Significance
The Notch pathway is a crucial metazoan cell–cell signaling pathway. The Notch receptor is regulated by O-glycosylation, a sugar modification that involves a series of enzyme-catalyzed additions to residues within EGF domains. Here, we demonstrate that the Fringe enzyme modification enhances the affinity of a receptor fragment for its ligand. X-ray crystallographic analysis demonstrates that the backbone structure does not change as a consequence of the modification, suggesting that the Fringe addition directly enhances ligand binding, although indirect effects cannot be ruled out. These data help to explain the Notch–Delta signaling increase seen in the presence of Fringe, but suggest that the inhibitory effects observed with the Jagged/Serrate ligand class are mediated by other regions of modified Notch.
Keywords: glycosylation, mass spectrometry, X-ray
Abstract
The Notch signaling pathway is essential for many aspects of development, cell fate determination, and tissue homeostasis. Notch signaling can be modulated by posttranslational modifications to the Notch receptor, which are known to alter both ligand binding and receptor activation. We have modified the ligand-binding region (EGF domains 11–13) of human Notch1 (hN1) with O-fucose and O-glucose glycans and shown by flow cytometry and surface plasmon resonance that the Fringe-catalyzed addition of GlcNAc to the O-fucose at T466 in EGF12 substantially increases binding to Jagged1 and Delta-like 1 (DLL1) ligands. We have subsequently determined the crystal structures of EGF domains 11–13 of hN1 modified with either the O-fucose monosaccharide or the GlcNAc–fucose disaccharide at T466 of EGF12 and observed no change in backbone structure for each variant. Collectively, these data demonstrate a role for GlcNAc in modulating the ligand-binding site in hN1 EGF12, resulting in an increased affinity of this region for ligands Jagged1 and DLL1. We propose that this finding explains the Fringe-catalyzed enhancement of Notch–Delta signaling observed in flies and humans, but suggest that the inhibitory effect of Fringe on Jagged/Serrate mediated signaling involves other regions of Notch.
The Notch signaling pathway is universally conserved among all metazoan species and is involved in many aspects of cell fate determination, tissue patterning, and homeostasis (1). Notch (N) receptors are large single-pass transmembrane glycoproteins that undergo cleavage by a furin-like convertase before being targeted to the cell surface as a heterodimer (2, 3). The Notch extracellular domain (NECD) contains up to 36 tandem epidermal growth factor-like (EGF) modules, and EGF 11–12 in Drosophila (d)Notch and human (h)N1 (4, 5) are required for Notch to bind to the Delta, Serrate, Lag-2 (DSL) domain of a ligand molecule on an adjacent cell (6, 7). Drosophila has a single Notch receptor and two ligands, Delta and Serrate. Mammalian species contain four Notch homologs (Notch1–4) and five ligands [Jagged1 and 2 and Delta-like ligand (DLL) 1, 3, and 4], which are also expressed as large single-pass transmembrane proteins (8). Endocytosis of the receptor–ligand complex induces a conformational change in the negative regulatory region of the Notch receptor, which is cleaved by ADAM 10 (9) and the γ-secretase complex (10, 11). This process removes the NECD from the surface of the Notch-expressing cell and releases the Notch intracellular domain (NICD) from the membrane (12). NICD translocates to the nucleus, where it can activate expression of the HES/HEY family of genes (13).
Many of the EGF modules in the NECD are modified by O-fucose and/or O-glucose glycans (14–16). O-fucose is added to Ser or Thr residues in the consensus sequence C2-X-X-X-X-(S/T)-C3 (14, 17) by protein O-fucosyltransferase (Pofut)-1 in mammals (Ofut1 in flies) and is essential for optimal Notch signaling in flies and mice (18, 19). Similarly, O-glucose can also be added to a Ser in the consensus C1-X-S-X-(P/A)-C2 by protein O-glucosyltransferase (Poglut). O-glucosylation is also essential for Notch signaling in mouse and Drosophila (20, 21). The molecular mechanism for the effects of these sugars on Notch activity is not yet clear. The presence of O-fucose can enhance ligand binding by mammalian Notch receptors (22, 23), whereas O-glucose does not appear to affect ligand binding but a subsequent step before receptor cleavage (20, 21).
Many of the O-fucose monosaccharides on Notch can be extended with GlcNAc by a single Fringe enzyme in Drosophila and by any of three Fringe enzymes—Lunatic fringe (Lfng), Manic fringe (Mfng), or Radical fringe (Rfng) in mammalian organisms (24), each of which affects Notch activity in slightly different ways (17, 25–27). The simplest case appears in flies, where Fringe modification has a positive effect on Delta-mediated signaling and a negative effect on Serrate-mediated signaling. Prior studies have reported variable, and sometimes contradictory, effects of mammalian Fringes on Notch signaling (22, 26–30) (SI Appendix, Table S1). This variability may be the result of the different cell systems used in these studies, which could cause cell-based variations in the extent of Notch glycosylation. For instance, it is known that the GlcNAc–fucose disaccharide can be further extended with galactose and sialic acid to produce the mature tetrasaccharide: Siaα2–3/6Galβ1–4GlcNAcβ1–3fucose, and this subsequent extension is known to further modulate Notch signaling (30, 31). The molecular basis for the effects of Fringe on Notch signaling is unknown.
The core ligand-binding site in hN1 is located on the central β-sheet of EGF12 (32), directly adjacent to the threonine residue that forms part of the O-fucosylation consensus sequence within this domain (T466). Sugar modifications at this position therefore have the potential to directly or indirectly affect formation of the Notch ligand complex through intermolecular or intramolecular effects. Based on this observation, we have modified prokaryotically expressed and refolded hN111–13 with various O-fucose and -glucose sugars in vitro, and assayed the effects of these modifications on binding to a variety of different mammalian Notch ligands using a flow cytometry-based assay and by surface plasmon resonance (SPR). In parallel, we have determined the crystal structures of hN111–13 modified with both the O-fucose monosaccharide and disaccharide at this position. Collectively, these data demonstrate that the GlcNAc addition by Fringe to O-fucosylated EGF12 substantially enhances the affinity of this region for the ligands Jagged1 and DLL1 without altering the backbone structure of EGF12. Importantly, these data provide a plausible molecular explanation for the positive effect of Fringe on Delta-mediated signaling.
Results
Prokaryotically Expressed and Refolded hN111–13 Can Be Stoichiometrically Modified with O-Fucose and O-Glucose Glycans.
