Abstract
Epithelial tissues have sparse stroma, in contrast to their corresponding tumours. The effect of cancer cells on stromal cells is well recognized. Increasingly, stromal components, such as endothelial and immune cells, are considered indispensable for cancer progression. The role of desmoplastic stroma, in contrast, is poorly understood. Targeting such cellular components within the tumour is attractive. Recent evidence strongly points towards a dynamic stromal cell participation in cancer progression that impacts patient prognosis. The role of specific desmoplastic stromal cells, such as stellate cells and myofibroblasts in pancreatic, oesophageal and skin cancers, was studied in bio-engineered, physiomimetic organotypic cultures and by regression analysis. For pancreatic cancer, the maximal effect on increasing cancer cell proliferation and invasion, as well as decreasing cancer cell apoptosis, occurs when stromal (pancreatic stellate cells) cells constitute the majority of the cellular population (maximal effect at a stromal cell proportion of 0.66–0.83), accompanied by change in expression of key molecules such as E-cadherin and β-catenin. Gene-expression microarrays, across three tumour types, indicate that stromal cells consistently and significantly alter global cancer cell functions such as cell cycle, cell–cell signalling, cell movement, cell death and inflammatory response. However, these changes are mediated through cancer type-specific alteration of expression, with very few common targets across tumour types. As highlighted by these in vitro data, the reciprocal relationship of E-cadherin and polymeric immunoglobulin receptor (PIGR) expression in cancer cells could be shown, in vivo, to be dependent on the stromal content of human pancreatic cancer. These studies demonstrate that context-specific cancer–stroma crosstalk requires to be precisely defined for effective therapeutic targeting. These data may be relevant to non-malignant processes where epithelial cells interact with stromal cells, such as chronic inflammatory and fibrotic conditions.
Keywords: proliferation, apoptosis, invasion, organotypic model, gene-expression microarray
Introduction
Epithelial tissues have sparse stroma, in contrast to their corresponding tumours. The effect of cancer cells on stromal cells is well recognized. The stroma-rich tumour has recently been acknowledged to negatively impact patient prognosis in a variety of cancers, such as pancreatic [1,2], oesophageal [3] and skin [4] cancers. Increasing recognition for biological rationalization as well as therapeutic targeting is being paid to stromal cells such as endothelial cells (angiogenesis or hypoxia) and immune cells (cancer-promoting inflammation as well as, contrarily, evasion of immune surveillance), such that they are considered as emerging hallmarks indispensable for cancer progression [5]. The specific biological role of the innocuous-appearing desmoplastic stroma has received relatively little attention, primarily because of a lack of vital function being attributed to this cellular (myofibroblast [6]) and acellular [extracellular matrix (ECM)] [7] content, other than to provide a scaffold for the cancer. This is particularly bemusing, since hard cancers amongst soft organs have been a critical diagnostic feature taught to all medical students. Nowhere is this truer than pancreatic cancer, since the desmoplastic stroma may form ca. 70–80% of the tumour volume [8,9].
Targeting the desmoplastic stromal components within the tumour is attractive, as they are tumourspecific, in contrast to, for example, circulating immune cells. Since stromal cells are non-transformed, they are less likely to develop resistance if appropriately targeted [10]. Whilst conventional chemotherapeutics may not target stromal cells, recent advances, specifically in pancreatic cancer, in stroma-specific targeting has made substantial inroads [10–12]. The characterization and isolation of specialized stromal cells, such as cancer-associated fibroblasts [5], myofibroblasts [6] and activated stellate cells [8], and bio-engineering to construct in vitro, high-throughput, physiomimetic, organotypic cultures [10,13–16] has helped us dissect the cancer–stromal (desmoplasia) crosstalk in an unprecedented manner. We have harnessed this capability, with pancreatic cancer forming a prototype, to help us elaborate the interaction between cancer cells and stromal cells (activated stellate cells in the instance of pancreatic cancer [8]).
Materials and methods
Cell lines and organotypic culture models
Pancreatic cancer organotypic cultures were constructed as described previously [10,13,14]. Cancer cells (Capan1, well-differentiated; or AsPc1, poorly-differentiated: StR profiles elaborated in the Supplementary material, Table S1) or epithelial cells (DEChTERT [17]) and stellate (PS1 [13]) cells were mixed to keep a total count of 5 × 105 in cancer:stellate cell ratios of 1:0, 10:1, 5:1, 2:1, 1:1, 1:2, 1:5, 1:10 and 0:1, and plated on gels [52.5% collagen I (cat. no. 354249, BD Biosciences), 17.5% Matrigel (cat. no. 354234, BD Biosciences), 10% 10× Dulbecco's modified Eagle's medium (DMEM), 10% normal DMEM (cat. no. E15-843, PAA Laboratories)], lifted next day and fed on alternate days from below. Gels were harvested on day 10, fixed in formol saline, embedded in paraffin and cut into 4 μm sections. Submerged organotypic cultures were constructed by embedding 5 × 105 PS1 cells within the gel matrix and seeding 2.5 × 105 cancer cells on top after polymerization. The gels were harvested on day 10, frozen in embedding medium (cat. no. 6760096, Thermo Scientific) and stored at −80 °C for RNA extraction. Primary cancer-associated stellate cells were cultured in an ethically approved manner (Research Ethics Committee; Trent MREC 05/MRE04/82) by the outgrowth method [18], and verified using the standard markers [13] and ability to store retinol [10]. The relative paucity of primary stellate cells to conduct all the experiments in sufficient replicates restricted us to using mini-organotypics constructed in Transwell inserts, using fewer cells (total 1 × 105) per organotypic gel in a limited range of cancer:stellate cell ratios of 1:0, 1:2, 1:1 and 2:1 [14]. Squamous cell (skin) cancer [15] and oesophageal cancer [16] organotypic cultures were constructed as described before.
