Abstract
Purpose
Loss of mitochondrial DNA (mtDNA) has been described in whole blood samples from a small number of patients with sepsis, but the underlying mechanism and clinical implications of this observation are not clear. We have investigated the cellular basis of the mtDNA depletion in sepsis, and determined clinical correlates with mtDNA depletion.
Methods
Whole blood samples were obtained from 147 consecutive patients with severe sepsis admitted to a General Critical Care Unit in a University Hospital and 83 healthy controls. In a separate study of 13 patients with severe sepsis, blood was obtained for immediate cell sorting by flow cytometry. MtDNA content was determined in whole blood DNA by PCR methods, and subsequently in the 13 samples where white cell subtypes were separated.
Results
The mtDNA content of peripheral blood in human subjects was lower in patients with sepsis than controls (P < 0.0001). By studying leukocyte subsets in a subgroup of 13 patients, we showed that this was largely due to an increase in the proportion of circulating neutrophils, which contained ~3-fold less mtDNA than mononuclear leukocytes. However, isolated monocytes (P = 0.041) and lymphocytes (P = 0.021) from septic patients showed clear evidence of mtDNA depletion, which correlated with the APACHE II score (P = 0.015).
Conclusions
In severe sepsis much of the apparent whole blood mtDNA depletion is due to a change in the differential leukocyte count. However mtDNA depletion in mononuclear cells occurs in patients with sepsis and correlates with disease severity.
Keywords: Sepsis, Mitochondria, Copy number, Neutrophil, Monocyte, Lymphocyte
Introduction
Sepsis is the inflammatory and metabolic host response to infectious pathogens which results in multiple organ failure with a mortality rate in excess of 40% [1]. Increasing evidence suggests that the organ dysfunction in sepsis is ultimately due to a failure of mitochondrial energy metabolism [2, 3]. Failure of cellular oxygen (O2) utilization appears to be central to the development of multiple organ dysfunction in severe sepsis [4]. Over 98% of O2 delivered to tissues is used by the mitochondrial respiratory chain to produce adenosine triphosphate (ATP), the principal intracellular energy source. Mitochondria contain their own DNA (mtDNA) which codes for 13 essential respiratory chain polypeptides, and the 24 RNA genes required for intra-mitochondrial protein synthesis. In a growing group of inherited diseases, mutations and depletion (loss) of mtDNA cause a respiratory chain defect, and compromise ATP synthesis leading to widespread organ dysfunction [5]. It is therefore of great interest that in several animal models of sepsis, organ dysfunction was accompanied by mtDNA depletion [6–8], and low levels of mtDNA were reported in 28 patients admitted to an intensive care unit (ICU), with the level increasing in survivors [9]. Sepsis now causes 47% of hospital mortality and is responsible for 45% of hospital critical care bed occupancy [10]. Understanding the underlying mechanisms is therefore of great importance, including the role of mtDNA depletion, which could serve as a prognostic marker in the acute situation, or be directly relevant to the mechanisms of survival.
Materials and methods
Patients and subjects
The studies had ethical approval from the Newcastle and North Tyneside Local Research Ethics Committee. We initially studied 147 patients with severe sepsis, as defined by the European/North American consensus criteria [11], recording basic demographic information, the APACHE II (Acute Physiology And Chronic Health Evaluation) severity of illness data set, and outcome in ICU, at hospital discharge and at 180 days. Patients were defined as “survivors” if they were alive at 180 days after admission to the ICU. Eighty-three healthy community controls were recruited from a geographically and ethnically matched population. Subsequent studies were carried out on a subgroup of 13 prospectively recruited sepsis patients and 14 matched healthy controls.