By using purified recombinant Pofut-1 and GDP–fucose, it was possible to stoichiometrically modify hN111–13 protein in vitro with the O-fucose monosaccharide at T466. The O-fucose was then elongated to the GlcNAc–fucose disaccharide by using purified recombinant Lfng and UDP–GlcNAc and further elongated to produce the mature O-fucose tetrasaccharide by using the β4-galactosyltransferase and α3-sialyltransferase and the appropriate nucleotide sugars. Protein modified with the O-fucose monosaccharide (mono), GlcNAc–fucose disaccharide (di), Gal–GlcNAc–fucose trisaccharide (tri), and Sia–Gal–GlcNAc–fucose tetrasaccharide (tetra) were purified by HPLC, and the attachment site and molecular weight of each sugar was confirmed by nano-liquid chromatography (LC)-electrospray ionization (ESI)-tandem MS (MS/MS) (Fig. 1 and SI Appendix, Fig. S1). We also stoichiometrically modified S458 at EGF12 and S496 at EGF13 with O-glucose using recombinant Poglut (33), extended the O-glucose with xylose using purified recombinant Gxylt2, purified the protein, and demonstrated that it is modified with the O-glucose monosaccharides or xylose–glucose disaccharides (SI Appendix, Figs. S2–S4).
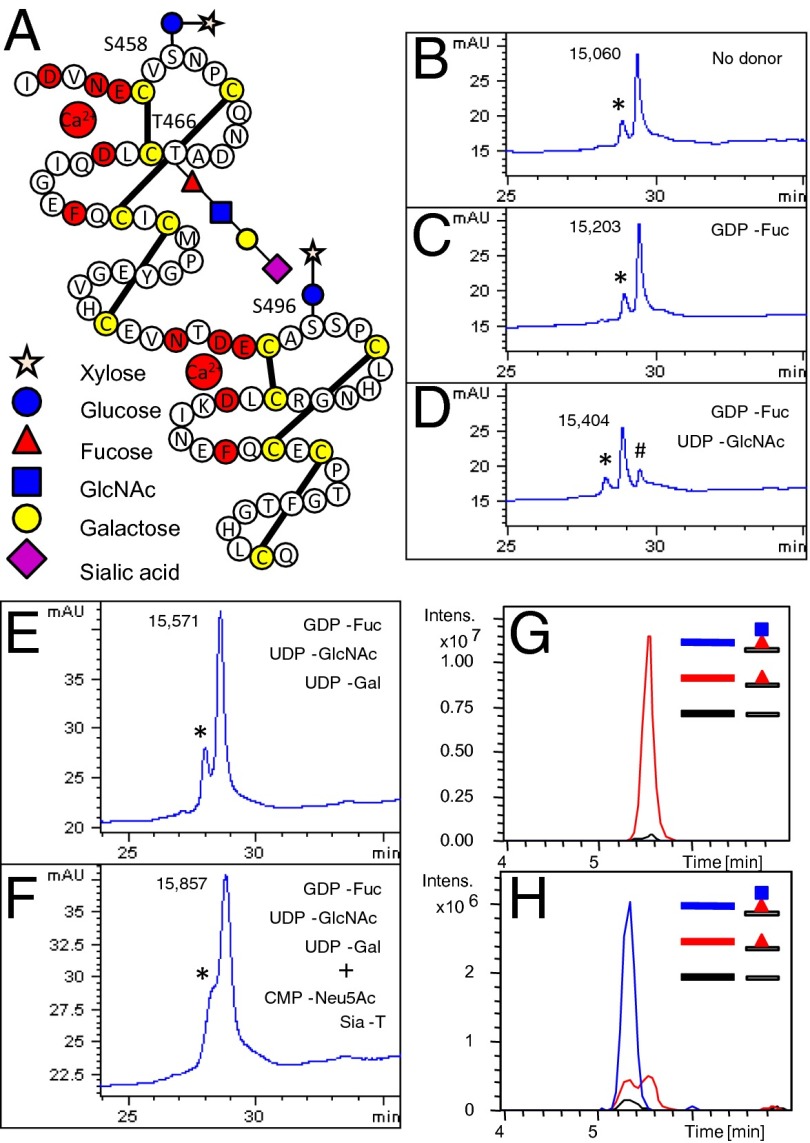
Fig. 1.
Stoichiometric modification of T466 with O-fucose glycans. (A) Diagramatic representation of EGF domains 12 and 13 of hN1, with cysteines highlighted in yellow and residues responsible for coordination of calcium highlighted in red. O-fucose and -glucose glycans added in this study are indicated at the appropriate sites. EGF11 is not shown because it lacks O-glycosylation consensus sites. (B–F) Synthesis of O-fucose monosaccharide, disaccharide, trisaccharide, and tetrasaccharide forms of hN111–13. Representative HPLC profiles of hN111–13 incubated with the appropriate enzymes and donor substrates as indicated are shown. The masses of the species in each peak as determined by mass spectrometry are shown, indicating addition of the appropriate sugar. mAU, milli-absorbance units. * indicates oxidized species; # indicates an unmodified substrate. (B) No donor, Pofut1, and Lfng. (C) GDP–fucose, Pofut1, and Lfng. (D) GDP–fucose, UDP–GlcNAc, Pofut1, and Lfng. (E) GDP–fucose, UDP–GlcNAc, UDP–galactose, Pofut1, Lfng, and β4-galactosyltransferase. (F) After overnight incubation of E, CMP–sialic acid and α3-(N)sialyltransferase were added to an aliquot and incubated for an additional 6 h. (G) Extracted ion chromatograms (EICs) of the ions corresponding to unmodified (black line), O-fucose monosaccharide (red line), and disaccharide glycoforms (blue line) of the peptide containing the O-fucose consensus sequence from EGF12, 452DVNECVSNPCQNDATCL468, derived from Asp-N digests of hN111–13 modified with O-fucose monosaccharide (from C). See SI Appendix, Fig. S1A for description of the relevant ions. (H) EICs of the ions corresponding to the unmodified (black line), O-fucose monosaccharide (red line), and disaccharide (blue line) glycoforms of the same peptide from Asp-N digests of hN111–13 modified with O-fucose disaccharide (from D). See SI Appendix, Fig. S1B for description of the relevant ions.
