Significance
It is known that the gastrointestinal microbiota influences adiposity and weight gain in the host. However the mechanisms by which gut microorganisms coordinate host physiological processes are currently unclear. We demonstrate that a single, widely distributed function of the gut microbiota, bile salt hydrolase (BSH) activity, significantly influences lipid metabolism, weight gain, and cholesterol levels in the host. In our study microbial BSH activity was shown to direct expression of host signalling pathways with known roles in lipid metabolism, circadian rhythm, and epithelial cell function. The work defines the significant impact of in situ bile hydrolysis on host metabolism and indicates how this finding may be exploited as a potential intervention strategy for the control of obesity and metabolic syndrome.
Keywords: FXR, host response, Lactobacillus salivarius, adiponectin, barrier function
Abstract
Alterations in the gastrointestinal microbiota have been implicated in obesity in mice and humans, but the key microbial functions influencing host energy metabolism and adiposity remain to be determined. Despite an increased understanding of the genetic content of the gastrointestinal microbiome, functional analyses of common microbial gene sets are required. We established a controlled expression system for the parallel functional analysis of microbial alleles in the murine gut. Using this approach we show that bacterial bile salt hydrolase (BSH) mediates a microbe–host dialogue that functionally regulates host lipid metabolism and plays a profound role in cholesterol metabolism and weight gain in the host. Expression of cloned BSH enzymes in the gastrointestinal tract of gnotobiotic or conventionally raised mice significantly altered plasma bile acid signatures and regulated transcription of key genes involved in lipid metabolism (Pparγ, Angptl4), cholesterol metabolism (Abcg5/8), gastrointestinal homeostasis (RegIIIγ), and circadian rhythm (Dbp, Per1/2) in the liver or small intestine. High-level expression of BSH in conventionally raised mice resulted in a significant reduction in host weight gain, plasma cholesterol, and liver triglycerides, demonstrating the overall impact of elevated BSH activity on host physiology. In addition, BSH activity in vivo varied according to BSH allele group, indicating that subtle differences in activity can have significant effects on the host. In summary, we demonstrate that bacterial BSH activity significantly impacts the systemic metabolic processes and adiposity in the host and represents a key mechanistic target for the control of obesity and hypercholesterolemia.
The gastrointestinal microbiota exerts a major influence on host energy metabolism and adiposity (1–5), but the precise microbial activities that influence lipid metabolism in the host remain largely unidentified. Large-scale sequencing studies have cataloged the genetic composition of the human gut microbiota (the microbiome), providing insights into the core microbial genes whose products are predicted to influence host metabolism (6, 7). There is a significant challenge in categorizing both gut-specific and gut-enriched microbial activities functionally to determine the relevance of specific gene sets in a physiological or pathological context.
Bile acids are the main functional components of bile secretions that play a role in the emulsification of dietary lipids (8) and also act as signaling molecules in the host, triggering host responses mediated by both cellular farnesoid X receptor (FXR) and G protein-coupled receptor (9–11). Bile acids influence the composition of the gastrointestinal microbiota and in turn are chemically modified by bacterial enzymes in the gut. Bile acids can be considered as mediators of reciprocal microbe–host crosstalk with the ability to influence host metabolic pathways (10, 12, 13) and the potential to influence microbial community structure (12, 14). Bile acids are synthesized in hepatocytes as cholesterol moieties conjugated to either a taurine or a glycine amino acid and are stored in the gallbladder before secretion into the duodenum via the common bile duct. Bacterial enzymes in the gut significantly modify bile acids, a process that in turn influences host bile acid synthesis through a feedback mechanism in which the hepatic cholesterol hydroxylase enzymes involved in bile acid synthesis (including Cyp7A1 and Cyp8B1) are regulated (10, 13). Recent evidence suggests that this feedback mechanism involves activation of the nuclear bile acid receptor FXR in enterocytes, leading to the production of FGF15/19, which regulates hepatic Cyp7A1 (13). The existence of additional regulatory networks that influence bile acid biosynthesis also has been demonstrated (15).
Bacterial bile salt hydrolase (BSH) enzymes in the gut catalyze an essential gateway reaction in the metabolism of bile acids. BSH enzymes cleave the amino acid side chain of glyco- or tauro-conjugated bile acids to generate unconjugated bile acids (cholic and chenodeoxycholic acids), which then are amenable to further bacterial modification to yield secondary bile acids (deoxycholic and lithocholic acid) (8). We have shown previously that functional BSH activity is a conserved microbial adaptation that is unique to the gut-associated microbiota and is distributed across the major bacterial divisions and archaeal species in the gastrointestinal tract (12). We demonstrated that BSH contributes to bile tolerance in gut bacteria and hypothesized that the evolution of BSH activity is governed by host-driven selection (12).
Here, using a controlled experimental system, we significantly extend our previous analysis of BSH activities in the host to analyze the impact of in situ bile salt hydrolysis on host metabolism and weight gain. We demonstrate that expression of cloned bacterial BSH enzymes in the gastrointestinal tract significantly modifies plasma bile acid profiles in gnotobiotic mice and influences both local and systemic gene-expression profiles in pathways governing lipid metabolism, metabolic signaling events, circadian rhythm, and immune function. We show that by elevating BSH activity in conventionally raised mice (animals raised with a normal microbiota) we can significantly reduce weight gain, serum cholesterol, and liver triglycerides in these animals. This work definitively identifies BSH as a mechanism through which the microbiota modulates host lipid metabolism and demonstrates that this mechanism represents a key target for the development of intervention strategies for the control of obesity and metabolic syndrome.
Results
Significant Alteration of Bile Acid Profiles in Gnotobiotic Mice Through Gastrointestinal BSH Activity.
