Abstract
Mitotic cell division is the most fundamental task of all living cells. Cells have intricate and tightly regulated machinery to ensure that mitosis occurs with appropriate frequency and high fidelity. A core element of this machinery is the kinesin-5 motor protein, which plays essential roles in spindle formation and maintenance. In this review, we discuss how the structural and mechanical properties of kinesin-5 motors uniquely suit them to their mitotic role. We describe some of the small molecule inhibitors and regulatory proteins that act on kinesin-5, and discuss how these regulators may influence the process of cell division. Finally, we touch on some more recently described functions of kinesin-5 motors in non-dividing cells. Throughout, we highlight a number of open questions that impede our understanding of both this motor's function and the potential utility of kinesin-5 inhibitors.
Keywords: kinesin, mitosis, microtubule, tpx2, phosphorylation
I. Kinesin-5 plays a critical role in the mitotic spindle
The mitotic spindle is a complex multi-protein machine made up of microtubule (MT) filaments, motor proteins that walk along and organize these filaments, nonmotor microtubule-associated proteins (MAPs) and various non-structural signaling molecules. MTs act as the major structural scaffold of the mitotic spindle, while the dynamic properties of the mitotic spindle depend upon the variety of proteins that attach to MTs in different regions of the mitotic spindle.
While hundreds of components have been shown to associate with the mitotic spindle (Sauer et al., 2005), theoretical and experimental evidence both point toward a central role for kinesin-5. In simulations, only four mechanical activities are required to establish and maintain a stable MT-based spindle: 1) extension and retraction of MTs, 2) a pole cohesion factor that pulls the minus-ends of MTs together, 3) the MT cross-linking force of a minus-end directed motor protein, dynein, and 4) an outward-directed force between interpolar MTs, generated by kinesin-5 motors (Loughlin et al., 2010) (Figure 1A). An RNAi screen of all the MT-based motor proteins identified only three that were absolutely required for completion of mitosis: kinesin-5, kinesin-6 (which is involved in separating the two daughter cells) and kinesin-8 (which acts to shorten MTs) (Goshima , Vale, 2003). Thus, determining how kinesin-5 functions is a critical task as we seek to understand how mitosis works and how it can be altered for therapeutic interventions.
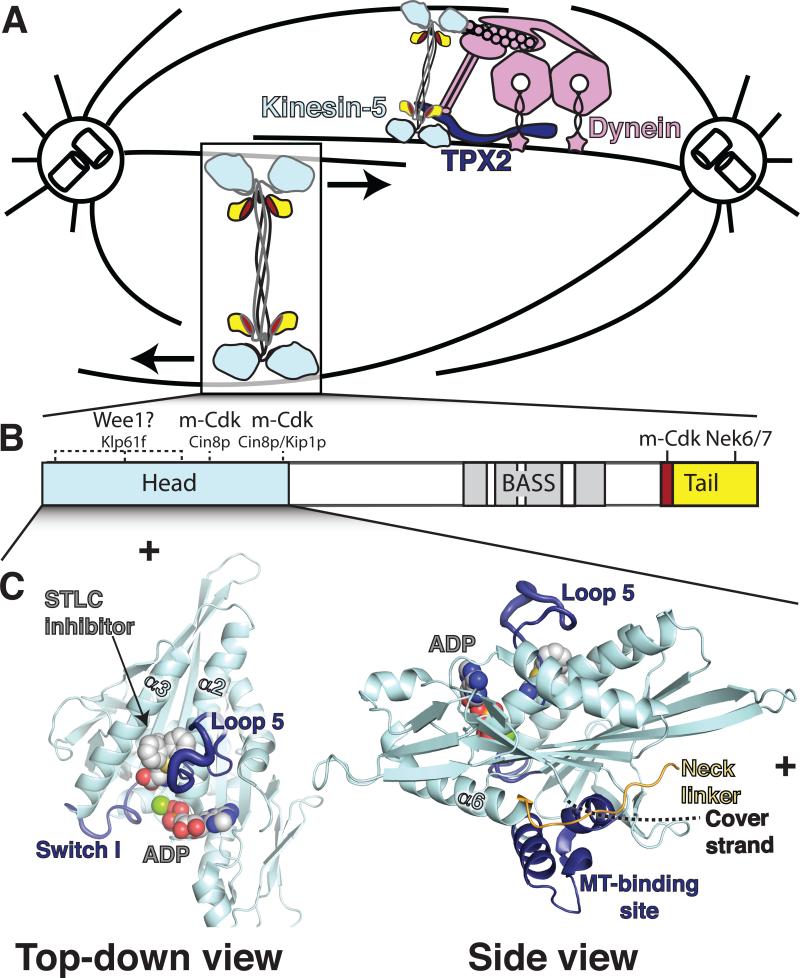
Figure 1. Kinesin-5 structure, function, and regulation.
A. Kinesin-5 slides spindle poles apart during mitosis. The motor associates with dynein (pink) and TPX2 (dark blue) for proper localization. The Eg5 tail interacts with the p150 glued subunit of dynactin (Blangy et al., 1997). It is not which domain(s) of Eg5 interact with TPX2. B. Bar diagram of kinesin-5, showing conserved domains and known phosphorylation sites. Coloring is as in A with the BimC box in red. The BASS domain has only been identified in the D. melanogaster homolog Klp61F and is shown in gray and white (*Acar et al., 2013). Phosphorylation sites in the heads are only known to occur in non-mammalian kinesin-5 homologs. These are indicated with dashed lines along with the homolog. Tail phosphorylation sites are conserved among metazoan kinesin-5 motors and are shown in solid lines. C. Left, Eg5 head structure (PDB ID#3KEN, (Kim et al., 2010) showing Loop 5 and the binding site for allosteric inhibitors. This view would be top-down for a MT-bound Eg5 head, with the MT plus end up as indicated. Right, side view, with MT plus end to the right. The MT-binding site is at the bottom of the molecule. The neck linker is shown in orange. The cover strand is not ordered in this structure, but its approximate position is shown in dashed lines, based on the work of Goulet and Hesse (*Goulet et al., 2012; Hesse et al., 2013).
Despite the core function shared by all eukaryotic kinesin-5 homologs, each protein seems to play a somewhat different role in its organism's mitotic process. In S. cerevisiae and Drosophila embryos, overexpression of kinesin-5 increases the metaphase spindle length (Brust-Mascher et al., 2009; Saunders et al., 1997; Straight et al., 1997); while in other cell types, including Drosphila S2 cells, spindle length is unaffected by overexpression of kinesin-5 (Goshima, Vale, 2005). In both S. cerevisiae and D. melanogaster embryos, kinesin-5 is responsible for pushing spindle poles apart in anaphase (Brust-Mascher et al., 2009; Straight et al., 1997). In contrast, mammalian cells require kinesin-5 in early mitosis for centrosome separation (Blangy et al., 1995; Tanenbaum et al., 2008). Unlike the other kinesin-5 family members, which are essential for mitosis and generate outward force to power spindle separation, the C. elegans kinesin-5 BMK-1 does not appear to be required for mitosis and acts to slow the rate of spindle extension (Saunders et al., 2007). The kinesin-5 motor homologs in these organisms are generally similar, based on primary sequence. Determining how these motors fulfill their basic role of pushing mitotic spindle poles apart and identifying what variations on this theme occur in different organisms remain major outstanding questions for the field. In our view, addressing these questions requires a focus on the mechanism and regulation of kinesin-5 motors, as well as on their interplay with other mitotic proteins.
