Abstract
Recent research suggests that in addition to their role as soluble electron carriers, pyridine nucleotides [NAD(P)(H)] also regulate ion transport mechanisms. This mode of regulation seems to have been conserved through evolution. Several bacterial ion–transporting proteins or their auxiliary subunits possess nucleotide-binding domains. In eukaryotes, the Kv1 and Kv4 channels interact with pyridine nucleotide–binding β-subunits that belong to the aldo-keto reductase superfamily. Binding of NADP+ to Kvβ removes N-type inactivation of Kv currents, whereas NADPH stabilizes channel inactivation. Pyridine nucleotides also regulate Slo channels by interacting with their cytosolic regulator of potassium conductance domains that show high sequence homology to the bacterial TrkA family of K+ transporters. These nucleotides also have been shown to modify the activity of the plasma membrane KATP channels, the cystic fibrosis transmembrane conductance regulator, the transient receptor potential M2 channel, and the intracellular ryanodine receptor calcium release channels. In addition, pyridine nucleotides also modulate the voltage-gated sodium channel by supporting the activity of its ancillary subunit—the glycerol-3-phosphate dehydrogenase-like protein. Moreover, the NADP+ metabolite, NAADP+, regulates intracellular calcium homeostasis via the 2-pore channel, ryanodine receptor, or transient receptor potential M2 channels. Regulation of ion channels by pyridine nucleotides may be required for integrating cell ion transport to energetics and for sensing oxygen levels or metabolite availability. This mechanism also may be an important component of hypoxic pulmonary vasoconstriction, memory, and circadian rhythms, and disruption of this regulatory axis may be linked to dysregulation of calcium homeostasis and cardiac arrhythmias.
Keywords: KATP channels, NAADP+, potassium channels, ryanodine receptor calcium release channel, Slo channels, TRPM2 channels, TPC channels
The capacity to perform controlled oxidation–reduction reactions is one of the distinguishing features of life. All living organisms use such reactions to trap or liberate energy or to synthesize essential cell constituents and metabolites. This is accomplished by specialized biomolecules that package and shuttle energy between different processes. Reactions that need energy are fueled by ATP, which is the universal energy currency of cells, but to generate ATP energy must be extracted from nutrients by a series of coupled catabolic reactions. This process requires specialized electron carriers that can deliver energy from nutrients to the mitochondrial electron-transport chain. Electrons derived from nutrients also are needed for the biosynthesis of new material and for the removal of toxins and reactive metabolites. This shuttling of electrons between different reactions is accomplished by soluble electron carriers—the pyridine nucleotide coenzymes.
Both prokaryotic and eukaryotic cells synthesize 2 sets of specialized pyridine nucleotides (NAD+/NADH and NADP+/NADPH). The structure of these nucleotides is optimized for ready exchange of electrons and for recognition by specialized nucleotide-binding domains (NBDs) located at the active sites of >200 different oxidoreductases. The business end of these nucleotides is a nitrogenous, nicotinamide ring that donates hydride ions. This transfer of electrons is accompanied by a large negative free energy change that leads to a more stable arrangement of electrons. The ring is also optimized for accepting electrons during exergonic oxidation reactions and is linked to adenosine via a pyrophosphate bridge (Figure 1). The unique structure of pyridine nucleotides imparts specificity of function. The NBD of most oxidoreductases interacts with both the nicotinamide and the adenosyl moieties of pyridine nucleotides and, therefore, these enzymes can discriminate between pyridine nucleotides and their precursors or metabolites. Moreover, these enzymes also can distinguish between reduced and oxidized states of the nucleotide and by specifically recognizing the phosphorylation state of the ribose ring, between NAD+ and NADP+.
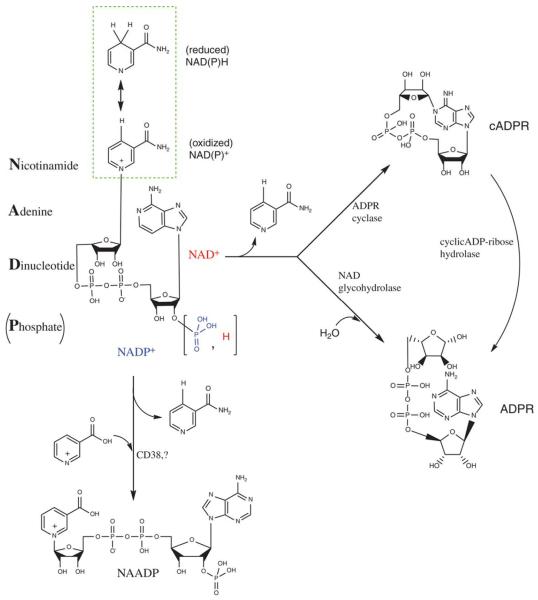
Figure 1. Pyridine nucleotides and their metabolites.
Chemical structure of pyridine nucleotides and their metabolites. NAD+ and NADP+ have different substituents at 2′ position of the ribose ring. The highlighted inset shows the pyridine ring in its oxidized and reduced forms. cADPR indicates cyclic adenosine diphosphate ribose.
Specific interactions between pyridine nucleotides and oxidoreductases support a diverse range of metabolic reactions. NADP(H) is recognized by enzymes that are involved primarily in anabolic (reductive) reactions, such as lipid or cholesterol synthesis or fatty acid chain elongation, whereas NAD(H) is recognized by enzymes that catalyze catabolic reactions of glycolysis and by components of the electron-transport chain. This differential recognition of NADP(H) and NAD(H) enables the cell to maintain the supply of electrons for 2 contrasting purposes, independent of each other. A high NAD+:NADH ratio is maintained to readily accept electrons generated by catabolic reactions, whereas the low NADP+:NADPH ratio reflects a state of readiness to donate electrons to biosynthetic reactions or antioxidant defense. As a result, the availability and the redox state of pyridine nucleotides regulate the activity of the processes involved in intermediary metabolism,1 biosynthesis, antioxidant defense, and xenobiotic detoxification. Moreover, because the pyridine nucleotide couples have one of the lowest standard reduction potentials in the cell (−0.315 V for NAD[H] and −0.320 V for NADP[H]), their redox poise (the NAD[P]H:NAD[P] ratio) dictates the overall redox state of the cell.
Although the principal role of pyridine coenzymes is to donate or accept electrons without undergoing any other transformation, these nucleotides also are used by some enzymes as substrates and are converted to reactive metabolites that regulate cell signaling. For instance, NAD+ is used by poly-ADP ribose polymerases to construct polymers of ADP ribose to form scaffolding that recruits DNA repair enzymes for repairing single-strand DNA breaks. NAD+ also is consumed during lysine deacetylation of proteins by sirtuins. Some mitochondrial and nuclear sirtuins use NAD+ to support their ADP-ribosyl transferase activity. In addition, pyridine nucleotides are metabolically converted to specialized signaling molecules, such as NAADP+, ADP-ribose, and cyclic adenosine diphosphate ribose (cADPR) (Figure 1). The use of NAD+ by these reactions affects a wide array of signaling and gene-transcription events (see other review articles in this series).2,3 Thus, acting as essential cofactors in redox reactions, or substrates in ribosyl transfer reactions or precursors of signaling molecules, pyridine nucleotides directly or indirectly regulate a large network of metabolic reactions and link the activity of several enzymes to the redox status of the cell. Moreover, in addition to these regulatory functions, recent work has shown that pyridine nucleotides also regulate the activity of ion channels, by acting either as ligands or as substrates of accessory subunits that modify channel gating. Further understanding how pyridine nucleotides regulate ion channels therefore might shed new light on the mechanisms by which cells respond to environmental variations in oxygen and nutrient availability and those underlying the metabolic regulation of ion transport, excitability, apoptosis, and osmoregulation.
In this article, we review evidence implicating pyridine nucleotides and their metabolites as direct or indirect regulators of ion channels. Several ion channels, such as the voltage-gated potassium and sodium channels, have been shown to be regulated by pyridine nucleotides. We discuss each of these channels individually. Although in some cases direct evidence is lacking, we have included speculative discussion on these channels to stimulate future research in this area. To underscore the importance of this relationship, we highlight the evolutionary conservation of this link from bacterium to human. Regulation of bacterial transporters represents the most direct link between pyridine nucleotides and ion transport, and the simple regulatory modes seen in bacteria recur in eukaryotic systems in more elaborate forms. In addition, the structural bases and the biophysical mechanisms of this regulation in bacteria seem to be prototypes that have been conserved during evolution or have been developed to regulate ion channels by other ligands, such as calcium or ATP. Moreover, recognition of the similarities between bacterial and eukaryotic modes of regulation might help in investigating conserved functional mechanisms.
Regulation of Potassium Transport by Pyridine Nucleotides
Bacterial Potassium Transporters
It is currently believed that life originated in an aqueous environment in which negatively charged biomolecules, such as proteins and nucleic acids, were trapped in a semipermeable membrane. The high osmotic pressure exerted by charged biomolecules was counterbalanced by a high concentration of positively charged K+ ions within the membrane-delimited cell. This intracellular accumulation of K+ and exclusion of the more abundant sea water cation, Na+ was probably used to energize the cell membrane. As a result, all living cells tightly regulate K+ transport and use K+ as the major solute to control osmolarity. The regulation of K+ transport is critically important not only for survival and growth but also for maintaining cytosolic pH and for transmitting information from the extracellular to the intracellular environment. Although it is unknown how archaic cells regulated K+ transport, modern bacteria, such as Escherichia coli, maintain separate systems for K+ uptake and efflux. Transport systems, such as Trk, Ktr, and T2M channels, mediate active uptake of K+ ions, whereas K+ efflux is effected by the Kef system. Remarkably, all 4 of these transport systems possess a nucleotide-binding potassium transport nucleotide–binding domain (KTNBD)4,5 (Figure 2). In uptake systems, this domain is in the cytosolic subunits (TrKA or KtrA) that assemble with the membrane-spanning subunits (TrkG/H or KtrB/D), whereas in systems mediating K+ efflux (KefC/B), the KTNBD is covalently linked to the cytosolic C-terminus of the ion transporter. In both instances, the cytosolic location of the NBD suggests a sensing mechanism in which ligand binding to the intracellular domains of the transporter could alter K+ flux through the ion-conducting pore. This possibility is reinforced by the invariant proximity of the KTNBDs to the base of the innermost pore-forming helices,4 suggesting that conformational changes in the KTNBD could readily alter the ion transport properties of the pore. The KTNBDs of bacteria form a well-conserved Rossmann fold, which is a stable NAD+-binding motif composed of 6 parallel β strands linked to 2 pairs of α-helices. This fold is commonly found in several bacterial and eukaryotic dehydrogenases. The Rossmann fold of KtrA binds to both NAD+ and NADH. Binding of these ligands is essential for the maintenance of the tetrameric state of KTNBD and ligand-mediated changes. This could impart conformational changes in the ion-transporting subunit to alter its conducting properties,4 although this has not been directly demonstrated.
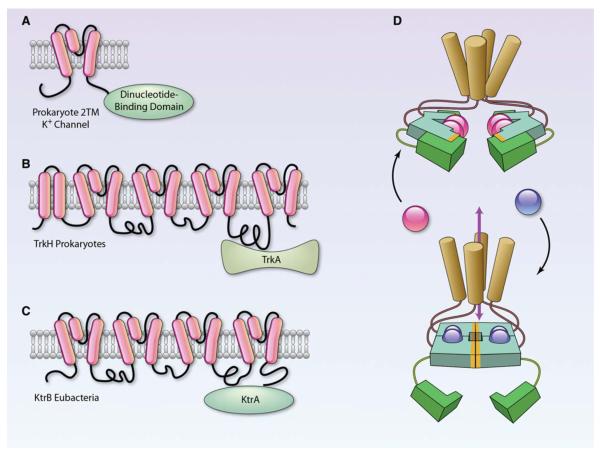
Figure 2. Association of pyridine nucleotide-binding proteins with bacterial potassium transporters.
A, The ancestral 2-transmembrane-type of prokaryotic K+ channel contains a dinucleotide-binding domain covalently attached to the C-terminus of the channel, whereas both the TrkH (B) and the KtrB (C) transporter associate with cytosolic proteins (TrkA and KtrA) that bind nucleotides. TrkA contains 2 nucleotide-binding domains. Adapted with permission from Biophys J, Durrell et al.5D, Model of channel gating in which alternative potassium transport nucleotide–binding domain (KTNBD) conformations can impart an asymmetrical twist on the inner pore-lining helices of the channel, which could affect ion passage. KTNBD is colored with hydrophobic interaction surfaces highlighted in orange. These conformational changes can be either ligand-mediated (blue and red balls represent small cytoplasmic molecules, such as NADH and NAD+) or stabilized by the binding or the release of auxillary protein subunits (green). Adapted with permission from Cell, Roosild et al4 (Illustration: Ben Smith).
Like KtrA, the K+ uptake proteins TrkG/H also interact with subunits (TrkA) containing the KTNBD. The TrkA subunit of TrkG/H has 2 distinct dinucleotide-binding sites in each of 2 similar subdomains and, in addition to TrkG/H, TrkA also interacts with several other proteins, such as TrkE, to form functional channels. Each half of the protein sequence of TrkA aligns with NAD+-dependent dehydrogenases, such as lactate, malate, and alanine dehydrogenase, and purified TrkA binds NAD+ and NADH with much higher affinity than ATP.6 Because TrkA lacks the C-terminal catalytic domain of dehydrogenases, it is considered unlikely that the protein has enzyme activity. Nevertheless, it has been proposed that binding of NAD(H) to TrkA regulates the transport activity of the TrkG and the TrkH systems,4 but the functional regulation of Trk transporters by NAD(H) has not been directly demonstrated to date. However, the presence of a pyridine NBD in TrkA suggests potential coupling between energy expensive import of K+ ions and active metabolism. This coupling might be particularly important during cell growth. A high NADH:NAD+ ratio is a prerequisite for cell growth, and activation of K+ import by pyridine nucleotides may be required to maintain cytoplasmic K+ levels and turgor pressure during cell expansion.4
In contrast to the K+ uptake systems, which associate with nucleotide-binding proteins, the K+-efflux system, KefC, has a KTNBD that is covalently linked to its ion-transporting subunits. The KefC transport system is inactivated by reduced glutathione, and it is activated by glutathione-S-conjugates.7 Activation of this transport by glutathione conjugates leads to acidification of the cytosol. Because thiol reactivity is decreased at low pH, this might be a strategy for preventing the modification of cytosolic protein thiols, and thereby for protecting the bacteria from electrophilic stress. The C-terminal KTNBD of KefC is similar in structure to the Rossmann fold of dihydrofolate reductase and it binds glutathione. Inhibition of KefC by glutathione is enhanced by NADH, but not NAD+, indicating that NADH:NAD+ ratio could regulate the antiporter activity of KefC.8
In addition to glutathione and NADH, the KTNBD of KefC also binds to the auxiliary subunits, KefF and KefG, which are required for the full activation of KefB/C.9 The primary sequences of KefC and KefG show striking resemblance to human quinone reductases and in the crystal structure of KefF, FMN is bound to the KTNBD of the protein10 (Figure 2). Recently, it has been shown that the KefF is a bonafide oxidoreductase in which NADH and NADPH act as electron donors and quinones and ferricyanide act as acceptors.11 Although enzyme activity was not found to be required for KefC activation, it was suggested that by catalyzing the reduction of quinones, KefF protects KefC from the toxicity of electrophilic quinones.11
As discussed, the link between K+ channels and nucleotide-binding proteins in bacteria is conserved in eukaryotic K+ channels. Like the bacteria efflux systems (Kef), some of the eukaryotic channels, such as the Slo channels, possess a nucleotide-binding site in their cytosolic domain, whereas others, such as Kv channels, associate with auxiliary subunits that bind pyridine nucleotides in a manner reminiscent of the bacterial K+ uptake systems (Trk/Ktr). Moreover, like bacterial channels, the mammalian K+ channels also are regulated by pyridine nucleotides. It is likely that this mode of regulation is conserved during evolution because it plays a critical, nonredundant role in linking K+ transport to the metabolic state of the cell, thereby enabling the cell to sense and respond to changes in the external environment.
