Abstract
A surface protein of Lactobacillus reuteri, mucus adhesion-promoting protein (MapA), is considered to be an adhesion factor. MapA is expressed in L. reuteri strains and adheres to piglet gastric mucus, collagen type I, and human intestinal epithelial cells such as Caco-2. The aim of this study was to identify molecules that mediate the attachment of MapA from L. reuteri to the intestinal epithelial cell surface by investigating the adhesion of MapA to receptor-like molecules on Caco-2 cells. MapA-binding receptor-like molecules were detected in Caco-2 cell lysates by 2D-PAGE. Two proteins, annexin A13 (ANXA13) and paralemmin (PALM), were identified by MALDI TOF-MS. The results of a pull-down assay showed that MapA bound directly to ANXA13 and PALM. Fluorescence microscopy studies confirmed that MapA binding to ANXA13 and PALM was colocalized on the Caco-2 cell membrane. To evaluate whether ANXA13 and PALM are important for MapA adhesion, ANXA13 and PALM knockdown cell lines were established. The adhesion of MapA to the abovementioned cell lines was reduced compared with that to wild-type Caco-2 cells. These knockdown experiments established the importance of these receptor-like molecules in MapA adhesion.
Keywords: Lactobacillus reuteri, adhesion, Caco-2, receptor-like molecules
INTRODUCTION
The gastrointestinal tracts (GITs) of animals are colonized by vast numbers of microorganisms living in harmony with their hosts [1]. The host acquires metabolites as stimulating factors from these microorganisms, and changes in the intestinal bacterial flora can affect the health of the host. Gibson and Fuller suggested a definition of a “probiotic” as a live bacterial cell that beneficially affects the health of the host by improving the microorganism balance [2].
Lactic acid bacteria (LAB) meet this definition of “probiotic” and have attracted interest because of their beneficial effects such as immunostimulation and competitive inhibition of pathogens. LAB must adhere to the GIT in order to exert their effects on the tissues and intestinal flora of the host. Bacteria that enter the GIT from the external environment are usually rejected by the host’s defense mechanisms and eliminated in the feces by intestinal peristalsis. Normal microbiota in GIT can resist this elimination. However, unlike infectious pathogens, the mechanisms by which LAB adhere to the gut remain unknown. For successful research and development of probiotics, we need to have a basic understanding of their interactions with the host.
Lactobacillus reuteri strains are LAB that are found in the GITs of various animals and live symbiotically with other bacteria and the host [3]. These strains have beneficial effects, including cholesterol-lowering activity, immunomodulation and reduction of the viability of pathogens such as Escherichia coli O157 via the actions of reuterin [4,5,6,7,8,9]. One of the adhesion factors of L. reuteri DSMZ 20016T is mucus adhesion-promoting protein, MapA. MapA has been isolated from the surfaces of L. reuteri 104R; this protein adheres to porcine gastric mucus [10,11,12]. It is widely distributed in L. reuteri strains and is a component of the ABC transporter. This protein shares amino acid sequence homology with a collagen-binding protein (CnBP) of L. reuteri NCIB 11951 [13, 14].
Several types of LAB are known to adhere to the GIT in conjunction with extracellular matrix components such as mucin and collagen. These molecules or their constituents can mediate the affinity between bacterial extracellular molecules and mucus or epithelial cells [15]. Therefore, these extracellular matrix components are useful for screening for LAB adhesion factors. However, to understand the mechanism of LAB adhesion, we considered that binding of adhesion factors of LAB to receptor-like molecules on intestinal epithelial cells must also be studied.
We previously showed that MapA binds directly to the surfaces of human intestinal epithelial Caco-2 cells and mediates the adhesion of L. reuteri [16]. In this study, we investigated MapA binding to receptor-like molecules on Caco-2 cells.
MATERIALS AND METHODS
Cell culture
Caco-2 (RCB0988) cells were purchased from the Riken Cell Bank (Tsukuba, Japan) and maintained in a cell culture flask (Greiner Bio-One) in minimal essential medium (MEM; Sigma-Aldrich) supplemented with 1% MEM nonessential amino acids (Gibco), 1% MEM sodium pyruvate (Gibco), penicillin/streptomycin (100 U/100 mg; Gibco), and 10% fetal bovine serum (Lot SFBM30-1894; Equitech-Bio Inc.). The cells were cultured for 5–7 days at 37°C in a humidified atmosphere of 5% CO2 and 95% air.
MapA purification
We used QIAexpress® (Qiagen) to purify MapA. Protein expression and purification were performed according to the manufacturer’s instructions. Purified MapA was dialyzed against phosphate-buffered saline (PBS) and stored at –80°C [16].
Preparation of Caco-2 cell lysates
Caco-2 cells grown to confluence in a 90-mm culture dish were washed two times with 2 ml of ice-cold PBS. A cell scraper was used to harvest the cells, and the cells were then suspended in 1 ml of radioimmunopre-cipitation assay (RIPA) buffer (20 mM HEPES-NaOH (pH 7.4), 150 mM NaCl, 1% NP-40, 0.1% SDS, and 0.1% sodium deoxycholate) supplemented with 50 mg/ml DNase I and 25 mg/ml RNase A (Sigma). The cells were homogenized by drawing them through a 25-gauge needle 10 times. This mixture was kept on ice for 2 hr. The suspension was centrifuged at 20,000 ×g for 30 min, and the supernatant was collected and used as a Caco-2 cell lysate.
Preparation of MapA-immobilized resin
HisLink Protein Purification Resin (10 µl; Promega) was added to a new tube. The resin was washed, suspended in 40 μl of 250 mM imidazole, and incubated on ice for 30 min. The resin was then washed two times with 40 μl of PBS. MapA (20 μg) was bound to the resin by affinity capture. The resin mix was then incubated on ice for 30 min, and the resin was then washed two times with 40 μl PBS. As a negative control, resin was incubated without MapA.