Transwell migration assay
Cancer cells (5 × 104 Capan1 or AsPC1) seeded in serum-free medium were allowed to migrate through 8 μm Transwell membranes (Corning, NY, USA) over 24 h towards conditioned media from activated (ethanol) or quiescent (ATRA) stellate cells, as described previously [10]. Percentages of cells migrating through the membranes were counted using a Casy Counter (Schärfe System, Reutlingen, Germany). Fresh medium was used as control.
Laser-capture microdissection
For pancreatic and skin organotypic cultures, frozen sections were obtained on polyethylene naphthalate membrane-coated slides (cat. no. 415190–9041, Carl Zeiss) pretreated with RNase Zap (cat. no. AM9780, Ambion) and rendered sterile and hydrophilic with UV light irradiation (Stratalinker®, Stratagene). The sections were fixed (ice-cold 70% ethanol), stained (haematoxylin) and the non-invaded cancer cell layer on top of the gel matrix was captured by laser microdissection (PALM MicroBeam, Carl Zeiss). For oesophageal organotypic cultures, frozen sections were taken onto membrane-mounted metal frame slides (cat. no. 50103, MMI) and were immediately fixed, stained and dehydrated before laser microdis-section (Nikon Eclipse TE 2000–5). For pancreatic cancer organotypic cultures, cancer cells captured by laser microdissection were incubated in 100 μl RNA lysis buffer at 42 °C for 30 min, followed by RNA extraction using the RNAqeous micro kit (cat. no. AM1931, Ambion). RNA from oesophageal and skin organotypic cultures was isolated using the PicoPure RNA isolation kit (cat. no. KIT0204, Arcturus, Invitrogen) and Qiagen RNAeasy Micro Kit (cat. no. 74004, Qiagen) respectively.
Microarray analysis
For pancreatic cancer organotypic cultures, 250 ng extracted RNA was labelled using an Illumina TotalPrep™ RNA Amplification Kit, according to the manufacturer's instructions. 1.5 μg labelled cRNA was hybridized onto Illumina Human-12 v. 4 Expression BeadChips (47 231 probes). Quality Control and normalization were performed using GenomeStudio v. 2011.1 (Illumina). Statistical analyses were performed using Bioconductor [19] packages within the open source R statistical environment [20]. We applied a filter using the standard deviation (SD) of gene expression values to select the top 10 000 probes. We used the Limma package for differential expression analysis [21] and Empirical Bayes was used to borrow information across genes for stable analyses. We set a double threshold for significant changes in gene expression with a false discovery rate (FDR) ≤ 0.05 and an absolute fold change > 2.
For oesophageal and skin organotypic cultures, purified total RNA was Biotin-labelled and hybridized onto Affymetrix GeneChip U133 Plus 2.0 oligonucleotide array. The microarray data were analysed using the Microarray Suite v. 5.0 (MAS 5.0) algorithm, using Affymetrix default parameter settings and global scaling as the normalization method. The Limma package was used on the log2-transformed MAS 5.0 scaled data for the two-group (skin) and multiple-group (oesophageal) ANOVA differential expression analysis. Since the number of replicates was low (2) for each group, a significance threshold of raw p value < 0.01 was applied [22] and an absolute fold change of > 2 was applied for two-group comparisons. Hierarchical clustering analysis was subsequently performed. The heat maps were generated using ‘heatmap.2’ from the R package gplots, and the Venn diagram generated using the R code [23]. The raw data of the MIAME-compliant microarray experiments are deposited on the Gene Expression Omnibus [24], Accession Nos GSE36775 (pancreatic), GSE36776 (skin) and GSE19472 (oesophageal).
Pathway analysis
Ingenuity Pathway analysis was used to dissect out cellular and biological functions [10]. The global functional analysis feature calculated the significance using the right-tailed Fisher's exact test, with p < 0.05 taken as significant, after applying the Benjamini–Hochberg approach for multiple testing [22].
qRT – PCR
Primers were designed using the online primer design tool [25] (see Supplementary material, Table S2). SensiFAST SYBR Hi-ROX One-Step Kit™ (cat. no. 73005, Bioline), formulated for first-strand cDNA synthesis and subsequent real-time PCR in a single tube, was used. 25 ng RNA was added to the master mix, consisting of SensiFAST SYBR Hi-ROX One-Step mix (2×), reverse transcriptase, RNase inhibitor, forward and reverse primers (400 nM final concentration) and DEPC-treated water, made up to a final volume of 10 μl. PCR was performed in the Applied Biosystems Step One™ real-time PCR systems. Fold change was calculated using the comparative Ct (ΔΔCt) method [26] after normalizing for endogenous 18 s expression.