Laboratory methods
Whole blood mtDNA analysis
Whole blood samples were collected from 147 sepsis patients within 24 h of fulfilling the Joint Consensus definition of severe sepsis and shock [11]. Total genomic DNA was extracted from whole blood from the 147 sepsis patients and 83 controls using a Nucleon DNA Extraction Kit (Tepnel Lifescience). The relative amount of mtDNA per cell was determined by real-time PCR using iQ Sybr Green on a BioRad ICycler iQ5 (BioRad, Hercules, CA, USA) to a target template spanning from nt 3459 to nt 3569 of the MTND1 gene [12]. This was normalized using a nuclear-encoded (nDNA) template for the glyceraldehyde-3-phosphate dehydrogenase (GAPDH [MIM *138400]) gene spanning from nt 804 to nt 903 (NCBI accession number, NM_002046.2). Each reaction was optimized and confirmed linear over an appropriate concentration range. Each assay was performed in separate wells of the same microtitre plate. The same standards were used on each run to normalize between experiments. Samples and standards were analyzed in triplicate for both assays. The inter- and intra-assay coefficient of variation (CV) was calculated from 12 assays, each carried out in triplicate using the same genomic DNA standard. The intra-assay CV was 0.23% for MTND1, and 0.97% for GAPDH; and the inter-assay CV was 0.22% for MTND1, and 0.72% for GAPDH. The mtDNA content was expressed as a relative mtDNA/nDNA ratio.
Cell sorting
Fluorescence activated cell sorting (FACS) was used to identify the major blood cell components, using forward and side scatter based on size and morphology. Pacific Blue (CD14) was used to differentiate monocytes and lymphocytes that may be overlapping. Differential cell counts on whole blood were performed using Pacific Blue (CD14) and Pacific Orange (CD45). Platelets were collected from whole blood. To collect monocytes, lymphocytes and granulocytes, whole blood red cell lysis was performed by hypotonic lysis using Pharmlyse (Pharmingen, CA, USA) ammonium chloride (1×) with 20 min incubation at room temperature. Cells were then washed three times using a BD FACS™ Lyse/Wash Assistant (Becton–Dickinson, NJ, USA). Cell types were collected in sets of 10, 50, 100 and 1,000 from both patients and controls; they were unidirectionally sorted into single wells of a 96-well plate using a BD FACSAria (Becton–Dickinson, NJ, USA) using 100-mW sapphire laser, 100-μm nozzle at a sort rate of 5,000 events per second. Quality control and stability was proven daily by the use of Spherotech single peak Ultra Rainbow beads. Plates were stored at −20°C until required.
MtDNA analysis in specific cell types
A real-time PCR assay was used to quantify the absolute number of mtDNA molecules in each cell type using the standard curve method [13]. A PCR template was generated spanning the mtDNA genome between nt 3017 and nt 4057. The template was purified by gel extraction (Qiagen, Hilden, Germany), quantified by UV absorbance at 260 nm, and serially diluted to generate a standard curve for absolute quantification of mtDNA content in samples. To analyze the samples, cells were lysed for 16 h in 50 mM Tris–HCl, pH 8.5, with 0.5% Tween 20 and 100 mg proteinase K, at 55°C, followed by heat inactivation at 95°C for 10 min. All samples and standards were analyzed in triplicate. The intra- and inter-assay variation for the single cell analysis was similar to the whole blood genomic DNA analysis, with CVs ranging from 0.11–0.73% for the MTND1 assay on fluorescently sorted single cells. There was no difference in the CV between control subjects and sepsis patients.
Statistical analysis
Data was analyzed using GraphPad Prism v3 software (GraphPad, La Jolla, CA, USA). Quantitative variables were compared using Student’s t test and linear regression analysis was performed where appropriate.
Results
Patients and controls
The median age for the patients with sepsis was 63 (range 22–87), 57% being male. The median APACHE II score on recruitment was 26 (range 10–46). Mortality in ITU was 27%, at hospital discharge 45% and at 180 days 48%. There was no significant difference between the survivor and non-survivor groups in terms of ethnicity, gender, mtDNA copy number, co-morbidity and cause of the sepsis. The median age of the control volunteers was 69 (range 41–92), 36% being male.
