Abstract
While recent studies indicated roles of long non-coding RNAs (lncRNAs) in physiologic aspects of cell-type determination and tissue homeostasis1 yet their potential involvement in regulated gene transcription programs remain rather poorly understood. Androgen receptor (AR) regulates a large repertoire of genes central to the identity and behavior of prostate cancer cells2, and functions in a ligand-independent fashion in many prostate cancers when they become hormone refractory after initial androgen deprivation therapy3. Here, we report that two lncRNAs highly overexpressed in aggressive prostate cancer, PRNCR1 and PCGEM1, bind successively to the AR and strongly enhance both ligand-dependent and ligand-independent AR-mediated gene activation programs and proliferation in prostate cancer cells. Binding of PRNCR1 to the C-terminally acetylated AR on enhancers and its association with DOT1L appear to be required for recruitment of the second lncRNA, PCGEM1, to the DOT1L-mediated methylated AR N-terminus. Unexpectedly, recognition of specific protein marks by PCGEM1-recruited Pygopus2 PHD domain proves to enhance selective looping of AR-bound enhancers to target gene promoters in these cells. In “resistant” prostate cancer cells, these overexpressed lncRNAs can interact with, and are required for, the robust activation of both truncated and full length AR, causing ligand-independent activation of the AR transcriptional program and cell proliferation. Conditionally-expressed short hairpin RNA (shRNA) targeting of these lncRNAs in castration-resistant prostate cancer (CRPC) cell lines strongly suppressed tumor xenograft growth in vivo. Together, these results suggest that these overexpressed lncRNAs can potentially serve as a required component of castration-resistance in prostatic tumors.
Keywords: Nuclear Receptor, Non-coding RNA, Enhancer, Pygopus2, Transcription Activation, Castration-resistant Prostate Cancer
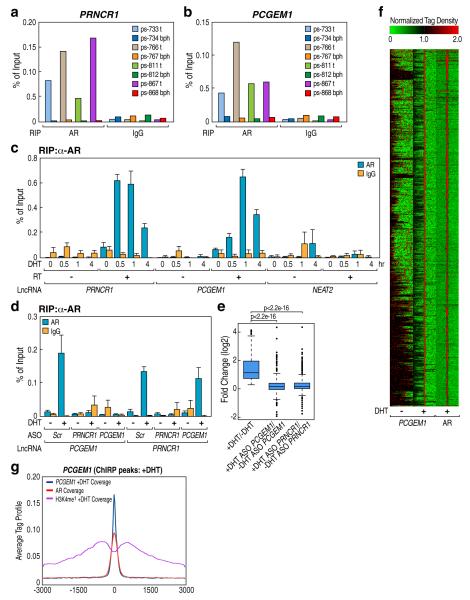
One of the overexpressed lncRNAs in prostate cancer, PCGEM1, is tissue-specific and correlated with high-risk prostate cancer patients, including African-American men4, while a second highly expressed lncRNA, PRNCR1 (PCNCR1), is pervasively transcribed from the 8q24 “gene desert” region in strong association with susceptibility of prostate cancer5. Paired benign prostatic hyperplasia (BPT) and aggressive tumor specimens (Gleason scores 3+3) derived from three individual prostate cancer patients exhibited >100-fold upregulation of PRNCR1 and PCGEM1 expression (Supplementary Fig. 1a, b). Native RNA-Immunoprecipitation (RIP) experiments in paired prostate tumor and BPH tissues (Gleason scores 2+3 to 4+3), revealed a specific association of full-length AR with both PRNCR1 and PCGEM1 in prostate tumor tissues (Fig. 1a, b; Supplementary Fig. 1a, c) compared to minimal interactions with glucocorticoid receptor (GR) (Supplementary Fig. 1c and data not shown). In LNCaP cells, DHT treatment induced association of AR with both PRNCR1 and PCGEM1 (Fig. 1c), but not with NEAT2 (Fig. 1c). Antisense oligonucleotides (ASO)-based knock-down of PRNCR1 abolished both AR-PRNCR1 and AR-PCGEM1 interactions while knock-down of PCGEM1 did not affect the AR-PRNCR1 interaction (Fig. 1d; Supplementary Fig. 2a), suggesting the PRNCR1-dependent recruitment of PCGEM1.
Figure 1. Signal-Dependent Interaction between AR and Prostate Specific LncRNAs.
a and b, RIP assay performed in paired benign prostatic hyperplasia (BPH) and prostate tumor (T) tissues. c, RIP assay in DHT-treated LNCaP cells (100 nM) for indicated time points. d, RIP assay in LNCaP cells transfected with indicated ASO followed by DHT treatment (100 nM). e, Global changes in DHT-induced AR target genes in PCGEM1 or PRNCR1 depleted LNCaP cells. f, Heatmap showing the distribution of PCGEM1 and AR binding sites in DHT-stimulated LNCaP cells. g, Average tag profile analysis of the aligned 2,142 PCGEM1 ChIRP peaks. Mean ± SEM for panel c and e (n=3).
Knock-down of either PRNCR1 or PCGEM1 resulted in a significantly decrease in DHT-induced activation of canonical AR target genes while not affecting AR levels (Supplementary Fig. 2a-c). Global run-on sequencing (GRO-Seq) confirmed that knock-down of either PRNCR1 or PCGEM1 significantly decreased the induction of 617 DHT-upregulated genes (n=617, edgeR FDR < 0.01, and read density RD > 2) with AR-bound enhancers within 200kb (Fig. 1e), but had no effect on DHT-unresponsive genes located >200kb away from AR-bound enhancers (Supplementary Fig. 2d), verified by randomly extracting sets of 1,000 genes (data not shown).
Using Chromatin Isolation by RNA Purification (ChIRP)6 with biotin-labeled DNA probes (40-60 nt) tiling PRNCR1 and PCGEM1 RNAs (Supplementary Fig. 3a-c), we identified 2,142 high-confidence PCGEM1 occupancy sites genome-wide (Supplementary Fig. 3d, e) and motif analysis revealed a very significantly enriched AR response element (ARE) DNA motif (Supplementary Fig. 3f), revealing that ~82% of PGCEM1 co-localized with AR-bound sites (−3kb/+3kb relative to the center of PCGEM1 peak), of which ~70% corresponded to AR bound, H3K4me1-marked enhancers (Fig. 1f, g and Supplementary Fig. 3g), independently confirmed by qPCR analyses (Supplementary Fig. 3h, i) and ChIRP-Seq using even-numbered and odd-numbered probe sets (Supplementary Fig. 3j and data not shown). These data suggest a stoichiometry of PCGEM1 sufficient to account for the recruitment to AR DNA regulatory binding sites on enhancers. Levels of PRNCR1 in LNCaP cells are estimated as ~400-600 copies/cell (data not shown). The ability of these lncRNAs to read enhancer-associated histone marks might account for their preferential presence at AR-bound enhancers (vide infra).
