in the phase plane) remain the same when alternative bounding functions are used; see SI Text, section B.7 for details. We note that without the bounds, runaway evolution (i.e., evolution toward arbitrarily large trait values) could occur under disruptive selection. However, we expect the trait values to have bounds in natural systems. For example, prey cannot invest less in defense when completely vulnerable to predation and prey cannot invest more in defense once they are completely invulnerable to predation.
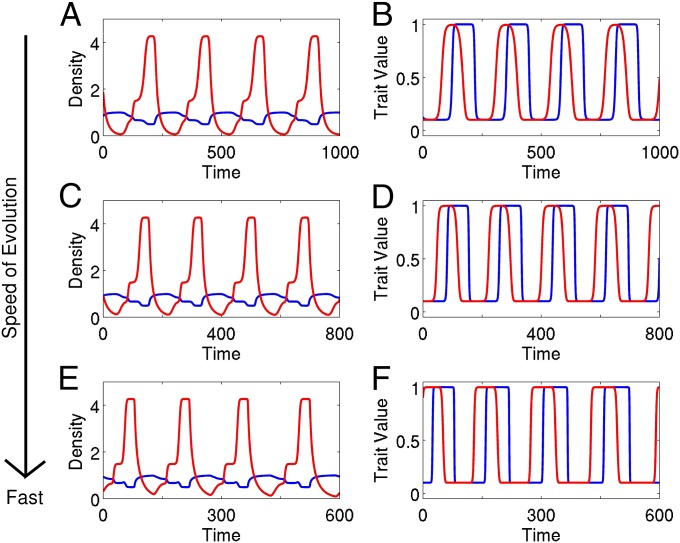
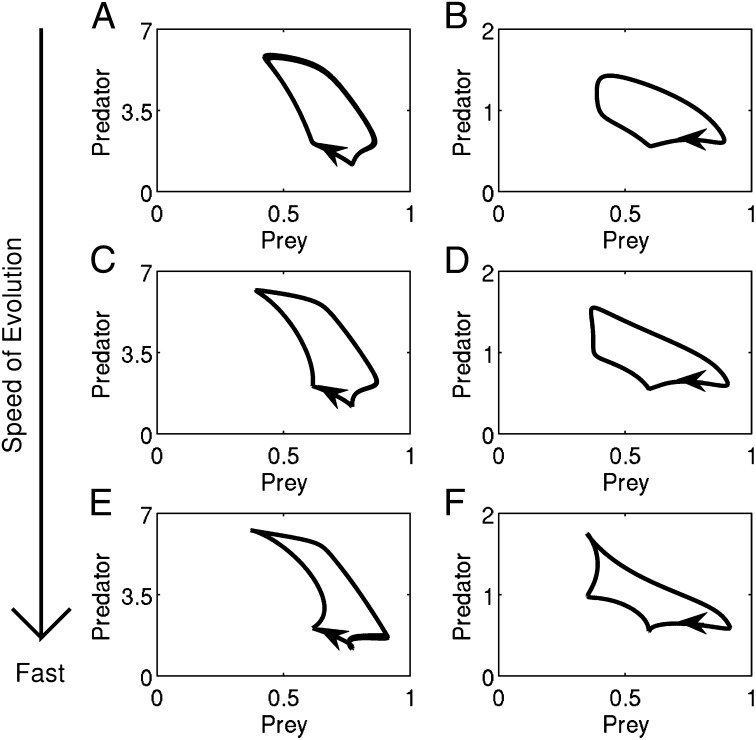
Changing the amount of genetic variation (V) in the continuous trait model 2 changes the speed of the evolutionary dynamics. The rates of ecological and evolutionary change are comparable when . To study and understand the dynamics that arise when , we artificially increase the amount of genetic variation in system 2 until the speed of evolution is nearly instantaneous with respect to the population dynamics of the system. Mathematically this is done by studying the dynamics of system 2 when V is large and positive . In studying this fast evolution limit, we are not positing that nearly instantaneous evolution can occur in nature. Instead, as seen in Fig. 2, while numerical differences arise, the essential qualitative features of the time series, e.g., the phase relations between the population densities, do not change as the speed of evolution is increased by a factor of 2 (Fig. 2 C and D) and 5 (Fig. 2 E and F). Furthermore, as the speed of evolution increases, the ecological dynamics of the system do not become increasingly unrealistic. Thus, as has been done previously in eco-evolutionary models with a single evolving species (28), by studying the dynamics where evolution is nearly instantaneous, we can understand how coevolution alters the ecological dynamics of natural predator–prey communities.