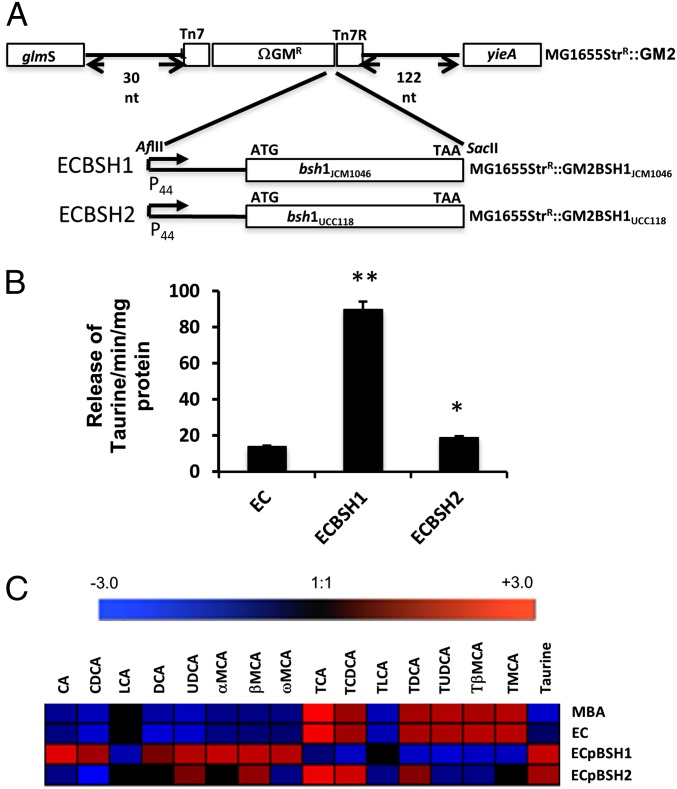
The wide variation in BSH enzymes within the gut microbiota suggests that different BSH alleles may have differing impacts on in vivo bile metabolism and downstream responses (12). To compare different BSH enzymes using an isogenic delivery system, we expressed bsh genes in Escherichia coli MG1655, a K-12 strain that lacks BSH activity and colonizes both conventionally raised (16) and germ-free (this study) mice at high levels. To achieve stable expression in long-term colonization experiments, we used the miniTn7 transposon system for the cloning of bsh genes in single copy into the region downstream of glmS in the E. coli host (Fig. 1A). We cloned and expressed bsh genes from Lactobacillus salivarius JCM1046 (herein designated as BSH1) and L. salivarius UCC118 (herein designated BSH2), which display defined structural differences (17). Both BSHs can deconjugate tauroconjugated bile acids in vitro, as determined by the ninhydrin release assay (Fig. 1B), with BSH1 demonstrating greater efficiency in catalyzing the release of taurine. We also exposed BSH-negative E. coli MG1655 (EC) or E. coli clones expressing BSH1 (ECBSH1) or BSH2 (ECBSH2) to ex vivo murine gallbladder bile and then examined individual bile acid profiles using a sensitive ultra-performance liquid chromatography mass spectronomy (UPLC-MS) protocol. BSH1 exhibited the greatest efficacy in generating deconjugated bile acids and free taurine when measured in this in vitro system (Fig. 1C).
Fig. 1.
Expression of cloned BSH in E. coli MG1655 and activity in murine gallbladder bile in vitro. (A) Cloning strategy for expression of BSH enzymes in E. coli MG1655. (B) Ninhydrin assay showing the release of taurine from conjugated bile acids as an index of BSH activity. *P < 0.05; **P < 0.005, (n = 5 per group). (C) Heat maps summarizing UPLC-MS analysis of individual bile acids in murine bile in vitro following 90-min exposure to EC, to plasmid-borne ECpBSH1 or ECpBSH2, or to the empty vector control, ECpNZ44. Results represent analysis of three biological replicates. MBA, murine bile acids.
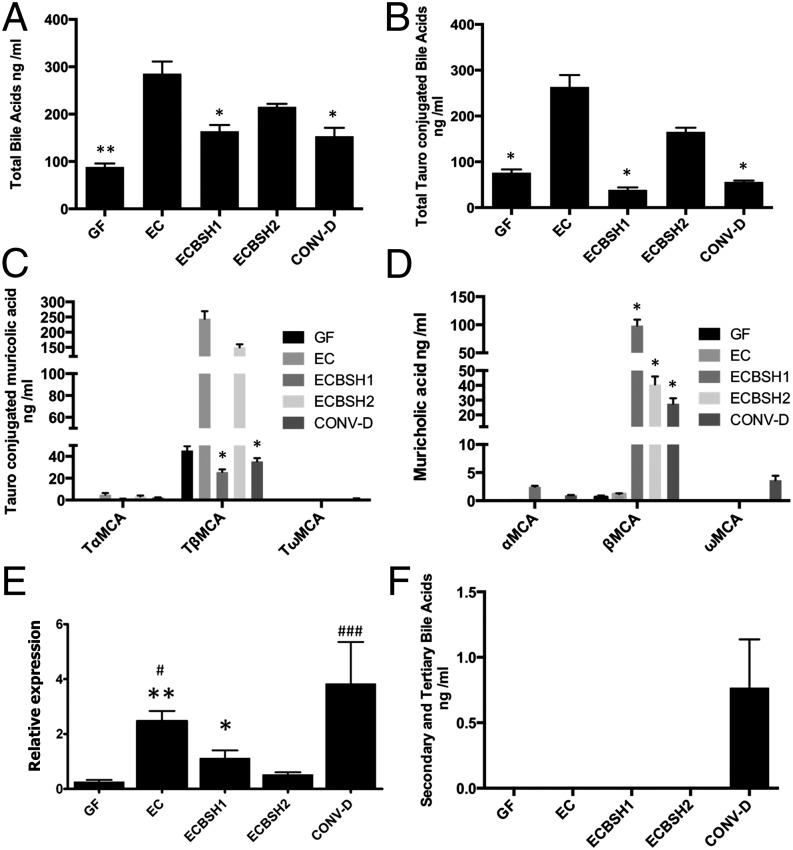
To analyze the physiological effects of bile hydrolysis in a controlled system that lacks extant bile-modification systems, we monocolonized gnotobiotic mice with our E. coli strains expressing BSH activity (ECBSH1 or ECBSH2). Colonization of germ-free mice with EC resulted in a significant elevation of total plasma bile acids (Fig. 2A), indicating that bacterial colonization influences bile metabolism regardless of BSH status. In our system in situ BSH activity resulted in a significant reduction of total plasma bile acids and a specific reduction in tauroconjugated bile acids relative to levels in EC-colonized mice, demonstrating the effects of in vivo deconjugation of bile acids (Fig. 2 A and B). In particular we noted a reduction in the levels of the potent FXR antagonist tauro-β-muricholic acid (TβMCA) (13) relative to levels in EC-colonized gnotobiotic mice as a result of in situ BSH expression (Fig. 2 C and D). Levels of taurocholic acid (TCA) also were reduced by in vivo BSH1 expression in our system with a resultant elevation in cholic acid (CA) (Fig. S1). The findings may reflect poor enterohepatic uptake of deconjugated bile acids relative to conjugated bile acids in the ileum (18). However, gastrointestinal BSH activity significantly reduced the expression of the hepatic gene encoding an enzyme that is rate limiting in the synthesis of bile acids (Cyp7a1), as is consistent with reduced de novo synthesis of bile acids (Fig. 2E). This finding indicates that it is possible to manipulate a bile acid feedback mechanism [potentially mediated via TβMCA (13)] in the host through gut expression of BSH. As expected, we failed to detect significant levels of secondary or tertiary bile acids in gnotobiotic mice, although these bile acids were abundant in mice with a reconstituted microbiota [conventionalized (CONV-D) mice] (Fig. 2F).
