Abstract
Despite being essential for successful infection, the molecular cues involved in host recognition and genome transfer of viruses are not completely understood. Bacterial outer membrane proteins A and C co-purify in lipid vesicles with bacteriophage Sf6, implicating both outer membrane proteins as potential host receptors. We determined that outer membrane proteins A and C mediate Sf6 infection by dramatically increasing its rate and efficiency. We performed a combination of in vivo studies with three omp null mutants of Shigella flexneri, including classic phage plaque assays and time-lapse fluorescence microscopy to monitor genome ejection at the single virion level. Cryo-electron tomography of phage “infecting” outer membrane vesicles shows the tail needle contacting and indenting the outer membrane. Lastly, in vitro ejection studies reveal that lipopolysaccharide and outer membrane proteins are both required for Sf6 genome release. We conclude that Sf6 phage entry utilizes either outer membrane proteins A or C, with outer membrane protein A being the preferred receptor.
Keywords: bacteriophage, virus, host receptor, outer membrane proteins, Shigella flexneri, fluorescence time-lapse, cryo-electron tomography, in vitro genome ejection
Introduction
Non-enveloped viruses include the vast majority of bacteriophages and many human pathogens. Despite their widespread occurrence and medical relevance, the molecular mechanisms that govern their genome delivery are not completely understood. Productive infection requires precise docking to a designated location on the cell surface to enable delivery of the viral genome through several steps that may include: initial specific recognition of the cell surface through interaction with a “primary” host receptor, “triggering” the process of nucleic acid release from the virion (e.g. by receptor recognition), followed by coordinated conformational changes in viral proteins, and ultimately genome transfer into the target cell. Such transfer is a universal phenomenon among viruses and must be highly regulated since evolutionary pressures demand that errors leading to premature or inappropriate release be avoided. Although the genomes of viruses must breach or transit through a wide variety of obstacles such as bacterial cell walls, some common strategies and mechanisms are utilized (Poranen et al., 2002). Receptor-binding proteins in viruses are needed to attach to the outside of the host cell, and these can have conserved structures. For example, cell receptor-binding proteins of PRD-1 (infects a range of Gram negative bacteria such as Pseudomonas aeruginosa, Escherichia coli, Vibrio cholera, and Salmonella typhimurium) and adenoviruses (infects humans) share a common polypeptide fold (Bamford et al., 2005). Another common theme is that ion-permeable pores facilitate genome translocation as occurs in diverse viral systems such as tailed bacteriophage and poliovirus (Poranen et al., 2002).
Bacteriophage Sf6 is a short-tailed dsDNA virus (family Podoviridae) that infects Shigella flexneri, a causative agent of bacillary dysentery. Shigella, the third most prevalent bacterial foodborne pathogen (Warren et al., 2006), is responsible for over 165 million human cases of dysentery worldwide each year (Kotloff et al., 1999). Although most Shigella infections are reported in underdeveloped countries, the disease has become more widespread in recent years (Feil, 2012; Holt et al., 2012). Interactions between phage Sf6 and its host can result in altered host pathogenicity by changing its surface characteristics, such as via an acetylase that affects the O-antigen polysaccharide of lipopolysaccharide (LPS) (Clark et al., 1991; Verma et al., 1991). Therefore, a more detailed understanding of host recognition and infection mechanisms is needed (Allison and Verma, 2000; Banks et al., 2002; Broadbent et al., 2010). In many tailed phages, short, thick fibers (“tailspike proteins”) are responsible for initial interaction with LPS (the “primary” receptor) and mediate reversible cell binding (Figure 1, step 1). Sf6 is a member of the P22-like group of tailed phages, which includes over 150 phages and prophages that have similar sets of virion assembly genes (Casjens and Thuman-Commike, 2011), and tailspikes of this phage group bind to and cleave the surface O-antigen of their host LPS. Atomic-resolution structures are available for P22 and Sf6 tailspike proteins (Muller et al., 2008; Steinbacher et al., 1997), and these are strikingly similar despite having no recognizable similarity in amino acid sequence (Baxa et al., 1996; Chua et al., 1999). Purified LPS of Salmonella enterica serovar Typhimurium is sufficient to cause a slow release of the P22 genome in vitro (Andres et al., 2010), but it is unclear if LPS alone is sufficient to induce rapid and accurate genome release in vivo. Secondary receptors (i.e. those that often mediate irreversible cell binding) are necessary for infection by a number of other phages and the presence of secondary receptors in Podoviridae including N4 has been reported (Kiino et al., 1993; McPartland and Rothman-Denes, 2009).
Figure 1. Schematic diagram of steps in dsDNA genome ejection for members of the family Podoviridae.
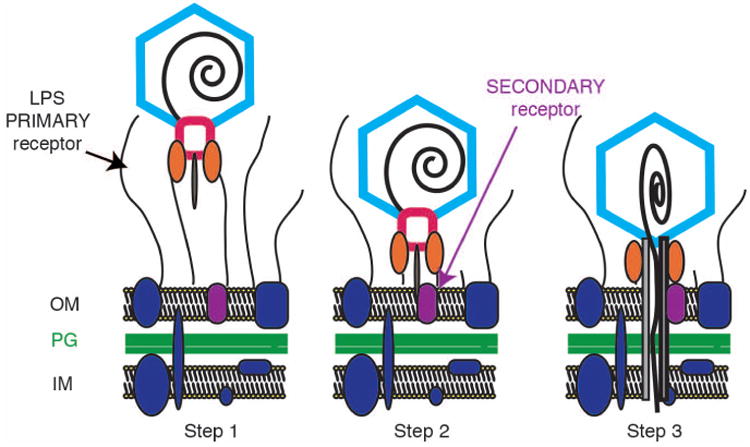
Step 1: A virion binds to its cell surface primary receptor. In the P22-like phages, the phage tailspike proteins (orange ovals) bind the lipopolysaccharide (LPS) through the O-antigen. Step 2: Tailspike proteins hydrolyze the LPS and the virion moves closer to the cell surface where it can now bind a putative secondary receptor. Step 3: dsDNA likely enters the cell through a channel formed by phage “ejection” proteins (P22 releases three proteins, gp7, gp16, and gp20 during DNA release). OM; outer membrane, PG; peptidoglycan, IM; inner membrane. This schematic was modified from (Casjens and Molineux, 2012).
Many diverse Gram negative bacteria, including Shigella, secrete outer membrane vesicles (OMVs) (Berlanda Scorza et al., 2012), and OMV production can increase in response to attack by lytic phages (Kulp and Kuehn, 2010). Previously, host-derived outer membrane proteins (Omps) A and C were reported to co-purify with Sf6, even after CsCl purification, through OMVs attached to the Sf6 tail machinery (Parent et al., 2012; Zhao et al., 2011). The tight association of Sf6 with OMVs indicates that Sf6 and P22 may differ in their binding to hosts since OMVs do not co-purify with P22. In the present study we investigated the roles of S. flexneri OmpA and OmpC during Sf6 infection, and demonstrate that both Omps act as secondary receptors and increase Sf6 genome ejection efficiency. Our results suggest that host recognition and genome transfer follow diverse pathways even within rather closely related phages.
Results
Sf6 plating efficiency is reduced in the absence of host OmpA and OmpC
Three null mutants of S. flexneri were created to investigate the role of Omps A and C during the initial stages of Sf6 infection. Two of the mutants contain single gene knockouts (ompA− and ompC−) as described previously (Parent et al., 2012), and the third strain lacks both genes (ompA−C−, constructed as described in Supplementary Materials). We compared Sf6 growth on and infection of wild-type (WT) Shigella with these three null mutants to explore how OmpA and OmpC affect phage growth. For all experiments described herein, the Sf6 phage carried a clear mutation that blocks lysogen formation (Casjens et al., 2004). Sf6 plaque morphology differed between the WT and each of the three omp knockout strains (Figure S1A). The efficiency of Sf6 plating was determined on the omp defective hosts relative to the WT host at a range of temperatures between 20 and 42 °C. Sf6 plating efficiency was essentially the same on WT and the ompA− and ompC− single knockout hosts. However, at all temperatures it was ∼10-fold or more lower when Sf6 was plated on the ompA−C− strain (Figure S1B).