Fig. 2.
The qualitative characteristics of the ecological time series from continuous trait model 2 remain the same as the speed of evolution increases. (A, C, and E) Predator (red) and prey (blue) densities. (B, D, and F) Mean predator (offense, red) and mean prey (vulnerability, blue) traits. The speed of evolution is (A and B) as fast, (C and D) two times as fast, and (E and F) five times as fast as the ecological dynamics of the system. In A–F, prey exhibit logistic growth in the absence of predation, predation rates are Type II functional responses, and predators have a linear death rate; see SI Text, section D for equations and parameters.
Results
Clonal Model Yields Clockwise Cycles.
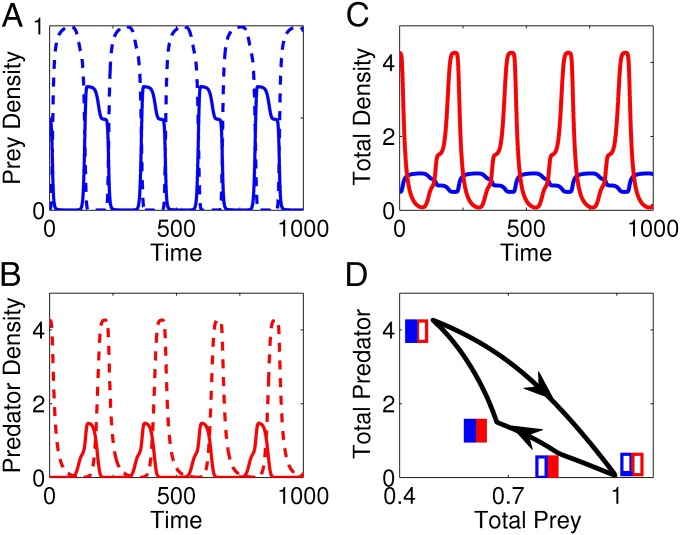
In the clonal model 1, clockwise oscillations can occur between the total prey density and the total predator density . These cycles arise when the defense of low-vulnerability prey is effective against low-offense predators and offense is costly. As an example, consider Fig. 3 with low- (dashed blue, Fig. 3A) and high-vulnerability (solid blue, Fig. 3A) prey clones and low- (dashed red, Fig. 3B) and high-offense (solid red, Fig. 3B) predator clones. In Fig. 3 A and B, the densities and frequencies of the clones fluctuate over time. Summing the clonal densities to obtain the total predator and prey population densities yields the predator–prey oscillations in Fig. 3C. These cycles have a clockwise orientation in the phase plane (Fig. 3D), the opposite of what is predicted by classical predator–prey models.
Fig. 3.
Example of a clonal model exhibiting clockwise predator–prey cycles. (A) Low- (dashed blue) and high- (solid blue) vulnerability prey densities. (B) Low- (dashed red) and high- (solid red) offense predator densities. (C) Total prey (blue) and total predator (red) densities. (D) Total prey and predator densities exhibit clockwise cycles in the phase plane; arrows denote the flow of time. Blue and red rectangles denote the frequencies of the prey and predator clonal types along the cycle, respectively. In the rectangles, open areas correspond to the frequency of low-vulnerability or low-offense clones and filled areas correspond to the frequency of high-vulnerability or high-offense clones. Simulations are of clonal model 1 with logistic growth of the prey clones, Type II functional responses, and linear predator mortality rates; see SI Text, section D for equations and parameters.
Biologically, the clockwise cycles are driven by fluctuations in the densities and fitnesses of the clonal types. To understand how, consider a small prey population dominated by low-vulnerability clones interacting with a large predator population dominated by low-offense clones. The defended prey drive the predators to low abundance, allowing the prey to increase. Here, a prey peak follows the predator peak. The increase in low-vulnerability prey increases the fitness of high-offense predators, allowing high-offense predators to increase and begin to drive the prey population down. Due to high costs for offense, the abundance of high-offense predators remains low, resulting in a selective advantage for the high-vulnerability clone. As the prey population becomes dominated by high-vulnerability clones, the high-offense predators increase and the prey population continues to decrease. Since relatively few prey are present and low-offense predators pay a lesser cost, selection favors low-offense predators. The low-offense predators replace the high-offense predators, resulting in increased predator abundance and a further decrease in the prey population. With many predators present, selection favors low-vulnerability prey and the cycle repeats.
Continuous Trait Models Can Approximate Clonal Models.
