Significance
Noncoding transcripts are found in high complexity but low abundance in most genomes, but their functional relevance is unclear. We investigate the function of a set of antisense transcripts expressed from an important floral repressor gene in Arabidopsis. Different polyadenylated forms of the antisense transcripts correlate with high or low expression states of the floral repressor gene. We now identify a mutation in a conserved transcription elongation factor that specifically disrupts the antisense transcription. The direct reduction of antisense transcription releases a repression mechanism that indirectly increases expression of both the floral repressor gene and antisense expression. This study reveals tight interplay between sense and antisense transcription and a mechanism that could have a widespread role in quantitative gene regulation.
Keywords: lncRNA, autonomous pathway, transcriptional regulation, chromatin silencing
Abstract
The functional significance of noncoding transcripts is currently a major question in biology. We have been studying the function of a set of antisense transcripts called COOLAIR that encompass the whole transcription unit of the Arabidopsis floral repressor FLOWERING LOCUS C (FLC). Alternative polyadenylation of COOLAIR transcripts correlates with different FLC sense expression states. Suppressor mutagenesis aimed at understanding the importance of this sense–antisense transcriptional circuitry has identified a role for Arabidopsis cyclin-dependent kinase C (CDKC;2) in FLC repression. CDKC;2 functions in an Arabidopsis positive transcription elongation factor b (P-TEFb) complex and influences global RNA polymerase II (Pol II) Ser2 phosphorylation levels. CDKC;2 activity directly promotes COOLAIR transcription but does not affect an FLC transgene missing the COOLAIR promoter. In the endogenous gene context, however, the reduction of COOLAIR transcription by cdkc;2 disrupts a COOLAIR-mediated repression mechanism that increases FLC expression. This disruption then feeds back to indirectly increase COOLAIR expression. This tight interconnection between sense and antisense transcription, together with differential promoter sensitivity to P-TEFb, is central to quantitative regulation of this important floral repressor gene.
We are investigating the role of specific antisense transcripts in gene regulation through our analysis of the regulation of expression of Arabidopsis thaliana FLOWERING LOCUS C (FLC), an important regulator of flowering time (1, 2). Multiple genetic pathways have been defined that regulate FLC expression; some function in parallel whereas others function antagonistically. FRIGIDA (FRI) up-regulates FLC expression, causing plants to overwinter vegetatively (3). FRI function is antagonized by vernalization, a process through which prolonged cold epigenetically silences FLC (4–7). Acting in parallel with vernalization to repress FLC expression is the autonomous pathway. The autonomous pathway was initially characterized through mutations specifically affecting flowering time but has subsequently been shown to regulate many other targets in the A. thaliana genome (8–10). The autonomous pathway is composed of a number of factors: RNA-binding proteins FCA (11), FPA (12), and FLK (13, 14), RNA 3′ processing factors FY (15–17), CstF64, and CstF77 (18), a histone 3 lysine 4 (H3K4) demethylase FLD (19, 20), a homolog of MSI1 (multicopy suppressor of ira1) FVE (21), and a homeodomain protein LUMINIDEPENDENS (LD) (22). These factors link alternative processing of a set of antisense transcripts produced at the FLC locus, collectively termed COOLAIR, with chromatin modifications in the FLC gene body (8). FCA, FY, and FPA promote proximal polyadenylation of COOLAIR transcripts (18), FLD-dependent H3K4me2 demethylation across the body of the gene, and low FLC transcription (23). Their loss results in distal polyadenylation in COOLAIR, increased H3K4 methylation across the gene, and high expression (24), but how COOLAIR processing is linked to FLC transcription is still not fully understood. COOLAIR is also regulated transcriptionally via an extensive R-loop that covers the COOLAIR promoter and first exon (25).
To gain a better understanding of the sense–antisense mechanism regulating FLC, we have undertaken suppressor mutagenesis and have identified a requirement for cyclin-dependent kinase C (CDKC;2). This protein is an Arabidopsis ortholog of a component of the positive transcription elongation factor b (P-TEFb) (26–29). P-TEFb regulates transcription elongation and integrates mRNA synthesis with histone modification, pre-mRNA processing, and mRNA export (30). Arabidopsis CDKC;2 has previously been shown to be important for flowering time control and plant virus infection (26) and to colocalize with spliceosome components in nuclear bodies (31). Here, we show that CDKC;2 functions globally in the A. thaliana genome to influence the phosphorylation status of RNA polymerase II (Pol II), as anticipated from a P-TEFb function. We also investigate CDKC;2 function on sense FLC and COOLAIR transcription through analysis of transgenic lines, where both are expressed independently of each other. This analysis established that cdkc;2 specifically reduces transcription of COOLAIR, which indirectly up-regulates FLC expression through disruption of a COOLAIR-mediated repression mechanism. The feedback mechanisms that link sense and antisense transcription in the endogenous gene context (18) then indirectly increase COOLAIR expression. This sensitivity of the COOLAIR promoter to P-TEFb function suggests that the antisense transcription is the driver quantitatively regulating expression levels at FLC.
Results
sof77, a Mutation in CDKC;2, Increases FLC Expression.