Fig. 2.
Alterations of host bile acid signatures through gastrointestinal expression of cloned BSH in gnotobiotic mice. (A) Total plasma bile acids (assessed by UPLC-MS) in germ-free (GF) mice, mice monocolonized with EC, ECBSH1, or ECBSH2, and conventionalized mice (CONV-D). (B) Total tauroconjugated bile acids (assessed by UPLC-MS) in plasma of GF mice, mice monocolonized with EC, ECBSH1, or ECBSH2, and CONV-D mice. (C) Total tauroconjugated muricholic acid moieties in plasma of GF mice, mice monocolonized with EC, ECBSH1, or ECBSH2, and CONV-D mice. (D) Total unconjugated muricholic acid moieties in plasma of GF mice, mice monocolonized with ESC, ECBSH1, or ECBSH2, and CONV-D mice. In all cases statistical comparisons were made relative to E. coli controls using the Student t test and were corrected for false discovery using the Benjamini–Hochberg procedure. *P < 0.05; **P < 0.005 (n = 5 per group). (E) Influence of gastrointestinal BSH expression upon Cyp7a expression in the livers of monocolonized mice. Cyp7a transcript was measured by qRT-PCR (n = 5 per group). Data are presented as means ± SEM; *P < 0.05 vs. GF; **P < 0.01 vs. GF; #P < 0.05 vs. ECBSH1; ###P < 0.001 vs. ECBSH1. (F) Total levels of secondary and tertiary bile acids in plasma of GF mice, mice monocolonized with EC, ECBSH1, or ECBSH2, and CONV-D mice. (n = 5 per group).
Overall, the data indicate that the induction of in situ BSH activity in our model system significantly redirected the murine bile acid signature in a number of tissue compartments, with BSH1 in particular altering the ratio of conjugated to unconjugated bile acids in plasma (Fig. S2A), liver (Fig. S2B), and feces (Fig. S2C). Principal component analysis (PCA) of plasma-extracted samples indicated that all groups could be delimited on the basis of bile acid signature. The germ-free animals showed tight clustering and clear separation from all of the other groups. Mice colonized by EC and ECBSH2 were more similar, whereas animals treated with ECBSH1 and CONV-D animals were clearly separated (Fig. S2D).
Impact of Gastrointestinal Bile Salt Hydrolysis on Local and Systemic Gene-Expression Patterns in the Host.
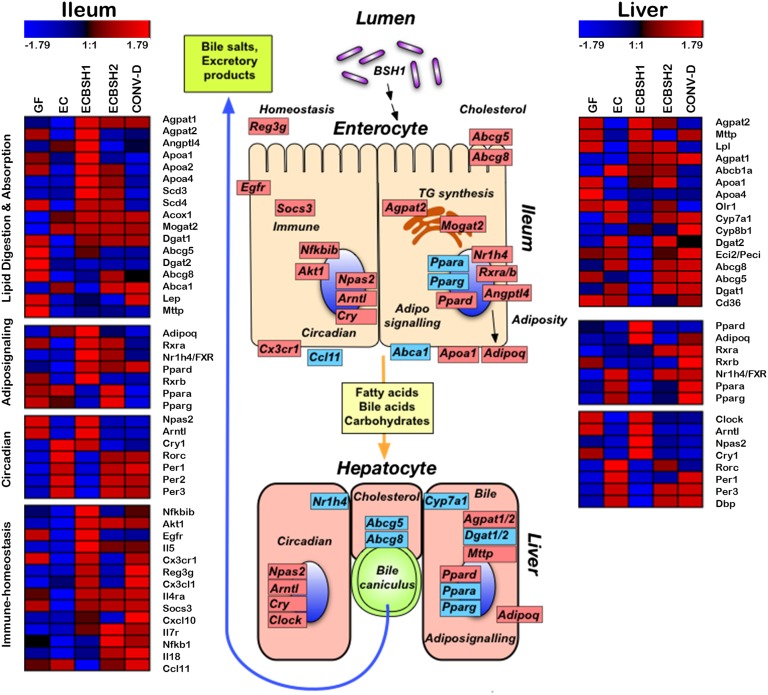
We examined the expression patterns of more than 23,000 genes in the liver and ileum in germ-free mice monocolonized with EC, ECBSH1, or ECBSH2 and in CONV-D mice. Overall significant changes in host gene-expression patterns in both the ileum and liver were induced in BSH1- and BSH2-colonized mice relative to EC-colonized mice. We used gene annotation and pathway mapping using Subio software to examine the primary functional groups of host genes regulated through in situ expression of BSH enzymes in the host gastrointestinal tract. Because of the potent activity of BSH1 in vitro and in vivo, we focus here primarily on the influence of BSH1 expression in our system. However, many of the loci influenced by BSH1 also are influenced by BSH2 activity (Fig. 3). The figure outlines selected genes in which BSH activity significantly modulated expression levels relative to the EC-colonized control. In the ileum BSH1 activity altered the expression of loci associated with immune function, cholesterol transport, and lipid transport and synthesis (Fig. 3). Gene expression also was significantly altered in the livers of mice following gastrointestinal colonization by ECBSH1, with the regulation of major metabolic pathways involved in triglyceride biosynthesis, bile synthesis, and fatty acid transport and synthesis. Gene-expression profiles for a number of target genes were validated using quantitative RT-PCR (qRT-PCR) (Fig. S3).