Assaying the Effect of O-Linked Sugars on Binding to Notch Ligands by Flow Cytometry.
hN111–13 modified with each of the glycans outlined above was tested for binding to B16 cells stably transfected with either full-length murine Jagged1 or murine DLL4 in a flow cytometry-based assay, and the level of binding was compared with the unmodified protein. Protein modified with the O-fucose monosaccharide at T466 showed slightly enhanced binding to B16 Jagged1 cells, but binding was substantially enhanced by the extension of the O-fucose with GlcNAc (Fig. 2A). Extension toward the trisaccharide and tetrasaccharide had no further effect on binding (Fig. 2A). In contrast, these modifications had no detectable effect on binding to B16 DLL4 cells (Fig. 2B). The effects of the O-fucose monosaccharide and the GlcNAc–fucose disaccharide on binding were subsequently tested by using CHO-K1 cells stably transfected with full-length human DLL1. As had been observed for Jagged1, hN111–13 modified with the monosaccharide showed a slight enhancement in binding compared with the unmodified protein, whereas the GlcNAc–fucose disaccharide showed a far larger increase in binding (Fig. 2C). None of the positive effects on binding were due to a lectin-like interaction between proteins on the cell surface and O-fucose glycans, because untransfected cells did not bind any of the Notch variants (Fig. 2D). Quantification of these data showed the increased binding of the disaccharide-, trisaccharide-, and tetrasaccharide-modified protein to Jagged1 and the binding of the disaccharide-modified protein to DLL1 to be statistically significant compared with binding of the unmodified form using Tukey’s multiple comparison test (P ≤ 0.0220) (Fig. 2E). None of the cell lines used in this study showed any change in binding when tested with hN111–13 modified with the O-glucose glycans (SI Appendix, Fig. S5).
Fig. 2.
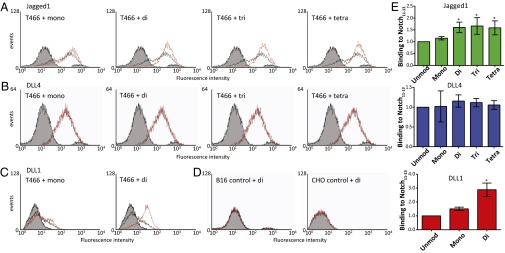
Flow-cytometry analysis of the effect of the addition of sugars to T466 on binding to Notch ligands. (A–C) Flow cytometry of B16 cells expressing Jagged1 (A), B16 cells expressing DLL4 (B), or CHO cells expressing DLL1 (C) after interaction with biotinylated hN111–13 modified with various sugars at T466 (red line) bound to avidin-coated fluorescent beads. In each case, a representative trace is shown. Binding was compared with a negative control (fibrillin-1 cbEGF12–14; gray shading) and positive control (hN111–13 WT; black line). A bimodal distribution is observed for the negative control in the presence of B16 cells due to fluorescence from unbound beads (Materials and Methods). (A) The effect on binding of each of the four separate sugar modifications is shown; the addition of the O-fucose monosaccharide (+mono) causes a small increase in binding compared with unmodified protein (indicated by the small rightward shift compared with the positive control), and the addition of the GlcNAc–fucose disaccharide (+di) causes a much larger increase in binding (indicated by a further shift to the right). Binding of hN111–13 modified with the trisaccharide (+tri) or tetrasaccharide (+tetra) is indistinguishable from the disaccharide. (B) None of the sugar modifications had any apparent effect on the interaction between hN111–13 and DLL4. (C) The addition of the O-fucose monosaccharide causes a slight increase in binding to DLL1, and the addition of the GlcNAc–fucose causes a much larger increase in binding. (D) Untransfected B16 and CHO cells do not bind unmodified or disaccharide (+di) hN111–13. (E) FACS binding data normalized to unmodified Notch111–13 with SD shown. For Jagged1, increased binding for the disaccharide, trisaccharide, and tetrasaccharide was found to be significant (*) by Tukey’s multiple comparison test (P = 0.0028, 0.0074, and 0.0220), as was increased binding of the disaccharide to DLL1 (P = 0.0001). None of the sugar modifications was found to significantly alter binding to DLL4.
Studying the Notch–Ligand Interaction Using SPR.
Initially, an Fc-tagged eukaryotically expressed Notch protein containing EGF domains 1–14 (hN11–14 Fc) was immobilized, and a titration was performed by using monomeric human Jagged1 protein spanning the N terminus to EGF3 domains (Jagged1NE3) as analyte. This construct contains the Notch-binding DSL domain in a native context. It was possible to calculate the Kd of this interaction as 7.1 ± 0.1 μM (SI Appendix, Fig. S6). Yields of this Notch construct are very low, and, because it contains multiple possible glycosylation sites in addition to containing both the subsidiary ligand binding site for Serrate/Jagged around EGF8 (34) as well as the major site in EGF12 (32), it cannot be used to dissect the effects of Fringe modifications within the primary ligand-binding site. Coupling the shorter, prokaryotic hN111–13 to the chip surface did not allow ligand binding to be detected—presumably due to steric interference of the chip matrix with the binding site. We therefore inverted the experiment to use Fc-fused ligands on the surface with the hN111–13 in the solution phase. In this orientation, binding could reliably be detected to Jagged1NE3, DLL1NE3, and DLL4NE3 (Fig. 3A). The interaction of the unmodified hN11–13 with Jagged1NE3 and DLL1NE3 was sufficiently weak (KD > 50 μM) that solubility of hN111–13 precluded a reliable quantification, whereas the interaction with DLL4NE3 was seen to be tighter but could not be satisfactorily fit using a simple 1:1 Langmuir model to derive a KD.
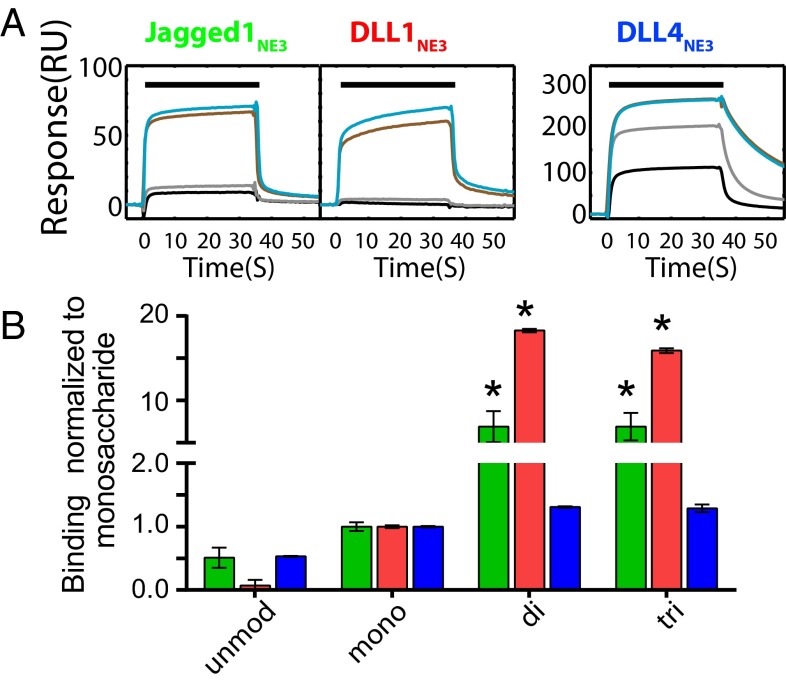
Fig. 3.