The total prey and total predator population dynamics in Fig. 3C are quantitatively similar to the population dynamics in Fig. 2A. This suggests that the continuous trait model 2 can approximate the dynamics of the discrete trait clonal model 1. In SI Text, section A, we show that the continuous trait model can approximate the dynamics of the clonal model by focusing on the dynamics of the total prey density , the total predator density , the mean prey trait , and the mean predator trait . In SI Text, section A we also quantify the error that arises in using this approximation, identify cases in which the approximation is exact, and discuss extensions to systems with more than two clonal types per species. The time series in Figs. 3 and 2 A and B are examples of when the clonal and continuous trait models exhibit identical dynamics. In total, via this approximation, we can explore how clockwise cycles arise in systems with continuous or discrete traits by studying the continuous trait model 2.
Continuous Trait Model Yields Clockwise Cycles.
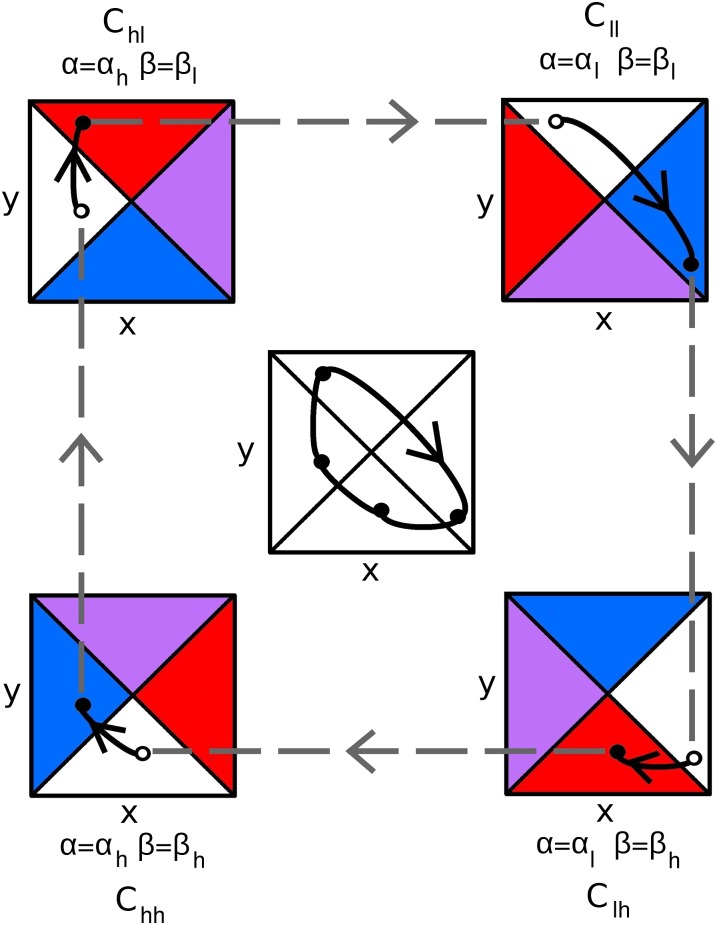
To understand when clockwise cycles arise in system 2, we focus on the dynamics where evolution is much faster than ecology . In the fast evolution limit, the eco-evolutionary dynamics of the system decompose into fast evolutionary jumps and slow changes in population densities. These fast and slow dynamics can be understood, via fast–slow dynamical system theory (37) by studying the four 2D planes called critical manifolds, as shown in Fig. 4. Each critical manifold defines the population dynamics that occur when the traits are fixed at their lowest or highest values. The white regions of the planes are stable and the colored regions are unstable with respect to the evolutionary dynamics of the system. When a solution to system 2 is near a critical manifold, the populations slowly change as if stuck to that critical manifold (solid black curves in Fig. 4). During this time, the traits remain essentially fixed. After the population densities cross into a colored region of a plane, the solution jumps away and lands in the white region of another critical manifold (dashed gray lines in Fig. 4). During the jump, one trait evolves while the other trait and the population densities remain fixed. Repeating this process yields an eco-coevolutionary cycle. Concatenating the population dynamics on the critical manifolds results in the predator–prey cycle shown in the middle plane of Fig. 4. See SI Text, section B for additional details.
Fig. 4.