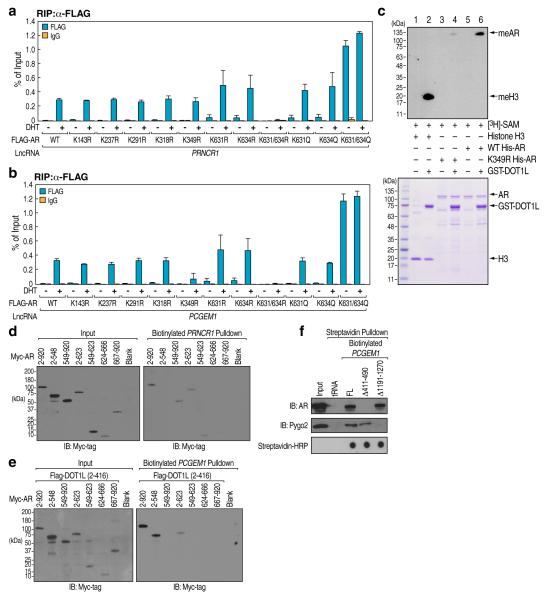
By imposing a high stringency wash condition, we identified that DOT1L, CARM1, GADD45α, and AR specifically associated with in vitro-transcribed biotinylated PRNCR1 by Mass-spectrometry analysis, while AR, β-Catenin, and Pygopus2 (Pygo2) preferentially associated with in vitro-transcribed biotinylated PCGEM1 (Supplementary Fig. 4a-c; Supplementary Tables 1-3). β-Catenin, CARM1 and GADD45α have been suggested to play important roles in AR signaling7. Knock-down of AR, Dot1L, β-catenin, and Pygo2 by specific siRNAs impaired DHT-induced activation of AR-target genes, TMPRSS2, PSA, and FKBP5 (Supplementary Fig. 4d). Mass-spectrometry analysis revealed that the lncRNA-bound AR contains several post-translational modifications, including K631/634 acetylation and K349 methylation (Supplementary Fig. 4e; Supplementary Tables 1-3). Consistent with the proposed importance of acetylation of AR in activation of an AR target gene8, a K631/634R mutation on AR inhibited its interaction with PRNCR1 and PCGEM1 (Fig. 2a; Supplementary Fig. 5a) and DHT-induced expression of AR target genes (Supplementary Fig. 5b), while overexpression of a AR K631/634Q mutant resulted in enhanced DHT-dependent interactions with PRNCR1 and PCGEM (Fig. 2a, b; Supplementary Fig. 5c). These data suggest that PRNCR1 and PCGEM1 interact with AR in a K631/634 acetylation- and K349 methylation-dependent manner, respectively.
Figure 2. Mechanistic Study of LncRNA with Associated Transcription Factors/Co-activators.
a and b, RIP assay in DHT-treated LNCaP cells (100 nM) expressing indicated plasmids. c. In vitro methylation assay for AR. d and e, LNCaP cells expressing Myc-tagged AR fragments (d) or co-transfect with Flag-tagged DOT1L (e) were subjected to RNA pulldown assay. f, In vitro transcribed PCGEM1 full-length, Δ411-490, or Δ1191-1270 were incubated with cell lysates extracted from DHT-treated LNCaP cell (100 nM, 1 hr) for in vitro RNA pulldown assay. Mean ± SEM for panel a and b (n=3).
Because effective AR-PCGEM1 interaction requires the methylation of AR at K349 (Fig. 2b), we confirmed DOT1L-mediated AR methylation at K349 using in vitro methylation assays, finding that a K349R point mutation significantly inhibited AR methylation (Supplementary 6a; Fig. 2c). Specific DOT1L knock-down impaired the interaction between AR and PCGEM1, but not that with PRNCR1 (Supplementary Fig. 6b, c), suggesting that AR K349 methylation, mediated by PRNCR1-bound DOT1L, is critical for the recruitment of PCGEM1 to AR. Indeed, overexpression of an AR K349R mutant significantly reduced DHT-induced gene activation in LNCaP cells (Supplementary Fig. 5b).
In vitro binding studies demonstrated that PRNCR1 bound to the region aa 549-623 of AR and PCGEM1 bound to AR N terminal region when methylated at K349 by overexpressing DOT1L (Fig. 2d, e; Supplementary Fig. 7a). By incubating in vitro transcribed PCGEM1 with nuclear lysate from cells overexpressing Myc-tagged Pygo2 proteins, including full-length, N- or C-terminally truncated proteins, we identified strong interactions between PCGEM1 and the Pygo2 C-terminus (Supplementary Fig. 7b).
To map the sequence motif of PCGEM1 responsible for AR or Pygo2 binding, we performed modified in vitro RNA pulldown followed by dot-blot assay (Supplementary Fig. 8a), using two regions of NEAT2-bound by unmethylated Pc2 as a control for the CLIP assay9 (Supplementary Fig. 8b). Methylated AR bound/protected PCGEM1 sequence was identified to encompass 421GAT…TCC480 (Supplementary Fig. 8c) with unmethylated AR or the unrelated protein, His-tagged MURF1, not showing specific binding to any region of PCGEM1 (Supplementary Fig. 8c). A sequence motif of PCGEM1 encompassing 1201TGT…ATT1260, distinct from the AR binding region, was identified as the Pygo2 binding motif, with deletion of this motif (Δ1191-1270) abolishing Pygo2 binding with no effect on AR binding (Supplementary Fig. 8c; Fig. 2f). Similarly, deletion of AR binding site of PCGEM1 (Δ411-490) abolished AR-PCGEM1 interaction, with minimal effect on Pygo2-PCGEM1 interaction (Fig. 2f).
MODified Histone Peptide Array experiments using in vitro transcribed biotinylated PCGEM1 or PRNCR1 revealed that they selectively recognize H3K4me1 and H4K16ac histone marks indicative of enhancers10,11, respectively (Supplementary Fig. 9a-c). Therefore, it is likely that these histone tail associations of PRNCR1 and PCGEM1 serve as a functional component of their preferred recruitment to enhancers of AR-regulated transcription units.
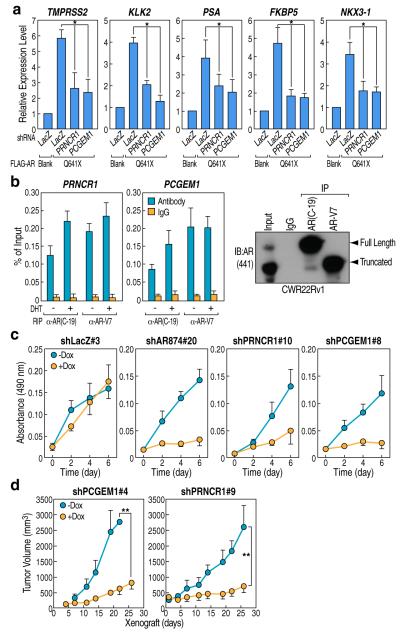
PCGEM1 and PRNCR1 were highly upregulated in the LNCaP-cds2, and CWR22Rv1 castration-resistant prostate cancer cell line models compared with immortalized “normal” prostate epithelial cell lines, WPE and RWPE, or even LNCaP cells (Supplementary Fig. 10a, b). The AR antagonist, bicalutamide (Casodex), reduced the DHT-induced PSA expression in LNCaP cells but failed to act as an antagonist in LNCaP-cds2 cells (Supplementary Fig. 10c). Transduction of LNCaP-cds2 cells with lentivirus expressing shRNAs against PRNCR1 or PCGEM1, but not a non-specific (LacZ) shRNA, significantly reduced the activation of several canonical AR target genes while having no effect on AR expression levels (Supplementary Fig. 10d-f and 11a). Because truncated forms of AR that exhibit ligand-independent transcriptional activation are frequently detected in castration-resistant prostate cancer and may often reflect alterations in AR gene structure, we investigated the potential roles of PRNCR1 and PCGEM1 in AR-mediated basal transcription activity in CRPC cells. RT-PCR using primers specific for one AR “splicing” variant, AR-V712 confirmed the presence of this variant in LNCaP-cds2 cells (Supplementary Fig. 11b). Western blot analysis using N-terminal AR-specific antibody (441), revealed that the AR-V7 variant (~75kDa) represents ~1-2% of total AR in LNCaP-cds2 cells, although it is the predominant form in CWR22Rv1 cells (Supplementary Fig. 10g). Because overexpression of truncated AR can constitutively activate androgen-responsive genes in the absence of ligand12, we transfected LNCaP cells with AR Q641X mutant, with activation of canonical androgen-responsive genes, including TMPRSS2, PSA, KLK2, FKBP5, and NKX3-1 in the absence of added androgen (Fig. 3a; Supplementary Fig. 12a, b). This constitutive effect of AR Q641X was highly reduced upon knock-down of either PRNCR1 or PCGEM1 (Fig. 3a). RIP assay in CWR22Rv1 cells demonstrated that both PRNCR1 and PCGEM1 associated constitutively with truncated AR (AR-V7) with or without ligand (Fig. 3b). By immunoblotting the AR-V7 immunoprecipitates with N-terminal AR-specific antibody (441), we did not observe any residual pull-down indicative of interaction between full length and truncated AR (Fig. 3b, right panel), arguing against an indirect association of PRNCR1 and PCGEM1 with truncated AR consequent to hetrodimerization with full length AR. Using an antibody specific for the C-terminal ligand binding domain of AR (C-19) to selectively recognize full length AR, we observed interactions between these lncRNAs and full length AR in the absence of added ligand (Fig. 3b), possibly due to the relative higher level of basal acetylation and methylation of full-length AR in CWR22Rv1 cells (Supplementary Fig. 12c).
