Abstract
The ABO histo-blood group, first discovered over a century ago, is found not only in humans but also in many other primate species, with the same genetic variants maintained for at least 20 million years. Polymorphisms in ABO have been associated with susceptibility to a large number of human diseases, from gastric cancers to immune or artery diseases, but the adaptive phenotypes to which the polymorphism contributes remain unclear. We suggest that variation in ABO has been maintained by frequency-dependent or fluctuating selection pressures, potentially arising from co-evolution with gut pathogens. We further hypothesize that the histo-blood group labels A, B, AB, and O do not offer a full description of variants maintained by natural selection, implying that there are unrecognized, functionally important, antigens beyond the ABO group in humans and other primates.
Keywords: ABO, balancing selection, evolution, host-pathogen interaction, population genetics, primates
Long-term maintenance of the ABO histo-blood group in primates
The ABO histo-blood groups, encoded by the A, B, and O alleles at the ABO gene 1, was the first polymorphism to be discovered in humans. Genetic diversity at the ABO gene is unusually high, suggesting that distinct blood groups have persisted due to balancing selection, a form of adaptation that maintains diversity in a species in the face of genetic drift (the chance fluctuations in allele frequencies that occur in finite populations). Why ABO blood groups might be under balancing selection has been debated for close to a century 2.
Strikingly, A and B are both found in at least 17 other primate species (see Fig. 1A), and the genetic differences between the A and B alleles consist of the same two amino acid changes in exon 7 of ABO 3,4. In contrast, there are a number of distinct loss-of-function (O) alleles, which are not shared among species 5. We recently showed that the A/B polymorphism emerged at least around 20 millions years ago and persisted in some primate species until the present 6. Notably, humans and gibbons inherited A and B types from a common ancestor at the origin of apes 6. The maintenance of a polymorphism for that long is exceedingly unlikely by chance alone, providing compelling evidence that variants in ABO have been maintained by ancient balancing selection and thus must have important effects on individual fitness 7.
Figure 1.
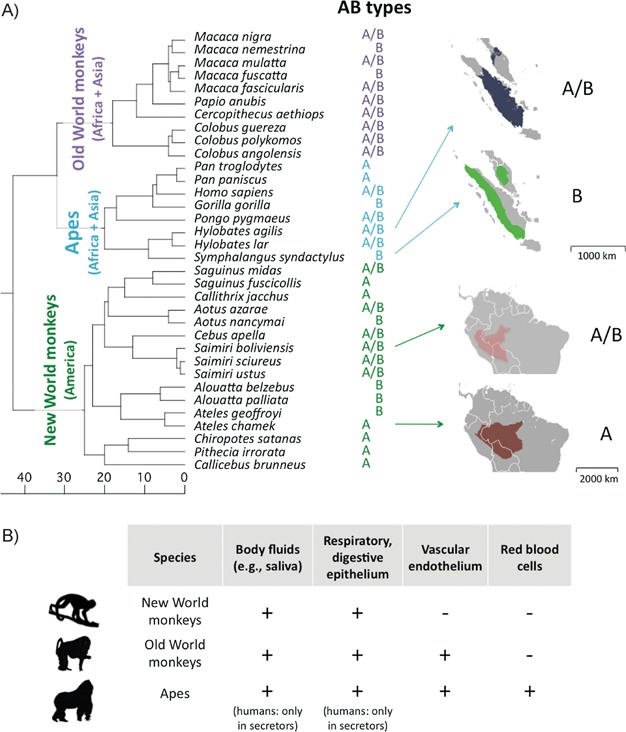
A: Phylogenetic information about the A/B polymorphism for primate species in which it has been characterized (see 6 and references therein), along side two examples of overlapping geographical ranges for pairs of species that differ in their ABO phenotype. The scale is in Millions of years. Geographical ranges are from the IUCN Red List maps (http://www.iucnredlist.org/). B: Expression pattern of ABO in different tissues and primate species 22.