Mammalian Channels
Slo K+ Channels
The Slo family comprises high-to-intermediate conductance channels with a C-terminal domain that bears close resem blance to TrkA and other NAD+-binding prokaryotic K+ transporters.12 These channels are widely distributed across Linnaean borders and are expressed in many types of cells, including cardiac myocytes and smooth muscle cells. The 4 mammalian Slo genes, Slo1 (BKCa), Slo2.1 (Slick), Slo2.2 (Slack), and Slo3, encode proteins that form K+-selective homotetrameric channels.13 The core region of these channels resembles the canonical Kv channels, but their cytoplasmic domain shows unusually high structural diversity. Variations in the cytosolic domain enable these channels to respond to a wide range of intracellular ions and metabolites (Figure 3). The cytosolic region of Slo1 binds to calcium via the calcium bowl located at the distal end of its hydrophilic tail; therefore, these channels are sensitive to changes in both membrane potential and [Ca2+]. The Slo2.1 channel is regulated by Cl−, but it contains an ATP-binding domain as well. The Slo2.2 channel is insensitive to Cl− and it does not bind ATP. Nevertheless, both Slo2 channels respond to elevated [Na+]i levels, giving rise to the KNa current.15
Figure 3. Structure of the Slo channels and their regulation by pyridine nucleotides.
Organization of the structural subunits of Slo1 (A) and Slo 2 family of channels (B). Adapted with permission from Nat Rev Neurosci, Salkoff et al.13C, Inside-out patch-clamp recordings from pulmonary artery smooth muscle cells; horizontal bars represent application of 2 mmol/L NADH or NAD+. Current was recorded with a holding potential of +50 mV. Reprinted with permission from Exp Physiol, Park et al130; D, Inside-out patch-clamp recordings from rat dorsal root ganglion neurons in the absence and presence of 1 mmol/L NAD+ made from a holding potential of −80 mV. Adapted with permission from J Neurosci, Tamsett et al12 (Illustration: Ben Smith).
Initial work showed that the KNa channels are sensitive to sodium only at supraphysiological levels (50–80 mmol/L), making it doubtful whether they could be regulated by physiological changes in [Na+]i that usually vary between 5 and 15 mmol/L.16 Channel run-down after initial excision also was frequently observed. These discrepancies remained unresolved until Tamsett et al12 found that the cytoplasmic domain of both Slo2.1 and Slo2.2 contains NBDs similar to TrkA. This site includes a canonical NAD+-binding βαβαβ motif that was required for nucleotide binding.12 They also found that application of physiological levels of NAD+ to inside-out patches of rat dorsal root ganglion neurons led to a 2- to 2.5-fold increase in the open probability of KNa channels (Figure 3) and a decrease in EC of Na+ 50 from 50 to 20 mmol/L. The specificity of this interaction was reinforced by the finding that other cytosolic factors (cAMP, cGMP, and ATP) were without effect.17 NAD+, but not NADH, was effective in altering the gating properties of the channel, but only in the presence of Na+. Moreover, like native KNa channels, the current generated by Slack channels also was increased by NAD+ or NADP+. Site-directed mutations at the NAD+-binding site of the Slo2 channel abolished this response, suggesting that direct nucleotide binding to the cytosolic region of the channel is required for these channels to respond to changes in pyridine nucleotide levels.
The regulation of KNA/Slo2 channels by NAD(P)+ suggests that the activity of these channels may be coupled to the metabolic state of the cell. This mode of regulation may be particularly important during ischemia–reperfusion and other conditions in which accumulation of NAD(P)+ could increase K+ efflux via these channels. High levels of intracellular NAD(P)+ also would increase the sensitivity of these channels to intracellular sodium. Indeed, it has been suggested that in ischemic cardiac myocytes, elevated [Na+]i levels activate KNa, and an increase in this current shortens the action potential duration and induces calcium overload.18,19 Therefore, regulation by pyridine nucleotides would allow these channels to adapt simultaneously to both the metabolic and the ionic conditions prevalent in the ischemic heart. Interestingly, even though the evidence is indirect, it has been proposed that the Slo2 channels are present in cardiac mitochondria.20 If present, the regulation of these channels by pyridine nucleotides might represent conservation of the link between metabolism and ion transport in modern mitochondria and their prokaryotic ancestors.
The NAD+-binding site of Slo2 resembles the calcium binding site of the cytoplasmic domain of Slo1 channels, which contain 2 regulators of potassium conductance domains.13 The regulator of potassium conductance domain is similar to the KTNBD of bacterial channels4 found in 6-transmembrane K+ channels, except that in the KTNBD the nucleotide binding Rossmann fold is not conserved. The N-terminus of the regulator of potassium conductance domain of Slo1 forms a Rossmann fold, which is similar to that seen in the structure of the cytoplasmic region of Slo2.221; however, amino acid replacements in this domain during evolution have recruited the structure to bind calcium ions in case of Slo1 and sodium in case of Slo2.2. Although no direct pyridine nucleotide binding to the regulator of potassium conductance domain of Slo1 channels has been demonstrated, Lee et al22 have reported that application of 2 mmol/L NAD+ to the internal face of excised patches from small (<300 μm) pulmonary arteries reduces the open probability of BKCa channels, whereas NADH has the opposite effect (Figure 3). However, pyridine nucleotides had no effect on steady-state BKCa conductance in ear arterial smooth muscle cells22 or in large (>300 μm) intralobar pulmonary arteries,23 although in large arteries, application of NADH did lead to a voltage-dependent block of the channel.23 Although these observations are intriguing, further studies are required to understand how intracellular changes in pyridine nucleotides regulate the activity and the physiological role of Slo channels.
Voltage-Gated Potassium Channels
Like the bacterial KefC K+ transporters, several eukaryotic voltage-gated potassium (Kv) channels also associate with pyridine nucleotide–binding proteins. These channels play a fundamental role in many physiological processes. They control the membrane potential of excitable cells and they modulate the waveform and the frequency of the action potential. These channels also regulate neurotransmitter release, as well as cell volume,24,25 proliferation,26 and apoptosis,27 and they play a key role in T-cell differentiation, activation, and cytokine production.28 Also, the activity of these channels controls basal-stimulated and agonist-stimulated vasomotor tone, and membrane hyperpolarization caused by the activation of Kv channels regulates vasodilation.29 In small resistance arteries, oxygen-sensitive changes in Kv channel activity mediate hypoxic pulmonary vasoconstriction (HPV).30,31 Therefore, altered Kv channel activity is associated with cardiac arrhythmias,32 pulmonary hypertension,33 epilepsy, and abnormal immune responses.28
The diverse functions of Kv channels relate to their varied structure. The ion-conducting pore of Kv channels is formed by 4 membrane-spanning α-subunits in homotetrameric or heterotetrameric assembly. To date, 12 different Kv channel proteins have been described.24,34,35 The pore-forming subunits of several Kv families associate in situ with ancillary subunits that aid channel assembly and modify channel function24 (Figure 4), such as the proteins of the Kvβ family, which interact with the cytosolic domains of Kv1 and Kv4 channel proteins.37 In heterologous expression systems, cloned Kv1 α-subunits generate slowly inactivating (delayed rectifiers, eg, Kv1.1) or rapidly inactivating (A-type channels, eg, Kv1.4) outward currents. However, coexpression with Kvβ1 confers rapid inactivation to Kv1.1 currents38 and accelerates Kv1.4 inactivation. The inactivation of A-type Kv currents has been shown to be mediated by the N-terminal domain (ball-and-chain) of the α-subunits, which blocks the internal mouth of the channel on membrane depolarization (N-type inactivation). Inactivation of Kv1.4 is lost when the N-terminal ball of the channel is removed, but it could be restored by coexpressing Kvβ1, indicating that the Kvβ1 proteins can provide the inactivating ball in a manner analogous to the inactivating N-terminus of Kv1.4. Consistent with this model, Kvβ2, which has a shorter N-terminal domain, does not induce N-type inactivation in delayed rectifying channels,38 although when assembled with Kv1.5, Kvβ2 causes a −10-mV shift in the activation threshold and accelerates channel activation.39 Kvβ2 also has been shown to modestly accelerate inactivation of Kv1.4 currents.39 The Kvβ3, which possesses an inactivation ball with 90% sequence identity to that of Kvβ1.1, has been found to impart inactivation to slowly inactivating Kv1.5 channels.40
Figure 4. Model of physiological regulation of voltage-gated potassium (Kv) channel by pyridine nucleotides.
A, Composite representation of the Kvα-Kvβ complex. The membrane-spanning domains of Kvα are shown in blue. The T1 domain, which docks with Kvβ, is shown in light blue. The N-terminus of Kvβ forms the inactivating ball and chain assembly. In the NADPH-bound state of the Kvβ subunit, the N-terminal domain of Kvβ inactivates the channel by plugging the internal opening of the ion-permeation pathway, resulting in N-type inactivation (right). Binding of NADP+ to Kvβ prevents inactivation. For clarity, only 2 of the 4 subunits of Kvα and β are shown. B, Schematic showing the regulation of channel function by NADPH. The noninactivated (open) state of the channel is stabilized by NADP+, whereas NADPH binding induces inactivation. The transition between the inactivated and noninactivated state of the channel is mediated either by pyridine nucleotide exchange or by catalytic turnover involving the substrate-dependent conversion of NADPH to NADP+. C, Effect of NAD+ on single channel Kv activity in inside-out patches recorded in COS-7 cells expressing Kvα1.5 and Kvβ1.3 before and after exposure to 1 mmol/L NAD+. Currents recorded in response to a +50-mV depolarizing pulse. Reprinted with permission from Am J Physiol Cell Physiol, Tipparaju et al.42D, Stereoview of a ribbon representation showing the side view of the channel complex containing the transmembrane (TM) domain, the T1 scaffolding, and the auxiliary β-subunits. The NADP+ cofactor bound to each β-subunit is shown as indicated.36 (Illustration: Ben Smith.)
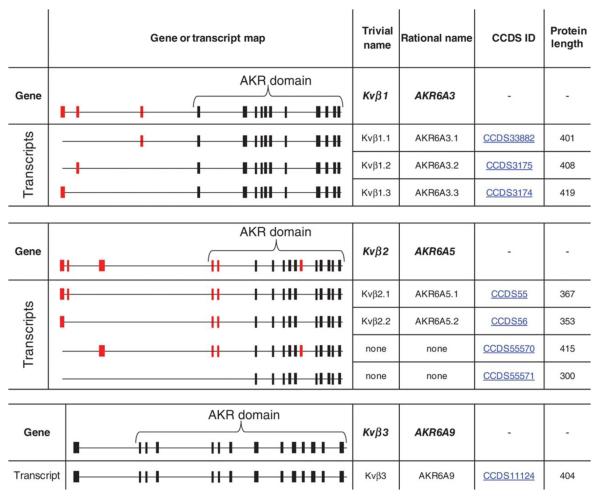
The Kvβ proteins are members of the aldo-keto reductase (AKR) superfamily that includes 15 individual families of oxidoreductases involved in carbonyl metabolism. Members of individual families share at least 40% sequence homology with each other and <40% homology with other AKRs. All AKRs share a (β/α)8 triosephosphateisomerase-barrel structural fold in which the active site is located at the C-terminus. These proteins do not contain a Rossmann fold, but they bind pyridine nucleotides with high affinity via a unique AKR NBD. Mammalian AKRs belong to 3 families, AKR 1, 6, and 7, and the Kvβ proteins belong to the AKR6A family. The hyperkinetic locus of Drosophila melanogaster is the founding member of the AKR6B family, whereas the homologous β-subunit proteins from plants constitute AKR6C family. In humans, there are 3 Kvβ genes, systematically designated as AKR6A3 (Kvβ1), AKR6A5 (Kvβ2), and AKR6A9 (Kvβ3). In addition, the AKR6A3 gene undergoes alternative splicing to give rise to Kvβ1.1, 1.2, and 1.3, and the 6A5 gene is alternatively spliced to generate Kvβ2.1 and 2.2. Figure 5 lists Kvβ proteins identified to date. To avoid confusion caused by variable names in current databases, we have included a revised nomenclature for human Kvβ proteins and their splice variants, which is consistent with the AKR structure of these proteins.
Figure 5. Structure and classification of human Kvβ genes.
Schematic showing the structure of Kvβ genes with alternative splicing sites based on the Ensembl database (http://www.ensembl.org). Common exons are shown in black and alternatively spliced exons are in red. Only splice variants resulting in different protein sequences and coding exons are shown. Trivial names refer to the name most widely used in literature. The rational nomenclature uses a gene name accepted by the aldo-keto reductase (AKR) superfamily database (http://www.med.upenn.edu/akr/) to reflect the AKR nature of the Kvβ proteins.
The AKR nature of Kvβ proteins was first identified by the significant homology between the amino acid sequence of the Shaker β-subunit and proteins of the AKR superfamily.41 Amino acid alignments also showed that the AKR residues involved in cofactor binding are conserved in the Shaker β-subunit.42 In accordance with these predictions, we found that purified Kvβ proteins bind to pyridine nucleotides with high affinity (Kd values between 0.1 and 6 μmol/L).43 In vitro these proteins bind NADP(H) with higher affinity than NAD(H). However, the nature of the cofactor bound to Kvβ in vivo is unclear, because NAD(H) levels in excitable tissues are 2- to 7-fold higher than that of NADP(H); therefore, it is possible that the lower affinity of Kvβ for NAD(H) is offset by higher NAD(H) levels in the cell. Hence, the extent and the nature of the nucleotide bound to Kvβ in vivo may depend on the prevailing concentration of all 4 nucleotides in a cell. We believe that this might allow the Kvβ to respond to a wide range of metabolic conditions by being sensitive to changes in both NADP(H) and NAD(H) levels.