Immunostaining
Paraffin-embedded organotypic sections were dewaxed and rehydrated. Heat-induced epitope retrieval in citrate buffer, pH 6, was used for all antibodies. For immunohistochemistry, endogenous peroxidase was blocked with 3% H2O2 in methanol. Primary antibodies (see Supplementary material, Table S3) were incubated at 4 °C overnight, followed by 1 h incubation with biotinylated secondary antibody. Peroxidase-labelled Avidin–Biotin complex (cat. no. PK4000, Vectastain ABC kit™, Vector Laboratories) was added and visualized, using 3,3-diaminobenzidine tetrahydrochloride followed by counterstaining with haematoxylin. For immunofluorescence, sections were permeabilized with 0.2% Triton X-100 and blocked with 2% bovine serum albumin (BSA; cat. no. K45-001, PAA Laboratories), 0.02% fish skin gelatin (cat. no. G7765, Sigma), 10% FBS (cat. no. A15-104, PAA Laboratories). Primary antibodies were incubated at 4 °C overnight. Fluorescent-labelled appropriate secondary antibodies (AlexaFluor® 488, 546) were incubated at room temperature for 1 h and nuclei were counterstained with DAPI. Controls were uniformly negative with appropriate isotype-specific immunoglobulin at matching dilutions.
Quantification
Organotypic culture length and thickness was measured by summating the length of serial low-power (×50) fields across the gel from end to end, limiting to within the area of cellularity to avoid edge artefacts (Figure 1b–i). Cancer cell counts were obtained in six random high-power fields/gel: L2 h/lt = estimated total cancer cell count, where L = gel length, h = number of cells per high-power field, l = length of one high-power field and t = thickness of the section (usually 4 μm). Fold change was arrived at by calculating the ratio between the cancer cell counts at the end of the experiment to those at the start. The proliferation (Ki67), apoptosis (cleaved-caspase-3) and invasive (cytokeratin or dual stains) indices were calculated as described previously [10]. Clumps of more than two cells were counted as a cohort.
Figure 1.
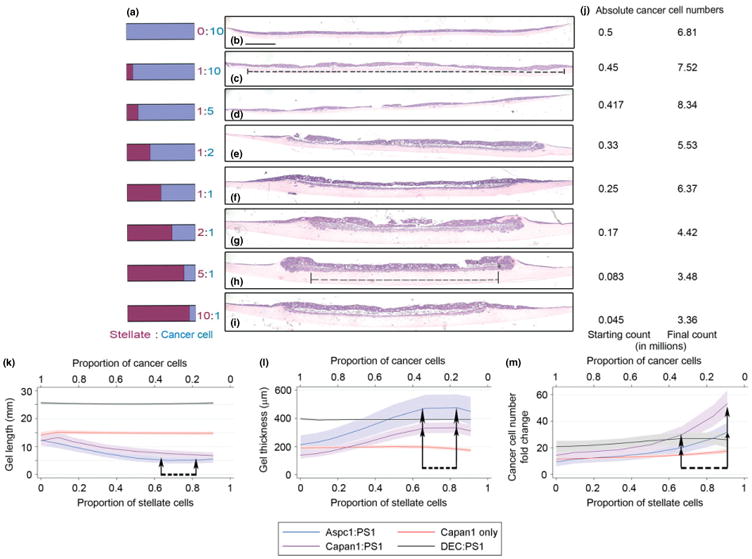
Effects of stellate cells on ECM gel contraction and cancer cell number. Organotypic culture gels were constructed by seeding specific ratios (a) of cancer cells [either Capan1 (b–i) or AsPc1] and stellate cells (PS1). As the proportion of stellate cells increased, reduction in length and increase in thickness of the ECM gels were observed (b–i), associated with increased ratios of absolute final cancer cell count to starting cell numbers (j). Broken lines (c, h) show the extent of measurement of organotypic ECM ‘cellular’ length. Bold lines in (c, h) indicate gel thickness; scale bar = 1000 μm. Fractional polynomial regression lines [bold; 95% confidence intervals (shaded areas) demonstrate contractions of ECM gels, length (k), thickness (l)] in cancer–stellate cell organotypic culture gels (Capan1:PS1, AsPC1:PS1) as opposed to epithelial–stellate (DEC:PS1) or cancer cells alone (Capan1) gels. Maximum contraction was observed when stellate cells formed 66% or 83% of the starting cell number (arrows). A marked increase in the fold-change of epithelial cells (m) with a steep upward trend was noted from starting stellate proportion of 0.66 onwards (arrows) (see Supplementary material, Figure S1, for raw data). Statistical significance of the specific points of change determined by Friedman's test with Dunn's multiple comparison test. Details of regression models used are summarized in Table S4 (see Supplementary material).
Human tissue microarrays
Previously described human tissue microarrays [13], constructed with ethical approval (Research Ethics Committee, East London & the City REC3 07/H0705/87) were used to quantify desmoplastic stromal content [cytokeratin/α-smooth muscle actin (αSMA) co-stain] and correlate with expression of specific molecules (PIGR/E-cadherin co-stain) in sequential sections.
Fluorescent intensity
Random high-power fields/gel were captured for analysis of E-cadherin, β-catenin and PIGR expression in the non-invaded cancer cell layer. The intensity of fluorescence in the green/red channel for respective molecules was calculated using ImageJ software [27]. The threshold and normalization was set according to the intensity of the green/red channel in the cancer cell-only organotypic cultures. Cellularity was ascertained by counting cells in the DAPI channel and the fluorescent intensity/cell in the non-invaded cancer cell layer was calculated.
