The zebrafish is emerging as a model organism for the safety assessment and hazard ranking of engineered nanomaterials. In this communication, we demonstrate the implementation of a roboticized high throughput screening (HTS) platform with automated image analysis to assess the impact of dissolvable oxide nanoparticles on embryo hatching. We further demonstrate that this hatching interference is mechanistically linked to an effect on the metalloprotease, ZHE 1, which is responsible for degradation of the chorionic membrane. Our data indicate that 4 of 24 metal oxide nanoparticles (CuO, ZnO, Cr2O3 and NiO) could interfere in embryo hatching by a chelator-sensitive mechanism that involves ligation of critical histidines in the ZHE1 center by the shed metal ions. We established a recombinant ZHE1 enzymatic assay to demonstrate that the dialysates from the same materials responsible for hatching interference also inhibit ZHE1 activity, in a dose-dependent fashion. A peptide-based BLAST search identified several additional aquatic species that express enzymes with homologous histidine-based catalytic centers, suggesting that the ZHE1 mechanistic paradigm could be used to predict the toxicity of a large number of oxide nanoparticles that pose a hazard to aquatic species. In summary, we have developed a mechanisms-based high throughput screening paradigm in zebrafish that can be used for hazard ranking of soluble metal oxide nanoparticles.
1. Introduction
Zebrafish is emerging as a model organism for safety assessment and hazard ranking of engineered nanomaterials.[1-9] Because the traditional toxicological evaluation of the different developmental stages involves time-consuming and labor-intensive technical procedures such as embryo selection, imaging, and phenotype assessment, the number of materials that can be assessed in any comparative analysis is restricted, thereby limiting the statistical power of the analysis as well as hampering the establishment of quantitative structure-activity relationships (QSARs). In order to take full advantage of this animal model, high throughput screening (HTS) platforms are required to increase the number of materials that can be evaluated in each comparative analysis.[10-13] We have previously demonstrated the utility of automated image analysis to speed up the hazard ranking of a limited number of transition metal oxide nanoparticles, but could not achieve HTS status because of the limitations imposed by the number of embryos that could be analyzed during manual embryo selection and plating.[14-15]
In addition to the need to increase the number of embryos to be analyzed, HTS also requires the use of robust toxicological mechanisms that can be used for the screening and for the development of SARs.[16-18] We have previously demonstrated that interference in zebrafish embryo hatching by a limited number of metal oxide nanoparticles is due to the shedding of metal ions and that the hatching interference could be reversed by chelators.[14, 19] The release of metal ions such as Cu2+ and Ni2+ to the chorion prompted us to hypothesize their interaction with metal-sensitive binding sites in the active center of the zebrafish hatching enzyme, ZHE1.[14] If proven to be correct, this molecular hypothesis should allow the introduction of a robust screening assay that can be used for the safety analysis of dissolvable oxide nanoparticles that are used by the semiconductor, catalyst and energy sectors. However, this hypothesis needs to be proven and requires predictive in vitro and in vivo assays that can be performed in a high throughput mode.
In this communication, we investigated whether recombinant ZHE1 (rec. ZHE1) could be used as a robust abiotic platform for the toxicological analysis of 24 metal oxide nanoparticles, as well as demonstrating that this paradigm is predictive of the materials’ effects on zebrafish embryos, in our newly established HTS platform. First, we developed an enzymatic assay that utilizes rec. ZHE1 and a fluorogenic substrate to show that selected metal oxides can interfere in enzyme activity based on their solubility. Second, we constructed and implemented a robotic pick and plate system for rapid suspension of zebrafish embryos, which could then be further analyzed by automated imaging. We demonstrate that interference in ZHE1 activity could correctly predict the oxide nanoparticles that lead to hatching interference, as assessed in the HTS assay. Moreover, a peptide-based homology search reveals that the evolutionary conserved histidine residues in the ZHE1 active center is also present in metalloproteases expressed in a large number of aquatic species. This study demonstrates the utility of HTS and a mechanisms-based toxicological approach to speed up safety assessment of dissolvable metal oxide nanoparticles.
2. Results
2.1. Use of recombinant ZHE1 to develop an abiotic assay that can be used for comparing interference in metalloprotease activity by metal oxide nanoparticles
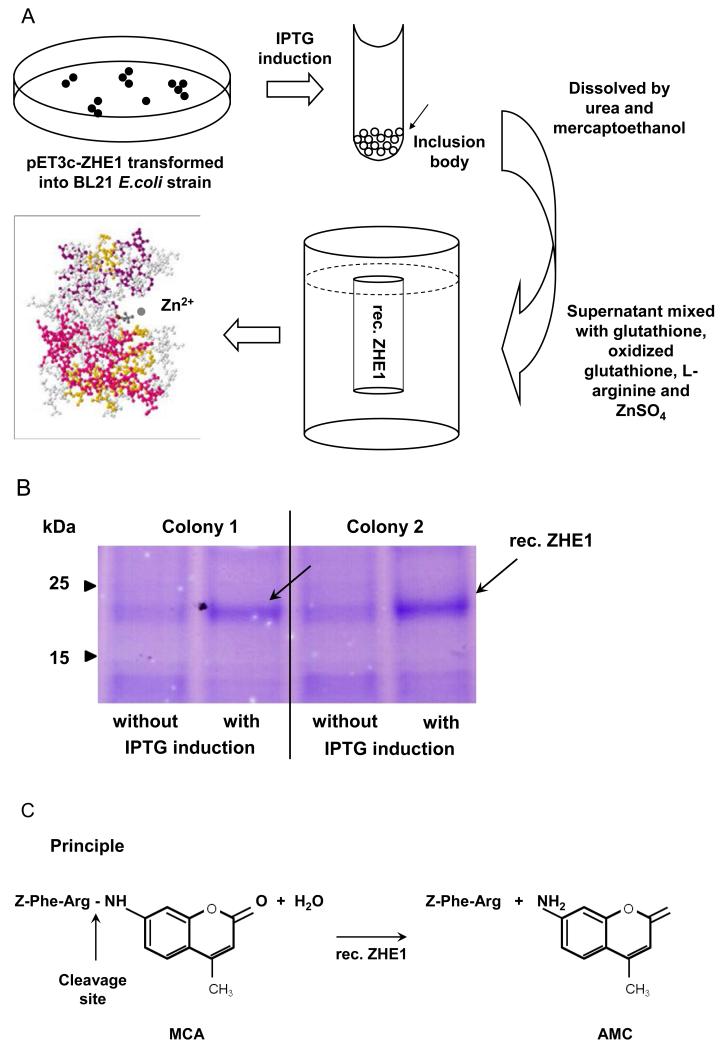
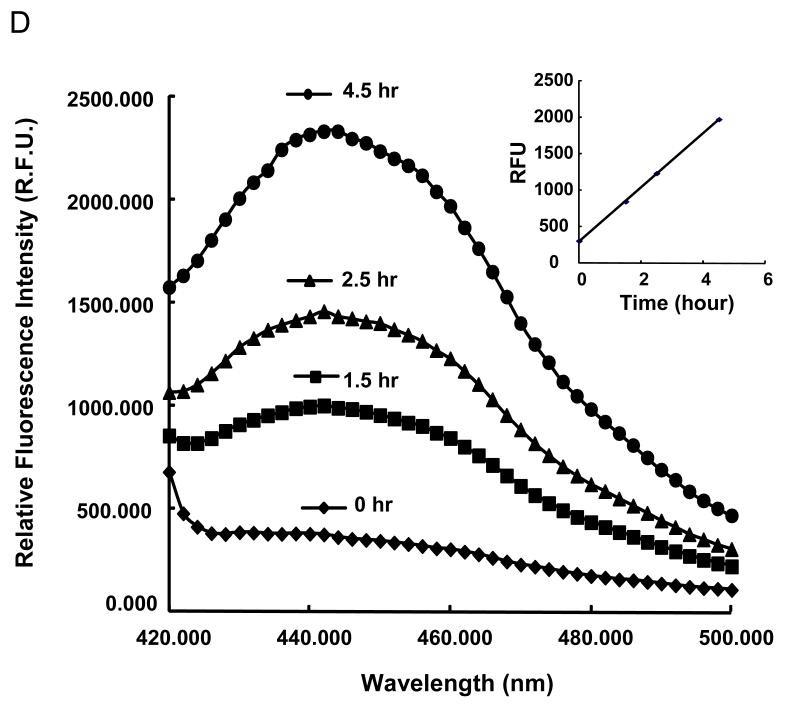
We have previously suggested that dissolution of ZnO nanoparticles and the concentration of Zn++ the chorionic sac constitutes the basis for interfering in embryo hatching due to an inhibitory effect on the metalloprotease, zebrafish hatching enzyme, ZHE1.[20-21] Purified rec. ZHE1 was prepared by expressing the coding sequence in a pET3c vector in the Escherichia coli strain, BL21. The stepwise approach for the purification of recombinant enzyme is summarized in Figure 1A and discussed in details in Experimental section. ZHE1 expression was induced by isopropyl thio-β-D-galactoside (IPTG) in randomly selected E coli. colonies. Following purification through Ni-NTA Superflow beads and elution by imidazole in a Tris–HCl buffer, the collected material was subjected to SDS/PAGE gel electrophoresis. The stained gel showed a single band at 23 kDa, which corresponds to the molecular weight of ZHE1 (Figure 1B).[21-22] The enzyme activity of the recombinant enzyme was further compared against natural ZHE1 by using a bioassay to look at the protein fragments generated during the proteolytic degradation of the chorion; this demonstrated the appearance of proteolytic products with the same molecular weight profile (not shown). To develop the abiotic assay, we used a peptidyl-4-methylcoumaryl-7-amides (Z-Phe-Arg-MCA) substrate, which is achieved by ZHE 1 (Figure 1C) to release the amino methyl cumarin (AMC) fragment that can be quantified by spectrophotometric analysis at 460 nm (Figure 1D).[23-24] This allowed us to demonstrate recombinant enzyme activities of 1.04 and 1.24 nmole/mg*minin Tris buffer and Holtfreter’s medium, respectively (Figure S1B). Holtfreter’s medium is used for the culture of intact zebrafish embryos.