Sf6 kills WT S. flexneri cells more efficiently than cells lacking OmpA and OmpC
A decrease in plating efficiency (Figure S1B) on the double S. flexneri ompA−C− null strain could result from one of two possibilities. Either the phage is unable to recognize and infect the mutant cells, or infection occurs but no progeny are produced (as was suggested by work that proposed OmpA and OmpC are incorporated into virions as a means of stabilization (Zhao et al., 2011)). To determine which of these possibilities accounts for the observed decrease, we performed a cell survival assay. In this experiment, mid-logarithmically growing cells at 2 ×108 cells/mL were incubated at increasing multiplicity of infection (MOI, the number of phage added per cell) of Sf6 at 30°C. Resulting colonies were counted and relative survival values were calculated as the number of colony forming units (CFU) at each MOI divided by the number of CFU present prior to infection (Figure S2). The colonies represent cells that have not been infected, since infected Shigella cells do not survive Sf6 clear mutant infection. For the WT strain, an MOI of two was sufficient to reduce the Shigella population by more than half, and the relative survival decreased as the MOI of Sf6 increased. These results are consistent with similar experiments performed using Salmonella enterica serovar Typhimurium infected by phage P22 (Gordon, 1993). The ompC− host demonstrated a very slight change in cell survival compared to WT, whereas the ompA− host showed a larger, and statistically significant increase. The ompA−C− host had the highest level of survival at all MOIs and required 4-5 times more phage to achieve the same killing as in the WT host.
Sf6 infection is slower in S. flexneri that lack both OmpA and OmpC
Single step growth curve experiments obtain information about the rates of phage absorption to the host as well as the rates and amounts (“burst size”) of phage produced per cell. Sf6 phage were added to mid-logarithmically growing cells at an MOI that ensured >95% of the cells were infected in each experiment. The phage/cell ratio at each time point was calculated by dividing the number of viable phage (an aliquot of the infected culture was titered post chloroform-induced lysis and immediate dilution) by the number of cells present in the initial culture. For WT Shigella, the observed pfu per cell decreases rapidly as the phage attach to the host cells, inject their genomes, and enter the eclipse phase. The pfu per cell subsequently increases as progeny phage are produced, until reaching steady-state at approximately 60 minutes post-infection. The growth curve kinetics and burst size for Sf6 in WT Shigella are similar to those reported for P22 in Salmonella (Aramli and Teschke, 1999; Botstein et al., 1973). Compared to the curves obtained for WT S. flexneri, Sf6 growth curves were altered for all three omp null mutant strains highlighting the relative changes in DNA delivery when Omps are absent. The rate of infection decreased slightly on ompC−, whereas infection dropped by an order of magnitude on ompA− (Figure 2A). For ompA−C−, infection was significantly slower and is several orders of magnitude lower (Figure 2A). Nevertheless, the relative rates of progeny phage formation and relative burst sizes were similar in all strains, indicating that Omps A and C affected the initial infection step but did not alter phage production once the cells were infected. In addition, we repeated the experiment above with the exception of not artificially rupturing the cells via chloroform to determine the minimum time to lysis as well as the average rise time for each strain. Minimum time to lysis from onset of Sf6 addition at 30 °C occurs as follows: WT = 63 min; ompA- = 76 min; ompC- = 65 min; ompA-C- = 78 min (data are averaged from three experiments and the error was +/- 3 min). The average rise time once lysis had initiated was similar for all strains (∼45 min).
Figure 2. Sf6 infection is slowest in ompA-C- Shigella.
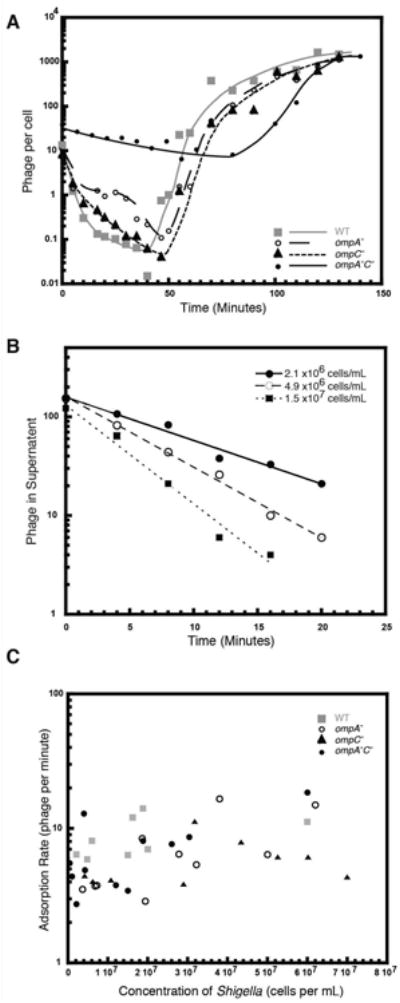
A) Exponentially growing Shigella were infected with Sf6 phage and at the designated times after infection, aliquots of each reaction were treated with chloroform and plated at 30 °C on WT Shigella. The number of phage per cell was calculated as the titer of phage at each time point divided by the total number of cells in the reaction. Each growth curve was repeated in triplicate, and representative plots are shown. B) Representative plots of reversible Sf6 adsorption rates to WT cells. C) Reversible adsorption rates of Sf6 to WT and the three omp null Shigella hosts are plotted as a function of cell concentration and do not differ significantly between WT and the omp null hosts.
Initial rates of Sf6 reversible adsorption to WT and omp null Shigella were monitored by adding phage to cells, taking aliquots at various times, spinning out the cells and attached phage, and titering the supernatant. This experiment allowed us to determine the number of unadsorbed phage as a function of time, and thus calculate the rate of Sf6 adsorption to LPS under a variety of conditions. Representative data are shown for Sf6 adsorbing to WT Shigella (Figure 2B). For each strain, adsorption rates were calculated from 8 to 11 different concentrations of cells and were plotted as a function of cell concentration for each strain (Figure 2C). There are no substantial differences between the strains, indicating that initial, reversible interaction with LPS is not varied as a result of the omp deletions. Therefore, the relative differences that we observed in the eclipse phase in our single step growth curves (Figure 2A) is independent of LPS adsorption rate, and is most likely owing to a slower irreversible adsorption step from the lack of Omps A and C.
Why is the eclipse phase slightly altered in ompC- and ompA- and dramatically altered on the ompA-C- host? Removal of OmpA and OmpC may have a pleiotropic or negative effect on cells, and slower eclipse phases could be unrelated to secondary receptor binding, but instead influenced by the overall health of the omp mutant strains. For example, if the omp deletions alter production of the primary receptor (O-antigen portion of LPS), the phage would also show changes in adsorption (Gemski et al., 1975). To test this possibility, we performed LPS extractions of each S. flexneri strain and found no discernable differences in the amount of LPS present or in O-antigen length as measured by silver staining tris tricine gels displaying the isolated LPS (data not shown). This result is consistent with a previous LPS analysis of S. flexneri strain HND92, which carries a different ompA deletion mutant (Ambrosi et al., 2012). Therefore, phage likely bind the LPS of the omp null cells in a normal manner (Figure 1, step 1). A second possibility is that omp deletion may affect outer membrane integrity. We tested growth of all four strains on MacConkey agar, a bile-salt-rich media that selects for Gram negative bacteria with an intact outer membrane (Macconkey, 1905). All four strains grew on MacConkey plates (data not shown) indicating that the outer membranes were intact.
Time-lapse fluorescence microscopy shows that Sf6 infection is slower in omp null mutants
We used time-lapse fluorescence microscopy to monitor phage Sf6 infection of Shigella and visualize real time genome translocation at the single virion level at 20°C (Figure 3). Sf6 virions were stained with Sytox green since it penetrates capsids but not live cells (Langsrud and Sundheim, 1996; Roth et al., 1997). Images were recorded every three minutes after initiation of infection (see Experimental Procedures). Phage particles attached to cells appeared as bright green foci immediately adjacent to the cells (Figure 3A). Transfer of phage DNA into the host cell was observed directly as a decrease in fluorescence of the phage particle and the simultaneous emergence of diffuse fluorescence within the cell (Figure 3A, Movie S3). Fluorescence inside the cell was dispersed as the dye molecules were released from ejected phage DNA and then bound to the bacterial chromosome; this is similar to comparable experiments with phage λ (Van Valen et al., 2012) and P22 (Mosier-Boss et al., 2003). Quantitation of the fluorescence intensity at each time point is shown in the panels below each fluorescence micrograph (Figure 3A). We measured the average time elapsed after mixing phage and bacteria until DNA transfer occurred for Sf6 bound to cells (Figure 3B). Each data point in Figure 3B represents a single Sf6 DNA ejection event, and the data are grouped into five-minute bins. Phage infecting WT Shigella exhibited the shortest time between adsorption and DNA transfer (average of ∼25 minutes, n= 32). The average time to ejection increased for the omp null strains, with the double knockout showing the longest delay (ompA−, 36 minutes, n= 22; ompC−, 36 minutes n= 19; ompA−C−, 52 minutes, n = 17). There is a large distribution in the times to ejection for individual virions infecting each strain, a similar phenomenon was also observed during in vivo fluorescence imaging of phage λ infecting E. coli (Van Valen et al., 2012). We performed a T-test to validate that the observed average variation in ejection times was significant between strains. Injection times into the WT strain were significantly different than either of the single mutants as well as the double mutant. The resulting p values are as follows: WT vs. ompA-C-, p = <0.0001,(t(47) = -7.57); WT vs. ompA-, p = 0.002, (t(52) = -3.25); WT vs. ompC-, p=0.0031,(t(49) = -3.11); and. ompA- vs. ompC-, p = 0.96, t(39) = - 0.055). The average times to ejection are slower than the eclipse measured in burst experiments (Figure 2A), likely owing to differences in experimental design. For example, the fluorescence time-lapse experiments are performed at lower temperatures and with specimens immobilized on agar pads, versus higher temperatures and liquid culture as was used for the burst experiments.