Sepsis is associated with whole blood mtDNA depletion
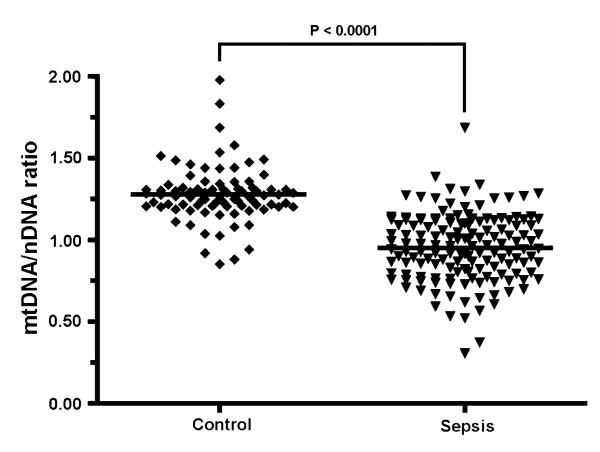
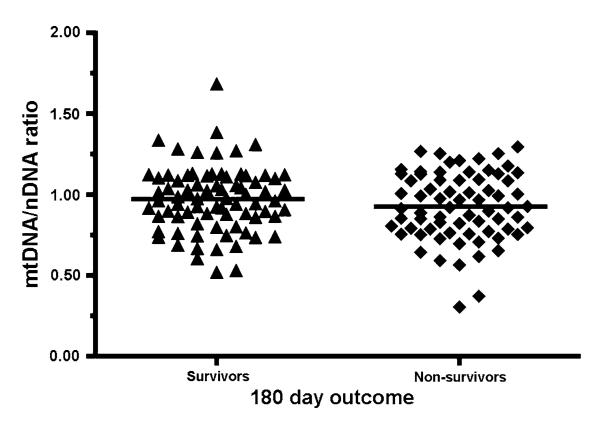
In keeping with the previous study of 28 patients [9], blood samples from the patients with sepsis contained significantly less mtDNA than controls (P = 1.18 × 10−26, Fig. 1). However, we saw no relationship between mtDNA content and survival outcome at 180 days in our cohort of 147 patients (P = 0.1705, Fig. 2). Although the septic group was 57% male while the control group was 36% male we found no gender influence on the blood cell mtDNA content.
Fig. 1.
Relative mtDNA/nDNA ratio in whole blood DNA from 147 sepsis patients and 83 controls. Horizontal lines represent mean value
Fig. 2.
Relative mtDNA/nDNA ratio in whole blood DNA from sepsis patient survivors (n = 77) and non-survivors (n = 70) 180 days after ICU admission. Horizontal lines represent mean value
MtDNA content of leukocyte sub-types in healthy control subjects
Acute sepsis is usually accompanied by a dramatic increase in total circulating blood leukocytes and an increase in the proportion of neutrophils. We speculated that the apparent decrease in whole blood mtDNA content could be due to an increase in the proportion of neutrophils in the blood sample.
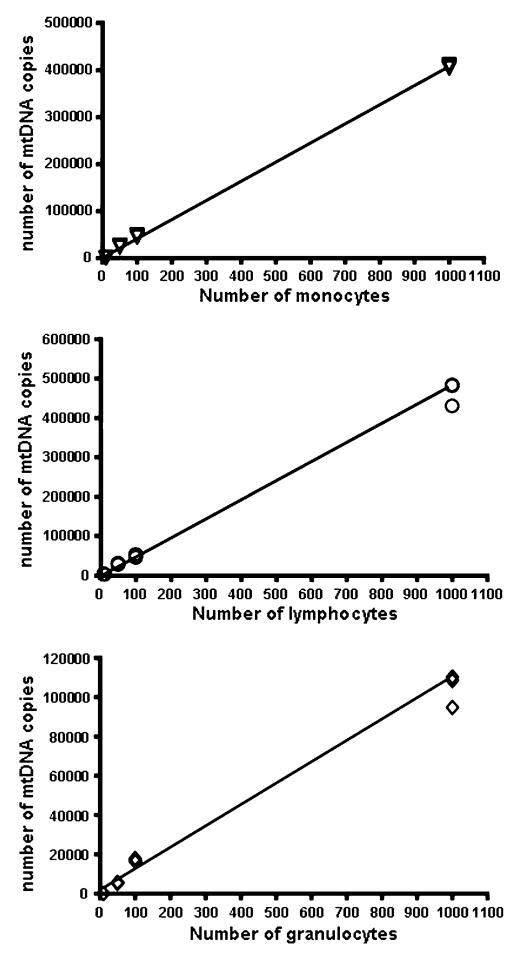
It is recognized that different cell types contain different amounts of mtDNA [14], and we initially determined the mtDNA content in healthy control subjects. MtDNA content was determined in wells containing 10, 50, 100, and 1,000 monocytes, lymphocytes and granulocytes (Fig. 3), confirming that the absolute quantification assay was linear over the required range (r2 = 0.99). Subsequent measurements were carried out on wells containing 1,000 cells in triplicate on 14 healthy control subjects.