Other examples of ancient balancing selection in primates include the major histocompatibility complex (MHC), which plays a critical role in immune response 8, and the opsin polymorphism in New World Monkeys that underlies trichromatic color vision 9. In contrast to these two canonical cases, the adaptive phenotype to which ABO contributes is less clear 10,11. It was originally suggested that ABO was under selection because of its protective role with regard to fetal-maternal Rhesus incompatibility 12. Since then, ABO variation has been associated with susceptibility to a large number of human diseases, from gastric cancers to immune or artery diseases 10,11,13. However, because these associations correspond to multiple, potentially unrelated phenotypes, it remains unknown which of them are responsible for the persistence of ABO types in multiple primate species. Here, we suggest that variation in ABO is maintained by frequency-dependent or fluctuating selection, possibly in response to gut pathogens, and that there exists functionally important cryptic variation in the gene yet to be uncovered.
Fluctuating selection in response to gut pathogens?
Balancing selection is often equated with heterozygote advantage, typified by the (evolutionarily young) sickle cell polymorphism in humans 14. ABO variation in primates is unlikely to be maintained by this mechanism, however, given that there exist haplotypes encoding the AB phenotype (“cis-AB” alleles), which are fixed in Mus musculus for instance 15, and yet these are found only at very low frequencies in humans and have not been reported in other primates 6. More generally, heterozygote advantage is thought to represent a transient solution that can be relatively rapidly resolved by the evolution of greater phenotypic plasticity or by duplication 16, as appears to have happened at least twice for the opsin polymorphism 9.
Genetic variation can also be maintained in the population by negative frequency-dependent selection, in which rare types have a fitness advantage (as in self-incompatibility loci in plants 17). One scenario by which this might occur, proposed for ABO 18, is when pathogens exploit specific host proteins to initiate infection and train on the more common types in the population. Host and pathogen co-evolution can also lead to the maintenance of variation when it induces temporally fluctuating selective pressures, as can arise when there is an interaction between the genotypes of the host and that of the pathogen and both virulence and resistance are costly 21. Consistent with these models, many of the well-characterized examples of long-term balancing selection are related to host immunity (e.g. 19).
For ABO specifically, multiple lines of evidence suggest that host-pathogen interactions are responsible for the maintenance of the polymorphism. First, variation in ABO antigens has been associated with susceptibility to a number of infectious diseases 10,13, and an interaction between ABO types and specificity of binding has been found in strains of Norwalk virus 20. Second, the composition of Helicobacter pylori appears to have evolved in response to changes in human ABO histo-blood group frequencies: the frequency of strains able to bind to the A blood group is greatly decreased in the Native Amerindian populations that are fixed for O 21. Thus, at least some of the conditions for frequency-dependent or fluctuating selection arising from host-pathogen co-evolution appear to be met.
The phylogenetic distribution of ABO provides additional hints about the source of balancing selection pressures. In apes, ABO antigens are expressed at the surface of red blood cells and on the vascular endothelium as well as in body fluids, mucus secretions and various epithelial tissues (in humans, only in “secretor” individuals who carry an intact FUT2; see Fig. 1B). In contrast, in Old World Monkeys, ABO antigens are absent on red blood cells, and in New World Monkeys, they are also absent from the vascular endothelium (see Fig. 1B 22). This observation strongly suggests that the balancing selection pressures did not arise from the presence of ABO antigens on blood cells alone, and for example, that the influence of ABO on rosetting 23, the binding of red blood cells infected by Plasmodium falciparum to uninfected cells, could not explain the ABO polymorphism outside of apes. The adaptive phenotype must be due instead, at least originally, to its more ancestral expression pattern on the surface of epithelial cells. Notably, in all primates, ABO antigens are present on the digestive tract, which is an important site of infection, e.g. for H. pylori and Norwalk virus. Interestingly, H. pylori is known to infect macaques and New World Monkeys 24,25. Thus, the interaction between variation at ABO and gut pathogens could impose a shared selective pressure among primates.
Also enlightening are findings about B4galnt2 in mice. A cis-regulatory region of this gene appears to be under long-term balancing selection, with the two highly diverged haplotypes controlling a tissue-specific switch between expression in gut and blood 26. Intriguingly, variation in this regulatory region is associated both to the presence of Helicobacter species in the mice gut 27 and to VWF levels in the blood (a protein involved in blood clotting), two phenotypes also associated with ABO histo-blood groups in humans 11,21. These parallels seem unlikely to be purely coincidental, and suggest that the association with Helicobacter species – or a trade-off between roles in different tissues – may be important in the maintenance of variation at ABO.