When evolution is nearly instantaneous, eco-evolutionary cycles can be decomposed into slow ecological dynamics and fast evolutionary jumps. Slow population dynamics (solid black curves) occur on four critical manifolds (x,y planes labeled “” in the corners). The critical manifolds are defined by fixing the mean prey (α) and mean predator (β) traits at their low (l) or high (h) values. White regions in the planes are evolutionary attractors and colored regions evolutionary repellers in the α-direction (red), the β-direction (blue), or both directions (purple). Fast evolutionary dynamics (dashed gray lines) occur as solutions jump between the critical manifolds. Solutions behave in the following way. A solution lands in the white region of a critical manifold (open circle) and then moves (solid black curves) toward the attracting equilibrium point on that manifold (filled circle). After crossing into a colored region of the manifold, the solution jumps to the white region of another critical manifold (dashed gray line). Repeating and concatenating the population dynamics on each critical manifold yields the eco-coevolutionary cycle in the center.
While clockwise cycles do not always arise in system 2, we find that clockwise cycles can arise when selection favors extreme trait values and intermediate trait values are never optimal (i.e., disruptive selection), the defense of the low-vulnerability prey is effective against low-offense predators, and offense is costly. These conditions come from our analysis of the general continuous trait model 2 in the fast evolution limit; see SI Text, section B.4 for details. The conditions and the species’ trade-offs ensure that the disadvantages of high vulnerability and low offense and the advantages of high offense and low vulnerability do not lead to evolutionary fixation. For a particular eco-coevolutionary model, these conditions will be realized through particular constraints on the parameter values of the model. In SI Text, section B.5, we provide a worked illustrative example of a coevolutionary Lotka–Volterra model and show that in the regions of parameter space where clockwise cycles arise, our general conditions also hold.
The sequence of slow ecological and fast evolutionary dynamics in Fig. 4 elucidates how clockwise cycles arise in the continuous trait model. Consider a small population of low-vulnerability prey and a large population of low-offense predators ( in Fig. 4). Due to the effective defense, the low-vulnerability prey drive the predators down while increasing in abundance. This results in selection for high-offense predators ( in Fig. 4). The high-offense predators decrease the prey population, however the predator population remains low due to high costs for offense. Low predation pressure selects for more competitive, more vulnerable prey ( in Fig. 4). Consequently, the predators increase and drive the prey population down further. Low-prey abundance selects for low-offense predators ( in Fig. 4). As predator density increases and prey density decreases, selection increases for low vulnerability and the cycle repeats.
While our conditions and analysis of clockwise cycles are general, we present three numerical examples in Figs. 2 and 5 where prey exhibit logistic growth and predation rates are Type II functional responses. In Fig. 2, peaks in predator abundance precede peaks in prey abundance. Since the dynamics of the continuous trait model in Fig. 2A and the clonal model in Fig. 3 are identical, the time series in Fig. 2A have a clockwise orientation when plotted in the predator–prey phase plane (Fig. 2D). Fig. 5 presents two additional examples of clockwise predator–prey cycles for varying evolutionary speeds. Note that the cycle orientation is preserved as the speed of evolution increases by a factor of 2 (Fig. 5 C and D) and 5 (Fig. 5 E and F).
Fig. 5.
Two examples of clockwise predator–prey cycles. The speed of evolution is (A and B) as fast, (C and D) two times as fast, and (E and F) five times as fast as the ecological dynamics of the system. Simulations are of continuous trait model 2 where the prey exhibit logistic growth in the absence of predation and predation rates follow a Type II functional response. In A, C, and E, the predators have a linear death rate and in B, D, and F the predators have a nonlinear death rate; see SI Text, section D for equations and parameters.
Empirical Datasets Exhibiting Clockwise Cycles.