SPR analysis of the effect of the addition of sugars to T466 on binding to Notch ligands. (A) Representative SPR traces for binding of 10 μM unmodified (black), monosaccharide (gray), disaccharide (cyan), and trisaccharide (brown) hN111–13 constructs over ∼3,000 response units (RU) Jagged1NE3-Fc (Left), DLL1NE3-Fc (Center), and DLL4NE3-Fc (Right). Traces are shown corrected for refractive index changes seen in the control channel (Materials and Methods). (B) SPR binding of unmodified and glycosylated hN111–13 constructs to NE3-Fc fusion constructs of human Jagged1 (green), DLL1 (red), and DLL4 (blue) normalized to the monosaccharide construct, with SD shown. Increased binding to the disaccharide and trisaccharide for Jagged1NE3-Fc and DLL1NE3-Fc was found to be significant by Tukey’s multiple comparison test. *P < 0.0001.
To investigate the effect of the O-fucose modifications on ligand binding, we then compared binding by hN111–13 proteins modified with the monosaccharide, disaccharide, or trisaccharide, as well as the unmodified protein. Experiments were performed at the maximum concentration for which sufficient trisaccharide-modified material could be obtained (10 μM). For Jagged1NE3 and DLL1NE3 surfaces, extension of sugars beyond the monosaccharide caused increases in the amount of binding seen (∼9- and 18-fold change respectively for the disaccharide), which were statistically significant, but there was no significant differences seen for DLL4NE3 (Fig. 3B). In this latter case, the level of binding seen with even the unmodified material was much higher than for the other ligand surfaces (presumably due to the higher affinity of this ligand) and did not significantly further increase with the addition of sugars. Further extension of the disaccharide had no additional effects on binding (Fig. 3) for any of the ligand surfaces. Raw SPR data are shown in SI Appendix, Fig. S7. Multiangle light-scattering (MALS) analysis demonstrating the monomeric status of disaccharide-modified and unmodified material is shown in SI Appendix, Fig. S8.
Structures of hN111–13 Modified with O-Fucose Sugars at T466.
X-ray crystallographic structures of proteins modified with the O-fucose monosaccharide and GlcNAc–fucose disaccharide on T466 were obtained (SI Appendix, Table S2). Structures were solved to <2 Å and were compared with the previously described structure of the unmodified protein (35) (Fig. 4A). The O-fucose sugar projects directly away from the central β-sheet region of EGF12 that has been implicated in ligand binding into a solvent-filled cavity within the crystal. The conformation of the threonine side chain is conserved in all of the structures with the ”m” rotamer (SI Appendix, Fig. S9) adopted in all (despite the fact that the solvent void would accommodate other orientations), and the T466 side chain and sugar modifications are well ordered, having thermal mobility (B factors) consistent with those of the other residues within the beta-hairpin. This stability presumably arises from the intramolecular contacts that are made between the modifications and EGF12—e.g., the C6 methyl group from the O-fucose sugar packs between residues I477 and M479—and these intramolecular interactions are likely to stabilize the position of the fucose sugar and account for the well-defined electron density seen in this region (Fig. 4 A and B and SI Appendix, Fig. S9). Attachment of GlcNAc to O-fucose via a β1–3 glycosidic linkage extends the sugar further away from the ligand-binding region of EGF12 (Fig. 4 A and B and SI Appendix, Fig. S9) and can be seen to extend the ligand-binding surface comprising residues L468, E473, Q475, and I477 identified in a previous mutagenesis study (Fig. 4C) (32). Additional contacts are formed between the GlcNAc sugar and residues D464 and M479. Despite the extensive contacts formed between the protein and the sugar, there is no evidence that the addition of sugars to T466 induces any conformational change in hN111–13. The backbone structure of EGF12 is unaffected, and the interdomain twist and tilt angles for hN111–13 modified with each of the sugars are within the level of experimental error of those for the unmodified protein (Fig. 4A).
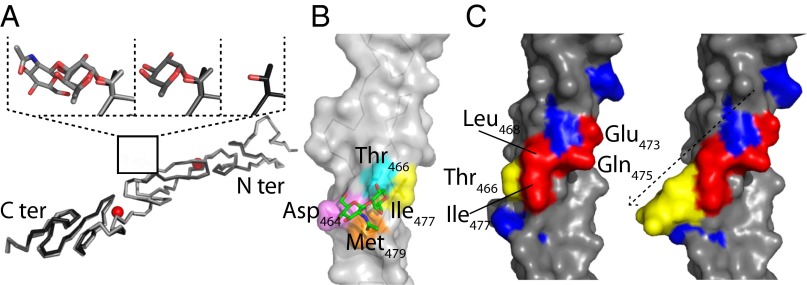
Fig. 4.
Structure of O-fucosylated variants of hN111–13. (A) Superimposed X-ray structures of the unmodified hN111–13 (PDB ID code 2VJ3) and the monosaccharide and disaccharide structures with the subsequent additions to the T466 region highlighted. Ca2+ ions are shown in red. (B) hN112 disaccharide X-ray structure highlighting contacts between the C6 methyl group of the O-fucose with Ile477 (yellow) and Met479 (orange), the C6 methoxy group of GlcNAc with Asp464 (pink), and the N-acetyl group of GlcNAc with Met479 (orange). Thr466 is highlighted in cyan. (C) hN112 unmodified and disaccharide X-ray structures highlighting residues shown to contribute to Jagged1 binding in a previous mutagenesis study (red); those residues that did not contribute to binding are also shown (blue) (32). T466 is highlighted in yellow, together with the disaccharide; the sugar may extend the known ligand-binding surface or act indirectly to promote complex formation.