Figure 1. Generation of rec. ZHE1 and development of the abiotic assay.
(A) Schematic to describe the essential steps in generating active rec. ZHE1. A full length ZHE1 cDNA copy was obtained by PCR amplification and subsequently cloned into the pET3 expression vector the E. coli expression system (BL21). After thio-ß-D-galactoside (IPTG) induction, the bacterial cells were harvested and the inclusion bodies dissolved in denaturing solution to collect the rec. ZHE1. rec. ZHE1 was obtained by subsequent refolding, dialysis and purification through Ni-NTA Superflow. (B) SDS/PAGE gel electrophoresis demonstrates the 23 kD rec. ZHE1 protein. The IPTG inducible expression was consistent for randomly chosen E. coli colonies, as indicated by black arrows. (C) Principle of the abiotic colorimetric assay based on a peptidyl-4-methylcoumaryl-7-amides (MCA) substrate (Z-Phe-Arg-MCA). During digestion, rec.ZHE1 cleaves the peptide bond between Arginine (Arg) and MCA, as indicated by the arrow. As a result, the non-fluorescent MCA motif is converted to fluorescent amino methyl cumarin (AMC). (D) The fluorescence spectrum was monitored real-time to reflect the kinetics of enzyme digestion. The enzymatic activity was subsequently calculated based on the rate of increase of the fluorescence intensity at 460 nm (insert).
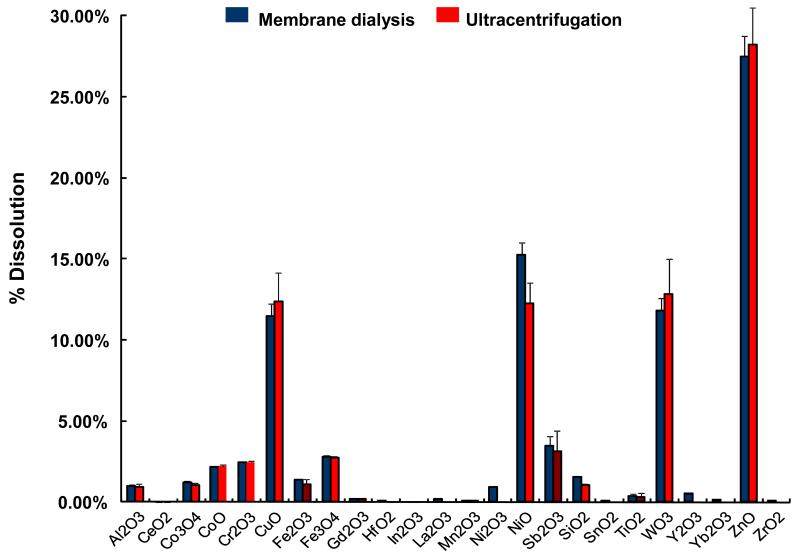
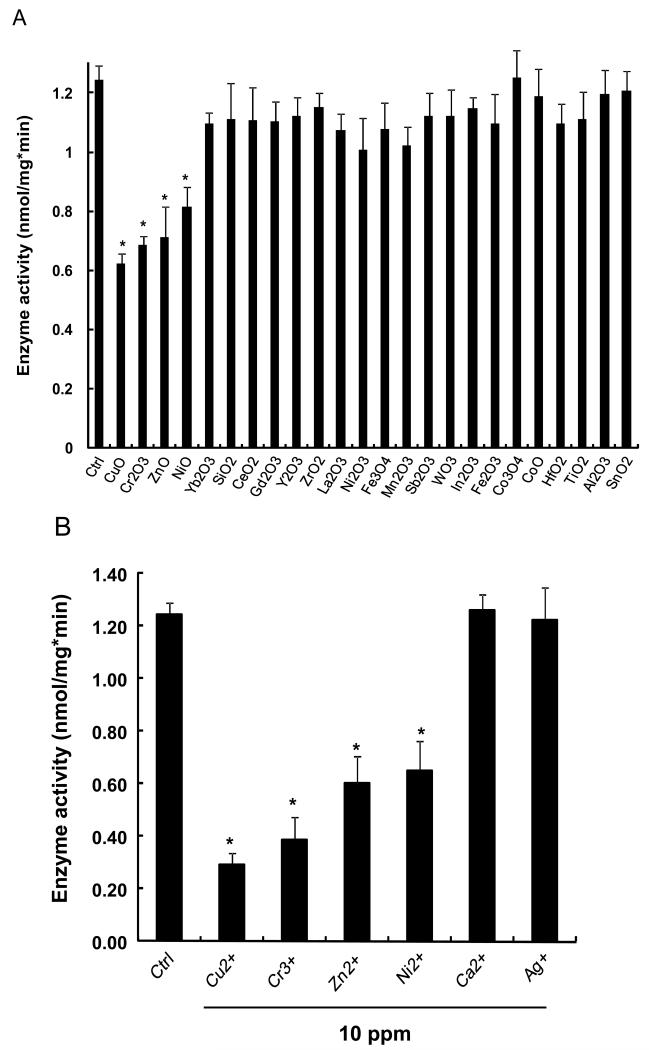
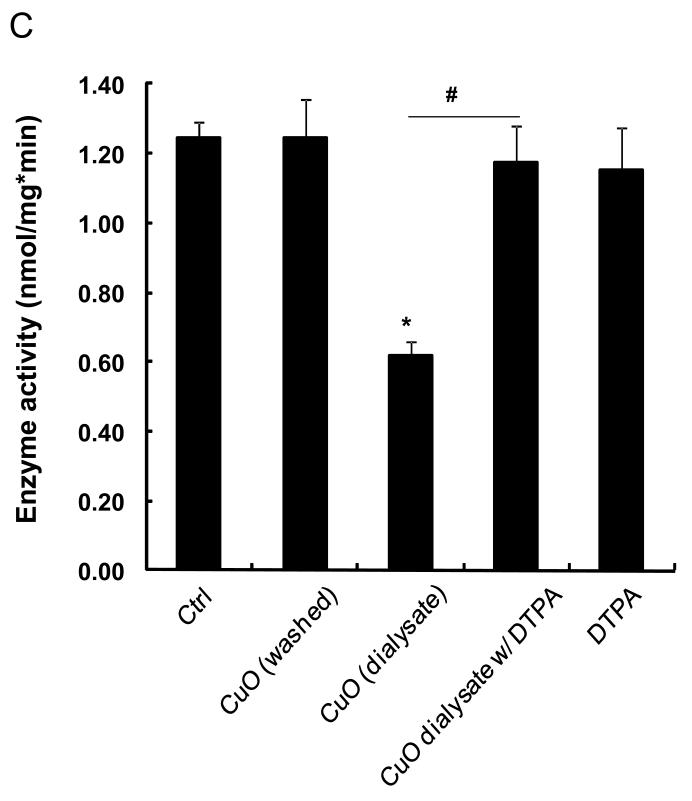
The abiotic assay was used subsequently to assess the effects of 24 metal oxide nanoparticles that are representative of commonly used materials in the semiconductor, catalysis and energy industries. The majority of these materials were acquired from commercial sources but a few (CuO, Co3O4, Fe3O4, Sb2O3, TiO2, WO3, and ZnO) were also synthesized in-house by flame spray pyrolysis.[25] The primary particle sizes, as determined by transmission electron microscopy (TEM), including their hydrodynamic size distribution and surface charge in Holtfreter’s medium (Table 1). The data demonstrate that the hydrodynamic size of these materials in Holtfreter’s medium (pH 7.0), supplemented with 100 μg/mL alginate to improve particle dispersal, was in the range of 200-500 nm, with a few materials (Al2O3, Fe3O4, Ni2O3, SnO2, Y2O3, Yb2O3) exhibiting a size of of 500 nm. The ζ-potentials of all the materials were negative (−20 to −30 mV) as a result of alginate coating. In the first analysis, we collected particle dialysates and supernatants (following ultracentrifugation) from the nanoparticle suspensions to quantify the % material dissolution over a 48 hour time period. Both methods resulted in comparable dissolution profiles for the materials as shown in Figure 2. The dialysates, suspended at a concentration of 50 ppm, were then used to conduct abiotic ZHE1 assays. As shown in Figure 3A, the metal ions released from four of these materials (CuO, ZnO, Cr2O3 and NiO) significantly interfered in ZHE1 activity acording to a ranking order CuO>Cr2O3≥ZnO>NiO. In order to confirm that the inhibition was due to shed metal ions, we also measured the effect of soluble metal salts on enzyme activity. This confirmed that ZHE1 activity was significantly affected by Cu2+, Zn2+, Cr3+ and Ni2+ salt solutions, but no inhibitory effect was seen with Ca2+ and Ag+ salt solutions (Figure 3B). Moreover, we demonstrated that metal chelation with DTPA could prevent the interference in ZHE1 activity. This is demonstrated for nano-Cu in Figure 3C. To rule out a direct effect of the nanoparticle on the enzyme activity, CuO nanoparticles were washed to remove shed ions prior to repeating the abiotic assay; this demonstrated that ZHE1 activity was not directly affected by the particles (Figure 3C). Other than the 4 specified materials, the rest of the oxide nanoparticles or their supernatants had no effect on ZHE1 activity. In summary, the above data provide direct demonstration that shedding of metal ions by dissolvable metal oxide nanoparticles interferes in rec. ZHE1 activity as previously suggested in our ZnO studies.[19] Based on this result, we predicted that CuO, ZnO, Cr2O3 and NiO nanoparticles should be the materials that interfere in embryo hatching.