Fig. 3.
BSH expression in the gastrointestinal tract of gnotobiotic mice significantly alters gene-expression patterns in ileal and hepatic tissue. Microarray analysis of ileal and liver tissue from GF mice, CONV-D mice, and mice monocolonized with EC, ECBSH1, or ECBSH2. Shown are heat maps representing gene-expression profiles of selected genes that were significantly (P < 0.05) altered through BSH1 expression in our system. Pathways related to lipid digestion and absorption, circadian rhythm, adipo-signaling, and immune homeostasis were most significantly affected, as determined by pathway analysis, and are shown here (n = 5 mice per group). The schematic indicates key transcriptional changes affected by BSH1 expression. Genes increased in ECBSH1-colonized mice relative to EC-colonized mice are indicated in red, genes decreased in ECBSH1-colonized mice relative to EC-colonized mice are indicated in blue.
Functional Consequences of Elevated Gastrointestinal BSH Activity in Conventionally Raised Mice.
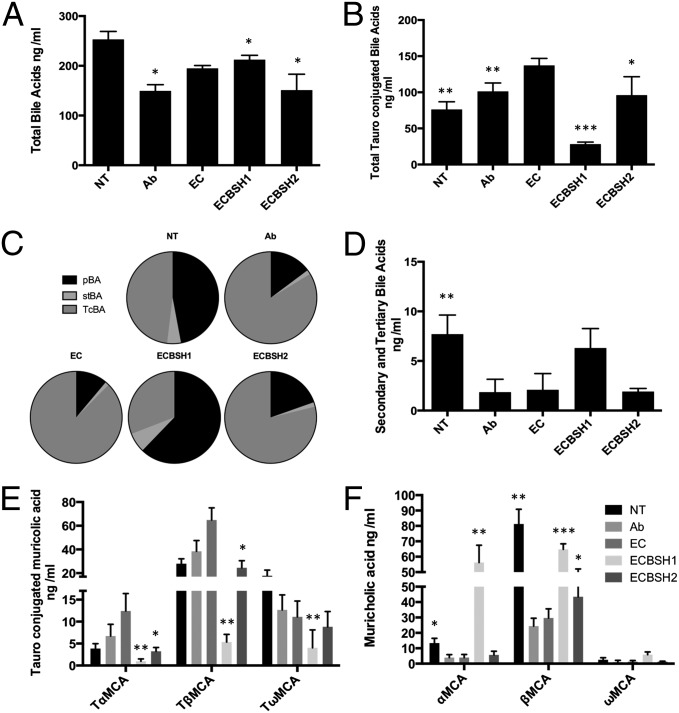
Given the influence of bacterial BSH on host energy pathways under controlled conditions in gnotobiotic mice, we examined whether modulation of gastrointestinal BSH activity could form the basis of an intervention strategy for the control of host weight gain and metabolic processes in conventionally raised animals. To obtain consistent, high-level expression of gastrointestinal BSH, we again used the E. coli MG1655 gut colonization model (16, 19) in which conventionally raised streptomycin-treated mice were significantly colonized for over 70 d with strepR EC, ECBSH1, or ECBSH2 (Fig. S4). Previous studies in our laboratory demonstrated efficient colonization of E. coli MG1655 within both the small bowel and large intestine in this model system (19). Expression of BSH1 in situ in the murine gut did not affect total plasma bile acids dramatically (Fig. 4A) but resulted in a fourfold reduction in tauroconjugated bile acids and a proportional increase in unconjugated primary bile acids in plasma (Fig. 4B and Fig. S5A) (Fig. 4C and Fig. S5A). Heightened BSH1 activity caused a moderate but not statistically significant increase in levels of secondary and tertiary bile acids (Fig. 4D). Notably, gastrointestinal expression of BSH in conventionally raised mice resulted in a dramatic reduction in plasma TβMCA (Fig. 4E) and a concomitant increase in levels of β-muricholic acid (Fig. 4F) relative to EC-colonized animals. Overall the effects of heightened deconjugation activity through BSH1 expression in the gastrointestinal tract were evident in the plasma (Fig. S5B), liver (Fig. S5C), and feces (Fig. S5D).
Fig. 4.
Gastrointestinal expression of cloned BSH in conventionally raised mice alters plasma bile acid profiles. Mice were provided with streptomycin (5 mg/mL) ad libitum in drinking water to promote stable high-level colonization of the host E. coli MG1655 StrepR strain as described previously (16). (A) Total plasma bile acids (assessed by UPLC-MS) in conventional mice (not treated, NT), conventional mice treated with antibiotic only (Ab), and mice colonized with EC, ECBSH1, or ECBSH2). (B) Total tauroconjugated plasma bile acids in uncolonized untreated or antibiotic-treated mice and mice colonized by EC, ECBSH1, or ECBSH2. (C) Relative proportions of primary bile acids (pBAs), secondary and tertiary bile acids (stBAs), and tauroconjugated bile acids (TcBAs) in the plasma of uncolonized untreated and antibiotic-treated mice and mice colonized by EC, ECBSH1, or ECBSH2. (D) Secondary and tertiary bile acid moieties in uncolonized, untreated and antibiotic-treated mice and mice colonized by EC, ECBSH1, or ECBSH2. (E) Total tauroconjugated muricholic acid moieties in plasma of conventionally raised untreated and antibiotic-treated mice and mice colonized by EC, ECBSH1, or ECBSH2. (F) Total unconjugated muricholic acid moieties in plasma of untreated mice, antibiotic-treated mice, and mice colonized by EC, ECBSH1, or ECBSH2. In all cases comparisons were made relative to E. coli controls using the Student t test and were corrected for false discovery using the Benjamini–Hochberg procedure. *P < 0.05; **P < 0.005; ***P < 0.0005 (n = 5 per group).