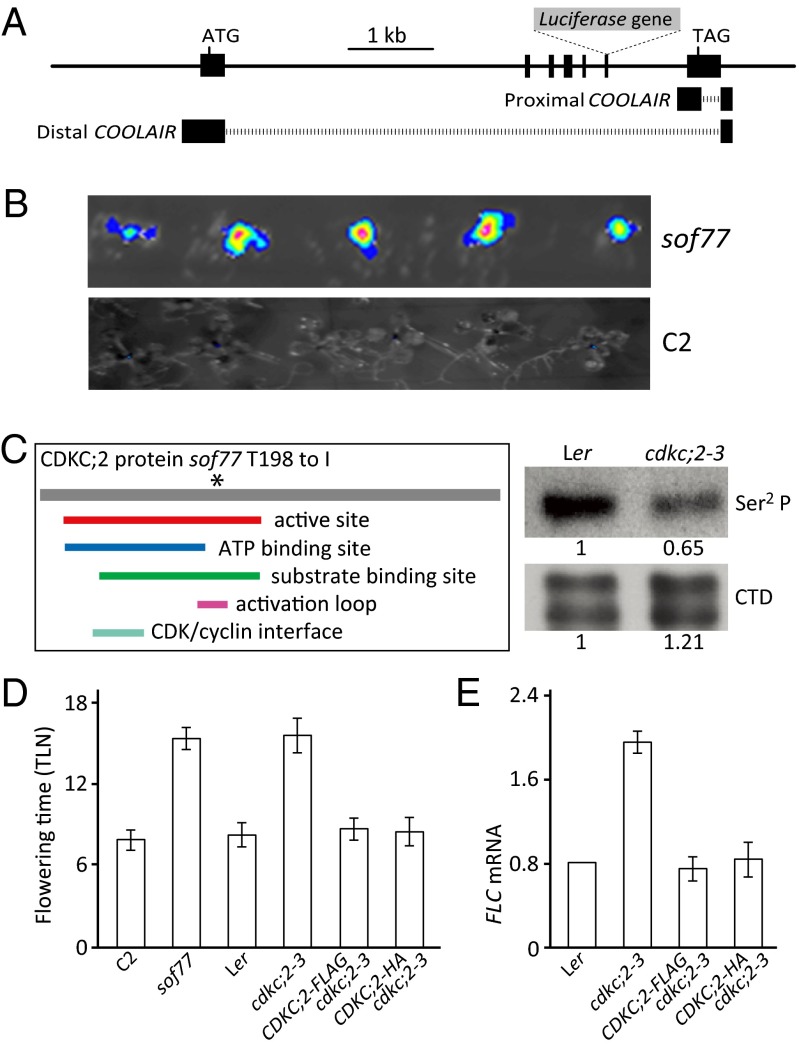
We used suppressor mutagenesis to identify components mediating repression of FLC expression. The Landsberg erecta (Ler) progenitor line expresses high levels of FCA in combination with FRI and an FLC (Col)::LUC transgene (18, 23). This line provides a system where the function of pathways both promoting and repressing FLC expression is enhanced relative to rapid-cycling genotypes normally used in Arabidopsis mutagenesis, so giving a sensitized state where even subtle effect mutations can easily be detected. A mutant called sof77 was identified that increased FLC::LUC expression and delayed flowering (Fig. 1 A, B, D, and E). Genetic mapping and sequencing revealed that sof77 carried a mutation that changes the ACA threonine codon (amino acid 198) into ATA encoding isoleucine in CDKC;2 (AT5G64960) (Fig. 1C). We named the mutation cdkc;2–3 because the two transferred DNA (T-DNA) insertion alleles in Col were named cdkc;2–1 and cdkc;2–2 (Fig. S1A) (26). CDKC;2 (AT5G64960) is one of two Arabidopsis orthologs of the CDK9 component of P-TEFb, the positive transcription elongation factor b, the other being CDKC;1 (AT5G10270) (26, 27, 29).
Fig. 1.
Characterization of cdkc;2. (A) Schematic illustration of the FLC::LUC construct. (B) sof77 shows high FLC::LUC in young seedlings compared with the C2 progenitor containing FRI, 35S::FCAγ, FLC::LUC. (C) Domains within CDKC;2. The * shows the position of the mutation in cdkc;2–3 plus Western blots of protein extracted from seedlings of cdkc;2–3 and Ler. Membranes were probed with antibodies against CTD Ser2 P Pol II (3E10) or total Pol II (8WG16). (D and E) The CDKC;2 HA or FLAG-tagged genomic fusions complemented the cdkc;2–3 mutant phenotype. Flowering time values in D are means ± SD (n = 20); FLC expression values in E are means ± SEM from three biological repeats.
We analyzed the involvement of CDKC;2 in FLC repression in a genotype not sensitized to FCA action by out-crossing the 35S::FCAγ and FRI transgenes. cdkc;2–3 elevated endogenous FLC mRNA levels by ∼2.5 fold and flowered later with approximately twice as many leaves compared with the Ler wild type (Fig. 1 D and E). This phenotype is much weaker than fca but demonstrates that CDKC;2 is involved in repressing FLC expression in wild-type Arabidopsis plants. Expression of HA- and FLAG-tagged CDKC;2 under its native regulatory sequences complemented the late flowering of the cdkc;2–3 mutant (Fig. 1 D and E and Fig. S1C). Allelic analysis using the T-DNA insertion mutant (SALK_029546, cdkc;2–2) also supported the enhanced FLC::LUC signal and later flowering time being caused by loss-of-function of the CDKC;2 gene.
As well as late flowering, cdkc;2–3 was associated with changed color of the leaves, slow growth, curved siliques, and aborted seeds, suggesting that CDKC;2 function is generally important for plant growth and development (Fig. S1 C–F). These mutant phenotypes reveal that CDKC;1 cannot cover loss of CDKC;2 function, perhaps because in silico gene expression analysis suggests that CDKC;1 expression could be much lower throughout development (Fig. S2).
CDKC;2 Functions in the FCA-Dependent Genetic Pathway to Down-Regulate FLC.
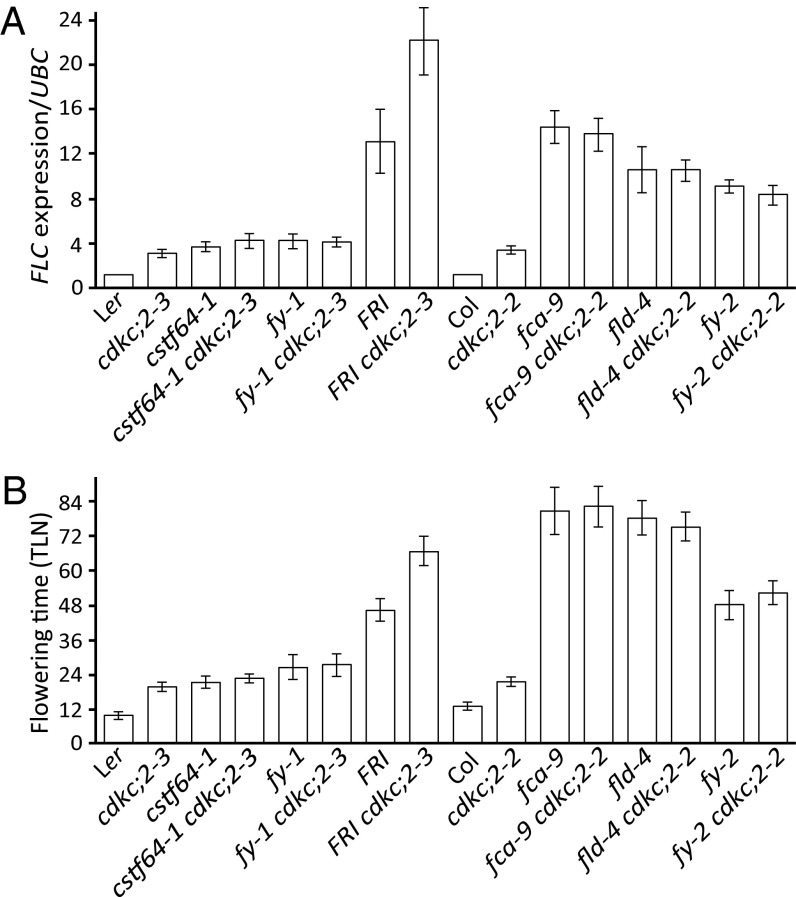
We first determined whether the cdkc;2 mutation affected expression of FLC regulators. We found no change in FCA, FY, FLD RNA expression and the feedback regulation of FCA on the processing of its own transcript, showing that the increase of FLC expression was not due to an effect on FCA function (Fig. S3). We undertook an epistasis analysis to test whether CDKC;2 functions in the same pathway as known autonomous pathway components, namely FCA, FY, CstF64, and FLD. We used both the cdkc;2–3 allele in Ler and the T-DNA insertion cdkc;2–2 allele in Col for the analysis. cdkc;2 was not additive with fca, fld, fy, or cstf64-1 mutant alleles but was additive with active FRI on the basis of analysis of both FLC expression levels and flowering time (Fig. 2). Thus, our analysis demonstrates that CDKC;2 functions in the same genetic pathway as FCA to limit FLC expression.