Statistical analysis
We conduct a form of trend test to analyse the association between the different proportions of stellate cells and the outcome variables. We investigated whether there was a linear association for the experiments; however, a constant for the controls. We fitted a linear regression model for each cell line and compared it to a null model. Since the data were measured in batches, all models are adjusted for batch variation. A formal test was used comparing the difference of the deviances. The p value was computed by referring the difference to a χ2 distribution on nine degrees of freedom (df) for the experiments and two df for the controls. The normality assumption of the residuals was tested by a Shapiro–Wilk test. The homogeneity of variance was analysed by a plot of the residuals versus the fitted values. A transformation of the outcome variable, either logarithm or square root, was used if the model fit was better regarding to normality assumption, diagnostics plots and R2. Additionally, we fitted a fractional polynomial (FP) regression model [28] to analyse whether the association was non-linear. The degree of FP to fit was set to two. Again, a formal test was used to contrast the FP with the linear model. The difference of the deviances was referred to a χ2 distribution on three df. For E-cadherin and β-catenin we included an extra dummy variable, which modulates the proportion of stellate cells equal to zero separately, resulting in a better model fit. Furthermore, we fitted a regression model including the data of the experiment and the control to examine whether the effect of stellate cells differed between the two groups. An interaction term between the proportion of stellate cells and a group dummy variable was included into the model. The model type was chosen by the most significant model for the experiment. A likelihood-ratio test was used to test the significance of the interaction term. The test statistic follows approximately a χ2 distribution, whereas df depend on the chosen model type (one df for the linear model, two df for FP). In the case of invasive cells, this part was missed out, since we had no control and a negative binomial regression model was considered, because count data were given and the variance was greater than the mean.
For all outcome variables, the fitted FP with two power terms was plotted, giving everyone the flexibility of a possible non-linear association. If a transformed outcome variable was considered, the plotted predicted values were transformed back to the normal scale. Scatter plots with fitted trend line were used to visualize the association between stromal content and the molecules PIGR and E-cadherin in human samples. Pearson's correlation coefficient is reported for the ratio PIGR:Ecadherin (log-transformed). A trend test for ordered groups was used for PIGR and E-cadherin scores [29]. In all tests, p ≤ 0.05 was considered statistically significant. The statistical analysis was performed using STATA 11.2 (StataCorp, 2009).
Results
Increasing stromal cell proportion causes ECM contraction
The most striking feature when pancreatic cancer cells (pancreatic ductal adenocarcinoma cells, PDACs) were admixed with varying proportions of stellate cells (PS1 [13]) on top of ECM gel [10] was the contraction of the ECM gels. This characteristic, accompanied by an obvious increase in the stiffness of the gels, was particularly prominent as the stellate cell proportion in the cellular admixture increased (maximal at a stellate cell proportion of 0.66–0.83; Figure 1a–i; see also Supplementary material, Figure S1a–k). We ensured that the cell–cell (heterotypic and homeotypic) and cell–matrix interactions, cellular organization and cellular (cancer and stellate) invasion into the matrix [10,13,14,30] and perfusion/diffusion gradient across acellular matrix proteins mimicked the in vivo scenario [31]. Artefacts due to differences in metabolic requirements were avoided by keeping the total number of cells constant. Strikingly, this visible shrinkage of ECM gels was absent when stellate cells were plated with normal epithelial cells (DEChTERT [17]), implying a distinction between cancer–stellate cell and epithelial–stellate cell interactions (Figure 1 k, l; see also Supplementary material, Figure S1f, j, q). Interestingly there was minimal de novo cancer cell or stellate cell effect when the respective cells were used alone at the same cell numbers as the above experiments (Figure 1 k, l; see also Supplementary material, Figure S1g, k, p, Table S4).
Increasing stromal cell proportion increases cancer cells' pro-survival and pro-invasive capacity
In addition, it was evident that the epithelial cell layer thickness, as well as invasion of cancer cells, was more prominent when stellate cells were the predominant cell type (see Supplementary material, Figure S1b, c). The fold change in cancer cell numbers (Figure 1j) over a 10 day period excluded any artefacts due to gel contraction-induced cellular crowding. We demonstrate a maximal fold change in cancer cell population in the presence of more stromal cells (maximal effect started at a stellate cell proportion of 0.66). In comparison, the control experiments (cancer cells alone, epithelial–stellate cell admixture) did not demonstrate a similar rise in fold change of cancerous or normal epithelial cells (Figure 1 m; see also Supplementary material, Figure S1l–o). This rise in cancer cell number was due to a combination of marked increase in proliferation (as determined by percentage of Ki-67-positive cancer cells: Figure 2a; see also Supplementary material, Figure S2a, c, d) and decrease in apoptotic fraction (percentage of cleaved caspase-3 positive cancer cells: Figure 2b; see also Supplementary material, Figure S2b, g, h). Surprisingly, control experiments showed a constant proliferative and apoptotic index in epithelial cells across all proportions (see Supplementary material, Figure S2e, f, i, j).
Figure 2.
