Abstract
Objective
The etiology of Parkinson disease (PD) is complex and multifactorial, with hereditary and environmental factors contributing. Monogenic forms have provided molecular clues to disease mechanisms but genetic modifiers of idiopathic PD are still to be determined.
Methods
We carried out whole-genome expression profiling of isolated human substantia nigra (SN) neurons from patients with PD vs. controls followed by association analysis of tagging single-nucleotide polymorphisms (SNPs) in differentially regulated genes. Association was investigated in a German PD sample and confirmed in Italian and British cohorts.
Results
We identified four differentially expressed genes located in PD candidate pathways, ie, MTND2 (mitochondrial, p = 7.14 × 10−7), PDXK (vitamin B6/dopamine metabolism, p = 3.27 × 10−6), SRGAP3 (axon guidance, p = 5.65 × 10−6), and TRAPPC4 (vesicle transport, p = 5.81 × 10−6). We identified a DNA variant (rs2010795) in PDXK associated with an increased risk of PD in the German cohort (p = 0.00032). This association was confirmed in the British (p = 0.028) and Italian (p = 0.0025) cohorts individually and reached a combined value of p = 1.2 × 10−7 (odds ratio [OR], 1.3; 95% confidence interval [CI], 1.18–1.44).
Interpretation
We provide an example of how microgenomic genome-wide expression studies in combination with association analysis can aid to identify genetic modifiers in neurodegenerative disorders. The detection of a genetic variant in PDXK, together with evidence accumulating from clinical studies, emphasize the impact of vitamin B6 status and metabolism on disease risk and therapy in PD.
Parkinson’s disease (PD) is a neurodegenerative disorder, characterized by age-related dysfunction and loss of dopaminergic neurons in the substantia nigra (SN) pars compacta (SNc). Hereditary forms of PD have provided evidence for linkage to 13 loci and nine causative genes, and their functional study has greatly helped to elucidate molecular disease mechanisms.1 For idiopathic PD, both genetic and environmental factors are believed to modulate disease risk, but the impact of genetic variants remains unclear.2 Genome-wide association (GWA) studies offer an unbiased approach for detecting effects in common variants, but large studies with sufficient power are still lacking. Restriction to candidate genes increases the a priori odds for phenotypic involvement and thus the chance of detecting significant associations. Here, we applied a functional genomic approach for the discovery of candidate genes and key regulatory pathways. We focused directly on the affected tissue and specifically on dopaminergic neurons of the SNc using single cell, whole-genome expression profiling followed by a genetic association study of differentially regulated genes.
Materials and Methods
Tissue Samples for Expression Profiling
Frozen human brain tissue was obtained from the Newcastle Brain Tissue Resource. Samples were available from patients with a clinical history of PD fulfilling UK PD Brain Bank criteria and from age-matched control individuals without neurological disease. The cases were clinically well documented and neuropathologically confirmed. Inclusion criteria comprised the neuropathological diagnosis of Lewy body disease with typical pathological features, including moderate to severe dopaminergic neuronal loss in the SNc and gliosis. The local research ethics committee approved all procedures.
Sectioning, Staining, Laser Microdissection, In Vitro Transcription, and Microarray
In order to isolate high-quality RNA from frozen human brain tissues, tissue pH and RNA integrity numbers (RIN) were measured in homogenates of frontal cortex and mid-brain sections from 41 suitable cases and 39 controls. This provided eight cases and nine age-matched controls of good RNA quality for laser microdissection (LMD). Postmortem data for matched cases/controls were as follows: age at death 78.6 ± 6.5/76.8 ± 9.8 years; postmortem delay 28.3 ± 11.9/20.5 ± 10.2 hours; pH 6.5 ± 0.1/6.61 ± 0.21; and RIN 7.7 ± 0.8/7.6 ± 1.1 (Supporting Table 1; Supporting Fig 1).All procedures were carried out under RNAse-free conditions. Frozen 20μm midbrain sections were mounted on Leica PEN-slides (Leica, Wetzlar, Germany), rapidly stained with toluidine blue, dehydrated in an ethanol series, and processed on a Leica LMD microscope. A total of 100 neurons per case/control were collected and RNA was extracted with the PicoPure® Kit (Molecular Devices, Sunnyvale, CA) according to the manufacturer’s protocol. In two cases and two controls sampling and experiments were repeated to enable analysis of reproducibility. In vitro transcription (IVT) comprised one round of linear amplification with MessageAmpII (Applied Biosystems/Ambion, Austin, TX), followed by a second round of IVT with the Illumina® TotalPrep™ RNA Amplification Kit (Ambion). Second-round IVT yielded >3μg cRNA with an average length of 800bp and was used for hybridization on Illumina WG6v1 expression chips (Illumina, San Diego, CA). On average, more than 12,000 transcripts were detected (p < 0.01). For validation of microarray results, excess aRNA was reverse-transcribed using SuperScript™ III First-Strand Synthesis SuperMix (Invitrogen, Carlsbad, CA) and assayed on a StepOnePlus™ Real-Time PCR System (Applied Biosystems, Foster City, CA) using TaqMan® Gene Expression Assays (Applied Biosystems, Foster City, CA) by following standard protocols. Beta-actin was used as a housekeeping gene and relative expression of the two most significant genes was determined. Data was analyzed using the comparative C(T) method.3