Fig. 2.
CDKC;2 functions together with autonomous pathway components to repress FLC expression and accelerate flowering. (A and B) cdkc;2 is not additive with cstf64, fy, fca, or fld in FLC repression. FLC expression values are means ± SEM from three biological repeats. Flowering time [total leaf number (TLN)] data are means ± SD (n = 20).
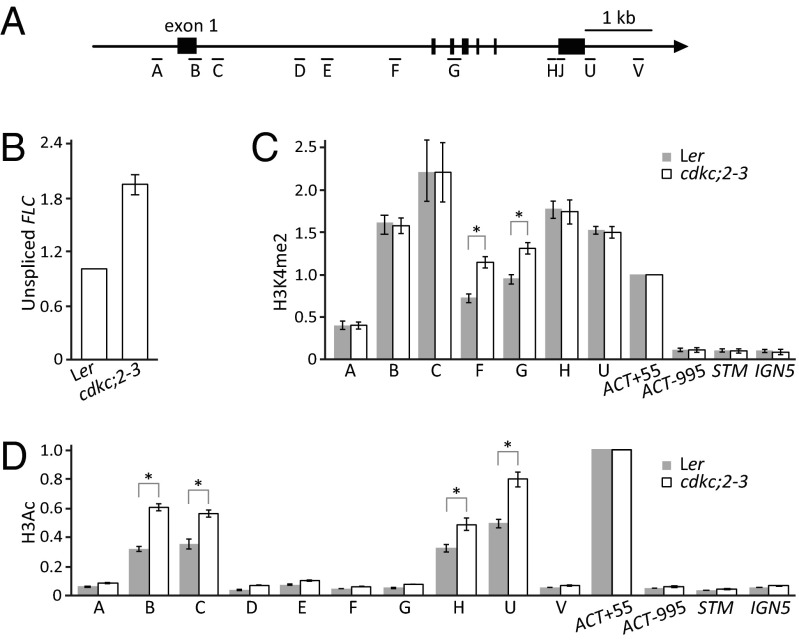
Our previous study had shown that that autonomous pathway mutants, including fld, cstf64, and cstf77, increase FLC transcriptionally (18, 23). Using similar assays, we found that cdkc;2–3 also caused increased FLC transcription. Unspliced transcript levels were higher (Fig. 3B), and H3K4me2 increased in the central region of the gene (regions F and G, Fig. 3C). H3Ac levels were higher in cdkc;2 near the sense and antisense promoter regions (regions B, C, H, and U, Fig. 3D). These data would suggest that loss of CDKC;2 perturbs aspects of the same repression mechanism as other autonomous pathway mutations.
Fig. 3.
cdkc;2 increases FLC transcription. (A) Schematic of FLC. Vertical bars show the exons. A, B, C, D, E, F, G, H, U, and V show regions analyzed by qPCR in ChIP. (B) Unspliced FLC transcript analysis by qRT-PCR. (C) H3K4me2 ChIP assay in FLC regions in cdkc;2–3 and Ler. (D) H3Ac ChIP assay in FLC regions in cdkc;2–3 and Ler. (B–D) Values are means ± SEM from three biological repeats; data were presented as ratio of (modified histone level at FLC/H3 FLC) to (modified histone level at Actin/H3 Actin). Actin, STM, and IGN5 were used as internal controls for the ChIP experiments. *P < 0.05.
The CDKC;2–CYCT Complex Forms an Arabidopsis P-TEFb Complex.
CDKC;2 is homologous to the CDK9 kinase subunit of human P-TEFb. P-TEFb phosphorylates the C-terminal domain (CTD) of Pol II at the Ser2 site within the heptad repeats (26, 29). This Ser2 posttranslational modification orchestrates the interplay between transcriptional elongation and processing of mRNA (32–35). This amino acid change in cdkc;2–3 is within the activation loop, known to be important for substrate binding, but not involved in interaction with the cyclin component of P-TEFb (Fig. 1C) (36). The specific kinase activity of CDKC;2 had not been previously explored so we assayed the phosphorylation status of Pol II CTD in wild-type and cdkc;2–3 plants. Western blot analysis using antibodies to CTD Ser2 phosphorylated Pol II (37) and hypophosphorylated CTD showed specific reduction in levels of CTD Ser2 phosphorylated Pol II in cdkc;2–3 (Fig. 1C). This result supports a functional role for CDKC;2 as part of an Arabidopsis P-TEFb complex.
Yeast two-hybrid and coimmunoprecipitation analyses have suggested that the gene products from the cyclin T genes CYCT1;3, CYCT1;4, and CYCT1;5 could be the cyclin partner of CDKC;2 (26). To definitively define the cyclin partner associated with CDKC;2 in P-TEFb in planta, we immunoprecipitated a complementing CDKC;2-FLAG stably expressed in cdkc;2–3 (Fig. 1 D and E and Fig. S1B) and identified associated proteins by mass spectrometry (MS). This experiment identified several peptides corresponding to CYCT1;4 and CYCT1;5, with most corresponding to CYCT1;5 (Table 1). These data suggest that the predominant partner in vivo is CYCT1;5 but that both can function in Arabidopsis P-TEFb activity in vivo. Consistent with a role in FLC regulation, a genotype deficient in CYCT1;5 and with reduced CYCT1;4 (cyct1;5/CYCT1;4RNAi) has been found to flower late with enhanced levels of FLC mRNA (26). We also detected phosphorylation of a conserved threonine in CDKC;2 by MS (Figs. S4A and S5). Phosphorylation of this threonine, T186, in mammals corresponding to T198 in Arabidopsis CDKC;2, on the activation loop in CDK9 is essential for kinase activity (36).
Table 1.
Summary of mass-spectral sequencing analysis of CDKC;2-FLAG purification IP-specific peptides
| Protein | Predicted molecular mass, Da | Unique (total) peptides | |
| Exp. 1 | Exp. 2 | ||
| CDKC;2-FLAG (AT5G64960) | 57,130 | 16 (19) | 20 (28) |
| CYCT1;5 (AT5G45190) | 65,168 | 5 (5) | 17 (21) |
| CYCT1;4 (AT4G19600) | 60,710 | 1 (1) | 4 (4) |
Minimum peptide identification probability value is 95%.
CDKC;2 Regulates FLC Expression via an Influence on COOLAIR Production.
