Abstract
Viral antibody–free BBDR and WF rats never develop spontaneous diabetes. BBDR rats, however, develop autoimmune diabetes after perturbation of the immune system, e.g., by viral infection. We previously identified a disease-susceptibility locus in the BBDR rat, iddm4, which is associated with the development of autoimmune diabetes after treatment with polyinosinic:polycytidylic acid and an antibody that depletes ART2+ regulatory cells. We have now developed lines of congenic WF.iddm4 rats and report that in an intercross of N5 generation WF.iddm4 rats, ∼70% of animals either homozygous or heterozygous for the BBDR origin allele of iddm4 became hyperglycemic after treatment to induce diabetes. Fewer than 20% of rats expressing the WF origin allele of iddm4 became diabetic. Testing the progeny of various recombinant N5 WF.iddm4 congenic rats for susceptibility to diabetes suggests that iddm4 is centered on a small segment of chromosome 4 bounded by the proximal marker D4Rat135 and the distal marker D4Got51, an interval of <2.8 cM. The allele at iddm4 has 79% sensitivity and 80% specificity in prediction of diabetes in rats that are segregating for this locus. These characteristics suggest that iddm4 is one of the most powerful non–major histocompatibility complex determinants of susceptibility to autoimmune diabetes described to date.
Type 1A autoimmune diabetes is heritable, but the mode of inheritance is complex and non-Mendelian (1). Intensive analysis has identified several high-risk class II major histocompatibility complex (MHC) haplotypes associated with the disease, but none are necessary or sufficient for diabetes expression. More than 16 additional non-MHC loci have also been linked to the disease, but the analysis of such a complex trait in outbred populations is extremely difficult (2). The relevant genes and their mechanism of action have not been identified.
The inbred NOD mouse also develops a heritable type 1A–like diabetic syndrome that affects up to 90% of female mice. As in humans, diabetes in NOD mice is associated with a high-risk class II MHC haplotype (I-Ag7) and at least 17 non-MHC loci, but only one gene (β 2 microglobulin) has been identified with absolute certainty (3).
The genetic control of type 1A diabetes in the BB rat model has been studied less extensively, but new data suggest that it may now provide important advantages for the analysis of this disorder (4). Approximately 90% of inbred viral antibody–free (VAF) diabetes-prone BBDP rats develop spontaneous autoimmune hyperglycemia (5,6). Unlike humans with type 1 diabetes, however, BBDP rats are lymphopenic. In contrast, the inbred diabetes-resistant BBDR rat was developed from diabetes-prone forebears selected for normoglycemia (4) and is nonlymphopenic and immunocompetent (5). Under VAF conditions, no BBDR rats develop autoimmunity spontaneously (7), but type 1A–like diabetes can be induced by several interventions. These include low-dose irradiation, cyclophosphamide, high-dose polyinosinic:polycytidylic acid (poly I:C), viral infection, and in vivo depletion of ART2+ regulatory cells in combination with low-dose poly I:C (5). Arguably, the BBDR rat models type 1A diabetes in humans with considerable fidelity; the predisposition to disease is clearly heritable but requires the kind of interaction with the environment that is often believed to be critical for disease expression in humans (8,9).
As is the case for human type 1A diabetes, inheritance of disease in BB rats is associated with a permissive MHC locus, designated iddm2 in the rat. The BB rat expresses the class I RT1Au/RT1Cu, class II RT1B/Du MHC haplotype (10). Expression of diabetes is independent of class I haplotype but requires the presence of at least one class II RT1B/Du allele (11-15). The RT1u allele of the BB rat does not appear to be a unique diabetogenic variant (11), and diabetes can be induced by immunological perturbation in several class II RT1u rat strains (16).
The autosomal recessive locus lyp (or iddm1) causes T-cell lymphopenia in BBDP rats (17-19). The lyp/iddm1 locus has been mapped to chromosome 4 (RNO4) (19,20). Multiple genetic crosses have shown that deficiency in peripheral T-cells is necessary, but not sufficient, for the expression of spontaneous type 1 diabetes in BB rats (12,17-19,21). Studies of (BBDR × WF) × WF backcross animals, all of which are lyp+/+, have demonstrated that lyp is not required for the latent predisposition to autoimmune diabetes in BBDR rats that develop disease in response to appropriate immunomodulatory and environmental perturbants (22). The lyp/iddm1 gene has recently been identified as Ian4, a mitochondrial membrane protein (23).
Additional susceptibility loci have been mapped in backcrosses of (BBDP × WF)F1 and (BBDR × WF)F1 rats to the WF rat. Neither F1 rat develops spontaneous diabetes, but the disease was inducible in >95% of F1 animals after combined treatment with low-dose poly I:C and depletion of the regulatory ART2+ cell population (24). In both backcrosses, we mapped a locus on chromosome 4 (iddm4) with significant linkage to insulitis and type 1A–like diabetes expression. The iddm4 locus is linked to, but different from, lyp/iddm1/Ian4. It has been mapped in two separate (DP × WF)F1 × WF backcrosses, and identity-by-descent analysis suggested that the iddm4 locus is likely to play a role in disease initiation in both BBDP and BBDR rats (22).
Using a marker-assisted breeding strategy, we have now created WF.iddm4 congenic rat lines as a first step toward identifying the iddm4 gene and to confirm our hypothesis that iddm4 participates in generating the latent autoreactivity that is revealed when animals are exposed to appropriate environmental perturbation. We also generated a control WF.ART2a congenic rat, which, unlike the standard ART2b WF rat, can be depleted of ART2+ regulatory cells using available reagents. We now report that diabetes segregates in a genetically dominant manner with iddm4 in an N5 intercross of WF.iddm4 congenic rats and that thymocytes from WF.iddm4d/− rats transfer diabetes to adoptive recipients with high efficiency. Finally, in a progeny testing analysis of N6 WF.iddm4 congenic rats, we have established likely boundaries for the iddm4 interval on chromosome 4.
RESEARCH DESIGN AND METHODS
1. Animals
BBDR/Wor rats (RT1u/u, ART2a) were obtained from a VAF colony originally maintained at the University of Massachusetts Medical School and now at BRM (Worcester, MA). Wistar Furth (WF) rats (RT1u/u, ART2b) were purchased from Harlan Sprague Dawley (Indianapolis, IN). (BBDR/Wor × WF) × WF animals were bred in our facilities, as described previously (22), and backcrossed repetitively to generate WF.iddm4 congenic lines, as described below. (BBDR/Wor × WF) × WF animals were also backcrossed repetitively to generate a WF.ART2a congenic line, as described below. Athymic WAG rnu/rnu (RT1u/u) rats were obtained from BRM. Animals in these studies were maintained under VAF conditions, tested monthly, and consistently confirmed to be serologically free of Sendai virus, pneumonia virus of mice, sialodacryoadenitis virus, rat corona virus, Kilham’s rat virus, H-1 (Toolan’s virus), GD7, Reo-3, Mycoplasma pulmonis, lymphocytic choriomeningitis virus, mouse adenovirus, Hantaan virus, and encephalitozöon cuniculi. All animals were maintained in accordance with recommendations in the Guide for the Care and Use of Laboratory Animals (Institute of Laboratory Animal Resources, National Research Council, National Academy of Sciences, 1996) and the guidelines of the Institutional Animal Care and Use Committee of the University of Massachusetts Medical School.
2. Marker-assisted congenic rat production
Congenic rats were bred using a marker-assisted selection protocol (25). Rats from each backcross generation were treated as follows. DNA samples from all progeny were prepared from tail snips and screened using microsatellite markers that define the iddm4 and ART2 intervals plus a selected panel of 194 additional microsatellite loci chosen to be distributed as evenly as possible over all the autosomes. Typed rats found suitable for breeding were mated with diabetes-resistant rats to create the next generation. Initially, the diabetes-resistant rats were standard ART2b WF rats, and later in the breeding program they were WF.ART2a congenic rats (see below). Rats for breeding were selected according to the following criteria: 1) they were ART2a/b (so their progeny could be depleted with anti-ART2.1 antibody); 2) they had DR-derived alleles for the iddm4 interval, as determined conservatively by heterozygosity for markers between D4Rat16 and D4Rat44; and 3) they had the fewest DR-derived alleles at all other typed loci. At the next round, the same strategy was used for iddm4 and ART2, and so on for several generations. Congenic parental rats though the N5 generation were males. Thereafter, due to periodic unavailability of males, female congenic rats were used as parents when needed. Induced diabetes in BBDR rats is observed equally in both sexes (4), and at no time in the breeding program was an effect of sex on susceptibility to diabetes noted.