To examine the functional effects of pyridine nucleotide binding, we studied Kvβ1.3-mediated inactivation of Kv1.5 expressed in COS-7 cells.44 We found that oxidized nucleotides (NAD[P]+) prevent inactivation, whereas reduced nucleotides have no effect. These findings have been corroborated and expanded by subsequent investigations, which have shown that NAD(P)+ removes the inactivation of Kv1.1 by Kvβ145 and Kv1.5 inactivation by Kvβ3.46 NAD(P)+ also has been shown to prevent the Kvβ2-mediated hyperpolarizing shift in Kv1.5 activation46 and the acceleration of Kv1.4 current inactivation.47 The results of whole-cell patch-clamp have been substantiated by excised inside-out single-channel experiments in which it was found that the mean open time and the open probability of Kv1.5+Kvβ1.344 and Kv1.1+Kvβ145 currents are increased by adding NAD+ to the perfusate. The specificity of these interactions has been established by site-directed mutagenesis studies, which indicate that active site mutations that prevent nucleotide binding abolish the effects of NAD(P)+ on current inactivation.48 Additional mutagenesis and domain-deletion experiments have shown that oxidized nucleotides promote the binding of Kvβ N-terminus to the core of the protein and thereby remove inactivation by preventing the N-terminus from blocking the channel.49 This interaction also may be regulated by the cytosolic C-terminus of the membrane-spanning channel protein. For instance, it has been reported that deletion of Kv1.5 C-terminus prevents NAD(P)+-mediated removal of inactivation by Kvβ3.46 Moreover, even though NAD(P)H does not affect Kv1.5 inactivation by Kvβ1.3,44 it accelerates Kv1.5 inactivation by Kvβ3.46 Collectively, these observations indicate that pyridine nucleotides bind directly to the Kvβ active site and that NAD(P)+ binding induces a specific conformational change that prevents Kvβ-induced inactivation, whereas NADP(H) binding preserves or promotes inactivation (Figure 4).
The gating of the Kv-Kvβ assembly could be regulated by changes in the extent of cofactor binding that passively mirrors the cellular levels of these nucleotides. Thus, high intracellular levels of NAD(P)H would promote inactivation, whereas an increase in NAD(P)+ would remove inactivation. However, the effects of pyridine nucleotides also could be altered by catalysis. Substrate-supported catalytic conversion could alter the redox state of the pyridine nucleotide bound to Kvβ, and therefore modify Kv gating. Tertiary structure analysis shows that the 3-dimensional orientation of the residues involved in hydride transfer in other AKRs is conserved in Kvβ proteins,42 suggesting that like other AKRs, these proteins might be catalytically active. Both Kvβ1 and Kvβ2 proteins display measurable catalytic activity with model aldehydes, such as 4-cyanobenzaldehyde (4-CY) and 4-carboxybenzaldehyde. They also display catalytic activity with endogenous aldehydes, such as 1-palmitoyl-2-(5'-oxo-valeroyl)-sn-glycero-3-phosphocholine and 4-oxononenal, 12-lipoxygenase products, such as prostaglandin J2 (PGJ2) and 12-oxo-eicosatetraenoic acid, and advanced glycation end-product precursors, 3-deoxyglucosone, and methylglyoxal.50,51 Enzyme kinetic studies have shown that like other AKRs, Kvβ uses the active site tyrosine residue (Tyr-90) for acid-base catalysis and that the catalytic cycle follows an ordered reaction scheme in which the bond-breaking step is rate-limiting.50 These results support the notion that Kvβ proteins are bonafide oxidoreductases. Nevertheless, it is unclear how catalysis regulates Kvβ function in vivo.
There is no evidence that any of the substrates identified to date are endogenous regulators of Kv channels or whether these carbonyls are generated in vivo to levels required to regulate Kv gating. Moreover, mice expressing catalytically inactive Kvβ2 have no overt phenotype,52 making it difficult to postulate a major role of Kvβ catalysis in regulating Kv channel gating. However, data from in vitro studies support the concept that, at least in principle, the catalytic activity of Kvβ could regulate Kv activity. Zhou et al47 have demonstrated that in inside-out patches excised from oocytes expressing Kv1.4 and Kvβ2, perfusion with 4-CY decreases the rate of channel inactivation. Perfusion with 4-CY also decreased the rate of inactivation of Kv1.1 by Kvβ1. Because 4-CY is a Kvβ substrate, it could remove Kv inactivation by oxidizing NADPH to NADP+ at the active site of Kvβ. This possibility is supported by the observation that NADPH addition could reverse the effects of 4-CY47 and that mutation of the active site lysine (Lys-152) to methionine, which diminishes the catalytic rate, slows inactivation of Kv1.1-Kvβ1 currents.45 Interestingly, the rate of channel modulation was higher than the rate of catalysis measured with the pure enzyme,47 suggesting that the hydride transfer step could be accelerated by the coassembly of Kvβ with Kv1 in the cell. Furthermore, it was found that the effects of both 4-CY and NADP+ were faster at more depolarized potentials, suggesting that NADPH oxidation or exchange could be regulated by the membrane potential. Thus, catalysis could affect Kv currents, and changes in membrane potential could in turn affect the catalytic rate. This is an exciting observation because it suggests an entirely new mechanism by which membrane voltage could regulate cell chemistry.
Membrane voltage could affect catalysis by altering the interaction of Kvβ with the cytosolic T1 domain or the C-terminus of Kv channels. Electron microscopic single particle analysis of the Shaker channel bound to Kvβ2 shows that the C-terminus of the channel is in close contact with Kvβ active site.53 This analysis indicates that the C-terminus of the Kv channel is connected directly to the inner helices of the channel, which are expected to undergo large movements during the opening and closing of the gate, thus the conformation and the orientation of Kv C-terminus relative to the β-subunits could change as function of the membrane voltage. Although the role of Kv C-terminus in imparting voltage-sensitivity to Kvβ catalysis has not been studied, it has been recently reported that in vitro the C-terminal peptide of Kv1.5 interacts more avidly with NADPH than NADP+-bound Kvβ2 and that its deletion prevents differential regulation of Kv1.5+Kvβ2 and Kv1.5+Kvβ3 currents by reduced and oxidized nucleotides.46 Despite these findings, the general physiological role of Kv C-terminus remains unclear. Unlike the C-terminus of Kv1.5, the C-terminal domain of Kv1.1 does not affect channel regulation by Kvβ1 bound to pyridine nucleotides.45 Moreover, in addition to Kvβ, Kv channels bind to several other ancillary proteins,37 which could potentially modify α–β interactions and the interaction of the C-terminal domain with the β-subunit. In the hippocampus, leucine-rich glioma inactivated gene 1 (LGI1) protein coassembles with Kv1.4 and Kv1.1 and prevents N-type inactivation of these channels imparted by Kvβ1.54 Moreover, the inactivation of Kv1.1 by Kvβ1.1 is prevented by phosphoinositides (eg, PIP2), which immobilize the ball-and-chain inactivation domain of both Kvα and β.55 The inactivation of Kv channels by Kvβ is also modified by the subunit assembly of Kv- subunits. For instance, the N-type inactivation prevention domain of the N-terminus of Kv1.6 channels prevents β-mediated inactivation in heteromeric Kvα complexes.56 However, the role of individual components of native Kv channel complexes in regulating Kv α–β interactions and their dependence on pyridine nucleotides are unclear and merit additional investigation.
Although the regulation of Kv currents by pyridine nucleotides has been studied in heterologous systems, the physiological significance of this regulatory axis has not been assessed. Kv channels participate in several physiological processes and, therefore, their activity is extensively regulated by posttranslational modification and subunit assembly. Regulation by pyridine nucleotides could provide additional control by coupling the activity of these channels to the changes in metabolic activity or the redox state of the cell. Previous work has shown that Kv channels are sensitive to changes in cell metabolism and oxygen levels. For instance, hypoxic depolarization of pulmonary artery smooth muscle cells (PASMCs), which underlies the phenomenon of HPV, has been shown to be attributable to the inhibition of Kv1.5.57 Moreover, hypoxic inhibition of IKv in PASMC as well as HPV in isolated lungs and pulmonary arteries is reduced in Kv1.5-deficient mice,31 and adenoviral transfection with Kv1.5 restores oxygen-sensitive Kv currents in PASMCs and normalizes HPV in isolated lungs of chronically hypoxic rats.58 However, Kv1.5 has been found to be sensitive to oxygen when expressed in PASMCs, but not other cell types,59 indicating that factors other than the pore-forming subunits of the channels may be required for the oxygen sensitivity of the channel.
The pyridine nucleotide–binding Kvβ proteins with their ability to regulate Kv current could in principle impart oxygen sensitivity to Kv1.5 channels. Kvβ is highly expressed in PASMCs, and its expression is much higher in the distal than the proximal bovine pulmonary artery,60 consistent with its potential role in oxygen sensing and HPV. Moreover, the Kv1.5-Kvβ1.3 channels are the major components of Ikv in PASMC,57 and in COS-7 cells, these channels are differentially regulated by oxidized and reduced pyridine nucleotides.44 Therefore, it is conceivable that an increase in NADPH:NADP+ ratio during hypoxia activates Kv1.5-Kvβ1.3 currents at more negative values of the membrane potential, whereas the current is inhibited at more positive membrane potential, when inactivation becomes more prominent. Such behavior has been observed in hypoxic canine PASMC,61 but not in other species. In rat PASMCs hypoxia inhibits Kv currents, and this inhibition is associated with shift in activation threshold to more positive membrane potential.62 This species difference may be attributable to differences in Kvβ expression. Activation of Kv currents by hypoxia may be attributable to Kvβ1.3, whereas inhibition may be related to Kvβ2, which does not affect Kv inactivation but causes a shift in voltage-dependence of activation, although an increase in NADPH:NADP+ ratio would be expected to cause a hyperpolarizing rather than depolarizing shift in activation threshold.46 Clearly, further work is required to implicate Kvβ in HPV and to delineate the contribution of different Kvβ subunits in regulating the oxygen sensitivity of Kv channels.
In addition to regulating Kv1 channels, Kvβ proteins could affect Kv4 activity as well. In heterologous expression systems, Kvβ interacts with both Kv4.2 and Kv4.3 channels. Experiments of Pérez-García et al63 show that when expressed in HEK-293 cells, Kvβ1.2 does not affect the rate of inactivation of Kv4.2 currents, but it makes these currents sensitive to thiol-modifying reagents. They also found that the Kv4.2 channels when expressed alone do not respond to hypoxia, but when coexpressed with Kvβ1.2 the Kv4.2 current is inhibited by hypoxia, suggesting that Kvβ imparts oxygen sensitivity to Kv4.2 currents. Nevertheless, the mechanism underlying Kvβ1.2-dependent oxygen sensitivity of Kv4.2 remains unclear. The activity of Kv4.2-Kvβ1.2 channels was affected by hypoxia even in excised patches and based on this observation, Pérez-García et al63 concluded that oxygen sensor must be located in the membrane and that it affects Kv currents only in the presence of Kvβ. Although, the identity of this sensor remains a mystery, its sensitivity to CO suggests that it might be a heme-containing protein that could alter pyridine nucleotides bound to the Kvα-β pair or change the nature of the cofactor bound to Kvβ by generating a Kvβ substrate. At present, such mechanisms remain speculative, and further investigations are required to determine whether oxygen-sensitive changes in Kvα–β current are reflective of metabolism-driven changes in the redox ratio of pyridine nucleotides or the catalysis of an oxygen-sensitive Kvβ substrate.
Kv2.1 Channels
Like members of the Kv1 and Kv4 family, the Kv2.1 channels also have been shown to be sensitive to the redox ratio of pyridine nucleotides. MacDonald et al64 have reported that in whole-cell recordings of Kv2.1 from pancreatic β-cells, increasing the NADPH:NADP+ ratio in the patch pipette from 1:10 to 10:1 increased the contribution of fast inactivation to total inactivation from 40% to 60%. The effects, however, were modest and could not be duplicated by Yoshida et al.65 Moreover, it is unclear how Kv2.1 is regulated by NADP(H). The Kv2.1 protein does not associate with pyridine-binding subunit, such as Kvβ, and direct binding of pyridine nucleotide to Kv2.1 has not been demonstrated. Although it is possible that changes in NADPH/NADP+ levels could also affect Kv2.1 currents indirectly by changing cell metabolism or kinase activation, there is no evidence to support this possibility. Moreover, the physiological significance of the redox sensitivity of Kv2.1 in insulin secretion is unclear, because it has been reported that changes in Kv2.1 channels do not affect the levels of the critical pool of subplasma membrane calcium that regulates exocytosis.66 Because NADPH facilitates insulin exocytosis,67 it has been speculated that the binding of NADPH increases the association of Kv2.1 with SNARE proteins, which facilitates granule docking or priming.68 Although this is an interesting possibility, additional investigations are required to fully elucidate the relationship between NADP(H) and Kv2.1 and to assess its importance in regulating insulin secretion or other physiological phenomena.