Table 1. Primary Size, Hydrodynamic Size and Zeta-potential of 24 Metal Oxide Nanoparticles.
| Number | MOx | Primary Size (nm)a |
Hydrodynamic Size (nm)b |
Zeta-potential (mV)c |
|---|---|---|---|---|
| 1 | Al2O3 | 14.7±5.2 | 524.8 ± 32.8 | −24.0 ± 0.5 |
| 2 | CeO2 | 12.8 ± 3.4 | 321.3 ± 8.6 | −28.9 ± 3.3 |
| 3 | CoO | 18.3 ± 6.8 | 378.3 ± 16.4 | −25.5 ± 1.3 |
| 4 | Co3O4 | 10.0 ± 2.4 | 247.6 ± 16.9 | −29.0 ± 2.2 |
| 5 | Cr2O3 | 71.8 ±16.2 | 478.5 ± 7.2 | −26.2 ± 3.1 |
| 6 | CuO | 193.0 ± 90.0 | 289.5 ± 31.0 | −26.9 ± 0.8 |
| 7 | Fe2O3 | 12.3 ± 2.9 | 385.2 ± 6.3 | −24.1 ± 2.0 |
| 8 | Fe3O4 | 12.0 ± 3.2 | 831.7 ± 41.8 | −27.0 ± 2.3 |
| 9 | Gd2O3 | 43.8 ± 15.8 | 726.7 ± 54.8 | −34.7 ± 0.7 |
| 10 | HfO2 | 28.4 ± 7.3 | 349.9 ± 5.2 | −24.3 ± 2.1 |
| 11 | In2O3 | 59.6 ± 19.0 | 303.2 ± 5.2 | −35.5 ± 2.4 |
| 12 | La2O3 | 24.6 ± 5.3 | 471.2 ± 20.9 | −27.8 ± 0.6 |
| 13 | Mn2O3 | 51.5±7.3 | 525.9 ± 7.8 | −30.9 ± 0.4 |
| 14 | NiO | 13.1 ± 5.9 | 277.5 ± 23.0 | −23.1 ± 2.0 |
| 15 | Ni2O3 | 140.6 ± 52.5 | 665.8 ± 46.4 | −24.4 ± 2.2 |
| 16 | Sb2O3 | 11.8 ± 3.3 | 459.9 ± 22.7 | −25.8 ± 0.9 |
| 17 | SiO2 | 13.5 ± 4.2 | 374.9 ± 29.0 | -16.8 ± 2.0 |
| 18 | SnO2 | 62.4 ± 13.2 | 635.0 ± 52.0 | −26.4 ± 0.3 |
| 19 | TiO2 | 12.6 ± 4.3 | 497.0 ± 17.1 | −31.5 ± 1.4 |
| 20 | WO2 | 16.6 ±4.3 | 511.9 ± 19.4 | −23.3 ± 1.1 |
| 21 | Y2O3 | 32.7 ±8.1 | 594.5 ± 33.0 | −27.6 ± 0.4 |
| 22 | Yb2O3 | 61.7 ± 11.3 | 682.6 ± 56.2 | −29.7 ± 0.5 |
| 23 | ZnO | 22.6 ± 5.1 | 379.0 ± 11.0 | −27.0 ± 1.1 |
| 24 | ZrO2 | 40.1 ± 12.6 | 384.4 ± 25.0 | -19.7 ±3.6 |
Primary size of the particles in their dry state was obtained by transmission electron microscopy (JEOL, 1200 EX).
Hydrodynamic size was determined by high throughput dynamic light scattering (HT-DLS, Dynapro Plate Reader, Wyatt Tech).
Particle ζ-potential was measured using ZetaPALS (Brookhaven Instruments, Holtsville, NY). Introduction of the nanoparticles in Holtfreter’s medium (pH 7.0) did not significantly change the medium pH in spite of the dissolution of metal oxide nanoparticles.
Figure 2. Dissolution characteristics of 24 metal oxide nanoparticles using membrane dialysis and ultracentrifugation methods.
Nanoparticle suspensions (50 ppm) in Holtfreter’s medium supplemented with 100 μg/mL were used for dissolution characterization. For membrane dialysis, nanoparticle suspensions were dialyzed in a dialysis membrane with 3500 Dalton MWCO pore size against Holtfreter’s medium over 48 hours at 28.5 °C. The dialysates were quantified by ICP-MS. For ultracentrifugation, the nanoparticle suspensions were kept at 28.5 °C for 48 hours and followed by centrifugation at 20,000g for 1 hour. The supernatants were quantified by ICP-MS. Both methods resulted in comparable weight percentage dissolution for all the nanoparticles.
Figure 3. Enzyme activity of rec. ZHE1 measured by abiotic colorimetric assay.
(A) ZHE1 activity was significantly decreased by the dialysates from CuO, Cr2O3, ZnO and NiO nanoparticles suspended at 50 ppm in the dialysis bag. Dialysates from the rest of the materials had no effect. The dialysates were mixed with 100 μM of MCA substrate and 310 ng/mL rec. ZHE1 at 28.5 °C, followed by real-time measurement of the increase in fluorescence intensity at 460 nm (ex: 380 nm) for 1 hour. The enzyme activity was subsequently calculated based on rate of increase in fluorescence intensity. (B) Change of ZHE1 activity was assessed by incubating the recombinant enzyme in 10 ppm Cu2+, Cr3+, Zn2+, Ni2+, Ca2+ and Ag+ salt solution, showing that while Cu2+, Cr3+, Zn2+, Ni2+ had an inhibitory effect (p<0.05), Ca2+ and Ag+ had no effect. (C) Enzyme activity of rec. ZHE1 in response to washed CuO, CuO dialysate, CuO co-exposed with DTPA, and DTPA alone. The decreased enzymatic activity was only observed for the CuO dialysates (p<0.05). Co-exposure with DTPA prevented the decline in enzyme activity.