Unrecognized variation of functional relevance?
Another potentially informative phylogenetic pattern is the loss of ABO histo-blood groups in some species. Among 41 primate species for which data are available, 10 species do not present the B allele/phenotype and 11 do not present the A allele/phenotype 6. In apes, notably, chimpanzees and bonobos lack B, while gorillas lack A. The differences among species could reflect the loss of A, B, or O by chance (i.e. genetic drift) if they have undergone a marked reduction in population size 28. For example, although the variants in the MHC have been maintained for millions of years in mammals (and other vertebrates), MHC variability is greatly decreased in species that have experienced strong, recent bottlenecks 29. To test this possibility, we examined whether primates with smaller effective population sizes (as measured by putatively neutral diversity levels) tend to have lost A or B. For the 11 species for which reliable genetic diversity estimates were available 30, there is no discernable correlation (phylogenetic least-square regression, p-value = 0.28).
Another explanation for the loss of ABO types might be that species face different selective pressures, for example because of differences in pathogen community composition. While this hypothesis seems sensible, the loss of allelic classes occurred in locations in which other species have maintained all ABO histo-blood groups: for example, Symphalangus syndactylus and Hylobates agilis are in sympatry on the Sumatra island and the Malay peninsula, yet one is fixed for B and the second presents both A and B (see Fig. 1A). A similar observation holds for Ateles chamek and Saimiri boliviensis, which are both found in parts of Brazil, Bolivia, and Peru (with the important caveat that they may occupy different ecological niches within those geographic areas; Fig. 1A).
A third (not mutually exclusive) hypothesis is that there are more allelic classes at ABO than the three commonly defined A, B, and O, so that natural selection might actually be maintaining a larger number of variants as part of a multi-allelic balanced polymorphism. In that regard, we note that the A, B, AB, and O blood groups are categories defined based on hemaglutination patterns after mixing of blood. Given that shared selective pressures among primate species cannot be the result of the presence of ABO on red blood cells, there is no reason to assume that the A, B, and O labels fully describe the spectrum of variants distinguished by natural selection. Thus, species apparently monomorphic for one category, e.g. for the A class, may actually be harboring variation among A alleles of functional importance. If so, we would misclassify these species as monomorphic and underestimate the number of relevant functional classes. In support of this hypothesis, functional variation is known to exist within histo-blood types in humans: for instance, A1 and A2 alleles, while equivalent for transfusion purposes, differ in quality and quantity of antigens 31. These sub-groups have been shown to have an effect on levels of VWF 11, but tend not to have been tested systematically in studies of disease phenotypes, so that we know little about their effects on immune or other phenotypes. Intriguingly, these phenotypic sub-groups have also been observed in chimpanzees, gibbons, and orangutans 32.
Additional support for the third hypothesis comes from population genetic analyses: surveys of variation at ABO in humans have revealed unusually old variants not only in exon 7, where changes distinguish A and B types, but also in exon 4 and intron 1 (see Fig. 2); these polymorphisms are not in linkage disequilibrium with those in exon 7, raising the possibility of additional targets of ancient balancing selection along the ABO gene 33. Moreover, we recently discovered that two polymorphisms around intron 4 of ABO are found in both humans and chimpanzees 34 (see Fig. 2) and appear to be old 33,34 and unrelated to the balancing selection acting on exon 7 (unpublished simulation results). This sharing between humans and chimpanzees is unexpected if the only functionally important variation distinguishes A and B types, as chimpanzees lack the B type and therefore should not share ancestral polymorphisms with humans. Similarly, in exon 7, there is a non-synonymous variant (position 703, Gly235Ser) shared between humans, orangutans, and gibbons, species that are all polymorphic for A and B, as well as with gorillas, which lack A 6 (see Fig. 2). In humans, alleles with the Gly at this site on a B background have reduced B activity and small amounts of A activity (B(A) allele) 31, suggesting that some gorillas may in fact have small levels of A activity rather than being fixed for B. Regulatory variation near ABO may also be important: notably, differences in the number of repeats bound by the CCAAT-binding factor NF-Y have been associated with ABO expression differences in humans 31 and polymorphisms for the number of binding motifs are also found in chimpanzees (Thompson and Ober, personal communication). Thus, the variation patterns across species point to currently unrecognized polymorphisms of selective (and hence functional) importance in ABO.