Voltage-gated Sodium Channel
The voltage-gated sodium channel (Nav) conducts the fast inward sodium current that gives rise to the upstroke of the action potential and regulates the action potential duration. Therefore, small changes in sodium current profoundly impact myocardial excitability and conductance and such changes attributable to genetic mutations increase myocardial susceptibility to arrhythmias. Several gain-of-function and loss-of-function mutations in the cardiac channel (SCN5A) and its auxiliary subunits (Nav β1–β4 subunits) have been linked to arrhythmic syndromes, such as the long Q-T (LQTS type 3), the Brugada, the sick sinus, and the sudden infant death syndromes.69 In a large multigenerational family of Italian descent with Brugada syndrome70 and in several cases of sudden infant death syndrome,71 the pathogenic cause has been identified to mutations in the glycerol-3-phosphate dehydrogenase-1-like protein (GPD1-L), which decrease surface trafficking of SCN5A and the peak sodium current. The importance of GPD1-L is further underscored by recent evidence showing that common variations in or near GPD1-L are associated with increased risk of sudden cardiac death in patients with coronary artery disease.72
GPD1-L is a 40-kDa protein that shares 84% sequence homology with GPD, an oxidoreductase that converts dihydroxyacetone phosphate to glycerol-3-phosphate. Because glycerol-3-phosphate is required for lipid synthesis, the activity of GPD connects carbohydrate metabolism to lipid synthesis. GPD also contributes electrons to the mitochondrial electron transport system and maintains the redox status of the pyridine nucleotide levels in the mitochondria by participating in glycerol phosphate shuttle. GPD1-L displays glycerol phosphate dehydrogenase activity, although its catalytic activity is slower than that of GPD.73,74 Results of GST pull-down assays in a heterologous expression system suggest that GPD1-L is directly or closely associated with the pore-forming α-subunit of SCN5A.73 GPDL-1 mutations that have been linked to Brugada syndrome (A280V) and sudden infant death syndrome (E83K) decrease GPDL-1 activity, but do not alter its association with SCN5A.73 However, these mutations decrease the surface expression of SCN5A, thereby reducing total INa.70,73 This phenomenon is reminiscent of the behavior of Kvβ, which is also an oxidoreductase that regulates the surface expression of its pore-forming partner (Kv1). As seen with GPDL-1, loss-of-function mutations also decrease the effects of Kvβ on Kv1 trafficking.75
The close association between Nav and GPD1-L suggests that the sodium current may be sensitive to redox chemistry. Patch-clamp experiments of Dudley et al76 show that in rat neonatal cardiac myocytes and in HEK cells expressing SCN5A, intracellular dialysis with 20 to 100 μmol/L NADH directly inhibits INa (Figure 6) and that this is antagonized by incubating the cells with NAD+. This inhibition of INa was linked to NADH-dependent protein kinase C (PKC) activation or mitochondrial superoxide generation.76 However, the processes by which NADH could stimulate PKC have not been identified and it was unclear how activated PKC could increase mitochondrial reactive oxygen species production. Moreover, NADH-mediated INa suppression was not accompanied by changes in the inactivation of the channel or the induction of window current or a late sodium current, usually seen in cardiac myocytes exposed to oxidants.78–80 Valdivia et al73 suggest that at least some of the effects of NADH on sodium current may be attributable to changes in GPD1-L activity, because they were completely abolished by inhibiting PKC, indicating that NADH has no direct effects on Nav. They link PKC activation to GPD1-L activity, suggesting that NADH increased the production of glycerol-3-phosphate by GPD1-L, which increases diacylglycerol formation. This increase in diacylglycerol stimulates PKC activity and results in greater SCN5A phosphorylation. Moreover, they found that PKC activation acutely decreases the surface expression of SCN5A and this effect is prevented by the NADPH oxidase inhibitor apocynin, suggesting that both channel phosphorylation and reactive oxygen species production are required for PKC-mediated regulation of SCN5A trafficking.81 However, these signaling pathways have been delineated mostly in heterologous systems and, therefore, additional experiments are required to determine endogenous regulation of INa by pyridine nucleotide in cardiac myocytes (or neurons) and to determine whether GPD1-L and pyridine nucleotide–dependent changes in PKC activation and reactive oxygen species generation affect only sodium channels or other ion transport mechanisms as well.
Figure 6. Structural representation of Nav channel.
A, Schematic representation of the α-subunits of the cardiac sodium channel consisting of 4 serially linked homologous domains each containing 6 transmembrane segments. Glycerol-3-phosphate dehydrogenase-1-like protein is shown as a protein interacting with the cytosolic domain of the channel subunits. Adapted with permission from Circ Res, Wilde et al.67B, Representative traces of whole-cell sodium currents recorded from cardiac myocytes with patch pipettes containing 0 or 100 μmol/L NADH. Reprinted with permission from Circ Res, Liu et al.77 (Illustration: Ben Smith.)
ATP-regulated K+ Channels
The ATP-regulated K+ channels represent another class of K+ channels that are regulated by nucleotides. Although these channels are primary regulated by ATP, they also have been found to be sensitive to pyridine nucleotides as well. The effects of pyridine nucleotides on the KATP channels were first described by Dunne et al,82 who reported that in excised patches of insulin-secreting cells low (100 μmol/L) concentrations of NAD(P)+ and NAD(P)H promoted channel opening, whereas high concentrations (500 μmol/L) led to channel closure. These effects were modified by ATP and ADP, indicating that pyridine coenzymes compete with adenine nucleotides for the NBD of the channel.
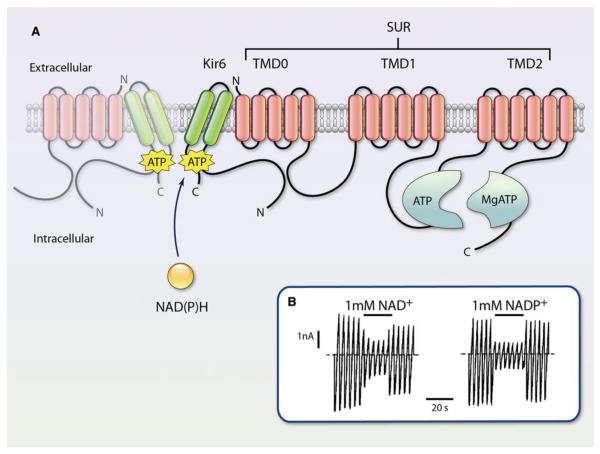
The KATP currents are generated by a large conductance channel present in the plasma membrane of several tissues, including the heart, smooth muscle, and pancreatic β-cells.83,84 An ATP-dependent potassium conductance has also been identified in the mitochondria,85,86 which has recently been found to be attributable to a channel related to the ROMK (Kir1.1) channel of the renal outer medulla.87 The sarcolemmal KATP channels open when the cellular concentrations of ATP are low and are blocked at high ATP levels. These channels are formed by the 4 pore-forming Kir6.2 subunits and 4 regulatory sulfonylurea receptor (SUR2A) subunits. The current is inhibited by the binding of adenine nucleotides to Kir6.2. Moreover, the NBD of SUR interacts specifically with Mg–nucleotide complexes, resulting in channel opening.83 Therefore, in any given metabolic state, the activity of the channel is a balance between the stimulatory and inhibitory effects of adenine nucleotide binding. Experiments with Kir6.2/SUR1 expressed in Xenopus oocytes have shown that inhibition of these currents by NAD+ and NADP+ is mediated via binding to the Kir6.2 NBD, but not to the SUR1 NBD (Figure 7). The affinity of Kir6.2 for NADP(H) is markedly enhanced on interaction with SUR1, perhaps because modification of the nucleotide-binding pocket of Kir6.2 by SUR1 facilitates the attachment of molecules bulkier than adenine mononucleotides, such as NADP(H).88 Nevertheless, the physiological significance of this interaction has not been assessed, and it is not clear whether the cardiac SUR2A isoform responds similarly. Because pyridine nucleotides are much less potent in inhibiting IKATP, it is likely that these nucleotides make only a small contribution to inhibition of the channel. Moreover, because the Kir6.2 displays no specificity for oxidized or reduced species but responds only to bulk nucleotide concentrations, it cannot participate in modulation of membrane potential by the redox state of pyridine nucleotides. Additionally, the ability of low concentrations of pyridine nucleotides to increase the activity of native KATP channels in pancreatic cells82 was not observed in oocytes expressing Kir6.2/SUR1.88 Clearly, additional work is required to elucidate the mechanism of binding of low levels of pyridine nucleotides and to assess the significance of binding to physiological levels of NADP(H). Nevertheless, because pyridine nucleotides levels change under conditions that affect ATP levels, it is likely that native KATP channels are sensitive to both adenine and pyridine nucleotides.
Figure 7. Structure and functional regulation of sulfonylurea receptor (SUR) and KATP channel.
A, The KATP channel is a heterooctamer composed of 4 Kir6 subunits and 4 SUR subunits. The Kir6 subunits form the pore, whereas the SUR subunits play a regulatory role. Pyridine nucleotides interact with the Kir domain of the channel as indicated. This site is distinct from the ATP and MgATP binding site with the cytosolic domains of SUR. TMD indicates transmembrane domain. Adapted with permission from Circ Res, Burke et al.84B, Macroscopic currents recorded from inside-out patches in response to a series of voltage ramps from −110 to +110 mV from oocytes expressing Kir 6.2. NAD+ or NADP+ was added as indicated by horizontal bars. The dashed line indicates zero current potential. Reprinted with permission from J Physiol, Dabrowski et al.88 (Illustration: Ben Smith.)
Like the sarcolemmal KATP channel, the mitochondrial KATP currents also respond to pyridine nucleotides. Measurements of mitochondrial KATP activation by osmotic swelling indicate that the channel activity could be inhibited by NADPH.89 The inhibition of the channel could not be related to reduction of mitochondrial thiols and, therefore, was ascribed to direct regulation of the channel activity by NADPH. These observations suggest that the regulation of mitoKATP channels by NADPH may be a physiological mechanism for sensing changes in energy metabolism and the redox status of mitochondria, but extensive work will be required to determine whether pyri-dine nucleotides are endogenous regulators of mitochondrial K+ transport.
Cystic Fibrosis Transmembrane Conductance Regulator
The cystic fibrosis transmembrane conductance regulator is an ATP-binding cassette ion exchanger responsible for moving chloride and thiocyanate ions across epithelial cell membranes. Mutations in this gene create a nonfunctional protein that does not transport chloride and water in and out of cells that line the lungs, the pancreas, the liver, and the reproductive and digestive tracts. This disruption of osmotic gradients results in the production of abnormally viscous mucous, causing the obstruction of the respiratory tract characteristic of cystic fibrosis, as well as chronic dysfunction of other affected organs. The cystic fibrosis transmembrane conductance regulator acts as a cAMP-activated ATP-gated ion channel that allows Cl− ions to flow down their electrochemical gradient and exit the cell. It has been reported that pyridine nucleotides can interact with the NBD of cystic fibrosis transmembrane conductance regulator and that the redox potential of pyridine nucleotides regulates the Cl− conductance of the channel.90 It was found that when ATP levels were clamped, there was a marked increase in Cl− conductance on dialysis of the cell with NADP+. In contrast, dialysis with NADPH inhibited Cl− conductance. Although these studies point to an intriguing link between the redox state of pyridine nucleotides and Cl− conductance, no studies seem to have followed-up on these initial findings.
Transient Receptor Potential (TRP) M2 Channel
Pyridine nucleotides and their metabolites also regulate calcium transport and homeostasis. Although the effects of pyridine nucleotides on the voltage-dependent calcium channels have not been reported, recent work has shown that NAD+ regulates calcium homeostasis by modifying the activity of TRPM2 channel (TRPM2). The transient receptor potential (TRP) M2 channel belongs to a large TRP superfamily which comprises several 6 transmembrane cation channels involved in a variety of processes ranging from sensation of touch, smell, taste, pain, temperature, osmotic pressure, and apoptosis.91 A distinguishing feature of the TRPM2 channel is the presence of a cytosolic nudix hydrolase domain in the C-terminus of the channel that is highly homologous to the ADP pyrophosphatase NUDT9 and is therefore named the NUDT9-homologous domain (NUDT9-H). The NUDT9 domain of ADP pyrophosphatase displays 39% sequence identity with TRPM2 and is a member of the Nudix family of proteins, such as 8-oxo-dGTP hydrolase (MutT) and diadenosine tetraphosphate pyrophosphatase (AP4A hydrolase).92 TRPM2 is highly abundant in the brain, but it is also expressed in other tissues, including, spleen, liver, lung, heart, and myeloid cells.93 The channel is nonselective for cations and displays a nearly linear current–voltage relationship with a reversal potential near 0 mV.94 The physiological functions of TRPM2 have not been completely elucidated, but there is evidence showing that the channel is involved in monocyte chemotaxis,95 insulin secretion by pancreatic β-cells,96 and lysosomal calcium release.97 Studies with TRPM2-null mice suggest that the channel controls the production of chemokines in monocytes and the infiltration of neutrophils during inflammation.98 Because TRPM2 is highly responsive to oxidative stress, it is likely that this channel could function as a redox sensor99 by directly binding to NAD+ or its metabolite, ADPR (Figure 1).
Data from whole-cell patch-clamp experiments show that intracellular dialysis with NAD+100 evokes a large inward current in cells expressing TRPM2 channels. In excised patches, application of NAD+ lead to instantaneous activation of the channel, suggesting that NAD+ directly activates the channel without the involvement of cytoplasmic or membrane components. However, regulation of the channel by NAD+ remains controversial. Some investigators suggest that stimulation of the channel by NAD+ may be attributable to contamination of the commercially available NAD+ preparations that contain trace levels of ADPR, which is the natural ligand of the channel.101 Nevertheless, direct binding of 32P-NAD+ to a GST fusion protein of the C-terminal domain of TRPM296,99 suggests that the channel protein interacts with NAD+, presumably as it does with ADPR. Moreover, even though trace contamination by ADPR could account for the effects of 1 mmol/L NAD+, channel activation at higher temperature also has been observed at 300 μmol/L NAD+96 and only minimal channel activation was observed with 10 μmol/L ADPR100 (EC50 ≈100 μmol/L),94 suggesting that contamination with >10% ADPR would be required to fully account for pronounced channel activation by commercial NAD+ preparations.
The binding of 32P-NAD+ to the C-terminus of TRPM2 channels indicates that NAD+ interacts with ADPR binding site of the Nudix domain. The importance of this domain has been confirmed by experiments showing that deletion of the C-terminus abolishes the activation of the channel by both ADPR and NAD+.102 Hara et al99 have suggested that H2O2 activates TRPM2 by increasing the intracellular NAD+, which precipitates cell death by inducing calcium and sodium overload. A similar NAD+-activated conductance, reminiscent of the TRPM2 activity, also has been implicated in rat striatal neuron cell death induced by cell depolarization and calcium influx.103 Additionally, it has been reported that a similar NAD+-gated nonselective cation channel is activated in CRI-G1 rat insulinoma cells treated with H2O2.104 The findings of these studies suggest that stimulation of TRPM2 by NAD+ could activate cation influx and trigger cell death. Similarly, in cardiac myocytes, activation of TRPM2 and the resultant sodium and calcium overload have been proposed to be obligatory steps in H2O2-mediated apoptosis.105 Nonetheless, it remains unclear whether myocardial oxidative injury in vivo during ischemia–reperfusion could be attributed to TRPM2 activation and whether this is mediated by NAD+ binding to the channel. Further experiments with TRPM2-null mice are warranted to rigorously delineate the role of TRPM2 in myocardial ischemic injury and heart failure and to determine whether the activity of the myocardial channel is regulated by NAD+.