A total of seven rats with smaller recombinant iddm4 intervals were identified at generation N5 and used to generate multiple litters of subcongenic progeny, which are hereafter designated as “types” 5–11, Rats studied for susceptibility to diabetes induction in the present report were either these seven types of N6 progeny (n = 124) or the progeny (n = 58) of an intercross between N5 congenic siblings bearing the full iddm4 interval. The breeding schemes were as follows.
iddm4d/d homozygote production
By the N5 generation, much of the WF.iddm4 congenic rat genome was fixed for WF alleles (∼98–99%, see below), and we intercrossed selected iddm4d/w N5 animals to create N5F1 rats with iddm4d/d, iddm4d/w, and iddm4w/w on the WF background.
WF.ART2a congenic production
Using similar procedures, we selected an N3 (BBDR × WF) × WF iddm4w/w male having >95% WF origin genome to create a congenic WF rat that expresses the ART2.1 allotype. The male was analyzed using a genome-wide scan of the available polymorphic microsatellite markers and flow cytometry to document the presence of ART2.1+ T lymphocytes. Screening for ART2 allotype in subsequent generations was done by direct typing for ART2 using conformation-sensitive gel electrophoresis (CSGE) as described below. This line is now at the N8 generation and is >99% WF, The BBDR origin ART2 interval on chromosome 1, as determined by typing for flanking markers, is <5.6 cM.
Residual DR origin chromosomal segments and ART2 genotype
In the rats studied, there were eight segregating DR origin chromosomal segments outside the iddm4 and ART2 intervals. These were located on chromosome 10 at 0–34 cM (N5F1; types 6 and 7), 12 at 27–54 cM (N5F1; types 5, 6, 7, 9, and 11), 16 at 34–45 cM (N5F1; types 5, 6, 7, and 9), 19 at 20–43 cM (N5F1; types 5 and 6), 20 at 0–23 cM (N5F1; types 5, 6,7,10, and 11) 5 at 76–105 cM (types 7, 8, and 10), 6 at 76–85 cM (types 6–10), and 13 at 23–38 cM (N5F1; types 5, 6, 7, 9, and 10). It should be noted that none of the WF.iddm4 animals used in these studies, including the N5F1 series and the seven congenic subtypes, carried all eight of the residual DR origin intervals.
The protocol for induction of diabetes in these studies requires the depletion of ART2+ regulatory T-cells. Until the creation of the animals described here, all analyses of iddm4 in the rat (22,26) were carried out using animals heterozygous for ART2a (which encodes ART2.1) and ART2b (which encodes ART2.2) (27). These genes are codominant (28,29) but expressed somewhat asymmetrically, with the ART2b gene product being favored (30). The generation of the N5F1 animals and the mating scheme used to generate the N6 congenic types led to the generation of rats homozygous for ART2a. All statistical analyses included an adjustment for ART2 genotype. Because there are no available reagents that deplete ART2.2+ T-cells, no ART2b homozygotes have been studied. The flanking markers of the ART2 region in the WF.iddm4 rats in this study were D1Rat29 and D1Rat287.
3. DNA samples
Genomic DNA was prepared using one of two protocols. Snap-frozen livers were ground on dry ice, and the dispersed tissue was treated with proteinase K in the presence of 10% sarkosyl and 0.5 mol/1 EDTA (pH 8.0). DNA was purified from these digests by phenol-chloroform extraction and dialysis against Tris EDTA (0.01 mol/1 Tris/0.001 mol/l EDTA, pH 7.4). Alternatively, genomic DNA was extracted from rat tail snips using the QIAamp Tissue kit (Qiagen, Stanford, CA). DNA was extracted according to the manufacturer’s instructions.
4. Microsatellite and mapping analysis
All microsatellite primers used in this study are available from Research Genetics (Huntsville, AL). The general map location of these microsatellites was taken from our own segregating backcrosses and from maps published by the Rat Genome Database (http://www.rgd.mcw.edu) and by Dr. R. Wilder and Dr. E. Remmers (www.nih.gov/niams/scientific/ratgbase/index.htm). Those primers found to be polymorphic between parental strains were then used in a genome-wide screen of the progeny. Primers were end-labeled using [γ-32P]ATP, used in a PCR, and resolved by polyacrylamide gel electrophoresis as described (26). For detection of polymorphisms within expressed sequence tags (ESTs), appropriate primers were chosen from the EST database (www.ratest.uiowa.edu) and were added unlabeled to a PCR. The products were resolved using CSGE as described (31). Autoradiographic films of CSGE gels were also used to resolve the two alleles of the ART2 gene, amplified using radiolabeled primers that flank the polymorphic nucleotides (forward: 5′-CCAATGCATTTGATGACCAG-3′; reverse: 5′-TCCCAGTGTAGGCAACTAAAGC-3′). The position of markers on the genetic map was established by inspection of the dataset and conventional calculation methods to establish meiotic map distances, which are expressed in centiMorgans.
5. Induction of diabetes
For the induction of autoimmune diabetes, all congenic animals were screened for expression of the ART2.1 allotype as described above. It was necessary to do so because, as noted earlier, there is no depleting anti-ART2.2 monoclonal antibody (mAb) comparable to the T-cell–depleting DS4.23 anti-ART2.1 mAb used in all prior analyses of diabetes induction in the BBDR rat. When 28–32 days of age, ART2.1+ rats were treated with the cytotoxic DS4.23 anti-ART2.1 mAb (1 ml of 2× hybridoma supernatant five times per week) and poly I:C, a nonspecific immune system activator (2.5 μg/g, three times per week), as described (32). The combination regimen administered to VAF BBDR/Wor rats induces type 1 diabetes in >90% of animals (32). Treatment with both mAb and poly I:C was stopped when diabetes was diagnosed or after 40 days of treatment. All experimental animals were screened three times weekly for the presence of glycosuria (Tes-Tape; Eli Lilly, Indianapolis, IN). The presence of diabetes in glycosuric rats was established on the basis of a plasma glucose concentration >250 mg/dl (Glucose Analyzer 2; Beckman Instruments, Fullerton, CA).
6. Histological evaluation of insulitis
After the diagnosis of diabetes or at the conclusion of the experiment, rats were killed and pancreata were removed and fixed in 10% buffered formalin. Paraffin-embedded sections of pancreas were prepared for histologic analysis and stained with hematoxylin and eosin. An evaluator who was not informed of the donor’s glycemic status scored the tissues. On the basis of histological appearance, pancreatic insulitis was scored as both a qualitative (present or absent) phenotype and a quantitative trait locus. Pancreata were graded on a scale of 0–4+ as follows: 0, no inflammatory mononuclear cell infiltration of any islets; 1+, islet mononuclear cell infiltration only at the periphery of the islet (“peri-insulitis”); 2+, small numbers of mononuclear cells infiltrating into islets with preservation of islet architecture; 3+, large numbers of mononuclear cells within most islets affected and some distortion of islet architecture; and 4+, florid infiltration of mononuclear cells or classical end-stage islets.
7. Thymocyte adoptive transfer
Thymocytes for use in adoptive transfer studies were obtained from the N5F1 WF.iddm4 donor animals described above. These rats had all been treated with poly I:C and anti-ART2.1 mAb, and thymi were recovered from diabetic donors at the time of disease onset and from nondiabetic donors immediately after the 40th day of treatment. Recovered thymi from each individual donor were processed as described (33) and adoptively transferred at a dose 200 × 106 cells i.v. into a single athymic WAG recipient. The recipients were treated with anti-ART2.1 mAb plus poly I:C as described (33). Neither treatment of the thymocyte donor with anti-ART2 mAb plus poly I:C nor the presence of diabetes in the donor influences the ability of BBDR rat thymocytes to adoptively transfer disease (33,34).
8. Data analysis
Differences among groups of rats with respect to diabetes-free survival were analyzed by the method of Kaplan and Meier using the log-rank statistic (35); when necessary, analyses were stratified by ART2 genotype (36). Parametric data are shown as arithmetic means ± SD. Insulitis scores were analyzed nonparametrically using Mann Whitney U tests (37). Analyses of proportions used χ2 analysis or the Fisher’s exact statistic (37), and P values <0.05 (two tailed) were considered statistically significant.