Statistical Analysis of Microarray Data
Raw data were exported from the Illumina Beadstudio software to R (http://cran.r-project.org), log-scale transformed (log2) and normalized (nonlinear transformation employing the LOESS smoother).4 A total of 8,491 transcripts were detected in all samples. Statistical analysis (t test) revealed no significant difference for age, postmortem delay, tissue pH, and RIN number of postmortem samples, and no associated effect was seen on gene expression. To select significant signals in the transcriptome-wide differential (t test) screening, conservative Bonferroni thresholds were used, which corresponded to a nominal level of 0.05.
Single-Nucleotide Polymorphism Analysis and German Cohort
Single-nucleotide polymorphism (SNP) selection was performed with the Tagger algorithm in Haploview 4.1 (http://www.broad.mit.edu/mpg/tagger) using pairwise tagging only, an r2 threshold of 0.8, and a minor allele frequency (MAF) of 0.1. Genotype data were taken from the CEU population in HapMap (data Rel 23a/phaseII Mar08). Selection of SNPs was restricted to those that were covered by the Illumina Hap550 array, since genotyping data from this platform in universal German controls was used for analysis. The genomic regions of both genes were defined as the transcriptional unit plus 10kb upstream and downstream. Twelve tagging SNPs for PDXK and five tagging SNPs for TRAPPC4 were selected. German PD patients were recruited by participating institutions in Munich and Tuebingen. Specialists in movement disorders examined the patients. Diagnosis was established according to UK PD Brain Bank criteria. Geno-typing of 676 German PD patients were performed on a Sequenom MassArray system with the iPlex Gold assay (Sequenom, San Diego, CA). A total of 15 assays had call rates of >98%. With the exception of one SNP (rs743463; Pearson p = 0.04) no deviations from Hardy-Weinberg equilibrium (HWE) were observed. The Armitage Trend Test was performed for association using our genotype data for the 676 PD patients and available Illumina Hap550 genotyping data from 485 KORA and 487 POPGEN universal German controls.5,6
British Cases and Controls
For the British cohort, a group of community-based cases with PD fulfilling UK PD Brain Bank criteria (n = 203) were compared to a group of ethnically age- and gender-matched controls (n = 142), with no clinical evidence of PD. All were of UK Caucasian origin. Two additional UK control groups were included to increase statistical power: The Orkney Complex Disease Study (ORCADES) is an ongoing, family-based, cross-sectional study in the isolated Scottish archipelago of Orkney. Data for participants aged 18 to 100 years, from a subgroup of 10 islands, were used for this analysis. The mean age was 53 years and 53% were female. Genetic diversity in this population is decreased compared to Mainland Scotland, consistent with the historically high levels of endogamy. Therefore, from 719 available samples, first-, second-, and third-degree relatives were excluded, resulting in 483 samples used for this study. All participants gave informed consent and the Research Ethics Committees in Orkney and Aberdeen approved the study. In the Study of Colon Cancer Survivors (SOCCS), 1,012 colorectal cancer cases were recruited from throughout mainland Scotland; 1,012 age- and gender-matched cancer-free population controls were selected at random according to matching criteria from a population-based register (518 males, 494 females; age 51.0 ± 5.9 years, mean ± standard deviation [SD]). From these, 503 samples were randomly selected for our study. Genotyping was conducted using Illumina HumanHap300 and Illumina Human-Hap240S arrays according to established protocols.7
Italian Cases and Controls
The Italian cohort comprised 356 unrelated, nonfamilial PD patients, fulfilling the criteria for PD reported in Ghezzi et al.8 The study was in agreement with the UK PD Brain Bank criteria. The male/female ratio was 1.72; the age range was 50 to 75 years (67 ± 6.6). The control group included 171 unrelated, ethnically- and age-matched controls (male/female ratio 0.4; age range 50–75 years; age 59 ± 16.3 years) with no clinical evidence of PD. All were Italians of Caucasian origin. All of the participants gave informed consent. One additional control group from Italy was included to increase statistical power. The Val Borbera Project is an ongoing family-based, cross-sectional study in the isolated population of the Val Borbera in Northwest Italy. Data for participants aged 18 to 102 years, from the seven villages of the valley, were used for this analysis. The mean age was 55 years and 56% were female. The population presents a high level of endogamy until around 1950. From the 1,436 samples genotyped, individuals presenting a Ks coefficient ≥ 0.05 (first-, second-, and third-degree relatives) were excluded, resulting in 532 samples used for this study. All participants gave informed consent and the Ethics Committees of the San Raffaele Hospital and of the Piemonte Region approved the study.