Fig. 3.
Absolute amount of mtDNA measured in fluorescence activated (FACS) sorted cells taken from whole blood. Monocytes (triangles), lymphocytes (circles), and granulocytes (diamonds) were FACS sorted from whole blood taken from three control subjects. MtDNA content was determined by real time PCR in 10, 50, 100 and 1,000 cells of each subset. Each symbol represents the mean value of one control sample taken from a triplicate real time measurement. Each sample was measured in triplicate. Lines represent linear regression (r2 = 0.99, P < 0.001 in each case)
The mean mtDNA content was greatest in lymphoctyes (481.2, SD = 48.1) followed by monocytes (445.0, SD = 56.9, Fig. 3). Granulocytes contained approximately threefold less mtDNA (132.9, SD = 31.5), and platelets substantially less (5.6, SD = 2.9). Based on these values, we estimate that the normal range of granulocyte count in healthy subjects (50–70% of total leukocytes) will be associated with ~1.3-fold difference in mtDNA content of the peripheral blood. This range is comparable to the range of values we observed in healthy subjects (Fig. 1), and suggests that a large proportion of the variation in blood mtDNA content between controls actually arises through differences in the proportion of neutrophils to other cell types.
MtDNA content of leukocyte sub-types in sepsis patients
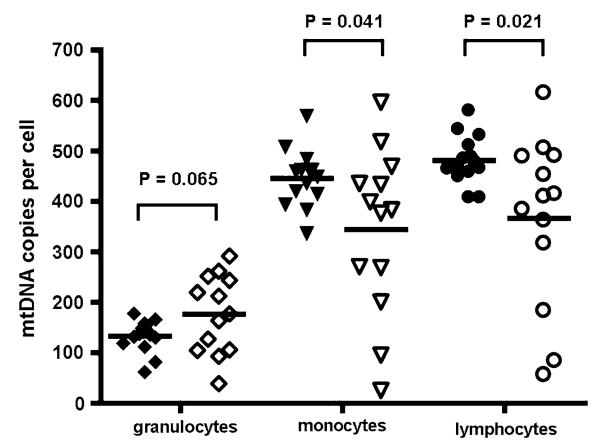
To determine whether the observed difference in whole blood mtDNA content in the sepsis patients was simply due to the associated neutrophil leukocytosis, we determined the absolute mtDNA content in FACS sorted leukocytes from 13 sepsis patients, taken within 12 h of fulfilling the Joint Consensus severe sepsis criteria [11], and compared this to 14 healthy control subjects (Fig. 4). Although there was no difference in the mean granulocyte mtDNA content (176.4, SD = 77.6, P = 0.065), the mean mtDNA content was significantly less in both monocyte (344.0, SD = 164.9, P = 0.041), and lymphocyte populations (366.7, SD = 167.1, P = 0.021).
Fig. 4.