The relative positions of markers on chromosome 4 were determined by examining the rate of recombination between them in both the N5F1 intercross and the progeny of the N4 and N5 congenic strains. In most cases, these positions were found to be very similar to the positions of the same markers, as determined by radiation hybrid panel analysis (rgd.mcw.edu).
RESULTS
1. Diabetes segregates with iddm4 in an N5 intercross of WF.iddm4 congenic rats
A panel of informative microsatellite markers on RNO4 was used to verify the retention of the iddm4 interval in each generation of marker-assisted congenics. From among the N5 WF.iddm4 progeny scored for markers linked to iddm4 (22,26), two males and two females, each bearing a full-length nonrecombinant iddm4 interval, were selected to generate intercross progeny. This interval was flanked by D4Rat16 and D4Rat44 (22). A total of 58 offspring were generated and treated with poly I:C and anti-ART2 mAb to induce diabetes beginning at 28–32 days of age and continuing for 40 days or until the onset of diabetes.
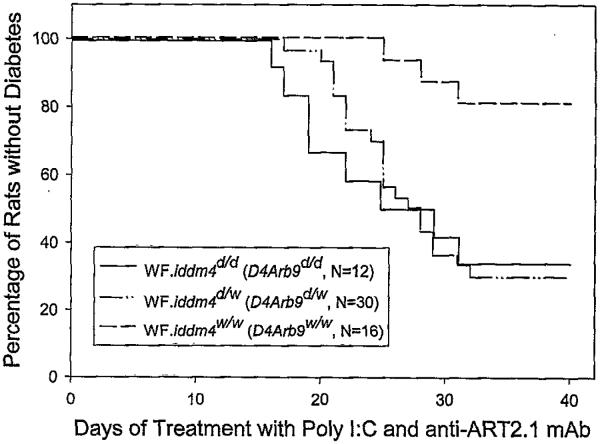
Confirming our original results (22,26), diabetes segregated significantly with markers in the iddm4 interval; 29/32 diabetic rats carried BBDR-derived alleles at these markers. As before, linkage analysis revealed that the diabetic phenotype correlated best with inheritance of BBDR-derived alleles at D4Arb9/D4Arb38/D4Mgh32 (P = 0.0011). We therefore used these markers to distinguish iddm4d/d, iddm4d/w, and iddm4w/w animals. Among iddm4d/d rats (n = 12), 67% became diabetic, with a mean latency of 22 days. Among iddm4d/w animals (n = 30), 70% became diabetic, with a mean latency of 25 days. In contrast, the cumulative frequency of diabetes in iddm4w/w progeny (n = 16) was significantly less (18%, P = 0.007 vs. iddm4d/d and P = 0.003 vs. iddm4d/w), with a mean latency of 28 days in these three rats (Fig. 1). There was no statistically significant difference in the cumulative frequency or latency to onset of diabetes in the homozygous iddm4d/d rats when compared with the heterozygous iddm4d/w animals (Fig. 1). Each of these analyses was adjusted for ART2 genotype because this experimental cohort of N5F1 rats included animals homozygous for ART2a. The overall life table analysis, considering all three iddm4 groups and adjusting for ART2 genotype, was significant at the P = 0.0027 level.
FIG. 1.
Kaplan Meier analysis of diabetes in WF.iddm4 N5F1 rats. From among N5 WF.iddm4 progeny scored for markers linked to iddm4, suitable rats were selected to generate intercross progeny as described in RESEARCH DESIGN AND METHODS. In this figure, progeny were typed as homozygous or heterozygous for the presence WF or BBDR origin alleles of the microsatellite marker D4Arb9. A total of 58 of the progeny expressed the ART2.1 regulatory T-cell alloantigen and could therefore be tested for susceptibility to the induction of diabetes by administration of poly I:C and anti-ART2.1 mAb. Among WF.iddm4d/d rats, 67% became diabetic, with a median latency of 25 days. Among WF.iddm4d/w animals, 70% became diabetic, with a median latency of 27 days. The cumulative frequency of diabetes in iddm4w/w progeny was statistically significantly less (18%, P = 0.007 vs. iddm4d/d and P =0.003 vs. iddm4d/w). The cumulative frequency of diabetes in homozygous iddm4d/d rats and heterozygous iddm4d/w rats was statistically similar (P = 0.57). Significance levels were determined by log-rank statistic with adjustment for ART2 genotype (see RESULTS).
The analysis also revealed that the ART2 genotype was a determinant of the degree of penetrance of iddm4. Overall, 94% of N5F1 WF.iddm4d/− rats homozygous for ART2a became diabetic (n = 16 of 17), compared with 52% of rats that were WF.iddm4d/−ART2a/b heterozygotes (n = 13 of 25, P < 0.004, Fisher’s exact test). Among animals that were WF.iddm4w/w, two of four rats homozygous for ART2a became diabetic, compared with 1 of 12 that were ART2a/b heterozygotes (P = 0.047).
2. Progeny analysis
Analysis of candidate parents for the N5 generation of WF.iddm4 rats revealed several recombinants carrying various segments of the interval bounded by D4Rat16 and D4Rat44, which we had used to define the original iddm4 locus (22). To narrow the boundaries of the iddm4 interval, we analyzed the offspring of recombinant progeny bearing BBDR origin markers in seven selected subcongenic intervals, each of a different length, on chromosome 4. It was necessary to adopt this strategy because the diabetes phenotype is incompletely penetrant and the resistant phenotype is incompletely resistant (22,26). The approximate map locations of these intervals, together with intermarker distances in centiMorgans, are shown in Fig. 2. Mapping analysis of the progeny in this study (based on >500 meioses) places the typed markers at positions that were generally consistent with the radiation hybrid and intercross maps (rgd.mcw.edu).
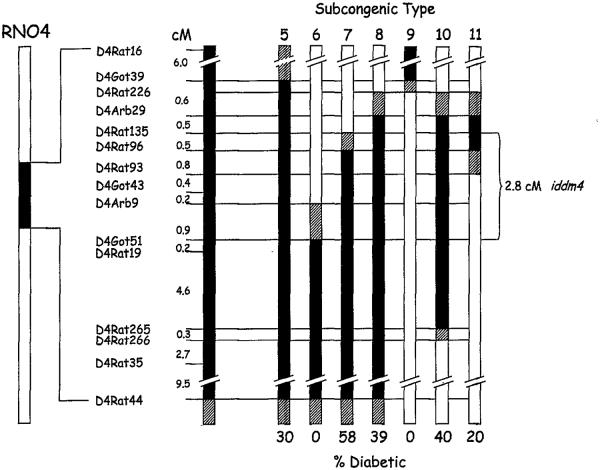
FIG. 2.
Map of diabetes susceptibility in heterozygous WF.iddm4 congenic rats. The left-most bar represents the entire length of rat chromosome 4 (RN04). The solid region of the bar indicates the iddm4 interval present in the N5 parents of the N5F1 rats shown in Fig. 1 and the N6 congenic progeny analyzed in Table 1. A magnified view of the iddm4 interval with the relevant microsatellite markers is shown in the adjacent bar, with the intermarker distances indicated in centiMorgans. The remaining bars represent the segment of this interval present in the parent of each of the seven subcongenic types, with the type number indicated on the top of each bar. At the bottom of each bar is the percentage of progeny of these animals that developed diabetes after treatment, as explained in Table 1 and RESULTS. Areas of the bars with no fill indicate WF origin alleles, and areas with solid fill indicate BBDR-derived alleles. Diagonal fill indicates transition intervals for which the strain of origin could not be determined with available markers. Note that these chromosomal segments are those of the parents and do not take into account recombinations that may be present in their progeny. Considering only the definitively typed regions (solid fill) and omitting from consideration type 11, which is indeterminate in its susceptibility to diabetes, the data from progeny testing define a 2.8-cM interval for iddm4 bounded by D4Rat135 and D4Got51.
The susceptibility to diabetes induction of rats bearing at least one DR-derived allele within the interval was compared with that of their WF.iddm4w/w littermates. As shown in Table 1 (bottom row), the susceptibility of N6 WF.iddm4w/w animals to diabetes induction was low; only 3 of 53 animals became diabetic. Among the seven subcongenic types tested, the frequency of diabetes in chromosome 4 heterozygotes ranged from 0 to 58% (Table 1).