Figure 3. Time-lapse fluorescence microscopy shows that Sf6 infection is slower on omp null strains.
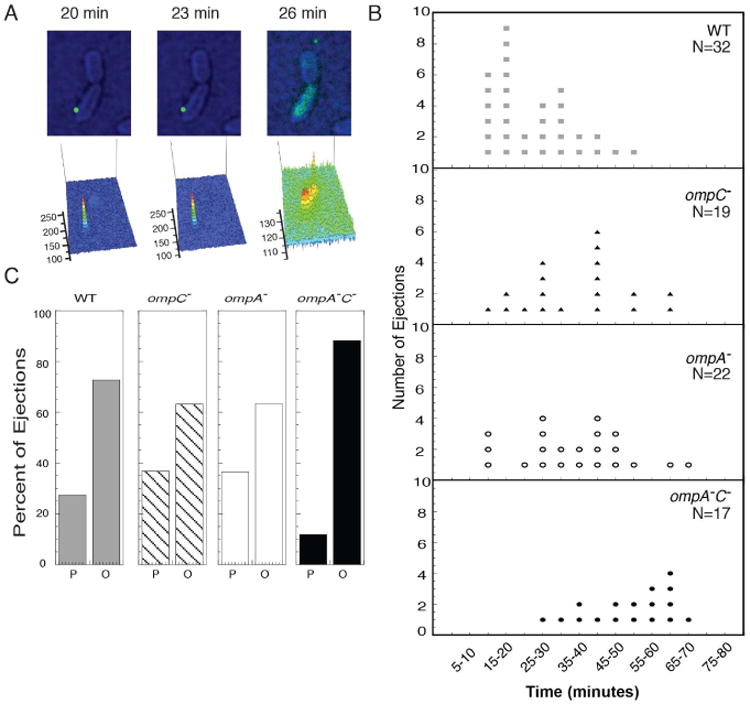
A) Still frames of a time-lapse experiment showing Sf6 (labeled with Sytox green dye) attached to unstained, live WT Shigella (artificially colored to enhance visibility). Below each still image is a schematic showing the pattern of fluorescence intensity of the Sytox signal in the frame. B) Distribution of single particle results showing time of genome ejection post initiation of infection. Each dot represents a single phage-infection event. C) Distribution of phage infection location. P (“at the pole”), is defined as phage infection observed in the quadrant nearest either pole and O (“regions other than the pole”) is defined as the middle two quadrants of the cells that are not near the poles.
We also examined the data to determine if there is a preferred site of infection on the cell surface, since phages SPP1, λ, T4, T7, P1, KVP40, and ϕA1122 are reported to preferentially infect cells at the poles (Jakutyte et al., 2011) (Edgar et al., 2008). Sf6 showed no obvious preference for infections to initiate near cell poles for any of the strains tested (Figure 3C). In instances involving infection of the null mutants of S. flexneri, adsorbed Sf6 virions appeared to diffuse along or “scan” the surface until a suitable locus for infection was found (Movie S4). In many cases the phage were observed to continually scan the cell surface for the entire duration of the fluorescence microscopy experiments. However, a small fraction of phage were observed to infect the ompA-C- cells, and the times until infection for these phage are reported in Figure 3B. Our results are consistent with experiments performed using phage λ that demonstrate a target-finding process (Rothenberg et al., 2011). As a negative control, a mixture of Sf6 and E. coli MG1655 cells (lacking O-antigen and are not hosts for Sf6) was imaged; as expected, Sf6 did not absorb to or infect these cells (data not shown).
OMVs co-purify with Sf6 when grown on WT, ompA−, and ompC− Shigella strains, but not on an ompA−C− strain
As established in the above experiments, Sf6 infection kinetics are altered when OmpA and OmpC are absent. Previously, it was shown that OmpA and OmpC proteins co-purify with Sf6 virions, even after CsCl purification (Casjens et al.; Zhao et al., 2011), and these proteins were subsequently shown be present in OMVs that appeared to be attached to the phage tails (Parent et al., 2012). It was unclear whether this association resulted from specific interactions between the Sf6 tail and the Omps or from other interactions with the lipid membrane. Therefore, we purified Sf6 virions propagated on WT, ompA−, ompC−, and ompA−C− S. flexneri, by CsCl step gradient density centrifugation, and analyzed the composition of each sample by cryo transmission electron microscopy (cryo-TEM) at low magnification (Figure S5) and determined the relative amounts of co-purified OMVs when phage were grown on WT and on omp null strains. We inspected images of Sf6 purified from the different hosts and quantified the number of virions and the number of OMVs present in several preparations of each particle type (data not shown). This measure of the relative amounts of OMVs and phage is imprecise because it doesn't account for heterogeneity in OMV diameters, but it does provide a reasonable estimate of the relative differences. Phage propagated on ompA-C- had significantly decreased levels (>5-fold) of associated OMVs, compared to the WT, ompA-, and ompC- strains. As negative controls, we analyzed similar preparations of P22 and CUS-3 virions, which are not expected to co-purify with OMVs, and indeed we did not observe OMVs in these samples (data not shown). These results demonstrate that OmpA or OmpC is required for efficient physical association of OMVs with Sf6. Although virions could in theory associate through tailspike interaction with OMV LPS, the fact that few OMVs bind in the absence of both OmpA and OmpC suggests that this is not sufficient and that direct virion-Omp protein contact may be required.
Cryo-electron tomography of Sf6 bound to OMVs implies that the tail needle knob may interact with OmpA and OmpC
To gain direct visual evidence concerning the interactions between phage and host-derived membranes, we analyzed Sf6 co-purified with OMVs from WT S. flexneri by cryo-electron tomography (Figure 4). In all 54 tomographic volumes that were reconstructed, we observed a mixed population of particles among a total of 537 phage particles. The majority of these particles (N=312), identified as “free” virions, clearly had no interaction with any OMV. The others appeared to lie in close proximity to the vesicles, each with its tail apparatus interposed between the capsid and the host outer membrane, which suggests that Sf6 associates with the OMVs via their tail machinery. A subpopulation of these adsorbed particles (N=170) contained high density inside the capsid (“full” virions), and the rest (N=55) showed low density inside the capsid (“empty virions”), indicating they had released their genome. In the raw (i.e. unaveraged) tomograms (Movie S6), it was not possible to determine accurately the conformation of the tail apparatus. This is likely owing to the high level of noise and the resolution anisotropy caused by the “missing wedge” effect (Frank, 2006), an artifact intrinsic to tomographic reconstructions because samples cannot be tilted a full ±90 degrees in the microscope, and thus are only observed from a limited range of views. Therefore, we computed a sub-tomogram average for each subpopulation by extracting individual particle sub-volumes from the tomograms and aligning them to a common reference. As a control for assessing the quality of the reconstructions, we compared each of them to an asymmetric map of the Sf6 virion (EMDB-5730, Figure 4B), which was previously reconstructed by single particle methods to 16-Å resolution (Parent et al., 2012). Aside from differences in resolution, the averaged “free” virion sub-tomogram (Figure 4C) compares quite well with the published model. Both capsids share the same structural features, and the tail apparatus adopts the same overall conformation. The sub-tomographic averages of the “full” (Figure 4D) and “empty” (Figure 4E) virions do not reveal any dramatic conformational differences, indicating that cell attachment and genome transfer can occur without extensive rearrangements of the phage tail. Furthermore, all the components of the tail apparatus are visible in the sub-tomogram averages, including the needle and its distal knob, which appears to insert into and indent the membrane.
Figure 4. Cryo-electron tomography and sub-tomogram averages of Sf6 “infecting” OMVs from S. flexneri.
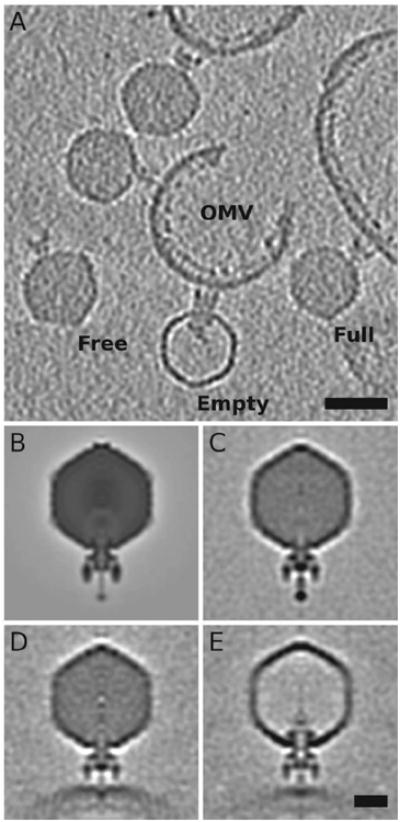
A) A 40Å thick slice from a representative 3D tomogram, showing isolated, unattached phage (“Free”), genome-containing phage that are attached to OMVs (“Full”), and genome-lacking phage attached to OMVs (“Empty”). Scale bar = 500Å. B) A planar, 5.85-Å thick, central section from a density map of Sf6 (filtered to 50-Å resolution, EMDB-5730) calculated by means of single particle, asymmetric reconstruction methods (Parent et al., 2012). C-E) Same as panel (B) but for sub-tomogram averages of “Free” (C), “Full” (D), and “Empty” (E) particles. Scale bar for B-E = 200Å.