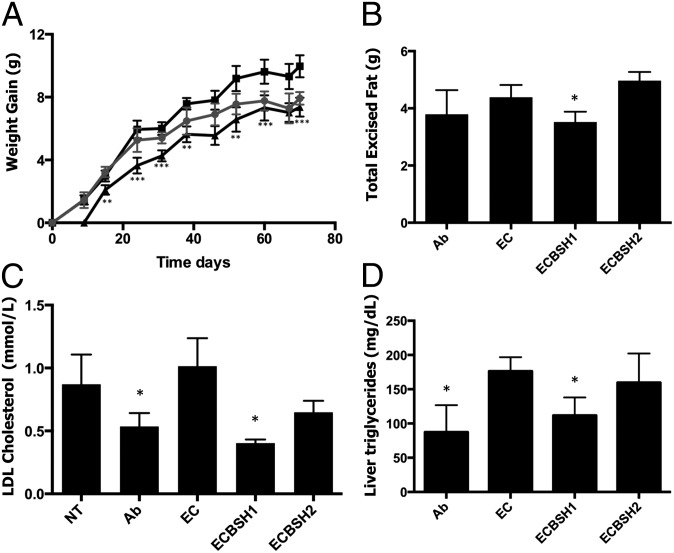
Colonization of conventionally raised mice by ECBSH1 resulted in significantly diminished weight gain (46% reduction) relative to mice colonized by EC in animals fed either a normal (Fig. 5A) or a high-fat diet (Fig. S6A). Food intake was not affected. Diminished weight gain was associated with reduced fat deposition in these animals (Fig. 5B and Fig. S6B). BSH1 expression also was capable of lowering serum LDL cholesterol (by 60.6%) and liver triglycerides (by 36.5%) relative to mice colonized by EC (Fig. 5 C and D). Reductions of this magnitude are likely to be physiologically relevant for the mammalian host (20). Similar results were seen in mice fed a high-fat diet (Fig. S6 C and D). We noted that colonization of mice with EC resulted in increased weight gain, supporting recent studies that link increases in body mass to increases in Proteobacteria (21), including E. coli (22). In our system BSH1 activity reversed this increase in weight gain. Significantly, we did not see an increase in systemic inflammation in our model (Fig. S7).
Fig. 5.
Gastrointestinal expression of cloned BSH in conventionally raised mice reduces weight gain, serum cholesterol, and liver triglycerides. (A) Average weight gain over time measured in grams following colonization of mice with EC or ECBSH1. Data represent antibiotic-treated mice (solid circles), EC-colonized mice (solid squares), and ECBSH1-colonized mice (solid triangles) with weight gain monitored over 10 wk. **P < 0.005 and ***P < 0.0005 for ECBSH1 mice relative to EC-colonized controls. (B) Weight of total excised fat from mice undergoing antibiotic treatment alone and mice colonized by EC, ECBSH1, or ECBSH2. *P < 0.05 relative to the EC-colonized control (n = 5 per group). (C) Levels of LDL cholesterol in plasma in mice colonized by EC, ECBSH1, or ECBSH2 and control uncolonized mice. *P < 0.05 relative to controls (n = 5 per group). (D) Levels of liver triglycerides in mice colonized by EC, ECBSH1, or ECBSH2 and in antibiotic-treated controls (Ab). *P < 0.05. (n = 5 per group). All analyses used the Student t test.
Effects of Elevated BSH Activity upon Local and Systemic Transcriptional Patterns in the Host.
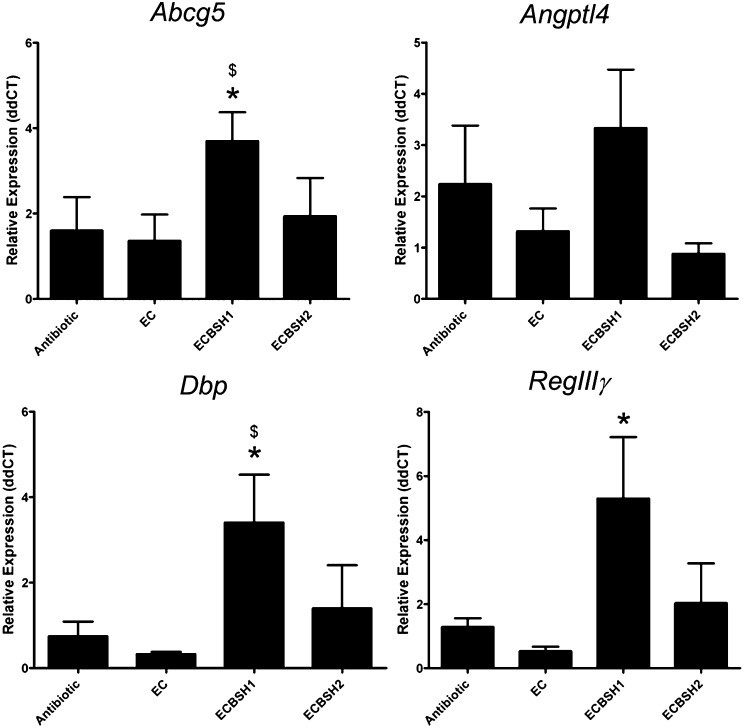
The data from monocolonized gnotobiotic mice identified a number of host pathways that are clearly affected by gastrointestinal BSH activity (Fig. 3). Given the phenotypic changes in host physiology seen in conventionally raised animals, we also examined the gene-expression profiles of a number of key genes in conventionally raised mice colonized by ECBSH1 or ECBSH2 (Fig. 6). The expression of these selected target genes was analyzed using qRT-PCR. In particular we detected an increase in intestinal gene expression of ATP-binding cassette sub-family G member (Abcg5) (23), angiopoietin-like 4 (Angptl4) (24), albumin D-box binding protein (Dbp) (25), and regenerating islet-derived protein 3 gamma (RegIIIγ) (26) (Fig. 6). Expression of Cdkn1a, a gene encoding a regulator of cell cycle (p21) (27), peroxisome proliferator-activated receptor gamma (Pparγ), and Abcg8 also were influenced by BSH1 in conventionally raised mice (Fig. S8).
Fig. 6.
Gastrointestinal expression of BSH influences gene-expression patterns in ileal tissue in conventionally raised mice. Mice given streptomycin were colonized by E. coli MG1655 StrepR as outlined in our model system. Gene-expression patterns of selected genes were examined using qRT-PCR in ileal tissue of mice colonized by EC, ECBSH1, or ECBSH2 and of uncolonized animals (n = 5 per group). Statistical significance was determined using ANOVA. Data are presented as means ± SEM; *P < 0.05 vs. GF; $P < 0.05 vs. EC.