TABLE 1.
Analysis of WF.iddm4 N6 progeny
| Progeny type | n |
n (%) diabetic |
Mean latency to diabetes (days) |
P * | Number of diabetic rats with D4Arb9d allele |
|---|---|---|---|---|---|
| Congenic rats with BBDR-derived chromosome 4 alleles |
|||||
| 5 | 10 | 3 (30) | 17 | <0.01 | 3/3 |
| 6 | 12 | 0 (0) | — | NS | 0/0 |
| 7 | 12 | 7 (58) | 24 | <0.001 | 7/7 |
| 8 | 18 | 7 (39) | 20 | <0.001 | 7/7 |
| 9 | 4 | 0 (0) | — | NS | 0/0 |
| 10 | 5 | 2 (40) | 30 | <0.01 | 2/2 |
| 11 | 10 | 2 (20) | 30 | NS | 0/2 |
| Congenic rats homozygous for WF-derived chromosome 4 alleles |
|||||
| Pooled from progeny types 5–11 | 53 | 3 (5) | 29 | — | 0/3 |
Data are the results of WF.iddm4 congenic progeny testing. N5 generation WF.iddm4 congenic rats with recombinant iddm4 intervals were identified and bred to WF.Art2a rats. Their N6 progeny were tested for susceptibility to the induction of diabetes as described in RESEARCH DESIGN AND METHODS. Seven subcongenic types were generated in this way. The approximate location and size of the BBDR rat-origin interval carried by each line is shown in Fig. 2.
P values were calculated by 2 × 2 χ2 analysis. Indicated P values are with respect to the pooled group having all WF-derived chromosome 4 markers. Progeny of types 6, 9, and 11 could not inherit D4Arb9d alleles, which were not present in their N5 congenic parents. A fraction of the progeny from types 5, 7, 8, and 10 were recombinants that did not inherit D4Arb9d. The number of diabetic rats in each type of congenic that carried D4Arb9d is shown in the right-most column. NS, not significant.
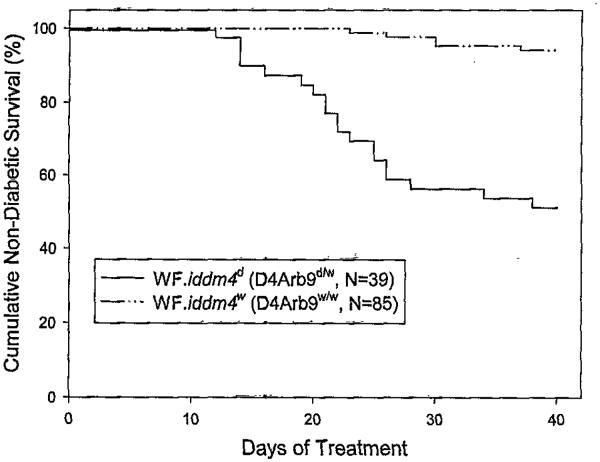
The linkage of diabetes in the congenic progeny to markers in the iddm4 interval is shown in Table 2. As was true for the N5F1 analysis, susceptibility to diabetes was best predicted by inheritance of a set of markers in the middle of the iddm4 interval, including D4Arb38, D4Arb9, and D4Mgh32. Inheritance of DR-derived alleles at these loci provided a surrogate for the iddm4 locus itself. There were no recombinants detected in the congenic population studied between D4Rat96 and D4Arb9 (Fig. 2), We therefore classified all of the N6 progeny as either D4Arb9d/− or D4Arb9w/w. Using this peak marker, we calculated the cumulative frequency and kinetics of induced diabetes in WF.iddm4w/w and WF.iddm4d/W rats, and the result demonstrates a striking difference in diabetes susceptibility between the two groups (Fig. 3). As was done for the N5F1 analysis, the analysis was stratified by ART2 genotype. After adjusting for ART2, the effect of D4Arb9 was highly statistically significant (P < 0.0001). The analysis confirmed that the ART2 genotype was an important determinant of the penetrance of iddm4. Overall, 78% (14 of 18) of WF.iddm4d/w rats that were homozygous for ART2a became diabetic, compared with 24% (5 of 21) of rats that were WF.iddm4d/w ART2a/b heterozygotes (P < 0.002, Fisher’s exact test). Among WF.iddm4w/w rats, very few developed diabetes, and there was no statistically significant difference between ART2a/a homozygotes (3 of 25) and ART2a/b heterozygotes (2 of 28, P = 0.15, Fisher’s exact test).
TABLE 2.
Linkage of markers near D4Arb9 to insulitis and diabetes
| Trait | Insulitis score P value |
Insulitis χ2 |
Insulitis P value |
Diabetes χ2 |
Diabetes P value |
Genomic exclusions determined by subcongenic types* |
|---|---|---|---|---|---|---|
| D4Rat16 | NS | 0.1 | NS | 0.46 | NS | 7, 10 |
| D4Rat134 | NS | 0.1 | NS | 0.00 | NS | 7, 10 |
| D4Mit9 | 0.03 | 4.0 | NS | 0.19 | NS | 7, 10 |
| D4Wox10 | 0.03 | 1.9 | NS | 0.19 | NS | 7, 10 |
| D4Arb11 | 0.03 | 4.6 | 0.03 | 0.06 | NS | 7 |
| D4Rat226 | 0.02 | 7.1 | 0.008 | 1.22 | NS | 7 |
| D4Arb29 | 0.01 | 7.1 | 0.008 | 9.26 | 0.0023 | 7 |
| D4Rat122 | 0.01 | 5.7 | 0.02 | 9.98 | 0.0016 | 7 |
| D4Rat135 | 0.01 | 5.7 | 0.02 | 9.98 | 0.0016 | 7 |
| D4Got48 | <0.003 | 8.6 | 0.003 | 28.70 | <0.0001 | None |
| D4Rat96 | <0.003 | 8.6 | 0.003 | 28.70 | <0.0001 | None |
| D4Rat93 | <0.003 | 8.6 | 0.003 | 31.43 | <0.0001 | None |
| EST tva | <0.003 | 8.6 | 0.003 | 31.43 | <0.0001 | None |
| D4Arb9 | <0.003 | 8.6 | 0.003 | 31.43 | <0.0001 | None |
| D4Got51 | NS | 3.6 | NS | 24.12 | <.0001 | 6 |
| D4Got52 | NS | 3.6 | NS | 24.12 | <.0001 | 6 |
| D4Rat19 | NS | 3.6 | NS | 24.12 | <.0001 | 6 |
| D4Mit5 | NS | 1.7 | NS | 20.53 | <.0001 | 6 |
| D4Rat228 | NS | 1.1 | NS | 19.58 | <.0001 | 6 |
| D4Arb30 | NS | 1.1 | NS | 19.58 | <.0001 | 6 |
| D4Rat265 | NS | 0.9 | NS | 9.18 | 0.002 | 6 |
| D4Rat266 | NS | 1.7 | NS | 6.79 | 0.009 | 6 |
| D4Rat35 | NS | 1.9 | NS | 3.33 | NS | 6 |
| D4Rat38 | NS | 1.0 | NS | 4.15 | NS | 6 |
| D4Rat44 | NS | 1.6 | NS | 3.18 | NS | 6 |
Data are linkage of diabetes and insulitis to markers near D4Arb9d in congenic rats. Microsatellite markers in the left-most column were linked to insulitis severity or the presence of insulitis in the 67 progeny that did not develop overt diabetes, as shown in Table 3. ANOVA was used to determine significance of linkage to the insulitis score, and the 2 × 2 χ2 test was used to determine the significance of the frequency of insulitis and diabetes, scored as binary traits. The frequency of diabetes with respect to inheritance of the DR-derived allele at each of the markers was calculated for the entire congenic cohort shown in Table 1 (N = 124 rats).
The right-most column identifies congenic subtypes with either a high (types 7 and 10) or a zero (type 6) frequency of diabetes (see Table 1) that were used to exclude regions of RN04 from the iddm4 interval. At the indicated markers, types 7 and 10 are “w” but are diabetes-susceptible and type 6 has the “d” allele but is diabetes-resistant. This information was used to construct the map shown in Fig. 2. NS, not significant (P > 0.05).
FIG. 3.