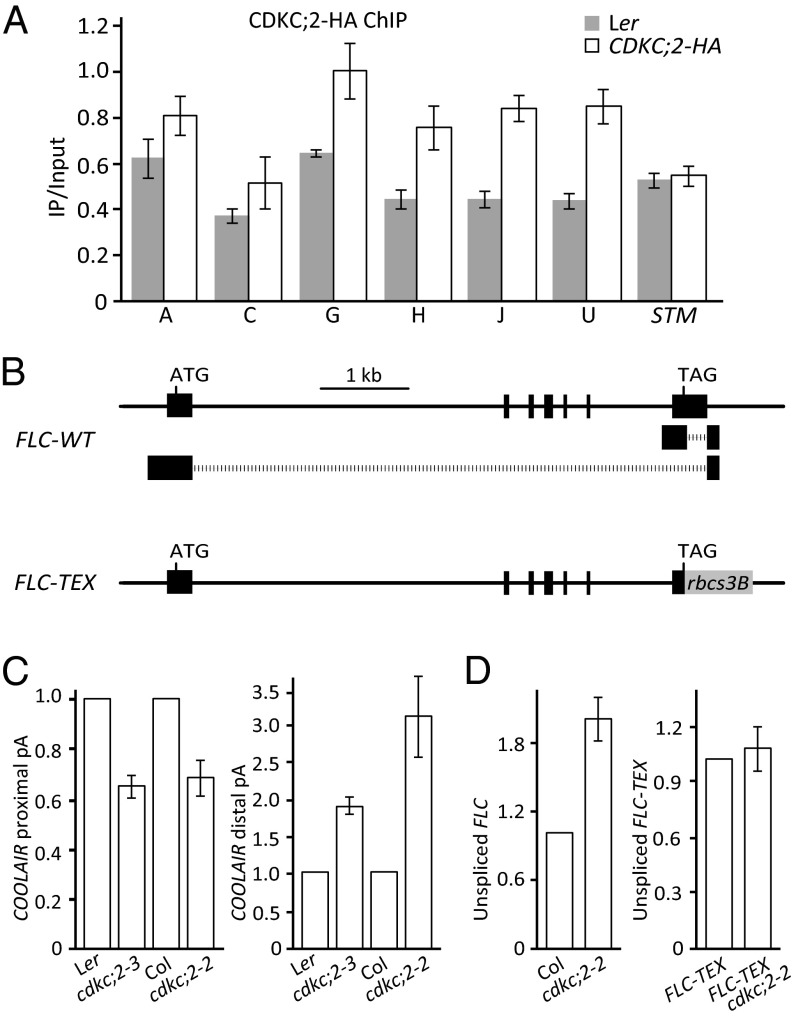
The requirement of CDKC;2 to repress expression of the FLC gene runs counter to the role of P-TEFb as a positive transcriptional elongation factor. We therefore used chromatin immunoprecipitation experiments using the lines carrying the complementing CDKC;2-HA fusion to test that CDKC;2 function at the FLC locus was direct (Fig. 1 D and E and Fig. S1B). There was a low level of enrichment across the whole locus, with the highest at the 3′ region of FLC near the COOLAIR promoter (Fig. 4A). The increase in COOLAIR expression in cdkc;2 was associated with changed COOLAIR processing; relative to total levels of COOLAIR, proximal polyadenylation was reduced and distal polyadenylation increased (Fig. 4C). This alteration parallels the situation in other mutants where FLC expression increased (18).
Fig. 4.
CDKC;2 associates directly with the FLC locus and requires COOLAIR to repress FLC expression. (A) ChIP experiments showing enrichment of FLC regions in by CDKC;2-HA. The regions A, C, G, H, J, and U analyzed by qPCR are shown in the schematic in Fig. 3A. (B) Schematic diagram showing the replacement of the FLC 3′ region with that from the Arabidopsis rbcs3B gene to form the FLC-terminator exchange construct or FLC-TEX. (C) Relative use of the proximal polyadenylation site of COOLAIR is reduced in cdkc;2–3 (the Ler allele identified in this study) and cdkc;2–2 (the Columbia T-DNA insertion allele), and relative use of the distal site increased. The proximally and distally polyadenylated COOLAIR transcript levels are normalized to total COOLAIR transcript level and given as fold change compared with the parental lines. (D) Unspliced FLC or FLC-TEX transcript analysis by qRT-PCR. Values are means ± SEM from three biological repeats.
Our previous work had demonstrated a role of COOLAIR in FLC regulation (38). To investigate whether the effect of CDKC;2 on FLC expression was mediated via COOLAIR, we generated an Arabidopsis line expressing an FLC transgene, where the native FLC 3′ region from the translation stop codon to past the poly (A) site (which corresponds to the COOLAIR promoter, the COOLAIR first intron, and the beginning of the COOLAIR second exon) was replaced by the 3′ untranslated region from the translation stop codon to past the poly (A) site of the Arabidopsis rbcs3B gene (Fig. 4B). The transgene, called FLC-TEX, complemented the flc-2 mutation; the plants flowered late and produced even higher levels of FLC unspliced transcript than the Col wild type (Fig. S6A) (39). FLC-TEX produced low levels of proximally polyadenylated COOLAIR approximately the same as the flc-2 parent (39) [the representative FLC-TEX line (no. 577) used in this analysis differed from that used previously (39) and is shown in Fig. S6C] and approximately half as much total antisense transcripts as wild-type plants (Fig. S6B), likely to result from Pol II firing from the efficient rbcs3B terminator (40). The FLC-TEX line was crossed to a cdkc;2 mutation (using the Col cdkc;2–2 allele to avoid mixing genetic backgrounds, selecting for fri individuals, but maintaining the endogenous Columbia FLC allele). We found that the transgene FLC-TEX expression in FLC-TEX cdkc;2 was unaffected by cdkc;2–2, indicating that CDKC;2 repression of FLC expression depends on COOLAIR (Fig. 4D). This result was obtained in a background where the expression of the endogenous FLC locus expressing native COOLAIR increased (Fig. 4D).