In addition to its direct effects on the channel, NAD+ could affect the regulation of TRPM2 by its endogenous ligand, ADPR, or inhibit the catalytic activity of the C-terminal domain of the channel. It is currently believed that TRPM2 is activated by ADPR generated from the cleavage of NAD+ by CD38.106 Thus, NAD+ levels could indirectly affect TRPM2 activity by regulating the supply of ADPR. NAD+ also could compete with ADPR binding and catalysis. The NUDT9 domain of the channel has low levels of ADPR pyrophosphatase activity,94 which could be inhibited by NAD+ and other pyridine nucleotides and their metabolites, although this possibility has not been directly tested. Regardless, the presence of a catalytic domain within the channel is fascinating because it indicates that as seen with the bacterial Kef system, the TRPM2 channels belong to a distinct class of channel proteins that possess catalytic activity. Even though the enzymatic activity of NUDT9 is considerably lower than in the ADPR pyrophosphatases, this may be an evolutionary adaptation to increase the dwell time of ADPR at the channel.102 Moreover, mutations of the catalytic domain that increase enzymatic activity of NUDT9-H decreased channel activity, suggesting that nucleotide binding, not catalysis, activates the channel. Notably, the interesting possibility that channel gating or ion flow modulates the catalytic activity of NUDT9-H has not been tested.
Ryanodine Receptor (RyR) Calcium Release Channel
The RyRs represent another class of ion channels that may be regulated by pyridine nucleotides. Sequence analysis and homology modeling studies show that the RyR of the skeletal muscle (RyR1) contains several dehydrogenase and NAD+/NADH oxidoreductase domains.107 This region is located near the N-terminus of the RyR1 and it shows significant structural homology to isocitrate dehydrogenase and isopropylmalate dehydrogenase. It also contains additional motifs related to the alcohol dehydrogenase. Notably, several of the residues that participate in NADP+ binding in isocitrate dehydrogenase are conserved in RyR1, suggesting that the channel may be capable of binding to pyridine nucleotides. Indeed, equilibrium-binding studies indicate low-affinity binding of [3H] NAD+ to the sarcoplasmic reticulum membrane (Kd=10 μmol/L), although kinetic studies indicate a much higher affinity (Kd=50 nmol/L). On the basis of these studies, it has been estimated that nearly 10 molecules of NAD+ bind to each subunit of RyR1.107 Sequence alignment demonstrates that the cardiac RyR2 protein shares ≈82% homology with RyR1 between amino acids 41 and 1200, and that this region of the protein contains multiple nucleotide-binding sites with significant structural and sequence homology to phosphorylated isocitrate dehydrogenase. The same region also encompasses both catalytic and binding sequences common to dehydrogenases and oxidoreductases.
The results of structural and biochemical studies are consistent with functional measurements. In permeabilized ventricular myocytes, addition of NADH decreases the frequency of calcium sparks,108 and this inhibitory effect of NADH is partially reversed by NAD+, although NAD+ by itself has no effect on calcium spark frequency. These findings suggest that an increase in NADH/NAD+ (eg, during ischemia) could inhibit spontaneous sarcoplasmic reticulum calcium release. Nevertheless, the biochemical basis and the physiological significance of these findings are yet to be established. Particularly, it is unclear whether these effects are because of direct binding of NAD+ to RyR or because of some other indirect NAD+-dependent changes, such as increased superoxide generation by NADH oxidase.109
Although NAD+ does not activate calcium sparks in permeabilized myocytes, addition of 1 μmol/L NAD+ increases the open probability of single RyR2 cardiac calcium release channels incorporated into planar phospholipid bilayers.110 A similar increase has been reported for skeletal muscle RyR1 channels; in which case, addition of 1 to 10 μmol/L NAD+ led to a 7- to 80-fold increase in open probability.111 These observations suggest that NAD+ can directly activate calcium release channels; however, additional investigations are required to fully assess the role of pyridine nucleotides in regulating the calcium release channels, to determine whether the oxidoreductase domains of the RyR are catalytically active, and whether this catalysis regulates calcium release by the channel.
Regulation of Calcium Signaling by NAADP+
In addition to directly regulating ion transport, pyridine nucleotides also generate specialized metabolites that regulate cell signaling, particularly calcium fluxes. The most potent of these metabolites is nicotinic acid adenine dinucleotide phosphate (NAADP+), which stimulates calcium release in different cell types at concentrations as low as 5 to 10 nmol/L. Activation of calcium release by NAADP+ has been found to regulate several physiological processes, including fertilization, neurite outgrowth, synaptic function,112 and insulin secretion.113 NAADP+ also mobilizes calcium stores in smooth muscle cells114 and cardiac myocytes.115 In endothelial cells, NAADP+ has been recognized as an essential mediator of histamine-induced secretion of von Willebrand factor116 and a regulator of nitric oxide production.117 On the basis of the observation that intravenous administration of a cell permanent NAADPester lowers blood pressure in rats, it has been suggested that NAADP+ regulates systemic blood pressure.117
NAADP+ is an NADP+ derivative in which the nicotinamide ring is replaced by nictonic acid (Figure 1). This structural difference may be sufficient in preventing NAADP+ binding to most NADP+-binding proteins, and in promoting specific recognition of NAADP+ by its cognate receptors. The biochemical processes involved in NAADP+ synthesis have not been completely identified. In vitro NAADP+ is synthesized from NADP+ by both ADP-ribosyl cyclases and CD38,118 but it is not clear whether these enzymes synthesize NAADP+ in vivo. Measurements of basal NAADP+ levels in several tissues show that CD38-null mice maintain normal NAADP+ levels.119 Moreover, increases in NAADP+ levels in histamine-stimulated myometrial cells119 and in glucose-stimulated islet cells113 are preserved in the absence of CD38, although NAADP+ generation in response to CCK stimulation in pancreatic acinar cells120 is attenuated. Thus, at least in some cells, NAADP+ synthesis seems to be CD38-independent and it is likely that there are additional enzyme(s) involved in generating basal and agonist-evoked NAADP+.
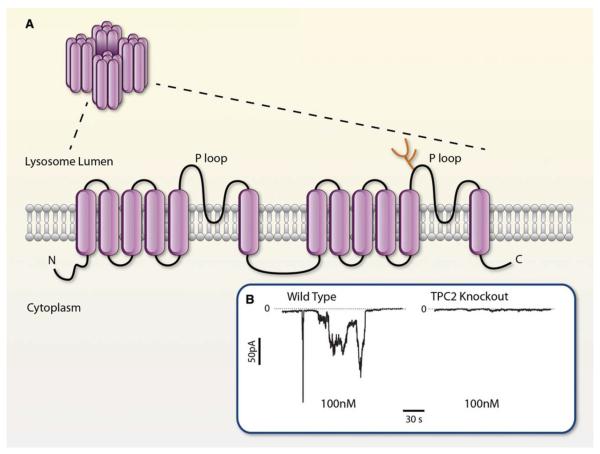
Agonist-stimulated increase in NAADP+ levels is associated with release of calcium from intracellular stores that are different from those mobilized by IP3 or cyclic ADPR.121 It is currently believed that NAADP+ targets lysosome-related stores as some of its effects are inhibited by depletion of acidic calcium stores, but not by inhibitors of sarco/plasmic or endoplasmic reticulum Ca2+-ATPase. The ability of NAADP+ to release calcium from lysosomes has been related to the activation of 2-pore channels (TPCs).122,123 These channels contain 2 putative pore-forming repeats and their transmembrane regions are similar to that of other channels, such as the Nav or TRP channels (Figure 8) however, instead of the plasma membrane, these channels are located in the endolysosomes and lysosomes or the ER. To date, 3 genes encoding TPCs (TPCN1-3) have been identified, of which TPCN2 is the predominant form expressed in primates and humans. Cells expressing TPC2 show a marked increase in calcium release on intracellular dialysis with 10 nmol/L NAADP+. Conversely, genetic knockdown of these channels abolishes NAADP+- induced calcium release, indicating that TPCs are endogenous targets of NAADP+.121 However, in addition to TPCs, NAADP+ also activates RyR124 and TRP subtype mucolipin 1 (TRP-ML1)125 and, at high concentrations, the TRPM2126 channels. The role of each of these channels in shaping the overall calcium response to NAADP+ is not clear, but it has been suggested that responses of multiple NAADP+ targets are integrated such that the small localized release of calcium by NAADP+ via TPCs is amplified by neighboring receptors to generate well-orchestrated calcium oscillations.
Figure 8. Membrane organization and NAADP+ regulation of two-pore channel (TPC).
A, Transmembrane organization of TPCs based on hydrophobicity analysis and membrane orientation of the voltage-gated Ca2+ channel and the transient potential channels. Adapted with permission from Am J Physiol Cell Physiol, Zhu et al.121B, Cation currents at −70 mV evoked by intracellular dialysis with 100 nmol/L NAADP+ in pancreatic β-cells isolated from wild-type (left) and TPC2-null mice (right). Reprinted with permission from Nature, Calcraft et al.122 (Illustration: Ben Smith).
The molecular mechanisms by which NAADP+ regulates TPCs remain to be fully elucidated. Data from HEK-293 cells show that relative to wild-type cells, cells stably overexpressing TPC2 display increased [32P] NAADP+ binding at high-affinity (Kd=5 nmol/L) and low-affinity (Kd=10 μmol/L) sites.122 However, the results of photoaffinity studies using radioactive 5-azido NAADP+ show no direct binding to the TPC protein. These studies, however, did show that some unknown low-molecular-weight proteins were labeled by [32P] NAADP+ and that the labeling of these proteins was preserved in TPC-null cells.127 These observations suggest that similar to what has been observed with other pyridine coenzyme-regulated channels (eg, Kv channels), there might be ancillary proteins within the larger TPC complex, which impart NAADP+ sensitivity to TPCs.
Because NAADP+ is synthesized from NADP+, it is possible that this synthesis is sensitive to prevailing intra-cellular levels of pyridine nucleotide as well as cellular redox state. However, CD38-ribose and ADP-ribose cyclase- dependent NAADP+ synthesis requires nicotinic acid, which binds to these enzyme with low affinity (half maximal effective concentration, 5 mmol/L).118 Therefore, under most conditions, the availability of nicotinic acid, rather than NADP+, is likely to be the limiting factor. NAADP+ signaling could, however, be coupled to the cellular redox state by enzymatic reduction of NAADP+ to NAADPH. NAADP+ is structurally related to NADP+ and it binds to NADP+-linked enzymes, such as glucose-6-phosphate dehydrogenase and 6-phospho gluconate dehydrogenase.128 The reduction of NAADP+ by glucose-6-phosphate dehydrogenase generates NAADPH, which does not induce calcium release. Hence, it is possible that enzymatic reduction is an off signal that limits the actions of NAADP+, and that this reductive process couples NAADP+ signaling to the overall redox state of the cell. In this regard, it is interesting to point out that several processes that involve NAADP+ signaling, for example, fertilization,129 are also associated with dramatic changes in the redox state; therefore, the redox sensitivity of NAADP+ may be the missing link between calcium-mediated signaling and cell metabolism.
Summary and Perspective
In classical biochemistry, pyridine nucleotides are most frequently viewed as soluble electron carriers. As coenzymes, they are known to support oxidation–reduction reactions and to control cell metabolism. However, recent research suggests that pyridine nucleotides can also regulate cell signaling, gene transcription, and ion transport by acting as electron donors, enzyme substrates, or ligands of specific receptors. Unlike most signaling molecules, pyridine nucleotides also impart redox sensitivity to regulatory processes. By doing so, these nucleotides control a large network of reactions, and therefore they can effectively integrate metabolism, cell signaling, gene transcription, proliferation, and cell death. Many of these processes depend on ion transport and homeostasis and, thus the ability to regulate ion channels may be a fundamental feature of the biological role of pyridine nucleotides.
Although research in this area is still maturing, several ion-transporting proteins have been shown to either contain NBD motifs or assemble with auxiliary subunits that bind pyridine nucleotides (Figure 9). The association between nucleotides and ion transport has been conserved during evolution, and NBD-containing ion transport systems have been found in organisms ranging from bacterium to human. Although during evolution, some of these domains have been recruited to provide structural stability to proteins or to bind other ligands, most are still capable of high-affinity pyridine nucleotide binding. In addition, recent studies have shown that some NBD motifs of ion transport proteins are functional and that the activity of several ion transporters is modified by exogenous addition of pyridine nucleotides. There is also evidence to suggest that pyridine nucleotides regulate ion fluxes by binding directly to ion transport proteins or their ancillary subunits. Yet, there is little direct evidence showing that pyridine nucleotides are endogenous regulators of ion transport or that physiological or pathological changes in pyridine nucleotide levels have any significant effect on ion transport. Additional research is needed to establish cause-and-effect relationships between pyridine nucleotides and ion transport. To delineate these relationships, it may be necessary to develop new methods for simultaneously measuring free nucleotide levels and ion transport in living cells and to determine how pyridine nucleotides regulate ion transport in vivo.
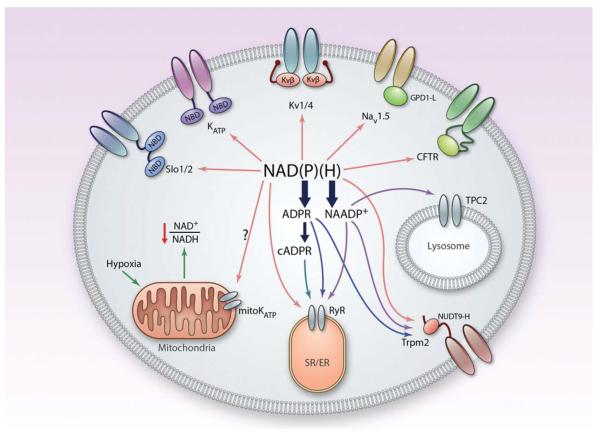
Figure 9. Integrated regulation of ion transport by pyridine nucleotides.
Hypothetical scheme showing the regulation of plasma membrane and intracellular ion channels by NAD(P)(H) and their products, NAADP+, adenosine diphosphate ribose (ADPR), and cyclic ADPR. The scheme shows all ion transport systems that have been shown to be regulated by pyridine nucleotides. Individual cells may or may not express the entire complement of ion channels shown in the figure. cADPR indicates cyclic adenosine diphosphate ribose; NBD, nucleotide-binding domain; and TPC, two-pore channel. (Illustration: Ben Smith.)
As discussed, recent research has uncovered several potential mechanisms by which pyridine nucleotides can regulate ion fluxes. In bacterial K+ transporters, such as KtrA, for example, nucleotide binding induces specific changes in channel conformation—changes that could possibly alter the ion-conducting properties of the channel pore.4 Similarly, in eukaryotic channels, such as the Slo K+ channels, the binding of pyridine nucleotides to the cytosolic domain of the channel alters channel gating, whereas in Kv1 complex nucleotide binding to Kvβ affects inactivation of the current. In addition, as shown for Kv175 and SCN5A73 channels, NBD proteins could also facilitate channel trafficking and localization. Moreover, as in the bacterial KefC channels, catalytically active NBD proteins could help protect channel proteins from oxidative injury. Such proteins could also provide the channel privileged access to metabolites that regulate channel activity—as in case of the Nav-associated protein GPD1-L, which regulates selective PKC phosphorylation of the channel. Although the general applicability of this function is unclear, other channel proteins, like Kvβ, are also constitutively associated with PKC,130 suggesting that association with other signaling proteins may be required to support local channel-specific regulation. In most cases described in the literature, however, the speculated roles of pyridine nucleotides in regulating ion fluxes remain unsubstantiated. Additional research is required to delineate the specific roles of pyridine nucleotides and their metabolites in the regulation of channel activity, localization, and posttranslational modification.