2.2. HTS in intact embryos confirms the prediction of the abiotic assay
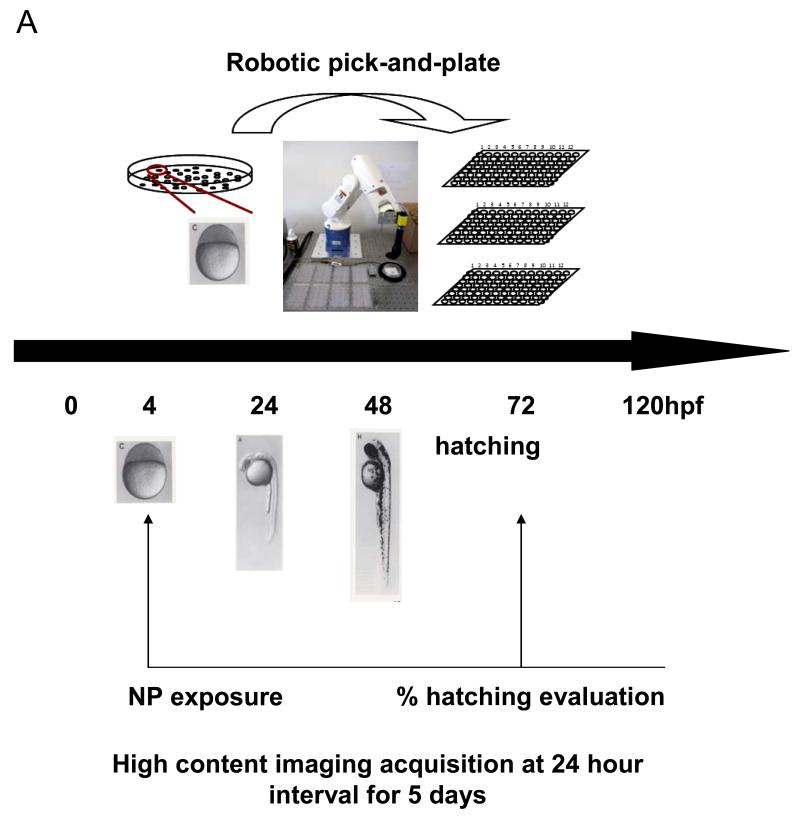
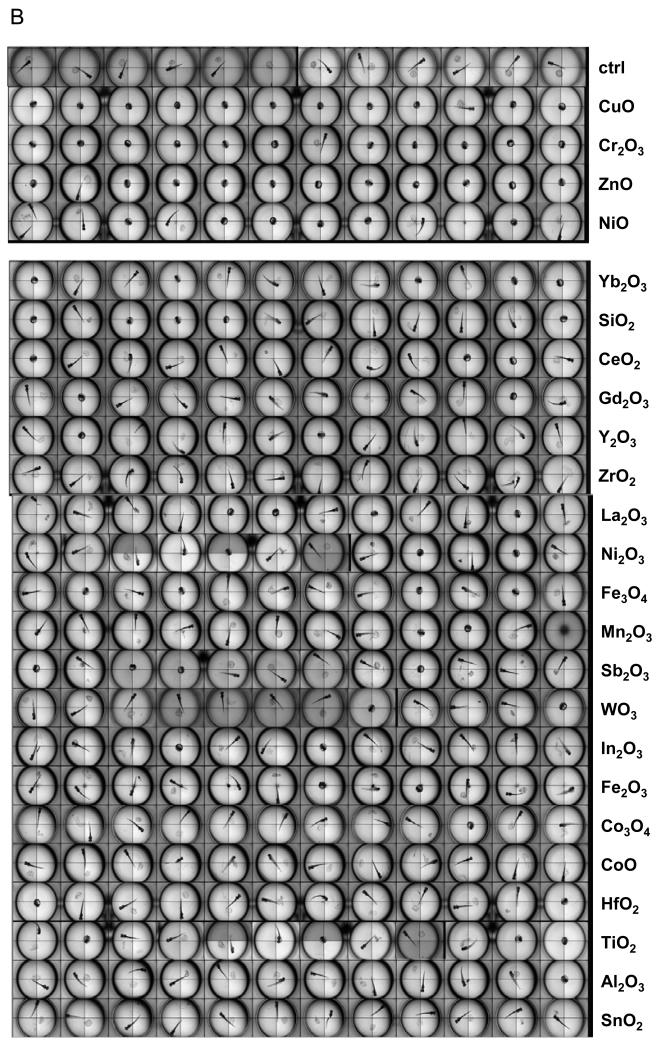
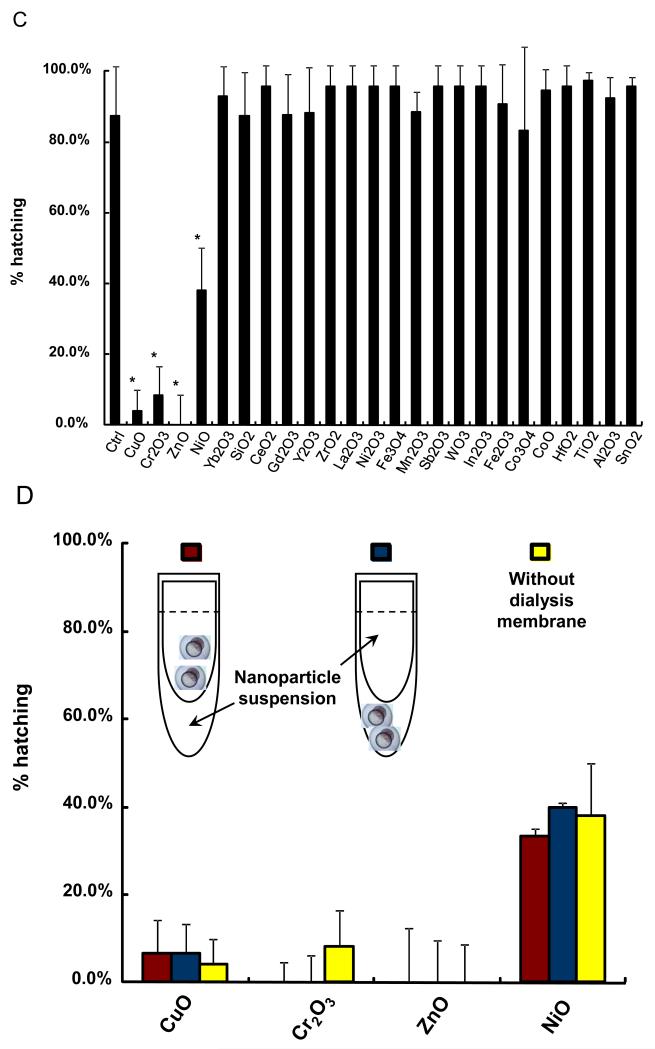
In order to demonstrate the predictive value of the abiotic assay for each of the metal oxides, it was necessary to develop a zebrafish HTS assay to handle the screening of 24 materials in a single experiment. A major limitation of our existing automated image analysis system has been the number of embryos (and incubation plates) that could be processed in a single experiment due to the labor intensity of handpicking and transferring the embryos. We therefore assembled a vision-guided robotic system to perform the pick-and plate-process, thereby speeding up embryo selection. Full details of the system appear online and a picture of the system is provided in Figure 4A and S2. High content bright field image analysis was added to the automated picking process, allowing the embryos to be screened every 24 hours following the introduction of the metal oxide nanoparticles in Holtfreter’s medium (Figure 4A). The particles, which were added at final concentrations of 0.05, 0.5, 5, 25, 50, 100, and 200 ppm, aggregated to a similar extent as described in Table 1. The fully integrated HTS platform allowed us to screen all the materials in a single experiment. Representative four-quadrant images of the 96 well plates demonstrating the across-the-board interference in embryo hatching by CuO, ZnO, Cr2O3 and NiO but not other nanoparticles (Figure 4B). In contrast, the rest of the materials had no effect. Statistical analysis of 5 replicate experiments confirmed that the hatching interference by the aforementioned materials was highly significant (p<0.05). Moreover, the in vivo results agree with the abiotic data, showing correlation coefficent of 0.92 (Pearson’s correlation test, p-value: 1.433e-10). Confirmation of the role of metal ions in the hatching interference was provided by introducing the nanoparticles to the culture system in dialysis membranes. This demonstrated that in spite of physical separation from the target, the dissolving materials still exerted effects on embryo hatching (Fig. 4D). We also demonstrated that hatching interference could be restored by including DTPA in the culture medium (not shown).