Figure 3. PCGEM1 and PRNCR1 Promote Hormone-independent Activation of the AR Transcriptional Program in Castration Resistant Prostate Cancer (CRPC).
a, qRT-PCR analyses of AR targets in LNCaP cells co-transfected with indicated vectors followed by Doxycycline induction (160 ng/ml, 2d). b, RIP assay in CWR22Rv1 cells treated with or without DHT (100 nM, 1hr) using indicated antibodies. Detection of immunoprecipitated full-length AR and AR-V7 were shown. c, Cell proliferation assay in CWR22Rv1 cells stably expressing indicated shRNAs followed by Doxycycline induction (160 ng/ml) for indicated times. d, Xenografts of CWR22Rv1 cell lines harboring Doxycycline-induced shRNAs were monitored for tumor growth for the indicated time, with or without Doxycycline intake (4 mice/group). Mean ± SEM (n=6, *p<0.05 and **p<0.01).
To study the biological roles of PRNCR1 or PCGEM1, we generated stable cell lines derived from CWR22Rv1 harboring Doxycycline (Dox)-induced shRNA against LacZ, PRNCR1 or PCGEM1 (Supplementary Fig. 13a). Dox-induced either PCGEM1 or PRNCR1 knock-down significantly reduced the expression of canonical AR target genes, with no noticeable effect on AR expression level (Supplementary Fig. 11a, 13b). Dox-induced knock-down of either PRNCR1 or PCGEM1 also inhibited the growth of CWR22Rv1 cells comparable to the effect of AR knock-down13, without affecting AR expression levels (Fig. 3c; Supplementary Fig. 11a and 13c). Remarkably, conditional shRNA-mediated inhibition of either PRNCR1 or PCGEM1 robustly inhibited in vivo tumor growth in a CWR22Rv1 prostate cancer xenograft mice model (Fig. 3d), indicative of an lncRNA-dependent regulatory network that critically regulates growth of castration-resistant prostate cancer cells in vivo.
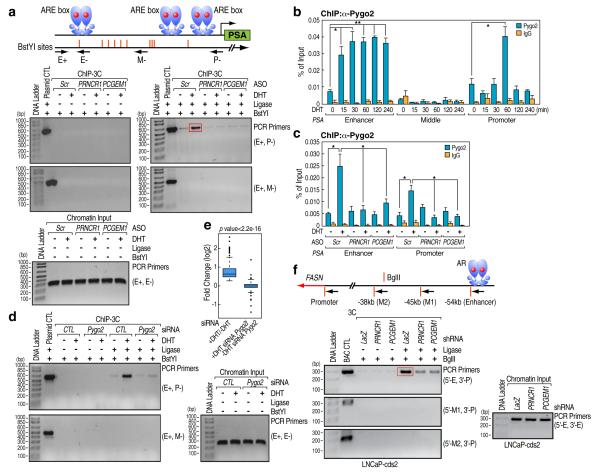
While knock-down of either PRNCR1 or PCGEM1 did not affect the recruitment of AR on PSA and KLK2 enhancers (Supplementary Fig. 14a, b, left panels), knock-down of PCGEM1 inhibited SMC1 recruitment on PSA and KLK2 promoters, with only minimal effects on SMC1 levels on enhancers (Supplementary Fig. 14a, b, right panels), consistent with proposed Cohesin-dependent14 formation of chromatin loops between enhancers and promoters. We further demonstrated ligand-induced enhancer: promoter interactions in the PSA transcription unit by ChIP-3C assay15 and found that these interactions were impaired by depletion of either PRNCR1 or PCGEM1 (Fig. 4a; Supplementary Fig. 14c).
Figure 4. Regulation of Enhancer: Promoter Interaction by PRNCR1 and PCGEM1.
a, ChIP-3C in LNCaP cells with indicated ASO transfection and treatment. b, ChIP–qPCR showing Pygo2 occupancy in DHT-treated LNCaP cells (100 nM) at indicated times. c, ChIP-qPCR indicate the Pygo2 recruitment in LNCaP cells with indicated transfection and treatment. d, ChIP-3C in LNCaP cells with indicated siRNA transfection and treatment. e, Global changes in DHT-induced AR targets in LNCaP cells with indicated siRNA transfection and treatment. f, 3C assay on FASN locus in LNCaP-cds2 cells transduced with indicated shRNA harboring lentivirus. Red box indicate PCR product sequenced in a and f. Mean ± SEM (n=3, *p<0.05 and **p<0.01).
The ability of Pygo2, associated with PCGEM1 (Supplementary Fig. 2a), to recognize a canonical promoter histone mark16, H3K4me3, raised the possibility that Pygo2 might be involved, at least quantitatively, in enhancer: promoter looping. ChIP assays revealed that Pygo2 was efficiently recruited to enhancer and promoter regions of the PSA, KLK2 and TMPRSS2 transcription units in a DHT-dependent manner, exhibiting relatively higher and earlier association with the enhancer regions (Fig. 4b; Supplementary Fig. 15a). Knock-down of either PRNCR1 or PCGEM1 in LNCaP cells inhibited Pygo2 recruitment to AR-dependent enhancers/promoters (Fig. 4c; Supplementary Fig. 15b). On knock-down of Pygo2 , AR and SMC1 failed to effectively associate with the PSA, KLK2 or TMPRSS2 promoters despite unperturbed DHT-dependent AR or SMC1 recruitment to their enhancers (Supplementary 16a-d) and DHT-induced enhancer: promoter looping in the PSA transcription unit was inhibited (Fig. 4d). GRO-Seq analysis revealed a broad inhibition of AR-dependent transcriptional program under the condition of Pygo2 knock-down (n=290, edgeR FDR < 0.01), which did not affect AR expression (Fig. 4e; Supplementary Fig. 16a).
Depleting PRNCR1 or PCGEM1 from LNCaP-cds2 cells also inhibited enhancer: promoter looping in FASN and NDRG1 (Supplementary Fig. 17a-d; Fig. 4f), previously shown to be activated following FoxA1 knock-down in LNCaP cells17. Again, knock-down of either PRNCR1 or PCGEM1 had no effect on recruitment of AR to enhancer regions of the FASN and NDRG1 transcription units or on Pygo2 expression levels (Supplementary Figs. 16a, 17e).