Assay of MapA binding to receptor-like molecules on Caco-2 cells
A Caco-2 cell lysate (250 μl) was diluted with 750 μl HNS buffer (20 mM HEPES-NaOH (pH 7.4), 1% NP-40, and 150 mM NaCl) and added to a 1/100 volume of 1 M imidazole stock (0.1 M HEPES, 1 M imidazole, pH adjusted to 7.4 with HCl). The resin sample in each tube was suspended in the diluted Caco-2 cell lysate mixture, incubated at 4°C for 16 hr with rotation, and centrifuged at 20,000 × g for 1 min. The resin was washed two times with 250 µl wash buffer containing increasing concentrations of imidazole (10, 25, 50, and 100 mM imidazole in RIPA buffer) and then centrifuged at 20,000 × g for 1 min. It was then suspended in elution buffer (0.5 M imidazole in RIPA buffer) and centrifuged at 20,000 ×g for 1 min. The clear supernatant fluid was used for two-dimensional polyacrylamide gel electrophoresis (2D-PAGE). 2D-PAGE gels were stained with SYPRO Ruby Protein Gel Stain (Bio-Rad). The PharosFX Plus System for molecular imaging (Bio-Rad) was used for sample analysis.
In-gel digestion
A gel slice with a receptor-like protein from 2D-PAGE was added to 50% acetonitrile in a centrifuge tube for 10 min at room temperature. The solution was removed, and 100% acetonitrile was added to the tube for 10 min. The acetonitrile was discarded, and the gel slice was dried. Reducing buffer (10 mg/ml of DTT in 0.1 M NH4HCO3) was then added to the dried gel slice and incubated for 90 min at 55 °C. Carboxymethylation buffer (100 mg/ml sodium iodoacetic acid in 0.1 M NH4HCO3) was added to the gel slice, and the gel slice was incubated at room temperature for 60 min in the dark. Excess solution was removed, 0.1 M NH4HCO3 was added, and the sample was then kept at room temperature for 10 min. The solution was then removed, 50% acetonitrile was added, and the sample was kept at room temperature for 10 min. The solution was removed, 100% acetonitrile was added, and the sample was again kept at room temperature for 10 min. The supernatant was then discarded, and the gel was dried. Trypsin solution (50 μg/ml trypsin stock solution* in 0.1 M NH4HCO3) was added to the dried gel, and the sample was incubated at 37°C overnight. The solution was placed in a new tube to which 50% acetonitrile with 0.1% trifluoroacetic acid was added; the sample was then kept at room temperature for 10 min. This step was performed two times. The solution was removed and placed in a new tube to which 100% acetonitrile with 0.1% trifluoroacetic acid was added; this solution was then incubated at room temperature for 10 min. Acetonitrile was removed from the sample by vacuum concentration. The substantially dried sample was desalted using ZipTip® pipette tips (Millipore). The sample was analyzed by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry.
Pull-down assay
To prepare receptor-like molecules, the fractionated receptor-like molecules were pulled down using an antibody directed against the molecules and a Catch and Release Reversible Immunoprecipitation System v2.0 (Millipore) according to the manufacturer’s protocol. In brief, the column resin was washed with the buffer provided. The following were then added to each column: 125 μl Caco-2 cell lysate and 4 μg anti-annexin A13 (ANXA13) antibody (Aviva Systems Biology) or 4 μg anti-paralemmin (PALM) antibody (Abnova Corporation). The columns were incubated overnight on a rotator at 4°C. MapA (1 μg) was then added to the columns with wash buffer, and the columns were incubated overnight on a rotator at 4 °C. The columns were eluted with varying concentrations of non-denaturing elution buffer contained in the kit. The eluted protein concentrations were determined, and the proteins were then used for Western blot analysis.
Fluorescence microscopy and image analysis
Caco-2 cells grown on Lab-Tek II Chamber Slide System 4-well glass slides (Nunc) were washed three times with PBS. The cells in the chambers were incubated with 10 μg/ml MapA in PBS at room temperature for 1 hr and then washed four times with PBS. As a negative control, 10 μg/ml of BSA instead of MapA was incubated with the Caco-2 cells. The cells in the chamber were fixed by adding 4% p-formaldehyde. MapA on Caco-2 cell surfaces was detected using anti-MapA antiserum from rabbit and anti-rabbit IgG-FITC (Sigma) or using anti-His (Qiagen) and anti-mouse IgG-FITC (Sigma). ANXA13 was detected using anti-ANXA13 antibody and anti-rabbit IgG-Cy3 conjugate from sheep (Sigma). PALM was detected using anti-PALM antibody and anti-mouse IgG-Cy3 conjugate from sheep (Sigma). Caveolin was detected using anti-caveolin-1 antibody (Cell Signaling Technology or Sigma) and anti-rabbit IgG-FITC. Fluorescence was analyzed using a confocal laser-scanning microscope (FV1000; Olympus). Images were processed using the software supplied with the Olympus FluoView Viewer (ver.2.1c).
Constructs for inducible short hairpin RNAs
Specific short hairpin RNAs (shRNAs) targeting the ANX A13 or PALM sequences (Table 1) were cloned into a pENTR/H1/TO entry vector using a BLOCK-iT Inducible H1 RNAi Entry Vector Kit (Invitrogen). These shRNAs were designed using a web-based software provided by Invitrogen. The constructed vectors were transferred into pLenti4/BLOCK-iT-DEST expression vectors by targeted recombination according to the manufacturer’s protocol. The pLenti expression constructs were cotransfected with packaging plasmids with Lipofectamine 2000 (Invitrogen) in the 293FT producer cell line using a ViraPower Lentiviral Expression System (Invitrogen) according to the manufacturer’s protocols. Lentivirus-containing supernatants of transfected 293FT cells were collected at 48 hr and stored at –80°C.