Absolute amount of mtDNA measured in fluorescence activated (FACS) sorted monocytes (triangles), lymphocytes (circles), and granulocytes (diamonds) from 14 control subjects (filled symbols) and 13 sepsis patients (open symbols). Horizontal lines represent mean values
Clinical implications of the mtDNA depletion
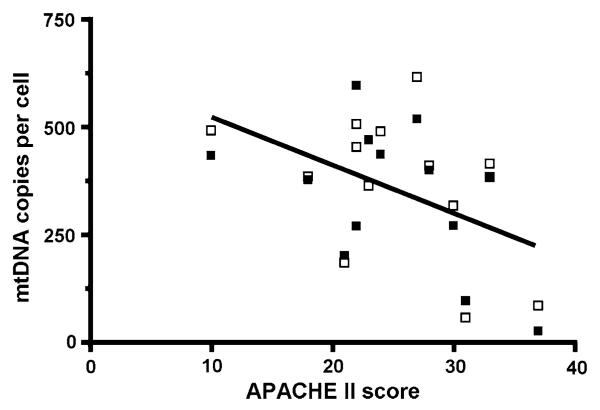
If there is a causal relationship between sepsis and the amount of mtDNA in leukocytes, then the degree of mtDNA depletion should correlate with the severity of the sepsis. In keeping with this, we found a significant linear relationship between illness severity (as assessed by the APACHE II score recorded at the time of sampling) and mtDNA copy number in the FACS sorted mononuclear sub-sets (pooled monocytes and lymphocytes) (P = 0.015; r2 = 0.222, Fig. 5). Part of the variation in the reduction in mtDNA copy number may therefore be the result of differing illness severity. This in turn is likely to reflect differences in the intensity of the inflammatory response between patients.
Fig. 5.
Correlation of APACHE II score of severity of illness in 13 sepsis patients and absolute mtDNA copy number from FACS sorted mononuclear sub-sets. Line represents linear regression (r2 = 0.222, P = 0.015). Open squares represent lymphocytes and filled squares represent monocytes
Discussion
Our observations in 147 patients confirm that sepsis is associated with a reduction of mtDNA content in peripheral blood. However, a large proportion of this change is actually attributable to an increased proportion of peripheral blood neutrophils. A previous study of 28 critically ill patients (14 of which were septic), demonstrated a longitudinal increase in blood mtDNA levels over a short period of time (less than 28 days) in ICU. Our study design differs as we observed mtDNA content cross-sectionally in septic survivors and non-survivors (at 180 days) in samples taken within 24 h of fulfilling joint consensus criteria. It is therefore possible that the previously observed increase in mtDNA content that accompanies recovery [9] following sepsis could simply reflect falling neutrophil counts after the acute phase. By studying cell subtypes in a limited group of patients with sepsis, we show for the first time that the mtDNA content of circulating monocytes and lymphocytes is decreased in the acute phase of sepsis. How can we explain this finding, and what are the implications for our understanding of sepsis?
Sepsis has been shown to be associated with a disturbance of toll-like receptor 4 (TLR4) expression in Kupffer cells [15], and the hepatic mtDNA damage shown in a mouse model of bacterial sepsis appeared to be mediated through TLR4 activation [16]. Based on these observations, one hypothesis is that TLR4 activation in human sepsis leads to mtDNA damage in circulating blood mononuclear cells, with the subsequent removal of damaged organelles by autophagy. Given their longer circulating half-life, monoctyes and lymphocytes would be more susceptible to the cumulative effects of TLR4 activation than neutrophils which can circulate for less than 12 h during septicaemia. This could explain why these cell types show the most obvious mtDNA depletion in our study. The global transcriptional downregulation of mitochondrial genes observed in acute systemic inflammation includes key genes required for mtDNA replication [15]. This would compromise the situation even further, leading to persistently low mtDNA levels. On the other hand, one could also speculate that the loss of mtDNA may not be a passive consequence of sepsis-induced damage, but actually contribute to the host defense mechanism, as has recently been demonstrated in eosinophils exposed to bacteria [17].
Could the depletion of mtDNA in the lymphocytes and monocytes contribute to the pathophysiology of sepsis? Mononuclear cells have important regulatory functions that influence the acute (innate) immune response to infections [18]. Monocytes and some lymphocyte sub-sets express pattern recognition receptors to micro-organisms and their breakdown products [19]. Binding of these ligands to the receptors results in cell activation with both initiation and amplification of the innate immune response. Recovery from infection also involves a successful transition to adaptive immunity. Monocytes and lymphoctes are again central to this process in terms of antigen processing, presentation and amplification of response. Alteration in mtDNA copy number is increasingly recognized as important in cell growth and differentiation [20]. T cell activation, for example, requires an acute increase in mtDNA copy number [21] as does the differentiation of some stem cell lines [22]. It is therefore tempting to speculate that the fall in mtDNA copy number in mononuclear cells will impair the immune response to infection [23].