Results
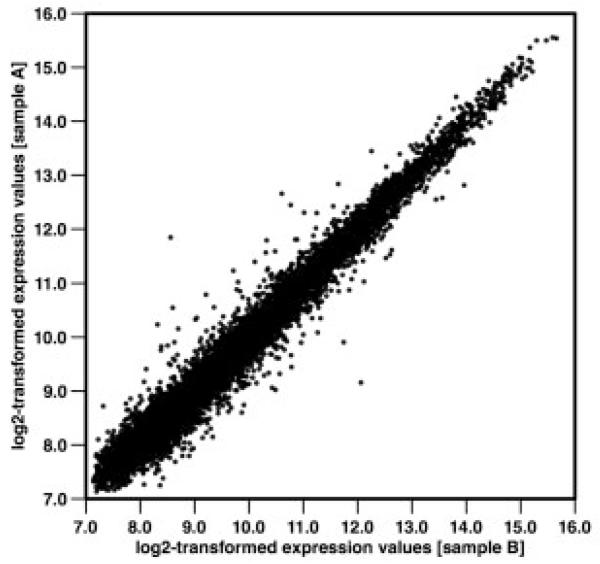
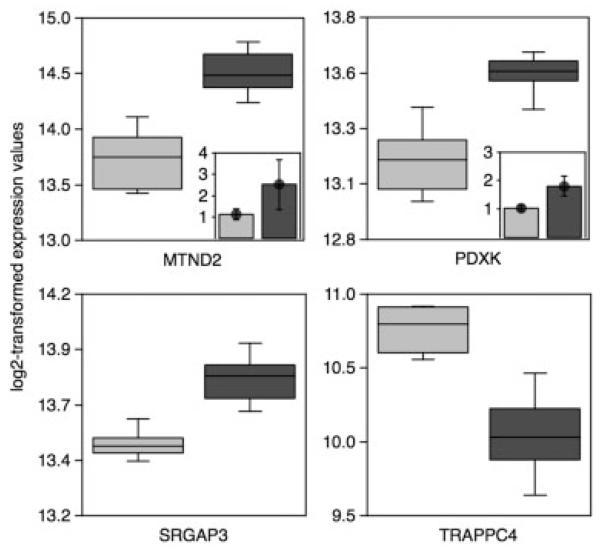
Gene expression profiles of 100 SNc neurons collected from the same brain sample, which were carried through LMD, IVT, and array hybridization individually, displayed an extremely high correlation (Fig 1). This degree of reproducibility strengthened confidence in the method used. Statistical analysis of the genome-wide expression profiles using stringent Bonferroni correction revealed four transcripts to be significantly altered between PD and control samples: the mitochondrial encoded complex I subunit gene ND2 (mtND2), pyridoxal (pyridoxine, vitamin B6) kinase (PDXK), SLIT-ROBO Rho guanosine triphosphatase (GTPase) activating protein 3 (SRGAP3), and trafficking protein particle complex 4 (TRAPPC4) (Fig 2; Table 1). Upregulation of ND2 and PDXK was confirmed by real-time analysis (ND2: +2.5-fold; PDXK: +1.8-fold). In addition to ND2, a further 17 genes coding for subunits of the mitochondrial respiratory chain were among the top 320 differentially regulated transcripts (significantly changed applying a false discovery rate, p < 0.025).9 The Database for Annotation, Visualization, and Integrated Discovery (DAVID) gene functional classification tool revealed a highly significant enrichment in the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway “oxidative phosphorylation” (p = 5.6 × 10−9).10 Interestingly, the mtDNA-encoded genes were all found to be upregulated: ND1 (p = 0.0002; +1.65-fold), ND2 (p = 7.14 × 10−7; +1.7-fold), ND4 (p = 0.0005; +1.27-fold), ND5 (p = 0.0002; +1.69-fold), and COXII (p = 0.00039; +1.31-fold). In contrast, the 13 nuclear genes coding for different members of the five respiratory chain complexes were robustly downregulated. Mitochondrial dysfunction in the pathogenesis of PD is clearly established,11 but our findings demonstrate an inverse regulation of mtDNA and nuclear-encoded subunits of the respiratory chain on a single-cell basis, which has been reported by Noureddine et al.12 and Duke et al.13 in gross dissections of SN tissue.