Table 1.
| Used applications | Primer Name | Primer sequence (5’ to 3’) |
| anx A13b-XhoI-F | CCGCTCGAGGTGGCAATCGTCATAGCCA | |
| anx A13b-NotI-R | ATAGTTTAGCGGCCGCTCAGTGCAAGAGGGCTACTAGCAG | |
| palm-XhoI-F | CCGCTCGAGGTGAGGTCCTGGCGGCAGAGA | |
| palm-NotI-R | ATAGTTTAGCGGCCGCTCACATGATGGAGCAGCATTTACA | |
| Construct of RNAi vector | anx A13-sh Top | CACCGCAGTTACGAGCCACCTTTCACGAATGAAAGGTGGCTCGTAACTGC |
| anx A13-sh Bottom | AAAAGCAGTTACGAGCCACCTTTCATTCGTGAAAGGTGGCTCGTAACTGC | |
| palm-sh Top | CACCGGACGAACTCATCCACAAAGCCGAAGCTTTGTGGATGAGTTCGTCC | |
| palm-sh Bottom | AAAAGGACGAACTCATCCACAAAGCTTCGGCTTTGTGGATGAGTTCGTCC | |
| Real Time qPCR | actin-RT-F | GTGTGACGTGGACATCCG |
| actin-RT-R | CGGCAATGCCAGGGTACAT | |
| anx A13-RT-F | GCAGTTACGAGCCACCTTTC | |
| anx A13-RT-R | GCAAGTCGCCTGATGTTTCT | |
| palm-RT-F | GTCCATGCTGTGGACGG | |
| palm-RT-R | GTCCGCTTTGTGGATGAGTT | |
Construction of stable cell lines
To generate inducible Caco-2 cell lines, we used pLent6/TR virus infection for establishing tetracycline repressor (tetR)-expressing cell lines. The virus-infected Caco-2 cells were treated with blasticidin (5–10 µg/ml; Invitrogen) for 2 weeks. Blasticidin-resistant Caco-2 cells were then selected, and their tetR mRNA levels were determined by RT-PCR. Next, we infected the tetR-expressing Caco-2 cell lines with shRNA-expressing lentivirus. During this step, tetR-expressing Caco-2 cells were treated with Zeocin (400 µg/ml; Invitrogen) for 2 weeks, and stable knockdown Caco-2 cell lines were established.
Analysis of ANXA13 or PALM mRNA expression by real-time PCR
The constructed knockdown Caco-2 cell lines were cultured for 1 week with or without 10 µg/ml tetracycline. Each cell line was then dissolved in Isogen, and partial total RNA was isolated. The partial total RNAs were reacted with DNase (RT-Grade; Wako) for 120 min at 37°C. The DNase-reacted total RNA was purified using a FastPure RNA Kit (Takara). Real-time reverse transcription PCR was performed using One Step SYBR PrimeScript RT-PCR Kit II (Takara), 100 ng of total RNA, and an primer set in a 20-μl reaction volume. The mRNA expression values of these proteins were analyzed by the 2-ΔΔCt method; the β-actin gene (NM_001101) was used as a reference. Real-time quantitative PCR was performed using a LightCycler 2.0 real-time PCR system (Roche). The amplification steps were as follows: reverse transcription at 42°C for 5 min, followed by 40 cycles of 95°C for 5 sec, 58°C for 15 sec, and 72°C for 10 sec, with a temperature transition rate of 20°C/sec.
Assay of MapA binding to Caco-2 cell lines
The amount of MapA binding was confirmed in an immunofluorescence assay. Caco-2 cell lines (RNAi-inducible Caco-2 cell line and RNAi-uninducible Caco-2 cell line) were cultured on 96-well plates (Nunc) for 15 days. The cells on each plate well were incubated with 100 µl of MapA (final concentrations of 25, 50 or 100 µg/ml in PBS) at 37°C for 1 hr. As a negative baseline, MapA was not added to 8 wells for each Caco-2 cell line. MapA bound to the Caco-2 cell surfaces was detected using anti-MapA antibody and anti-rabbit IgG-Cy3. Fluorescence intensity was determined using a CytoFluor Series 4000 Fluorescence Multi-Well Plate Reader (Applied Biosystems); this was used as a measure of the amount of MapA bound to Caco-2 cells per well.
Adhesion of L. reuteri DSMZ 20016T to Caco-2 cell lines
Monolayers of wild-type and knockdown Caco-2 cell lines were prepared on 96-well plates (Nunc) and then washed two times with prewarmed PBS. L. reuteri DSMZ 20016T was grown overnight at 37°C in Man–Rogosa–Sharpe (MRS) broth (Difco). The bacterial culture was centrifuged at 5,000 × g for 5 min, and the pellet was washed two times in sterile PBS. The pellet was then adjusted to a cell concentration of 2 × 108 cells/ml. The resuspended L. reuteri DSMZ 20016T cells were added to each well and then incubated at 37°C for 1 hr with gentle rocking. After incubation, the cell monolayers were washed gently three times with PBS, fixed with methanol, and stained with 0.1% methylene blue. Bacterial cells adhering to the Caco-2 cell lines were counted under a light microscope. The number of bacteria adhering to 10 cells in each of 50 randomly selected fields was counted.
RESULTS
Isolation and identification of receptor-like molecules
To investigate the presence of MapA-binding molecules on Caco-2 cells, lysed Caco-2 cells were bound to MapA in a pull-down assay. Proteins in eluted samples were separated by SDS-PAGE and visualized by SYPRO Ruby staining. We confirmed that binding had occurred between MapA and receptor-like molecules (data not show). We tested the ability of 1% NP-40 buffer to lyse Caco-2 cells but were unable to detect MapA binding (data not shown). However, binding between MapA and receptor-like molecules was observed when cells were lysed with RIPA buffer containing NP-40. Therefore, we suspected that the MapA-binding receptor-like molecules isolated were hydrophobic proteins. Moreover, binding between MapA and the receptor-like molecules was inhibited by SDS.
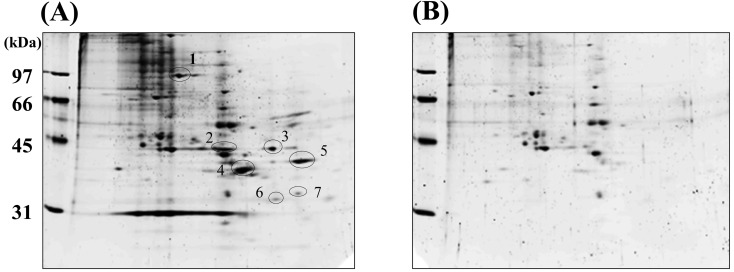
To identify these receptor-like molecules, we used 2D-PAGE; 7 spots were isolated from the Caco-2 cell membrane, which represented MapA-binding receptor-like molecules (Fig. 1). These spots were eluted from the MapA-bound resin. Thus, we verified that the 7 spots were MapA-binding receptor-like molecules.
Fig. 1.