II. Molecular anatomy of kinesin-5
Kinesin-5 is a homotetrameric protein, with each subunit containing an N-terminal kinesin motor domain, a central stalk domain and a C-terminal tail domain (Figure 1B. These subunits are thought to arrange themselves into bipolar homotetramers, with a pair of motor domains on either end. While only the D. melanogaster kinesin-5 Klp61F and the S. cerevisiae kinesin-5 Kip1 have been directly observed to form bipolar homotetramers, vertebrate kinesin-5s slide two MTs antiparallel to each other in microscopy assays, consistent with such an organization (*Acar et al., 2013; Gordon , Roof, 1999; *Kapitein et al., 2005; Kashina et al., 1996).
The N-terminal kinesin-5 motor domain is ~350 residues long and has all of the conserved structural elements that form the engine of every kinesin (Vale , Milligan, 2000). All kinesin motor domains, myosin motors and small GTPases have a central beta-sheet flanked by alpha helices. These proteins also share several critical structural elements that translate nucleotide hydrolysis into conformational changes. Several of these structural features are of particular interest for kinesin-5 motors and are discussed in detail in this review. They are identified here from the N-terminus to the C-terminus of the motor domain and are shown in Figure 1C
Cover strand: The cover strand is a short, ~ 5-10 residue segment at the N-terminus of the motor domain. This element is not often visualized in kinesin crystal structures, but has been shown to be critical for forward movement in kinesin-1 motors (Khalil et al., 2008). The cover strand forms a short beta-sheet segment, referred to as the cover neck bundle, with the neck linker during forward movement by both kinesin-1 and the human kinesin-5, Eg5 (*Goulet et al., 2012; Hesse et al., 2013).
Loop 5: The alpha 2 helix of all kinesin motors breaks into a short loop, called loop 5, near the nucleotide pocket. The sequence of this loop is kinesin family-specific, and it is unclear whether loop 5 has a conserved function for all kinesins. In kinesin 5 motors, loop 5 has been implicated in ADP release (Waitzman et al. 2011) and in communicating changes in nucleotide state to the neck linker and microtubule-binding regions of the motor domain (*Goulet et al., 2012).
Switch I and switch II: Switch I and Switch II are conserved nucleotide-sensing elements found in all kinesins, myosins, and small GTPases. These elements have been shown to play a role in nucleotide-induced conformational changes. Switch I plays a large role in mediating nucleotide release, which is stimulated by exchange factors in all of these enzymes (Kull , Endow, 2002; Vale , Fletterick, 1997). Several allosteric inhibitors of Eg5, like the STLC molecule shown in Figure 1C, bind in a groove between loop 5, alpha 2, alpha 3, and switch I and strongly affect microtubule-stimulated ADP release (Cochran et al. 2005). Switch II plays a role in binding to exchange factors, namely MTs for kinesins. Switch II communicates changes in motor nucleotide state to the relay helix within the MT-binding site and to neck linker, which drives forward motility (Kull , Endow, 2002; Sindelar , Downing, 2010).
Neck linker: The neck linker element undergoes nucleotide- and microtubule-dependent conformational changes to result in the directed movement of all kinesin motors (Endres et al., 2006; *Goulet et al., 2012; Rice et al., , 1999; Vinogradova , 2004). This directed movement is thought to be a coordinated, hand-over-hand movement of the motor domains for both kinesin-1 and the human kinesin-5 motor Eg5 (Vale et al., 1996; *Valentine et al., 2006), and the neck linker is critical to this coordination between motor heads (Jiang et al., 1997; Yildiz et al., 2008).
Given the importance of the human kinesin-5, Eg5 as a drug target, the kinesin-5 motor domain has been the subject of many crystallization screens and its mechanochemical mechanism has been intensively studied. The useful trove of structural and mechanistic data on kinesin-5 has informed our understanding of both its mechanism in particular and the enzymology of kinesins and ATPases in general. An X-ray crystal structure of Eg5 bound to AMPPNP (PDB ID#3HQD) revealed that kinesins, and possibly several other ATPases as well, hydrolyze nucleotide by coordinating the positions of two critical water molecules (*Parke et al., 2010). Structure-based molecular dynamics studies and cryo-EM studies on kinesin-5 motors have demonstrated that many kinesin families are likely to initiate forward movement by a similar mechanism (*Goulet et al., 2012; Hesse et al., 2013). While these reports enriched our general understanding of the kinesin superfamily, others have revealed significant differences between the enzymatic and kinematic properties of kinesin-5s vs. other kinesin families, suiting kinesin-5 family motors to their unique role in mitosis. These properties are discussed in detail in the “Mechanism of kinesin-5” section below and in Table 1.
Table 1.
ADP release, ATPase, and motility rates of human Eg5 motor proteins.
| Eg5-367 | Kinesin-1 monomer | Eg5-513 | Kinesin-1 dimer | |
|---|---|---|---|---|
| Basal ADP release (s−1) | 0.27 | 0.02 | ||
| Basal ATPase (s1 site−1) | 0.25 | 0.006 | ||
| Maximal MT-stimulated ADP release (s−1) | 42.3 | >100 | 28.2 1.0 |
95 |
| MT-stimulated ATPase (s−1 site−1) | 6.25 | 60 | 0.48 (steady-state) 5-10 (while stepping) |
31 |
| Motility rate (nm/s) | 20.5 | 88 | 32.9 | 450 |
Data are taken from these references: (Case et al., 2000; Krzysiak , Gilbert, 2006; Ma , Taylor, 1995; *Valentine et al., 2006; Waitzman et al., 2011). Steady-state microtubule-stimulated ATPase of Eg5-513 includes both engagement onto MTs and active stepping. Engagement is rate-limiting in this case. The rate of active stepping after engagement was estimated based on optical trapping and calculation from other rate constants at 5-10 s−1. At saturating MTs, Eg5-513 has two different rates for the MT-stimulated ADP release rates of the first and second heads; the second head rate is italicized.
Remarkably, kinesin-5 requires not just its enzymatic heads, but also its nonmotor stalk and tail domains to cross-link and slide microtubules. The central stalk domain of kinesin-5 orients the motor subunits relative to one other. Robust dimers of motor domains can be formed using the N-terminal motor domain and half of the stalk (residues 1-513 in human Eg5; Figure 1B). Using Electron Microscopy and electron paramagnetic resonance (EPR) spectroscopy on Drosophila Klp61F, Acar et al. identified a bipolar assembly (“BASS”) domain toward the C-terminal end of the stalk (residues 671-791) that allows two dimers to associate in an antiparallel fashion to form bipolar homotetramers (*Acar et al., 2013).
The C-terminal tail of kinesin-5 appears to have two major functions: to aid the motor's localization in mitosis and to increase the motor's affinity for MTs (Weinger et al., 2011). In early work on kinesin-5, it was noted that the motor localized to the mitotic spindle. This localization depends upon the BimC box, a 20-residue stretch in the C-terminal tail of the motor, which contains a consensus site for the cell-cycle kinase M-CDK at residue Thr926 of human Eg5/Thr937 of Xenopus Eg5 (Blangy et al., 1995; Sawin, Mitchison, 1995) (Figure 1B). The tails of all known kinesin-5 motors except for S. pombe Cut7 appear to be phosphoregulated by M-CDK (Drummond , Hagan, 1998). This is discussed in detail below, along with phosphoregulation of kinesin-5 heads.
III. Mechanism of Kinesin-5
While vertebrate Eg5 has been the primary focus of in vitro studies to determine the mechanism of kinesin-5 motors, recent work on yeast kinesin-5 motors suggests remarkably divergent behavior that challenges the field's thinking on how directionality is determined for kinesin superfamily motors.
In general, kinesins whose motor domain is located at the protein's N-terminus move toward the microtubule's plus end, while those whose motor domain is located at the C-terminus move toward the microtubule's minus end. This notion is supported by structural and computational data showing that the repositioning of the neck linker and alpha 6 helices in C-terminal vs. N-terminal motors accounts for the directionality difference (Jana et al., 2012; Vinogradova et al., 2004). However, new results on the directionality of the S. cerevisiae kinesin-5 Cin8 have called this idea into question.