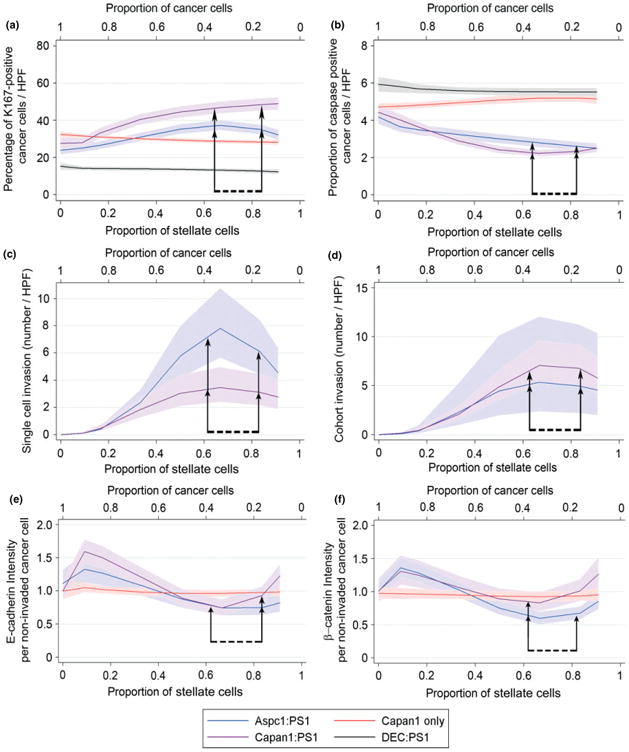
Stellate cell effects on survival, invasion and E-cadherin and β-catenin expression. Fractional polynomial regression lines (bold), along with 95% confidence intervals (shaded areas), demonstrate that increasing the proportion of stellate cells caused an increase in the percentage of proliferating cancer cells (a), concomitant with a decrease in the percentage of apoptotic cancer cells (b), when the starting stellate cell proportion was 0.66–0.83 (seen in both Capan1:PS1 and AsPc1:PS1 organotypic cultures). Control gels (DEC:PS1 or cancer cells alone) demonstrated no change in the proliferative or apoptotic fractions. The number of invading single cancer cells (c) and invading cohorts of cancer cells (d) (for raw data, see Supplementary material, Figure S3). The number of invading cohorts and single cancer cells increased as the proportion of stellate cells increased with maximal effect at stellate cell proportions of 0.66–0.83 (arrows). The expression of previously described proteins [10,13] for cell–cell adhesion (E-cadherin, e) and pro-survival (β-catenin, f) ‘per non-invaded cancer cell’ (for raw data, see Supplementary material, Figure S7). Stellate cells at a proportion of 0.66–0.83 (arrows) decreased the cancer cell expression of E-cadherin and β-catenin maximally. The statistical significance of the specific points of change was determined by Friedman's test with Dunn's multiple comparison test. Details of regression models are summarized in Table S4 (see Supplementary material).
Besides the marked pro-survival and anti-apoptotic effect of stromal cells on the cancer cell, an increasing proportion of stromal cells also increased single-cell invasion as well as cohort invasion [15,32] (Figure 2c, d; see also Supplementary material, Figure S3a–e) of cancer cells into the underlying matrix, whilst stellate cells alone, cancer cells alone and the normal epithelial–stellate cell admixture failed to demonstrate any invasion into the underlying ECM gel (see Supplementary material, Figure S1p, q). In agreement with the data on other observed changes, the maximal effect on cancer cell invasion was observed when the range of proportion of stellate cells was 0.66–0.83. Similarly, the pro-proliferative, pro-invasive and anti-apoptotic effect upon cancer cells could be induced upon co-culture in three dimensions (3D; mini-organotypics over the lesser range of ratios of cancer:stellate cells to accommodate the paucity of primary stellate cells) with primary cancer-associated stellate cells (see Supplementary material, Figures S4, S5). All the stellate cells used in this study represent activated stellate cells, since they were continuously in culture on plastic (PS1) or were derived from cancer-associated tissues (primary PSC) and were cultured in the presence of relatively high sugar content in the medium (3.15 g/l dextrose) [8]. Therefore, these stellate cells represented those found in human PDACs. We have recently demonstrated that these activated stellate cells could be rendered quiescent by restoring retinol depots [10]. Thus, using conditioned media from quiescent and activated stellate cells, we could demonstrate that the activation of stellate cells enhances the migration of cancer cells (see Supplementary material, Figure S6). These data, taken together, demonstrate that the presence of stellate cells mediates cancer cell pro-survival and pro-invasive capability, perhaps in conjunction with, or due to, matrix remodelling [15,32]. This effect is maximal when the stromal cells constitute the predominant component of the cellular mix. This extent of dissection of cell–cell interaction is possible only in these 3D organotypic cultures and is unachievable in vivo, due to the collaborative or conflicting effects of other stromal cell types, such as immune cells and endothelial cells, amongst others.
Increasing stromal cell proportion alters expression profile of cancer cells
Based on our [10,13] and other laboratories' [33] experience in the involvement of key survival (β-catenin [10,13]) and cell–cell adhesion (E-cadherin [13,33–35]) molecules, which are interlinked during wnt signalling [33], epithelial–mesenchymal transition [34] (EMT) as well as the pro-migratory [33,35–37] phenotype of cancer cells, we further explored these two molecules specifically in the context of cancer cell–stellate cell organotypic cultures. In order to avoid the confounding effect of heterogeneous cellular migration (single cell and collective), as well as possible EMT, we assessed the non-invaded sheet of epithelial cells above the epithelial cell–stromal cell–gel interface. Our data demonstrate a biphasic E-cadherin (Figure 2e; see also Supplementary material, Figure S7a, c–e) and β-catenin (Figure 2f; see also Supplementary material, Figure S7b, f–h) expression per cancer cell (initial increase in expression followed by significant reduction when the stellate cell proportion was 0.66, and a return of expression to basal levels corresponding to further increases in stellate cells). Importantly, the maximal reduction of expression of these molecules was demonstrable when the cancer cell proportion was less than that of the stellate cells, and this change was notably absent in control gels. These data offer explanations of many of the conflicting observations about E-cadherin and β-catenin expression in distinct cancers, both in vivo [10,13,38] and in vitro [33,39].