Kaplan Meier analysis of diabetes in WF.iddm4d (D4Arb9d/w) and WF.iddm4w (D4Arb9w/w) N6 congenic rats. Congenic N6 generation WF.idddm4 rats were bred and treated with poly I:C and anti-ART2.1 mAb as described in RESEARCH DESIGN AND METHODS. All animals were genotyped and grouped according to the presence of the WF and BBDR alleles of the microsatellite marker D4Arb9, as described in RESULTS and in the legend to Table 1. One WF.iddm4d rat that died on day 17 with unknown glycemic status has been omitted. The cumulative frequency of diabetes was 49% in the WF.iddm4d rats and 6% in the WF.iddm4w group (P < 0.0001, log-rank statistic after adjustment for ART2 genotype).
The linkage of diabetes in the various types of congenic progeny to markers in the iddm4 interval, as shown in Table 2, establishes the current working boundaries for iddm4. The key observations in Table 2 are 1) type 7 congenics (58% frequency of diabetes) had a WF-derived allele at D4Rat135 but a DR-derived allele at D4Rat96, and 2) type 6 congenics (0% frequency of diabetes) had WF-derived alleles at D4Arb9, D4Arb38, and D4Mgh32 and a DR-derived allele of D4Got51. Our mapping of the two flanking markers (D4Got51 and D4Rat135) places them in the central region of rat chromosome 4, ∼40–45 cM distal to the centromere. These observations, together with the map locations shown in Fig. 2, allowed us to narrow the iddm4 interval to a <2.8-cM segment of chromosome 4 in the vicinity of the trypsin and T-cell receptor β-chain loci.
We next analyzed the frequency and intensity of insulitis, the pathological substrate of autoimmune diabetes, in a sample of the congenic rats shown in Table 1 that were still nondiabetic at the conclusion of the period of observation on day 40. In previous reports (22,26), we noted an association between the presence of the BBDR origin allele of iddm4 and higher intensity insulitis scores. As shown in Table 3, the intensity of insulitis in our congenic rats with the WF origin allele of D4Arb9 was low. In contrast, insulitis was both more common and more intense in the nondiabetic rats with the DR origin allele. A complete assessment of the effect of ART2 genotype on insulitis was not possible because insulitis scores were available for only two rats that were both iddm4d and ART2a/b. The overall mean insulitis score in nondiabetic ART2a/a rats (2.3 ± 1.5, n = 18) was slightly higher than in ART2a/b animals (1.2 ± 1.5, n = 49, P < 0.025).
TABLE 3.
Insulitis in nondiapetic WF.iddm4 congenic rats
| Group | n | Mean insulitis score* |
n (%) with 0 or 1 + insulitis |
n (%) with 2+, 3+, or 4+ insulitis |
|---|---|---|---|---|
| Congenic WF.iddm4
rats that inherited D4Arb9d/w |
16 | 2.50 ± 1.41 | 4 (25)† | 12 (75)† |
| Congenic WF.iddm4
rats homozygous for D4Arb9w/w |
51 | 1.22 ± 1.46 | 34 (67) | 17 (33) |
A sample of pancreata from animals described in Table 1 that were nondiabetic at the conclusion of the experiment was scored for the presence of insulitis on a scale of 0–4 as described in RESEARCH DESIGN AND METHODS. Insulitis scores are shown as the arithmetic means ± SD.
P < 0.005 (Mann-Whitney U test).
P = 0.004 (Fisher’s exact statistic). Comparisons used nonparametric statistics to avoid making assumptions about the distribution of scores. Not all pancreata from the rats shown in Table 1 were analyzed because some of the nondiabetic animals were needed for subsequent breeding (N = 23), one rat died before harvest of the pancreas, and some histology specimens were unsatisfactory and could not be interpreted (N = 9).
We also analyzed insulitis in a sample of diabetic rats of all types to determine whether diabetes was induced at varying degree of insulitis in different groups. We observed, however, that for both the N5F1 intercross and the congenics, the mean insulitis scores were almost uniformly 4+. (Thirty-five of 36 scored pancreata from diabetic rats showed 4+ insulitis and the one exception was scored 2+.)
The linkage of insulitis scores in the nondiabetic congenic progeny to markers in the interval was assessed by ANOVA and a 2 × 2 χ2 analysis (Table 2). Once again, the most highly significant linkage occured between insulitis and the iddm4-associated markers within the larger interval. These include D4Arb9, D4Arb38, and the EST UI-R-C3-tv-a-09-0-UI (hereafter abbreviated tva) (http://www.ratest.uiowa.edu). Each of these genetic markers identifies one of the set of pancreatic trypsin genes on chromosome 4. Table 2 also demonstrates that the linkage maps for insulitis and diabetes do not coincide for the region of chromosome 4 distal to tva.
Analysis by ANOVA documented that there was no independent segregating contribution of those chromosomal segments outside the iddm4 and ART regions that still carry BBDR-derived alleles (see RESEARCH DESIGN AND METHODS) to susceptibility to diabetes or insulitis.
3. Thymocyte transfer
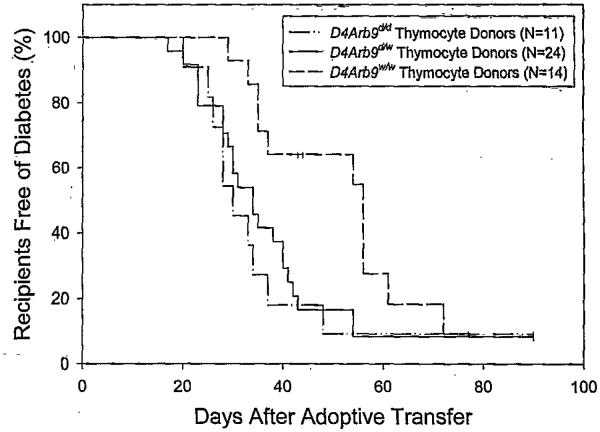
It is known that thymocytes from BBDR rats can adoptively transfer diabetes to athymic recipients (33,38), and we next sought to determine whether the ability to transfer diabetes was related to the presence of DR origin alleles at the iddm4 locus. Thymocytes for adoptive transfer were obtained from a sample of the WF.iddm4d/w, WF.iddm4d/d, and WF.iddm4w/w N5F1 animals described in Fig. 1. As shown in Fig. 4, the majority of adoptive recipients of thymocytes became diabetic within 90 days, irrespective of the genotype of the donor. However, diabetes appeared earlier when thymocytes were obtained from either WF.iddm4d/d N5F1 (n = 11, 10 diabetic, median latency 30 days) or WF.iddm4d/w N5F1 donors (n = 24, 3 diabetic, median latency 34 days) rather than WF.iddm4w/w donors (n = 14, 11 diabetic, median latency 56 days). Life table analysis, after adjustment for the ART2 genotype of the thymocyte donor, confirmed a statistically significant difference in the behavior of the three groups at the P < 0.005 level. Between-group analyses revealed that thymocytes from WF.iddm4d/d and WF.iddm4d/w were equally effective in transferring diabetes (P = 0.5), and both of these groups differed significantly from the WF.iddm4w/w thymocyte donors (P < 0.01). Overall, there was no difference in latency to onset of diabetes when the thymocyte donor was ART2a/a versus ART2a/b (P = 0.36).
FIG. 4.
Kaplan Meier analysis of diabetes in adoptive recipients of thymocytes. Thymocytes were obtained from the N5F1 WF.iddm4 rats described in Fig. 1 and prepared for adoptive transfer to WAG rnu/rnu rats, as described in RESEARCH DESIGN AND METHODS. Thymi were obtained from diabetic donors within 1 day of diagnosis; additional thymi were obtained from animals still nondiabetic at the conclusion of the protocol on day 40. Recipients were monitored for the development of diabetes for 90 days after transfer. Overall life table analysis, after adjustment for the ART2 genotype of the thymocyte donor, confirmed a statistically significant difference in the behavior of the three groups at the P < 0.005 level. Thymocytes from WF.iddm4d/d and WF.iddm4d/w were equally effective in transferring diabetes (P = 0.5), and both of these groups differed significantly from the WF.iddm4w/w thymocyte donors (P < 0.01). Recipients of thymocytes from donors bearing the WF allele of D4Arb9 became diabetic, with a median latency to onset of 56 days, whereas recipients of thymocytes from donors bearing a BBDR origin allele of D4Arb9 became diabetic, with a median latency of 33 days. There was no statistically significant independent effect attributable to the presence or absence of diabetes in the thymocyte donor. N indicates the number of rats entered into the study, and the small vertical bars indicate censored data, i.e., rats that either died during the study (n = 4) or were still normoglycemic at the end of the period of observation.