Effect of Substituting T466.
Several studies that have investigated the role of O-glycosylation in regulating Notch signaling have removed the O-fucosylation site in EGF12 by using site-directed mutagenesis (36–38). Given that T466 lies directly adjacent to the ligand-binding site in hN1, it is possible that these studies could have affected binding independently from the loss of glycosylation at this position. Therefore, a series of mutants of hN111–13 was produced, with T466 substituted by alanine, valine, or serine, and binding to Jagged1 was measured. Binding was initially assessed via flow cytometry using B16 cells expressing full-length Jagged1, as described above. hN111–13 mutants containing either the T466A or the T466S substitutions had reduced binding compared with hN111–13 WT (SI Appendix, Fig. S10). By contrast, hN111–13 containing the T466V substitution bound to Jagged1 similarly to hN111–13 WT (SI Appendix, Fig. S10). Quantification of these data showed the reduced binding of T466A and T466S to Jagged1 to be statistically significant, although a small increase in binding of T466V was also observed (SI Appendix, Fig. S10), albeit at a lower level of significance. Unsurprisingly, none of the mutants affected binding to DLL4 in our flow cytometry assay, presumably due to the higher affinity of even the unmodified protein (Fig. 3). These effects were confirmed in an SPR binding assay using the shorter construct Jagged1NE3 (SI Appendix, Fig. S10). Structures were obtained for each mutant (SI Appendix, Table S2) and showed that none of these substitutions altered the native fold of EGF12. These data indicate that the changes in binding to Jagged1 observed in the flow cytometry assay arise from changes in the affinity of the interaction, consistent with the proximity of T466 to the ligand-binding region.
Discussion
We have demonstrated that Fringe-mediated elongation of O-linked fucose to the GlcNAc disaccharide at T466 of EGF12 results in a substantial increase in binding to members of the Jagged (Jagged1) and Delta (DLL1) classes of mammalian Notch ligands. Trisaccharide and tetrasaccharide additions had no further effect on binding, and addition of O-glucose glycans had no detectable effects. Structural analysis demonstrated that neither the addition of fucose nor GlcNAc resulted in an alteration of the polypeptide backbone or relative orientation of adjacent EGF domains. MALS analysis showed that there was no difference in the oligomerization state of the disaccharide-modified hN111–13 compared with unmodified protein (SI Appendix, Fig. S8). These data demonstrate that the Fringe modification increases the affinity of the ligand-binding region for Jagged1 and DLL1. Furthermore, because of its location within EGF12 in the vicinity of the ligand-binding patch, substitution of T466 by serine or alanine may have implications for ligand binding, independently from the loss of O-glycosylation at this position reported in previous studies (36–38).
Using flow cytometry and SPR in tandem provides compelling evidence that, following O-fucosylation of EGF12, the subsequent extension with GlcNAc leads to an enhancement of binding of this region to various Notch ligands. The flow cytometry assay uses full-length ligand and provides a cellular context for the interaction between hN1 and its ligands, whereas the SPR assay allows precise measurement of this interaction and provides a more quantitative measure of the effects of O-glycosylation on binding to Jagged1, DLL1, and DLL4. Our SPR data also provide an explanation for why O-glycosylation has no observable effect on binding of hN111–13 to DLL4 in our flow cytometry assay: Binding of unmodified hN111–13 is already at a maximum level because of its higher affinity for DLL4 than either of the other two ligands. This high inherent affinity of the DLL4/Notch interaction in the absence of any Fringe modification was recently observed by Andrawes et al. (39).
An increase in affinity of the Notch/DLL1 complex resulting from Fringe-mediated elongation of O-fucose on EGF12, coupled with potential avidity effects due to increased concentrations of receptor and ligand at the cell surface, provides a convincing explanation for the known biological effects of the Fringes on the ability of Delta ligands to activate Notch (SI Appendix, Table S1). In contrast, the increase in Jagged1 binding was unexpected. Fringe is generally observed as a negative regulator of Jagged/Serrate activation of Notch, although its effects on binding have been mixed (SI Appendix, Table S1). In light of our data, a possible explanation for these observations is that, whereas Fringe-mediated elongation of O-fucose on EGF12 enhances interactions between Notch and Jagged/Serrate ligands, elongation of O-fucose on other EGF repeats causes the reduction in activation by Jagged1 typically associated with the biological effects of Fringe. Interestingly, prior studies have demonstrated that the O-fucose modifications on EGF26 and 27 are both elongated by Fringes and play important roles in ligand-mediated Notch activation (38).
A published solution structure of EGF12 modified with O-fucose sugars (40) indicated a stabilizing intramolecular effect of the sugar on the major β-hairpin. However, this finding is likely to be a nonphysiological effect specific to the experimental conditions used. This study was conducted under nonnative conditions (without covalent attachment of neighboring domains 11 and 13) and in the absence of Ca2+, which is required to stabilize the major β-hairpin of EGF12 in solution in the absence of any sugar addition (SI Appendix, Fig. S11). By contrast, another NMR study (41), performed on hN111–13 at physiologically relevant Ca2+ concentrations, demonstrated that EGF12 residues are characterized by order parameters >0.8, suggesting that this domain does not contain backbone segments that are dynamic on a fast timescale (42, 43). Our crystal structures, obtained in the presence of Ca2+, demonstrate that the addition of sugars to T466 does not appear to induce a conformational change either in the orientation of the Thr side chain or more globally in the overall structure of the EGF12 domain, or alter the packing interactions it makes with adjacent cbEGF domains. Furthermore, the Fringe addition almost doubles the surface area of the adjacent ligand-binding region. Collectively, these data suggest that the most likely mechanism to explain the effect of the disaccharide modification on affinity of the complex is by direct involvement of the sugar moiety in ligand binding. However, in the absence of a structure for the Notch/ligand complex, other explanations remain possible, such as an effect on side-chain dynamics or long-range electrostatics at the binding site or intramolecular Ca2+ binding.