Figure 4. High throughput screening in intact zebrafish embryos.
(A) Schematic to depict the approach to high throughput screening in zebrafish embryos. Zebrafish embryos at 2 hpf were washed and placed in a petri-dish. A robotic system was used to pick and dispense individual embryos into the wells of 96-well transparent, U-bottom plates. The exposure of nanoparticles suspensions started at 4 hpf and was carried out up to 5 days. The observation on hatching interference, morphological abnormalities and mortality was facilitated by high content imaging that automatically captures the bright-field image for each well at 24 hour intervals. (B) The representative four-quadrant high content images of zebrafish embryos exposed to 50 ppm for each of the 24 metal oxide nanoparticles. Compared to control embryos (first row), four (ZnO, CuO, Cr2O3 and NiO) of 24 materials resulted in significant hatching interference. (C) Statistical analysis of the hatching rate interference in response to the metal oxide nanoparticles. The hatching interference exerted by ZnO, CuO, Cr2O3 and NiO was statistically significant (p<0.05) compared to control embryos. (D) The hatching interference was not altered by physically separating the nanoparticles from embryos using a dialysis membrane. Zebrafish embryos were indirectly exposed to CuO, Cr2O3, ZnO and Cr2O3 by including the nanoparticle suspensions in dialysis bags made from a membrane with 3500 Dalton MWCO size (~5 nm). There was no difference at the level of hatching interference with and without the dialysis membrane.
The ability to perform HTS allowed us to analyze the minimum CuO, ZnO, Cr2O3 and NiO concentrations that could generate hatching interference. Thus, calculation of the lowest observed hatching interference effect demonstrated and exposure amount of 0.5 ppm for CuO, 5 ppm for ZnO and Cr2O3, and 50 ppm for NiO, respectively (Figure S3). Moreover, the in vivo ranking of the material effects agreed with the ranking obtained in the abiotic assay. In summary, the statistical agreement of in vitro with in vivo screening results establishes a predictive toxicological paradigm that can be used to quantitatively assess the toxicity of soluble metal oxide nanoparticles.
2.3. Peptide homology search for related metalloproteases in aquatic organisms
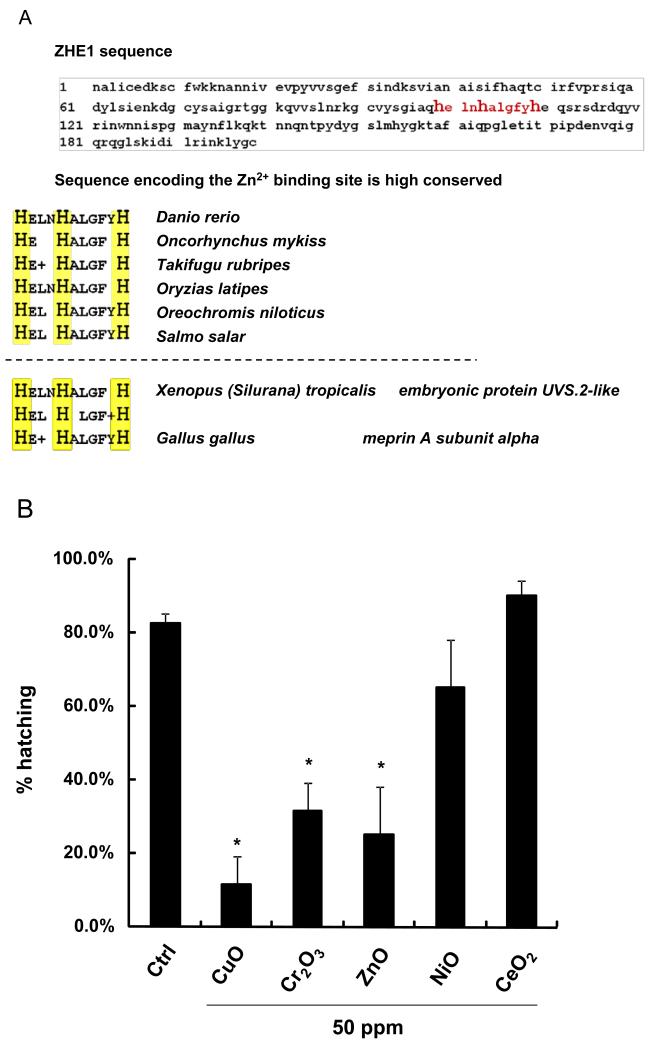
In addition to the utility of ZHE1 to be used as a predictive platform for the effect of soluble oxide nanoparticles in zebrafish embryos, the metal-sensitive catalytic center of the enzyme could be useful for the safety analysis of other aquatic species. Performance of a BLAST homology search, using the primary peptide sequence of ZHE1, demonstrated > 500 hits for proteins containing an identical histidine-based core sequence sequence in the catalytic center (Figure 5A). Most of these homologs could be classified as zinc metalloproteases or belonging to the astacin protein family. Importantly, the histidine-based metal ion binding site is highly conserved in a number of fish hatching enzymes, including in the Japanese medaka (Oryzias latipes), Atlantic salmon (Salmo salar) and Rainbow trout (Oncorhynchus mykiss) (Figure 5A). This could signify that dissolving metal nanoparticles entering the habitat of these fish could also impact their hatching. This prediction was confirmed by screening Japanese medaka embryos, in which we could confirm that CuO, ZnO and Cr2O3 nanoparticles (but not NiO) interfered in embryo hatching (Figure 5B). Interference in medaka hatching required significantly higher nanoparticle concentrations than zebrafish embryos, suggesting that different species may have different sensitivities to dissolving metal oxides.[26] Nonetheless, the results of our BLAST search demonstrate the potential utility of the ZHE1 toxicological paradigm for assessing the environmental impact of dissolving oxide nanoparticles.
Figure 5. The evolutionary conserved sequence responsible for Zn2+ ligation in the ZHE1 active center is also expressed in a wide range of environmental species.
(A) An excerpt from the BLAST search for homologous peptide sequences corresponding to the active center of ZHE1. The search resulted more than 500 species (such as Oncorhynchus mykiss and Oryzias latipes) sharing similar histidine liganding domains (highlighted in yellow). (B) The hatching rates of medaka embryos exposed to metal oxide nanoparticles (CuO, Cr2O3, ZnO, NiO and CeO2) at 50 ppm. Statistically significant hatching interference was observed for CuO, Cr2O3 and ZnO (p<0.05). Five replicates of twelve medaka embryos were exposed to nanoparticles suspension at concentrations of 0.05, 0.5, 5, and 50 ppm. The hatching rate of medaka embryos was assessed at 7 dpf.
3. Discussion
In this communication, we demonstrate the development of a predictive toxicological paradigm in which we used an abiotic colorimetric ZHE1 assay to forecast which among a number of dissolvable metal oxide nanoparticles could lead to hatching interference in zebrafish embryos. Moreover, we demonstrate the development of a HTS approach that allowed us to assess 24 metal oxide nanoparticles in a single zebrafish experiment. The integrated data show that while ZnO, CuO, Cr2O3 and NiO lead to ZHE1 inactivation and hatching interference, the rest of the materials did not exert similar effects, either as a result of low solubility or the inability of the shed metal ions to interact with the metal-binding histidines in the enzyme center. The expression of homologous metalloproteases or members of the astacin family of proteins in other aquatic species suggests that this predictive toxicological paradigm could be useful to perform hazard ranking on metal oxides in several exposed environments.
Predictive toxicology approaches have been implemented to identify potential hazardous nanomaterials from a human health perspective. For instance, the assessment of nanomaterials to generate oxidative stress or pro-fibrogenic effects in vitro has proven to be predictive for the in vivo toxicological outcome in the lung, including the generation of pulmonary inflammation and fibrosis.[27-30] The emergence of similar predictive toxicity platforms for aquatic organisms remains scarce, however. Thus, this study makes an important contribution in demonstrating that interference in the function of ZHE1 constitutes a predictive, mechanistic platform to assess the effect dissolving metal oxide nanoparticles on zebrafish embryo hatching. The value of this predictive platform was further enhanced by our ability to develop this into a HTS assay that can be used to confirm the results of the abiotic assay in vivo. This constitutes one of the earliest attempts to establish a predictive toxicological paradigm for HTS analysis of nanomaterials in an aquatic organism.