Figure 2.
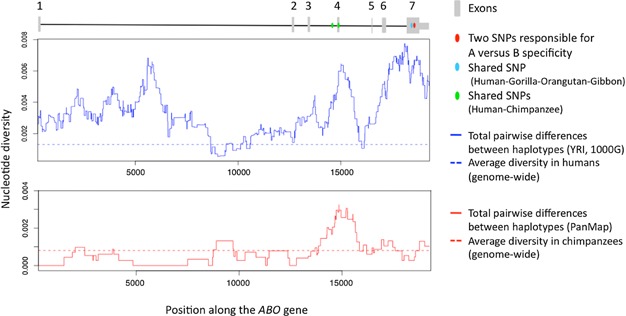
Structure of ABO and nucleotide diversity in humans (top) and chimpanzees (bottom) for sliding windows of 1 kb, using data for Yoruban individuals in the 1000 Genomes Project 35 and data for Western chimpanzees from the PanMap project 36. The location of the molecular changes distinguishing A and B types are indicated, as are a subset of the polymorphisms shared between ape species. The average genome-wide diversity is shown for YRI and for Western chimpanzees 30, respectively.
Future directions
The evolutionary history of ABO indicates that balancing selection has maintained a polymorphism at this locus for many millions of years, and hence that these variants are important to the fitness of humans and other primates. The mechanism of balancing selection is yet still unknown, but more likely to be fluctuating or frequency-dependent selection than heterozygote advantage. The adaptive phenotypes to which ABO contributes are also unclear, but its phylogenetic distribution strongly suggests that they do not stem from its role in blood alone but rather could be due to shared gut pathogens. This consideration, in turn, implies that the histo-blood group categories (A, B, AB, and O) may not fully describe the variation in ABO antigens, and raises the possibility of a larger number of allelic classes of relevance for natural selection. The case of ABO thus illustrates how the analysis of evolutionary pressures can help to reveal variation of biological importance.
The evolutionary analyses also serve to motivate further functional studies. For example, genetic variation data for the entire ABO gene in additional primates (notably New World Monkeys) would allow one to test whether regions with unusually high diversity are observed outside of exon 7, and could lead to the identification of additional targets of ancient balancing selection. Such variants could then be examined for their effects on enzymatic activity. To evaluate whether there is cryptic variation of functional importance in ABO, it may be particularly interesting to focus on activity levels of ABO in species thought to be lacking one of the main histo-blood groups (e.g. gorillas or chimpanzees). In parallel, phenotypic associations might be conducted to test the effect of histo-blood type subgroups and secretor status on susceptibility to infectious diseases and other plausible phenotypes. This information could then be integrated with data on population frequencies at ABO and local pathogen community composition to learn more about the selection mechanism underlying the remarkable evolution of this gene.
Acknowledgments
We thank Emma E. Thompson and Carole Ober for permission to cite their unpublished data and for sparking our interest in ABO, Joachim Hermisson, Ellen Leffler and Guy Sella for helpful discussions, and two anonymous reviewers and the editor for comments. This work was supported by NIH GM72861 to M. P. M. P. is a Howard Hughes Early Career Scientist.