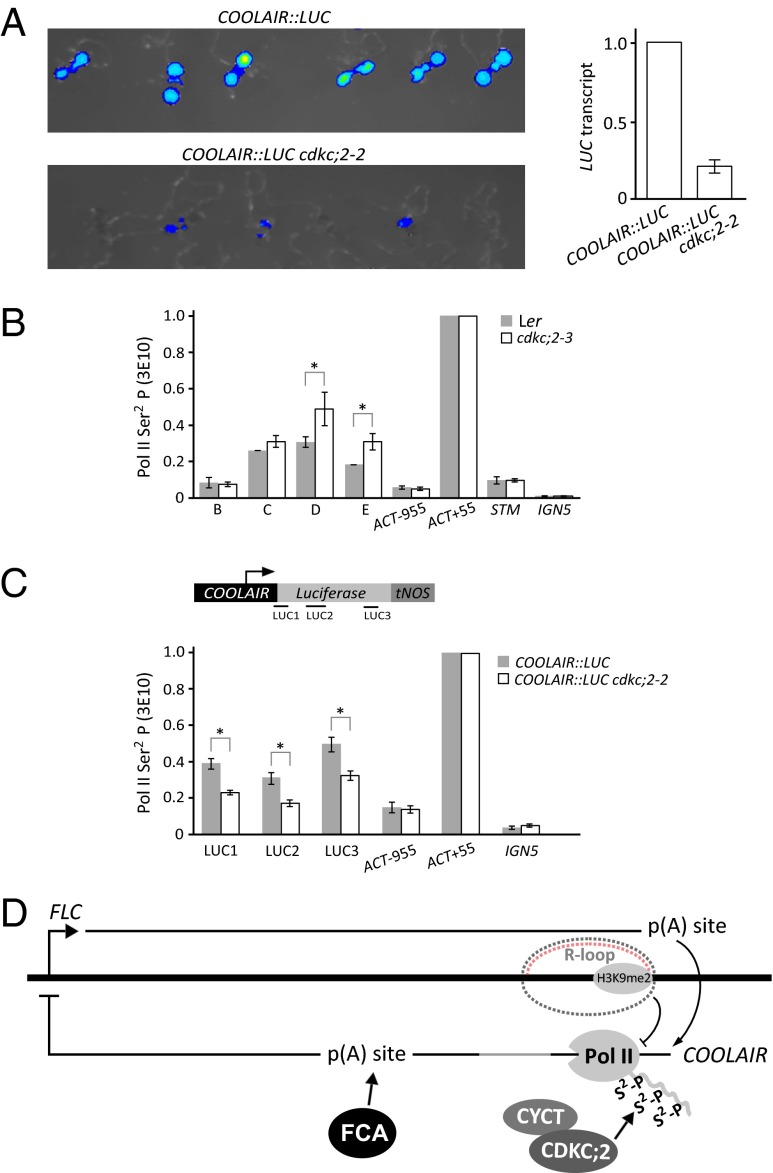
We used a COOLAIR::LUC fusion (25, 38), where the COOLAIR promoter, first exon, and first intron had been fused with an internal ribosome entry site to the firefly luciferase-coding region (LUC) to study the affect of cdkc;2, in the absence of FLC sense expression. This COOLAIR::LUC transgene mimics the behavior of the endogenous COOLAIR transcripts (25, 38). When combined with cdkc;2–2, the COOLAIR::LUC expression decreased significantly, and corresponded to reduced levels of LUC RNA transcript (Fig. 5A). Levels of CTD Ser2 phosphorylated Pol II were significantly reduced across this COOLAIR::LUC transgene in the cdkc;2–2 mutant (Fig. 5C and Fig. S7A). This result is consistent with CDKC;2 association near the COOLAIR promoter and its role as a positive transcriptional elongation factor. Interestingly, the CTD Ser2 phosphorylated Pol II levels are slightly increased in the endogenous gene in the context of the linked sense–antisense transcription circuitry (Fig. 5B and Fig. S7B). Together, these data suggest that, in wild-type plants, CDKC;2 promotes transcription of COOLAIR, which in turn indirectly decreases FLC expression (Fig. 5D). The counterintuitive outcome is that, through feedback mechanisms likely involving the FLC gene loop (41), when CDKC;2 is lost and FLC transcription increased, there is a secondary effect to increase COOLAIR transcription, thus masking the direct effect of the cdkc;2 mutations.
Fig. 5.
CDKC;2 is required for efficient COOLAIR transcription. (A) Effects of cdkc;2–2 on a COOLAIR::LUC fusion, expressed separately from the endogenous FLC. Luciferase activity and COOLAIR::LUC transcript analysis by qRT-PCR are shown. Values are means ± SEM from three biological repeats. (B) ChIP experiments assaying Pol II (using anti-Ser2 P-CTD-3E10) in FLC regions in cdkc;2–3 and Ler. (C) Schematic showing the COOLAIR::LUC fusion and primers used in ChIP analysis plus ChIP experiments assaying RNA Pol II (using anti-Ser2 P-CTD-3E10) in COOLAIR::LUC regions in cdkc;2–2 and Col. (B and C) Values are means ± SEM from two biological repeats; data were presented as ratio of (Pol II FLC/input FLC) to (Pol II Actin/input Actin). Actin, STM, and IGN5 were used as internal controls for the ChIP experiments. (D) Working model for how CDKC;2 functions in the FLC sense–antisense transcriptional circuitry. CDKC;2 is a component of P-TEFb required for COOLAIR transcription. It is required to produce the proximally polyadenylated COOLAIR transcript in the FCA-dependent pathway. The proximally polyadenylated COOLAIR transcript represses FLC transcription. Feedback mechanisms link sense and antisense transcription levels so, when autonomous pathway function is compromised and FLC transcription increases, COOLAIR transcription is increased (indicated by the positive arrow on the right of the schematic). The R-loop that covers the COOLAIR promoter (dashed line with the RNA:DNA heteroduplex shown as a red/black pair) could make COOLAIR transcription more sensitive to CDKC;2 function.
Discussion
The prevalence of antisense transcripts in many genomes suggests that they may have a widespread functional role (42, 43). Studies of specific loci in Saccharomyces cerevisiae (44) have demonstrated how sense/antisense pairs can function as genetic toggles switching expression states, but the general mechanisms in higher eukaryotes are yet to be fully established. We have been studying the sense/antisense transcriptional circuitry at the FLC/COOLAIR locus in the Arabidopsis thaliana genome. FLC encodes a floral repressor so quantitative changes in gene expression cause altered flowering time, an important adaptive trait in plants.
Our study showed that Arabidopsis P-TEFb transcription elongation complex promotes antisense COOLAIR transcription. The primary defect caused by the cdkc;2 mutation was revealed by analysis of the expression of a COOLAIR::LUC fusion, which no longer expressed the FLC sense transcription unit. The clear reduction of COOLAIR::LUC expression and reduced CTD Ser2 phosphorylated RNA Pol II occupancy at the COOLAIR promoter in cdkc;2 contrasted with the lack of effect of cdkc;2 mutations on an FLC transgene engineered to lack COOLAIR production. Thus, we conclude that, in the endogenous gene context, cdkc;2 indirectly up-regulates FLC expression through disruption of a COOLAIR-mediated repression mechanism. Complex feedback mechanisms central to FLC regulation then lead to the counterintuitive outcome that, as a secondary effect, expression of COOLAIR increases. The feedback mechanisms that lead to the positive correlation of FLC and COOLAIR transcription remain to be fully established. However, the presence of an FLC gene loop involving the physical interaction of the 5′ and 3′ regions (41), and/or the antisense transcription unit fully encompassing the sense transcription unit (38), could both be involved in efficient recycling of RNA polymerase between strands.