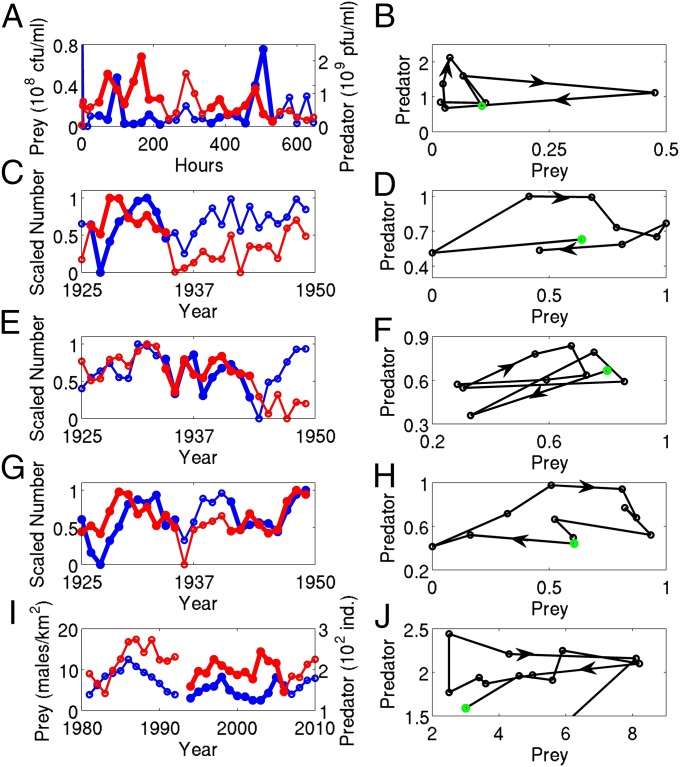
We have shown that predator–prey coevolution can, in theory, drive clockwise cycles. To what extent are clockwise cycles found in ecological time series data? We revisited long-term population time series and identified three cases of interest: phage–cholera chemostat experiments (38), gyrflacon–rock ptarmigan census data (39), and mink and muskrat trapping data from the Hudson’s Bay Company (40). In each case, we reexamined the time series and identified regions where predators (or analogs thereof) and their prey exhibit clockwise cycles; see SI Text, section C for a discussion of how cycle type was identified. In the thickened regions of the time series in Fig. 6 A, C, E, G, and I, peaks in predator density (or number) precede those of the prey. When the thickened segments are plotted in the predator–prey phase plane (Fig. 6 B, D, F, H, and J), the cycles have a clockwise orientation. These characteristics suggest that coevolution could be influencing the dynamics of those systems.
Fig. 6.
Examples of clockwise predator–prey cycles from (A and B) phage–cholera chemostat experiments (38), (C–H) mink and muskrat trapping data from the Hudson’s Bay Company (40), and (I and J) gyrfalcon and rock ptarmigan census data (39). (A, C, E, G, and I) Predator (red) and prey (blue) time series. (B, D, F, H, and J) Thickened segments of time series plotted in the phase plane. Arrows denote the flow of time and green circles denote the first time point. Only the first thickened segments of A and G are plotted in B and H; see SI Text, section C for the second segments. CFU, colony-forming unit; PFU, plaque-forming unit. See SI Text, section C for data sources.
Additional evidence supporting this idea is available for the phage–cholera system. First, in Fig. 6A the predator peaks always occur with or before the prey peaks. Second, in the Wei et al. study (38), the authors observed that the host and phage populations were each composed of two types (but did not measure the density of each type). Our results suggest that the clockwise phage–cholera cycles in Fig. 6B are driven by changes in the frequencies of phage and cholera types, similar to the dynamics of the clonal model 1.
Discussion
Our results add to a growing body of work showing that evolution can alter the dynamics of predator–prey systems and potentially mask classical signatures of predator–prey interactions. Phase relations between predator and prey oscillations have been used previously to infer the existence and strength of predator–prey interactions (41, 42). Based on ecological theory, large positive phase differences where prey peaks precede predator peaks are interpreted as strong predatory interactions. Negative phase differences where prey peaks follow predator peaks are interpreted as very weak interactions or the absence of predatory interactions (41). Previous work has shown that evolution in one or both species can drive cryptic cycles where one species oscillates while the other remains effectively constant (9, 14, 28, 29). Because the density of one species does not oscillate, phase relations cannot be measured or used to identify predator–prey interactions.
Coevolution, via clockwise cycles, can also mask classical signatures of predatory interactions. Because peaks in predator density precede peaks in prey density (i.e., there is a negative phase difference), one might conclude that the prey are eating the predator. Indeed, while later shown to be due to recording errors (3), clockwise cycles were observed in the classic lynx–hare time series and led Gilpin (43) to ask, “Do hares eat lynx?” Our results suggest that predator–prey coevolution is an alternative explanation for negative phase differences. This difference in interpretation points to important challenges in identifying species interactions from ecological time series data. In particular, information about phenotypic variation may be necessary to distinguish weak predator–prey interactions and predator–prey interactions mediated by predator–prey coevolution.
One underlying assumption of the continuous trait model is that there is standing genetic (and phenotypic) variation in the predator and prey populations. Because of this, we expect clockwise cycles to arise in systems where genotypes with extreme phenotypes are present (although possibly at low density) and oscillate in abundance over time, e.g., clonal systems. However, we do advise caution when making inferences about predator–prey coevolution from time series data like the mink–muskrat and gyrflacon–rock ptarmigan data in Fig. 6. Theoretical models similar to system 2 have been used to model the effects of phenotypic plasticity on community dynamics (36, 44) and to model species turnover and succession in multispecies communities (45). Plasticity and species diversity are two alternative mechanisms through which standing phenotypic diversity can be realized. Thus, while coevolution is one mechanism through which clockwise cycles arise, clockwise cycles may also arise via plastic adaptation (e.g., adaptively foraging predators) or in communities with multiple prey and predator species without evolution. An important area of future research is understanding if and when clockwise cycles can arise via these other mechanisms.