Fig 1.
Correlation of individual genome-wide expression profiles. Plot shows correlation of two genome-wide expression profiles after individual LMD and IVT of 100 neurons collected from the same SNc. Axes show normalized expression values (log2) with a range of expression levels nearing three orders of magnitude. The resulting Pearson product-moment correlation coefficient is r = 0.986.
Fig 2.
Differentially regulated genes identified by whole-genome expression analysis. Four significant genes after stringent Bonferroni correction: Box plots are showing normalized expression (log2) of significant genes in PD samples (light gray) compared to controls (black). Insets show fold change of gene expression as measured by real-time PCR.
Table 1. Description of Significant Genes Identified by Whole Genome Expression.
| Symbol | Definition | Fold change | p-Value | KEGG pathway/reactome event | Biological process |
|---|---|---|---|---|---|
| MTND2 | Homo sapiens NADH dehydrogenase, subunit 2 (complex I) | up 1.70 | 7.14 × 10−7 | Oxidative phosphorylation/electron transport chain | ATP synthesis coupled electron transport |
| PDXK | Homo sapiens pyridoxal (pyridoxine, vitamin B6) kinase | up 1.32 | 3.27 × 10−6 | Vitamin B6/dopamine metabolism | Pyridoxine biosynthetic process |
| SRGPA3 | Homo sapiens SLIT-ROBO Rho GTPase activating protein 3 | up 1.23 | 5.65 × 10−6 | Axon guidance/signaling by Rho GTPases | Signal transduction |
| TRAPPC4 | Homo sapiens trafficking protein particle complex 4 | down 1.69 | 5.8 × 10−6 | Vesicle tethering and fusion | ER to Golgi vesiclemediated transport |
KEGG = Kyoto Encyclopedia of Genes and Genomes; NADH = reduced nicotinamide adenine dinucleotide; ATP = adenosine triphosphate; SLIT-ROBO = SLIT-roundabout; GTPase = guanosine triphosphatase; ER = endoplasmic reticulum.
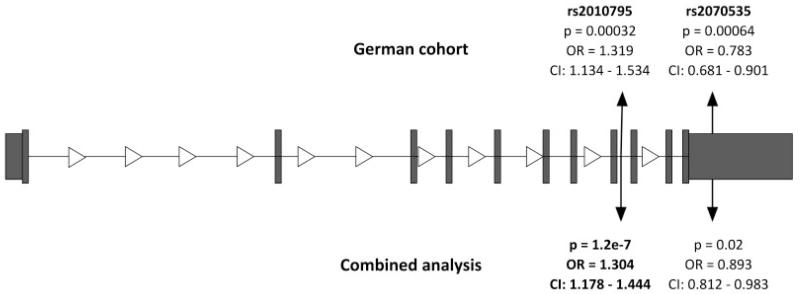
In order to validate a causal involvement of the new candidates in the pathogenesis of PD, we decided to follow a genetic approach. Since mtDNA variants and polymorphisms in the axon guidance pathway have already been shown to be involved in prior studies, our investigation was restricted to PDXK and TRAPPC4.14,15 No association was seen for tagging SNPs in the TRAPPC4 gene. By contrast, in PDXK, two SNPs with r2 = 0.29 reached significant p values for association with disease status: rs2010795 (p = 3.2 × 10−4; odds ratio [OR], 1.319; 95% confidence interval [CI], 1.134–1.534) lies in intron 8 and rs2070535 (p = 6.4 × 10−4; OR, 0.783; 95% CI, 0.681–0.901) in the 3′ untranslated region of the PDXK gene (Fig 3). Replication genotyping of the two lead SNPs in 203 PD and 1,126 control samples from the UK cohort confirmed the association of rs2010795 with PD (p = 0.028) giving a similar MAF in PD cases (0.34) and controls (0.29), and resulting in a similar OR of 1.3 (95% CI, 1.03–1.61). In the Italian cohort, association for rs2010795 was confirmed in 353 cases and 704 controls with a p value of 0.00252, again with a similar MAF (PD = 0.35, controls = 0.29) and OR (1.35; 95% CI, 1.11–1.63). In the combined analysis of 1,232 PD and 2,802 control individuals, the association of SNP rs2010795 reached a p value of 1.18 × 10−7 (combined OR, 1.304; 95% CI, 1.178–1.444). Details on allele frequency are given in Table 2. The geographic distribution of PD and control sampling sites is shown in Supporting Figure 2.