Both the T466A and T466S substitutions cause a large decrease in binding to Jagged1, whereas the T466V substitution appears to enhance binding. This finding is consistent with the earlier finding that the core ligand-binding site within EGF12 uses a hydrophobic patch to initiate ligand binding and suggests that the ligand-binding site extends to include T466. It has been reported that mutation of the O-fucosylation site within EGF12 to alanine causes a large drop in Notch signaling induced by Jagged1 and DLL1 (38). Our results suggest that the basis for this reduction in signaling is likely to be the result of a large decrease in binding to both classes of ligands—not just through a loss of O-glycosylation but through altering the major ligand-binding site. It is interesting that, whereas in our study prokaryotically expressed hN111–13 containing either the T466A or the T466S substitutions both show reduced binding to Jagged1, in previous coculture assays, full-length murine Notch1 containing the T466A substitution showed no activity in the presence of cells expressing DLL1, whereas the T466S mutant was both O-fucosylated and behaved similarly to WT protein (38, 44). In addition, T466 from mouse Notch1 is modified at high stoichiometry with O-fucose in cells (16), suggesting that O-fucose is present on this site in vivo. In light of these studies, our data highlight the importance of the presence of O-fucose glycans in this region in increasing the affinity of the interaction between Notch and its ligands and provide an explanation for the observation that the Notch receptor needs to be O-fucosylated in order for optimal signaling to occur (28, 30).
In summary, our work has demonstrated that Fringe-mediated extension of O-linked fucose sugars in EGF12 of the ligand-binding region can substantially enhance binding to both Jagged and DLLs. In addition to identifying a role for sugars in modulating a defined protein–protein interaction, these data provide a plausible affinity-based explanation for the observed enhancement of DLL-mediated signaling by Fringe and suggest that the inhibitory effect on Jagged/Serrate-mediated signaling involves other domains outside the ligand-binding region of Notch.
Materials and Methods
Protein Expression.
Purification and refolding of prokaryotic hN111–13 variants were performed as described (32). Sequences corresponding to hJagged1NE3-Fc, hDll-1 NE3-Fc, hDll-4 NE3-Fc, and hN11–14-Fc were cloned into a modified pLEXm eukaryotic expression plasmid (45). Sequence corresponding to hJagged1NE3 lacking Fc was cloned in pSecTag. HEK 293T cells were transiently transfected with ligand constructs by using polyethylenimine, and proteins were purified by Ni2+ column and Superdex 200 size-exclusion chromatography.
Purification of Glycosyltransferase Enzymes.
Recombinant human Pofut-1, mouse Lfng, human Poglut, and mouse Gxylt2 proteins were expressed in HEK293T cells and purified by Ni-NTA affinity chromatography (33). β4-galactosyltransferase from bovine milk (Sigma-Aldrich) and recombinant rat α3-(N)sialyltransferase (Calbiochem) were purchased.
In Vitro Glycosylation.
Addition of O-fucose monosaccharides or disaccharides to unmodified hN111–13 was performed as described (33). hN111–13 at a concentration of ∼10 μM was incubated with Pofut-1, Lfng, and 200 μM appropriate donor substrates at 37 °C overnight. For further elongation to the O-fucose trisaccharide or tetrasaccharide, UDP–galactose (final concentration, 200 μM; Sigma-Aldrich) and β4-galactosyltransferase (0.5 mU/μL) were added to the reaction. After overnight incubation, CMP–sialic acid (200 μM; Sigma-Aldrich) and α3-(N)sialyltransferase [1/20% (vol/vol)] were added to the reaction mixture and incubated for another 6 h. All products were HPLC purified, and molecular weights were confirmed by nano-LC-ESI-MS/MS on an Agilent ion trap mass spectrometer with a CHIP–Cube interface (33). For site mapping of O-fucose glycans, hN111–13 modified with an O-fucose monosaccharide or disaccharide was digested with Asp-N protease (Sigma-Aldrich), and glycopeptides were analyzed by nano-LC-ESI-MS/MS (46). Relative levels of the different glycoforms of the relevant peptides were compared by generating extracted ion chromatograms of the ions corresponding to the appropriate glycoform (20).
Flow Cytometry Binding Assay.
Biotinylated hN1 11–13 or control fibrillin triple EGF domain constructs were coupled to fluorescent avidin beads and, after washing, mixed with ligand-expressing cells (32). Following incubation, samples were analyzed directly by flow cytometry without removal of unbound beads. For qualitative evaluation, binding can be observed directly by fluorescence microscopy (SI Appendix, Fig. S12).
SPR.
hJagged1NE3-Fc and hDLL1NE3-Fc or hJagged1NE3-Fc and hDLL4NE3-Fc were immobilized on a CM5 chip surface through primary amine coupling, and data were collected by multiple injections of 10 μM hN111–13 (unmodified, monosaccharide, disaccharide, and trisaccharide) over the coupled chip surface. Traces were corrected for refractive index changes by subtraction of a control trace that was recorded simultaneously and analyzed by using the program BIA evaluation (Version 3.2) and GraphPad Prism (Version 6.0). All experiments were carried out at 25 °C in Hepes buffered saline (BIACore) supplemented with 1 mM Ca2+ on a T-3000 BiaCore instrument.
hN111–13 Crystallization and Structural Determination.
Glycosylated forms of hN111–13 were crystallized at a concentration of ∼18 mg/mL by using sitting drops and vapor diffusion with commercial screens (Molecular Dimensions) in H2O with 10 mM BaCl2 and 30% (vol/vol) mother liquor, 100 mM Mes (pH 6.0), 200 mM CaCl2, and 20% (wt/vol) PEG 6,000. Data were collected at the Diamond facility (beamline I041). Mutant forms of hN111–13 were crystallized at a concentration of 13.1 mg/mL for T466V, 17.0 mg/mL for T466S, and 15.3 mg/mL for T466A in H2O with 10 mM CaCl2 and 25% (vol/vol) mother liquor, 100 mM ammonium acetate (pH 4.5), and 22.5% (wt/vol) PEG 10,000 for T466A; 100 mM sodium acetate (pH 5.5) and 12% (wt/vol) PEG 5000 monomethylether for T466V; and 200 mM sodium bromide, 100 mM bis-Tris propane (pH 6.5), and 20% (wt/vol) PEG 3350 for T466S. Data for both T446V and T466S were collected at Diamond facility, beamline I02 and T466A on beamline I03. All datasets were indexed and scaled by using xia2, and phases were determined through molecular replacement [Protein Data Bank (PDB) ID code 2VJ3] by using phaser (47). Autobuster was used in conjunction with COOT to refine all structures (48). Molprobity was used to gauge structural quality (49).