The genetic functioning of CDKC;2 within the autonomous pathway supports the view that the repression of FLC expression involves a COOLAIR-mediated repression mechanism, in many respects rather similar to antisense-mediated repression of yeast genes (43–45). Loss of this mechanism leads to increased levels of distally polyadenylated COOLAIR transcripts and increased FLC transcription. The positive correlation between distal polyadenylation of COOLAIR with increased FLC expression has led to the suggestion that the process of antisense transcription facilitates access of FLC promoter sequences by the transcription machinery, so enhancing expression (24). This view was reinforced because of the concern that the absolute amount of proximal polyadenylation did not change in some autonomous pathway mutants (46). However, the higher FLC expression of the FLC-TEX transgene, which lacks the COOLAIR promoter and proximally polyadenylated COOLAIR transcript and has reduced antisense transcription (Fig. S6), is consistent with proximal polyadenylation of COOLAIR actively repressing FLC expression. The feedback mechanisms that so tightly connect sense and antisense transcription at FLC mean that the absolute changes in transcript level need to be interpreted carefully; COOLAIR transcription can drop as a primary defect in cdkc;2 but increase in the endogenous gene context due to release of the locus-wide repression mechanism.
P-TEFb is required for transcription of most genes via its kinase activity, leading to Ser2 phosphorylation of the RNA Pol II CTD. This posttranslational modification is associated with transcriptional elongation and intimately linked with splicing, polyadenylation, and nuclear export (30). A surprise therefore was that COOLAIR transcription but not sense FLC transcription was sensitive to loss of CDKC;2. Indeed, the differential sensitivity of sense and antisense transcription to this P-TEFb function appears to be an important component of the cotranscriptional mechanism limiting FLC expression. What causes this differential sensitivity is an interesting question. COOLAIR transcription initiates from a noncanonical promoter within a genomic region carrying termination sequences for the sense transcript, a feature frequently found in yeast (40). Small RNAs (24- and 30-mers) can be detected homologous to the COOLAIR promoter, and these are required to maintain a small patch of H3K9me2-modified chromatin just upstream of the major COOLAIR start site (47). This noncanonical promoter could be particularly sensitive to CDKC;2 function to ensure Pol II promoter clearance and entry into the elongating phase of transcription and cotranscriptional maturation of the transcripts. There may therefore be a greater dependence on P-TEFb function for recruitment of the 3′ processing machinery (including CstF and CPSF complexes) to the COOLAIR transcript (48, 49). Any change in CstF or CPSF recruitment would influence poly (A) site choice in the antisense transcript (50) so loss of CDKC;2 might be functionally analogous to a cstf64 or cstf77 mutation, previously isolated in the same suppressor screen (18). It is also possible that the 500- to 700-nucleotide R-loop, which covers the COOLAIR promoter and first exon and is stabilized by a homeodomain protein, AtNDX (25), could make COOLAIR transcription more sensitive to P-TEFb function. Enhanced cotranscriptional recruitment of splicing factors to the nascent transcript has been shown to resolve R-loops, thus aiding transcription (51).
Because P-TEFb is such a central component of transcriptional regulation, the viability of cdkc;2 loss-of-function mutants suggests some genetic redundancy with the second copy of CDKC, the CDKC;1 gene in the A. thaliana genome (Fig. S5B). However, the observed phenotypes show that the two copies cannot be fully redundant (Fig. S1 C–F). A division of labor in the transcriptional elongation function of P-TEFb components also seems to be the case in S. cerevisiae (52) and Schizosaccharomyces pombe (53). Recent studies demonstrating a second metazoan CTD Ser2 kinase (54, 55) suggest that the paradigm of CTD Ser2 kinase pairs might be retained throughout higher eukaryotes. The A. thaliana proteins CDKC;2 and CDKC;1 display CTD kinase activity in vitro (26), are associated with cyclin T (CycT), and have a conserved CTD kinase domain so it is likely that expression differences determine their division of labor.
In conclusion, our data reveal the complexity in the sense–antisense transcriptional circuitry at the Arabidopsis FLC locus. Sensitivity of COOLAIR transcription to P-TEFb function is an important component of the cotranscriptional mechanism limiting FLC expression. Loss of CDKC;2 reduces COOLAIR production, which disrupts the restraint on sense transcription. Through mechanisms that link sense–antisense transcription, probably involving kinetic coupling of chromatin state, transcription elongation rate, and RNA processing, in combination with gene loops, this increased FLC transcription then feeds back to increase COOLAIR transcription. These kinds of cotranscriptional mechanisms, which work in cis only, are clearly not limited to plant genomes (56, 57). Modulation of antisense transcription by P-TEFb linked to gene body chromatin modification could be a general mechanism regulating gene expression in eukaryotes.
Materials and Methods
The progenitor line C2 is described in ref. 18. For the luciferase screening, Arabidopsis seedlings were grown on germination medium agar plates at 23–25 °C with a 16-h photoperiod for 12 d. Seedlings were sprayed with 1mM of luciferin (Promega) solution and incubated in the dark at room temperature for 25 min. The LUC bioluminescence signal of the seedlings was assayed using a Nightowl light-sensitive CCD camera (Berthold Technologies). Arabidopsis CDKC;2 T-DNA insertion line (SALK_029546, cdkc;2–2) was obtained from the Nottingham Arabidopsis Stock Centre.
The COOLAIR::LUC line has been described previously and is in the FRI flc-2 background (25). Here, the COOLAIR::LUC gene was introgressed into cdkc;2–2 by crossing and genotyping. The resulting COOLAIR::LUC cdkc;2–2 genotype contains the endogenous active FLC allele.
Complementation analysis, FLC-TEX cloning, expression analysis using real-time RT-PCR, measurement of the COOLAIR levels in different genetic backgrounds, ChIP assays, Western blotting, CDKC;2 purification for proteomic analysis, and liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis are all described in SI Materials and Methods. Primers are listed in Table S1.
Supplementary Material
Acknowledgments
We thank Gerhard Saalbach for proteomic analysis and C.D. laboratory members for technical assistance and useful discussions, especially Fuquan Liu for advice. This work was supported by United Kingdom (UK) Biotechnology and Biological Sciences Research Council (BBSRC) Institute Strategic Programme Grant BB/J004588/1 (to the John Innes Centre), UK BBSRC Grant BB/D010799/1 (to C.D.), and a European Research Council Advanced Investigator grant (ENVGENE) (to C.D.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1406635111/-/DCSupplemental.