Discussion
Here we determined that a single, widely distributed, and gut-specific bacterial enzyme, BSH, is capable of directing local and systemic gene-expression profiles in metabolic pathways and influencing weight gain and adiposity in the host. We definitively establish a role for bacterial BSH as a major regulator of host physiological processes and a mediator of a microbe–host interaction that has functional consequences for the host.
Recent work has demonstrated the influence of the collective microbiota upon bile acid signatures and metabolic pathways in the host (10, 13). However, the functional impact of individual bacterial bile acid modifications upon the host has not been established (28). Such information is necessary to gain an appreciation of the physiological consequences that arise from dysbiosis of the microbiota and resultant perturbations of microbe-associated functions. BSH activity is one such function that is widely distributed within the microbiota and has been predicted, based upon in vitro analyses, to influence bile metabolism in the host (12, 29). BSH activity often is associated with bacteria used as probiotics in humans and animals and is considered likely to contribute to survival of probiotic organisms in the gastrointestinal tract (12, 29). However, the potential effects of probiotic BSH activity on the physiology of the host are currently unclear.
We established E. coli MG1655 as a delivery vector capable of expressing cloned bsh genes and colonizing the murine gastrointestinal tract to high levels (16). This isogenic system allowed the direct comparison of different bsh alleles both in vitro and in vivo. Expression of two different bsh alleles from L. salivarius strains was used here because they have been analyzed previously in vitro and are active against tauroconjugated bile acids, which are abundant in mice (17). In particular we demonstrate that both BSH enzymes are active against TβMCA, with the enzyme designated BSH1 demonstrating greater in vitro and in vivo deconjugating activity and more profound downstream consequences in the host. TβMCA was shown recently to be a potent antagonist of the host bile acid receptor FXR, a transcriptional repressor of genes involved in bile acid synthesis (including Cyp7a1) (13). BSH activity additionally releases free taurine, a compound that also may have physiological implications for the host. Using a gnotobiotic mouse model, we demonstrate that gastrointestinal BSH activity significantly shifts overall bile acid profiles in the host, decreases plasma TβMCA, and reduces expression of Cyp7a1 in the liver. Because bile acid profiles in the host can, in turn, influence the composition of the microbiota (11, 14, 29), we suggest that this activity provides a mechanism for crosstalk between microbe and host that has the potential to influence microbial community structure in health and disease. The allelic variation in BSH activities indicates that subtle differences in community BSH profiles may have physiological implications for the host, as suggested recently through metagenomic analysis of bsh alleles in the human gut microbiome (30).
To obtain a global overview of how BSH activity impacts host metabolic processes, we used whole-genome microarray analysis of tissues isolated from ECBSH1- and ECBSH2-monocolonized gnotobiotic mice and appropriate controls. BSH1 activity, in particular, significantly modified the expression of key regulators of lipid metabolism in both the ileum and liver. These regulatory elements included FXR, the hormone adiponectin (AdipoQ), the nuclear retinoid X receptors RXRα and RXRβ, and the PPARs. PPARα is known to be regulated diurnally and interacts with regulators of circadian rhythm (Clock/Bmal1) (31). We observed that gastrointestinal BSH1 activity also promotes a significant shift in the expression pattern of genes normally regulated by circadian rhythm. Along with modulation of major host adipo-signaling regulators, we see a parallel regulation of genes encoding effectors of lipid and cholesterol metabolic pathways (for example Dgat1, Abca1, Acox1, and ApoA1) (32) indicative of significant local and systemic alterations of lipid metabolism following bile acid deconjugation in the gastrointestinal tract. We also note the differential regulation of genes involved in epithelial differentiation and immune homeostasis in the host (Egfr, RegIIIγ, Nfkbib, Il5). This finding indicates that BSH activity may influence cellular homeostasis and supports previous observations documenting an immune-regulatory function for bile acids (11, 33).
The observed impact of BSH activity upon pathways regulating lipid and cholesterol metabolism in gnotobiotic mice prompted us to examine the physiological effects of this enzymatic activity in conventionally raised animals. Although previous studies have attempted to correlate BSH activity with weight gain in animals (29, 34), the physiological effects of gastrointestinal BSH enzymes have not been studied previously using a controlled experimental system. We demonstrate that BSH1 significantly reduces weight gain in mice fed normal or high-fat diets and also reduces serum LDL cholesterol and liver triglycerides in these animals. The small intestine is the major location of lipid metabolism and absorption in mammals (32, 35). We suggest that the ability of ECBSH1 to colonize this location in our model system (19) influences bile acid profiles and therefore lipid metabolism locally with effects on weight gain and energy metabolism that are further regulated through the systemic signaling properties of bile acids (9).
We demonstrated that many of the key gene systems regulated by BSH1 activity in germ-free mice also were regulated in conventionally raised animals, thus suggesting potential mechanisms underpinning the observed physiological effects. In particular we determined an increase in intestinal gene expression of Abcg5/8 loci encoding a cholesterol transport system known to contribute to cholesterol efflux into the lumen (32). This system is thought to provide a mechanistic basis for the systemic reduction of cholesterol by plant stanol esters (36). BSH1 also induced local expression of Angptl4 (also known as fasting-induced adipose factor, FIAF) in conventionally raised mice in our system. Induction of FIAF expression has been shown to be associated with a reduction in obesity in mouse models (37, 38).
BSH1 activity also induced ileal expression of RegIIIγ, which encodes a secreted antibacterial lectin that promotes host–microbial mutualism and immune homeostasis (26). This finding suggests that BSH activity and bile acid signaling may have broader implications for the development of host–microbe mutualism in the gut. BSH1 regulation of genes involved in circadian rhythm in conventionally raised mice (Dbp) and gnotobiotic mice (Clock, Arntl, Per1, Per2, Per3) suggests that microbial metabolism of bile may influence diurnal gene-expression patterns in the host. This influence is potentially an important means of microbe–host crosstalk, given that circadian desynchronizations are clearly linked to alterations in energy metabolism and weight regulation (39).
We appreciate that there are significant differences between the bile acid profiles of mice and humans. In particular the murine conjugated bile acid profile comprises a predominance of tauroconjugated bile acids (including tauromuricholic acids) and a substantial deficiency in glycoconjugated moieties (8, 10). Although bile acids common to mice and humans, including TCA and CA, were affected in our study, further work will be necessary to determine the relevance of our findings in humans.