Gene-expression microarray analysis confirmed the transcriptomic change, targeting multiple intracellular pathways, within cancer cells upon exposure to stellate cells or myofibroblasts [Figure 3; see also Supplementary material, Figures S8, S9, Table S5, and Gene Expression Omnibus website (Accession No. GSE 36776)]. To prevent artefacts, laser capture microdissection of the overlying epithelial [pancreatic [14] and squamous [15] (GSE 36775), oesophageal [35,36] (GSE 19472)] cell layer from organotypic culture gels, with or without stromal cells, was carried out in triplicate (pancreatic) or duplicate (squamous and oesophageal). To prevent contamination of cancer cell gene expression due to minute quantities of stellate or myofibroblast gene signature, we embedded stromal cells within the ECM gel and harvested only the non-invading cancer cells (see Supplementary material, Figure S8) [30]. Unsupervised hierarchical clustering confirmed experimental homogeneity (not shown).
Figure 3.
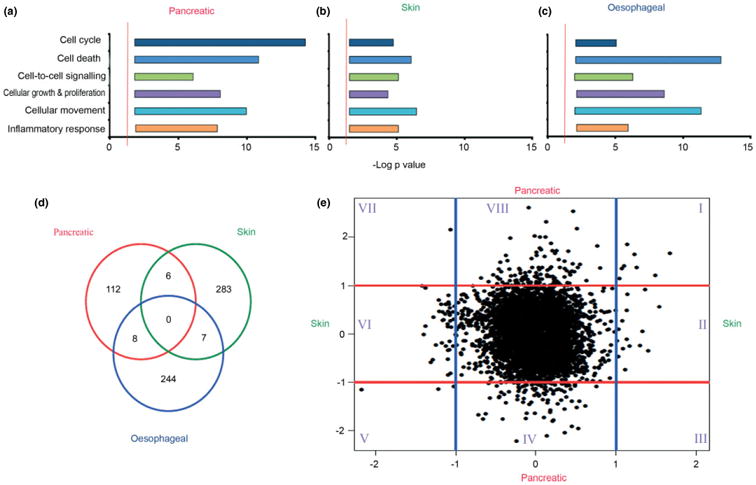
Cancer-specific stromal cell alterations in gene expression. Ingenuity Pathway analysis of gene expression microarray data demonstrated that similar pathways were affected in cancer cells upon signalling from stromal cells in pancreatic (a), skin (b) and oesophageal (c) cancer. Graphs represent the negative log of a range of p values (x axis) of various subfunctions of genes involved in cellular functions (y axis). The red line denotes significance, determined by right-tailed Fisher's exact test, with a p value of < 0.05 after applying Benjamini–Hochberg approach for multiple testing [22]. There is considerable heterogeneity in the specific genes affected, as demonstrated by the Venn diagram (d). Specific genes in pancreatic and skin cancer models influenced by stromal cells were plotted as log-fold change (e), demonstrating no consistent pattern across the two gene sets. Specific gene sets in the different subgroups (I–VIII) are listed in Table S6 (see Supplementary material).
Stromal cells significantly alter multiple signalling cascades within cancer cells
Ingenuity Pathway Analysis, a valid tool for obtaining an overview of changes in gene expression [10], highlighted statistically significant changes in gene expression mediating key cellular functions, such as cell cycle and proliferation, cell movement and death, cell–cell signalling and inflammatory response mediated through multiple signalling cascades, for both pancreatic (Figure 3a) and skin squamous (Figure 3b) cancer cells. Furthermore in a well-established and characterized oesophageal cancer model, we investigated the effect of the two most significant molecular targets within cancer cells of cancer–stroma signalling, E-cadherin and transforming growth factor (TGFβ), by genetically inactivating expression of E-cadherin and TGFβ-receptor II (TGFβRII) sequentially [36]. Whilst wild-type E-cadherin expressing hTERT-immortalized normal oesophageal cells demonstrated no invasion, knockdown of E-cadherin and both E-cadherin and TGFβRII demonstrated pronounced increase in cancer cell invasion [36], associated with marked changes in gene expression in cancer cells (Figure 3c; see also Supplementary material, Figure S9a) affecting similar cellular functions. Further interrogation of these datasets demonstrates changes in gene expression which are markedly different between pancreatic and squamous cancer cells upon exposure to their respective stromal cells (Figure 3d, e). These data suggest organ-specific changes influenced by respective stromal cells with very few genes, which are similarly regulated.