Fig 3.
Gene structure of PDXK and position of SNPs. The gene structure of PDXK and position of SNP rs2010795 in intron 8 and of rs2070535 in the 3′ untranslated region. OR = odds ratio; CI = confidence interval.
Table 2. Allele Frequency in All Cohorts.
| SNP rs2010795 | n | AA | f(AA) | AG | f(AG) | GG | f(GG) | MAF (A) | Allelic p | OR (95% CI) |
|---|---|---|---|---|---|---|---|---|---|---|
| German | 0.00032 | 1.319 (1.134–1.534) | ||||||||
| German PD | 676 | 77 | 0.11 | 294 | 0.43 | 305 | 0.45 | 0.33 | ||
| KORA | 485 | 29 | 0.06 | 192 | 0.40 | 264 | 0.54 | 0.26 | ||
| POPGEN | 487 | 33 | 0.07 | 215 | 0.44 | 239 | 0.49 | 0.29 | ||
| All German controls | 972 | 62 | 0.06 | 407 | 0.42 | 503 | 0.52 | 0.27 | ||
| British | 0.02815 | 1.286 | (1.027–1.610) | |||||||
| British PD | 203 | 24 | 0.12 | 90 | 0.44 | 89 | 0.44 | 0.34 | ||
| British controls | 142 | 15 | 0.11 | 46 | 0.32 | 81 | 0.57 | 0.27 | ||
| ORCADES | 483 | 27 | 0.06 | 200 | 0.41 | 256 | 0.53 | 0.26 | ||
| SOCCS | 501 | 43 | 0.08 | 228 | 0.46 | 230 | 0.46 | 0.31 | ||
| All UK controls | 1126 | 85 | 0.08 | 474 | 0.42 | 567 | 0.50 | 0.29 | ||
| Italian | 0.00252 | 1.346 | (1.110–1.632) | |||||||
| Italian PD | 353 | 47 | 0.13 | 154 | 0.44 | 152 | 0.43 | 0.35 | ||
| Italian controls | 172 | 18 | 0.10 | 69 | 0.40 | 85 | 0.49 | 0.31 | ||
| Val Borbera | 532 | 40 | 0.08 | 219 | 0.41 | 273 | 0.51 | 0.28 | ||
| All Italian controls | 704 | 58 | 0.08 | 288 | 0.41 | 358 | 0.51 | 0.29 | ||
| Combined analysis | 1.18E–07 | 1.304 (1.178–1.444) | ||||||||
| All PD | 1232 | 148 | 0.12 | 538 | 0.44 | 546 | 0.44 | 0.34 | ||
| All controls | 2802 | 205 | 0.07 | 1169 | 0.42 | 1428 | 0.51 | 0.28 |
SNP = single-nucleotide polymorphism; OR = odds ratio; CI = confidence interval; MAF = minor allele frequency; PD = Parkinson’s disease; KORA = Kooperative Gesundheits forschung in der Region Augsburg; POPGEN = Populations genetisches Forschungsprojekt des Nationalen; ORCADES = Orkney Complex Disease Study; SOCCS = Study of Colon Cancer Survivors.