References
- Yamamoto F, Clausen H, White T, Marken J, et al. Molecular genetic basis of the histo-blood group ABO system. Nature. 1990;345:229–33. doi: 10.1038/345229a0. [DOI] [PubMed] [Google Scholar]
- Livingstone FB. Natural selection, disease, and ongoing human evolution, as illustrated by the ABO blood groups. Hum Biol. 1960;32:17–27. [PubMed] [Google Scholar]
- Saitou N, Yamamoto F. Evolution of primate ABO blood group genes and their homologous genes. Mol Biol Evol. 1997;14:399–411. doi: 10.1093/oxfordjournals.molbev.a025776. [DOI] [PubMed] [Google Scholar]
- Kitano T, Noda R, Takenaka O, Saitou N. Relic of ancient recombinations in gibbon ABO blood group genes deciphered through phylogenetic network analysis. Mol Phylogenet Evol. 2009;51:465–71. doi: 10.1016/j.ympev.2009.02.023. [DOI] [PubMed] [Google Scholar]
- Kermarrec N, Roubinet F, Apoil PA, Blancher A. Comparison of allele O sequences of the human and non-human primate ABO system. Immunogenetics. 1999;49:517–26. doi: 10.1007/s002510050529. [DOI] [PubMed] [Google Scholar]
- Segurel L, Thompson EE, Flutre T, Lovstad J, et al. The ABO blood group is a trans-species polymorphism in primates. Proc Natl Acad Sci USA. 2012;109:18493–8. doi: 10.1073/pnas.1210603109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth D. Balancing selection and its effects on sequences in nearby genome regions. PLoS Genet. 2006;2:e64. doi: 10.1371/journal.pgen.0020064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spurgin LG, Richardson DS. How pathogens drive genetic diversity: MHC, mechanisms and misunderstandings. Proc Biol Sci. 2010;277:979–88. doi: 10.1098/rspb.2009.2084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surridge AK, Osorio D, Mundy NI. Evolution and selection of trichromatic vision in primates. Trends Ecol Evol. 2003;18:198–205. [Google Scholar]
- Garratty G. Relationship of blood groups to disease: do blood group antigens have a biological role. Rev Med Inst Mex Seguro Soc. 2005;43:113–21. [Google Scholar]
- Jenkins PV, O'Donnell JS. ABO blood group determines plasma von Willebrand factor levels: a biologic function after all. Transfusion. 2006;46:1836–44. doi: 10.1111/j.1537-2995.2006.00975.x. [DOI] [PubMed] [Google Scholar]
- Vos GH. The frequency of ABO-incompatible combinations in relation to maternal Rhesus antibody values in Rh immunized women. Am J Hum Genet. 1965;17:202–11. [PMC free article] [PubMed] [Google Scholar]
- Yamamoto F, Cid E, Yamamoto M, Blancher A. ABO research in the modern era of genomics. Transfus Med Rev. 2012;26:103–18. doi: 10.1016/j.tmrv.2011.08.002. [DOI] [PubMed] [Google Scholar]
- Allison AC. Protection afforded by sickle-cell trait against subtertian malareal infection. Br Med J. 1954;1:290–4. doi: 10.1136/bmj.1.4857.290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto M, Lin XH, Kominato Y, Hata Y, et al. Murine equivalent of the human histo-blood group ABO gene is a cis-AB gene and encodes a glycosyltransferase with both A and B transferase activity. J Biol Chem. 2001;276:13701–8. doi: 10.1074/jbc.M010805200. [DOI] [PubMed] [Google Scholar]
- Proulx SR, Phillips PC. Allelic divergence precedes and promotes gene duplication. Evolution. 2006;60:881–92. [PubMed] [Google Scholar]
- Castric V, Vekemans X. Plant self-incompatibility in natural populations: a critical assessment of recent theoretical and empirical advances. Mol Ecol. 2004;13:2873–89. doi: 10.1111/j.1365-294X.2004.02267.x. [DOI] [PubMed] [Google Scholar]
- Gagneux P, Varki A. Evolutionary considerations in relating oligosaccharide diversity to biological function. Glycobiology. 1999;9:747–55. doi: 10.1093/glycob/9.8.747. [DOI] [PubMed] [Google Scholar]
- Stahl EA, Dwyer G, Mauricio R, Kreitman M, et al. Dynamics of disease resistance polymorphism at the Rpm1 locus of Arabidopsis. Nature. 1999;400:667–71. doi: 10.1038/23260. [DOI] [PubMed] [Google Scholar]