Quantitative real-time RT–PCR in independent experiments (Figure 4), confirmed several of the most up-regulated and down-regulated genes across two pancreatic cancer cell lines (Capan1, AsPC1) and clustered them into four groups affecting one or more key cellular functions. We next investigated polymeric immunoglobulin receptor (PIGR) as an exemplar candidate protein, a significantly up-regulated gene in cancer cells upon exposure to stellate cells. PIGR expression has recently been linked to poor outcome as well as EMT in hepatocellular cancer [34,40], where hepatic stellate cells play a dominant role in the underlying fibrotic reaction. PIGR is involved in transcytosis of molecules in epithelial cells and, thus, may provide the missing link between inflammation, EMT and metastasis [40]. We found that PIGR expression was predominantly observed in cancer cells when stellate cells were most predominant (stellate proportion of 0.66–0.83; Figure 5a, b) and was inversely correlated with E-cadherin expression. We confirmed the reciprocal changes in E-cadherin and PIGR changes in human PDAC samples. This reciprocal relation of E-cadherin and PIGR expression correlated with the stromal predominance in human samples (Figure 5; see also Supplementary material, Figures S10, S11). Furthermore, the data indicated the causality of many of the genetic changes highlighted recently in pancreatic cancer, specifically such as those related to α2-zinc glycoprotein (AZGP1 [41]) and anterior gradient 2 (AGR2 [42]), amongst others, such as S100A8 [43,44] and p120-catenin [45], which have been implicated in both skin and oesophageal cancers. Thus, using PIGR as one of the many examples we could have chosen from, we provide evidence for the first time of the ‘dose-dependent’ influence of (desmoplasia-inducing) stromal cells on the cancer cell pro-invasive and pro-survival phenotype.
Figure 4.
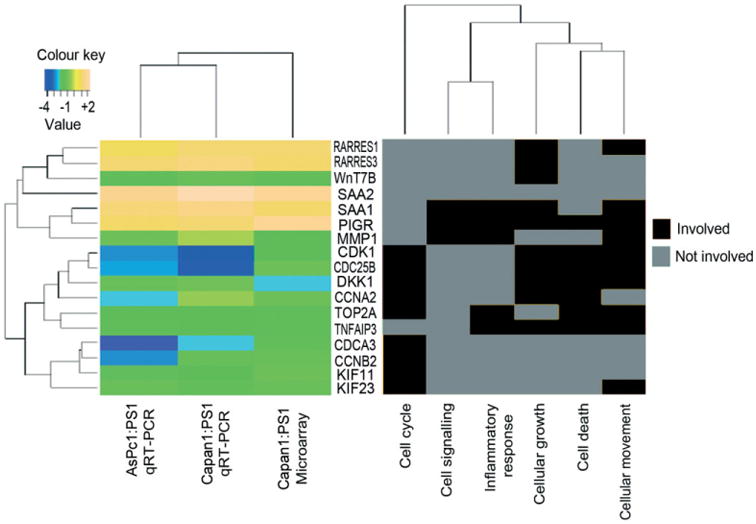
Confirmation of gene expression changes by qRT–PCR. The topmost up- or down-regulated genes for pancreatic cancer upon exposure to activated stellate cells were selected and validated by qRT–PCR of independent organotypic cultures. Heat map of log fold changes, which ranged from −4 (most down-regulated) to +2 (most up-regulated). Further, key cancer-related biological functions were identified and the involvement of each gene in a particular cellular function is depicted in the heat map on the right. The genes cluster in hierarchy into two major sets with four minor groups, depending on cellular function (binary coding) and log2-fold change in expression detected by gene-expression microarray and qRT–PCR. PIGR is a gene involved in most cellular functions, such as cell signalling, inflammatory response, cell growth, death and movement.
Figure 5.
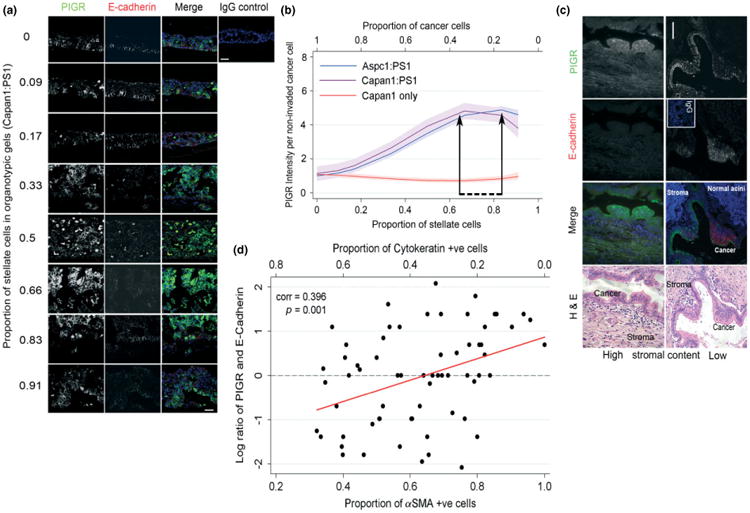
Confirmation of changes in protein expression in organotypic cultures and human tissue samples. Intense PIGR staining was noted in cancer cells when the stellate cell proportion was 0.66–0.83, which correlated inversely to expression of E-cadherin (a) in organotypic cultures; fractional polynomial regression lines (bold), along with 95% confidence intervals (shaded areas) of PIGR staining intensity (b). Inverse relation of E-cadherin expression to PIGR expression and the stromal context in vivo in human PDAC (c), validating the observations made in the physiomimetic in vitro organotypic cultures; scale bar = 100 μm. Scatter plot with fitted trend line shows a significant positive correlation between PIGR:E-cadherin staining and the proportion of stellate cells (d). Log-transformed PIGR:E-cadherin ratio was used.