To address whether the Pygo2 PHD domain might itself be instrumental for its function in mediating chromatin looping, we first depleted Pygo2 by shRNA followed by overexpression in LNCaP-cds2 cells of either shRNA-resistant wild-type Pygo218 or a W352A mutant defective for H3K4me3 recognition16. In 3C assays, knock-down of Pygo2 reduced FASN enhancer: promoter interactions, which could be rescued by overexpression of wild-type, but not W352A, Pygo2 (Supplementary Fig. 18a, 19a), even though there was equal recruitment of wild-type Pygo2 or the W352A mutant to enhancers, and no altered promoter H3K4me3 levels (Supplementary Fig. 18b-d). Knock-down of Pygo2 curtailed the expression of canonical of AR target genes TMPRSS2, KLK2, PSA, FKBP5, and NKX3-1, and overexpression of wild-type, but not W352A, Pygo2 was able to robustly rescue the induction of these genes (Supplementary Fig. 19b). These data suggest that Pygo2 exerts a quantitatively-important role in DHT-dependent enhancer: promoter interactions and coding target gene activation. For 220 AR-regulated coding gene promoters under regulation of an enhancer exhibiting ligand-dependent recruitment of Pygo2 also exhibiting recruitment to the coding gene promoter by ChIP-seq, we did not observe ligand-induced increase in the next adjacent, non-AR-regulated transcription unit (~204 promoters) (Supplementary Fig. 19c), indicating that the H3K4me3 mark cannot alone be sufficient to effectively recruit Pygo2, suggesting a role of other similarly-modified proteins in prostate cancer cell gene activation events.
In the present study, we have found a mechanistic link between prostate cancer-upregulated lncRNAs and AR transcriptional activity, revealing the biological importance of the lncRNAs, PRNCR1 and PCGEM1, in licensing C-terminally truncated, as well as full length, AR-dependent gene activation events in prostate cancer cells (Supplementary Fig. 19d). Considering the regulatory potential of enhancer RNAs (eRNAs) identified in recent studies19,20, lncRNAs may also be part of a broad transcription regulatory network.
Methods
Tissue Samples and Processing
Experiments using paired benign prostatic hyperplasia (BPT) and tumor (T) were performed from unidentified individual prostate cancer patients, which were obtained from Dr. Ralph W. deVere White, UC Davis Comprehensive Cancer Center. The informed consent was obtained from all subjects. The tissue samples used in manuscript were received as the de-identified samples without any PHI attached. The Gleason score or tumor/BPH status was considered pathological information, not patient information. We did not know the names or birth dates. The tissues were homogenized by Precellys®24 tissue homogenizer followed by downstream assays.
Cell Culture and Transfection
Prostate cancer LNCaP cells were obtained from ATCC and cultured in RPMI1640 containing 10% (vol/vol) FBS. The benign immortalized prostate cell line RWPE, WPE and the castration-resistant prostate cancer cell lines LNCaP-cds1, LNCaP-cds2, CWR22Rv1 were kindly provided by Dr. Christopher Evans (Department of Urology, University of California Davis). RWPE and WEP cells were grown in KGM media and bulletkit from Lonza supplemented with L-Glutamine. CWR22Rv1, LNCaP-cds1 and LNCaP-cds2 cells were grown in RPMI1640 media containing 5% final volume of Charcoal Stripped Serum. LNCaP cells were grown to 30-50% confluence and siRNA/ASO transfections were carried out using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. Transfection of LNCaP cells with DNA plasmids was performed using Amaxa™ Nucleofector™ kit R from Lonza. shRNAs specific for LacZ, PRNCR1, PCGEM1 or AR were delivered, by lentiviral transduction, to LNCaP-cds2 and CWR22Rv1 cells.
Cloning Procedures
The full-length AR expression vector has been previously described17. Human PCGEM1 (14-1556) and PRNCR1 fragments (1-3240, 3156-6428, 6331-9670 and 9531-12710) were amplified from cDNAs generated from LNCaP cells and cloned into pSTBlue-1 vector (Novagen) for in vitro transcription assay. PCGEM1 gene sequence with 411-490 and 1191-1270 deletion were synthesized by GeneScript Inc. and cloned into pcDNA3.1 vector (Invitrogen). Lentiviral vector pLKO.1 containing the shRNA against scrambled sequence, Pygo2 and pHIV vector containing RNAi-resistant Pygo2 cDNA were obtained from Dr. Xing Dai (Department of Biological Chemistry, University of California at Irvine)18. A 4.8 kb genomic sequence upstream of PSA promoter was amplified from LNCaP cells and subcloned into pSTBlue-1 vector (Novagen). Bacterial expression vectors for AR and K349R mutant were constructed by subcloning the gene sequences into pET-28a backbone (Novagen). Nuclear expression vectors for AR2-920, AR2-548, AR549-920, AR2-623, AR549-623, AR624-666, AR667-920 and Pygo22-406, Pygo22-266, Pygo2267-406 were constructed by subcloning the cDNA sequences into pCMV/myc/nuc backbone (Invitrogen). The expression vector of FLAG-DOT1L was kindly provided by Dr. Yi Zhang’s laboratory. All mutants were generated using QuikChange™ Lightning Site-Directed Mutagenesis Kit (Agilent Technologies). Detailed oligonucleotide sequences were listed in Oligonucleotide Sequences and Primers section.
Antisense DNA Oligonucleotide, siRNA and Lentiviral shRNA
2′-O-methyl phosphor-othioate oligonucleotides were designed and synthesized by Integrated DNA Technologies, Inc. Commercially available FlexiTube siRNA targeting AR (SI02757258) and CARM1 (SI02663815) from Qiagen, ON-TARGETplus SMARTpool siRNA targeting DOT1L (L-014900-01-0005) and GADD45α (L-003893-00-0005) from Dharmacon, MISSION® siRNA targeting β-Catenin (SASI_Hs01_00117958), Pygopus2 (SASI_Hs01_00059018) from Sigma-Aldrich were used in this study. The knockdown efficiency and specificity of all siRNAs were validated either by vendors or ourselves (Supplementary Fig. 6b, 16a). Oligonucleotides for shRNA targeting PRNCR1, PCGEM1 and AR were designed athttp://biosettia.com/support/shrna-designer and cloned into pLV-H1TetO-GFP-Puro vector according to manufacturer’s instructions (Biosettia). We tested the efficacy and specificity of 3 ASOs (see Supplementary Fig. 2a) and 2 shRNAs (see Supplementary Fig. 10d) in LNCaP and LNCa-cds2 cells, respectively, for both PRNCR1 and PCGEM1. For functional assays, the specific ASO/shRNA giving the best knockdown efficiency was used. Detailed ASO/shRNA sequences were listed in Oligonucleotide Sequences and Primers section.
Antibodies
Specific antibodies were purchased from the following commercial sources: anti-AR (N-20), anti-AR (C-19), anti-AR (441), anti-GR (E-20), anti-β-Catenin (D-10), and anti-GAPDH (6C5) from Santa Cruz Biotechnology; anti-CARM1 (4438), anti-GADD45α (3518) and anti-pan acetylated-lysine (#9441) from Cell Signaling Technology; anti-FLAG® M2, anti-γ-Tubulin (T5326) and anti-Pygo2 from Sigma-Aldrich Prestige Antibodies®; anti-Pygo2 (GTX119726) from GeneTex Inc.; Anti-DOT1L (39954) and anti-Myc tag (clone 4E12) from Active Motif; anti-SMC1 (A300-055A) from Bethyl Laboratories Inc.; anti-pan methylated lysine (7315) from Abcam and anti-AR-V7 from Precision Antibody™.
Protein Recombination and Purification
Recombinant His-AR proteins were expressed in E.coli strain BL21-CodonPlus® (DE3)-RIPL (Agilent Technologies) and purified using TALON® Metal Affinity Resin (Clontech). Recombinant Histone H3 was purchased from Active Motif. Human DOT1L (amino acids 2-416) was obtained from Sigma-Aldrich. Human Pygo2 was purchased from BioClone Inc. His-tagged MURF1 was purchased from BostonBiochem®.