Additional research also is required to evaluate the physiological and the pathological implications of this regulatory axis. For instance, even though circumstantial evidence suggests that pyridine nucleotides play an important role in the regulation of HPV, insulin secretion, oxygen sensing, or even circadian rhythms,131 there is no clear evidence to actually implicate pyridine nucleotides in these phenomena. It is similarly unclear whether the ischemic dysfunction of myocardial ion conductances is related to changes in pyridine nucleotide signaling. Finally, the exciting possibility that, in addition to being regulated by NBD proteins, ion transport proteins in turn can regulate the activity of pyridine nucleotide-dependent proteins has yet to be tested. As mentioned, several channel-associated proteins, such as KefF, Kvβ, GPD1-L, and NUDT9-H, are catalytically active; therefore, changes in membrane potential could affect the activities of these enzymes. Further exploration of this possibility could reveal new mechanisms by which membrane potential regulates cell chemistry and metabolism. In the brain, such processes might be the basic molecular units of memory and learning; in non-neuronal cells, this mechanism could, perhaps, impart metabolic memory or contribute to the epigenetic regulation of gene expression. To understand these and other complex relationships between ion transport and pyridine nucleotides, however, we would first need to identify and characterize specific components of the individual ions channels that are regulated by pyridine nucleotides. From these findings, we would have to develop an integrated systemwide view—a view that would detail how exactly different ion transport mechanisms are synchronously regulated to support basic cell function or to mount a well-orchestrated unified response to environmental cues. Thus, further elucidation of this link between pyridine nucleotides and ion transport might provide a new understanding of the mechanisms underlying several physiological processes and disease states.
Table 1.
Pyridine Nucleotide–Regulated Mammalian Ion Channels
| Channels* | Ligand-Binding Units | Ligands | Functions |
|---|---|---|---|
| Slo2.1, Slo2.2 | Cytosolic TrkA-like domain in the C-terminus | NAD+, NADP+ | Increased Na+ sensitivity, open probability |
| Slo1 | Cytosolic RCK domain (?) in the C-terminus | NAD+ | Reduces open probability |
| KATP | Cytosolic domain of Kir6.2 | NADP(H) | Channel stimulation at low concentrations; channel block at high concentrations |
| TRPM2 | Cytosolic NUDT9-H domain | NAD+, NAADP+ | Channel activation |
| Kv | Ancillary subunit-Kvβ | NAD(P)H, NAD(P+) | N-type inactivation by NADPH, removal of inactivation by NADP+, membrane trafficking |
| Nav | Ancillary subunit-GPD1-L | NADH | Phosphorylation (PKC), membrane trafficking |
| TPC2 | Ancillary subunits (?) | NAADP+ | Increase in calcium release |
| TRP-ML1 | ? | NAADP+ | Increase in calcium release |
| CFR | ? | NADP(H) | Current inhibition (NADPH) and stimulation (NADP+) |
Channels reported in the literature to be sensitive to pyridine nucleotides are listed. GPD1-L indicates glycerol phosphate dehydrogenase-1-like protein; PKC, protein kinase C; ?, unknown; and RCK, regulator of potassium conductance.
Acknowledgments
Sources of Funding This work was partly supported by National Institutes of Health grants GM103492, HL-78825, HL-55477, HL-59378 (to A. Bhatnagar), HL-89372 (to O.A. Barski), and HL-102171 and USF College of Pharmacy also provided funding (to S.M. Tipparaju).
Nonstandard Abbreviations and Acronyms
- 4-CY
4-cyanobenzaldehyde
- ADPR
adenosine diphosphate ribose
- AKR
aldo-keto reductase
- GPD1-L
glycerol-3-phosphate dehydrogenase-1-like protein
- HPV
hypoxic pulmonary vasoconstriction
- KTNBD
potassium transport nucleotide binding domain
- NBD
nucleotide-binding domains
- PASMC
pulmonary artery smooth muscle cell
- PKC
protein kinase C
- RyR
ryanodine receptor
- SUR
sulfonylurea receptor
- TPC
two-pore channel
- TRP
transient receptor potential cation channel
Footnotes
Disclosures None.
References
- 1.Ussher JR, Jaswal JS, Lopaschuk GD. Pyridine nucleotide regulation of cardiac intermediary metabolism. Circ Res. 2012;111:628–641. doi: 10.1161/CIRCRESAHA.111.246371. [DOI] [PubMed] [Google Scholar]
- 2.Nakamura M, Bhatnagar A, Sadoshima J. Overview of pyridine nucleotides review series. Circ Res. 2012;111:604–610. doi: 10.1161/CIRCRESAHA.111.247924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Oka S, Hsu CP, Sadoshima J. Regulation of cell survival and death by pyridine nucleotides. Circ Res. 2012;111:611–627. doi: 10.1161/CIRCRESAHA.111.247932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Roosild TP, Miller S, Booth IR, Choe S. A mechanism of regulating transmembrane potassium flux through a ligand-mediated conformational switch. Cell. 2002;109:781–791. doi: 10.1016/s0092-8674(02)00768-7. [DOI] [PubMed] [Google Scholar]
- 5.Durell SR, Hao Y, Nakamura T, Bakker EP, Guy HR. Evolutionary relationship between K(+) channels and symporters. Biophys J. 1999;77:775–788. doi: 10.1016/S0006-3495(99)76931-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schlösser A, Hamann A, Bossemeyer D, Schneider E, Bakker EP. NAD+ binding to the Escherichia coli K(+)-uptake protein TrkA and sequence similarity between TrkA and domains of a family of dehydrogenases suggest a role for NAD+ in bacterial transport. Mol Microbiol. 1993;9:533–543. doi: 10.1111/j.1365-2958.1993.tb01714.x. [DOI] [PubMed] [Google Scholar]
- 7.Roosild TP, Castronovo S, Healy J, Miller S, Pliotas C, Rasmussen T, Bartlett W, Conway SJ, Booth IR. Mechanism of ligand-gated potassium efflux in bacterial pathogens. Proc Natl Acad Sci USA. 2010;107:19784–19789. doi: 10.1073/pnas.1012716107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fujisawa M, Ito M, Krulwich TA. Three two-component transporters with channel-like properties have monovalent cation/proton antiport activity. Proc Natl Acad Sci USA. 2007;104:13289–13294. doi: 10.1073/pnas.0703709104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Miller S, Ness LS, Wood CM, Fox BC, Booth IR. Identification of an ancillary protein, YabF, required for activity of the KefC glutathione-gated potassium efflux system in Escherichia coli. J Bacteriol. 2000;182:6536–6540. doi: 10.1128/jb.182.22.6536-6540.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Roosild TP, Castronovo S, Miller S, Li C, Rasmussen T, Bartlett W, Gunasekera B, Choe S, Booth IR. KTN (RCK) domains regulate K+ channels and transporters by controlling the dimer-hinge conformation. Structure. 2009;17:893–903. doi: 10.1016/j.str.2009.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lyngberg L, Healy J, Bartlett W, Miller S, Conway SJ, Booth IR, Rasmussen T. KefF, the regulatory subunit of the potassium efflux system KefC, shows quinone oxidoreductase activity. J Bacteriol. 2011;193:4925–4932. doi: 10.1128/JB.05272-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tamsett TJ, Picchione KE, Bhattacharjee A. NAD+ activates KNa channels in dorsal root ganglion neurons. J Neurosci. 2009;29:5127–5134. doi: 10.1523/JNEUROSCI.0859-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Salkoff L, Butler A, Ferreira G, Santi C, Wei A. High-conductance potassium channels of the SLO family. Nat Rev Neurosci. 2006;7:921–931. doi: 10.1038/nrn1992. [DOI] [PubMed] [Google Scholar]
- 14.Park MK, Lee SH, Ho WK, Earm YE. Redox agents as a link between hypoxia and the responses of ionic channels in rabbit pulmonary vascular smooth muscle. Exp Physiol. 1995;80:835–842. doi: 10.1113/expphysiol.1995.sp003891. [DOI] [PubMed] [Google Scholar]
- 15.Yuan A, Santi CM, Wei A, Wang ZW, Pollak K, Nonet M, Kaczmarek L, Crowder CM, Salkoff L. The sodium-activated potassium channel is encoded by a member of the Slo gene family. Neuron. 2003;37:765–773. doi: 10.1016/s0896-6273(03)00096-5. [DOI] [PubMed] [Google Scholar]
- 16.Bhattacharjee A, Joiner WJ, Wu M, Yang Y, Sigworth FJ, Kaczmarek LK. Slick (Slo2.1), a rapidly-gating sodium-activated potassium channel inhibited by ATP. J Neurosci. 2003;23:11681–11691. doi: 10.1523/JNEUROSCI.23-37-11681.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Egan TM, Dagan D, Kupper J, Levitan IB. Properties and rundown of sodium-activated potassium channels in rat olfactory bulb neurons. J Neurosci. 1992;12:1964–1976. doi: 10.1523/JNEUROSCI.12-05-01964.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mitani A, Shattock MJ. Role of Na-activated K channel, Na-K-Cl co-transport, and Na-K pump in [K]e changes during ischemia in rat heart. Am J Physiol. 1992;263:H333–H340. doi: 10.1152/ajpheart.1992.263.2.H333. [DOI] [PubMed] [Google Scholar]
- 19.Lawrence CL, Rodrigo GC. The Na+-activated K+ channel contributes to K+ efflux in Na+-loaded guinea-pig but not rat ventricular myocytes. Pflugers Arch. 2001;442:595–602. doi: 10.1007/s004240100569. [DOI] [PubMed] [Google Scholar]
- 20.Wojtovich AP, Sherman TA, Nadtochiy SM, Urciuoli WR, Brookes PS, Nehrke K. SLO-2 is cytoprotective and contributes to mitochondrial potassium transport. PLoS ONE. 2011;6:e28287. doi: 10.1371/journal.pone.0028287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yuan P, Leonetti MD, Pico AR, Hsiung Y, MacKinnon R. Structure of the human BK channel Ca2+-activation apparatus at 3.0 A resolution. Science. 2010;329:182–186. doi: 10.1126/science.1190414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee S, Park M, So I, Earm YE. NADH and NAD modulates Ca(2+)-activated K+ channels in small pulmonary arterial smooth muscle cells of the rabbit. Pflugers Arch. 1994;427:378–380. doi: 10.1007/BF00374548. [DOI] [PubMed] [Google Scholar]
- 23.Thuringer D, Findlay I. Contrasting effects of intracellular redox couples on the regulation of maxi-K channels in isolated myocytes from rabbit pulmonary artery. J Physiol (Lond) 1997;500(Pt 3):583–592. doi: 10.1113/jphysiol.1997.sp022044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yellen G. The voltage-gated potassium channels and their relatives. Nature. 2002;419:35–42. doi: 10.1038/nature00978. [DOI] [PubMed] [Google Scholar]
- 25.Nerbonne JM. Molecular basis of functional voltage-gated K+ channel diversity in the mammalian myocardium. J Physiol (Lond) 2000;525(Pt 2):285–298. doi: 10.1111/j.1469-7793.2000.t01-1-00285.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dubois JM, Rouzaire-Dubois B. Role of potassium channels in mitogenesis. Prog Biophys Mol Biol. 1993;59:1–21. doi: 10.1016/0079-6107(93)90005-5. [DOI] [PubMed] [Google Scholar]
- 27.Krick S, Platoshyn O, Sweeney M, Kim H, Yuan JX. Activation of K+ channels induces apoptosis in vascular smooth muscle cells. Am J Physiol Cell Physiol. 2001;280:C970–C979. doi: 10.1152/ajpcell.2001.280.4.C970. [DOI] [PubMed] [Google Scholar]
- 28.Chandy KG, Wulff H, Beeton C, Pennington M, Gutman GA, Cahalan MD. K+ channels as targets for specific immunomodulation. Trends Pharmacol Sci. 2004;25:280–289. doi: 10.1016/j.tips.2004.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gutterman DD, Miura H, Liu Y. Redox modulation of vascular tone: focus of potassium channel mechanisms of dilation. Arterioscler Thromb Vasc Biol. 2005;25:671–678. doi: 10.1161/01.ATV.0000158497.09626.3b. [DOI] [PubMed] [Google Scholar]
- 30.Archer SL, Souil E, Dinh-Xuan AT, Schremmer B, Mercier JC, El Yaagoubi A, Nguyen-Huu L, Reeve HL, Hampl V. Molecular identification of the role of voltage-gated K+ channels, Kv1.5 and Kv2.1, in hypoxic pulmonary vasoconstriction and control of resting membrane potential in rat pulmonary artery myocytes. J Clin Invest. 1998;101:2319–2330. doi: 10.1172/JCI333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Archer SL, London B, Hampl V, Wu X, Nsair A, Puttagunta L, Hashimoto K, Waite RE, Michelakis ED. Impairment of hypoxic pulmonary vasoconstriction in mice lacking the voltage-gated potassium channel Kv1.5. FASEB J. 2001;15:1801–1803. doi: 10.1096/fj.00-0649fje. [DOI] [PubMed] [Google Scholar]
- 32.Tristani-Firouzi M, Chen J, Mitcheson JS, Sanguinetti MC. Molecular biology of K(+) channels and their role in cardiac arrhythmias. Am J Med. 2001;110:50–59. doi: 10.1016/s0002-9343(00)00623-9. [DOI] [PubMed] [Google Scholar]
- 33.Mandegar M, Yuan JX. Role of K+ channels in pulmonary hypertension. Vascul Pharmacol. 2002;38:25–33. doi: 10.1016/s1537-1891(02)00123-4. [DOI] [PubMed] [Google Scholar]
- 34.Hille B. Ionic channels of excitable membranes. Sinauer Associates; Sunderland, MA: 1992. [Google Scholar]
- 35.Choe S. Potassium channel structures. Nat Rev Neurosci. 2002;3:115–121. doi: 10.1038/nrn727. [DOI] [PubMed] [Google Scholar]