The current study demonstrates, for the first time, a definitive molecular mechanism for hatching interference by metal oxide nanoparticles, i.e. the inactivation of the ZHE1 hatching enzyme by shed metal ions. We show that there is an excellent correlation between the ability of CuO, ZnO, Cr2O3 and NiO to inhibit ZHE1 activity and exert hatching interference at the organism level. Moreover, the use of ZHE1 activity allows us to establish a property-activity relationship that can use the solubility of the material as well as the chemistry of the shed ions to understand how histidine ligation in the enzyme center could lead to toxicological outcomes. Interestingly, dissolution alone did not define toxicity among the materials with moderate (CoO, Cr2O3, Fe3O4 and Sb2O3) or high (CuO, NiO, WO3 and ZnO) dissolution potential, indicating that the chemistry of the metal ions is important role in the structure-activity relationship. ZHE1 is a zinc metalloprotease in which the active Zn2+ binding site includes three histidine residues.[20-22] This ligand configuration also has the ability to bind to Cu2+, Ni2+, and Cr3+.[31-33] Therefore, it is reasonable to hypothesize that the shed metal ions from CuO, NiO and Cr2O3 nanoparticles inhibit ZHE1 activity through substitution of the Zn2+ ion in the active enzyme center. The ionic radii of these metal ions as well as their ion-ligand coordination chemistry and binding constants could determine Zn2+ substitution in the enzyme center. This could involve a conformational change that interferes in enzymatic activity. This notion is further supported by studies showing that overdosing of Zn2+ or substituting Zn2+ with Cu2+ or Ni2+ in zinc metalloproteases can reduce/diminish the enzyme activity.[34-37]
In addition to predicting the impact of metal oxide nanoparticles in zebrafish embryo hatching, the ZHE1 platform may have wider implications based on the widespread presence of homologous metalloproteases across fish, aquatic organisms and even humans. As demonstrated in another fish model, Japanese medaka, which contains an evolutionary conserved ZHE1-like hatching enzyme, embryo hatching could be affected by CuO, ZnO and Cr2O3 nanoparticles. Although this hatching interference required higher concentrations of the materials than in zebrafish embryos, the results indicate that the inhibitory effect of shed metal ions also apply to homologous metalloproteases that contained a Zn2+ ligation site in their center. Taken together, the ZHE1 paradigm could be applied to other aquatic species and therefore shows promise as a screening tool for metal oxide toxicity and regulatory decision-making.
It is important to clarify that the ZHE1 predictive platform is specific to the metal ion shedding characteristics of the nanoparticles under study and is not intended to address the possibility that metal oxide nanoparticles could also exert toxicological effects through their surface characteristics or mechanisms other than ion shedding. In contrast to metal oxide nanoparticles, Ag nanoparticles cause a moderate degree of hatching interference in zebrafish embryos, which is accompanied by a high rate of mortality.[5, 7, 38] Interestingly, Ag ions do not interfere in rec. ZHE1 activity (Figure 3B), suggesting that the hatching interference by nano-Ag involves another injury mechanism, as further demonstrated by the high rate of morphological abnormalities and mortality in response to this material.[5, 7, 38] Thus, the ability of metal oxide nanoparticles to interfere in ZHE1 activity is only appropriate for the development of property-activity relationships that relate to interference in embryo hatching secondly to nanoparticle dissolution and ion release to the chorionic sac. It is possible that additional mechanisms exist by which ions and non-dissolved metal oxide nanoparticles could lead to toxicological outcomes. One example is the activation of stress-related pathways as demonstrated by the use of transgenic hsp70:eGFP zebrafish larvae.[14]
The development of a HTS platform for zebrafish embryos further advances safety screening of engineered nanomaterials. By making use of robotics, vision-recognition equipment and precise liquid handing tools for the automation of embryo pick-and-placement, our screening capabilities now allows the screening of large number of nanomaterials, which is in step with the rate of growth of nanotechnology. The increased use of automation in developing zebrafish HTS for related research purposes further demonstrated the utility of this organism for studying important biological problems.[39-41]
Finally, it is important to address the shortcomings of our study. First, the exposure scenario in this study does not reflect the complexity of real-life aquatic exposures. Environmental media contain a large number of components that could influence the agglomeration, fate and transport as well as dissolution of oxide nanoparticles. While our study attempted to obtain the best possible particle dispersion in the experimental Holtfreter’s medium, Table 1 demonstrates that some nanoparticle aggregation still occurred and could influence the dissolution rate and the dynamics of metal ion accumulation in the chorionic sac. Second, only pristine nanoparticles were studied and do not address the more complicated physicochemical state of commercial nano-enabled products as well as the chemical speciation of the shed metal ions in the environment. Nonetheless, we envisage that our HTS analysis will be useful for the initial hazard ranking of potentially dissolvable metal oxide nanoparticles and our study methods could then be used to assess and supplement real-life exposures, such as the ecosystems studies being carried out in freshwater and estuarine ecosystems in the University of California Center for the Environmental Implications of Nanomaterials (UC CEIN). Moreover, we also envisage that already exposed aquatic samples obtained from the real-life exposures can be introduced into our abiotic and zebrafish HTS analysis to make predictions about the possible contribution of shed metal ions in those environments. The current HTS paradigm for the toxicological analysis of metal oxide nanoparticles premised on dissolution characteristics supplements the recent development of a toxicological paradigm for the same class of materials in which material bandgap have been shown to play a role in pulmonary toxicity.[28] While it is also possible that oxidative stress effects might develop in the zebrafish embryo, we did not screen for this possibility in the current study.
4. Conclusion
We have successfully demonstrated a predictive toxicological paradigm that can be used for the safety assessment of dissolving metal oxide nanoparticles in an aquatic environment. The excellent correlation between ZHE1 inactivation and hatching interference in intact embryos delineate the molecular mechanism of hatching interference exerted by CuO, ZnO, Cr2O3 and NiO nanoparticles. This predictive toxicological paradigm, based on ligation of the ZHE1 enzyme center by specific metal ions, has wider implications for safety assessment of engineered nanomaterials across a wide spectrum of aquatic organisms. The emergence of a zebrafish HTS platform could speed up environmental and regulatory decision-making.
5. Experimental section
Materials and Reagents
The MCA substrate (Cat # 24096) was from AnaSpec, Inc. and was dissolved in DMSO at 10 mM before aliquoting and storing at −20 °C. The metal oxide nanoparticles library was assembled by commercial acquisition as well as in-house flame spray pyrolysis (CuO, Co3O4, Fe3O4, Sb2O3, TiO2, WO3, and ZnO) as previously described by us.[27]
Physicochemical characterizations of nanoparticles
Nanoparticles, originally obtained in powder form, were dispersed in Holtfreter’s medium (supplemented with 100 ppm alginate) by sonication to yield a stock solution of 5×103 ppm as previously described.[19] The primary particles sizes were determined by transmission electron microscopy (TEM), while the suspended particles were also used for assessment of hydrodynamic size as well as measurement of the ζ-potential. TEM samples were prepared by placing a drop of nanoparticle dispersion (25 ppm) on a carbon-coated TEM grid and waiting for the water to evaporate. Images were acquired at 80 kV accelerating voltage (JEOL 1200 EX) and the primary size was analyzed by NIH ImageJ software. The hydrodynamic size of the nanoparticles, dispersed in Holtfreter’s medium, was determined by high throughput dynamic light scattering (HT-DLS, Dynapro Plate Reader, Wyatt Technology) as described by Ji et al.[42] A ZetaPALS instrument (Brookhaven Instruments, Holtsville, NY) was used to measure the electrophoretic mobility of nanoparticles from which the ζ-potential was derived using the Helmholtz-Smoluchowski equation. The dissolution characteristics of these nanoparticle suspensions were analyzed by inductively coupled plasma mass spectroscopy (ICP-MS), employing two methods, membrane dialysis and ultracentrifugation. For membrane dialysis, the nanoparticle suspensions were added to dialysis membranes with 3500 Da molecular weight cutoff (MWCO) pore size (equivalent to 5 nm) and dialyzed against Holtfreter’s medium for 48 hours at 28.5 °C. The dialysates were collected for ICP-MS analysis. For ultracentrifugation, the nanoparticle suspensions were kept at 28.5 °C for 48 hours and centrifuged for 1 hour at 20,000g as previously described. The supernatants were collected for ICP-MS analysis.