RNA Immunoprecipitation (RIP)
RIP was performed in native conditions as described21. Briefly, 1×107 LNCaP cell nuclei were pelleted and lysed in 1mL ice-cold Polysomal Lysis Buffer (100mM KCl, 5mM MgCl2, 10mM HEPES [pH 7.0], 0.5% NP-40, 1mM DTT) supplemented with Anti-RNase, Protease Inhibitor Cocktail, Phosphatase Inhibitor Cocktail, Panobinostat, and Methylstat. The lysate were passed through a 27.5 gauge needle 4 times to promote nuclear lysis. Turbo™ DNase (400 U) was then added to the lysate and incubated on ice for 30 min. The cell lysate was diluted in the NT2 buffer (50mM Tris-HCl [pH 7.4], 150mM NaCl, 1mM MgCl2, 0.05% NP-40) and 50 ul of the supernatant was saved as input for PCR analysis. 500 ul of the supernatant was incubated with 4 μg of AR (441) antibodies-IgG magnetic beads (pre-blocked by 1×PBS+5 mg/ml BSA) at 4 °C overnight. The RNA/antibody complex was washed four times (1 ml wash, 5 minutes each) by NT2 buffer supplemented with Anti-RNase, Protease Inhibitor Cocktail, Phosphatase Inhibitor Cocktail, Panobinostat and Methylstat. The RNA was extracted using Acid-Phenol: Chloroform, pH 4.5 (with IAA, 125:24:1) (Invitrogen) according to the manufacturer’s protocol and subjected to RT-qPCR analysis.
Chromatin Isolation by RNA Purification (ChIRP)
ChIRP was performed as described6 with minor modifications. Briefly, 60-mer antisense DNA probes targeting PRNCR1 and PCGEM1 full-length sequences were designed at http://www.singlemoleculefish.com/designer.html. A set of probes targeting LacZ RNA was also designed as the negative control. All probes were biotinylated using Label I® Nucleic Acid Biotin Labeling Kit from Mirus Biotechnology. LNCaP cells were fixed with 1% formaldehyde for 10 min at room temperature. Crosslinking was then quenched with 125mM glycine for 5 min. The chromatin preparation, hybridization/elution, deep sequencing steps were essentially performed as described4 except that wash was conducted at 50 °C and 65°C. The image analysis and base calling were performed using Illumina’s computational analysis pipeline. The sequencing reads were aligned to hg18 human genome by using Bowtie222 and only 1 read/position was kept for downstream analyses. Peak finding was performed by using HOMER suite23 and the peaks within 1kb from each other were merged. Peak intersection was computed by using intersectBed in BedTools24, after extending the peaks with 1kb. In order to call reliable peaks, we have excluded from analysis the peaks that overlapped the satellite repeats or LacZ ChIRP peaks. The annotation of the peaks on the human genome and the tag density profiles were computed in HOMER, and the display of the heatmap were carried in MeV25. Sequenced motif enrichment analysis was carried in HOMER. For ChIP-Seq data (AR and H3K4me1), peak finding was performed by using HOMER or SICER26.
Data Processing Procedure for Overlapping ChIP-Seq and ChIRP-Seq
We used the standard ChIP-Seq peak finding tools in the processing of ChIRP-Seq data. We chose HOMER software suite (http://biowhat.ucsd.edu) for the analysis of both AR ChIP-Seq and PCGEM1 ChIRP-Seq data, using the same program routine (i.e. findPeaks command) and the same default parameters that calls only the robust peaks (these parameters are outlined below): 1) fold enrichment over input tag count, default: 4.0; 2) poisson p-value threshold relative to input tag count, default: 0.0001; 3) fold enrichment over local tag count, default: 4.0; 4) poisson p-value threshold relative to local tag count, default: 0.0001; 5) fdr <#> false discovery rate, default = 0.001; 6) size of region used for local filtering = 10000; 7) fold over local region required = 4.00. Peak finding procedure: 1) typically, the tag distribution along the genome could be modeled by a Poisson distribution and the peak finding algorithm slides windows of fixed size across the genome to find candidate peaks with a significant tag enrichment (Poisson distribution p-value default 10-4 to 10-5); 2) we use very strict parameters in HOMER to call a peak: a very low FDR (i.e. 0.001), and a high fold enrichment over input tag count (i.e. 4). By default, HOMER also requires the tag density at peaks to be 4-fold greater than in the surrounding 10 kb region. Therefore, we ensure that only the sharp peaks with low local background are called and considered for downstream analyses. Both ChIP-seq and ChIRP-seq data were processed precisely in the same way using the same default settings of HOMER.
For the analysis of overlapping between ChIP-Seq and ChIRP-Seq data, we used the following samples: PCGEM1ChIRP-Seq (−DHT), PCGEM1 ChIRP-Seq (+DHT), and AR ChIP-Seq (at a higher sequencing depth). The heatmaps were generated in 2 steps: 1) we used HOMER and the command [(annotatePeaks.pl <peak file> <genome> −size <#> −hist <#> −ghist) that specifies the list of peaks (<peak file>), and the tag density in a region that covers <size> kb around the peak center; the tag density is specified in a bin size that is specified by the parameter <hist>)] to generate a matrix of tag densities across the samples, for each peak. Typically the tag densities are normalized to 10 mil reads for each sample; 2) we used MeV package (http://www.tm4.org/mev/) to display the heatmap and to color code it on a scale from 0 to 2: a difference in the colors from 2 (red) to 0 (green) may reflect a fold enrichment over local tag count higher than 4 (4 is typically the default value, used for peak finding, as we described above).
GRO-Seq
The image analysis and base calling were performed by using Illumina’s standard computational analysis pipeline. Bowtie222 was used to align the sequencing reads to hg18 human genome; when multiple reads aligned to the same genomic position, only 1 read/position was kept for downstream analyses. We have used RefSeq annotations in order to estimate the gene expression level by counting the sequencing reads over the gene body (from 400bp downstream of TSS to TTS) on the sense strand with respect to the gene transcription, by using intersectBed in BedTools24. In order to call statistically significant differentially expressed genes, we have used edgeR27 and a FDR < 0.01. Additional criteria, including read density (normalized number of reads/kb) were imposed in order to filter the differentially expressed genes. Wilcoxon tests for sample comparisons were computed in R.
Chromatin immunoprecipitation (ChIP) assay, ChIP-Seq and ChIP-3C assay
ChIP assays and ChIP-Seq was performed as described28. ChIP-3C assays were performed as described15 with minor modifications. Briefly, LNCaP cells were cross-linked with 1% formaldehyde for 10 min followed by incubation with 125mM glycine for 5 min. The cross-linked chromatin was sonicated and digested with restriction enzyme BstYI (New England Lab) followed by immunoprecipitated with specific antibody coupled to Protein A Dynabeads® (Invitrogen). The beads bound chromatin was ligated with T4 DNA ligase (Promega), eluted and reverse-cross-linked. The ChIP-3C material was purified by QIAquick Gel Extraction Kit (Qiagen) and subjected to PCR analysis with sets of primers as described15.
3C Assay
LNCaP-cds2 cells were cross-linked with 1% formaldehyde for 10 min at room temperature and processed according to the procedures as described17 with minor modification. Briefly, BAC clones (Empire Genomics) for FASN (RP11-1033I, RP11-1033J9) and NDRG1 (RP11-671M3, RP11-125I19) loci were used to generate control templates for the positive controls. 15 μg of the BAC clone for each locus were mixed and digested with 300 units of corresponding restriction enzyme overnight at 37oC. DNA fragments were extracted and ligated with T4 DNA ligase at a DNA concentration of 300 ng/μl. The primers for the fragments on the FASN and NDRG1 loci were used as previously described17.