- 36.Long SB, Campbell EB, Mackinnon R. Crystal structure of a mammalian voltage-dependent Shaker family K+ channel. Science. 2005;309:897–903. doi: 10.1126/science.1116269. [DOI] [PubMed] [Google Scholar]
- 37.Pongs O, Schwarz JR. Ancillary subunits associated with voltage-dependent K+ channels. Physiol Rev. 2010;90:755–796. doi: 10.1152/physrev.00020.2009. [DOI] [PubMed] [Google Scholar]
- 38.Rettig J, Heinemann SH, Wunder F, Lorra C, Parcej DN, Dolly JO, Pongs O. Inactivation properties of voltage-gated K+ channels altered by presence of beta-subunit. Nature. 1994;369:289–294. doi: 10.1038/369289a0. [DOI] [PubMed] [Google Scholar]
- 39.Heinemann SH, Rettig J, Graack HR, Pongs O. Functional characterization of Kv channel beta-subunits from rat brain. J Physiol (Lond) 1996;493(Pt 3):625–633. doi: 10.1113/jphysiol.1996.sp021409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Leicher T, Bähring R, Isbrandt D, Pongs O. Coexpression of the KCNA3B gene product with Kv1.5 leads to a novel A-type potassium channel. J Biol Chem. 1998;273:35095–35101. doi: 10.1074/jbc.273.52.35095. [DOI] [PubMed] [Google Scholar]
- 41.McCormack T, McCormack K. Shaker K+ channel beta subunits belong to an NAD(P)H-dependent oxidoreductase superfamily. Cell. 1994;79:1133–1135. doi: 10.1016/0092-8674(94)90004-3. [DOI] [PubMed] [Google Scholar]
- 42.Gulbis JM, Mann S, MacKinnon R. Structure of a voltage-dependent K+ channel beta subunit. Cell. 1999;97:943–952. doi: 10.1016/s0092-8674(00)80805-3. [DOI] [PubMed] [Google Scholar]
- 43.Liu SQ, Jin H, Zacarias A, Srivastava S, Bhatnagar A. Binding of pyridine nucleotide coenzymes to the beta-subunit of the voltage-sensitive K + channel. J Biol Chem. 2001;276:11812–11820. doi: 10.1074/jbc.M008259200. [DOI] [PubMed] [Google Scholar]
- 44.Tipparaju SM, Saxena N, Liu SQ, Kumar R, Bhatnagar A. Differential regulation of voltage-gated K+ channels by oxidized and reduced pyridine nucleotide coenzymes. Am J Physiol Cell Physiol. 2005;288:C366–C376. doi: 10.1152/ajpcell.00354.2004. [DOI] [PubMed] [Google Scholar]
- 45.Pan Y, Weng J, Cao Y, Bhosle RC, Zhou M. Functional coupling between the Kv1.1 channel and aldoketoreductase Kvbeta1. J Biol Chem. 2008;283:8634–8642. doi: 10.1074/jbc.M709304200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tipparaju SM, Li XP, Kilfoil PJ, Xue B, Uversky VN, Bhatnagar A, Barski OA. Interactions between the C-terminus of Kv1.5 and Kvβ regulate pyridine nucleotide-dependent changes in channel gating. Pflugers Arch. 2012;463:799–818. doi: 10.1007/s00424-012-1093-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Weng J, Cao Y, Moss N, Zhou M. Modulation of voltage-dependent Shaker family potassium channels by an aldo-keto reductase. J Biol Chem. 2006;281:15194–15200. doi: 10.1074/jbc.M513809200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tipparaju SM, Liu SQ, Barski OA, Bhatnagar A. NADPH binding to beta-subunit regulates inactivation of voltage-gated K(+) channels. Biochem Biophys Res Commun. 2007;359:269–276. doi: 10.1016/j.bbrc.2007.05.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pan Y, Weng J, Levin EJ, Zhou M. Oxidation of NADPH on Kvbeta1 inhibits ball-and-chain type inactivation by restraining the chain. Proc Natl Acad Sci USA. 2011;108:5885–5890. doi: 10.1073/pnas.1100316108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tipparaju SM, Barski OA, Srivastava S, Bhatnagar A. Catalytic mechanism and substrate specificity of the beta-subunit of the voltage-gated potassium channel. Biochemistry. 2008;47:8840–8854. doi: 10.1021/bi800301b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xie Z, Barski OA, Cai J, Bhatnagar A, Tipparaju SM. Catalytic reduction of carbonyl groups in oxidized PAPC by Kvβ2 (AKR6) Chem Biol Interact. 2011;191:255–260. doi: 10.1016/j.cbi.2011.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.McCormack K, Connor JX, Zhou L, Ho LL, Ganetzky B, Chiu SY, Messing A. Genetic analysis of the mammalian K+ channel beta subunit Kvbeta 2 (Kcnab2) J Biol Chem. 2002;277:13219–13228. doi: 10.1074/jbc.M111465200. [DOI] [PubMed] [Google Scholar]
- 53.Sokolova O, Accardi A, Gutierrez D, Lau A, Rigney M, Grigorieff N. Conformational changes in the C terminus of Shaker K+ channel bound to the rat Kvbeta2-subunit. Proc Natl Acad Sci USA. 2003;100:12607–12612. doi: 10.1073/pnas.2235650100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schulte U, Thumfart JO, Klöcker N, Sailer CA, Bildl W, Biniossek M, Dehn D, Deller T, Eble S, Abbass K, Wangler T, Knaus HG, Fakler B. The epilepsy-linked Lgi1 protein assembles into presynaptic Kv1 channels and inhibits inactivation by Kvbeta1. Neuron. 2006;49:697–706. doi: 10.1016/j.neuron.2006.01.033. [DOI] [PubMed] [Google Scholar]
- 55.Oliver D, Lien CC, Soom M, Baukrowitz T, Jonas P, Fakler B. Functional conversion between A-type and delayed rectifier K+ channels by membrane lipids. Science. 2004;304:265–270. doi: 10.1126/science.1094113. [DOI] [PubMed] [Google Scholar]
- 56.Roeper J, Sewing S, Zhang Y, Sommer T, Wanner SG, Pongs O. NIP domain prevents N-type inactivation in voltage-gated potassium channels. Nature. 1998;391:390–393. doi: 10.1038/34916. [DOI] [PubMed] [Google Scholar]
- 57.Sylvester JT, Shimoda LA, Aaronson PI, Ward JP. Hypoxic pulmonary vasoconstriction. Physiol Rev. 2012;92:367–520. doi: 10.1152/physrev.00041.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pozeg ZI, Michelakis ED, McMurtry MS, Thébaud B, Wu XC, Dyck JR, Hashimoto K, Wang S, Moudgil R, Harry G, Sultanian R, Koshal A, Archer SL. In vivo gene transfer of the O2-sensitive potassium channel Kv1.5 reduces pulmonary hypertension and restores hypoxic pulmonary vasoconstriction in chronically hypoxic rats. Circulation. 2003;107:2037–2044. doi: 10.1161/01.CIR.0000062688.76508.B3. [DOI] [PubMed] [Google Scholar]
- 59.Platoshyn O, Brevnova EE, Burg ED, Yu Y, Remillard CV, Yuan JX. Acute hypoxia selectively inhibits KCNA5 channels in pulmonary artery smooth muscle cells. Am J Physiol Cell Physiol. 2006;290:C907–C916. doi: 10.1152/ajpcell.00028.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Coppock EA, Tamkun MM. Differential expression of K(V) channel alpha- and beta-subunits in the bovine pulmonary arterial circulation. Am J Physiol Lung Cell Mol Physiol. 2001;281:L1350–L1360. doi: 10.1152/ajplung.2001.281.6.L1350. [DOI] [PubMed] [Google Scholar]
- 61.Post JM, Hume JR, Archer SL, Weir EK. Direct role for potassium channel inhibition in hypoxic pulmonary vasoconstriction. Am J Physiol. 1992;262:C882–C890. doi: 10.1152/ajpcell.1992.262.4.C882. [DOI] [PubMed] [Google Scholar]
- 62.Olschewski A, Hong Z, Nelson DP, Weir EK. Graded response of K+ current, membrane potential, and [Ca2+]i to hypoxia in pulmonary arterial smooth muscle. Am J Physiol Lung Cell Mol Physiol. 2002;283:L1143–L1150. doi: 10.1152/ajplung.00104.2002. [DOI] [PubMed] [Google Scholar]
- 63.Pérez-García MT, López-López JR, González C. Kvbeta1.2 subunit co-expression in HEK293 cells confers O2 sensitivity to kv4.2 but not to Shaker channels. J Gen Physiol. 1999;113:897–907. doi: 10.1085/jgp.113.6.897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.MacDonald PE, Salapatek AM, Wheeler MB. Temperature and redox state dependence of native Kv2.1 currents in rat pancreatic beta-cells. J Physiol (Lond) 2003;546:647–653. doi: 10.1113/jphysiol.2002.035709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yoshida M, Dezaki K, Yamato S, Aoki A, Sugawara H, Toyoshima H, Ishikawa SE, Kawakami M, Nakata M, Yada T, Kakei M. Regulation of voltage-gated K+ channels by glucose metabolism in pancreatic beta-cells. FEBS Lett. 2009;583:2225–2230. doi: 10.1016/j.febslet.2009.05.050. [DOI] [PubMed] [Google Scholar]
- 66.Ravier MA, Cheng-Xue R, Palmer AE, Henquin JC, Gilon P. Subplasmalemmal Ca(2+) measurements in mouse pancreatic beta cells support the existence of an amplifying effect of glucose on insulin secretion. Diabetologia. 2010;53:1947–1957. doi: 10.1007/s00125-010-1775-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ivarsson R, Quintens R, Dejonghe S, Tsukamoto K, `t Veld P, Renström E, Schuit FC. Redox control of exocytosis: regulatory role of NADPH, thioredoxin, and glutaredoxin. Diabetes. 2005;54:2132–2142. doi: 10.2337/diabetes.54.7.2132. [DOI] [PubMed] [Google Scholar]
- 68.MacDonald PE. Signal integration at the level of ion channel and exocytotic function in pancreatic β-cells. Am J Physiol Endocrinol Metab. 2011;301:E1065–E1069. doi: 10.1152/ajpendo.00426.2011. [DOI] [PubMed] [Google Scholar]
- 69.Wilde AA, Brugada R. Phenotypical manifestations of mutations in the genes encoding subunits of the cardiac sodium channel. Circ Res. 2011;108:884–897. doi: 10.1161/CIRCRESAHA.110.238469. [DOI] [PubMed] [Google Scholar]
- 70.London B, Michalec M, Mehdi H, et al. Mutation in glycerol-3-phosphate dehydrogenase 1 like gene (GPD1-L) decreases cardiac Na+ current and causes inherited arrhythmias. Circulation. 2007;116:2260–2268. doi: 10.1161/CIRCULATIONAHA.107.703330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Van Norstrand DW, Valdivia CR, Tester DJ, Ueda K, London B, Makielski JC, Ackerman MJ. Molecular and functional characterization of novel glycerol-3-phosphate dehydrogenase 1 like gene (GPD1-L) mutations in sudden infant death syndrome. Circulation. 2007;116:2253–2259. doi: 10.1161/CIRCULATIONAHA.107.704627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Westaway SK, Reinier K, Huertas-Vazquez A, Evanado A, Teodorescu C, Navarro J, Sinner MF, Gunson K, Jui J, Spooner P, Kaab S, Chugh SS. Common variants in CASQ2, GPD1L, and NOS1AP are significantly associated with risk of sudden death in patients with coronary artery disease. Circ Cardiovasc Genet. 2011;4:397–402. doi: 10.1161/CIRCGENETICS.111.959916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Valdivia CR, Ueda K, Ackerman MJ, Makielski JC. GPD1L links redox state to cardiac excitability by PKC-dependent phosphorylation of the sodium channel SCN5A. Am J Physiol Heart Circ Physiol. 2009;297:H1446–H1452. doi: 10.1152/ajpheart.00513.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kelly TJ, Souza AL, Clish CB, Puigserver P. A hypoxia-induced positive feedback loop promotes hypoxia-inducible factor 1alpha stability through miR-210 suppression of glycerol-3-phosphate dehydrogenase 1-like. Mol Cell Biol. 2011;31:2696–2706. doi: 10.1128/MCB.01242-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Campomanes CR, Carroll KI, Manganas LN, Hershberger ME, Gong B, Antonucci DE, Rhodes KJ, Trimmer JS. Kv beta subunit oxidoreductase activity and Kv1 potassium channel trafficking. J Biol Chem. 2002;277:8298–8305. doi: 10.1074/jbc.M110276200. [DOI] [PubMed] [Google Scholar]
- 76.Liu M, Liu H, Dudley SC., Jr Reactive oxygen species originating from mitochondria regulate the cardiac sodium channel. Circ Res. 2010;107:967–974. doi: 10.1161/CIRCRESAHA.110.220673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Liu M, Sanyal S, Gao G, Gurung IS, Zhu X, Gaconnet G, Kerchner LJ, Shang LL, Huang CL, Grace A, London B, Dudley SC., Jr Cardiac Na+ current regulation by pyridine nucleotides. Circ Res. 2009;105:737–745. doi: 10.1161/CIRCRESAHA.109.197277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bhatnagar A, Srivastava SK, Szabo G. Oxidative stress alters specific membrane currents in isolated cardiac myocytes. Circ Res. 1990;67:535–549. doi: 10.1161/01.res.67.3.535. [DOI] [PubMed] [Google Scholar]
- 79.Bhatnagar A. Electrophysiological effects of 4-hydroxynonenal, an aldehydic product of lipid peroxidation, on isolated rat ventricular myocytes. Circ Res. 1995;76:293–304. doi: 10.1161/01.res.76.2.293. [DOI] [PubMed] [Google Scholar]
- 80.Wagner S, Ruff HM, Weber SL, Bellmann S, Sowa T, Schulte T, Anderson ME, Grandi E, Bers DM, Backs J, Belardinelli L, Maier LS. Reactive oxygen species-activated Ca/calmodulin kinase IIδ is required for late I(Na) augmentation leading to cellular Na and Ca overload. Circ Res. 2011;108:555–565. doi: 10.1161/CIRCRESAHA.110.221911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hallaq H, Wang DW, Kunic JD, George AL, Jr, Wells KS, Murray KT. Activation of protein kinase C alters the intracellular distribution and mobility of cardiac Na+ channels. Am J Physiol Heart Circ Physiol. 2012;302:H782–H789. doi: 10.1152/ajpheart.00817.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Dunne MJ, Findlay I, Petersen OH. Effects of pyridine nucleotides on the gating of ATP-sensitive potassium channels in insulin-secreting cells. J Membr Biol. 1988;102:205–216. doi: 10.1007/BF01925714. [DOI] [PubMed] [Google Scholar]