Supplementary Material
Acknowledgments
P.T. was supported by the Biotechnology and Biological Sciences Research Council. P.A.H., S.M.L., D.S., and C.C. were supported by Wellcome Trust Grant 3097928. S.M.L. is supported by Wellcome Senior Investigator Award 100298. This work was supported in part by National Institutes of Health Grant GM061126 (to R.S.H.). We thank Prof. A. Harris for B16 cell lines; P. Whiteman and S. Liang for help with FACS and microscopy; C. Redfield for assistance with NMR analysis; S. Johnson for MALS analysis; and R.S.H. laboratory members for helpful discussion. We acknowledge Diamond Light Source for time on beamlines I02 and I03 (MX9306).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
Data deposition: The crystallography, atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 4cue, 4cuf, 4d0f, 4cud, and 4d0e).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1319683111/-/DCSupplemental.
References
- 1.Artavanis-Tsakonas S, Rand MD, Lake RJ. Notch signaling: Cell fate control and signal integration in development. Science. 1999;284(5415):770–776. doi: 10.1126/science.284.5415.770. [DOI] [PubMed] [Google Scholar]
- 2.Blaumueller CM, Qi H, Zagouras P, Artavanis-Tsakonas S. Intracellular cleavage of Notch leads to a heterodimeric receptor on the plasma membrane. Cell. 1997;90(2):281–291. doi: 10.1016/s0092-8674(00)80336-0. [DOI] [PubMed] [Google Scholar]
- 3.Sanchez-Irizarry C, et al. Notch subunit heterodimerization and prevention of ligand-independent proteolytic activation depend, respectively, on a novel domain and the LNR repeats. Mol Cell Biol. 2004;24(21):9265–9273. doi: 10.1128/MCB.24.21.9265-9273.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fehon RG, et al. Molecular interactions between the protein products of the neurogenic loci Notch and Delta, two EGF-homologous genes in Drosophila. Cell. 1990;61(3):523–534. doi: 10.1016/0092-8674(90)90534-l. [DOI] [PubMed] [Google Scholar]
- 5.Rebay I, et al. Specific EGF repeats of Notch mediate interactions with Delta and Serrate: Implications for Notch as a multifunctional receptor. Cell. 1991;67(4):687–699. doi: 10.1016/0092-8674(91)90064-6. [DOI] [PubMed] [Google Scholar]
- 6.Shimizu K, et al. Mouse jagged1 physically interacts with notch2 and other notch receptors. Assessment by quantitative methods. J Biol Chem. 1999;274(46):32961–32969. doi: 10.1074/jbc.274.46.32961. [DOI] [PubMed] [Google Scholar]
- 7.Fleming RJ. Structural conservation of Notch receptors and ligands. Semin Cell Dev Biol. 1998;9(6):599–607. doi: 10.1006/scdb.1998.0260. [DOI] [PubMed] [Google Scholar]
- 8.Kopan R, Ilagan MX. The canonical Notch signaling pathway: Unfolding the activation mechanism. Cell. 2009;137(2):216–233. doi: 10.1016/j.cell.2009.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brou C, et al. A novel proteolytic cleavage involved in Notch signaling: The role of the disintegrin-metalloprotease TACE. Mol Cell. 2000;5(2):207–216. doi: 10.1016/s1097-2765(00)80417-7. [DOI] [PubMed] [Google Scholar]
- 10.De Strooper B, et al. A presenilin-1-dependent gamma-secretase-like protease mediates release of Notch intracellular domain. Nature. 1999;398(6727):518–522. doi: 10.1038/19083. [DOI] [PubMed] [Google Scholar]
- 11.Mumm JS, et al. A ligand-induced extracellular cleavage regulates gamma-secretase-like proteolytic activation of Notch1. Mol Cell. 2000;5(2):197–206. doi: 10.1016/s1097-2765(00)80416-5. [DOI] [PubMed] [Google Scholar]
- 12.Schroeter EH, Kisslinger JA, Kopan R. Notch-1 signaling requires ligand-induced proteolytic release of intracellular domain. Nature. 1998;393(6683):382–386. doi: 10.1038/30756. [DOI] [PubMed] [Google Scholar]
- 13.Jarriault S, et al. Signaling downstream of activated mammalian Notch. Nature. 1995;377(6547):355–358. doi: 10.1038/377355a0. [DOI] [PubMed] [Google Scholar]
- 14.Moloney DJ, et al. Mammalian Notch1 is modified with two unusual forms of O-linked glycosylation found on epidermal growth factor-like modules. J Biol Chem. 2000;275(13):9604–9611. doi: 10.1074/jbc.275.13.9604. [DOI] [PubMed] [Google Scholar]
- 15.Rana NA, Haltiwanger RS. Fringe benefits: Functional and structural impacts of O-glycosylation on the extracellular domain of Notch receptors. Curr Opin Struct Biol. 2011;21(5):583–589. doi: 10.1016/j.sbi.2011.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rana NA, et al. O-glucose trisaccharide is present at high but variable stoichiometry at multiple sites on mouse Notch1. J Biol Chem. 2011;286(36):31623–31637. doi: 10.1074/jbc.M111.268243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shao L, Moloney DJ, Haltiwanger R. Fringe modifies O-fucose on mouse Notch1 at epidermal growth factor-like repeats within the ligand-binding site and the Abruptex region. J Biol Chem. 2003;278(10):7775–7782. doi: 10.1074/jbc.M212221200. [DOI] [PubMed] [Google Scholar]
- 18.Okajima T, Irvine KD. Regulation of notch signaling by o-linked fucose. Cell. 2002;111(6):893–904. doi: 10.1016/s0092-8674(02)01114-5. [DOI] [PubMed] [Google Scholar]
- 19.Shi S, Stanley P. Protein O-fucosyltransferase 1 is an essential component of Notch signaling pathways. Proc Natl Acad Sci USA. 2003;100(9):5234–5239. doi: 10.1073/pnas.0831126100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Acar M, et al. Rumi is a CAP10 domain glycosyltransferase that modifies Notch and is required for Notch signaling. Cell. 2008;132(2):247–258. doi: 10.1016/j.cell.2007.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fernandez-Valdivia R, et al. Regulation of mammalian Notch signaling and embryonic development by the protein O-glucosyltransferase Rumi. Development. 2011;138(10):1925–1934. doi: 10.1242/dev.060020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stahl M, et al. Roles of Pofut1 and O-fucose in mammalian Notch signaling. J Biol Chem. 2008;283(20):13638–13651. doi: 10.1074/jbc.M802027200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yao D, et al. Protein O-fucosyltransferase 1 (Pofut1) regulates lymphoid and myeloid homeostasis through modulation of Notch receptor ligand interactions. Blood. 2011;117(21):5652–5662. doi: 10.1182/blood-2010-12-326074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Johnston SH, et al. A family of mammalian Fringe genes implicated in boundary determination and the Notch pathway. Development. 1997;124(11):2245–2254. doi: 10.1242/dev.124.11.2245. [DOI] [PubMed] [Google Scholar]