In Vitro RNA pulldown Coupled with Dot-Blot Assay
In vitro transcribed biotinylated RNA was formed secondary structure as described29 and incubated with recombinant His-tagged or GST-tagged proteins in gel shift protein binding buffer (Promega) on ice for 1 hour. The reactions were ultraviolet (UV) irradiated (150 mJ/cm2) to crosslink RNA-protein complexes. After UV irradiation, the RNA was partially digested by RNase I (Ambion) at 1:50 and 1: 500 dilutions for 5 min, allowing a small fragment to remain attached to protein. RNA-protein complexes of interest were then partially purified by His tag or GST tag magnetic beads and the purified RNA-protein complexes were treated with proteinase K, which removes protein but leaves intact RNAs. The recovered RNAs were hybridized to BrightStar®-Plus Positively Charged Nylon Membrane spotted with 60-mer antisense DNA oligonucleotides tiling along the indicated lncRNA sequence at 37 °C overnight. The anti-sense oligonucleotides corresponding to the indicated lncRNAs were spotted on membrane as following order (left to right in each row): A1 is corresponding to the oligonucleotide sequence nt 1-60 of indicated lncRNA, A2 is corresponding to oligonucleotide sequence nt 61-120 of indicated lncRNA and so on till the end of the lncRNA sequences. The hybridized membrane was washed as described at sequential 37°C, 50 °C and 65°C. The protein-bound RNA sequence was visualized by detection of Streptavidin-HRP signals. The positions and sequences of antisense DNA oligonucleotides tiling along PCGEM1 is available upon request.
In Vitro Methylation Assay
In vitro methylation assays were conducted in a total volume of 30 μl using 2 μg of substrate, and 1 μg of recombinant GST-DOT1L in methylation buffer (50 mM HEPES [pH 8.0], 0.01% (v/v) NP-40, 10 mM NaCl, 1 mM DTT and 1 mM PMSF) supplemented with either 40 μM S-Adenosyl-methionine (SAM, Sigma-Aldrich; for nonradioactive methylation) or 300 nM S-Adenosyl-L-[methyl-[3H]methionine ([3H]-SAM, Perkin Elmer; 0.55 μCi per reaction for radioactive methylation). Reactions were carried out at 37°C for 3 hours and the reactions were separation on a 4-12% SDS-PAGE gel. The resulting protein bands were visualized by Coomassie blue staining, immunoblotting or autoradiography using EN3HANCE™ spray (Pelkin Elmer). Purification of methylated protein was carried out as described9.
Cell Lysis, Protein Immunoprecipitation, and Immunoblotting
Cells were homogenized in 1×RIPA buffer with protease inhibitor and Anti-RNase (Invitrogen). Lysates were cleared by centrifugation at 13,000 rpm for 15 min at 4°C. The protein concentration of the extracts was determined by Bradford assay (Bio-Rad). Immunoprecipitation experiments and immunoblotting analyses were performed as described in previous studies9. The blotting signals were detected using SuperSignal West Dura Extended Duration Substrate (Pierce).
Lentivirus Packaging and Transduction
Production of Lentiviral shRNA stocks were performed in 293LTV cell line according to manufacturer’s instruction (Cell Biolabs). The lentivirus was further purified and concentrated by ViraBind™ Lentivirus Concentration and Purification Kit (Cell Biolabs). LNCaP-cds2 and CWR22Rv1 cells were transduced using ViraDuctin™ Lentivirus Transduction Kit (Cell Biolabs) and purified with 0.6 and 0.4 μg/ml puromycin, respectively. To establish stable cell line expressing shRNA against LacZ, PRNCR1, PCGEM1 or AR, CWR22Rv1 cells were transduced as described above and stable transduced clones were generated following selection with 0.4 μg/ml puromycin. Single cell clones were then isolated by three rounds of single cell dilution, and the efficiency of Doxycycline-induced target gene knockdown was screened by RT- qPCR with 24 individual clones for each target gene (see Supplementary Fig. 13a).
RNA Pulldown Assay and Mass Spectrometric Analysis
RNA pulldown assay was performed as previously described9 with minor modifications. Biotin-labeled PCGEM1 RNA (14-1556) and PRNCR1 RNA fragments (1-3240, 3156-6428, 6331-9670 and 9531-12710) were in vitro transcribed with the Biotin RNA Labeling Mix (Roche) and T7 or SP6 RNA polymerase (Promega) and purified with RNeasy® Mini Kit (QIAGEN). RNAs were incubated with nuclear extract from LNCaP cells in the presence of anti-RNase, Protease Inhibitor Cocktail, Phosphatase Inhibitor Cocktail, Panobinostat, and Methylstat. Proteins pulled down by biotinylated PRNCR1 and PCGEM1 were first eluted, denatured, reduced, alkylated and then digested with immobilized trypsin (Promega). The digested peptides were batch purified on a reversed-phase ZipTip® (Millipore), and resulting peptide pools were then subjected to mass spectrometric analysis at UCSD Biomolecular and Proteomics Mass Spectrometry Facility. Data were analyzed using ProteinPilot™ software (Applied Biosystems).
Modified Histone Peptide Array
The interaction between in vitro transcribed PRNCR1 and PCGEM1 with Histone was analyzed by MODified™ Histone Peptide Array (Active Motif) according to manufacturer’s instruction. The specificity of interactions was quantified by Array Analyses Software (Active Motif).
Histone Peptide Pulldown Assay
Histone peptide pulldown assays were performed using SensoLyte® Methylated Histone H3 kit from AnaSpec. Briefly, 1μg of recombinant Pygo2 protein was incubated with 1μg of biotinylated histone H3 peptides (unmodified, K4me1, K4me2, or K4me3) in the presence of yeast tRNA or in vitro transcribed PCGEM1 in binding buffer (50 mM Tris-HCl, pH7.5, 150 mM NaCl, 0.05% NP-40) for 2hrs at 4°C with rotation. After 1hr incubation with streptavidin magnetic beads and extensive washing, the bound protein was analyzed by SDS-gel electrophoresis and western blotting with anti-Pygo2 antibodies. For this assay, PCGEM1 was in vitro transcribed using unlabeled UTP.
Cell Proliferation Assay
Cell proliferation assay was performed using CellTiter 96® AQueous One Solution Cell Proliferation Assay (MTS) (Promega). Briefly, cells were distributed in 96-well plate with 100 μl media each. After removing media and rinsed once by PBS, cells were supplied with 100 μl PBS mixed by 4 μl MTS reagent, followed by 1hr incubation at 37°C 5% CO2 incubator. After incubation, 490nm absorption of each well was measured by light absorbance reader. Every group has 6 replicates.
Xenografts and Animals
CWR22Rv1 stable cell lines with Doxycycline-inducible shRNA against LacZ, PRNCR1 or PCGEM1 were injected into male athymic Nu/Nu mice, 4-5 weeks old. Mice arrived in our facility were randomly put into cages with four mice each. They were implanted with respective tumor cells in the unit of cages. Three million of cells were suspended with matrigel in the ratio of 2:1 for subcutaneous injection. The experiment was set up to use eight animals per group to detect a 2-fold increase in tumor size with power of 80% and at the significance level of 0.05 by a two-sided test for significant studies. In these experiments, the tumor take rate was often about 50%, so we often ended up with about 4 animals per group, with or without doxycycline treatment. Tumor progression was monitored by caliper measurement twice a week and the tumor volume was calculated according to the equation, v=length × width2 ×1/2. When the tumor size reached between 150 – 200 mm3, animals were randomly distributed to groups receiving 1 mg/ml of doxycycline water or regular water with continuous tumor monitor until tumor burden exceeded the limit of IACUC humane endpoints (less than 20 mm in one dimension), for 3 to 4 weeks. The investigators were blinded to the group allocation during the experiment and/or when assessing the outcome. This study was carried out in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. Animals were housed in the UC Davis Medical Center Animal Facility (Sacramento, CA) under pathogen-free conditions; protocol approved by the Institutional Animal Care and Use Committee.