Purification of MapA-associated proteins by 2D-PAGE. The first-dimension separation was based on isoelectric focusing (pH: 4 to 6), and the second-dimension separation was performed using 10% SDS-PAGE. Gels were stained with SYPRO Ruby (Bio-Rad). (A) Elution of MapA-immobilized resin. (B) Elution of non-immobilized resin as a negative control. Arrows indicate MapA-associated proteins from Caco-2 cell lysates.
To identify the proteins, these spots were analyzed by MALDI-TOF. To analyze the MALDI-TOF data, we performed a database search using the PeptIdent tool of the ExPASy Proteomics Server. We were only able to identify spot numbers 4 and 5. Spot 4 was identified as ANXA13, and spot 5 was PALM. Therefore, we considered that ANXA13 and PALM were MapA-binding receptor-like molecules.
mRNA expression of receptor-like molecules on Caco-2 cells
We examined Caco-2 cells for mRNA expression of MapA-binding receptor-like molecules (ANXA13 and PALM). The amplified bands were excised from a 1% agarose gel and sequenced (data not show). The DNA sequences matched those of ANXA13 (NM_004306) and PALM (NM_002579, NM_001040134). PALM was expressed as two transcript variants by Caco-2 cells.
ANXA13 and PALM interact with MapA
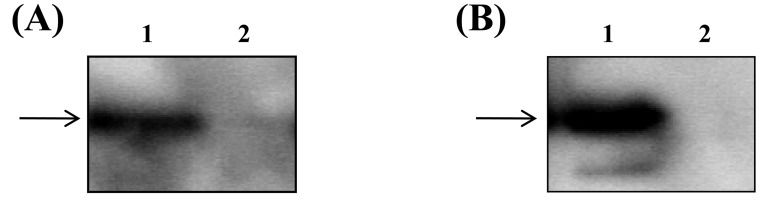
To confirm the interactions between MapA and the receptor-like molecules, we purified ANXA13 and PALM and used them in a co-immunoprecipitation assay. The receptor-like molecules in Caco-2 cell lysates were purified on resin using specific antibodies. MapA was then added to the purified ANXA13 and PALM resins. Resin without specific antibodies to the receptor-like molecules was used as a negative control. In Western blot analysis, MapA was detected only in the presence of the ANXA13 or PALM fraction (Fig. 2). This indicates that MapA binds directly to both ANXA13 and PALM.
Fig. 2.
(A) Pull-down assay. Lane 1: Annexin A13 added; Lane 2: Same as lane1 but without annexin A13 (lane 2). Samples were subjected to immunoprecipitation, and the resulting precipitates were subjected to immunoblot analysis using anti-MapA antibody. (B) Pull-down assay. Lane 1: Paralemmin added as a concentration gradient. Lane 2: Same as lane1 but without paralemmin. Samples were subjected to immunoprecipitation, and the resulting precipitates were subjected to immunoblot analysis with anti-MapA antibody.
MapA and receptor-like molecules colocalize on Caco-2 cells
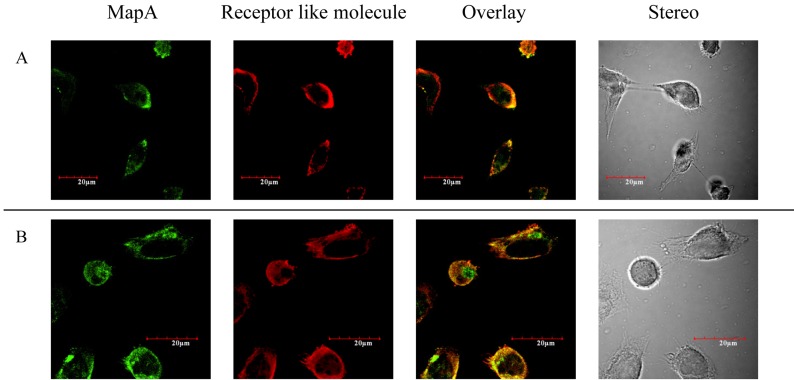
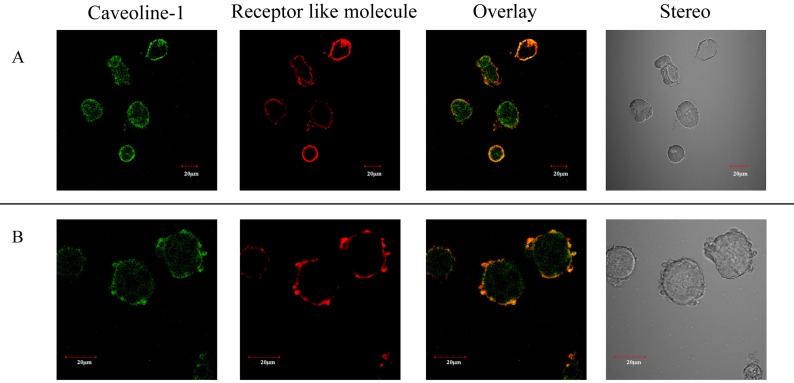
Binding between MapA and the receptor-like molecules was observed by CLSM. Nonspecific background fluorescence due to nonselective adsorption to surface antigens on Caco-2 cells was not detected under the same excitation intensity (data not shown). This suggests that ANXA13 and PALM are expressed on the Caco-2 cell membrane (Fig. 3). ANXA13 is reportedly involved in vesicular transport [17], and our observations with Caco-2 cells indicated that ANXA13 is localized in the Golgi network, which supports this hypothesis. PALM was also observed on the Caco-2 cell membrane. Therefore, MapA is localized on the Caco-2 cell membrane and is colocalized with ANXA13 and PALM (Fig. 3A, B). These receptor-like molecules have also been reported to be located within membrane microdomains [18]. Thus, we investigated the localization of caveolin as an indicator of the locations of membrane microdomains, and thereby determined the locations of the receptor-like molecules. Caveolin and MapA-binding receptor-like molecules were colocalized on the Caco-2 cell membrane (Fig. 4A, B).
Fig. 3.
(A) MapA was immunostained with anti-His antibody (green). Localization of anti-annexin A13 immunostaining (red) is shown in Caco-2 cells. Annexin A13 and MapA are colocalized (yellow). (B) MapA was immunostained with anti-MapA antibody (green). Localization of anti-paralemmin immunostaining (red) is shown in Caco-2 cells. Paralemmin and MapA are colocalized (yellow). Bars = 20 μm.