Sf6 in vitro genome ejection requires both LPS and an outer membrane protein
OmpA and OmpC clearly influence Sf6 infection in vivo. Hence, we hypothesized that genome ejection in vitro might also require an Omp protein. For phage P22, purified LPS alone is sufficient to induce genome release in vitro (Andres et al., 2010; Andres et al., 2012). Therefore, we determined whether this is also true for Sf6. LPS was extracted from S. enterica serovar Typhimurium strain DB7136 and from S. flexneri serotype Y strain PE577 (see Experimental Procedures). In vitro genome ejection was initiated by mixing phage particles and purified LPS (P22 was incubated with S. enterica serovar Typhimurium LPS, and Sf6 was incubated with S. flexneri serotype Y LPS), followed by incubation at 37°C for 120 min, as described (Andres et al., 2010; Andres et al., 2012). Phage genome release was confirmed by titering the remaining phage (Figure 5A), negative stain electron microscopy (data not shown), and agarose electrophoresis gels that separate intact capsids and ejected genome stained with ethidium bromide (data not shown).
Figure 5. In vitro genome ejection of Sf6 and P22.
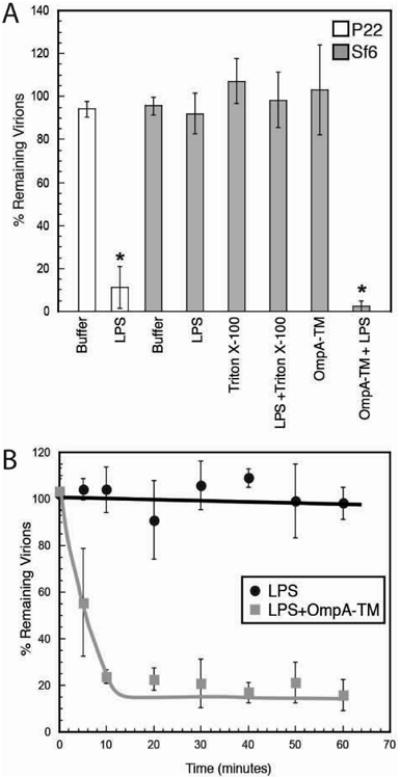
A) “% Remaining Virions” was calculated as the number of PFU remaining after incubation with detergent, LPS, OmpA-TM, or LPS and OmpA-TM, divided by the number of PFU when incubated with buffer. B) “% Remaining Virions” was calculated at each time point as the number of PFUs remaining after incubation with LPS or LPs and OmpA-TM, divided by the number of PFUs when incubated with buffer at t=0 minutes. Each data point is an average of at least three experiments, and the error bars signify one standard deviation.
As was previously reported (Andres et al., 2010; Andres et al., 2012), purified Salmonella LPS was found to be sufficient for phage P22 genome release for the vast majority of virus particles as less than 10% intact P22 virions remained. In contrast, essentially all Sf6 virions remained intact after incubation with Shigella LPS, indicating that LPS alone is not sufficient for Sf6 genome release. To determine the contribution of OmpA to Sf6 in vitro ejection, we expressed and purified the transmembrane domain of S. flexneri OmpA (“OmpA-TM”, residues 1-175, an 8-stranded β-barrel; this protein is missing its periplasmic long C-terminal domain) from E. coli inclusion bodies, and refolded the truncated protein in detergent micelles (see Supplementary Material). Similar to what was observed for LPS, the OmpA-TM domain alone was not sufficient for Sf6 genome ejection in vitro. However, in the presence of both LPS and OmpA-TM, ∼95% of Sf6 virions released their genomes (Figure 5A). To track Sf6 ejection rates, intact virions remaining in the Sf6 in vitro reactions were monitored for number of plaque-forming units. In the presence of OmpA-TM and LPS, functional virions are lost within 10 minutes, which is a physiologically relevant time scale. This suggests that, for Sf6, in vitro DNA release accurately reflects the natural infection (Figure 5B). These experiments showed no difference when performed with LPS extracted from WT or the three omp null Shigella strains (data not shown).
Discussion
Primary and secondary phage receptors
A large number of specific protein-protein interactions must occur between virus-encoded and host-encoded proteins during intracellular multiplication. Phage virions recognize surface features that are diagnostic of each host cell in order to avoid nonproductive infections. For tailed phages this recognition usually occurs in two stages: an initial and reversible stage in which the virion interacts with its primary receptor and a second, usually irreversible, stage since the virion must avoid nonproductive ejection before it reaches the surface of the cell's outer membrane (Casjens and Molineux, 2012; Davidson et al., 2012; Leiman and Shneider, 2012). It is often found that different virion proteins interact with primary and secondary receptors. T4 long tail fibers recognize the LPS core and/or OmpC protein as a primary receptor and its short tail fibers subsequently also recognize part of the core region of LPS (Riede, 1987; Thomassen et al., 2003) as the secondary receptor. Phage O×2 (a T4-like phage) tail fiber mutations that cause the fibers to bind to several different alternative primary receptor molecules do not affect the use of the secondary receptor (Drexler et al., 1991), so these two interactions do not apparently need to be precisely spatially coupled. On the other hand, for phages λ, T5, and SPP1, interaction between the long tail fiber and primary receptor is not essential in the laboratory (Baptista et al., 2008; Heller and Braun, 1979; Hendrix and Duda, 1992; Saigo, 1978). DNA delivery by podoviruses is particularly poorly understood, and in no case are both the primary and secondary receptors known.
Sf6 displays different receptor binding requirements from P22
Sf6 and P22 are related members of the Podoviridae family, and their virions are built from similar structural proteins (Casjens et al., 2004). However, our data suggest that they have different receptor requirements for productive infection. Recent work has identified LPS as the sole requirement for in vitro genome release for P22 (Andres et al., 2010) as well as siphovirus 9NA (Andres et al., 2012). For P22, this release rate in vitro is quite slow (>60 minutes). Phage 9NA demonstrates faster release rates that match values for in vitro ejection by other phages such as λ and T5, where ejection is complete in a few minutes when triggered with purified LamB (Evilevitch et al., 2003; Grayson et al., 2007; Novick and Baldeschwieler, 1988) or FhuA (Chiaruttini et al., 2010; Mangenot et al., 2005), respectively. The release rates reported for P22 reveal a discrepancy, as phage genome ejection happens in a few minutes, not several hours, in vivo. We have confirmed the slow P22 genome release results here as a comparison for our Sf6 studies. LPS alone is not sufficient for genome release in Sf6. In addition, Sf6 interactions with OmpA-TM alone are not enough to trigger genome release, but DNA is released in the presence of both LPS and OmpA-TM. The in vitro rates under these conditions are similar to preliminary observations of in vivo ejection rates (faster than 10 minutes by time-lapse fluorescence microscopy, data not shown). We propose that primary hydrolysis of LPS followed by a secondary interaction with Omps are needed to promote a productive Sf6 infection. The combination of the data presented here indicates that Sf6 requires interactions with a secondary receptor (OmpA or OmpC) for genome ejection.
OmpA and OmpC are receptors for many phages, including Sf6
OmpA is a major component of enterobacterial outer membranes, with ∼100,000 copies presented on the surface of each cell (Koebnik et al., 2000). OmpA has ∼99.6% identity between E. coli and S. flexneri (Power et al., 2006) and may present an alternative or accessory cell recognition mechanism for some members of the P22-like phages. OmpC has also been implicated as an essential receptor for bacteriophages PP01 (Mizoguchi et al., 2003), Gifsy-1/Gifsy-2 (Ho and Slauch, 2001b), and AR1 (Yu et al., 2000). In other instances, deletion of ompC decreases but does not eliminate phage sensitivity, indicating that OmpC may be one of several possible receptors, or may function as a co-receptor (Montag et al., 1989; Tanji et al., 2008). As reported here, it appears that OmpA is the preferred receptor for Sf6.