Two groups have observed Cin8 walking toward the minus-end of the MT; as the motor contains an N-terminal motor domain, this result was entirely unexpected (Gerson-Gurwitz et al., 2011; Roostalu et al., 2011). In Roostalu et al, the authors observe that tetramers of Cin8 walk toward the minus-end of the MT when attached to a single MT, but switch direction when bound to two MTs such that both pairs of motor domains walk toward the plus-ends of the MTs to which they are attached. The authors attribute this switch to a collective behavior of the motor, as the motor acts with a minus-end-directionality at very low concentrations. Gerson-Gurwitz and colleagues note that this directionality preference is biased by the presence of M-CDK phosphorylation sites in Loop 8 and the ionic strength of buffers used. Recently the same lab has reported similar direction-switching behavior in Kip1, another S. cerevisiae kinesin-5 motor (Fridman et al., 2013). The biophysical basis of this switching behavior and the role that it plays in mitosis are not yet established, and it clearly stands in contrast to the “N-terminal motors walk toward the plus-end” paradigm.
Unlike the yeast kinesin-5 motors mentioned above, human Eg5 does follow the kinesin directionality paradigm. Eg5 tetramers walk toward the plus-ends of both MTs to which they are attached, allowing the motor protein to generate an outward force on the mitotic spindle. This point was elegantly demonstrated in the work of Kapitein et al. (Kapitein et al., 2008; *Kapitein et al., 2005). The authors observed that, when attached to a surface-bound MT bundle, fluorescent Eg5 diffused randomly along the MT's length; when additional fluorescent MTs were flowed in, the Eg5 acted to crosslink between the surface and free MTs and slide them antiparallel to each other by walking processively to the MT plus ends. Recent work by Thiede et al on a kinesin-1/Eg5 chimera shows that the diffusive and processive behaviors of the motor are separated by an energetic barrier, and propose that this switch is mediated by an interaction between the heads and tails of Eg5 (Thiede et al., 2013). While a head-tail interaction in Eg5 has never been directly observed, understanding the nature of the diffusive/processive switching behavior of this motor would greatly inform the field's view of its role in mitosis and provide a potential window for the development of specific inhibitors.
Given the size of the kinesin-5 tetramer and the difficulty of studying four identical but non-synchronized catalytic sites, much effort has focused on determining the mechanism of monomers and dimers of motor domains. The amino acid sequence of the human Eg5 motor domain is 45% identical to that of the well-studied kinesin-1, and the use of stable dimers of human Eg5 in a majority of these studies allows for a direct comparison of Eg5 activity to that of kinesin-1. While both motors hydrolyze ATP to perform mechanical work, there are several features of the Eg5 mechanism that distinguish it from kinesin-1 (Table 1).
While the core structural elements that enable ATP hydrolysis and MT motility are conserved between kinesin-5 and kinesin-1, there are substantial differences between the motors’ sequences that translate to differences in enzymatic properties and function. The largest difference between kinesin-1 and Eg5 is an 8-residue insertion in Loop 5 of Eg5 (residues 125-132; see Figure 1c). Loop 5 is a break in the α2 helix that occurs in all kinesin family members; however, the length of the loop is longer in kinesin-5 than any other family member. Eg5 also has a noticeably longer Loop 1 and neck linker than kinesin-1.
Within a single Eg5 head, allosteric communication between Loop 5, the nucleotide-sensing elements (the P-loop, Switch I and Switch II), and the neck linker accounts for the motor's distinct kinetics and functional properties. This communication has been revealed by kinetic studies of monomeric kinesin motor heads, which show substantial, and at least to some extent, unexplained differences between Eg5 and kinesin-1. The ADP release rate and ATPase rates of wild-type Eg5 are far less activated by the addition of MTs than kinesin-1 (25-fold v. 3000-fold; Table 1). Deletion of seven residues in Loop 5 in Eg5 abolishes the MT stimulation of ADP release upon initial engagement, but has only modest effects on the ATPase activity and motility of Eg5 along MTs after this slow engagement step (Waitzman et al., 2011). The reason why Loop 5 is critical for Eg5 ADP release is unclear. Whether Loop 5, which is found in all kinesins, has a conserved function in the superfamily is also unknown.
Eg5 dimers have curious properties as well. Eg5 dimers release ADP from one head quickly (28 s-1), but then release their second ADP after a slow, ~ s-1 isomerization event. This isomerization event appears to be unique to Eg5, and this isomerization event only occurs during Eg5's initial engagement with the MT. Subsequent steps and coupled ATP hydrolysis events occur much more rapidly than this first step, at about 13 s1 1). (Krzysiak et al., 2008; *Valentine et al., 2006). These data suggest that the Eg5 dimer releases ADP very slowly as it takes its first step on the MT. However in taking subsequent steps coupled to ATP hydrolysis events, Eg5 releases both ADP and phosphate simultaneously and far more repidly than the initial microtubule-stimulated ADP release event (Wiatzman et al., 2011). This mechanism may help establish an energy barrier kinesin-5’s diffusing and walking states, or create a rate-limiting step in the motor's engagement that can be targeted by exogenous regulators. The structural features that are responsible for this unique property of Eg5 are unknown. However, a clue has emerged from recent cryo-EM data suggesting that binding to two adjacent sites on the microtubule places constraints on the neck linker conformations of the two heads. These constraints may influence Loop 5 conformation and the ADP•Pi or ADP release properties of Eg5 through allosteric connections (*Goulet et al., 2012).
Like dimers of kinesin-1 heads, Eg5 dimers move towards the plus ends of microtubules, alternating their heads in a hand-over-hand fashion (*Valentine et al., 2006). Both kinesin-1 and Eg5 dimers are processive, meaning that they can take multiple coupled mechanical and enzymatic steps along microtubules. However, Eg5 dimers are much more weakly processive than kinesin-1. Eg5 dimers are capable of taking 8-10 processive steps along a single MT (*Valentine et al., 2006). In contrast, kinesin-1 can take one hundred or more steps along the MT without detaching (Vale et al., 1996). The specific mechanical and kinetic properties responsible for Eg5's low processivity are not yet known, but several ideas have been proposed. Kinetic studies on Eg5 dimers indicate that the dimer initiates a processive run when one of its heads releases ADP and engages the MT (Krzysiak et al., 2008). The same head then binds ATP, sending the second head forward to its binding site. If the second head remains ADP-bound, or prematurely binds ADP from the APO state, it will not be tightly bound to the MT when the first head hydrolyzes ATP and detaches (Valentine , Block, 2009). Eg5 has a longer neck linker than kinesin-1, but studies differ as to whether Eg5's longer neck linker may be responsible for its reduced processivity (Duselder et al., 2012; Shastry, Hancock, 2011). Another possible explanation is that Eg5 remains MT-bound during processive stepping as long as it releases ADP from the ADP †Pi state, but if it releases Pi before ADP or binds ADP with its other head, it detaches. In this manner, Eg5's mechanochemical properties may be the result of its unique kinetics, which certainly involve Loop 5.
IV. Loop 5 of the human kinesin-5, Eg5 is a target for inhibitor development
Aside from its significant role in Eg5's mechanochemical cycle, Loop 5 is the target of a structurally diverse class of chemical inhibitors that shut down the activity of human Eg5. The first of the Eg5 inhibitors was identified in a screen performed by Tim Mitchison's group in 1999, and named “monastrol” for the mono-astral spindle phenotype it induced in treated U2OS cells (Kapoor et al., 2000; *Mayer et al., 1999). Further studies have identified a number of drugs that inhibit Eg5 activity, including ispinesib, enastron and S-Trityl-L-Cysteine (STLC). These agents require residues in Loop 5 and the α3 helix to bind the motor and inhibit it (Brier et al., 2006; Maliga , Mitchison, 2006) (Figure 1C Kinetic characterization of these agents indicates that they inhibit the motor via an allosteric mechanism that reduces the affinity of the motor for MTs and inhibits its ATPase activity. Interestingly, these agents appear to bind to Eg5 primarily while the motor is in solution, as MT-bound Eg5 has low affinity for monastrol, and monastrol-bound Eg5 has low affinity for the MT (Cochran et al., 2005; Maliga , 2002). These properties of Loop 5-directed inhibitors are consistent with the kinetic data described above, which suggest that Loop 5 is required for Eg5 engagement on MTs.