Discussion
Integrating gene expression with association analysis data is a promising approach for the study of disease biology. This concept, termed genomic convergence, was first proposed by Hauser et al.16 and aims to narrow down a large pool of candidate genes to a few selected choices. With four biologically plausible candidates being differentially expressed in SNc neurons of PD samples, we aimed to evaluate their contribution in idiopathic PD. If differential regulation of these genes is involved in the pathogenesis of PD, their genetic variants might affect the risk for disease.
mtDNA variants and polymorphisms in the axon guidance pathway have been investigated in prior genetic studies. For example, the mitochondrial UKJT haplogroup was shown to be associated with a decreased risk of PD.2,8,14 We also demonstrate that multiple mtDNA genes are upregulated, supporting previous observations that mitochondrial abnormalities are present in SN neurons from patients with PD.11 The axon guidance pathway was recently implicated in the pathogenesis of PD by a systematic analysis of GWA PD studies.15,17 Ephrin, netrin, semaphorin, and slit proteins are the main effectors of the axon guidance pathway and are important for brain development, dopaminergic axonal maintenance, regeneration, and target recognition. Lesnick et al.15 used the combined SNP information, rather than individual variants, to predict PD susceptibility, survival free of PD, and PD age at onset with extremely high power.15 Together with subsequent studies, two out of four GWA PD studies show this significant association.18,19 With the identification of differential regulation of SRGAP3 in dopaminergic neurons of the SNc, we provide functional data supporting the importance of the axon guidance pathway in the development of PD.
TRAPPC4 appears to be a good candidate as it is localized in neuronal synapses and is part of the human transport protein particle complex I that is required for tethering endoplasmic reticulum (ER)-derived vesicles to Golgi membranes.20,21 Evidence for impaired synaptic vesicle endocytosis as well as disrupted ER-to-Golgi transport in PD comes from recent studies into the role of the PD proteins alpha-synuclein and LRRK2.22,23 Since no SNP in TRAPPC4 was significantly associated with disease in our study based on 676 cases, the causal involvement could not be shown. Either the differential expression pattern of TRAPPC4 is a consequence of the PD pathology or the effect size is to small to be shown in a study of this size.
PDXK catalyzes the conversion of vitamin B6 (pyridoxine, pyridoxal, and pyridoxamine) to pyridoxal 5′-phosphate (PLP), which is thought to be a cofactor in multiple eukaryotic enzyme-catalyzed reactions. Of direct relevance to PD, the second step in the biosynthesis of dopamine by the enzyme aromatic L-amino acid decarboxylase (dopa decarboxylase [DDC]) is dependent on PLP as a cofactor and becomes rate-limiting in patients receiving L-DOPA therapy (L-DOPA + pyridoxal phosphate = dopamine + pyridoxal phosphate + CO2).24 A recent clinical phase I trial showed promising results of intrastriatal induction of DDC gene expression in PD patients.25 Upregulation of PDXK in dopaminergic neurons may be explained by an adaptive mechanism to increased dopamine metabolism in the remaining functional dopaminergic neurons of the SNc or to L-DOPA therapy. Treatment strategies of PD with pyridoxine were published as early as 1954, but abandoned because of metabolic interaction with the therapeutically more potent L-DOPA in the periphery.26 Today, this effect can be negated in the presence of a peripheral DDC inhibitor and a recent study showed that high doses of pyridoxine therapy (at 400mg/day) might be beneficial for the treatment of PD.24 Independent of L-DOPA therapy, a prospective, population-based cohort study showed that dietary vitamin B6 might decrease the risk of PD, although this effect was restricted to smokers.27
SNP analysis showed a significant influence of the marker rs2010795 on disease status in our study. Our data is the first to indicate an effect of polymorphisms in the PDXK gene on the risk for PD, possibly by influencing enzyme amount or activity in dopaminergic neurons of the SNc. The effect size of this marker is small and the power of current GWA studies would be to low for significant detection. Candidate gene selection for association studies was based on experimental data coming from microarray gene expression analysis, thus providing the link to a likely biological effect. We therefore introduce this promising approach for the study of genetic risk factors in complex neurodegenerative disorders.
Supplementary Material
Acknowledgments
This work was supported by the European Neurological Society (M.E.), the Wellcome Trust/UK (P.C.), the Impulse and Networking Fund of the Helmholtz Association in the framework of the Helmholtz Alliance for Mental Health in an Ageing Society (HA-215) (T.M. and H.P.), and the German National Genome Network of the German Ministry for Education and Research (NGFNplus) (T.M.). C.M.M. acknowledges funding from the Health Protection Agency UK and D.M.T. the Newcastle University Centre for Brain Ageing and Vitality.
T.K., T.M., and H.P. are members of the German network for mitochondrial disorders (mitoNET, 01GM0862), funded by the German ministry of education and research (BMBF, Bonn, Germany). Tissue for this study was provided by the Newcastle Brain Tissue Resource, which is funded in part by a grant from the UK Medical Research Council (G0400074). The Newcastle Brain Tissue Resource is part of the Brains for Dementia Research initiative and is funded by the Alzheimer’s Research Trust and the Alzheimer’s Society.