Fig. 4.
(A) Caveoline-1 was immunostained with anti- Caveoline-1 antibody (green). Localization of anti-annexin A13 immunostaining (red) is shown in Caco-2 cells. Annexin A13 and Caveoline-1 are colocalized (yellow). (B) Caveoline-1 was immunostained with anti- Caveoline-1 antibody (green). Localization of anti-paralemmin immunostaining (red) is shown in Caco-2 cells. Paralemmin and Caveoline-1 are colocalized (yellow). Bars = 20 μm.
Construction of knockdown Caco-2 cell lines
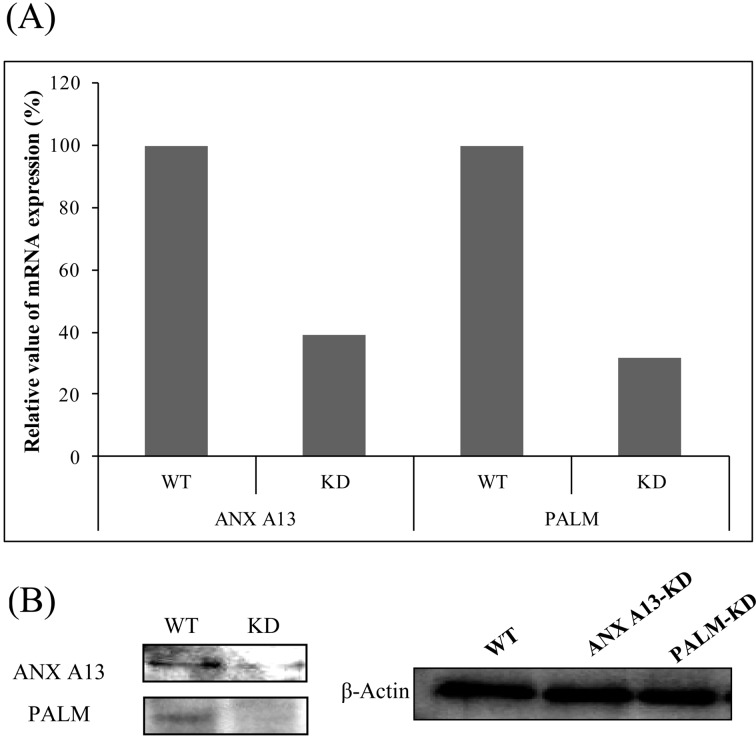
To examine the adhesion of MapA to Caco-2 cells in more detail, we used lentiviral delivery to create stable knockdown of ANXA13 and PALM. Compared with wild-type cells, the stable knockdown Caco-2 cell lines exhibited lower ANXA13 (60.8% mean reduction) and PALM (68.4% mean reduction) mRNA levels (Fig. 5A). We also performed Western blot analysis of ANXA13 and PALM protein expression levels; these levels were significantly reduced in ANXA13 and PALM knockdown cells compared with wild-type cells (Fig. 5B).
Fig. 5.
(A) Relative mRNA expression levels of annexin A13 (ANXA13) and paralemmin (PALM) in knockdown cells, as estimated by qPCR. As a reference, the level of mRNA expression in the wild-type Caco-2 cell line was set at 1.0. β-Actin was used as a reference (housekeeping) gene to determine ΔCT, and the results were expressed as 2-ΔΔCT for both RNAi Caco-2 cell lines. (B) Whole cell lysates were analyzed by Western blot analysis with antibodies specific for ANXA13, PALM, and β-actin. WT: wild type.
MapA adherence to Caco-2 cell lines
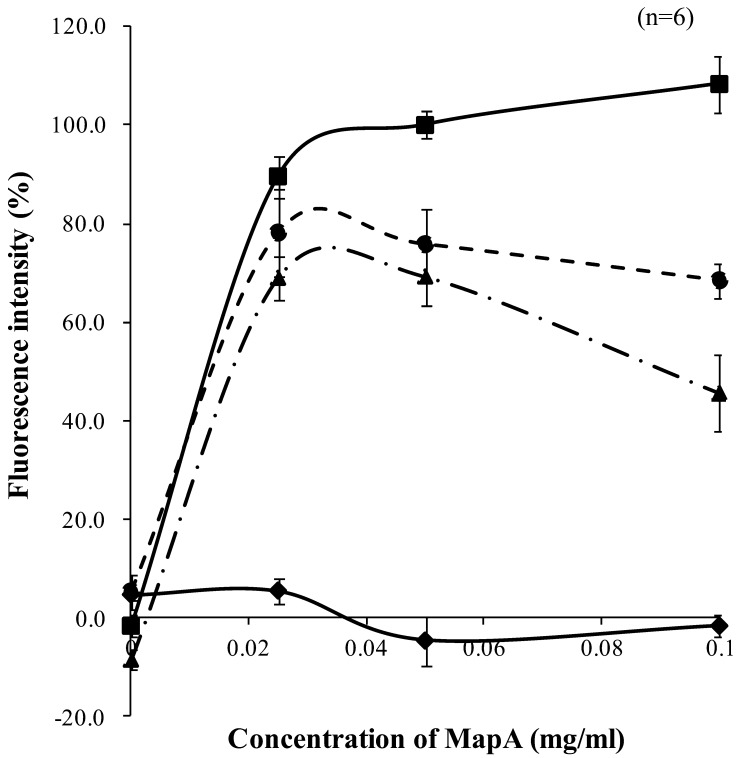
We investigated the effects of downregulation of ANXA13 and PALM expression on the adhesion of MapA to these knockdown cell lines. We used a fluorescent dye-labeled antibody to quantify the in vitro adhesion of MapA to Caco-2 cells. Fluorescence intensity increased in proportion to MapA concentration (Fig. 6). In contrast, compared with its adhesion to wild-type Caco-2 cells, adhesion of MapA to the ANXA13 and PALM knockdown cell lines was reduced by 24.2% and 30.7%, respectively. This suggests that MapA binding to Caco-2 cells requires ANXA13 and PALM on the Caco-2 cell membrane.