Loop 5 is clearly a “hot spot” for Eg5 inhibitor binding. For much of the history of Eg5 drug development, nearly all of the agents targeted Loop 5 (Brier et al., 2004). However, new classes of Eg5 inhibitors have been developed that appear to bind to other parts of the motor or have alternate catalytic effects. The chemical FCPT (2-(1-(4-fluorophenyl)cyclopropyl)-4-(pyridin-4-yl)thiazole) appears to bind at a site near Loop 5, but has ATP-competitive like effects on the kinetics of Eg5 (Groenet al., 2008). Additionally new work by the Kozielski group has identified BI8 (2-(3-fluoro-4-methoxyphenylamino)-1-((2-trifluoromethylbenzyl)-1H-benzo[d]imidazole-5-carboxylic acid), an agent that binds a novel pocket of Eg5 (Ulaganathan et al., 2013). While these agents may yet enter clinical trials and demonstrate efficacy, determining the role played by Loop 5 in the Eg5 motor mechanism remains a question of high basic and clinical interest.
Eg5 inhibitors targeting Loop 5 have been tested in several different Phase I and II clinical trials, where their efficacy has been poor. A good summary of these can be found in (Sarli , Giannis, 2008). This poor efficacy has been a disappointing development, as Eg5 inhibition very robustly arrests cell division in cultured cells. Eg5 inhibitors did work in selective conditions, such as xenograft tumor models (Sakowicz et al., 2004). There are several possible reasons for the failure of Eg5 inhibitors in these early trials. One reason is common to all antimitotics; human tumor cells undergo mitosis much less frequently than cells within tumor xenografts in mice, and cells have to enter mitosis for Eg5 inhibitors to cause apoptosis (Chan et al., 2012). Secondly, mutations in Eg5 can confer drug resistance to Eg5 inhibitors via either direct or allosteric mechanisms (Brier et al., 2006). Lastly, mitosis is an extremely robust process, and alterations in regulatory factors or other mitotic prote may potentially overcome Eg5 inhibition (Raaijmakers et al., 2012). It is therefore possible that agents acting on Eg5 or other mitotic target proteins may be more effective in combination than when used as monotherapies. That said, before we can understand what combination therapies with Eg5 would be effective, we must know how Eg5 is regulated and how it interacts with other potential drug targets in mitosis. What other targets would be effective in conjunction with Eg5? Are there kinases or other regulatory binding partnersupstream of Eg5 that might be better targets with similar cytotoxic effects?
V. Regulation of Kinesin-5 by binding partners
As one would expect given their critical role in mitosis, kinesin-5 motors are regulated by binding partners as well as by phosphorylation. Kinesin-5 localization changes throughout mitosis in a tightly regulated manner. During metaphase, kinesin-5 localizes along spindle microtubules and at spindle poles. The motor relocalizes to the spindle midzone in anaphase (Gable et al., 2012). Kinesin-5 has been reported to associate directly with NuMa (Nuclear Mitotic Apparatus protein). which is involved in spindle pole assembly. NuMa appears to require both kinesin-5 and dynein, but kinesin-5 does not require NuMA for localization to the spindle pole (Iwakiri et al., 2013).
While kinesin-5 does not require NuMA, it does require the minus-end directed motor dynein for proper targeting throughout mitosis. The human Eg5 tail interacts with dynein through the p150glued subunit of dynactin, and this interaction is regulated by a conserved phosphorylation site, Thr926, which is discussed in more detail below (Blangy et al., 1997). Nuclear envelope-associated dynein cooperates with Eg5 to separate spindle poles during prophase, and in fact, this dynein can separate spindle poles in cell lines that completely lack functional Eg5 (Raaijmakers et al., 2012). During metaphase, a fraction of Eg5 remains stationary at the spindle midzone, while the remainder is transported poleward. Inhibition of dynein by injection of the CC1 fragment of p150 blocks this movement, resulting in altered spindle length and organization (Gable et al., 2012).
Proper Eg5 targeting also requires the MAP TPX2 (Targeting Protein for Xenopus kinesin-like protein 2 (Xklp2)). Xklp2 is a kinesin-15 family motor protein that localizes to spindle poles and is essential for their separation. Because TPX2 is required for targeting both Eg5 and Xklp2 kinesins, it appears to play multiple roles in mitotic spindle assembly. TPX2 resides in the nucleus in interphase, and undergoes dyneindepenent relocalization to MTs near the spindle poles in early mitosis.
Wittmann, et al. showed that TPX2 is critical for establishing the localization of the Xklp2 motor protein during spindle pole separation. They also noted that adding an excess of TPX2 resulted in the formation of monopolar spindle structures (Wittmann et al., 2000). This finding was not followed up on until nearly a decade late, when Eckerdt et al showed that injection of Xenopus embryos with either the full-length or the C-terminal half of TPX2 led to cleavage arrest (Eckerdt et al., 2008). Based on an observation that both TPX2 and Eg5 had been isolated in a complex together (Koffa et a;., 2006), Eckerdt et al performed pull-downs between Eg5 and a variety of TPX2 constructs. The authors noted that TPX2 interacted with Eg5, and that this interaction required the C-terminal 35 residues of TPX2. could cause cleavage arrest in Xenopus Additionally, the only TPX2 proteins that embryos contained this C-terminal Eg5-interacting region (Eckerdt et al., 2008). Using LLC-Pk1 cells, Ma and colleagues demonstrated similarly that disrupting the TPX2-Eg5 interaction by deleting the C-terminal 35 residues of TPX2 led to mislocalization of TPX2 and poorly organized mitotic spindles (Ma et al., 2010). The findings that TPX2 appeared to directly interact with Eg5 and that excess amounts of the Eg5-interacting region of TPX2 induced monopolar spindle formation suggested that TPX2 might regulate Eg5 activity. Supporting this notion, TPX2 helps localize Eg5 to the mitotic spindle (*Ma et al., 2011), and TPX2 is required for Eg5 to transition from spindle pole localization to midbody (Gable et al., 2012).In vitro, TPX2 slows down the MT-sliding activity of Eg5 but not kinesin-1 (Ma et al., 2011). These data indicate that TPX2 may facilitate slower, but perhaps more persistent movement of Eg5 to the spindle midzone.
Previous studies proposed mechanisms by which MAPs could regulate kinesin motors through local MT lattice alteration and/or competition for MT binding sites (McVicker et al., 2011; Seitz et al., 2002). The TPX2/Eg5 interaction is certainly different, because the effects of TPX2 are specific to Eg5 and are not seen on kinesin-1 (*Ma et al., 2011). This suggests higher specificity than could be explained by either MAPs locally altering the MT lattice or competing for binding sites. It is intriguing that similar high-specificity regulation of kinesin-1 by the MAP ensconsin has been proposed (Barlan et al., 2013; Metzger et al., 2012). Therefore, TPX2 regulation of kinesin-5 may typify a new class of mechanisms for kinesin motor regulation that we do not yet fully understand.