DISCUSSION
These data confirm and extend our hypothesis that iddm4, now mapped to a <2.8-cM interval on chromosome 4, is an exceptionally strong non-MHC determinant of susceptibility to autoimmune diabetes in the rat. This hypothesis was developed in previous studies of (WF × BBDP) × WF and (WF × BBDR) × WF backcrosses. Those studies clearly established iddm4, localized initially to a 40-cM interval, as a major determinant of diabetes susceptibility that is common to both the DP and DR sublines of the BB rat (22,26). Our observation here that nearly 75% of N5 generation WF.iddm4d intercross rats developed autoimmune diabetes after immunological perturbation confirms the importance of iddm4 and the fidelity of our phenotype within the congenic breeding scheme. The intercross also confirmed that the diabetogenic BBDR origin allele of iddm4 is genetically dominant.
Testing the N6 progeny of various recombinant N5 WF.iddm4 congenic parents revealed that susceptibility to diabetes is controlled by a segment of RNO4 containing tva and bounded by the proximal marker D4Rat135 and the distal marker D4Got51. Analysis of the entire dataset generated in the progeny study suggests that the presence of the BB origin allele of iddm4 is 79% sensitive and 80% specific in the prediction of diabetes in rats that segregate for this locus. In the N5F1 cohort, the relative risk of developing diabetes in treated animals bearing the BBDR origin allele of iddm4 was 3.31 (95% CI 1.00–10.8). In the N6 congenic cohort, the relative risk was 8.60 (95% CI 2.90–25.6).
The experimental cohort of N5F1 and congenic rats presented here is the first to include animals homozygous for ART2a. In previous analyses of the iddm4 locus, all rats were ART2a/b heterozygotes. The present data reveal that the ART2 genotype is an important determinant of the penetrance of iddm4. There are several possible mechanisms by which homozygosity for ART2.1 could affect the penetrance of iddm4. 1) There may be differential efficiency of depletion of ART2+ regulatory T-cells in the peripheral lymphoid compartment of homozygotes versus heterozygotes. In at least one strain combination (BBDP × LEW.B6), ART2.1 and ART2.2 are expressed differentially on T-cells (30). In the heterozygous rats, not only was there more ART2.2 on cells expressing both it and ART2.1, but there also was noted a small population of T-cells that expressed only ART2.2 and would therefore not be depleted by the DS4.23 mAb used in these studies (30). We have confirmed this phenotype in a small sample of (BBDR × WF)F1 rats (J.P.M., unpublished observations). 2) A second possibility is that ART2.1+ and ART2.1+/ART2.2+ cells in the intestinal lymphoid compartment might be differentially affected by antibody treatment (39). ART2+ cells in the gut include a population of NK cells that could be important modulators of autoimmunity (39). 3) It is also possible that the effect on penetrance is due to genes within the ART2 interval on chromosome 1. The present study, designed solely to map iddm4, did not generate sufficient data to distinguish among these alternatives, the analysis of which will become part of future studies.
We also analyzed insulitis, the pathological precursor and substrate of autoimmune diabetes. In animals that were treated but not diabetic at the conclusion of the protocol, the frequency of insulitis was greater in WF.iddm4d rats than in controls. In addition, the quantitative trait locus controlling the intensity of insulitis was found within the same segment of RNO4 as susceptibility to diabetes. These data suggest that the iddm4 gene plays a role during the early stages of autoimmune diabetes pathogenesis in the rat.
We did observe that a small fraction of WF.iddm4 congenic rats homozygous for the WF origin allele of iddm4 became hyperglycemic after treatment to induce diabetes (8 of 101, considering both the N5F1 and congenic progeny testing analyses). This observation is consistent with previous reports that some degree of susceptibility to autoimmune diabetes is common among rats that express the class II RT1B/Du MHC haplotype (16). With respect specifically to the WF rat, Ellerman and Like (16) reported that 7% of these animals treated for 3 weeks with poly I:C alone could be induced to become diabetic. They also reported that spleen cells from treated WF rats were capable of the adoptive transfer of autoimmune diabetes. The simplest interpretation of these observations, given the common ancestry of the WF and BBDR rats, is that a mutation in the iddm4 gene has amplified the susceptibility to autoimmune diabetes that is inherent in the ancestral Wistar rat. These data can also be interpreted to suggest that iddm4w is a recessive non-MHC resistance gene for autoimmunity. Because diabetes in all of these animals was induced using poly I:C, it is interesting to speculate that differences in the metabolism of poly I:C could determine diabetes susceptibility. A corollary possibility is that the differences in the response to poly I:C (e.g., IL-18, IL-12, and interferons) could influence diabetes susceptibility. We have previously reported that rat strains differ in the amount of interferon produced in response to poly I:C (40), but that report did not measure responses in either BBDR or WF rats. There is no independently measured response to poly I:C in the present genetic analysis, but it remains our intention to test this and other immunophenotypes when the WF.iddm4 rat is fully congenic.
The region of rat chromosome 4 containing the iddm4 interval contains a number of interesting potential candidate genes. Among these are the T-cell receptor β-chain genes, the trypsin multigene family, ephrin B6, caspase 2, and others. None of these candidates has been formally excluded. As the sequence of the rat genome becomes available and bioinformatics tools become more sophisticated, the prospects for identifying iddm4 would appear to be good.
Our initial analyses of the iddm4 locus suggest that it is not syntenic with any of the known human iddm or mouse idd diabetes susceptibility loci. It is important to point out, however, that this does not imply that iddm4 is rat specific. To identify a locus as conferring susceptibility, it must be associated with a difference that segregates, and such unrecognized nonsegregating loci could be present in either humans or NOD mice.
Although our data clearly demonstrate that iddm4 is a major determinant of autoimmune diabetes in the RT1u rat, the data are also indicative of the action of additional disease-modifying genes. We show here that iddm4 is incompletely penetrant and that its penetrance is modified by ART2 genotype. In several reports, the presence of a diabetes resistance gene has been inferred from crosses between diabetic BBDP and resistant, nonlymphopenic non-BB rats. In two studies, the RT1u and lyp genes were placed on either the ACI (41) or the F344 (42) strain background, and these animals were found to be resistant to the development of diabetes. A third study reported results from F2 offspring of (DP × F344)F1 hybrids. The authors found six F2 rats homozygous for both RT1u and lyp (19). Because none of these became diabetic, they proposed that there was a third, diabetes-modifying locus that conferred resistance to diabetes. They designated this locus iddm3. Proof of the existence of iddm3 was sought by Klöting et al. (43) who, using the BB/OK variant of the BB rat, discovered a candidate location for iddm3 on chromosome 18. Using the BB/OK rat and crosses to several strains, including DA and SHR, Klöting and colleagues have also reported several other disease-modifying loci in the BB/OK rat on chromosomes 1, 6, 18, 20, and X (44-48). BB/OK rats exhibit several features not observed in the more common BB rats of Worcester origin (4), and the relationship of these loci to those observed in the BBDR rat of Worcester origin is uncertain.
With respect to the likelihood that multiple genes are responsible for diabetes susceptibility in the BB rat, it is important to point out that the frequency of diabetes in our various “susceptible” subcongenic lines expressing D4Arb9d varied from 30 to 70%. These data are consistent with the possibility that one or more additional genes important for the expression of autoimmunity may be present in the original 40-cM iddm4 interval. As noted previously (22), a number of genes important for autoimmunity in the rat map to this region of chromosome 4. These include loci associated with susceptibility to experimental autoimmune uveitis, adjuvant arthritis, and collagen-induced arthritis. Although our data are consistent with the possibility of a second gene, the sample size lacks the power to formally prove or disprove the hypothesis.