References
- 1.Michaels SD, Amasino RM. FLOWERING LOCUS C encodes a novel MADS domain protein that acts as a repressor of flowering. Plant Cell. 1999;11(5):949–956. doi: 10.1105/tpc.11.5.949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sheldon CC, et al. The FLF MADS box gene: A repressor of flowering in Arabidopsis regulated by vernalization and methylation. Plant Cell. 1999;11(3):445–458. doi: 10.1105/tpc.11.3.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Crevillén P, Dean C. Regulation of the floral repressor gene FLC: The complexity of transcription in a chromatin context. Curr Opin Plant Biol. 2011;14(1):38–44. doi: 10.1016/j.pbi.2010.08.015. [DOI] [PubMed] [Google Scholar]
- 4.Angel A, Song J, Dean C, Howard M. A Polycomb-based switch underlying quantitative epigenetic memory. Nature. 2011;476(7358):105–108. doi: 10.1038/nature10241. [DOI] [PubMed] [Google Scholar]
- 5.Gendall AR, Levy YY, Wilson A, Dean C. The VERNALIZATION 2 gene mediates the epigenetic regulation of vernalization in Arabidopsis. Cell. 2001;107(4):525–535. doi: 10.1016/s0092-8674(01)00573-6. [DOI] [PubMed] [Google Scholar]
- 6.Sung SB, Amasino RM. Vernalization in Arabidopsis thaliana is mediated by the PHD finger protein VIN3. Nature. 2004;427(6970):159–164. doi: 10.1038/nature02195. [DOI] [PubMed] [Google Scholar]
- 7.Greb T, et al. The PHD finger protein VRN5 functions in the epigenetic silencing of Arabidopsis FLC. Curr Biol. 2007;17(1):73–78. doi: 10.1016/j.cub.2006.11.052. [DOI] [PubMed] [Google Scholar]
- 8.Bäurle I, Dean C. The timing of developmental transitions in plants. Cell. 2006;125(4):655–664. doi: 10.1016/j.cell.2006.05.005. [DOI] [PubMed] [Google Scholar]
- 9.Sonmez C, et al. RNA 3′ processing functions of Arabidopsis FCA and FPA limit intergenic transcription. Proc Natl Acad Sci USA. 2011;108(20):8508–8513. doi: 10.1073/pnas.1105334108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Veley KM, Michaels SD. Functional redundancy and new roles for genes of the autonomous floral-promotion pathway. Plant Physiol. 2008;147(2):682–695. doi: 10.1104/pp.108.118927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Macknight R, et al. FCA, a gene controlling flowering time in Arabidopsis, encodes a protein containing RNA-binding domains. Cell. 1997;89(5):737–745. doi: 10.1016/s0092-8674(00)80256-1. [DOI] [PubMed] [Google Scholar]
- 12.Schomburg FM, Patton DA, Meinke DW, Amasino RM. FPA, a gene involved in floral induction in Arabidopsis, encodes a protein containing RNA-recognition motifs. Plant Cell. 2001;13(6):1427–1436. doi: 10.1105/tpc.13.6.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lim MH, et al. A new Arabidopsis gene, FLK, encodes an RNA binding protein with K homology motifs and regulates flowering time via FLOWERING LOCUS C. Plant Cell. 2004;16(3):731–740. doi: 10.1105/tpc.019331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mockler TC, et al. Regulation of flowering time in Arabidopsis by K homology domain proteins. Proc Natl Acad Sci USA. 2004;101(34):12759–12764. doi: 10.1073/pnas.0404552101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ohnacker M, Barabino SML, Preker PJ, Keller W. The WD-repeat protein pfs2p bridges two essential factors within the yeast pre-mRNA 3′-end-processing complex. EMBO J. 2000;19(1):37–47. doi: 10.1093/emboj/19.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shi YS, et al. Molecular architecture of the human pre-mRNA 3′ processing complex. Mol Cell. 2009;33(3):365–376. doi: 10.1016/j.molcel.2008.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Simpson GG, Dijkwel PP, Quesada V, Henderson I, Dean C. FY is an RNA 3′ end-processing factor that interacts with FCA to control the Arabidopsis floral transition. Cell. 2003;113(6):777–787. doi: 10.1016/s0092-8674(03)00425-2. [DOI] [PubMed] [Google Scholar]
- 18.Liu FQ, Marquardt S, Lister C, Swiezewski S, Dean C. Targeted 3′ processing of antisense transcripts triggers Arabidopsis FLC chromatin silencing. Science. 2010;327(5961):94–97. doi: 10.1126/science.1180278. [DOI] [PubMed] [Google Scholar]
- 19.He YH, Michaels SD, Amasino RM. Regulation of flowering time by histone acetylation in Arabidopsis. Science. 2003;302(5651):1751–1754. doi: 10.1126/science.1091109. [DOI] [PubMed] [Google Scholar]
- 20.Shi Y, et al. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell. 2004;119(7):941–953. doi: 10.1016/j.cell.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 21.Ausín I, Alonso-Blanco C, Jarillo JA, Ruiz-García L, Martínez-Zapater JM. Regulation of flowering time by FVE, a retinoblastoma-associated protein. Nat Genet. 2004;36(2):162–166. doi: 10.1038/ng1295. [DOI] [PubMed] [Google Scholar]
- 22.Aukerman MJ, Lee I, Weigel D, Amasino RM. The Arabidopsis flowering-time gene LUMINIDEPENDENS is expressed primarily in regions of cell proliferation and encodes a nuclear protein that regulates LEAFY expression. Plant J. 1999;18(2):195–203. doi: 10.1046/j.1365-313x.1999.00442.x. [DOI] [PubMed] [Google Scholar]
- 23.Liu FQ, et al. The Arabidopsis RNA-binding protein FCA requires a lysine-specific demethylase 1 homolog to downregulate FLC. Mol Cell. 2007;28(3):398–407. doi: 10.1016/j.molcel.2007.10.018. [DOI] [PubMed] [Google Scholar]
- 24.Hornyik C, Terzi LC, Simpson GG. The spen family protein FPA controls alternative cleavage and polyadenylation of RNA. Dev Cell. 2010;18(2):203–213. doi: 10.1016/j.devcel.2009.12.009. [DOI] [PubMed] [Google Scholar]
- 25.Sun Q, Csorba T, Skourti-Stathaki K, Proudfoot NJ, Dean C. R-loop stabilization represses antisense transcription at the Arabidopsis FLC locus. Science. 2013;340(6132):619–621. doi: 10.1126/science.1234848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cui XF, Fan BF, Scholz J, Chen ZX. Roles of Arabidopsis cyclin-dependent kinase C complexes in cauliflower mosaic virus infection, plant growth, and development. Plant Cell. 2007;19(4):1388–1402. doi: 10.1105/tpc.107.051375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Barrôco RM, et al. Novel complexes of cyclin-dependent kinases and a cyclin-like protein from Arabidopsis thaliana with a function unrelated to cell division. Cell Mol Life Sci. 2003;60(2):401–412. doi: 10.1007/s000180300033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fülöp K, et al. The Medicago CDKC;1-CYCLINT;1 kinase complex phosphorylates the carboxy-terminal domain of RNA polymerase II and promotes transcription. Plant J. 2005;42(6):810–820. doi: 10.1111/j.1365-313X.2005.02421.x. [DOI] [PubMed] [Google Scholar]