VI. Phosphoregulation of Kinesin-5
Phosphorylation of almost all kinesin-5 homologs at a co within the BimC box (Thr926 in human Eg5, Thr937 in Xenopus) is essential for targeting the motor to the mitotic spindle (Blangy et al., 1995; Sawin , Mitchison, 1995). The only known exception is the kinesin-5, Cut7 (Drummond , Hagan, 1998). A second phosphorylation site at residue Ser1033 the Eg5 tail, phosphorylated by Nek6/7, was shown to help localize around 3% of the total Eg5 protein to the spindle pole (Rapley et al., 2008). Phosphorylation of the BimC box threonine (Thr926 in human Eg5) also regulates the interaction of Eg5 with dynactin, suggesting that association with dynein and dynactin may help target Eg5 to MTs (Blangy et al., 1997). However,in Cahu, et al. showed that phosphorylation of the Xenopus Eg5 tail by M-CDK increases Eg5 affinity for MTs ~10 fold, and also increases the amount of time the motor spends on the MT, with no additional factors in the assay (Cahu et al., 2008). This suggests that phospho-regulation of the kinesin-5 tail exerts a direct effect on motor activity that is not mediated exclusively through binding partners. This increased MT affinity upon phosphorylation is counter-intuitive, given that the microtubule surface is negatively charged. It is not known how phosphorylation alters the structure of the Eg5 tail or the Eg5 holoenzyme to enhance MT binding.
In addition to work described above that identified phosphoregulatory mechanisms that control kinesin-5 localization through the motor's tail, recent studies have shown that phosphorylation of kinesin-5 enzymatic heads may alter their activity. In a 2010 paper, Chee and Haase identified three sites in the S. cerevisiae Cin8 motor domain and four sites in the Kip1 motor domain that met the minimal consensus sequence for the checkpoint kinase M-CDK (Chee , Haase, 2010). One of these sites (Ser388 in Kip1/Ser455 in Cin8) is conserved between the two motors and many other kinesin-5 family members. The authors found that deleting Cin8 and mutating Ser388 in Kip1 to alanine or deleting Kip1 and mutating Ser455 in Cin8 to alanine resulted in defects in spindle pole body separation.
Subsequently, Avunie-Masala et al found that the consensus sites for M-CDK in the Cin8 motor domain are phosphorylated and that phosphorylation of these sites alters Cin8'S motility. Cin8 was phosphorylated in anaphase, but mutation of the three consensus M-CDK sites described above abolished this phosphorylation (*Avunie-Masala et al., 2011). Two of the three identified sites, Ser239 and Thr247, are located in the yeast-specific Loop 8 insertion of the motor, while the third, Ser455, is the conserved site identified above. Cin8 with all three phosphorylated residues mutated to alanine localized primarily to the spindle poles in anaphase, as opposed to wild type motors that primarily occupy the midzone. A second study performed by the same group found that Cin8 proteins containing Ser239Ala and Thr247Ala mutations exhibited spindle-pole directed (or toward the minus-end of the MT) movements, suggesting that Loop 8 may help determine the directionality of the motor (Gerson-Gurwitz et al., 2011). In contrast, Cin8 with all three phosphorylated residues mutated to aspartate failed to localize to the spindle apparatus, and cells expressing this Cin8-3D mutant were not viable.
Garcia, et al. showed that the D. melanogaster kinesin-5 motor Klp61F is phosphorylated on up to three tyrosines in its motor domain by the cell cycle kinase dWee1 in vitro (Garcia et al., 2009) To determine the effects of this phosphorylation event, Garcia et al expressed Klp61F with the phosphoacceptor tyrosines mutated to phenylalanine in Drosophila embryos, and observed increases in lethality and spindle defects. This work suggests an interesting interplay between M-CDK and Wee1 at the onset of mitosis. Wee1 phosphorylates and deactivates M-CDK until the onset of mitosis (Russell , Nurse, 1987). In turn, M-CDK activation negatively regulates Wee1 (McGowan , Russell, 1995). As described above, M-CDK also phosphorylates and activates kinesin-5 tails. It is intriguing that a kinase that directly activates Eg5 is antagonized by a kinase that directly inhibits it. One could speculate that Eg5 is acting as a mechanical conduit between two antagonistic signaling mechanisms mediating mitosis, Wee1 and M-CDK. Together, the phosphorylation data on kinesin-5 motors tell two stories: first, a highly conserved mechanism by which M-CDK phosphorylation recruits kinesin-5 to microtubules and second, multiple, possibly species-specific, phosphoregulatory mechanisms (M-CDK in yeast, Wee1 in Drosophila, and perhaps others?) that fine-tune the activity of the motor domains.
Much work remains to be done regarding phosphoregulation of kinesin-5 and kinesin motors in general. The PhosphoSitePlus database has identified phosphorylation sites in nearly every human kinesin family member (Hornbeck, 2012). Many of these sites have been reproduced by multiple MS techniques, but very few studies have probed the mechanistic effects of these phosphorylation events on kinesin motor activity in vitro and in vivo. It is difficult to assess how much of this sea of data is an opportunity to discover specific, poorly understood regulatory mechanisms governing kinesins, and how much of it is simply artifact. However, intriguing mechanistic studies on kinesin motor phosphoregulatory mechanisms continue to emerge, such as those referenced above and the work of Mennella et al, who mechanistically link phosphorylation of the Drosophila kinesin-13 Klp10A to changes in MT depolymerization (Mennella et al., 2009).
VII. Kinesin-5 roles beyond mitosis
While kinesin-5 is considered a “mitotic” kinesin, several lines of evidence have suggested roles for the motor outside of mitosis. Peter Baas'S group has published several studies demonstrating that Eg5 regulates the length of axons and the migration of neurons in culture (Falnikar et al., 2011; *Myers , Baas, 2007). Recent work by the Baas group suggests that phosphorylation of the BimC box of Eg5 may act as a switch that turns on the motor'S activity in axonal growth cones, as it does in mitosis (Nadar et al., 2012).
Additionally, Bartoli et al have reported a global decrease in protein synthesis in cells treated with specific Eg5 inhibitors, and shown that Eg5 helps structure polysomes along MT scaffolds (Bartoli et al., 2011).
VIII. Conclusions: Directions for future study
Investigations of the mechanism and the biological role of kinesin-5 motors have generated several emerging and controversial storylines that force the entire kinesin field to re-examine many of its assumptions. Having highlighted many of these storylines above, we conclude here with a series of questions that, in our view, would make excellent topics for future research.
How and why is it that yeast kinesin-5s, which have N-terminal motor domains, switch directions? Is this direction switching the target of regulatory mechanisms that establish unique roles for yeast kinesin-5s?
Does loop 5 have a conserved function for all kinesins that is amplified for the kinesin-5 family? Is this structural element a natural regulatory target region for kinesins, and/or a “hot spot” for inhibitors?
How can a specific MAP, TPX2, regulate a specific kinesin, Eg5?
Can combination drug therapies be developed with a knowledge of how Eg5 interacts with regulatory proteins and mitotic kinases?
Does kinesin-5 require different binding partners to fulfill its conventional roles in mitosis vs. “moonlighting” roles in axonal length regulation and protein synthesis? Does kinesin-5 perform different mechanochemical activities in these settings?
Does kinesin-5 have other “moonlighting” roles besides axonal length regulation and protein synthesis?
Acknowledgments
The authors have no competing interests.
Funding: J.S.W. is supported by the Myhrvold Family Fellowship from the Fannie and John Hertz Foundation, the Malkin Scholars Program from the Robert H. Lurie Comprehensive Cancer Center of Northwestern University, a Medical Scientist Training Program training grant (GM08152) and a Cellular and Molecular Basis of Disease training grant (GM08061). S.E.R. is supported by National Institutes of Health under grant GM072656.
Abbreviations
- MT
microtubule
- MAPs
Microtubule-associated proteins
- EPR
electron paramagnetic resonance
- “BASS” domain
bipolar assembly domain
- STLC
S-trityl L-cysteine
- TPX2
Targeting Protein for Xenopus Kinesin-like protein 2
- MS
Mass spectrometry
Footnotes
Publisher's Disclaimer: This article has been accepted for publication and undergone full peer review but has not been through the copyediting, typesetting, pagination and proofreading process, which may lead to differences between this and the Version of Record. Please cite this article as DOI:10.1002/boc.201300054.