If OmpA and OmpC are the only secondary receptors for Sf6, the ompA−C− double mutant should be completely resistant to Sf6 infection, and Sf6 virion preparations (as described above) might be expected not to have even the <20% level of bound omp- OMVs. Why is Sf6 eventually able to infect ompA−C− Shigella, albeit much more slowly than WT Shigella, and why are some virion-bound vesicles present after an ompA−C− infection? Phage infection is a robust, dynamic process, and another membrane protein might be able to substitute for OmpA and OmpC. As described previously, OmpA and OmpC are the major protein components found in OMVs that co-purify with Sf6 virions, and this corresponds to the prominent bands seen in Coomassie-stained SDS-PAGE from “purified” virions (Parent et al., 2012; Zhao et al., 2011). Mass spectrometry of such Sf6 virion preparations (Parent et al., 2012), also identified other outer membrane proteins, including OmpX and OmpF, but at much lower abundance than OmpA or OmpC. It is also possible that Sf6 can infect cells at sites of membrane defects when OmpA and OmpC are absent, which would correspond to the much slower rate and efficiency as observed by the fluorescence microscopy experiments on the ompA-C- null host.
OmpA (Morona et al., 1984; Smith et al., 2007), OmpC (Ho and Slauch, 2001a; Mizoguchi et al., 2003; Yu and Mizushima, 1982), OmpX (Drexler et al., 1991), OmpF (Meyer et al., 2012), LamB (Randall-Hazelbauer and Schwartz, 1973), and FhuA (Braun and Wolff, 1973) have each been implicated as host receptors for various tailed phages. These proteins all have homologous folds made up of transmembrane β-barrels and surface-exposed loops, although their diameters and oligomeric states differ (Koebnik et al., 2000). Several studies have shown that phage have the ability to switch between a preferred receptor to an alternative one as a consequence of mutation (such as caused by a mutagen or culture environment) (Nguyen et al., 2012). A study of phage O×2 host range mutants of E. coli showed that O×2 can utilize OmpC for infection when OmpA is deleted (Morona and Henning, 1984). In a more recent study that monitored co-evolution of phage and hosts (Meyer et al., 2012), phage λ was able to recognize OmpF when LamB was absent and several mutations arose in the λ J protein. Therefore, given the prevalence and structural similarities of Omps, it is possible that, when OmpA is absent, Sf6 can recognize and bind to other Omps, such as OmpC. This apparent flexible use of receptors by phages should provide evolutionary advantages and adds another layer of complexity to the interactions between host and virus and could have implications for the use of phage in industrial and medical settings.
Cryo-electron tomograms indicate that Sf6 may bind OmpA and OmpC via the tail needle knob
Cryo-electron tomography has emerged as a very powerful tool for understanding complex phage architecture and binding to isolated membrane vesicles or hosts (Guerrero-Ferreira and Wright, 2013). Recent cryo-tomography work on Podoviridae interactions with hosts include studies of P-SSP7 (Liu et al., 2010) and T7 (Hu et al., 2013). These two phages are similar in structure, and both have an inner core and tail fibers (not tailspikes) that undergo extensive rearrangements upon docking to minicell surfaces. Alternatively, as presented here, Sf6 has large tailspike proteins that do not appear to undergo similar flexible and dramatic motions when docking to OMVs (Figure 4). It is possible that small-scale rearrangements do occur in Sf6 tailspike proteins upon interaction with OMVs, but at the current resolution limits of cryo-tomography (4-5 nm for these samples) we cannot detect such changes. However, at such low-resolution, we do observe an indentation in the OMVs that corresponds to the apparent point of contact of the Sf6 tail needle with the membrane, mediated by the distal tip (knob domain). The tail needle knob, which is present in Sf6 but is remarkably absent in other phages like P22, has a tertiary structure (Olia et al., 2007) with a fold that is similar to virus receptor proteins in adenovirus and PRD-1 (Bhardwaj et al., 2011). Therefore, we propose that the tail needle knob in Sf6 is a likely candidate for interacting with Shigella OmpA and OmpC proteins when the phage approaches the cell membrane.
The needle tips of the 164 known, P22-like phage genomes are present as one of two non-homologous types; 69 have tip domains that are homologous with the Sf6 needle and 95 are homologous to the P22 needle tip (Casjens and Thuman-Commike, 2011; Leavitt et al., 2013). The P22-type tips are rather diverse, with the more divergent pairs being about 50% identical in amino acid sequence, whereas the Sf6-type tips are all >95% identical to one another. The simplest explanation for this is that the P22-type tip is ancestral within this phage group, and the Sf6-type tip has much more recently introgressed into this population, but has not yet had time to diverge much since its arrival in the group (Leavitt et al., 2013). Such successful introgression implies that the Sf6-type tip confers an advantage to some phages over the P22-type tip under some circumstances. To date, the Sf6-type tip has only been found in phages that infect Shigella or E. coli, and not in phages that infect Salmonella, suggesting that it may give a special evolutionary advantage in the infection of the former species.
Experimental Procedures
Single step growth curves
S. flexneri (WT, ompA-, ompC-, and ompA-C-) were grown in LB at 30 °C to a concentration of 2×108 cells per mL. Phage infection was initiated at an MOI of 15 for the WT, ompA-, and ompC- strains, and an MOI of 30 was used for the ompA-C- strain. At times ranging from zero to three hours after infection was initiated, aliquots of infected cells were diluted with phage buffer containing saturating amounts of chloroform to lyse the cells. Lysates were plated and the number of phage per cell at each time point was calculated as described previously (Parent et al., 2004). Aliquots of the cells were assayed before infection and ten minutes post infection to ensure that >95% of the cells were infected, and therefore burst size determination was not affected by a second round of infected cells. These experiments were repeated in triplicate and representative plots are shown in Figure 2A.
Adsorption rates of Sf6 to Shigella
Initial Sf6 adsorption to Shigella was monitored by adding phage to cells at high MOI, taking aliquots at various times (ranging from zero to 20 minutes at 30 °C), spinning out the cells and attached phage for 30 seconds in a microfuge, and titering the supernatant. The concentration of phage was held constant in each experiment (∼1 × 109 phage per mL). We also accurately determined the cell concentration for each sample by plating and counting CFUs before the addition of phage. The fit of these data was done using Kaleidagraph software and only data sets that demonstrated linear kinetics were included in further analysis.
Fluorescence microscopy
Fluorescent Sf6 were made by incubating a high-titer lysate (>109 PFU/ml) with 1 μM of Sytox green nucleic acid stain at 4°C for 24 hours. Overnight cultures of Shigella flexneri (WT, ompA-, ompC-, and ompA-C-) were diluted in LB and grown to mid-log at 30°C with aeration. Cells (1 mL) were pelleted with 1 minute of centrifugation and resuspended in approximately 50μL of fresh LB. Five μL of a DNase I mixture (two units DNase I (New England Biolabs), in 1× supplied DNase I buffer, in ddH2O) were added to the cell re-suspension. A five μL aliquot of the cell suspension was mixed with one μL of Sytox-stained phage (at an MOI <10 phage phage per cell) and immediately spotted onto a thin, dried LB agar pad. Cells were incubated at room temperature for the duration of the experiment in order to increase the chances of capturing phage entry events. The earliest time point was taken within 3-5 minutes of phage addition and cells were imaged using DIC and FITC excitation for Sytox fluorescence, and images were recorded every three minutes for one hour with a GE/Applied Precision Deltavision Elite System with an Olympus IX71 microscope and a 100× 1.4 PlanApo lens.
High titer phage purification
S. flexneri (WT, ompA-, ompC-, and ompA-C-) were grown in LB at 37 °C to 1×108 cells/mL. Sf6 infection was initiated at a MOI of 0.1. The cultures were shaken until complete lysis occurred (∼3.5 hours at 37 °C). A saturating amount of chloroform was added, and cell debris was removed by centrifugation (Sorvall SS-34 rotor, for 10 minutes at 10,000 rpm, 4°C). Phage were then concentrated at 18,000 rpm for 90 minutes at 4°C. The resulting pellet was resuspended in phage buffer by nutation at 4°C overnight. Aggregated material was removed by centrifugation and the phage were further purified using a CsCl step gradient as described (Casjens et al., 2004), dialyzed against phage buffer (10 mM Tris pH=7.6, 10 mM MgCl2), and concentrated to ∼1×1014 phage/mL using a 30 MWCO micro concentrator (AMICON). Phage stocks were stored in phage buffer at 4 °C.
Cryo-TEM
Small (3.5 μL) aliquots of purified phage were vitrified and examined using established procedures (Baker et al., 1999). Briefly, samples were applied to holey (Quantifoil) grids that had been glow-discharged for ∼15 s in an Emitech K350 evaporation unit. Grids were then blotted with Whatman filter paper for ∼5 s, plunged into liquid ethane, and transferred into a precooled FEI Polara, multi-specimen holder, which maintained the vitrified specimen at liquid nitrogen temperature. Micrographs were recorded on a Gatan 4K2 CCD camera at 200 keV in an FEI Polara microscope at a nominal magnification of 19,542 × (7.68 Å per pixel).