Fig. 6.
Quantitative assay of MapA binding to Caco-2 cells. Puried MapA was incubated with Caco-2 monolayers (■), annexin A13 knockdown Caco-2 monolayers (●), or paralemmin knockdown Caco-2 monolayers (▲). The amount of MapA bound to Caco-2 cells was determined by uorescence intensity using a fluorescent dye-labeled anti-MapA antibody. Intensities are shown as percentages, with that from 0.05 mg/ml MapA set at 100%. Results are means + 1 standard deviation of 6 samples. (♦) Caco-2 monolayers incubated with bovine serum albumin.
Adhesion of L. reuteri to human intestinal epithelial cells
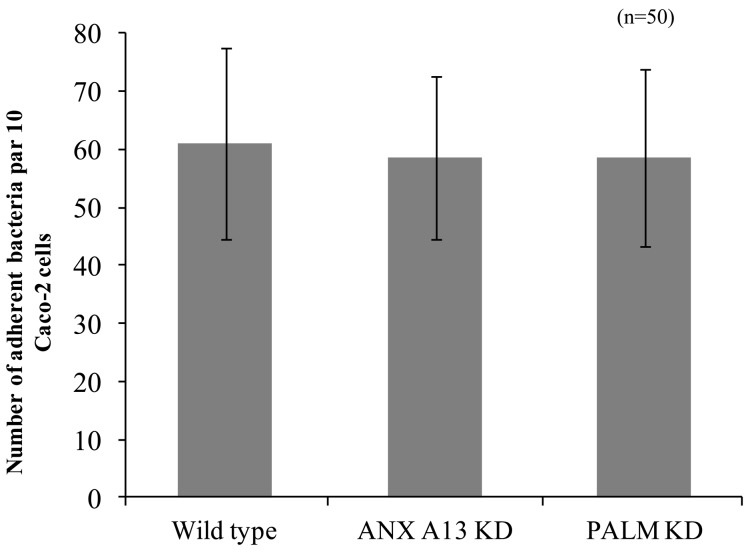
We compared the adherence of L. reuteri DSMZ 20016T to wild-type Caco-2 cells and ANXA13 and PALM knockdown Caco-2 cell lines. Microscopic observations showed that L. reuteri DSMZ 20016T adhered to Caco-2 cells [61.0 ± 16.4 (average ± standard deviation) bacteria per 10 Caco-2 cells]. The number of L. reuteri DSMZ 20016T cells adhering to the ANXA13 and PALM knockdown Caco-2 cell lines were 58.6 ± 14.0 and 58.5 ± 15.3 bacteria per 10 Caco-2 cells (Fig. 7).
Fig. 7.
Adhesion of L. reuteri to the two Caco-2 cell lines. Light microscopy was used to count the number of L. reuteri DSMZ 20016T that adhered to 10 cells for each of the Caco-2 cell lines. Results are means ± 1 standard deviation (error bars) of 50 samples.
DISCUSSION
L. reuteri is one of the dominant lactobacillus species that is a part of the normal microbiota in the GITs of numerous animals. It expresses MapA, which can bind to biogenic molecules. However, the mechanisms by which L. reuteri adheres to intestinal epithelial cells are unknown.
In this study, we used Caco-2 cells, which are widely used in adhesion studies of LAB [19]. We used a pull-down assay to investigate the MapA-binding receptor-like molecules obtained from Caco-2 cell lysates. These molecules could be lysed in the presence of SDS and sodium deoxycholate, although these surfactants interfered with the binding of these receptor-like molecules to MapA. However, these molecules could bind to MapA in diluted RIPA buffer containing NP buffer (1% NP-40). This indicates that the MapA-binding receptor-like molecules are hydrophobic. Several MapA-binding molecules were detected by 2D-PAGE; among them, ANXA13 and PALM were identified by MALDI TOF-MS.
ANXA13 and PALM were isolated from Caco-2 cells. The co-immunoprecipitation binding assay showed that MapA bound directly to ANXA13 and PALM. However, this assay was performed under specific conditions in which the Caco-2 cells were solubilized. Therefore, we used CFLM to confirm MapA binding to ANXA13 and PALM on the Caco-2 cell membrane. These results showed that MapA localized with ANXA13 and PALM on the membrane, which suggested that the binding occurred not only under lysis conditions but also under physiological conditions.
In addition, we investigated whether these receptor-like molecules were important for MapA adhesion to Caco-2 cells. Because ANXA13 and PALM were localized on the Caco-2 cell membrane, it was possible that the shapes of the Caco-2 cells would be changed if these receptor-like molecules were knocked down. Therefore, we used RNAi to inhibit mRNA expression of these receptor-like molecules.
To determine the role of these molecules in adhesion, we constructed ANXA13 and PALM knockdown cell lines and used them in an adhesion assay. Compared with the MapA adhesion to normal Caco-2 cells, ANXA13 or PALM downregulation resulted in reduced MapA adhesion to knocked down Caco-2 cells. This indicates that MapA is bound to ANXA13 and PALM on the Caco-2 cell membrane and that efficient adhesion of MapA to Caco-2 cells depends on the presence of these receptor-like molecules.
ANXA13 is expressed in the GIT and is localized in the intestinal epithelia cell membrane [17]. Lipid modification of the N terminus of ANXA13 by myristoylation suggests that ANXA13 regulates vesicular transport [20,21,22]. PALM plays a role in the central nervous system, as revealed by prenylation and palmitoylation of its C terminus [23, 24]. One study using an epithelial cell line showed that these proteins were located in membrane microdomains. Lipid rafts form part of these membrane microdomains, and floating lipid rafts on the cell membrane are involved in the transfer of various membrane proteins [25, 26]. The incorporation of receptor-like molecules into caveolae—specialized lipid rafts—is likely to promote signaling by these molecules. Caveolae are parts of the membrane microdomain that have abundant amounts of caveolin [27].
It has been suggested that these membrane microdomains are the points of cellular contact with pathogens such as E. coli O157 and influenza viruses [28,29,30,31]. Many pathogens use these lipid rafts and the endocytosis functions of caveolae to infect host cells [32, 33]. For example, Campylobacter jejuni uses this pathway [34]. Thus, we considered that L. reuteri probably adheres to these membrane microdomains via MapA. We observed the localization of receptor-like molecules for MapA with canavalin-1 on Caco-2 cell membranes. ANXA13 and CAV-1 reportedly colocalize on Madin–Darby canine kidney (MDCK) cells, and we obtained similar results with Caco-2 cells [35].