References
- Acar S, Carlson DB, Budamagunta MS, Yarov-Yarovoy V, Correia JJ, Ninonuevo MR, Jia W, Tao L, Leary JA, Voss JC, Evans JE, Scholey JM. The bipolar assembly domain of the mitotic motor kinesin-5. Nat Commun. 2013;4:1343. doi: 10.1038/ncomms2348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avunie-Masala R, Movshovich N, Nissenkorn Y, Gerson-Gurwitz A, Fridman V, Koivomagi M, Loog M, Hoyt MA, Zaritsky A, Gheber L. Phospho-regulation of kinesin-5 during anaphase spindle elongation. J Cell Sci. 2011;124:873–878. doi: 10.1242/jcs.077396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlan K, Lu W, Gelfand VI. The microtubule-binding protein ensconsin is an essential cofactor of kinesin-1. Curr Biol. 2013;23:317–322. doi: 10.1016/j.cub.2013.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartoli KM, Jakovljevic J, Woolford JL, Jr., Saunders WS. Kinesin molecular motor Eg5 functions during polypeptide synthesis. Mol Biol Cell. 2011;22:3420–3430. doi: 10.1091/mbc.E11-03-0211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blangy A, Arnaud L, Nigg EA. Phosphorylation by p34cdc2 protein kinase regulates binding of the kinesin-related motor HsEg5 to the dynactin subunit p150. J Biol Chem. 1997;272:19418–19424. doi: 10.1074/jbc.272.31.19418. [DOI] [PubMed] [Google Scholar]
- Blangy A, Lane HA, d'Herin P, Harper M, Kress M, Nigg EA. Phosphorylation by p34cdc2 regulates spindle association of human Eg5, a kinesin-related motor essential for bipolar spindle formation in vivo. Cell. 1995;83:1159–1169. doi: 10.1016/0092-8674(95)90142-6. [DOI] [PubMed] [Google Scholar]
- Brier S, Lemaire D, Debonis S, Forest E, Kozielski F. Identification of the protein binding region of S-trityl-L-cysteine, a new potent inhibitor of the mitotic kinesin Eg5. Biochemistry. 2004;43:13072–13082. doi: 10.1021/bi049264e. [DOI] [PubMed] [Google Scholar]
- Brier S, Lemaire D, DeBonis S, Forest E, Kozielski F. Molecular dissection of the inhibitor binding pocket of mitotic kinesin Eg5 reveals mutants that confer resistance to antimitotic agents. J Mol Biol. 2006;360:360–376. doi: 10.1016/j.jmb.2006.04.062. [DOI] [PubMed] [Google Scholar]
- Brust-Mascher I, Sommi P, Cheerambathur DK, Scholey JM. Kinesin-5-dependent poleward flux and spindle length control in Drosophila embryo mitosis. Mol Biol Cell. 2009;20:1749–1762. doi: 10.1091/mbc.E08-10-1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahu J, Olichon A, Hentrich C, Schek H, Drinjakovic J, Zhang C, Doherty-Kirby A, Lajoie G, Surrey T. Phosphorylation by Cdk1 increases the binding of Eg5 to microtubules in vitro and in Xenopus egg extract spindles. PLoS One. 2008;3:e3936. doi: 10.1371/journal.pone.0003936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Case RB, Rice S, Hart CL, Ly B, Vale RD. Role of the kinesin neck linker and catalytic core in microtubule-based motility. Curr Biol. 2000;10:157–160. doi: 10.1016/s0960-9822(00)00316-x. [DOI] [PubMed] [Google Scholar]
- Chan KS, Koh CG, Li HY. Mitosis-targeted anti-cancer therapies: where they stand. Cell Death Dis. 2012;3:e411. doi: 10.1038/cddis.2012.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chee MK, Haase SB. B-cyclin/CDKs regulate mitotic spindle assembly by phosphorylating kinesins-5 in budding yeast. PLoS Genet. 2010;6:e1000935. doi: 10.1371/journal.pgen.1000935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cochran JC, Gatial JE, 3rd, Kapoor TM, Gilbert SP. Monastrol inhibition of the mitotic kinesin Eg5. J Biol Chem. 2005;280:12658–12667. doi: 10.1074/jbc.M413140200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond DR, Hagan IM. Mutations in the bimC box of Cut7 indicate divergence of regulation within the bimC family of kinesin related proteins. J Cell Sci. 1998;111(Pt 7):853–865. doi: 10.1242/jcs.111.7.853. [DOI] [PubMed] [Google Scholar]
- Duselder A, Thiede C, Schmidt CF, Lakamper S. Neck-linker length dependence of processive Kinesin-5 motility. J Mol Biol. 2012;423:159–168. doi: 10.1016/j.jmb.2012.06.043. [DOI] [PubMed] [Google Scholar]
- Eckerdt F, Eyers PA, Lewellyn AL, Prigent C, Maller JL. Spindle pole regulation by a discrete Eg5-interacting domain in TPX2. Curr Biol. 2008;18:519–525. doi: 10.1016/j.cub.2008.02.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endres NF, Yoshioka C, Milligan RA, Vale RD. A lever-arm rotation drives motility of the minus-end-directed kinesin Ncd. Nature. 2006;439:875–878. doi: 10.1038/nature04320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falnikar A, Tole S, Baas PW. Kinesin-5, a mitotic microtubule-associated motor protein, modulates neuronal migration. Mol Biol Cell. 2011;22:1561–1574. doi: 10.1091/mbc.E10-11-0905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fridman V, Gerson-Gurwitz A, Shapira O, Movshovich N, Lakamper S, Schmidt CF, Gheber L. Kinesin-5 Kip1 is a bi-directional motor that stabilizes microtubules and tracks their plus-ends in vivo. J Cell Sci. 2013 doi: 10.1242/jcs.125153. [DOI] [PubMed] [Google Scholar]
- Gable A, Qiu M, Titus J, Balchand S, Ferenz NP, Ma N, Collins ES, Fagerstrom C, Ross JL, Yang G, Wadsworth P. Dynamic reorganization of Eg5 in the mammalian spindle throughout mitosis requires dynein and TPX2. Mol Biol Cell. 2012;23:1254–1266. doi: 10.1091/mbc.E11-09-0820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia K, Stumpff J, Duncan T, Su TT. Tyrosines in the kinesin-5 head domain are necessary for phosphorylation by Wee1 and for mitotic spindle integrity. Curr Biol. 2009;19:1670–1676. doi: 10.1016/j.cub.2009.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerson-Gurwitz A, Thiede C, Movshovich N, Fridman V, Podolskaya M, Danieli T, Lakamper S, Klopfenstein DR, Schmidt CF, Gheber L. Directionality of individual kinesin-5 Cin8 motors is modulated by loop 8, ionic strength and microtubule geometry. EMBO J. 2011;30:4942–4954. doi: 10.1038/emboj.2011.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon DM, Roof DM. The kinesin-related protein Kip1p of Saccharomyces cerevisiae is bipolar. J Biol Chem. 1999;274:28779–28786. doi: 10.1074/jbc.274.40.28779. [DOI] [PubMed] [Google Scholar]