Finally, we return to the fast evolution framework used to understand how coevolution drives clockwise cycles. The fast evolution limit reduces model complexity by using fast–slow dynamical systems theory. This results in insight into how evolutionary processes alter the ecological dynamics of communities. Previous studies, including those on adaptive dynamics (46, 47), have used the same body of theory to reduce model complexity and study eco-evolutionary dynamics in the limit where evolution is much slower than ecology (48, 49). The slow evolution limit yields insight into how ecological processes alter the evolutionary dynamics of species. Because the fast and the slow evolution limits assume a separation of time scales between the ecological and evolutionary dynamics, both limiting cases are approximations of the dynamics that occur in natural systems where the rates of ecological and evolutionary processes are comparable. Both limiting cases are useful because they provide complementary viewpoints from which to study eco-evolutionary dynamics. While these two approaches may not yet yield a complete picture, the fast and slow evolution limits can help identify how eco-evolutionary feedbacks shape the ecological and evolutionary dynamics of natural communities.
Supplementary Material
Acknowledgments
The authors thank Peter Abrams, Luis Jover, Bradford Taylor, and two anonymous reviewers for helpful comments. The authors thank Stephen Ellner, Bruce Levin, and Olafur Nielsen for sharing time series data. M.H.C. was supported by the National Science Foundation under Award DMS-1204401. J.S.W. holds a Career Award at the Scientific Interface from the Burroughs Wellcome Fund.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. A.H. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1317693111/-/DCSupplemental.
References
- 1.Elton CS. Periodic fluctuations in the number of animals: Their causes and effects. J Exp Biol. 1924;2:119–163. [Google Scholar]
- 2.Gause GF. Experimental demonstration of Volterra’s periodic oscillations in the numbers of animals. J Exp Biol. 1935;12(1):44–48. [Google Scholar]
- 3.Elton CS, Nicholson M. Fluctuations in numbers of the muskrat (Ondatra zibethica) in Canada. J Anim Ecol. 1942;11(1):96–126. [Google Scholar]
- 4.Rosenzweig ML, MacArthur RH. Graphical representation and stability conditions of predator-prey interactions. Am Nat. 1963;97(895):209–223. [Google Scholar]
- 5.Lotka AJ. 1934. Théorie analytique des associations biologiques, Hermann, Paris.
- 6.Volterra V. Variazioni e fluttuazioni del numero dindividui in specie animali conviventi. Mem R Accad Naz Lincei (Rome) 1926;2:31–113. [Google Scholar]
- 7.Bulmer MG. Phase relations in the ten-year cycle. J Anim Ecol. 1975;44(2):609–621. [Google Scholar]
- 8.Becks L, Ellner SP, Jones LE, Hairston NG., Jr Reduction of adaptive genetic diversity radically alters eco-evolutionary community dynamics. Ecol Lett. 2010;13(8):989–997. doi: 10.1111/j.1461-0248.2010.01490.x. [DOI] [PubMed] [Google Scholar]
- 9.Bohannan BJM, Lenski RE. Effect of prey heterogeneity on the response of a model food chain to resource enrichment. Am Nat. 1999;153(1):73–82. doi: 10.1086/303151. [DOI] [PubMed] [Google Scholar]
- 10.Hairston NG, Jr, Walton WE. Rapid evolution of a life history trait. Proc Natl Acad Sci USA. 1986;83(13):4831–4833. doi: 10.1073/pnas.83.13.4831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hairston NG, Jr, Dillon TA. Fluctuating selection and reponse in a population of freshwater copepods. Evolution. 1990;44(7):1796–1805. doi: 10.1111/j.1558-5646.1990.tb05250.x. [DOI] [PubMed] [Google Scholar]
- 12.Reznick DN, Shaw FH, Rodd FH, Shaw RG. Evaluation of the rate of evolution in natural populations of guppies (Poecilia reticulata) Science. 1997;275(5308):1934–1937. doi: 10.1126/science.275.5308.1934. [DOI] [PubMed] [Google Scholar]