Many studies suggest that autoimmune diabetes in the BB rat is T-cell–dependent and that cells capable of diabetes induction are present in the thymus. This was most clearly demonstrated in studies showing that both unmanipulated and cultured BB rat thymocytes can transfer disease to adoptive recipients (33) and that transfer can be prevented by coculture in the presence of islet tissue (38). We hypothesized, accordingly, that the diabetogenic action of iddm4d could be detected in populations of thymocytes. To test this hypothesis, we used an adoptive transfer protocol and showed that thymocytes from susceptible WF.iddm4d animals were capable of the adoptive transfer of diabetes at high efficiency to athymic histocompatible WAG nude recipients. Surprisingly, however, thymocytes from WF.iddm4w animals also transferred the disease, and the effect of iddm4 appeared to be related to the kinetics of diabetes onset. Given the multigenic nature of diabetes in the rat and the latent susceptibility of the WF and other RT1u rat strains to diabetes, we interpret our transfer data to suggest that iddm4d is expressed at the level of the thymus and in some way facilitates the action of other susceptibility genes that are otherwise masked in the context of a normal regulatory immune environment in vivo.
In conclusion, we propose that iddm4 on chromosome 4 is a critical determinant of susceptibility to the induction of diabetes in the rat after environmental perturbation of the immune system. With respect to its ability to predict disease, iddm4 is 79% sensitive and 80% specific. These characteristics suggest that iddm4 is a major determinant of susceptibility to autoimmune diabetes in the rat. As additional markers become available, it should be possible to narrow further the iddm4 interval. Identification of iddm4 and elucidation of the disease-determining pathways that it affects may eventually provide useful information for the understanding of human type 1A diabetes.
ACKNOWLEDGMENTS
Supported in part by grants DK49106 (to J.P.M., D.L.G., and E.P.B.), DK 36024 (to D.L.G.), DK25306 (to J.P.M.), and Center Grant DK32520 from the National Institutes of Health. We thank Drs. Aldo Rossini and Eugene Handler for review of the manuscript.
The contents of this publication are solely the responsibility of the authors and do not necessarily represent the official views of the National Institutes of Health.
Glossary
- CSGE
conformation-sensitive gel electrophoresis
- EST
expressed sequence tag
- mAb
monoclonal antibody
- MHC
major histocompatibility complex
- poly I:C
polyinosinic:polycytidylic acid
- VAF
viral antibody free
REFERENCES
- 1.Pietropaolo M, Trucco M. Major histocompatibility locus and other genes that determine the risk of development of type 1 diabetes mellitus. In: LeRoith D, Taylor SI, Olefsky JM, editors. Diabetes Mellitus: A Fundamental and Clinical Text. Lippincott Williams & Wilkins; Philadelphia: 2000. pp. 399–410. [Google Scholar]
- 2.Dahlman I, Eaves IA, Kosoy R, Morrison VA, Heward J, Gough SCL, Allahabadia A, Franklyn JA, Tuomilehto J, Tuomilehto-Wolf E, Cucca F, Giya C, Ionescu-Tirgoviste C, Stevens H, Carr P, Nutland S, McKinney P, Shield JP, Wang W, Cordell HJ, Walker N, Todd JA, Concarmon P. Parameters for reliable results in genetic association studies in common disease. Nat Genet. 2002;30:149–150. doi: 10.1038/ng825. [DOI] [PubMed] [Google Scholar]
- 3.Leiter E. Genetics and immunogenetics of NOD mice and related strains. In: Leiter E, Atkinson M, editors. NOD Mice and Related Strains: Research Applications in Diabetes, AIDS, Cancer, and Other Diseases. R.G. Landes; Austin, TX: 1998. pp. 37–69. [Google Scholar]
- 4.Mordes JP, BorteZI K, Groen H, Gubersld DL, Rossini AA, Greiner DL. Autoimmune diabetes mellitus in the BB rat. In: Sima AAF, Shafrir E, editors. Animal Models of Diabetes: A primer. Harwood Academic; Amsterdam: 2001. pp. 1–41. [Google Scholar]
- 5.Crisé L, Mordes JP, Rossini AA. Autoimmune diabetes mellitus in the BB rat (Review Article) Diabetes Metab Rev. 1992;8:4–37. [PubMed] [Google Scholar]
- 6.Mordes JP, Bortell R, Doukas J, Rigby MR, Whalen BJ, Zipris D, Greiner DL, Rossini AA. The BB/Wor rat and the balance hypothesis of autoimmunity. Diabetes Metab Rev. 1996;2:103–109. doi: 10.1002/(SICI)1099-0895(199607)12:2<103::AID-DMR161>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 7.Gubersld DL. Diabetes-prone and diabetes-resistant BB rate: animal models of spontaneous and virally induced diabetes mellitus, lymphocytic thyroiditis, and collagen-induced arthritis. ILAR News. 1994;35:29–37. [Google Scholar]
- 8.Knip M, Akerblom HK. Environmental factors in the pathogenesis of type 1 diabetes mellitus. Exp Clin Endocrinol Diabetes. 1999;107:S93–S100. doi: 10.1055/s-0029-1212160. [DOI] [PubMed] [Google Scholar]
- 9.Todd JA, Stemman L. Autoimmunity: the environment strikes back. Curr Opinion Immunol. 1993;5:863–865. doi: 10.1016/0952-7915(93)90097-c. [DOI] [PubMed] [Google Scholar]
- 10.Colle E, Guttmann RD, Seemayer TA. Spontaneous diabetes mellitus in the rat. I. Association with the major histocompatibility complex. J Exp Med. 1981;154:1237–1242. doi: 10.1084/jem.154.4.1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Colle E, Guttmann RD, Fuks A. Insulin-dependent diabetes mellitus is associated with genes that map to the right of the class 1 RT1.A locus of the major histocompatibility complex of the rat. Diabetes. 1986;35:454–458. doi: 10.2337/diab.35.4.454. [DOI] [PubMed] [Google Scholar]
- 12.Awata T, Guberski DL, Like AA. Genetics of the BB rat: Association of autoimmune disorders (diabetes, insulitis, and thyroiditis) with lymphopenia and major histocompatibility complex class II. Endocrinology. 1995;136:5731–5735. doi: 10.1210/endo.136.12.7588330. [DOI] [PubMed] [Google Scholar]
- 13.Colle E, Guttmann RD, Fuks A, Seemayer TA, Prud’homme GJ. Genetics of the spontaneous diabetic syndrome: interaction of MHC and non-MHC-associated factors. Mol Biol Med. 1986;3:13–23. [PubMed] [Google Scholar]
- 14.Colle E. Genetic susceptibility to the development of spontaneous insulin-dependent diabetes mellitus in the rat. Clin Immunol Immunopathol. 1990;57:1–9. doi: 10.1016/0090-1229(90)90017-k. [DOI] [PubMed] [Google Scholar]
- 15.Fuks A, Ono SJ, Colle E, Guttmann RD. A single dose of the MHC-linked susceptibility determinant associated with the RT1u haplotype is permissive of insulin-dependent diabetes mellitus in the BB rat. Expl Clin Immunogenet. 1990;7:162–169. [PubMed] [Google Scholar]
- 16.Ellerman KE, Like AA. Susceptibility to diabetes is widely distributed in normal class IIu haplotype rats. Diabetologia. 2000;43:890–898. doi: 10.1007/s001250051466. [DOI] [PubMed] [Google Scholar]
- 17.Guberski DL, Butler L, Kastem W, Like AA. Genetic studies in inbred BB/Wor rats: analysis of progeny produced by crossing lymphopenic diabetes-prone rats with nonlymphopenic diabetic rats. Diabetes. 1989;38:887–893. doi: 10.2337/diab.38.7.887. [DOI] [PubMed] [Google Scholar]
- 18.Markholst H, Eastman S, Wilson D, Andreasen BE, Lemmark Å . Diabetes segregates as a single locus in crosses between inbred BB rats prone or resistant to diabetes. J Exp Med. 1991;174:297–300. doi: 10.1084/jem.174.1.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jacob HJ, Pettersson A, Wilson D, Mao Y, Lemmark A, Lander ES. Genetic dissection of autoimmune type I diabetes in the BB rat. Nat Genet. 1992;2:56–60. doi: 10.1038/ng0992-56. [DOI] [PubMed] [Google Scholar]