- Goshima G, Vale RD. The roles of microtubule-based motor proteins in mitosis: comprehensive RNAi analysis in the Drosophila S2 cell line. J Cell Biol. 2003;162:1003–1016. doi: 10.1083/jcb.200303022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goshima G, Vale RD. Cell cycle-dependent dynamics and regulation of mitotic kinesins in Drosophila S2 cells. Mol Biol Cell. 2005;16:3896–3907. doi: 10.1091/mbc.E05-02-0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goulet A, Behnke-Parks WM, Sindelar CV, Major J, Rosenfeld SS, Moores CA. The structural basis of force generation by the mitotic motor kinesin-5. J Biol Chem. 2012;287:44654–44666. doi: 10.1074/jbc.M112.404228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groen AC, Needleman D, Brangwynne C, Gradinaru C, Fowler B, Mazitschek R, Mitchison TJ. A novel small-molecule inhibitor reveals a possible role of kinesin-5 in anastral spindle-pole assembly. J Cell Sci. 2008;121:2293–2300. doi: 10.1242/jcs.024018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hesse WR, Steiner M, Wohlever ML, Kamm RD, Hwang W, Lang MJ. Modular aspects of kinesin force generation machinery. Biophys J. 2013;104:1969–1978. doi: 10.1016/j.bpj.2013.03.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornbeck PV, Kornhauser JM, Tkachev S, Zhang B, Skrzypek E, Murray B, Latham V, Sullivan M. PhosphoSitePlus: a comprehensive resource for investigating the structure and function of experimentally determined post-translational modifications in man and mouse. Nucleic Acids Res. 2012;40:D261–270. doi: 10.1093/nar/gkr1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwakiri Y, Kamakura S, Hayase J, Sumimoto H. Interaction of NuMA protein with the kinesin Eg5: its possible role in bipolar spindle assembly and chromosome alignment. Biochem J. 2013;451:195–204. doi: 10.1042/BJ20121447. [DOI] [PubMed] [Google Scholar]
- Jana B, Hyeon C, Onuchic JN. The origin of minus-end directionality and mechanochemistry of Ncd motors. PLoS Comput Biol. 2012;8:e1002783. doi: 10.1371/journal.pcbi.1002783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W, Stock MF, Li X, Hackney DD. Influence of the kinesin neck domain on dimerization and ATPase kinetics. J Biol Chem. 1997;272:7626–7632. doi: 10.1074/jbc.272.12.7626. [DOI] [PubMed] [Google Scholar]
- Kapitein LC, Kwok BH, Weinger JS, Schmidt CF, Kapoor TM, Peterman EJ. Microtubule cross-linking triggers the directional motility of kinesin-5. J Cell Biol. 2008;182:421–428. doi: 10.1083/jcb.200801145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapitein LC, Peterman EJ, Kwok BH, Kim JH, Kapoor TM, Schmidt CF. The bipolar mitotic kinesin Eg5 moves on both microtubules that it crosslinks. Nature. 2005;435:114–118. doi: 10.1038/nature03503. [DOI] [PubMed] [Google Scholar]
- Kapoor TM, Mayer TU, Coughlin ML, Mitchison TJ. Probing spindle assembly mechanisms with monastrol, a small molecule inhibitor of the mitotic kinesin, Eg5. J Cell Biol. 2000;150:975–988. doi: 10.1083/jcb.150.5.975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashina AS, Baskin RJ, Cole DG, Wedaman KP, Saxton WM, Scholey JM. A bipolar kinesin. Nature. 1996;379:270–272. doi: 10.1038/379270a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalil AS, Appleyard DC, Labno AK, Georges A, Karplus M, Belcher AM, Hwang W, Lang MJ. Kinesin's cover-neck bundle folds forward to generate force. Proc Natl Acad Sci U S A. 2008;105:19247–19252. doi: 10.1073/pnas.0805147105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim ED, Buckley R, Learman S, Richard J, Parke C, Worthylake DK, Wojcik EJ, Walker RA, Kim S. Allosteric drug discrimination is coupled to mechanochemical changes in the kinesin-5 motor core. J Biol Chem. 2010;285:18650–18661. doi: 10.1074/jbc.M109.092072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koffa MD, Casanova CM, Santarella R, Kocher T, Wilm M, Mattaj IW. HURP is part of a Ran-dependent complex involved in spindle formation. Curr Biol. 2006;16:743–754. doi: 10.1016/j.cub.2006.03.056. [DOI] [PubMed] [Google Scholar]
- Krzysiak TC, Gilbert SP. Dimeric Eg5 maintains processivity through alternating-site catalysis with rate-limiting ATP hydrolysis. J Biol Chem. 2006;281:39444–39454. doi: 10.1074/jbc.M608056200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krzysiak TC, Grabe M, Gilbert SP. Getting in sync with dimeric Eg5. Initiation and regulation of the processive run. J Biol Chem. 2008;283:2078–2087. doi: 10.1074/jbc.M708354200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kull FJ, Endow SA. Kinesin: switch I & II and the motor mechanism. J Cell Sci. 2002;115:15–23. doi: 10.1242/jcs.115.1.15. [DOI] [PubMed] [Google Scholar]
- Loughlin R, Heald R, Nedelec F. A computational model predicts Xenopus meiotic spindle organization. J Cell Biol. 2010;191:1239–1249. doi: 10.1083/jcb.201006076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma N, Titus J, Gable A, Ross JL, Wadsworth P. TPX2 regulates the localization and activity of Eg5 in the mammalian mitotic spindle. J Cell Biol. 2011;195:87–98. doi: 10.1083/jcb.201106149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma N, Tulu US, Ferenz NP, Fagerstrom C, Wilde A, Wadsworth P. Poleward transport of TPX2 in the mammalian mitotic spindle requires dynein, Eg5, and microtubule flux. Mol Biol Cell. 2010;21:979–988. doi: 10.1091/mbc.E09-07-0601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma YZ, Taylor EW. Mechanism of microtubule kinesin ATPase. Biochemistry. 1995;34:13242–13251. doi: 10.1021/bi00040a040. [DOI] [PubMed] [Google Scholar]
- Maliga Z, Kapoor TM, Mitchison TJ. Evidence that monastrol is an allosteric inhibitor of the mitotic kinesin Eg5. Chem Biol. 2002;9:989–996. doi: 10.1016/s1074-5521(02)00212-0. [DOI] [PubMed] [Google Scholar]
- Maliga Z, Mitchison TJ. Small-molecule and mutational analysis of allosteric Eg5 inhibition by monastrol. BMC Chem Biol. 2006;6:2. doi: 10.1186/1472-6769-6-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer TU, Kapoor TM, Haggarty SJ, King RW, Schreiber SL, Mitchison TJ. Small molecule inhibitor of mitotic spindle bipolarity identified in a phenotype-based screen. Science. 1999;286:971–974. doi: 10.1126/science.286.5441.971. [DOI] [PubMed] [Google Scholar]
- McGowan CH, Russell P. Cell cycle regulation of human WEE1. EMBO J. 1995;14:2166–2175. doi: 10.1002/j.1460-2075.1995.tb07210.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McVicker DP, Chrin LR, Berger CL. The nucleotide-binding state of microtubules modulates kinesin processivity and the ability of Tau to inhibit kinesin-mediated transport. J Biol Chem. 2011;286:42873–42880. doi: 10.1074/jbc.M111.292987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mennella V, Tan DY, Buster DW, Asenjo AB, Rath U, Ma A, Sosa HJ, Sharp DJ. Motor domain phosphorylation and regulation of the Drosophila kinesin 13, KLP10A. J Cell Biol. 2009;186:481–490. doi: 10.1083/jcb.200902113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzger T, Gache V, Xu M, Cadot B, Folker ES, Richardson BE, Gomes ER, Baylies MK. MAP and kinesin-dependent nuclear positioning is required for skeletal muscle function. Nature. 2012;484:120–124. doi: 10.1038/nature10914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers KA, Baas PW. Kinesin-5 regulates the growth of the axon by acting as a brake on its microtubule array. J Cell Biol. 2007;178:1081–1091. doi: 10.1083/jcb.200702074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadar VC, Lin S, Baas PW. Microtubule redistribution in growth cones elicited by focal inactivation of kinesin-5. J Neurosci. 