- 13.Yoshida T, Jones LE, Ellner SP, Fussmann GF, Hairston NG., Jr Rapid evolution drives ecological dynamics in a predator-prey system. Nature. 2003;424(6946):303–306. doi: 10.1038/nature01767. [DOI] [PubMed] [Google Scholar]
- 14.Yoshida T, et al. Cryptic population dynamics: Rapid evolution masks trophic interactions. PLoS Biol. 2007;5(9):e235. doi: 10.1371/journal.pbio.0050235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reznick DN, Ghalambor CK, Crooks K. Experimental studies of evolution in guppies: A model for understanding the evolutionary consequences of predator removal in natural communities. Mol Ecol. 2008;17(1):97–107. doi: 10.1111/j.1365-294X.2007.03474.x. [DOI] [PubMed] [Google Scholar]
- 16.Wei Y, Ocampo P, Levin BR. An experimental study of the population and evolutionary dynamics of Vibrio cholerae O1 and the bacteriophage JSF4. Proc Biol Sci. 2010;277(1698):3247–3254. doi: 10.1098/rspb.2010.0651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hairston NG, Jr, et al. Rapid evolution revealed by dormant eggs. Nature. 1999;401:446. [Google Scholar]
- 18.Grant PR, Grant BR. Unpredictable evolution in a 30-year study of Darwin’s finches. Science. 2002;296(5568):707–711. doi: 10.1126/science.1070315. [DOI] [PubMed] [Google Scholar]
- 19.Mizoguchi K, et al. Coevolution of bacteriophage PP01 and Escherichia coli O157:H7 in continuous culture. Appl Environ Microbiol. 2003;69(1):170–176. doi: 10.1128/AEM.69.1.170-176.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hall AR, Scanlan PD, Buckling A. Bacteria-phage coevolution and the emergence of generalist pathogens. Am Nat. 2011;177(1):44–53. doi: 10.1086/657441. [DOI] [PubMed] [Google Scholar]
- 21.Fussmann GF, Loreau M, Abrams PA. Eco-evolutionary dynamics of communities and ecosystems. Funct Ecol. 2007;21:465–477. [Google Scholar]
- 22.Kinnison MT, Hairston NG., Jr Eco-evolutionary conservation biology: Contemporary evolution and dynamics of persistence. Funct Ecol. 2007;21:444–454. [Google Scholar]
- 23.Horne MT. Coevolution of Escherichia coli and bacteriophages in chemostat culture. Science. 1970;168(3934):992–993. doi: 10.1126/science.168.3934.992-a. [DOI] [PubMed] [Google Scholar]
- 24.Jones LE, et al. Rapid contemporary evolution and clonal food web dynamics. Philos Trans R Soc Lond B Biol Sci. 2009;364(1523):1579–1591. doi: 10.1098/rstb.2009.0004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Palkovacs EP, et al. Experimental evaluation of evolution and coevolution as agents of ecosystem change in Trinidadian streams. Philos Trans R Soc Lond B Biol Sci. 2009;364(1523):1617–1628. doi: 10.1098/rstb.2009.0016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hall AR, Scanlan PD, Morgan AD, Buckling A. Host-parasite coevolutionary arms races give way to fluctuating selection. Ecol Lett. 2011;14(7):635–642. doi: 10.1111/j.1461-0248.2011.01624.x. [DOI] [PubMed] [Google Scholar]
- 27.Abrams PA, Matsuda H. Prey adaptation as a cause of predator-prey cycles. Evolution. 1997;51(6):1742–1750. doi: 10.1111/j.1558-5646.1997.tb05098.x. [DOI] [PubMed] [Google Scholar]
- 28.Cortez MH, Ellner SP. Understanding rapid evolution in predator‐prey interactions using the theory of fast‐slow dynamical systems. Am Nat. 2010;176(5):E109–E127. doi: 10.1086/656485. [DOI] [PubMed] [Google Scholar]
- 29.Mougi A. Predator-prey coevolution driven by size selective predation can cause anti-synchronized and cryptic population dynamics. Theor Popul Biol. 2012;81(2):113–118. doi: 10.1016/j.tpb.2011.12.005. [DOI] [PubMed] [Google Scholar]
- 30.Holling CS. Some characteristics of simple types of predation and parasitism. Can Entomol. 1959;91(7):385–398. [Google Scholar]