- 83.Flagg TP, Enkvetchakul D, Koster JC, Nichols CG. Muscle KATP channels: recent insights to energy sensing and myoprotection. Physiol Rev. 2010;90:799–829. doi: 10.1152/physrev.00027.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Burke MA, Mutharasan RK, Ardehali H. The sulfonylurea receptor, an atypical ATP-binding cassette protein, and its regulation of the KATP channel. Circ Res. 2008;102:164–176. doi: 10.1161/CIRCRESAHA.107.165324. [DOI] [PubMed] [Google Scholar]
- 85.Inoue I, Nagase H, Kishi K, Higuti T. ATP-sensitive K+ channel in the mitochondrial inner membrane. Nature. 1991;352:244–247. doi: 10.1038/352244a0. [DOI] [PubMed] [Google Scholar]
- 86.O'Rourke B. Myocardial K(ATP) channels in preconditioning. Circ Res. 2000;87:845–855. doi: 10.1161/01.res.87.10.845. [DOI] [PubMed] [Google Scholar]
- 87.Foster DB, Ho AS, Rucker J, Garlid AO, Chen L, Sidor A, Garlid KD, O'Rourke B. Mitochondrial ROMK channel is a molecular component of mitoK(ATP) Circ Res. 2012;111:446–454. doi: 10.1161/CIRCRESAHA.112.266445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Dabrowski M, Trapp S, Ashcroft FM. Pyridine nucleotide regulation of the KATP channel Kir6.2/SUR1 expressed in Xenopus oocytes. J Physiol (Lond) 2003;550:357–363. doi: 10.1113/jphysiol.2003.041715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Queliconi BB, Wojtovich AP, Nadtochiy SM, Kowaltowski AJ, Brookes PS. Redox regulation of the mitochondrial K(ATP) channel in cardioprotection. Biochim Biophys Acta. 2011;1813:1309–1315. doi: 10.1016/j.bbamcr.2010.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Stutts MJ, Gabriel SE, Price EM, Sarkadi B, Olsen JC, Boucher RC. Pyridine nucleotide redox potential modulates cystic fibrosis trans-membrane conductance regulator Cl− conductance. J Biol Chem. 1994;269:8667–8674. [PubMed] [Google Scholar]
- 91.Ramsey IS, Delling M, Clapham DE. An introduction to TRP channels. Annu Rev Physiol. 2006;68:619–647. doi: 10.1146/annurev.physiol.68.040204.100431. [DOI] [PubMed] [Google Scholar]
- 92.Shen BW, Perraud AL, Scharenberg A, Stoddard BL. The crystal structure and mutational analysis of human NUDT9. J Mol Biol. 2003;332:385–398. doi: 10.1016/s0022-2836(03)00954-9. [DOI] [PubMed] [Google Scholar]
- 93.Du J, Xie J, Yue L. Intracellular calcium activates TRPM2 and its alternative spliced isoforms. Proc Natl Acad Sci USA. 2009;106:7239–7244. doi: 10.1073/pnas.0811725106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Perraud AL, Fleig A, Dunn CA, Bagley LA, Launay P, Schmitz C, Stokes AJ, Zhu Q, Bessman MJ, Penner R, Kinet JP, Scharenberg AM. ADP-ribose gating of the calcium-permeable LTRPC2 channel revealed by Nudix motif homology. Nature. 2001;411:595–599. doi: 10.1038/35079100. [DOI] [PubMed] [Google Scholar]
- 95.Massullo P, Sumoza-Toledo A, Bhagat H, Partida-Sánchez S. TRPM channels, calcium and redox sensors during innate immune responses. Semin Cell Dev Biol. 2006;17:654–666. doi: 10.1016/j.semcdb.2006.11.006. [DOI] [PubMed] [Google Scholar]
- 96.Togashi K, Hara Y, Tominaga T, Higashi T, Konishi Y, Mori Y, Tominaga M. TRPM2 activation by cyclic ADP-ribose at body temperature is involved in insulin secretion. EMBO J. 2006;25:1804–1815. doi: 10.1038/sj.emboj.7601083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Lange I, Yamamoto S, Partida-Sanchez S, Mori Y, Fleig A, Penner R. TRPM2 functions as a lysosomal Ca2+-release channel in beta cells. Sci Signal. 2009;2:ra23. doi: 10.1126/scisignal.2000278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Yamamoto S, Shimizu S, Kiyonaka S, et al. TRPM2-mediated Ca2+ in flux induces chemokine production in monocytes that aggravates inflammatory neutrophil infiltration. Nat Med. 2008;14:738–747. doi: 10.1038/nm1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Hara Y, Wakamori M, Ishii M, Maeno E, Nishida M, Yoshida T, Yamada H, Shimizu S, Mori E, Kudoh J, Shimizu N, Kurose H, Okada Y, Imoto K, Mori Y. LTRPC2 Ca2+-permeable channel activated by changes in redox status confers susceptibility to cell death. Mol Cell. 2002;9:163–173. doi: 10.1016/s1097-2765(01)00438-5. [DOI] [PubMed] [Google Scholar]
- 100.Sano Y, Inamura K, Miyake A, Mochizuki S, Yokoi H, Matsushime H, Furuichi K. Immunocyte Ca2+ influx system mediated by LTRPC2. Science. 2001;293:1327–1330. doi: 10.1126/science.1062473. [DOI] [PubMed] [Google Scholar]
- 101.Kolisek M, Beck A, Fleig A, Penner R. Cyclic ADP-ribose and hydrogen peroxide synergize with ADP-ribose in the activation of TRPM2 channels. Mol Cell. 2005;18:61–69. doi: 10.1016/j.molcel.2005.02.033. [DOI] [PubMed] [Google Scholar]
- 102.Kühn FJ, Lückhoff A. Sites of the NUDT9-H domain critical for ADP-ribose activation of the cation channel TRPM2. J Biol Chem. 2004;279:46431–46437. doi: 10.1074/jbc.M407263200. [DOI] [PubMed] [Google Scholar]
- 103.Smith MA, Herson PS, Lee K, Pinnock RD, Ashford ML. Hydrogen-peroxide-induced toxicity of rat striatal neurones involves activation of a non-selective cation channel. J Physiol (Lond) 2003;547:417–425. doi: 10.1113/jphysiol.2002.034561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Herson PS, Dulock KA, Ashford ML. Characterization of a nicotin-amide-adenine dinucleotide-dependent cation channel in the CRI-G1 rat insulinoma cell line. J Physiol (Lond) 1997;505(Pt1):65–76. doi: 10.1111/j.1469-7793.1997.065bc.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Yang KT, Chang WL, Yang PC, Chien CL, Lai MS, Su MJ, Wu ML. Activation of the transient receptor potential M2 channel and poly(ADP-ribose) polymerase is involved in oxidative stress-induced cardiomyocyte death. Cell Death Differ. 2006;13:1815–1826. doi: 10.1038/sj.cdd.4401813. [DOI] [PubMed] [Google Scholar]
- 106.Heiner I, Eisfeld J, Lückhoff A. Role and regulation of TRP channels in neutrophil granulocytes. Cell Calcium. 2003;33:533–540. doi: 10.1016/s0143-4160(03)00058-7. [DOI] [PubMed] [Google Scholar]
- 107.Baker ML, Serysheva II, Sencer S, Wu Y, Ludtke SJ, Jiang W, Hamilton SL, Chiu W. The skeletal muscle Ca2+ release channel has an oxidoreductase-like domain. Proc Natl Acad Sci USA. 2002;99:12155–12160. doi: 10.1073/pnas.182058899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Zima AV, Copello JA, Blatter LA. Effects of cytosolic NADH/NAD(+) levels on sarcoplasmic reticulum Ca(2+) release in permeabilized rat ventricular myocytes. J Physiol (Lond) 2004;555:727–741. doi: 10.1113/jphysiol.2003.055848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Cherednichenko G, Zima AV, Feng W, Schaefer S, Blatter LA, Pessah IN. NADH oxidase activity of rat cardiac sarcoplasmic reticulum regulates calcium-induced calcium release. Circ Res. 2004;94:478–486. doi: 10.1161/01.RES.0000115554.65513.7C. [DOI] [PubMed] [Google Scholar]
- 110.Sitsapesan R, McGarry SJ, Williams AJ. Cyclic ADP-ribose competes with ATP for the adenine nucleotide binding site on the cardiac ryano-dine receptor Ca(2+)-release channel. Circ Res. 1994;75:596–600. doi: 10.1161/01.res.75.3.596. [DOI] [PubMed] [Google Scholar]
- 111.Sitsapesan R, Williams AJ. Cyclic ADP-ribose and related compounds activate sheep skeletal sarcoplasmic reticulum Ca2+ release channel. Am J Physiol. 1995;268:C1235–C1240. doi: 10.1152/ajpcell.1995.268.5.C1235. [DOI] [PubMed] [Google Scholar]
- 112.Lee HC. Nicotinic acid adenine dinucleotide phosphate (NAADP)-mediated calcium signaling. J Biol Chem. 2005;280:33693–33696. doi: 10.1074/jbc.R500012200. [DOI] [PubMed] [Google Scholar]
- 113.Kim BJ, Park KH, Yim CY, Takasawa S, Okamoto H, Im MJ, Kim UH. Generation of nicotinic acid adenine dinucleotide phosphate and cyclic ADP-ribose by glucagon-like peptide-1 evokes Ca2+ signal that is essential for insulin secretion in mouse pancreatic islets. Diabetes. 2008;57:868–878. doi: 10.2337/db07-0443. [DOI] [PubMed] [Google Scholar]
- 114.Tugba Durlu-Kandilci N, Ruas M, Chuang KT, Brading A, Parrington J, Galione A. TPC2 proteins mediate nicotinic acid adenine dinucleo-tide phosphate (NAADP)- and agonist-evoked contractions of smooth muscle. J Biol Chem. 2010;285:24925–24932. doi: 10.1074/jbc.M110.129833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Macgregor A, Yamasaki M, Rakovic S, Sanders L, Parkesh R, Churchill GC, Galione A, Terrar DA. NAADP controls cross-talk between distinct Ca2+ stores in the heart. J Biol Chem. 2007;282:15302–15311. doi: 10.1074/jbc.M611167200. [DOI] [PubMed] [Google Scholar]
- 116.Esposito B, Gambara G, Lewis AM, Palombi F, D'Alessio A, Taylor LX, Genazzani AA, Ziparo E, Galione A, Churchill GC, Filippini A. NAADP links histamine H1 receptors to secretion of von Willebrand factor in human endothelial cells. Blood. 2011;117:4968–4977. doi: 10.1182/blood-2010-02-266338. [DOI] [PubMed] [Google Scholar]
- 117.Brailoiu GC, Gurzu B, Gao X, Parkesh R, Aley PK, Trifa DI, Galione A, Dun NJ, Madesh M, Patel S, Churchill GC, Brailoiu E. AcidicNAADP-sensitive calcium stores in the endothelium: agonist-specific recruitment and role in regulating blood pressure. J Biol Chem. 2010;285:37133–37137. doi: 10.1074/jbc.C110.169763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Aarhus R, Graeff RM, Dickey DM, Walseth TF, Lee HC. ADP-ribosyl cyclase and CD38 catalyze the synthesis of a calcium-mobilizing metabolite from NADP. J Biol Chem. 1995;270:30327–30333. doi: 10.1074/jbc.270.51.30327. [DOI] [PubMed] [Google Scholar]
- 119.Soares S, Thompson M, White T, Isbell A, Yamasaki M, Prakash Y, Lund FE, Galione A, Chini EN. NAADP as a second messenger: neither CD38 nor base-exchange reaction are necessary for in vivo generation of NAADP in myometrial cells. Am J Physiol Cell Physiol. 2007;292:C227–C239. doi: 10.1152/ajpcell.00638.2005. [DOI] [PubMed] [Google Scholar]
- 120.Cosker F, Cheviron N, Yamasaki M, Menteyne A, Lund FE, Moutin MJ, Galione A, Cancela JM. The ecto-enzyme CD38 is a nicotinic acid adenine dinucleotide phosphate (NAADP) synthase that couples receptor activation to Ca2+ mobilization from lysosomes in pancreatic acinar cells. J Biol Chem. 2010;285:38251–38259. doi: 10.1074/jbc.M110.125864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Zhu MX, Ma J, Parrington J, Calcraft PJ, Galione A, Evans AM. Calcium signaling via two-pore channels: local or global, that is the question. Am J Physiol Cell Physiol. 2010;298:C430–C441. doi: 10.1152/ajpcell.00475.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Calcraft PJ, Ruas M, Pan Z, et al. NAADP mobilizes calcium from acidic organelles through two-pore channels. Nature. 2009;459:596–600. doi: 10.1038/nature08030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Brailoiu E, Churamani D, Cai X, Schrlau MG, Brailoiu GC, Gao X, Hooper R, Boulware MJ, Dun NJ, Marchant JS, Patel S. Essential requirement for two-pore channel 1 in NAADP-mediated calcium signaling. J Cell Biol. 2009;186:201–209. doi: 10.1083/jcb.200904073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Dammermann W, Guse AH. Functional ryanodine receptor expression is required for NAADP-mediated local Ca2+ signaling in T-lymphocytes. J Biol Chem. 2005;280:21394–21399. doi: 10.1074/jbc.M413085200. [DOI] [PubMed] [Google Scholar]
- 125.Zhang F, Xu M, Han WQ, Li PL. Reconstitution of lysosomal NAADP-TRP-ML1 signaling pathway and its function in TRP-ML1(−/−) cells. Am J Physiol Cell Physiol. 2011;301:C421–C430. doi: 10.1152/ajpcell.00393.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Sumoza-Toledo A, Penner R. TRPM2: a multifunctional ion channel for calcium signalling. J Physiol (Lond) 2011;589:1515–1525. doi: 10.1113/jphysiol.2010.201855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Lin-Moshier Y, Walseth TF, Churamani D, Davidson SM, Slama JT, Hooper R, Brailoiu E, Patel S, Marchant JS. Photoaffinity labeling of nicotinic acid adenine dinucleotide phosphate (NAADP) targets in mammalian cells. J Biol Chem. 2012;287:2296–2307. doi: 10.1074/jbc.M111.305813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Billington RA, Thuring JW, Conway SJ, Packman L, Holmes AB, Genazzani AA. Production and characterization of reduced NAADP (nicotinic acid-adenine dinucleotide phosphate) Biochem J. 2004;378:275–280. doi: 10.1042/BJ20031284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Schomer B, Epel D. Redox changes during fertilization and maturation of marine invertebrate eggs. Dev Biol. 1998;203:1–11. doi: 10.1006/dbio.1998.9044. [DOI] [PubMed] [Google Scholar]
- 130.Gong J, Xu J, Bezanilla M, van Huizen R, Derin R, Li M. Differential stimulation of PKC phosphorylation of potassium channels by ZIP1 and ZIP2. Science. 1999;285:1565–1569. doi: 10.1126/science.285.5433.1565. [DOI] [PubMed] [Google Scholar]
- 131.Wang TA, Yu YV, Govindaiah G, Ye X, Artinian L, Coleman TP, Sweedler JV, Cox CL, Gillette MU. Circadian rhythm of redox state regulates excitability in suprachiasmatic nucleus neurons. Science. 2012;337:839–842. doi: 10.1126/science.1222826. [DOI] [PMC free article] [PubMed] [Google Scholar]