2012;32:5783–5794. doi: 10.1523/JNEUROSCI.0144-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parke CL, Wojcik EJ, Kim S, Worthylake DK. ATP hydrolysis in Eg5 kinesin involves a catalytic two-water mechanism. J Biol Chem. 2010;285:5859–5867. doi: 10.1074/jbc.M109.071233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raaijmakers JA, van Heesbeen RG, Meaders JL, Geers EF, Fernandez-Garcia B, Medema RH, Tanenbaum ME. Nuclear envelope-associated dynein drives prophase centrosome separation and enables Eg5-independent bipolar spindle formation. EMBO J. 2012;31:4179–4190. doi: 10.1038/emboj.2012.272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapley J, Nicolas M, Groen A, Regue L, Bertran MT, Caelles C, Avruch J, Roig J. The NIMA-family kinase Nek6 phosphorylates the kinesin Eg5 at a novel site necessary for mitotic spindle formation. J Cell Sci. 2008;121:3912–3921. doi: 10.1242/jcs.035360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice S, Lin AW, Safer D, Hart CL, Naber N, Carragher BO, Cain SM, Pechatnikova E, Wilson-Kubalek EM, Whittaker M, Pate E, Cooke R, Taylor EW, Milligan RA, Vale RD. A structural change in the kinesin motor protein that drives motility. Nature. 1999;402:778–784. doi: 10.1038/45483. [DOI] [PubMed] [Google Scholar]
- Roostalu J, Hentrich C, Bieling P, Telley IA, Schiebel E, Surrey T. Directional switching of the kinesin Cin8 through motor coupling. Science. 2011;332:94–99. doi: 10.1126/science.1199945. [DOI] [PubMed] [Google Scholar]
- Russell P, Nurse P. Negative regulation of mitosis by wee1+, a gene encoding a protein kinase homolog. Cell. 1987;49:559–567. doi: 10.1016/0092-8674(87)90458-2. [DOI] [PubMed] [Google Scholar]
- Sakowicz R, Finer JT, Beraud C, Crompton A, Lewis E, Fritsch A, Lee Y, Mak J, Moody R, Turincio R, Chabala JC, Gonzales P, Roth S, Weitman S, Wood KW. Antitumor activity of a kinesin inhibitor. Cancer Res. 2004;64:3276–3280. doi: 10.1158/0008-5472.can-03-3839. [DOI] [PubMed] [Google Scholar]
- Sarli V, Giannis A. Targeting the kinesin spindle protein: basic principles and clinical implications. Clin Cancer Res. 2008;14:7583–7587. doi: 10.1158/1078-0432.CCR-08-0120. [DOI] [PubMed] [Google Scholar]
- Sauer G, Korner R, Hanisch A, Ries A, Nigg EA, Sillje HH. Proteome analysis of the human mitotic spindle. Mol Cell Proteomics. 2005;4:35–43. doi: 10.1074/mcp.M400158-MCP200. [DOI] [PubMed] [Google Scholar]
- Saunders AM, Powers J, Strome S, Saxton WM. Kinesin-5 acts as a brake in anaphase spindle elongation. Curr Biol. 2007;17:R453–454. doi: 10.1016/j.cub.2007.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders W, Lengyel V, Hoyt MA. Mitotic spindle function in Saccharomyces cerevisiae requires a balance between different types of kinesin-related motors. Mol Biol Cell. 1997;8:1025–1033. doi: 10.1091/mbc.8.6.1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawin KE, Mitchison TJ. Mutations in the kinesin-like protein Eg5 disrupting localization to the mitotic spindle. Proc Natl Acad Sci U S A. 1995;92:4289–4293. doi: 10.1073/pnas.92.10.4289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seitz A, Kojima H, Oiwa K, Mandelkow EM, Song YH, Mandelkow E. Single-molecule investigation of the interference between kinesin, tau and MAP2c. EMBO J. 2002;21:4896–4905. doi: 10.1093/emboj/cdf503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shastry S, Hancock WO. Interhead tension determines processivity across diverse N-terminal kinesins. Proc Natl Acad Sci U S A. 2011;108:16253–16258. doi: 10.1073/pnas.1102628108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sindelar CV, Downing KH. An atomic-level mechanism for activation of the kinesin molecular motors. Proc Natl Acad Sci U S A. 2010;107:4111–4116. doi: 10.1073/pnas.0911208107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straight AF, Marshall WF, Sedat JW, Murray AW. Mitosis in living budding yeast: anaphase A but no metaphase plate. Science. 1997;277:574–578. doi: 10.1126/science.277.5325.574. [DOI] [PubMed] [Google Scholar]
- Tanenbaum ME, Macurek L, Galjart N, Medema RH. Dynein, Lis1 and CLIP-170 counteract Eg5-dependent centrosome separation during bipolar spindle assembly. EMBO J. 2008;27:3235–3245. doi: 10.1038/emboj.2008.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiede C, Lakamper S, Wessel AD, Kramer S, Schmidt CF. A chimeric kinesin-1 head/kinesin-5 tail motor switches between diffusive and processive motility. Biophys J. 2013;104:432–441. doi: 10.1016/j.bpj.2012.11.3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulaganathan V, Talapatra SK, Rath O, Pannifer A, Hackney DD, Kozielski F. Structural insights into a unique inhibitor binding pocket in kinesin spindle protein. J Am Chem Soc. 2013;135:2263–2272. doi: 10.1021/ja310377d. [DOI] [PubMed] [Google Scholar]
- Vale RD, Fletterick RJ. The design plan of kinesin motors. Annu Rev Cell Dev Biol. 1997;13:745–777. doi: 10.1146/annurev.cellbio.13.1.745. [DOI] [PubMed] [Google Scholar]
- Vale RD, Funatsu T, Pierce DW, Romberg L, Harada Y, Yanagida T. Direct observation of single kinesin molecules moving along microtubules. Nature. 1996;380:451–453. doi: 10.1038/380451a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vale RD, Milligan RA. The way things move: looking under the hood of molecular motor proteins. Science. 2000;288:88–95. doi: 10.1126/science.288.5463.88. [DOI] [PubMed] [Google Scholar]
- Valentine MT, Block SM. Force and premature binding of ADP can regulate the processivity of individual Eg5 dimers. Biophys J. 2009;97:1671–1677. doi: 10.1016/j.bpj.2009.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valentine MT, Fordyce PM, Krzysiak TC, Gilbert SP, Block SM. Individual dimers of the mitotic kinesin motor Eg5 step processively and support substantial loads in vitro. Nat Cell Biol. 2006;8:470–476. doi: 10.1038/ncb1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinogradova MV, Reddy VS, Reddy AS, Sablin EP, Fletterick RJ. Crystal structure of kinesin regulated by Ca(2+)-calmodulin. J Biol Chem. 2004;279:23504–23509. doi: 10.1074/jbc.M400741200. [DOI] [PubMed] [Google Scholar]
- Waitzman JS, Larson AG, Cochran JC, Naber N, Cooke R, Jon Kull F, Pate E, Rice SE. The loop 5 element structurally and kinetically coordinates dimers of the human kinesin-5, Eg5. Biophys J. 2011;101:2760–2769. doi: 10.1016/j.bpj.2011.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinger JS, Qiu M, Yang G, Kapoor TM. A nonmotor microtubule binding site in kinesin-5 is required for filament crosslinking and sliding. Curr Biol. 2011;21:154–160. doi: 10.1016/j.cub.2010.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittmann T, Wilm M, Karsenti E, Vernos I. TPX2, A novel xenopus MAP involved in spindle pole organization. J Cell Biol. 2000;149:1405–1418. doi: 10.1083/jcb.149.7.1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yildiz A, Tomishige M, Gennerich A, Vale RD. Intramolecular strain coordinates kinesin stepping behavior along microtubules. Cell. 2008;134:1030–1041. doi: 10.1016/j.cell.2008.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]