- 31.Kraaijeveld AR, Hutcheson KA, Limentani EC, Godfray HCH. Costs of counterdefenses to host resistance in a parasitoid of Drosophila. Evolution. 2001;55(9):1815–1821. doi: 10.1111/j.0014-3820.2001.tb00830.x. [DOI] [PubMed] [Google Scholar]
- 32.Chao L, Levin BR, Stewart FM. A complex community in a simple habitat: An experimental study with bacteria and phage. Ecology. 1977;58(2):369–378. [Google Scholar]
- 33.Lenski RE. Experimental studies of pleiotrophy and epistasis in Escherichia coli ii. compensation for maladaptive effects associated with resistance to virus T4. Evolution. 1988;42(3):433–440. doi: 10.1111/j.1558-5646.1988.tb04150.x. [DOI] [PubMed] [Google Scholar]
- 34.Kraaijeveld AR, Ferrari J, Godfray HCJ. Costs of resistance in insect-parasite and insect-parasitoid interactions. Parasitology. 2002;125(Suppl):S71–S82. doi: 10.1017/s0031182002001750. [DOI] [PubMed] [Google Scholar]
- 35.Lande R. A quantitative genetic theory of life history evolution. Ecology. 1982;63(3):607–615. [Google Scholar]
- 36.Abrams PA, Matsuda H, Harada Y. Evolutionarily unstable fitness maxima and stable fitness minima of continuous traits. Evol Ecol. 1993;7:465–487. [Google Scholar]
- 37.Arnold L, Jones CKRT, Mischaikow K, Raugel G. 1995. Geometric singular perturbation theory. Dynamical Systems (Springer, Heidelberg), Vol 1609, pp. 44–118.
- 38.Wei Y, Kirby A, Levin BR. The population and evolutionary dynamics of Vibrio cholerae and its bacteriophage: Conditions for maintaining phage-limited communities. Am Nat. 2011;178(6):715–725. doi: 10.1086/662677. [DOI] [PubMed] [Google Scholar]
- 39.Nielsen OK. Gyrfalcon Population and Reproduction in Relation to Rock Ptarmigan Numbers in Iceland. Gyrfalcons and Ptarmigans in a Changing World. Boise, ID: The Peregrine Fund; 2011. [Google Scholar]
- 40.Haydon DT, Stenseth NC, Boyce MS, Greenwood PE. Phase coupling and synchrony in the spatiotemporal dynamics of muskrat and mink populations across Canada. Proc Natl Acad Sci USA. 2001;98(23):13149–13154. doi: 10.1073/pnas.221275198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Holmengen N, Seip KL. Cycle lengths and phase portrait characteristics as probes for predator-prey interactions: Comparing simulations and observed data. Can J Zool. 2009;87:20–30. [Google Scholar]
- 42.Holmengen N, Seip KL, Boyce M, Stenseth NC. Predator prey coupling: Interaction between mink mustela vison and muskrat ondatra zibethicus across canada. Oikos. 2009;118:440–448. [Google Scholar]
- 43.Gilpin ME. Do hares eat lynx? Am Nat. 1973;107(957):727–730. [Google Scholar]
- 44.Cortez MH. Comparing the qualitatively different effects rapidly evolving and rapidly induced defences have on predator-prey interactions. Ecol Lett. 2011;14(2):202–209. doi: 10.1111/j.1461-0248.2010.01572.x. [DOI] [PubMed] [Google Scholar]
- 45.Tirok K, Bauer B, Wirtz K, Gaedke U. Predator-prey dynamics driven by feedback between functionally diverse trophic levels. PLoS ONE. 2011;6(11):e27357. doi: 10.1371/journal.pone.0027357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dieckmann U, Marrow P, Law R. Evolutionary cycling in predator-prey interactions: Population dynamics and the red queen. J Theor Biol. 1995;176(1):91–102. doi: 10.1006/jtbi.1995.0179. [DOI] [PubMed] [Google Scholar]
- 47.Marrow P, Dieckmann U, Law R. Evolutionary dynamics of predator-prey systems: An ecological perspective. J Math Biol. 1996;34(5-6):556–578. doi: 10.1007/BF02409750. [DOI] [PubMed] [Google Scholar]
- 48.Khibnik AI, Kondrashov AS. Three mechanisms of Red Queen dynamics. Philos Trans R Soc Lond B Biol Sci. 1997;264:1049–1056. [Google Scholar]
- 49.Dercole F, Ferrière R, Gragnani A, Rinaldi S. Coevolution of slow-fast populations: eEvolutionary sliding, evolutionary pseudo-equilibria and complex Red Queen dynamics. Proc Biol Sci. 2006;273(1589):983–990. doi: 10.1098/rspb.2005.3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.