- 20.Homum L, Markholst H. Genetic map of the lymphopenia region on rat chromosome 4. Transplant Proc. 1999;31:1615–1617. doi: 10.1016/s0041-1345(99)00058-5. [DOI] [PubMed] [Google Scholar]
- 21.Fuks A, Colle E, Ono S, Prud’homme GJ, Seemayer T, Guttmann RD. Immunogenetic studies of insulin-dependent diabetes in the BB rat. In: Shafrir E, Renold AE, editors. Frontiers in Diabetes Research: Lessons From Animal Diabetes II. John Libbey; London: 1988. pp. 29–33. [Google Scholar]
- 22.Martin A-M, Maxson MN, Leif J, Mordes JP, Greiner DL, Blankenhom EP. Diabetes-prone and diabetes-resistant BB rats share a common major diabetes susceptibility locus, iddm4: additional evidence for a “universal autoimmunity locus” on rat chromosome 4. Diabetes. 1999;48:2138–2144. doi: 10.2337/diabetes.48.11.2138. [DOI] [PubMed] [Google Scholar]
- 23.Homum L, Rømer J, Markholst H. The diabetes-prone BB rat carries a frameshift mutation in Ian4, a positional candidate of iddml. Diabetes. 2002;51:1972–1979. doi: 10.2337/diabetes.51.6.1972. [DOI] [PubMed] [Google Scholar]
- 24.Doukas J, Mordes JP, Swymer C, Niedzwiecki D, Mason R, Rozing J, Rossini AA, Greiner DL. Thymic epithelial defects and predisposition to autoimmune disease in BB rats. Am J Pathol. 1994;145:1517–1525. [PMC free article] [PubMed] [Google Scholar]
- 25.Wakeland E, Morel L, Achey K, Yui M, Longmate J. Speed congenics: a classic technique in the fast lane (relatively speaking) Immunol Today. 1997;18:472–477. doi: 10.1016/s0167-5699(97)01126-2. [DOI] [PubMed] [Google Scholar]
- 26.Martin A-M, Blankenhom EP, Maxson MN, Zhao M, Leif J, Mordes JP, Greiner DL. Non-major histocompatibility complex-linked diabetes susceptibility loci on chromosomes 4 and 13 in a backcross of the DP BB/Wor rat to the WF rat. Diabetes. 1999;48:50–58. doi: 10.2337/diabetes.48.1.50. [DOI] [PubMed] [Google Scholar]
- 27.Bortell R, Kanaitsuka T, Stevens LA, Moss J, Mordes JP, Rossini AA, Greiner DL. The RT6 (Art2) family of ADP-ribosyltransferases in rat and mouse. Mol Cell Biochem. 1999;193:61–68. [PubMed] [Google Scholar]
- 28.Angelillo M, Greiner DL, Mordes JP, Handler ES, Nakamura N, McKeever U, Rossini AA. Absence of RT6+ T cells in diabetes-prone BioBreeding/Worcester rats is due to genetic and cell developmental defects. J Immunol. 1988;141:4146–4151. [PubMed] [Google Scholar]
- 29.Koch F, Kashan A, Thiele H-G. The rat T-cell differentiation marker RT6.1 is more polymorphic than its alloantigenic counterpart RT6.2. Immunology. 1988;65:259–265. [PMC free article] [PubMed] [Google Scholar]
- 30.Thiele H-G, Haag F, Nolte F. Asymmetric expression of RT6.1 and RT6.2 alloantigens in (RT6a × RT6b)F1 rats is due to a pretranslational mechanism. Transplant Proc. 1993;25:2786–2788. [PubMed] [Google Scholar]
- 31.Korkko J, Annunen S, Pihlajamaa T, Prockop DJ, Ala-Kokko L. Conformation sensitive gel electrophoresis for simple and accurate detection of mutations: comparison with denaturing gradient gel electrophoresis and nucleotide sequencing. Proc Natl Acad Sci USA. 1998;95:1681–1685. doi: 10.1073/pnas.95.4.1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thomas VA, Woda BA, Handler ES, Greiner DL, Mordes JP, Rossini AA. Altered expression of diabetes in BB/Wor rats by exposure to viral pathogens. Diabetes. 1991;40:255–258. doi: 10.2337/diab.40.2.255. [DOI] [PubMed] [Google Scholar]
- 33.Whalen BJ, Rossini AA, Mordes JP, Greiner DL. DR-BB rat thymus contains thymocyte populations predisposed to autoreactivity. Diabetes. 1995;44:963–967. doi: 10.2337/diab.44.8.963. [DOI] [PubMed] [Google Scholar]
- 34.Whalen BJ, Doukas J, Mordes JP, Rossini AA, Greiner DL. Induction of insulin-dependent diabetes mellitus in PVG.RTP1u rats. Transplant Proc. 1997;29:1684–1685. doi: 10.1016/s0041-1345(97)00015-8. [DOI] [PubMed] [Google Scholar]
- 35.Kaplan EL, Meier P. Nonparametric estimation from incomplete observations. J Am Statist Assn. 1958;53:457–481. [Google Scholar]
- 36.Nie NH, Hull CH, Jenkins JG, Steinbrenner K, Bent DH. Statistical Package for the Social Sciences. McGraw-Hill; New York: 1975. pp. 1–675. [Google Scholar]
- 37.Siegel S. Nonparametric Statistics. McGraw-Hill; New York: 1956. pp. 1–239. [Google Scholar]
- 38.Whalen BJ, Marounek J, Weiser P, Appel MC, Greiner DL, Mordes JP, Rossini AA. BB rat thymocytes cultured in the presence of islets lose their ability to transfer autoimmune diabetes. Diabetes. 2001;50:972–979. doi: 10.2337/diabetes.50.5.972. [DOI] [PubMed] [Google Scholar]
- 39.Todd DJ, Greiner DL, Rossini AA, Mordes JP, Bortell R. An atypical population of natural killer cells that spontaneously secrete IFN-γ and IL-4 is present in the intraepithelial lymphoid compartment of the rat. J Immunol. 2001;167:3600–3609. doi: 10.4049/jimmunol.167.7.3600. [DOI] [PubMed] [Google Scholar]
- 40.Davis CT, Blankenhom EP, Murasko DM. Genetic variation in the ability of several strains of rats to produce interferon in response to polyriboino-sinic-polyribocytodilic acid. Infect Immun. 1984;43:580–583. doi: 10.1128/iai.43.2.580-583.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Colle E, Fuks A, Poussier P, Edouard P, Guttmann RD. Polygenic nature of spontaneous diabetes in the rat: permissive MHC haplotype and presence of the lymphopenic trait of the BB rat are not sufficient to produce susceptibility. Diabetes. 1992;41:1617–1623. doi: 10.2337/diab.41.12.1617. [DOI] [PubMed] [Google Scholar]
- 42.Homum L, Lundsgaard D, Markholst H. An F344 rat congenic for BB/DP rat-derived diabetes susceptibility loci Iddm1 and Iddm2. Mamm Genome. 2001;12:867–868. doi: 10.1007/s00335-001-2065-3. [DOI] [PubMed] [Google Scholar]
- 43.Klöting I, Vogt L, Serikawa T. Locus on chromosome 18 cosegregates with diabetes in the BB/OK rat subline. Diabète Metab. 1995;21:338–344. [PubMed] [Google Scholar]
- 44.Klöting I, Van den Brandt J, Kuttler B. Genes of SHR rats protect spontaneously diabetic BB/OK rats from diabetes: lessons from congenic BB.SHR rat strains. Biochem Biophys Res Commun. 2001;283:399–405. doi: 10.1006/bbrc.2001.4798. [DOI] [PubMed] [Google Scholar]
- 45.Van den Brandt J, Kovécs P, Klöting I. Congenic diabetes-prone BB.Sa and BB.Xs rats differ from their progenitor strain BB/OK in frequency and severity of insulin-dependent diabetes mellitus. Biochem Biophys Res Commun. 1999;263:843–847. doi: 10.1006/bbrc.1999.1456. [DOI] [PubMed] [Google Scholar]
- 46.Klöting I, Kovacs P. Genes of the immune system cosegregate with the age at onset of diabetes in the BB/OK rat. Biochem Biophys Res Commun. 1998;242:461–463. doi: 10.1006/bbrc.1997.7985. [DOI] [PubMed] [Google Scholar]
- 47.Klöting I, Schmidt S, Kovacs P. Mapping of novel genes predisposing or protecting diabetes development in the BB/OK rat. Biochem Biophys Res Commun. 1998;245:483–486. doi: 10.1006/bbrc.1998.8464. [DOI] [PubMed] [Google Scholar]
- 48.Klöting I, Kovacs P. Phenotypic differences between diabetes-prone BB rat sublines cosegregate with loci on chromosomes X and 10. Biochem Mol Biol Int. 1998;45:865–870. doi: 10.1002/iub.7510450503. [DOI] [PubMed] [Google Scholar]