KORA and POPGEN were supported by the German Ministry of Education and Research through the National Genome Research Network (NGFN). ORCADES was supported by the Scottish Executive Health Department (Chief Scientist Office), the Royal Society and the European Union framework program 6 EURO-SPAN project (contract no. LSHG-CT-2006-018947). DNA extractions were performed at the Wellcome Trust Clinical Research Facility in Edinburgh. We acknowledge the invaluable contributions of L. Anderson and the research nurses in Orkney, the administrative team in Edinburgh, and the people of Orkney.
SOCCS was funded by grants Cancer Research UK (C348/A3758 and A8896, C48/A6361); Medical Research Council (G0000657-53203); Scottish Executive Chief Scientist’s Office (K/OPR/2/2/D333, CZB/4/449); Centre Grant from CORE as part of the Digestive Cancer Campaign (http://www.corecharity.org.uk). Samples, DNA extraction, and phenotype data were collected at the Wellcome Trust Clinical Research Facility, Edinburgh. We acknowledge the important role played by Dr. Susan Farrington, Dr. Albert Tenesa, and Dr. Evi Theodoratou. We also thank departments in central Scottish NHS, including the Family Practitioner Committee for population control recruitment.
The Strategic Research Grant of the Italian Ministry of Health, entitled “Analisi dei fattori di rischio e di potenziali elementi predittivi di danno neurodegenerativo nelle sindromi Parkinsoniane” supported the Italian contribution. Italian DNA samples contributed by the Parkinson’s Institute - Istituti Clinici di Perfezionamento were from the “Human genetic bank of patients affected by PD and Parkinsonisms” (http://www.Parkinson’s.it/dnabank.html), which is supported by the Italian Telethon (n. GTB07001) and by the “Fondazione Grigioni per il Morbo di Parkinson’s.” The Val Borbera Project was supported by Compagnia di San Paolo, Torino, by Cariplo Fundation, Milano, and by the Italian Ministry of Health, Grant Ricerca Finalizzata 2008.
Footnotes
Potential conflict of interest: Nothing to report.
Additional Supporting Information may be found in the online version of this article.
References
- 1.Lesage S, Brice A. Parkinson’s disease: from monogenic forms to genetic susceptibility factors. Hum Mol Genet. 2009;18:R48–R59. doi: 10.1093/hmg/ddp012. [DOI] [PubMed] [Google Scholar]
- 2.Tan EK. The role of common genetic risk variants in Parkinson disease. Clin Genet. 2007;72:387–393. doi: 10.1111/j.1399-0004.2007.00890.x. [DOI] [PubMed] [Google Scholar]
- 3.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 4.Cleveland WS. Robust locally weighted regression and smoothing scatterplots. J Am Stat Assoc. 1979;74:829–836. [Google Scholar]
- 5.Wichmann HE, Gieger C, Illig T, MONICA/KORA Study Group KORA-gen—resource for population genetics, controls and a broad spectrum of disease phenotypes. Gesundheitswesen. 2005;67(Suppl 1):S26–S30. doi: 10.1055/s-2005-858226. [DOI] [PubMed] [Google Scholar]
- 6.Krawczak M, Nikolaus S, von Eberstein H, et al. PopGen: population-based recruitment of patients and controls for the analysis of complex genotype-phenotype relationships. Community Genet. 2006;9:55–61. doi: 10.1159/000090694. [DOI] [PubMed] [Google Scholar]
- 7.Tenesa A, Farrington SM, Prendergast JG, et al. Genome-wide association scan identifies a colorectal cancer susceptibility locus on 11q23 and replicates risk loci at 8q24 and 18q21. Nat Genet. 2008;40:631–637. doi: 10.1038/ng.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ghezzi D, Marelli C, Achilli A, et al. Mitochondrial DNA haplogroup K is associated with a lower risk of Parkinson’s disease in Italians. Eur J Hum Genet. 2005;13:748–752. doi: 10.1038/sj.ejhg.5201425. [DOI] [PubMed] [Google Scholar]
- 9.Benjamini Y, Hochberg Y. Multiple hypothesis testing with weights. Scand J Stat. 1997;24:407–418. [Google Scholar]
- 10.Dennis G, Sherman BT, Hosack DA, et al. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4:P3. [PubMed] [Google Scholar]