In addition, toll-like receptor (TLR) 2 is expressed on the microdomains of epithelial and immune cells, and TLR9, which is expressed in the cytoplasm, recognizes the chromosomal CpG DNA motif of bacteria [36, 37]. Gram-positive bacteria such as Staphylococcus aureus and Streptococcus group A recognize the epithelial receptor asialo GM1 and induce inflammatory cytokine production via TLR2 [38, 39]. Moreover, asialo GM1 is a receptor-like molecule for L. reuteri [40]. These findings support the hypothesis that L. reuteri inhibits the adhesion of particular pathogens.
We also examined the effect of these receptor-like molecules on the adhesion of L. reuteri DSMZ 20016T to Caco-2 cell monolayers. The numbers of L. reuteri DSMZ 20016T cells that adhered to wild-type Caco-2 cells and receptor-like molecule knockdown Caco-2 cell lines were similar. On the knocked down Caco-2 cell membrane, other receptor-like molecules may bind to MapA, even if ANXA13 and PALM are knocked down (Fig. 7). On the surface layer of L. reuteri DSMZ 20016T, MapA may bind not as isolated receptor-like molecules, as shown in this study, but as other receptor-like molecules. MapA bound to a lot of acidic proteins as ANXA13 (pI 5.38) and PALM (pI 4.92) in receptor-like molecule isolated from Caco-2 cells (Fig. 1). Since MapA is a basic protein (pI 9.7), we think that binding with receptor-like molecules is electrostatic interaction under neutral condition. But if the binding with MapA and receptor-like molecules was only electrostatic interaction, MapA could have bound with a lot of acidic protein in this study (Fig.1). Taken together, we infer that the isolated receptor-like molecules have some sort of common denominator other than electrostatic interaction.
The binding between isolated receptor-like molecules and MapA was highly sensitive to the effects of surfactants. Our evaluations of the adhesion of LAB to host cells in vitro suggest empirically that strong washing could tear bacterial cells away from epithelial cell membranes. These results and our empirical observations suggest that intestinal adhesion of LAB does not result from specific binding mechanisms, as in the case of pathogenic organisms, but occurs by some weak binding mechanism(s). In future studies, we will examine in particular the weak binding mechanisms between MapA and the two receptor-like molecules identified here.
REFERENCES
- 1.Mitsuoka T. 1992. Intestinal flora and aging. Nutr Rev 50: 438–446 [DOI] [PubMed] [Google Scholar]
- 2.Gibson GR, Fuller R. 2000. Aspects of in vitro and in vivo research approaches directed toward identifying probiotics and prebiotics for human use. J Nutr 139: 391S–395S [DOI] [PubMed] [Google Scholar]
- 3.Casas IA, Dobrogosz WJ. 2000. Validation of the probiotic concept: Lactobacillus reuteri confers broad-spectrum protection against disease in humans and animals. Microb Ecol Health Dis 12: 247–285 [Google Scholar]
- 4.Chen CN, Sung HW, Liang HF, Chang WH. 2002. Feasibility study using a natural compound (reuterin) produced by Lactobacillus reuteri in sterilizing and crosslinking biological tissues. J Biomed Mater Res 61: 360–369 [DOI] [PubMed] [Google Scholar]
- 5.El-Ziney M, Debevere J. 1998. The effect of reuterin on Listeria monocytogenes and Escherichia coli O157:H7 in milk and cottage cheese. J Food Prot 61: 1275–1280 [DOI] [PubMed] [Google Scholar]
- 6.Maasen CBM, van Holten-Neelen C, Balk F, den Bak-Glashouer MJH, Leer RJ, Laman JD, Boersma WJA, Claassen E. 2000. Strain-dependent induction of cytokine profiles in the gut by orally administered Lactobacillus strains. Vaccine 18: 2613–2623 [DOI] [PubMed] [Google Scholar]
- 7.Taranto MP, Medici M, Perdigon G, Ruiz Holgado AP, Valdez GF. 1998. Evidence for hypocholesterolemic effect of Lactobacillus reuteri in hypercholesterolemic mice. J Dairy Sci 81: 2336–2340 [DOI] [PubMed] [Google Scholar]
- 8.Valeur N, Engel P, Carbajal N, Connolly E, Ladefoged K. 2004. Colonization and immunomodulation by Lactobacillus reuteri ATCC 55730 in the human gastrointestinal tract. Appl Environ Microbiol 70: 1176–1181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wolf BW, Wheeler KB, Ataya DG, Garleb KA. 1998. Safety and tolerance of Lactobacillus reuteri supplementation to a population infected with the human immunodeficiency virus. Food Chem Toxicol 36: 1085–1094 [DOI] [PubMed] [Google Scholar]
- 10.Henriksson A, Conway PL. 1992. Adhesion to porcine squamous epithelium of saccharide and protein moieties of Lactobacillus fermentum strain 104-S. J Gen Microbiol 138: 2657–2661 [DOI] [PubMed] [Google Scholar]
- 11.Henriksson A, Conway PL. 1996. Adhesion of Lactobacillus fermentum 104-S to porcine stomach mucus. Curr Microbiol 33: 31–34 [DOI] [PubMed] [Google Scholar]
- 12.Satoh E, Leer RJ, Conway PL, Pouwels PH. 1999. Mucus adhesion promoting protein of Lactobacillus reuteri 104R. In Sixth Symposium on Lactic Acid Bacteria; Genetics, Metabolism and Applications, Veldhoven, Book of Abstracts, J39.