Lymphocyte depletion in chronic sepsis is associated with a poor prognosis but the underlying mechanism is not clear [23]. For similar reasons, it is plausible that the observed mtDNA depletion contributes to this, through bioenergetic compromise operating on peripheral blood leukocytes or their precursors in the bone marrow. In addition, it is possible that the depletion we have observed in specific blood cell types could also be a marker of mtDNA depletion in other tissues such as vascular endothelium, skeletal muscle and myocardium, which are intimately involved in the pathophysiology of sepsis [1, 24]. Studying the underlying mechanisms of mtDNA depletion is likely to have direct relevance to the observed decrease in respiratory chain activity in these tissues, and thus advance our understanding of the pathophysiology of sepsis [3].
Finally, our observations also have more general relevance for the measurement of mtDNA in blood. For example, the differential white blood cell count should be considered in HIV patients treated with nucleoside analogues because a drop in the blood mtDNA content could simply reflect a neutrophil response to mild infection. Likewise, the “mild” mtDNA depletion measured in blood samples from patients with genetically determined mtDNA depletion disorders [25, 26] could also be partly due to a neutrophil leukocytosis—particularly on acute presentation which is often complicated by systemic infection. Previous studies have reported with either normal or low mtDNA values in both of these contexts [27, 28]. We have shown that the differential leukocyte count is a major confounding variable in this context, which provides a possible explanation for the discrepancies between previous studies. This raises the possibility that by incorporating blood count data, it might be feasible to diagnose mtDNA depletion disorders with a peripheral blood sample, and thus prevent invasive tissue biopsies in children who are often critically ill.
Conclusions
Mitochondrial content of whole blood falls in early human sepsis. Much of this fall is a result of the higher proportion of circulating neutrophil leukocytes which contain a lower mitochondrial copy number compared with mononuclear cells. However, there is a real fall in mitochondrial content in mononuclear cells. It is possible that this will have functional consequences in terms of immune depression.
Acknowledgments
PFC is a Wellcome Trust Senior Fellow in Clinical Science who also receives funding from the EU FP6 program, EUmitocombat and MITOCIRCLE, the Parkinson’s Disease Society (UK) and the Medical Research Council (UK). We thank Ian Dimmick and Rebecca Stewart for their assistance with the flow cytometry experiments. This work was principally funded by an unconditional research grant from the United States Army.
Contributor Information
Angela Pyle, Mitochondrial Research Group, Institute for Ageing and Health, The Medical School, Newcastle University, Framlington Place, Newcastle NE2 4HH, UK.
David J. Burn, Clinical Ageing Research Unit, Institute for Ageing and Health, The Medical School, Newcastle University, Newcastle NE4 5PL, UK
Charlotte Gordon, Institute of Cellular Medicine, Newcastle University, Newcastle NE2 4HH, UK; Critical Care Unit, Royal Victoria Infirmary, Newcastle NE1 4LP, UK.
Catherine Swan, Institute of Cellular Medicine, Newcastle University, Newcastle NE2 4HH, UK; Critical Care Unit, Royal Victoria Infirmary, Newcastle NE1 4LP, UK.