Generation of rec. ZHE1
Active rec. ZHE1 was generated using the pET3c as an expression vector in the Escherichia coli expression system (BL21).[20] First, a full length cDNA copy of ZHE1 was synthesized by PCR, using a forward primer, 5′-CATATGAATGCTCTCATCTGCGAGGACA-3′, containing a 5′ NdeI site and start codon, and a reverse primer, 5′-GGATCCTAGTGATGGTGATGGTGGCATCCATACAGCTTATTGATCC-3′, containing a tail encoding five histidine residues and a BamHI restriction site. The PCR product was subsequently cloned into the pET3c expression vector (Agilant Technologies) through the NdeI and BamHI sites. The pET3c-ZHE1 construct thus obtained was transformed into E. coli strain BL21 (DE3) (Agilant Technologies, CA). A single colony from the transformation was grown in 25 ml LB broth (containing 100 μg/ml ampicillin) overnight in a shaking incubator. The next morning, 12 ml of the overnight culture was added to 800 ml LB broth containing 100 μg/ml of ampicillin and the mixture incubated at 37 °C. Following growth expansion until a A600 of 0.6 was reached, isopropyl thio-ß-D-galactoside (IPTG) was added to the culture medium to a final concentration of 1 mM. Following incubation at 37 °C for an additional 4 hour, bacterial cells were harvested by centrifugation (3200 rpm for 10 minutes), transferred to a 50 ml tube, and frozen at −20 °C. The frozen cells were suspended in 10 mL lysis buffer (50 mM NaPO4, 100 mM NaCl, 10 mM imidazole, pH 7.0) supplemented with protease inhibitors. Triton X-100 was added to the lysates to a final concentration of 1%. The lysates were sonicated four times (20 seconds each) with a one-minute interval between each sonication, followed by centrifugation at 4 °C (12,500 rpm for 20 minutes). To recover the rec. ZHE1 protein from the inclusion bodies, the pellet was dissolved in 3 mL denaturing solution (8 M urea, 0.1 M 2-mercaptoethanol, 50 mM Tris–HCl buffer, pH 8.0) and incubated at 37 °C for 1 h. After centrifugation (12,500 rpm for 20 minutes), the supernatant was collected and 50% was added to 300 mL refolding solution (1 mM glutathione, 0.1 mM oxidized glutathione, 0.8 M L-Arginine, 5 μM ZnSO4, 50 mM Tris–HCl buffer, pH 8.0). The refolding mixture was kept at 4 °C for 48 hours and subsequently dialyzed twice (4 hours then overnight) with 4 L of cold 50 mM Tris–HCl buffer (pH 8.0) at 4 °C. After dialysis, the refolding mixture was filtered through presterilized Durapore 0.22 μm Filter (Millipore), distributed into six 50 mL tubes, and further purified using Ni-NTA Superflow (QIAGEN). In short, 250 μL of Ni-NTA Superflow beads were washed four times with 50 mM Tris–HCl buffer (pH 8.0), resuspended in 220 μL of 50 mM Tris–HCl buffer (pH 8.0), and added to each tube of filtered mixture. After a 1.5 hour-incubation at 4 °C, the mixture was applied to a poly-prep gravity column (Bio-Rad). Subsequently, rec. ZHE1 proteins were eluted from the beads with 10 mL of 50 mM Tris–HCl buffer (pH 8.0) containing 0.15 M NaCl and 0.4 M imidazole.
Digestion of zebrafish chorions by rec. ZHE1
Chorions were removed with tweezers from unhatched embryos at 4, 24 and 48 hpf and washed three times with 50 mM Tris–HCl buffer (pH 7.5). Twelve chorions from each group were incubated in 50 μl of 50 mM Tris–HCl (pH 7.5) containing 1 μg of rec. ZHE1 at 30 °C for 1 hour, 2 hours and 3 hours, respectively. For controls, twelve chorions from each group and twelve chorions collected immediately following natural hatching were incubated at 30 °C for 3 hours in 50 μl of 50 mM Tris–HCl (pH 7.5) without rec. ZHE1. The resulting mixtures were then subjected to SDS / PAGE analysis and the gel was stained with Coomassie Brilliant Blue R-250 (EM Science).
Abiotic enzyme assay
The abiotic enzyme assay was performed in 96-well plates in 100 uL of solution in 3 replicate wells. This solution consisted of nanoparticle dialysates (or metal salt solutions), MCA substrate (100 uM) and rec. ZHE1 (~310 ng/mL). Wells receiving substrate only, enzyme only or dialysates (or metal salt solution) only served as negative controls. The incremental increase in fluorescence intensity at 460 nm (excited at 380 nm) was measured every 2 minutes for 1 hour using a microplate spectrophotometer (SpectraMax M5, Molecular Probes). The enzyme activity was further calculated based on the increase in fluorescence intensity according to a calibration curve (Figure S5), which converts fluorescence intensity (relative fluorescence unit, RFU) to the amount of substrate consumed (uM). This calibration curve was constructed by plotting the fluorescence intensities against increasing amount of AMC (the fluorescent compound after enzyme digestion).
Zebrafish care and spawning
Wild type adult zebrafish (Danio rerio) were housed and maintained in the UCLA zebrafish facility on a 14:10D photoperiod. Two pairs of male/female fish were placed in a single cage a day ahead of time and released in the next morning to trigger spawning. The embryos were collected at 2 hpf and rinsed with Holtfreter’s solution to remove any residue on the embryo surface. The embryos were subsequently examined under a stereomicroscope (Zeiss, Stemi 2000) for viability and developmental stage before being selected and distributed by the robotic system. All procedures were carried out in accordance with the Animal Care and Use Committee guidelines at UCLA.
Embryo screening and high content imaging
Healthy embryos at the same developmental stage (2 hpf) were collected and washed three times in Holtfreter’s medium. The embryos were then placed in a petri-dish and robotically picked up and plated into 96-well transparent, U-bottom plates, dispersing one embryo into each well. One hundred microliters of each nanoparticle suspension at working concentrations of 0.05, 0.5, 5, 25, 50, 100, and 200 ppm was added to the wells at 4 hpf. To achieve robust statistical calculation, five replicate trials were carried out, each using 12 embryos. The observations on hatching interference, morphological abnormalities and mortality were carried out every 24 h for five consecutive days through the use of bright-field high content imaging (ImageXpress), as described previously.[14]
Supplementary Material
Acknowledgements
This work is supported by the National Science Foundation and the Environmental Protection Agency under Cooperative Agreement Number DBI 0830117. Any opinions, findings, conclusions or recommendations expressed herein are those of the author(s) and do not necessarily reflect the views of the National Science Foundation or the Environmental Protection Agency. This work has not been subjected to an EPA peer and policy review. Key support was provided by the US Public Health Service Grants (RO1 ES016746 and U19 ES019528). We acknowledge Dr. Shane Que Hee, School of Public Health, University of California, Los Angeles, for the ICP-MS analysis, and Linda Dong, Department of Molecular, Cell and Developmental Biology, University of California, Los Angeles, for the support on zebrafish care and spawning.
Footnotes
Additional figures describing the fluorescence spectra of MCA substrate, quantification of enzyme activity in different medium conditions, a picture of the robotic pick-and-plate platform, dose-dependent hatching interference, embryo survival rate and calibration curve for enzyme activity unit conversion as described in the text. This material is available from the Wiley Online Library or from the author.