- 25.Rampal R, et al. Lunatic fringe, manic fringe, and radical fringe recognize similar specificity determinants in O-fucosylated epidermal growth factor-like repeats. J Biol Chem. 2005;280(51):42454–42463. doi: 10.1074/jbc.M509552200. [DOI] [PubMed] [Google Scholar]
- 26.Shimizu K, et al. Manic fringe and lunatic fringe modify different sites of the Notch2 extracellular region, resulting in different signaling modulation. J Biol Chem. 2001;276(28):25753–25758. doi: 10.1074/jbc.M103473200. [DOI] [PubMed] [Google Scholar]
- 27.Yang LT, et al. Fringe glycosyltransferases differentially modulate Notch1 proteolysis induced by Delta1 and Jagged1. Mol Biol Cell. 2005;16(2):927–942. doi: 10.1091/mbc.E04-07-0614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moloney DJ, et al. Fringe is a glycosyltransferase that modifies Notch. Nature. 2000;406(6794):369–375. doi: 10.1038/35019000. [DOI] [PubMed] [Google Scholar]
- 29.Hicks C, et al. Fringe differentially modulates Jagged1 and Delta1 signaling through Notch1 and Notch2. Nat Cell Biol. 2000;2(8):515–520. doi: 10.1038/35019553. [DOI] [PubMed] [Google Scholar]
- 30.Chen J, Moloney DJ, Stanley P. Fringe modulation of Jagged1-induced Notch signaling requires the action of beta 4galactosyltransferase-1. Proc Natl Acad Sci USA. 2001;98(24):13716–13721. doi: 10.1073/pnas.241398098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hou X, Tashima Y, Stanley P. Galactose differentially modulates lunatic and manic fringe effects on Delta1-induced NOTCH signaling. J Biol Chem. 2012;287(1):474–483. doi: 10.1074/jbc.M111.317578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Whiteman P, et al. Molecular basis for Jagged-1/Serrate ligand recognition by the Notch receptor. J Biol Chem. 2013;288(10):7305–7312. doi: 10.1074/jbc.M112.428854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Takeuchi H, et al. Site-specific O-glucosylation of the epidermal growth factor-like (EGF) repeats of notch: Efficiency of glycosylation is affected by proper folding and amino acid sequence of individual EGF repeats. J Biol Chem. 2012;287(41):33934–33944. doi: 10.1074/jbc.M112.401315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yamamoto S, et al. A mutation in EGF repeat-8 of Notch discriminates between Serrate/Jagged and Delta family ligands. Science. 2012;338(6111):1229–1232. doi: 10.1126/science.1228745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cordle J, et al. A conserved face of the Jagged/Serrate DSL domain is involved in Notch trans-activation and cis-inhibition. Nat Struct Mol Biol. 2008;15(8):849–857. doi: 10.1038/nsmb.1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lei L, Xu A, Panin VM, Irvine KD. An O-fucose site in the ligand binding domain inhibits Notch activation. Development. 2003;130(26):6411–6421. doi: 10.1242/dev.00883. [DOI] [PubMed] [Google Scholar]
- 37.Ge C, Stanley P. The O-fucose glycan in the ligand-binding domain of Notch1 regulates embryogenesis and T cell development. Proc Natl Acad Sci USA. 2008;105(5):1539–1544. doi: 10.1073/pnas.0702846105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rampal R, Arboleda-Velasquez JF, Nita-Lazar A, Kosik KS, Haltiwanger RS. Highly conserved O-fucose sites have distinct effects on Notch1 function. J Biol Chem. 2005;280(37):32133–32140. doi: 10.1074/jbc.M506104200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Andrawes MB, et al. Intrinsic selectivity of Notch 1 for Delta-like 4 over Delta-like 1. J Biol Chem. 2013;288(35):25477–25489. doi: 10.1074/jbc.M113.454850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hiruma-Shimizu K, et al. Chemical synthesis, folding, and structural insights into O-fucosylated epidermal growth factor-like repeat 12 of mouse Notch-1 receptor. J Am Chem Soc. 2010;132(42):14857–14865. doi: 10.1021/ja105216u. [DOI] [PubMed] [Google Scholar]
- 41.Hambleton S, et al. Structural and functional properties of the human notch-1 ligand binding region. Structure. 2004;12(12):2173–2183. doi: 10.1016/j.str.2004.09.012. [DOI] [PubMed] [Google Scholar]
- 42.Yang D, Kay LE. Contributions to conformational entropy arising from bond vector fluctuations measured from NMR-derived order parameters: Application to protein folding. J Mol Biol. 1996;263(2):369–382. doi: 10.1006/jmbi.1996.0581. [DOI] [PubMed] [Google Scholar]
- 43.Akke M, Skelton NJ, Kördel J, Palmer AG, 3rd, Chazin WJ. Effects of ion binding on the backbone dynamics of calbindin D9k determined by 15N NMR relaxation. Biochemistry. 1993;32(37):9832–9844. doi: 10.1021/bi00088a039. [DOI] [PubMed] [Google Scholar]
- 44.Shi S, et al. The threonine that carries fucose, but not fucose, is required for Cripto to facilitate Nodal signaling. J Biol Chem. 2007;282(28):20133–20141. doi: 10.1074/jbc.M702593200. [DOI] [PubMed] [Google Scholar]
- 45.Aricescu AR, Lu W, Jones EY. A time- and cost-efficient system for high-level protein production in mammalian cells. Acta Crystallogr D Biol Crystallogr. 2006;62(Pt 10):1243–1250. doi: 10.1107/S0907444906029799. [DOI] [PubMed] [Google Scholar]
- 46.Leonhard-Melief C, Haltiwanger RS. O-fucosylation of thrombospondin type 1 repeats. Methods Enzymol. 2010;480:401–416. doi: 10.1016/S0076-6879(10)80018-7. [DOI] [PubMed] [Google Scholar]
- 47.Winter G. xia2: An expert system for macromolecular crystallography data reduction. J Appl Cryst. 2010;43:186–190. [Google Scholar]
- 48.Bricogne G, et al. BUSTER. Cambridge, U.K: Global Phasing Ltd; 2011. [Google Scholar]
- 49.Chen VB, et al. MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 1):12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.