Patrick F. Chinnery, Mitochondrial Research Group, Institute for Ageing and Health, The Medical School, Newcastle University, Framlington Place, Newcastle NE2 4HH, UK
Simon V. Baudouin, Institute of Cellular Medicine, Newcastle University, Newcastle NE2 4HH, UK; Critical Care Unit, Royal Victoria Infirmary, Newcastle NE1 4LP, UK
References
- 1.Hotchkiss RS, Karl IE. The pathophysiology and treatment of sepsis. N Engl J Med. 2003;348:138–150. doi: 10.1056/NEJMra021333. [DOI] [PubMed] [Google Scholar]
- 2.Singer M, Brealey D. Mitochondrial dysfunction in sepsis. Biochem Soc Symp. 1999;66:149–166. doi: 10.1042/bss0660149. [DOI] [PubMed] [Google Scholar]
- 3.Brealey D, Singer M. Mitochondrial dysfunction in sepsis. Curr Infect Dis Rep. 2003;5:365–371. doi: 10.1007/s11908-003-0015-9. [DOI] [PubMed] [Google Scholar]
- 4.Fink MP. Cytopathic hypoxia. Mitochondrial dysfunction as mechanism contributing to organ dysfunction in sepsis. Crit Care Clin. 2001;17:219–237. doi: 10.1016/s0749-0704(05)70161-5. [DOI] [PubMed] [Google Scholar]
- 5.Zeviani M, Di Donato S. Mitochondrial disorders. Brain. 2004;127:2153–2172. doi: 10.1093/brain/awh259. [DOI] [PubMed] [Google Scholar]
- 6.Watts JA, Kline JA, Thornton LR, Grattan RM, Brar SS. Metabolic dysfunction and depletion of mitochondria in hearts of septic rats. J Mol Cell Cardiol. 2004;36:141–150. doi: 10.1016/j.yjmcc.2003.10.015. [DOI] [PubMed] [Google Scholar]
- 7.Suliman HB, Carraway MS, Piantadosi CA. Postlipopolysaccharide oxidative damage of mitochondrial DNA. Am J Respir Crit Care Med. 2003;167:570–579. doi: 10.1164/rccm.200206-518OC. [DOI] [PubMed] [Google Scholar]
- 8.Crouser ED, Julian MW, Huff JE, Struck J, Cook CH. Carbamoyl phosphate synthase-1: a marker of mitochondrial damage and depletion in the liver during sepsis. Crit Care Med. 2006;34:2439–2446. doi: 10.1097/01.CCM.0000230240.02216.21. [DOI] [PubMed] [Google Scholar]
- 9.Cote HC, Day AG, Heyland DK. Longitudinal increases in mitochondrial DNA levels in blood cells are associated with survival in critically ill patients. Crit Care. 2007;11:R88. doi: 10.1186/cc6096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Martin GS, Mannino DM, Eaton S, Moss M. The epidemiology of sepsis in the United States from 1979 through 2000. N Engl J Med. 2003;348:1546–1554. doi: 10.1056/NEJMoa022139. [DOI] [PubMed] [Google Scholar]
- 11.Levy MM, Fink MP, Marshall JC, Abraham E, Angus D, Cook D, Cohen J, Opal SM, Vincent JL, Ramsay G. 2001 SCCM/ESICM/ACCP/ATS/SIS International Sepsis Definitions Conference. Intensive Care Med. 2003;29:530–538. doi: 10.1007/s00134-003-1662-x. [DOI] [PubMed] [Google Scholar]
- 12.Pyle A, Taylor RW, Durham SE, Deschauer M, Schaefer AM, Samuels DC, Chinnery PF. Depletion of mitochondrial DNA in leucocytes harbouring the 3243A->G mtDNA mutation. J Med Genet. 2007;44:69–74. doi: 10.1136/jmg.2006.043109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Durham SE, Bonilla E, Samuels DC, DiMauro S, Chinnery PF. Mitochondrial DNA copy number threshold in mtDNA depletion myopathy. Neurology. 2005;65:453–455. doi: 10.1212/01.wnl.0000171861.30277.88. [DOI] [PubMed] [Google Scholar]
- 14.Veltri KL, Espiritu M, Singh G. Distinct genomic copy number in mitochondria of different mammalian organs. J Cell Physiol. 1990;143:160–164. doi: 10.1002/jcp.1041430122. [DOI] [PubMed] [Google Scholar]