- 13.Aleljung P, Shen W, Rozalska B, Hellman U, Ljungh A, Wadstrom T. 1994. Purification of collagen-binding proteins of Lactobacillus reuteri NCIB 11951. Curr Microbiol 28: 231–236 [DOI] [PubMed] [Google Scholar]
- 14.Roos S, Aleljung P, Robert N, Lee B, Wadstrom T, Lindberg M, Jonsson H. 1996. A collagen binding protein from Lactobacillus reuteri is part of an ABC transporter system? FEMS Microbiol Lett 144: 33–38 [DOI] [PubMed] [Google Scholar]
- 15.Rojas M, Conway PL. 1996. Colonisation by Lactobacillus of piglet small intestinal mucus. J Appl Bacteriol 81: 474–480 [DOI] [PubMed] [Google Scholar]
- 16.Miyoshi Y, Okada S, Uchimura T, Satoh E. 2006. A mucus adhesion promoting protein, MapA, mediates the adhesion of Lactobacillus reuteri to Caco-2 human intestinal epithelial cells. Biosci Biotechnol Biochem 70: 1622–1628 [DOI] [PubMed] [Google Scholar]
- 17.Lafont F, Lecat S, Verkade P, Simons K. 1998. Annexin Xlllb associates with lipid microdomains to function in apical delivery. J Cell Biol 142: 1413–1427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kutzleb C, Petrasch-Parwez E, Kilimann MW. 2007. Cellular and subcellular localization of PALM-1, a protein involved in cell shape control, in the rat brain, adrenal gland and kidney. Histochem Cell Biol 127: 13–30 [DOI] [PubMed] [Google Scholar]
- 19.Greene JD, Klaenhammer TR. 1994. Factors involved in adherence of lactobacilli to human Caco-2 cells. Appl Environ Microbiol 60: 4487–4494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moss SE, Morgan RO. 2004. The annexins. Genome Biol 5: 219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Iglesias JM, Morgan RO, Jenkins NA, Copeland NG, Gilbert DJ, Fernandez MP. 2002. Comparative genetics and evolution of annexin A13 as the founder gene of vertebrate annexins. Mol Biol Evol 19: 608–618 [DOI] [PubMed] [Google Scholar]
- 22.Morgan RO, Martin-Almedina S, Iglesias JM, Gonzalez-Florez MI, Fernandez MP. 2004. Evolutionary perspective on annexin calcium-binding domains. Biochim Biophys Acta 1742: 133–140 [DOI] [PubMed] [Google Scholar]
- 23.Hu B, Copeland NG, Gilbert DJ, Jenkins NA, Kilimann MW. 2001. The PALM protein family: identification of PALM-2, an isoform differentially spliced to AKAP2/AKAP-KL, and of palmdelphin, a more distant cytosolic relative. Biochem Biophys Res Commun 285: 1369–1376 [DOI] [PubMed] [Google Scholar]
- 24.Kutzleb C, Sanders G, Yamamoto R, Wang X, Lichte B, Petrasch-Parwez E, Kilimann MW. 1998. Paralemmin, a prenyl-palmitoyl-anchored phosphoprotein abundant in neurons and implicated in plasma membrane dynamics and cell process formation. J Cell Biol 143: 795–813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nakahata N, Ohkubo S. 2003. Lipid rafts and their analytical methods. Nippon Yakurigaku Zasshi 122: 419–125 [DOI] [PubMed] [Google Scholar]
- 26.Simons K, Ikonen E. 1997. Functional rafts in cell membranes. Nature 387: 569–572 [DOI] [PubMed] [Google Scholar]
- 27.Williams TM, Lisanti MP. 2004. The caveolin proteins. Genome Biol 5: 214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fournier B, Philpott DJ. 2005. Recognition of Staphylococcus aureus by the innate immune system. Clin Microbiol Rev 18: 521–540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jin H, Subbarao K, Bagai S, Leser GP, Murphy BR, Lamb RA. 1996. Palmitylation of the influenza virus hemagglutinin (H3) is not essential for virus assembly or infectivity. J Virol 70: 1406–1414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mukai T, Kaneko S, Ohori H. 1998. Haemagglutination and glycolipid-binding activities of Lactobacillus reuteri. Lett Appl Microbiol 27: 130–134 [DOI] [PubMed] [Google Scholar]
- 31.Tohno M, Shimosato T, Moue M, Aso H, Watanabe K, Kawai Y, Yamaguchi T, Saito T, Kitazawa H. 2006. Toll-like receptor 2 and 9 are expressed and functional in gut-associated lymphoid tissues of presuckling newborn swine. Vet Res 37: 791–812> [DOI] [PubMed] [Google Scholar]
- 32.Lafont F, van der Goot FG. 2005. Bacterial invasion via lipid rafts. Cell Microbiol 7: 613–620 [DOI] [PubMed] [Google Scholar]
- 33.Zaas DW, Duncan M, Rae Wright J, Abraham SN. 2005. The role of lipid rafts in the pathogenesis of bacterial infections. Biochim Biophys Acta 1746: 305–313 [DOI] [PubMed] [Google Scholar]
- 34.Wooldridge KG, Williams PH, Ketley JM. 1996. Host signal transduction and endocytosis of Campylobacter jejuni. Microb Pathog 21: 299–305 [DOI] [PubMed] [Google Scholar]
- 35.Verkade P, Harder T, Lafont F, Simons K. 2000. Induction of caveolae in the apical plasma membrane of Madin-Darby canine kidney cells. J Cell Biol 148: 727–739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Soong G, Reddy B, Sokol S, Adamo R, Prince A. 2004. TLR2 is mobilized into an apical lipid raft receptor complex to signal infection in airway epithelial cells. J Clin Invest 113: 1482–1489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shimosato T, Kitazawa H, Saito T. 2005. Immunoactivity of DNA from immunobiotic lactic acid bacteria via Toll-like receptor 9 and potential applications. Milk Sci 54: 9–15 [Google Scholar]
- 38.Fournier B, Philpott D. 2005. Recognition of Staphylococcus aureus by the innate immune system. Clin Microbiol Rev 18: 521–540> [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Molinari G, Talay SR, Valentin-Weigand P, Rohde M, Chhatwal GS. 1997. The fibronectin-binding protein of Streptococcus pyogenes, SfbI, is involved in the internalization of group A streptococci by epithelial cells. Infect Immun 65: 1357–1363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mukai T, Asasaka T, Sato E, Mori K, Matsumoto M, Ohori H. 2002. Inhibition of binding of Helicobacter pylori to the glycolipid receptors by probiotic Lactobacillus reuteri. FEMS Immunol Med Microbiol 32: 105–110 [DOI] [PubMed] [Google Scholar]