Cryo-electron tomography and sub-tomogram averaging
OMVs that co-purified with Sf6 from WT Shigella lytic infections were vitrified as described above, except with the addition of 5-nm gold beads (Sigma). Images were recorded at ∼31,000× magnification at 200 keV in a tomographic, ± 60° tilt series using an FEI F20 microscope operated by Leginon (Potter et al., 1999), and images were collected at 2° tilt increments with a Gatan 4K2 CCD camera. Resulting tomograms were reconstructed using the IMOD program with the gold particles serving as fiducial markers (Kremer et al., 1996). We performed sub-tomogram averaging using routines previously implemented (Cardone et al., 2007), after some modifications tailored to this data set were made. Briefly, after computing 54 tomograms, we extracted from them sub volumes centered around “free” virions, “full” particles, and “empty” particles. Initial orientations of the particles were estimated manually using the visualization program 3dmod in IMOD, and these served to generate an initial template without using any external reference. For each sub-volume, a marker was placed on the phage tail machinery, and its position with respect to the center of the sub-volume was used to align the particles so that all tails pointed in the same direction. The alignment was refined by iterative template matching, after masking a region just encompassing the tail machinery, to eliminate any contribution from the density of the capsid, without assuming or imposing any symmetry. Several iterations of refinement were performed by narrowing progressively the range allowed for the orientation parameters search until no improvement was observed both by FSC calculations (van Heel and Schatz, 2005) and visual inspection. The final maps, showing both capsid and tail machinery, were obtained by averaging the entire sub-volumes, with no mask applied, after aligning them according to the orientation parameters determined for the corresponding tails.
LPS extraction and in vitro genome ejection
LPS was extracted from overnight cultures of S. typhimurium and S. flexneri using a kit (BULLDOG BIO). LPS purity and concentration were determined by analysis of silver-stained tris-tricine SDS gels, dry weight, and absorbance spectra (data not shown). Phage were incubated with either purified LPS (solubilized in 10mM Tris, pH=7.6) derived from their respective host cells according to previous experiments (Andres et al., 2010), 150 μg/mL OmpA-TM (refolded in 1% triton X-100), or both LPS and OmpA-TM. The percent remaining virions was calculated by dividing the PFU in each of the reactions by the PFU with only buffer added. For the time course experiments, Sf6 phage were incubated with LPS or both LPS and OmpA-TM (150 μg/mL) as described above, and an aliquot was taken at each time point. The percent remaining virions was calculated by dividing the PFU at each time point by the PFU with only buffer added. Plates were grown at 30 °C with either DB7136 (P22) or PE577 (Sf6). Each histogram represents an average of at least three experiments, and the error bars signify one standard deviation.
Supplementary Material
Acknowledgments
We thank N. Olson (UCSD) for expert guidance in cryo-TEM methods, K. Willingford (UCSD) for counting statistics of Sf6 and OMVs from cryo-electron micrographs, J. Fredlund (Institut Pasteur) for advice on construction of the expression vector, and H. Hong (MSU) for advice on OmpA purification and refolding. This work was supported in part by NIH grants R37 GM-033050 and 1S10 RR-020016 to TSB, AI074825 to SRC, R01 GM-073898 to JP, and support from UCSD and the Agouron Foundation to TSB to establish and support the UCSD cryo-TEM facilities. Some of the work presented here was conducted at the National Resource for Automated Molecular Microscopy, which is supported by the National Institutes of Health though the National Center for Research Resources' P41 program (RR017573).
Abbreviations
- WT
wild-type
- cryo-TEM
cryo transmission electron microscopy
- Omp
outer membrane protein
- LPS
lipopolysaccharide
- CFU
colony forming units
- PFU
plaque forming units
- MOI
multiplicity of infection
- OMVs
outer membrane vesicles
- TM
transmembrane
References
- Allison GE, Verma NK. Serotype-converting bacteriophages and O-antigen modification in Shigella flexneri. Trends Microbiol. 2000;8:17–23. doi: 10.1016/s0966-842x(99)01646-7. [DOI] [PubMed] [Google Scholar]
- Ambrosi C, Pompili M, Scribano D, Zagaglia C, Ripa S, Nicoletti M. Outer Membrane Protein A (OmpA): A New Player in Shigella flexneri Protrusion Formation and Inter-Cellular Spreading. PloS one. 2012;7:e49625. doi: 10.1371/journal.pone.0049625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andres D, Hanke C, Baxa U, Seul A, Barbirz S, Seckler R. Tailspike interactions with lipopolysaccharide effect DNA ejection from phage P22 particles in vitro. J Biol Chem. 2010;285:36768–36775. doi: 10.1074/jbc.M110.169003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andres D, Roske Y, Doering C, Heinemann U, Seckler R, Barbirz S. Tail morphology controls DNA release in two Salmonella phages with one lipopolysaccharide receptor recognition system. Molecular microbiology. 2012;83:1244–1253. doi: 10.1111/j.1365-2958.2012.08006.x. [DOI] [PubMed] [Google Scholar]
- Aramli LA, Teschke CM. Single amino acid substitutions globally suppress the folding defects of temperature-sensitive folding mutants of phage P22 coat protein. J Biol Chem. 1999;274:22217–22224. doi: 10.1074/jbc.274.32.22217. [DOI] [PubMed] [Google Scholar]
- Baker TS, Olson NH, Fuller SD. Adding the third dimension to virus life cycles: three-dimensional reconstruction of icosahedral viruses from cryo-electron micrographs. Microbiology & Molecular Biology Reviews. 1999;63:862–922. doi: 10.1128/mmbr.63.4.862-922.1999. erratum appears in Microbiol Mol Biol Rev 2000 Mar;64(1):237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bamford DH, Grimes JM, Stuart DI. What does structure tell us about virus evolution? Curr Opin Struct Biol. 2005;15:655–663. doi: 10.1016/j.sbi.2005.10.012. [DOI] [PubMed] [Google Scholar]
- Banks DJ, Beres SB, Musser JM. The fundamental contribution of phages to GAS evolution, genome diversification and strain emergence. Trends Microbiol. 2002;10:515–521. doi: 10.1016/s0966-842x(02)02461-7. [DOI] [PubMed] [Google Scholar]
- Baptista C, Santos MA, Sao-Jose C. Phage SPP1 reversible adsorption to Bacillus subtilis cell wall teichoic acids accelerates virus recognition of membrane receptor YueB. Journal of bacteriology. 2008;190:4989–4996. doi: 10.1128/JB.00349-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baxa U, Steinbacher S, Miller S, Weintraub A, Huber R, Seckler R. Interactions of phage P22 tails with their cellular receptor, Salmonella O-antigen polysaccharide. Biophysical journal. 1996;71:2040–2048. doi: 10.1016/S0006-3495(96)79402-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berlanda Scorza F, Colucci AM, Maggiore L, Sanzone S, Rossi O, Ferlenghi I, Pesce I, Caboni M, Norais N, Di Cioccio V, et al. High yield production process for Shigella outer membrane particles. PloS one. 2012;7:e35616. doi: 10.1371/journal.pone.0035616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhardwaj A, Molineux IJ, Casjens SR, Cingolani G. Atomic structure of bacteriophage Sf6 tail needle knob. J Biol Chem. 2011;286:30867–30877. doi: 10.1074/jbc.M111.260877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botstein D, Waddell CH, King J. Mechanism of head assembly and DNA encapsulation in Salmonella phage P22. I. Genes, proteins, structures and DNA maturation. J Mol Biol. 1973;80:669–695. doi: 10.1016/0022-2836(73)90204-0. [DOI] [PubMed] [Google Scholar]
- Braun V, Wolff H. Characterization of the receptor protein for phage T5 and colicin M in the outer membrane of E. coli B. FEBS letters. 1973;34:77–80. doi: 10.1016/0014-5793(73)80707-0. [DOI] [PubMed] [Google Scholar]
- Broadbent SE, Davies MR, van der Woude MW. Phase variation controls expression of Salmonella lipopolysaccharide modification genes by a DNA methylation-dependent mechanism. Molecular microbiology. 2010;77:337–353. doi: 10.1111/j.1365-2958.2010.07203.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardone G, Winkler DC, Trus BL, Cheng N, Heuser JE, Newcomb WW, Brown JC, Steven AC. Visualization of the herpes simplex virus portal in situ by cryo-electron tomography. Virology. 2007;361:426–434. doi: 10.1016/j.virol.2006.10.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casjens S, Molineux I. Short Noncontractile Tail Machines: Adsorption and DNA delivery by Podoviruses. In: Rossmann M, Rao V, editors. Viral Molecular Machines. New York: Springer; 2012. [DOI] [PubMed] [Google Scholar]