References
- [1].Fako VE, Furgeson DY. Adv. Drug Deliver. Rev. 2009;61:478–486. doi: 10.1016/j.addr.2009.03.008. [DOI] [PubMed] [Google Scholar]
- [2].Heiden TC, Dengler E, Kao WJ, Heideman W, Peterson RE. Toxicol. Appl. Pharmacol. 2007;225:70–79. doi: 10.1016/j.taap.2007.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Griffitt RJ, Luo J, Gao J, Bonzongo JC, Babber D. Environ. Toxicol. Chem. 2008;27:1972–1978. doi: 10.1897/08-002.1. [DOI] [PubMed] [Google Scholar]
- [4].Griffitt RJ, Weil R, Hyndman KA, Denslow ND, Powers K, Taylor D, Barber DS. Environ. Sci. Technol. 2007:8178–8186. doi: 10.1021/es071235e. [DOI] [PubMed] [Google Scholar]
- [5].George S, Xia T, Rallo R, Zhao Y, Ji Z, Lin S, Wang X, Zhang H, France B, Schoenfeld D, Damoiseaux R, Liu R, Lin S, Bradley KA, Cohen Y, Nel AE. ACS Nano. 2011;5:1805–1817. doi: 10.1021/nn102734s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Harper SL, Carriere JL, Miller JM, Hutchison JE, Maddux BLS, Tanguay RL. ACS Nano. 2011;5:4688–4697. doi: 10.1021/nn200546k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Asharani PV, Lian Wu Y, Gong Z, Valiyaveettil S. Nanotechnology. 2008;19:255102. doi: 10.1088/0957-4484/19/25/255102. [DOI] [PubMed] [Google Scholar]
- [8].Zhu X, Zhu L, Li Y, Duan Z, Chen W, Alvarez PJJ. Environ. Toxicol. Chem. 2007;26:976–979. doi: 10.1897/06-583.1. [DOI] [PubMed] [Google Scholar]
- [9].Browning LM, Lee KJ, Huang T, Nallathamby PD, Lowman JE, Xu XH. Nanoscale. 2009;1:138–152. doi: 10.1039/b9nr00053d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Vogt A, Codore H, Day BW, Hukriede NA, Tsang M. JoVE. 2010 doi: 10.3791/1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Vogt A, Cholewinski A, Shen X, Nelson SG, Lazo JS, Tsang M, Hukriede NA. Dev. Dyn. 2009;238:656–663. doi: 10.1002/dvdy.21892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Burns CG, Milan DJ, Grande EJ, Rottbauer W, MacRae CA, Fishman MC. Nat. Chem. Biol. 2005;1:263–264. doi: 10.1038/nchembio732. [DOI] [PubMed] [Google Scholar]
- [13].Love DR, Pichler FB, Dodd A, Copp BR, Greenwood DR. Curr. Opin Biotechnol. 2004;15:564–571. doi: 10.1016/j.copbio.2004.09.004. [DOI] [PubMed] [Google Scholar]
- [14].Lin S, Zhao Y, Xia T, Meng H, Ji ZX, Liu R, George S, Xiong SJ, Wang X, Zhang HY, Pokhrel S, Madler L, Damoiseaux R, Lin S, Nel AE. ACS Nano. 2011;5:7284–7295. doi: 10.1021/nn202116p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Liu R, Lin S, Rallo R, Zhao Y, Damoiseaux R, Xia T, Lin S, Nel A, Cohen Y. PloS One. 2012;7:e35014. doi: 10.1371/journal.pone.0035014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Meng H, Xia T, George S, Nel AE. ACS Nano. 2009;3:1620–1627. doi: 10.1021/nn9005973. [DOI] [PubMed] [Google Scholar]
- [17].Liu R, Rallo R, George S, Ji ZX, Nair S, Nel AE, Cohen Y. Small. 2011;7:1118–1126. doi: 10.1002/smll.201002366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Clark KA, White RH, Silbergeld EK. Regul. Toxicol. Pharmacol. 2011;59:361–363. doi: 10.1016/j.yrtph.2011.02.002. [DOI] [PubMed] [Google Scholar]
- [19].Xia T, Zhao Y, Sager T, George S, Pokhrel S, Li N, Schoenfeld D, Meng H, Lin S, Wang X, Wang M, Ji Z, Zink JI, Madler L, Castranova V, Lin S, Nel AE. ACS Nano. 2011;5:1223–1235. doi: 10.1021/nn1028482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Sano K, Inohaya K, Kawaguchi M, Yoshizaki N, Iuchi I, Yasumasu S. Febs J. 2008;275:5934–5946. doi: 10.1111/j.1742-4658.2008.06722.x. [DOI] [PubMed] [Google Scholar]
- [21].Okada A, Sano K, Nagata K, Yesumasu S, Ohtsuka J, Yamamura A, Kubota K, Iuchi I, Tanokura M. J Mol Biol. 2010;402:865–878. doi: 10.1016/j.jmb.2010.08.023. [DOI] [PubMed] [Google Scholar]
- [22].Okada A, Nagata K, Sano K, Yasumasu S, Kubota K, Ohtsuka J, Iuchi I, Tanokura M. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2009;65:1018–1020. doi: 10.1107/S1744309109033016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Morita T, Kato H, Iwanaga S, Takada K, Kimura T, Sakakibara S. J. Biochem. 1977;82:1495–1498. doi: 10.1093/oxfordjournals.jbchem.a131840. [DOI] [PubMed] [Google Scholar]
- [24].Lisk G, Pain M, Gluzman IY, Kambhampati S, Furuya T, Su XZ, Fay MP, Goldberg DE, Desai SA. Antimicrob. Agents Ch. 2008;52:2346–2354. doi: 10.1128/AAC.00057-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Teoh WY, Amal R, Madler L. Nanoscale. 2010;2:1324–1347. doi: 10.1039/c0nr00017e. [DOI] [PubMed] [Google Scholar]
- [26].Cameron IL, Hunter KE. J. Exp. Zool. 1984;231:447–454. doi: 10.1002/jez.1402310320. [DOI] [PubMed] [Google Scholar]
- [27].Xia T, Zhao Y, Sager T, George S, Pokhrel S, Li N, Schoenfeld D, Meng H, Lin S, Wang X, Wang M, Ji Z, Zink JI, Madler L, Castranova V, Lin S, Nel AE. ACS Nano. 2011;5:1223–1235. doi: 10.1021/nn1028482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Zhang H, Ji Z, Xia T, Meng H, Low-Kam C, Liu R, Pokhrel S, Lin S, Wang X, Liao Y, Wang M, Li L, Rallo R, Damoiseaux R, Telesca D, Madler L, Cohen Y, Zink JI, Nel AE. ACS Nano. 2012;6:4349–4368. doi: 10.1021/nn3010087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Wang X, Xia T, Addo Ntim S, Ji Z, Lin S, Meng H, Chung CH, George S, Zhang H, Wang M, Li N, Yang Y, Castranova V, Mitra S, Bonner JC, Nel AE. ACS Nano. 2011;5:9772–9787. doi: 10.1021/nn2033055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Wang X, Xia T, Duch MC, Ji Z, Zhang H, Li R, Sun B, Lin S, Meng H, Liao YP, Wang M, Song TB, Yang Y, Hersam MC, Nel AE. Nano Lett. 2012;12:3050–3061. doi: 10.1021/nl300895y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Remko M, Fitz D, Rode BM. Amino Acids. 2010;39:1309–1319. doi: 10.1007/s00726-010-0573-8. [DOI] [PubMed] [Google Scholar]
- [32].Hu P, Loo JA. J. Am. Chem. Soc. 1995;117:11314–11319. [Google Scholar]
- [33].Hoggard PE. Inorg. Chem. 1981;20:415–420. [Google Scholar]
- [34].Kooi C, Subsin B, Chen R, Pohorelic B, Sokol PA. Infect.Immun. 2006;74:4083–4093. doi: 10.1128/IAI.00297-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Gomis-Ruth FX, Grams F, Yiallouros I, Nar H, Kusthardt U, Zwilling R, Bode W, Stocker W. J. Biomed. Chem. 1994;269:17111–17117. [PubMed] [Google Scholar]
- [36].Fukasawa KM, Hata T, Ono Y, Hirose J. J. Amino Acids. 2011;2011:574816. doi: 10.4061/2011/574816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Abbasi SA, Nipaney PC, Soni R. Int. J. Environ. Stud. 1985;24:107–114. [Google Scholar]
- [38].Powers CM, Yen J, Linney EA, Seidler FJ, Slotkin TA. Neurotoxicol. Teratol. 2010;32:391–397. doi: 10.1016/j.ntt.2010.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Pardo-Martin C, Chang T-Y, Koo BK, Gilleland CL, Wasserman SC, Yanik MF. Nat. Methods. 2010;7:634. doi: 10.1038/nmeth.1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Peravali R, Gehrig J, Giselbrecht S, Lutjohann DS, Hadzhiev Y, Muller F, Liebel U. BioTechniques. 2011;50:319–324. doi: 10.2144/000113669. [DOI] [PubMed] [Google Scholar]
- [41].Wang WH, Liu XY, Sun Y. IEEE Int.Conf. Robot. Autom. 2008 [Google Scholar]
- [42].Ji Z, Jin X, George S, Xia T, Meng H, Wang X, Suarez E, Zhang H, Hoek EMV, Godwin H, Nel AE, Zink JI. Environ. Sci. Technol. 2010;44:7309–7314. doi: 10.1021/es100417s. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.