- 29.Guo Z, Stiller JW. Comparative genomics of cyclin-dependent kinases suggest co-evolution of the RNAP II C-terminal domain and CTD-directed CDKs. BMC Genomics. 2004;5:69. doi: 10.1186/1471-2164-5-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brès V, Yoh SM, Jones KA. The multi-tasking P-TEFb complex. Curr Opin Cell Biol. 2008;20(3):334–340. doi: 10.1016/j.ceb.2008.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kitsios G, Alexiou KG, Bush M, Shaw P, Doonan JH. A cyclin-dependent protein kinase, CDKC2, colocalizes with and modulates the distribution of spliceosomal components in Arabidopsis. Plant J. 2008;54(2):220–235. doi: 10.1111/j.1365-313X.2008.03414.x. [DOI] [PubMed] [Google Scholar]
- 32.Buratowski S. Progression through the RNA polymerase II CTD cycle. Mol Cell. 2009;36(4):541–546. doi: 10.1016/j.molcel.2009.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moore MJ, Proudfoot NJ. Pre-mRNA processing reaches back to transcription and ahead to translation. Cell. 2009;136(4):688–700. doi: 10.1016/j.cell.2009.02.001. [DOI] [PubMed] [Google Scholar]
- 34.Phatnani HP, Greenleaf AL. Phosphorylation and functions of the RNA polymerase II CTD. Genes Dev. 2006;20(21):2922–2936. doi: 10.1101/gad.1477006. [DOI] [PubMed] [Google Scholar]
- 35.Hajheidari M, Farrona S, Huettel B, Koncz Z, Koncz C. CDKF;1 and CDKD protein kinases regulate phosphorylation of serine residues in the C-terminal domain of Arabidopsis RNA polymerase II. Plant Cell. 2012;24(4):1626–1642. doi: 10.1105/tpc.112.096834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Baumli S, et al. The structure of P-TEFb (CDK9/cyclin T1), its complex with flavopiridol and regulation by phosphorylation. EMBO J. 2008;27(13):1907–1918. doi: 10.1038/emboj.2008.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chapman RD, et al. Transcribing RNA polymerase II is phosphorylated at CTD residue serine-7. Science. 2007;318(5857):1780–1782. doi: 10.1126/science.1145977. [DOI] [PubMed] [Google Scholar]
- 38.Swiezewski S, Liu FQ, Magusin A, Dean C. Cold-induced silencing by long antisense transcripts of an Arabidopsis Polycomb target. Nature. 2009;462(7274):799–802. doi: 10.1038/nature08618. [DOI] [PubMed] [Google Scholar]
- 39.Marquardt S, et al. Functional consequences of splicing of the antisense transcript COOLAIR on FLC transcription. Mol Cell. 2014;54(1):156–165. doi: 10.1016/j.molcel.2014.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Murray SC, et al. A pre-initiation complex at the 3′-end of genes drives antisense transcription independent of divergent sense transcription. Nucleic Acids Res. 2012;40(6):2432–2444. doi: 10.1093/nar/gkr1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Crevillén P, Sonmez C, Wu Z, Dean C. A gene loop containing the floral repressor FLC is disrupted in the early phase of vernalization. EMBO J. 2013;32(1):140–148. doi: 10.1038/emboj.2012.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Magistri M, Faghihi MA, St Laurent G, 3rd, Wahlestedt C. Regulation of chromatin structure by long noncoding RNAs: Focus on natural antisense transcripts. Trends Genet. 2012;28(8):389–396. doi: 10.1016/j.tig.2012.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.van Dijk EL, et al. XUTs are a class of Xrn1-sensitive antisense regulatory non-coding RNA in yeast. Nature. 2011;475(7354):114–117. doi: 10.1038/nature10118. [DOI] [PubMed] [Google Scholar]
- 44.Bumgarner SL, Dowell RD, Grisafi P, Gifford DK, Fink GR. Toggle involving cis-interfering noncoding RNAs controls variegated gene expression in yeast. Proc Natl Acad Sci USA. 2009;106(43):18321–18326. doi: 10.1073/pnas.0909641106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Castelnuovo M, et al. Bimodal expression of PHO84 is modulated by early termination of antisense transcription. Nat Struct Mol Biol. 2013;20(7):851–858. doi: 10.1038/nsmb.2598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Duc C, Sherstnev A, Cole C, Barton GJ, Simpson GG. Transcription termination and chimeric RNA formation controlled by Arabidopsis thaliana FPA. PLoS Genet. 2013;9(10):e1003867. doi: 10.1371/journal.pgen.1003867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Swiezewski S, et al. Small RNA-mediated chromatin silencing directed to the 3′ region of the Arabidopsis gene encoding the developmental regulator, FLC. Proc Natl Acad Sci USA. 2007;104(9):3633–3638. doi: 10.1073/pnas.0611459104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ahn SH, Kim M, Buratowski S. Phosphorylation of serine 2 within the RNA polymerase II C-terminal domain couples transcription and 3′ end processing. Mol Cell. 2004;13(1):67–76. doi: 10.1016/s1097-2765(03)00492-1. [DOI] [PubMed] [Google Scholar]
- 49.Ni ZY, Schwartz BE, Werner J, Suarez JR, Lis JT. Coordination of transcription, RNA processing, and surveillance by P-TEFb kinase on heat shock genes. Mol Cell. 2004;13(1):55–65. doi: 10.1016/s1097-2765(03)00526-4. [DOI] [PubMed] [Google Scholar]
- 50.Takagaki Y, Seipelt RL, Peterson ML, Manley JL. The polyadenylation factor CstF-64 regulates alternative processing of IgM heavy chain pre-mRNA during B cell differentiation. Cell. 1996;87(5):941–952. doi: 10.1016/s0092-8674(00)82000-0. [DOI] [PubMed] [Google Scholar]
- 51.Li X, Manley JL. Inactivation of the SR protein splicing factor ASF/SF2 results in genomic instability. Cell. 2005;122(3):365–378. doi: 10.1016/j.cell.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 52.Qiu H, Hu C, Hinnebusch AG. Phosphorylation of the Pol II CTD by KIN28 enhances BUR1/BUR2 recruitment and Ser2 CTD phosphorylation near promoters. Mol Cell. 2009;33(6):752–762. doi: 10.1016/j.molcel.2009.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Egloff S, Dienstbier M, Murphy S. Updating the RNA polymerase CTD code: Adding gene-specific layers. Trends Genet. 2012;28(7):333–341. doi: 10.1016/j.tig.2012.03.007. [DOI] [PubMed] [Google Scholar]
- 54.Bartkowiak B, et al. CDK12 is a transcription elongation-associated CTD kinase, the metazoan ortholog of yeast Ctk1. Genes Dev. 2010;24(20):2303–2316. doi: 10.1101/gad.1968210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Blazek D, et al. The Cyclin K/Cdk12 complex maintains genomic stability via regulation of expression of DNA damage response genes. Genes Dev. 2011;25(20):2158–2172. doi: 10.1101/gad.16962311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Faghihi MA, Wahlestedt C. Regulatory roles of natural antisense transcripts. Nat Rev Mol Cell Biol. 2009;10(9):637–643. doi: 10.1038/nrm2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Houseley J, Rubbi L, Grunstein M, Tollervey D, Vogelauer M. A ncRNA modulates histone modification and mRNA induction in the yeast GAL gene cluster. Mol Cell. 2008;32(5):685–695. doi: 10.1016/j.molcel.2008.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.