- Casjens S, Winn-Stapley DA, Gilcrease EB, Morona R, Kuhlewein C, Chua JE, Manning PA, Inwood W, Clark AJ. The chromosome of Shigella flexneri bacteriophage Sf6: complete nucleotide sequence, genetic mosaicism, and DNA packaging. Journal of molecular biology. 2004;339:379–394. doi: 10.1016/j.jmb.2004.03.068. [DOI] [PubMed] [Google Scholar]
- Casjens SR, Thuman-Commike PA. Evolution of mosaically related tailed bacteriophage genomes seen through the lens of phage P22 virion assembly. Virology. 2011;411:393–415. doi: 10.1016/j.virol.2010.12.046. [DOI] [PubMed] [Google Scholar]
- Chiaruttini N, de Frutos M, Augarde E, Boulanger P, Letellier L, Viasnoff V. Is the in vitro ejection of bacteriophage DNA quasistatic? A bulk to single virus study. Biophysical journal. 2010;99:447–455. doi: 10.1016/j.bpj.2010.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chua JE, Manning PA, Morona R. The Shigella flexneri bacteriophage Sf6 tailspike protein (TSP)/endorhamnosidase is related to the bacteriophage P22 TSP and has a motif common to exo- and endoglycanases, and C-5 epimerases. Microbiology. 1999;145(Pt 7):1649–1659. doi: 10.1099/13500872-145-7-1649. [DOI] [PubMed] [Google Scholar]
- Clark CA, Beltrame J, Manning PA. The oac gene encoding a lipopolysaccharide O-antigen acetylase maps adjacent to the integrase-encoding gene on the genome of Shigella flexneri bacteriophage Sf6. Gene. 1991;107:43–52. doi: 10.1016/0378-1119(91)90295-m. [DOI] [PubMed] [Google Scholar]
- Davidson AR, Cardarelli L, Pell LG, Radford DR, Maxwell KL. Long noncontractile tail machines of bacteriophages. Advances in experimental medicine and biology. 2012;726:115–142. doi: 10.1007/978-1-4614-0980-9_6. [DOI] [PubMed] [Google Scholar]
- Drexler K, Dannull J, Hindennach I, Mutschler B, Henning U. Single mutations in a gene for a tail fiber component of an Escherichia coli phage can cause an extension from a protein to a carbohydrate as a receptor. Journal of molecular biology. 1991;219:655–663. doi: 10.1016/0022-2836(91)90662-p. [DOI] [PubMed] [Google Scholar]
- Edgar R, Rokney A, Feeney M, Semsey S, Kessel M, Goldberg MB, Adhya S, Oppenheim AB. Bacteriophage infection is targeted to cellular poles. Molecular microbiology. 2008;68:1107–1116. doi: 10.1111/j.1365-2958.2008.06205.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evilevitch A, Lavelle L, Knobler CM, Raspaud E, Gelbart WM. Osmotic pressure inhibition of DNA ejection from phage. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:9292–9295. doi: 10.1073/pnas.1233721100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feil EJ. The emergence and spread of dysentery. Nature genetics. 2012;44:964–965. doi: 10.1038/ng.2389. [DOI] [PubMed] [Google Scholar]
- Frank J. Electron tomography: methods for three-dimensional visualization of structures in the cell. 2nd. New York; London: Springer; 2006. [Google Scholar]
- Gemski P, Jr, Koeltzow DE, Formal SB. Phage conversion of Shigella flexneri group antigens. Infection and immunity. 1975;11:685–691. doi: 10.1128/iai.11.4.685-691.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon CL. Biology. Cambridge, MA: Massachusettes Institute of Technology; 1993. Analysis of Conditional-Lethal Mutations of the Phage P22 Coat Protein. [Google Scholar]
- Grayson P, Han L, Winther T, Phillips R. Real-time observations of single bacteriophage lambda DNA ejections in vitro. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:14652–14657. doi: 10.1073/pnas.0703274104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerrero-Ferreira RC, Wright ER. Cryo-electron tomography of bacterial viruses. Virology. 2013;435:179–186. doi: 10.1016/j.virol.2012.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heller K, Braun V. Accelerated adsorption of bacteriophage T5 to Escherichia coli F, resulting from reversible tail fiber-lipopolysaccharide binding. Journal of bacteriology. 1979;139:32–38. doi: 10.1128/jb.139.1.32-38.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrix RW, Duda RL. Bacteriophage lambda PaPa: not the mother of all lambda phages. Science. 1992;258:1145–1148. doi: 10.1126/science.1439823. [DOI] [PubMed] [Google Scholar]
- Ho TD, Slauch JM. OmpC is the receptor for Gifsy-1 and Gifsy-2 bacteriophages of Salmonella. Journal of bacteriology. 2001a;183:1495–1498. doi: 10.1128/JB.183.4.1495-1498.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho TD, Slauch JM. OmpC is the receptor for Gifsy-1 and Gifsy-2 bacteriophages of Salmonella. Journal of bacteriology. 2001b;183:1495–1498. doi: 10.1128/JB.183.4.1495-1498.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt KE, Baker S, Weill FX, Holmes EC, Kitchen A, Yu J, Sangal V, Brown DJ, Coia JE, Kim DW, et al. Shigella sonnei genome sequencing and phylogenetic analysis indicate recent global dissemination from Europe. Nature genetics. 2012;44:1056–1059. doi: 10.1038/ng.2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu B, Margolin W, Molineux IJ, Liu J. The bacteriophage t7 virion undergoes extensive structural remodeling during infection. Science. 2013;339:576–579. doi: 10.1126/science.1231887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakutyte L, Baptista C, Sao-Jose C, Daugelavicius R, Carballido-Lopez R, Tavares P. Bacteriophage infection in rod-shaped gram-positive bacteria: evidence for a preferential polar route for phage SPP1 entry in Bacillus subtilis. Journal of bacteriology. 2011;193:4893–4903. doi: 10.1128/JB.05104-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiino DR, Singer MS, Rothman-Denes LB. Two overlapping genes encoding membrane proteins required for bacteriophage N4 adsorption. Journal of bacteriology. 1993;175:7081–7085. doi: 10.1128/jb.175.21.7081-7085.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koebnik R, Locher KP, Van Gelder P. Structure and function of bacterial outer membrane proteins: barrels in a nutshell. Molecular microbiology. 2000;37:239–253. doi: 10.1046/j.1365-2958.2000.01983.x. [DOI] [PubMed] [Google Scholar]
- Kotloff KL, Winickoff JP, Ivanoff B, Clemens JD, Swerdlow DL, Sansonetti PJ, Adak GK, Levine MM. Global burden of Shigella infections: implications for vaccine development and implementation of control strategies. Bull World Health Organ. 1999;77:651–666. [PMC free article] [PubMed] [Google Scholar]
- Kremer JR, Mastronarde DN, McIntosh JR. Computer visualization of three-dimensional image data using IMOD. J Struct Biol. 1996;116:71–76. doi: 10.1006/jsbi.1996.0013. [DOI] [PubMed] [Google Scholar]
- Kulp A, Kuehn MJ. Biological functions and biogenesis of secreted bacterial outer membrane vesicles. Annual review of microbiology. 2010;64:163–184. doi: 10.1146/annurev.micro.091208.073413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langsrud S, Sundheim G. Flow cytometry for rapid assessment of viability after exposure to a quaternary ammonium compound. The Journal of applied bacteriology. 1996;81:411–418. doi: 10.1111/j.1365-2672.1996.tb03527.x. [DOI] [PubMed] [Google Scholar]
- Leavitt JC, Gogokhia L, Gilcrease EB, Bhardwaj A, Cingolani G, Casjens SR. The tip of the tail needle affects the rate of DNA delivery by bacteriophage P22. PloS one. 2013;8:e70936. doi: 10.1371/journal.pone.0070936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leiman PG, Shneider MM. Contractile tail machines of bacteriophages. Advances in experimental medicine and biology. 2012;726:93–114. doi: 10.1007/978-1-4614-0980-9_5. [DOI] [PubMed] [Google Scholar]
- Liu X, Zhang Q, Murata K, Baker ML, Sullivan MB, Fu C, Dougherty MT, Schmid MF, Osburne MS, Chisholm SW, et al. Structural changes in a marine podovirus associated with release of its genome into Prochlorococcus. Nature structural & molecular biology. 2010;17:830–837. doi: 10.1038/nsmb.1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macconkey A. Lactose-Fermenting Bacteria in Faeces. The Journal of hygiene. 1905;5:333–379. doi: 10.1017/s002217240000259x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangenot S, Hochrein M, Radler J, Letellier L. Real-time imaging of DNA ejection from single phage particles. Current biology: CB. 2005;15:430–435. doi: 10.1016/j.cub.2004.12.080. [DOI] [PubMed] [Google Scholar]