RNA isolation and qRT–PCR
Total RNA was isolated from cells using RNeasy Plus Mini Kit (Qiagen) following the manufacturer’s protocol. First-strand cDNA synthesis from total RNA was carried out using iScript™ cDNA Synthesis Kit (Bio-Rad). Resulting cDNA was then analyzed by qPCR using SsoAdvanced™ SYBR® Green Supermix (Bio-Rad) on Stratagene Mx3000 or CFX Connect™ Real-Time PCR Detection System. Primers are specific for genes tested and their sequences are listed in Oligonucleotide Sequences and Primers section. All qPCRs were repeated at least three times.
Oligonucleotide Sequences (5′-3′) and Primers (forward and reverse)
Antisense Oligonucleotide
Scrambled (mA*mA*mG* mC*mG*C* G*C*A* C*C*A* G*C*G* mC*mC*mU* mC*mC)30, PRNCR1 #1 (mC*mC*mC* mU*mC*C* T*C*C* C*T*C* T*C*T* mC*mU*mU* mG*mC), PRNCR1 #2 (mA*mC*mU* mC*mU*C* C*T*T* C*T*C* C*A*C* mC*mU*mC* mC*mA), PRNCR1 #3 (mA*mC*mU* mC*mC*C* A*C*A* C*C*A* C*C*A* mC*mC*mA* mC*mC), PCGEM1 #1 (mU*mU*mC* mC*mC*T* C*T*G* C*T*T* G*C*C* mU*mG*mU* mU*mG), PCGEM1 #2 (mG*mC*mU* mU*mU*A* C*C*C* T*T*A* G*T*C* mC*mU*mC* mC*mA), and PCGEM1 #3 (mA*mG*mU* mC*mC*T* C*C*A* C*G*T* G*C*C* mU*mA*mC* mC*mC).
shRNA
Lac Z (AAA AGC AGT TAT CTG GAA GAT CAG GTT GGA TCC AAC CTG ATC TTC CAG ATA ACT GC) (Bioseitta); PRNCR1 #1 (AAA ACA GTT TGA TTA GGG AGG CAC ATT TAT TGG ATC CAA TAA ATG TGC CTC CCT AAT CAA ACT G); PRNCR1 #2 (AAA AAA GGA AGG ACT TTC CAG CAC CTT AAT TGG ATC CAA TTA AGG TGC TGG AAA GTC CTT CCT T); PCGEM1 #1 (AAA ACC TTT GCA GAG AGC ATG CTT TCC TAT TGG ATC CAA TAG GAA AGC ATG CTC TCT GCA AAG G); PCGEM1 #2 (AAA AAC AAC CTT TGC AGA GAG CAT GCT TTT TGG ATC CAA AAA GCA TGC TCT CTG CAA AGG TTG T); and AR (AAA AGG TTC TCT GCT AGA CGA CAT TGG ATC CAA TGT CGT CTA GCA GAG AAC C)13.
qPCR Primer for Gene Expression and RIP
Actin (GCT CGT CGT CGA CAA CGG CTC and CAA ACA TGA TCT GGG TCA TCT); PRNCR1 (CCA GAT TCC AAG GGC TGA TA and GAT GTT TGG AGG CAT CTG GT); PCGEM1 (GGT GCC TTT GCC AAT GTT AT and AGC ATG CTC TCT GCA AAG GT); NEAT2 (TGG GGG AGT TTC GTA CTG AG and TCT CCA GGA CTT GGC AGT CT); TMPRSS2 (CTG GTG GCT GAT AGG GGA T and GTC TGC CCT CAT TTG TCG AT); KLK2 (AGC CTG CCA AGA TCA CAG AT and GCA AGA ACT CCT CTG GTT CG); PSA (GAT GCT GTG AAG GTC ATG GA and TGG AGG TCC ACA CAC TGA AG); FKBP5 (TCC CTC GAA TGC AAC TCT CT and GCC ACA TCT CTG CAG TCA AA); NKX3-1 (GCC AAG AAC CTC AAG CTC AC and AGA AGG CCT CCT CTT TCA GG); PGC-1 (GAG TTC CTG AGG ACC CAC AA and AGG AAG TTC TGG GGT GGA GT); FASN (AGG ATC ACAG GGA CAA CCT G and ACT CCA CAG GTG GGA ACA AG); and NDRG1 (ACC TGC TAC AAC CCC CTC TT and TGA TCC ATG GAG GGG TAC AT).
qPCR Primer for ChIRP and ChIP
PSA enhancer (TGG GAC AAC TTG CAA ACC TG and CCA GAG TAG GTC TGT TTT CAA TCC A); PSA middle region (CAG TGG CCA TGA GTT TTG TTT G and AAC CAA TCC AAC TGC ATT ATA CAC A); PSA promoter (CCT AGA TGA AGT CTC CAT GAG CTA CA and GGG AGG GAG AGC TAG CAC TTG); KLK2 enhancer (GTT GAA AGC AGA CCT ACT CTG GA and GCA TAT TTG TAC AGC AGA TAG CC); KLK2 middle region (ATC TCA AGG ACT TCT GGG TGG A and TGG GTA GTC CCT GTT ACA AGA T); KLK2 promoter (GGG AAT GCC TCC AGA CTG ATC and CTT GCC CTG TTG GCA CCT AGA); TMPRSS2 enhancer (TGG TCC TGG ATG ATA AAA AAA GTT T and GAC ATA CGC CCC ACA ACA GA); TMPRSS2 middle region (CCA GAA GAA TAC AAT GAT TAA AAG GCT and TGG AAC TGA AGT ATT GGA AAA CCA); TMPRSS2 promoter (CTG AGC CCC CAC AAT TGC AAA AC and GGT GGG ACA CAC CTC AGC C); RBL1 promoter (CAG CGT GGG GCT TGT CCT CG and AGC GGA GGC AGA CGG TGG AT)9; MyoD1 enhancer (CAG CCA AGT ATC CTC CTC CA and AAG CTG AGC ACT CTG GGA GA)14; FASN enhancer (CTA CTT CTC CCG TGC CAC TC and TCT CTC CCC TTC GAT GTG TC); and NDRG1 enhancer (GGT CAC ATC CAA GTG GGA CT and AGA AGG TGG AGA GGG CAG TT).
PCR primer for PSA Enhancer
(ATA GGG TTG GGC ACT CAC AGC TGA AT and AAT GCT GGC AGA GTC CAT GAG ACT CC).
RT-PCR primer for AR variants
The full-length and truncated ARs were detected using primers as described12. F1: TGT CAC TAT GGA GCT CTC ACA TGT GG; R1: CAC CTC TCA AAT ATG CTA GAC GAA TCT GT; R2: GTA CTC ATT CAA GTA TCA GAT ATG CGG TAT CAT; F6: CCA TCT TGT CGT CTT CGG AAA TGT TAT GAA GC; R8: AGC TTC TGG GTT GTC TCC TCA GTG G
Supplementary Material
Acknowledgements
We are grateful to X. Dai for providing the Pygo2 shRNA and cDNA constructs and J. Hightower for assistance with figure presentation. This work was supported by NIH grants DK039949, DK18477, NS034934 to M.G.R. and from DoD and CA173903 to M.G.R. and initially by a grant from PCF to M.G.R.; by grants from the National Institutes of Health Pathway to Independence Award (1K99DK094981-01) to C.-R.L.; the US Army Medical Research and Material Command (W81XWH-08-1-0554), the National Institutes of Health Pathway to Independence Award (4R00CA166527-02) and Cancer Prevention Research Institute of Texas First-time Faculty Recruitment Award (R1218) to L.-Q.Y.; C.-Y.J. is the recipient of a Cancer Research Institute Postdoctoral Fellowship. M.G.R. is an Investigator of the Howard Hughes Medical Institute.