The influence of the gut microbiota upon energy metabolism is certainly multifactorial (4). Using a controlled biological system, we show that bacterial BSH represents a mechanism by which the microbiota influences cholesterol metabolism and weight gain in the host via a number of potential effector pathways (summarized in Fig. S9). We propose that the wide distribution of BSH enzymes across major divisions and phyla within the gut microbiota (12) reflects host-driven selection of this trait. Our data provide definitive support for previous observations that correlate reduced BSH function in antibiotic-treated animals with weight gain (29, 40). Importantly, in the context of a global obesity epidemic and a significant increase in the incidence of metabolic syndrome, we clearly demonstrate the feasibility of intervention strategies for disease states based on rational manipulation of bile metabolism by the gut microbiota. Because many probiotic bacteria generally regarded as safe express potent BSH activity (29), we propose that the current work provides a basis for the rational selection of probiotic bacteria for the control of obesity, hypercholesterolemia, and metabolic syndrome.
Materials and Methods
BSH Cloning.
BSHs from L. salivarius strains (17) were cloned independently into E. coli using pBKminiTn7GM2 under the control of the P44 promoter as outlined in SI Materials and Methods.
Bile Salt Activity Assay.
EC, ECBSH1, and ECBSH2 were examined for their ability to deconjugate bile in vitro using the ninhydrin assay for free taurine and by coincubation for 90 min in murine gall bladder bile acid followed by UPLC-MS analysis. Details are given in SI Materials and Methods.
Mice.
Experiments were approved by the University College Cork Animal Experimentation Ethics Committee. For experimental details see SI Materials and Methods.
UPLC-MS.
UPLC-MS analysis of bile acids (Table S1) was performed using the method of Swann et al., with modifications (10). Details can be found in SI Materials and Methods.
Microarrays.
Microarrays were carried out by the Almac Group, Craigavon, Northern Ireland using mouse Exon ST1.0 arrays (Affymetrix) and were analyzed using Subio Platform software (Subio Inc) and Genesis Software. Microarray data are deposited on the Gene Expression Omnibus website (accession no. GSE46952). Details are in SI Materials and Methods.
Statistical Analysis.
Data are presented as mean values with SD (unless otherwise indicated). Statistical analysis was performed by ANOVA and Student t test and where indicated were corrected for false discovery using the Benjamini–Hochberg procedure.
Supplementary Material
Acknowledgments
We thank Brian Jones (University of Brighton) for critical reading of the manuscript; Robert O'Doherty (University of Pittsburgh) and Paul O’Toole (Alimentary Pharmabiotic Centre, University College Cork) for helpful initial discussions; Adam Clooney for statistical analysis; and Frances O'Brien and colleagues for advice and input. The mass spectrometry unit was supported by an institutional grant from the Irish government via the Programme for Research in Third-Level Institutions. This work was funded by Grants 07/CE/B1368 and 12/RC/2273 from Science Foundation Ireland (to the Alimentary Pharmabiotic Centre).
Footnotes
Conflict of interest statement: S.A.J., C.H., and C.G.M.G. have filed a patent relating to a portion of the work outlined in this paper.
*This Direct Submission article had a prearranged editor.
Data deposition: The microarray data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE46952).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1323599111/-/DCSupplemental.
References
- 1.Murphy EF, et al. Composition and energy harvesting capacity of the gut microbiota: Relationship to diet, obesity and time in mouse models. Gut. 2010;59(12):1635–1642. doi: 10.1136/gut.2010.215665. [DOI] [PubMed] [Google Scholar]
- 2.Turnbaugh PJ, et al. A core gut microbiome in obese and lean twins. Nature. 2009;457(7228):480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Turnbaugh PJ, et al. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444(7122):1027–1031. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- 4.Clarke SF, et al. The gut microbiota and its relationship to diet and obesity: New insights. Gut Microbes. 2012;3(3):186–202. doi: 10.4161/gmic.20168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cho I, et al. Antibiotics in early life alter the murine colonic microbiome and adiposity. Nature. 2012;488(7413):621–626. doi: 10.1038/nature11400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lepage P, et al. A metagenomic insight into our gut’s microbiome. Gut. 2013;62(1):146–158. doi: 10.1136/gutjnl-2011-301805. [DOI] [PubMed] [Google Scholar]
- 7.Qin J, et al. MetaHIT Consortium A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464(7285):59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Begley M, Gahan CG, Hill C. The interaction between bacteria and bile. FEMS Microbiol Rev. 2005;29(4):625–651. doi: 10.1016/j.femsre.2004.09.003. [DOI] [PubMed] [Google Scholar]
- 9.Watanabe M, et al. Bile acids induce energy expenditure by promoting intracellular thyroid hormone activation. Nature. 2006;439(7075):484–489. doi: 10.1038/nature04330. [DOI] [PubMed] [Google Scholar]
- 10.Swann JR, et al. Systemic gut microbial modulation of bile acid metabolism in host tissue compartments. Proc Natl Acad Sci USA. 2011;108(Suppl 1):4523–4530. doi: 10.1073/pnas.1006734107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Inagaki T, et al. Regulation of antibacterial defense in the small intestine by the nuclear bile acid receptor. Proc Natl Acad Sci USA. 2006;103(10):3920–3925. doi: 10.1073/pnas.0509592103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jones BV, Begley M, Hill C, Gahan CG, Marchesi JR. Functional and comparative metagenomic analysis of bile salt hydrolase activity in the human gut microbiome. Proc Natl Acad Sci USA. 2008;105(36):13580–13585. doi: 10.1073/pnas.0804437105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sayin SI, et al. Gut microbiota regulates bile acid metabolism by reducing the levels of tauro-beta-muricholic acid, a naturally occurring FXR antagonist. Cell Metab. 2013;17(2):225–235. doi: 10.1016/j.cmet.2013.01.003. [DOI] [PubMed] [Google Scholar]