- 11.Bender A, Krishnan KJ, Morris CM, et al. High levels of mitochondrial DNA deletions in substantia nigra neurons in aging and Parkinson disease. Nat Genet. 2006;38:515–517. doi: 10.1038/ng1769. [DOI] [PubMed] [Google Scholar]
- 12.Noureddine MA, Li YJ, van der Walt JM, et al. Genomic convergence to identify candidate genes for Parkinson disease: SAGE analysis of the substantia nigra. Mov Disord. 2005;20:1299–1309. doi: 10.1002/mds.20573. [DOI] [PubMed] [Google Scholar]
- 13.Duke DC, Moran LB, Kalaitzakis ME, et al. Transcriptome analysis reveals link between proteasomal and mitochondrial pathways in Parkinson’s disease. Neurogenetics. 2006;7:139–148. doi: 10.1007/s10048-006-0033-5. [DOI] [PubMed] [Google Scholar]
- 14.Pyle A, Foltynie T, Tiangyou W, et al. Mitochondrial DNA haplogroup cluster UKJT reduces the risk of PD. Ann Neurol. 2005;57:564–567. doi: 10.1002/ana.20417. [DOI] [PubMed] [Google Scholar]
- 15.Lesnick TG, Papapetropoulos S, Mash DC, et al. A genomic pathway approach to a complex disease: axon guidance and Parkinson disease. PLoS Genet. 2007;3:e98. doi: 10.1371/journal.pgen.0030098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hauser MA, Li YJ, Takeuchi S, et al. Genomic convergence: identifying candidate genes for Parkinson’s disease by combining serial analysis of gene expression and genetic linkage. Hum Mol Genet. 2003;12:671–677. [PubMed] [Google Scholar]
- 17.Maraganore DM, de Andrade M, Lesnick TG, et al. High-resolution whole-genome association study of Parkinson disease. Am J Hum Genet. 2005;77:685–693. doi: 10.1086/496902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li Y, Rowland C, Xiromerisiou G, et al. Neither replication nor simulation supports a role for the axon guidance pathway in the genetics of Parkinson’s disease. PLoS ONE. 2008;3:e2707. doi: 10.1371/journal.pone.0002707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Srinivasan BS, Doostzadeh J, Absalan F, et al. Whole genome survey of coding SNPs reveals a reproducible pathway determinant of Parkinson disease. Hum Mutat. 2009;30:228–238. doi: 10.1002/humu.20840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim YG, Raunser S, Munger C, et al. The architecture of the multisubunit TRAPP I complex suggests a model for vesicle tethering. Cell. 2006;127:817–830. doi: 10.1016/j.cell.2006.09.029. [DOI] [PubMed] [Google Scholar]
- 21.Ethell IM, Hagihara K, Miura Y, et al. Synbindin, a novel syndecan-2-binding protein in neuronal dendritic spines. J Cell Biol. 2000;151:53–68. doi: 10.1083/jcb.151.1.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shin N, Jeong H, Kwon J, et al. LRRK2 regulates synaptic vesicle endocytosis. Exp Cell Res. 2008;314:2055–2065. doi: 10.1016/j.yexcr.2008.02.015. [DOI] [PubMed] [Google Scholar]
- 23.Cooper AA, Gitler AD, Cashikar A, et al. Alpha-synuclein blocks ER-Golgi traffic and Rab1 rescues neuron loss in Parkinson’s models. Science. 2006;313:324–328. doi: 10.1126/science.1129462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tan EK, Cheah SY, Fook-Chong S, et al. Functional COMT variant predicts response to high dose pyridoxine in Parkinson’s disease. Am J Med Genet B Neuropsychiatr Genet. 2005;137B:1–4. doi: 10.1002/ajmg.b.30198. [DOI] [PubMed] [Google Scholar]
- 25.Eberling JL, Jagust WJ, Christine CW, et al. Results from a phase I safety trial of hAADC gene therapy for Parkinson disease. Neurology. 2008;70:1980–1983. doi: 10.1212/01.wnl.0000312381.29287.ff. [DOI] [PubMed] [Google Scholar]
- 26.Yahr MD, Duvoisin RC. Pyridoxine and levodopa in the treatment of Parkinsonism. JAMA. 1972;220:861. [PubMed] [Google Scholar]
- 27.de Lau LM, Koudstaal PJ, Witteman JC, et al. Dietary folate, vitamin B12, and vitamin B6 and the risk of Parkinson disease. Neurology. 2006;67:315–318. doi: 10.1212/01.wnl.0000225050.57553.6d. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.