Footnotes
Author Contributions L.-Q.Y., C.-R.L. and M.G.R. designed the research, and L.-Q.Y. and C.-R.L. performed most of the experiments, with participation of C.-Y.J.; J.-C. Y., under supervision of C.E., performed in vivo tumor xenograft experiments. B.T. performed bioinformatics analyses on GRO-Seq, ChIP-seq and ChIRP-Seq data, W.-B.L., J.Z. and K.A.O. conducted high-throughput sequencing, and Da. M. helped with ChIRP assays, L.-Q.Y., C.-R.L. and M.G.R. wrote the manuscript.
Author Information The high-throughput sequencing data sets are deposited in the Gene Expression Omnibus database under accession GSE 47807. Reprints and permissions information is available at www.nature.com/reprints.
The authors declare no competing financial interests.
References
- 1.Gupta RA, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–1076. doi: 10.1038/nature08975. doi:nature08975 [pii] 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Heinlein CA, Chang C. Androgen receptor in prostate cancer. Endocr Rev. 2004;25:276–308. doi: 10.1210/er.2002-0032. [DOI] [PubMed] [Google Scholar]
- 3.Scher HI, Sawyers CL. Biology of progressive, castration-resistant prostate cancer: directed therapies targeting the androgen-receptor signaling axis. J Clin Oncol. 2005;23:8253–8261. doi: 10.1200/JCO.2005.03.4777. doi:23/32/8253 [pii] 10.1200/JCO.2005.03.4777. [DOI] [PubMed] [Google Scholar]
- 4.Petrovics G, et al. Elevated expression of PCGEM1, a prostate-specific gene with cell growth-promoting function, is associated with high-risk prostate cancer patients. Oncogene. 2004;23:605–611. doi: 10.1038/sj.onc.1207069. doi:10.1038/sj.onc.1207069 1207069 [pii] [DOI] [PubMed] [Google Scholar]
- 5.Chung S, et al. Association of a novel long non-coding RNA in 8q24 with prostate cancer susceptibility. Cancer Sci. 2011;102:245–252. doi: 10.1111/j.1349-7006.2010.01737.x. doi:10.1111/j.1349-7006.2010.01737.x. [DOI] [PubMed] [Google Scholar]
- 6.Chu C, Qu K, Zhong FL, Artandi SE, Chang HY. Genomic maps of long noncoding RNA occupancy reveal principles of RNA-chromatin interactions. Mol Cell. 2011;44:667–678. doi: 10.1016/j.molcel.2011.08.027. doi:S1097-2765(11)00680-0 [pii] 10.1016/j.molcel.2011.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kypta RM, Waxman J. Wnt/beta-catenin signalling in prostate cancer. Nature reviews. Urology. 2012 doi: 10.1038/nrurol.2012.116. doi:10.1038/nrurol.2012.116. [DOI] [PubMed] [Google Scholar]
- 8.Fu M, et al. Acetylation of androgen receptor enhances coactivator binding and promotes prostate cancer cell growth. Mol Cell Biol. 2003;23:8563–8575. doi: 10.1128/MCB.23.23.8563-8575.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang L, et al. ncRNA- and Pc2 methylation-dependent gene relocation between nuclear structures mediates gene activation programs. Cell. 2011;147:773–788. doi: 10.1016/j.cell.2011.08.054. doi:S0092-8674(11)01216-5 [pii] 10.1016/j.cell.2011.08.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zippo A, et al. Histone crosstalk between H3S10ph and H4K16ac generates a histone code that mediates transcription elongation. Cell. 2009;138:1122–1136. doi: 10.1016/j.cell.2009.07.031. doi:S0092-8674(09)00911-8 [pii] 10.1016/j.cell.2009.07.031. [DOI] [PubMed] [Google Scholar]
- 11.Heintzman ND, et al. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature. 2009;459:108–112. doi: 10.1038/nature07829. doi:nature07829 [pii] 10.1038/nature07829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hu R, et al. Ligand-independent androgen receptor variants derived from splicing of cryptic exons signify hormone-refractory prostate cancer. Cancer Res. 2009;69:16–22. doi: 10.1158/0008-5472.CAN-08-2764. doi:69/1/16 [pii] 10.1158/0008-5472.CAN-08-2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sun A, et al. Adeno-associated virus-delivered short hairpin-structured RNA for androgen receptor gene silencing induces tumor eradication of prostate cancer xenografts in nude mice: a preclinical study. Int J Cancer. 2010;126:764–774. doi: 10.1002/ijc.24778. doi:10.1002/ijc.24778. [DOI] [PubMed] [Google Scholar]
- 14.Taberlay PC, et al. Polycomb-repressed genes have permissive enhancers that initiate reprogramming. Cell. 2011;147:1283–1294. doi: 10.1016/j.cell.2011.10.040. doi:S0092-8674(11)01335-3 [pii] 10.1016/j.cell.2011.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang Q, Carroll JS, Brown M. Spatial and Temporal Recruitment of Androgen Receptor and Its Coactivators Involves Chromosomal Looping and Polymerase Tracking. Molecular cell. 2005;19:631–642. doi: 10.1016/j.molcel.2005.07.018. [DOI] [PubMed] [Google Scholar]
- 16.Gu B, et al. Pygo2 expands mammary progenitor cells by facilitating histone H3 K4 methylation. J Cell Biol. 2009;185:811–826. doi: 10.1083/jcb.200810133. doi:jcb.200810133 [pii] 10.1083/jcb.200810133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang D, et al. Reprogramming transcription by distinct classes of enhancers functionally defined by eRNA. Nature. 2011 doi: 10.1038/nature10006. doi:nature10006 [pii] 10.1038/nature10006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gu B, Watanabe K, Dai X. Pygo2 regulates histone gene expression and H3 K56 acetylation in human mammary epithelial cells. Cell Cycle. 2012;11:79–87. doi: 10.4161/cc.11.1.18402. doi:10.4161/cc.11.1.18402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lam MT, et al. Rev-Erbs repress macrophage gene expression by inhibiting enhancer-directed transcription. Nature. 2013 doi: 10.1038/nature12209. doi:10.1038/nature12209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li W, et al. Functional roles of enhancer RNAs for oestrogen-dependent transcriptional activation. Nature. 2013 doi: 10.1038/nature12210. doi:10.1038/nature12210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhao J, et al. Genome-wide identification of polycomb-associated RNAs by RIP-seq. Mol Cell. 2010;40:939–953. doi: 10.1016/j.molcel.2010.12.011. doi:10.1016/j.molcel.2010.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. doi:10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Heinz S, et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. doi:10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. doi:10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Saeed AI, et al. TM4 microarray software suite. Methods in enzymology. 2006;411:134–193. doi: 10.1016/S0076-6879(06)11009-5. doi:10.1016/S0076-6879(06)11009-5. [DOI] [PubMed] [Google Scholar]
- 26.Zang C, et al. A clustering approach for identification of enriched domains from histone modification ChIP-Seq data. Bioinformatics. 2009;25:1952–1958. doi: 10.1093/bioinformatics/btp340. doi:10.1093/bioinformatics/btp340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. doi:10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu W, et al. PHF8 mediates histone H4 lysine 20 demethylation events involved in cell cycle progression. Nature. 2010;466:508–512. doi: 10.1038/nature09272. doi:nature09272 [pii] 10.1038/nature09272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tsai MC, et al. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329:689–693. doi: 10.1126/science.1192002. doi:science.1192002 [pii] 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liao DF, Monia B, Dean N, Berk BC. Protein kinase C-zeta mediates angiotensin II activation of ERK1/2 in vascular smooth muscle cells. J Biol Chem. 1997;272:6146–6150. doi: 10.1074/jbc.272.10.6146. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.