- 14.Islam KB, et al. Bile acid is a host factor that regulates the composition of the cecal microbiota in rats. Gastroenterology. 2011;141(5):1773–1781. doi: 10.1053/j.gastro.2011.07.046. [DOI] [PubMed] [Google Scholar]
- 15.Kir S, Zhang Y, Gerard RD, Kliewer SA, Mangelsdorf DJ. Nuclear receptors HNF4α and LRH-1 cooperate in regulating Cyp7a1 in vivo. J Biol Chem. 2012;287(49):41334–41341. doi: 10.1074/jbc.M112.421834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chang DE, et al. Carbon nutrition of Escherichia coli in the mouse intestine. Proc Natl Acad Sci USA. 2004;101(19):7427–7432. doi: 10.1073/pnas.0307888101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fang F, et al. Allelic variation of bile salt hydrolase genes in Lactobacillus salivarius does not determine bile resistance levels. J Bacteriol. 2009;191(18):5743–5757. doi: 10.1128/JB.00506-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dawson PA, Lan T, Rao A. Bile acid transporters. J Lipid Res. 2009;50(12):2340–2357. doi: 10.1194/jlr.R900012-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cronin M, et al. High resolution in vivo bioluminescent imaging for the study of bacterial tumour targeting. PLoS ONE. 2012;7(1):e30940. doi: 10.1371/journal.pone.0030940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Klop B, Elte JW, Cabezas MC. Dyslipidemia in obesity: Mechanisms and potential targets. Nutrients. 2013;5(4):1218–1240. doi: 10.3390/nu5041218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fei N, Zhao L. An opportunistic pathogen isolated from the gut of an obese human causes obesity in germfree mice. ISME J. 2013;7(4):880–884. doi: 10.1038/ismej.2012.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Looft T, et al. In-feed antibiotic effects on the swine intestinal microbiome. Proc Natl Acad Sci USA. 2012;109(5):1691–1696. doi: 10.1073/pnas.1120238109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yu L, et al. Overexpression of ABCG5 and ABCG8 promotes biliary cholesterol secretion and reduces fractional absorption of dietary cholesterol. J Clin Invest. 2002;110(5):671–680. doi: 10.1172/JCI16001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bäckhed F, et al. The gut microbiota as an environmental factor that regulates fat storage. Proc Natl Acad Sci USA. 2004;101(44):15718–15723. doi: 10.1073/pnas.0407076101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ando H, et al. Impairment of peripheral circadian clocks precedes metabolic abnormalities in ob/ob mice. Endocrinology. 2011;152(4):1347–1354. doi: 10.1210/en.2010-1068. [DOI] [PubMed] [Google Scholar]
- 26.Vaishnava S, et al. The antibacterial lectin RegIIIgamma promotes the spatial segregation of microbiota and host in the intestine. Science. 2011;334(6053):255–258. doi: 10.1126/science.1209791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim WB, et al. Expression and clinical significance of cell cycle regulatory proteins in gallbladder and extrahepatic bile duct cancer. Ann Surg Oncol. 2009;16(1):23–34. doi: 10.1245/s10434-008-0182-x. [DOI] [PubMed] [Google Scholar]
- 28.Nicholson JK, et al. Host-gut microbiota metabolic interactions. Science. 2012;336(6086):1262–1267. doi: 10.1126/science.1223813. [DOI] [PubMed] [Google Scholar]
- 29.Begley M, Hill C, Gahan CG. Bile salt hydrolase activity in probiotics. Appl Environ Microbiol. 2006;72(3):1729–1738. doi: 10.1128/AEM.72.3.1729-1738.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ogilvie LA, Jones BV. Dysbiosis modulates capacity for bile acid modification in the gut microbiomes of patients with inflammatory bowel disease: A mechanism and marker of disease? Gut. 2012;61(11):1642–1643. doi: 10.1136/gutjnl-2012-302137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lin JD, Liu C, Li S. Integration of energy metabolism and the mammalian clock. Cell Cycle. 2008;7(4):453–457. doi: 10.4161/cc.7.4.5442. [DOI] [PubMed] [Google Scholar]
- 32.Abumrad NA, Davidson NO. Role of the gut in lipid homeostasis. Physiol Rev. 2012;92(3):1061–1085. doi: 10.1152/physrev.00019.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ichikawa R, et al. Bile acids induce monocyte differentiation toward interleukin-12 hypo-producing dendritic cells via a TGR5-dependent pathway. Immunology. 2012;136(2):153–162. doi: 10.1111/j.1365-2567.2012.03554.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Feighner SD, Dashkevicz MP. Subtherapeutic levels of antibiotics in poultry feeds and their effects on weight gain, feed efficiency, and bacterial cholyltaurine hydrolase activity. Appl Environ Microbiol. 1987;53(2):331–336. doi: 10.1128/aem.53.2.331-336.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Joyce SA, Gahan CG. The gut microbiota and the metabolic health of the host. Curr Opin Gastroenterol. 2014;30(2):120–127. doi: 10.1097/MOG.0000000000000039. [DOI] [PubMed] [Google Scholar]
- 36.Repa JJ, et al. Regulation of ATP-binding cassette sterol transporters ABCG5 and ABCG8 by the liver X receptors alpha and beta. J Biol Chem. 2002;277(21):18793–18800. doi: 10.1074/jbc.M109927200. [DOI] [PubMed] [Google Scholar]
- 37.Bäckhed F, Manchester JK, Semenkovich CF, Gordon JI. Mechanisms underlying the resistance to diet-induced obesity in germ-free mice. Proc Natl Acad Sci USA. 2007;104(3):979–984. doi: 10.1073/pnas.0605374104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Aronsson L, et al. Decreased fat storage by Lactobacillus paracasei is associated with increased levels of angiopoietin-like 4 protein (ANGPTL4) PLoS ONE. 2010;5(9):e13087. doi: 10.1371/journal.pone.0013087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ekmekcioglu C, Touitou Y. Chronobiological aspects of food intake and metabolism and their relevance on energy balance and weight regulation. Obes Rev. 2011;12(1):14–25. doi: 10.1111/j.1467-789X.2010.00716.x. [DOI] [PubMed] [Google Scholar]
- 40.Guban J, Korver DR, Allison GE, Tannock GW. Relationship of dietary antimicrobial drug administration with broiler performance, decreased population levels of Lactobacillus salivarius, and reduced bile salt deconjugation in the ileum of broiler chickens. Poult Sci. 2006;85(12):2186–2194. doi: 10.1093/ps/85.12.2186. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.