- 15.Hsieh YC, Frink M, Kawasaki T, Thobe BM, Choudhry MA, Schwacha MG, Bland KI, Chaudry IH. Downregulation of TLR4-dependent ATP production is critical for estrogen-mediated immunoprotection in Kupffer cells following trauma-hemorrhage. J Cell Physiol. 2007;211:364–370. doi: 10.1002/jcp.20943. [DOI] [PubMed] [Google Scholar]
- 16.Suliman HB, Welty-Wolf KE, Carraway MS, Schwartz DA, Hollingsworth JW, Piantadosi CA. Toll-like receptor 4 mediates mitochondrial DNA damage and biogenic responses after heat-inactivated E. coli. Faseb J. 2005;19:1531–1533. doi: 10.1096/fj.04-3500fje. [DOI] [PubMed] [Google Scholar]
- 17.Yousefi S, Gold JA, Andina N, Lee JJ, Kelly AM, Kozlowski E, Schmid I, Straumann A, Reichenbach J, Gleich GJ, Simon HU. Catapult-like release of mitochondrial DNA by eosinophils contributes to antibacterial defense. Nat Med. 2008;14:949–953. doi: 10.1038/nm.1855. [DOI] [PubMed] [Google Scholar]
- 18.Medzhitov R, Janeway C., Jr Innate immunity. N Engl J Med. 2000;343:338–344. doi: 10.1056/NEJM200008033430506. [DOI] [PubMed] [Google Scholar]
- 19.Kumagai Y, Takeuchi O, Akira S. Pathogen recognition by innate receptors. J Infect Chemother. 2008;14:86–92. doi: 10.1007/s10156-008-0596-1. [DOI] [PubMed] [Google Scholar]
- 20.Jeng JY, Yeh TS, Lee JW, Lin SH, Fong TH, Hsieh RH. Maintenance of mitochondrial DNA copy number and expression are essential for preservation of mitochondrial function and cell growth. J Cell Biochem. 2008;103:347–357. doi: 10.1002/jcb.21625. [DOI] [PubMed] [Google Scholar]
- 21.D’Souza AD, Parikh N, Kaech SM, Shadel GS. Convergence of multiple signaling pathways is required to coordinately up-regulate mtDNA and mitochondrial biogenesis during T cell activation. Mitochondrion. 2007;7:374–385. doi: 10.1016/j.mito.2007.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen CT, Shih YR, Kuo TK, Lee OK, Wei YH. Coordinated changes of mitochondrial biogenesis and antioxidant enzymes during osteogenic differentiation of human mesenchymal stem cells. Stem Cells. 2008;26:960–968. doi: 10.1634/stemcells.2007-0509. [DOI] [PubMed] [Google Scholar]
- 23.Wesche DE, Lomas-Neira JL, Perl M, Chung CS, Ayala A. Leukocyte apoptosis and its significance in sepsis and shock. J Leukoc Biol. 2005;78:325–337. doi: 10.1189/jlb.0105017. [DOI] [PubMed] [Google Scholar]
- 24.Annane D, Bellissant E, Cavaillon JM. Septic shock. Lancet. 2005;365:63–78. doi: 10.1016/S0140-6736(04)17667-8. [DOI] [PubMed] [Google Scholar]
- 25.Saada A, Shaag A, Mandel H, Nevo Y, Eriksson S, Elpeleg O. Mutant mitochondrial thymidine kinase in mitochondrial DNA depletion myopathy. Nat Genet. 2001;29:342–344. doi: 10.1038/ng751. [DOI] [PubMed] [Google Scholar]
- 26.Bourdon A, Minai L, Serre V, Jais JP, Sarzi E, Aubert S, Chretien D, de Lonlay P, Paquis-Flucklinger V, Arakawa H, Nakamura Y, Munnich A, Rotig A. Mutation of RRM2B, encoding p53-controlled ribonucleotide reductase (p53R2), causes severe mitochondrial DNA depletion. Nat Genet. 2007;39:776–780. doi: 10.1038/ng2040. [DOI] [PubMed] [Google Scholar]
- 27.Cote HC, Brumme ZL, Craib KJ, Alexander CS, Wynhoven B, Ting L, Wong H, Harris M, Harrigan PR, O’Shaughnessy MV, Montaner JS. Changes in mitochondrial DNA as a marker of nucleoside toxicity in HIV-infected patients. N Engl J Med. 2002;346:811–820. doi: 10.1056/NEJMoa012035. [DOI] [PubMed] [Google Scholar]
- 28.Zsurka G, Baron M, Stewart JD, Kornblum C, Bos M, Sassen R, Taylor RW, Elger CE, Chinnery PF, Kunz WS. Clonally expanded mitochondrial DNA mutations in epileptic individuals with mutated DNA polymerase gamma. J Neuropathol Exp Neurol. 2008;67:857–866. doi: 10.1097/NEN.0b013e3181839b2d. [DOI] [PubMed] [Google Scholar]