- McPartland J, Rothman-Denes LB. The tail sheath of bacteriophage N4 interacts with the Escherichia coli receptor. Journal of bacteriology. 2009;191:525–532. doi: 10.1128/JB.01423-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer JR, Dobias DT, Weitz JS, Barrick JE, Quick RT, Lenski RE. Repeatability and contingency in the evolution of a key innovation in phage lambda. Science. 2012;335:428–432. doi: 10.1126/science.1214449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizoguchi K, Morita M, Fischer CR, Yoichi M, Tanji Y, Unno H. Coevolution of bacteriophage PP01 and Escherichia coli O157:H7 in continuous culture. Applied and environmental microbiology. 2003;69:170–176. doi: 10.1128/AEM.69.1.170-176.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montag D, Schwarz H, Henning U. A component of the side tail fiber of Escherichia coli bacteriophage lambda can functionally replace the receptor-recognizing part of a long tail fiber protein of the unrelated bacteriophage T4. Journal of bacteriology. 1989;171:4378–4384. doi: 10.1128/jb.171.8.4378-4384.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morona R, Henning U. Host range mutants of bacteriophage Ox2 can use two different outer membrane proteins of Escherichia coli K-12 as receptors. Journal of bacteriology. 1984;159:579–582. doi: 10.1128/jb.159.2.579-582.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morona R, Klose M, Henning U. Escherichia coli K-12 outer membrane protein (OmpA) as a bacteriophage receptor: analysis of mutant genes expressing altered proteins. Journal of bacteriology. 1984;159:570–578. doi: 10.1128/jb.159.2.570-578.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosier-Boss PA, Lieberman SH, Andrews JM, Rohwer FL, Wegley LE, Breitbart M. Use of fluorescently labeled phage in the detection and identification of bacterial species. Applied spectroscopy. 2003;57:1138–1144. doi: 10.1366/00037020360696008. [DOI] [PubMed] [Google Scholar]
- Muller JJ, Barbirz S, Heinle K, Freiberg A, Seckler R, Heinemann U. An intersubunit active site between supercoiled parallel beta helices in the trimeric tailspike endorhamnosidase of Shigella flexneri Phage Sf6. Structure. 2008;16:766–775. doi: 10.1016/j.str.2008.01.019. [DOI] [PubMed] [Google Scholar]
- Nguyen AH, Molineux IJ, Springman R, Bull JJ. Multiple genetic pathways to similar fitness limits during viral adaptation to a new host. Evolution; international journal of organic evolution. 2012;66:363–374. doi: 10.1111/j.1558-5646.2011.01433.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novick SL, Baldeschwieler JD. Fluorescence measurement of the kinetics of DNA injection by bacteriophage lambda into liposomes. Biochemistry. 1988;27:7919–7924. doi: 10.1021/bi00420a050. [DOI] [PubMed] [Google Scholar]
- Olia AS, Casjens S, Cingolani G. Structure of phage P22 cell envelope-penetrating needle. Nat Struct Mol Biol. 2007 doi: 10.1038/nsmb1317. [DOI] [PubMed] [Google Scholar]
- Parent KN, Gilcrease EB, Casjens SR, Baker TS. Structural evolution of the P22-like phages: Comparison of Sf6 and P22 procapsid and virion architectures. Virology. 2012;427:177–188. doi: 10.1016/j.virol.2012.01.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parent KN, Ranaghan MJ, Teschke CM. A second site suppressor of a folding defect functions via interactions with a chaperone network to improve folding and assembly in vivo. Mol Microbiol. 2004;54:1036–1054. doi: 10.1111/j.1365-2958.2004.04326.x. [DOI] [PubMed] [Google Scholar]
- Poranen MM, Daugelavicius R, Bamford DH. Common principles in viral entry. Annual review of microbiology. 2002;56:521–538. doi: 10.1146/annurev.micro.56.012302.160643. [DOI] [PubMed] [Google Scholar]
- Potter CS, Chu H, Frey B, Green C, Kisseberth N, Madden TJ, Miller KL, Nahrstedt K, Pulokas J, Reilein A, et al. Leginon: a system for fully automated acquisition of 1000 electron micrographs a day. Ultramicroscopy. 1999;77:153–161. doi: 10.1016/s0304-3991(99)00043-1. [DOI] [PubMed] [Google Scholar]
- Power ML, Ferrari BC, Littlefield-Wyer J, Gordon DM, Slade MB, Veal DA. A naturally occurring novel allele of Escherichia coli outer membrane protein A reduces sensitivity to bacteriophage. Applied and environmental microbiology. 2006;72:7930–7932. doi: 10.1128/AEM.01040-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randall-Hazelbauer L, Schwartz M. Isolation of the bacteriophage lambda receptor from Escherichia coli. Journal of bacteriology. 1973;116:1436–1446. doi: 10.1128/jb.116.3.1436-1446.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riede I. Receptor specificity of the short tail fibres (gp12) of T-even type Escherichia coli phages. Molecular & general genetics: MGG. 1987;206:110–115. doi: 10.1007/BF00326544. [DOI] [PubMed] [Google Scholar]
- Roth BL, Poot M, Yue ST, Millard PJ. Bacterial viability and antibiotic susceptibility testing with SYTOX green nucleic acid stain. Applied and environmental microbiology. 1997;63:2421–2431. doi: 10.1128/aem.63.6.2421-2431.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothenberg E, Sepulveda LA, Skinner SO, Zeng L, Selvin PR, Golding I. Single-virus tracking reveals a spatial receptor-dependent search mechanism. Biophysical journal. 2011;100:2875–2882. doi: 10.1016/j.bpj.2011.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saigo K. Isolation of high-density mutants and identification of nonessential structural proteins in bacteriophage T5; dispensability of L-shaped tail fibers and a secondary major head protein. Virology. 1978;85:422–433. doi: 10.1016/0042-6822(78)90449-x. [DOI] [PubMed] [Google Scholar]
- Smith SG, Mahon V, Lambert MA, Fagan RP. A molecular Swiss army knife: OmpA structure, function and expression. FEMS Microbiol Lett. 2007;273:1–11. doi: 10.1111/j.1574-6968.2007.00778.x. [DOI] [PubMed] [Google Scholar]
- Steinbacher S, Miller S, Baxa U, Budisa N, Weintraub A, Seckler R, Huber R. Phage P22 tailspike protein: crystal structure of the head-binding domain at 2.3A, fully refined strucutre of the endorhamnosidase at 1.56A resolution, and the molecular basis of O-antigen recognition and cleavage. J Mol Biol. 1997;267:865–880. doi: 10.1006/jmbi.1997.0922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanji Y, Hattori K, Suzuki K, Miyanaga K. Spontaneous deletion of a 209-kilobase-pair fragment from the Escherichia coli genome occurs with acquisition of resistance to an assortment of infectious phages. Applied and environmental microbiology. 2008;74:4256–4263. doi: 10.1128/AEM.00243-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomassen E, Gielen G, Schutz M, Schoehn G, Abrahams JP, Miller S, van Raaij MJ. The structure of the receptor-binding domain of the bacteriophage T4 short tail fibre reveals a knitted trimeric metal-binding fold. Journal of molecular biology. 2003;331:361–373. doi: 10.1016/s0022-2836(03)00755-1. [DOI] [PubMed] [Google Scholar]
- van Heel M, Schatz M. Fourier shell correlation threshold criteria. J Struct Biol. 2005;151:250–262. doi: 10.1016/j.jsb.2005.05.009. [DOI] [PubMed] [Google Scholar]
- Van Valen D, Wu D, Chen YJ, Tuson H, Wiggins P, Phillips R. A single-molecule Hershey-Chase experiment. Current biology: CB. 2012;22:1339–1343. doi: 10.1016/j.cub.2012.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma NK, Brandt JM, Verma DJ, Lindberg AA. Molecular characterization of the O-acetyl transferase gene of converting bacteriophage SF6 that adds group antigen 6 to Shigella flexneri. Molecular microbiology. 1991;5:71–75. doi: 10.1111/j.1365-2958.1991.tb01827.x. [DOI] [PubMed] [Google Scholar]
- Warren BR, Parish ME, Schneider KR. Shigella as a foodborne pathogen and current methods for detection in food. Critical reviews in food science and nutrition. 2006;46:551–567. doi: 10.1080/10408390500295458. [DOI] [PubMed] [Google Scholar]
- Yu F, Mizushima S. Roles of lipopolysaccharide and outer membrane protein OmpC of Escherichia coli K-12 in the receptor function for bacteriophage T4. Journal of bacteriology. 1982;151:718–722. doi: 10.1128/jb.151.2.718-722.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu SL, Ko KL, Chen CS, Chang YC, Syu WJ. Characterization of the distal tail fiber locus and determination of the receptor for phage AR1, which specifically infects Escherichia coli O157:H7. Journal of bacteriology. 2000;182:5962–5968. doi: 10.1128/jb.182.21.5962-5968.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H, Sequeira RD, Galeva NA, Tang L. The host outer membrane proteins OmpA and OmpC are associated with the Shigella phage Sf6 virion. Virology. 2011;409:319–327. doi: 10.1016/j.virol.2010.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