- Huang P, Farkas T, Marionneau S, Zhong W, et al. Noroviruses bind to human ABO, Lewis, and secretor histo-blood group antigens: identification of 4 distinct strain-specific patterns. J Infect Dis. 2003;188:19–31. doi: 10.1086/375742. [DOI] [PubMed] [Google Scholar]
- Aspholm-Hurtig M, Dailide G, Lahmann M, Kalia A, et al. Functional adaptation of BabA, the H. pylori ABO blood group antigen binding adhesin. Science. 2004;305:519–22. doi: 10.1126/science.1098801. [DOI] [PubMed] [Google Scholar]
- Oriol R, Le Pendu J, Mollicone R. Genetics of ABO, H, Lewis, X and related antigens. Vox Sang. 1986;51:161–71. doi: 10.1111/j.1423-0410.1986.tb01946.x. [DOI] [PubMed] [Google Scholar]
- Vigan-Womas I, Guillotte M, Juillerat A, Hessel A, et al. Structural basis for the ABO blood-group dependence of Plasmodium falciparum rosetting. PLoS Pathog. 2012;8:e1002781. doi: 10.1371/journal.ppat.1002781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linden S, Mahdavi J, Semino-Mora C, Olsen C, et al. Role of ABO secretor status in mucosal innate immunity and H. pylori infection. PLoS Pathog. 2008;4:e2. doi: 10.1371/journal.ppat.0040002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguiar DC, Barros VL, Pereira WL, Loiola Rdo S, et al. Immunodetection of Helicobacter sp. and the associated expression of ABO blood group antigens in the gastric mucosa of captive and free-living New World primates in the Amazon region. Mem Inst Oswaldo Cruz. 2011;106:936–41. doi: 10.1590/s0074-02762011000800007. [DOI] [PubMed] [Google Scholar]
- Linnenbrink M, Johnsen JM, Montero I, Brzezinski CR, et al. Long-term balancing selection at the blood group-related gene B4galnt2 in the genus Mus (Rodentia; Muridae) Mol Biol Evol. 2011;28:2999–3003. doi: 10.1093/molbev/msr150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staubach F, Kunzel S, Baines AC, Yee A, et al. Expression of the blood-group-related glycosyltransferase B4galnt2 influences the intestinal microbiota in mice. ISME J. 2012;6:1345–55. doi: 10.1038/ismej.2011.204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahata N. A simple genealogical structure of strongly balanced allelic lines and trans-species evolution of polymorphism. Proc Natl Acad Sci USA. 1990;87:2419–23. doi: 10.1073/pnas.87.7.2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Brien SJ, Yuhki N. Comparative genome organization of the major histocompatibility complex: lessons from the Felidae. Immunol Rev. 1999;167:133–44. doi: 10.1111/j.1600-065X.1999.tb01387.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leffler EM, Bullaughey K, Matute DR, Meyer WK, et al. Revisiting an old riddle: what determines genetic diversity levels within species. PLoS Biol. 2012;10:e1001388. doi: 10.1371/journal.pbio.1001388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chester MA, Olsson ML. The ABO blood group gene: a locus of considerable genetic diversity. Transfus Med Rev. 2001;15:177–200. doi: 10.1053/tmrv.2001.24591. [DOI] [PubMed] [Google Scholar]
- Socha WW, Moor-Jankowski J. Blood groups of anthropoid apes and their relationship to human blood groups. J Hum Evol. 1979;8:453–65. [Google Scholar]
- Calafell F, Roubinet F, Ramirez-Soriano A, Saitou N, et al. Evolutionary dynamics of the human ABO gene. Hum Genet. 2008;124:123–35. doi: 10.1007/s00439-008-0530-8. [DOI] [PubMed] [Google Scholar]
- Leffler EM, Gao Z, Pfeifer S, Segurel L, et al. Multiple instances of ancient balancing selection shared between humans and chimpanzees. Science. 2013;339:1578–82. doi: 10.1126/science.1234070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1000 Genomes Project Consortium. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–73. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auton A, Fledel-Alon A, Pfeifer S, Venn O, et al. A fine-scale chimpanzee genetic map from population sequencing. Science. 2012;336:193–8. doi: 10.1126/science.1216872. [DOI] [PMC free article] [PubMed] [Google Scholar]