Discussion
Our current study demonstrates that the progressive accumulation of desmoplastic stromal cells (believed to be triggered by cancer-induced activation of fibroblasts/stellate cells to myofibroblasts) has a pro-survival and pro-invasive effect on tumour cells, in addition to stiffening (contraction) of the ECM gels. This is mediated through a number of signalling cascades and molecular targets. The strength of our experimental plan lies in the use of a range of cell lines from different cancers alongside primary cells in bio-engineered, in vitro, high-throughput, physiomimetic organotypic culture conditions [10,13–16,32], which reduces the noise from other stromal elements and thus focuses on the cancer-desmoplastic stromal crosstalk. This approach is validated by findings in human samples, such as those for PIGR expression in the present study as well as the work of others (examples include AZGP1 [41], AGR2 [42], S100A8 [43,44] and p120 catenin [45]).
This work highlights that understanding the complexity and identifying the key regulators of the multiple signalling cascades involved in the cancer–desmoplastic stromal crosstalk is necessary to effectively target them. As a corollary, we suggest that drugs targeting single pathways influencing the desmoplastic stroma–cancer crosstalk are unlikely to be efficacious, since such signalling networks harness redundancy in other pathways. Interestingly, pursuing this avenue, we have recently demonstrated that pleiotropic agents such as ATRA modulate or revert the activated stromal cells to their quiescent state, thereby altering a number of signalling cascades in the tumour–stroma crosstalk [10]. This in turn influences tumour behaviour by reducing cancer cell proliferation and invasion, as well as increasing cancer cell apoptosis. Indeed, the dependence of cancer progression and clinical prognosis on this tumour–stromal cell proportion has been highlighted in various studies of pancreatic cancer [1,2], oesophageal cancer [3] and skin cancer [4]. These data suggest that dampening the tumour-promoting interaction between cancer and stromal cells by ‘multitargeting’ agents may allow traditional chemo- and/or radiotherapy to be effective.
Supplementary Material
Figure S11. PIGR and E-cadherin expression in human PDACs.
Figure S1. Construction of organotypics and the influence of stellate cells on ECM matrix contraction and increasing cancer cell number.
Figure S2. Stellate cells have pro-survival effect on cancer cells.
Figure S3. Stellate cells enhance cancer cell invasion.
Figure S4. Primary pancreatic stellate cells in mini-organotypic 3D co-culture caused ECM gel contraction and an increase in cancer cell number.
Figure S5. Primary stellate cells have a pro-survival, pro-invasive effect on cancer cells.
Figure S6. Activated stellate cells have a pro-migratory effect on cancer cells.
Figure S7. Stellate cells alter the expression of key molecules.
Figure S8. Retrieval of mRNA from organotypic gels and gene-expression microarray analysis.
Figure S9. Gene-expression microarray analysis.
Figure S10. PIGR and E-cadherin expression in human PDACs.
Table S1. STR profile of cell lines used for the experiments.
Table S2. Primer sequences used for qRT–PCR.
Table S3. Antibodies used for immunostaining.
Table S4. Statistical summary analysis for individual outcomes in each experiment and corresponding n values.
Table S5. 146 probes demonstrating differentially expressed genes (n = 126).
Table S6. List of genes in specific subsets shown in the correlation plot in Figure 3e.
Acknowledgments
This study was supported by a Lewin International Surgical Oncology Fellow Award (to RK) and a pump-priming grant from the Pancreatic Society of Great Britain and Ireland (to HMK). CDA is the recipient of a Research Scholar Award from the American Gastroenterology Association / Foundation for Digestive Health and Nutrition (AGA/FDHN) and supported by the National Institutes of Health, USA (Grant No. DK075379). AKR is supported by the NIH/NCI (Grant No. P01-CA098101; Cell Culture, Molecular Biology and Microarray Core Facilities) and acknowledges an American Cancer Society Research Professorship (No. RP-10-033-01-CCE). ES and PS acknowledge the support of Cancer Research UK.
Footnotes
No conflicts of interest were declared.
Author contributions: RK did all the pancreatic organotypic culture experiments and staining with various antibodies, generated data for figures and co-wrote the manuscript, with help from FDM, ESo, MG, DB and FEMF under the supervision of HMK. HB did all the statistical analyses under the supervision of PS. JW did all the bioinformatics analyses under the supervision of CC. SH, CDA, ESa, SB and AKR contributed valuable data, performed critical analysis of the data and helped write the manuscript. HMK initiated the project, designed the experiments/analysis to generate the data in all figures, interpreted the data and wrote the manuscript. HMK acts as a guarantor for the data in the manuscript.
Transcript profiling: GEO website, Accession Nos GSE 19472, 36775, 36776:
http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?token=zluxvckucmowkjc&acc=GSE19472
http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?token=tnuvvokecawkmxi&acc=GSE36775
http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?token=lhghlsmysysagji&acc=GSE36776
Supporting Information on the Internet
The following supporting information may be found in the online version of this article
Supplementary materials and methods
References
- 1.Erkan M, Michalski CW, Rieder S, et al. The activated stroma index is a novel and independent prognostic marker in pancreatic ductal adenocarcinoma. Clin Gastroenterol Hepatol. 2008;6:1155–1161. doi: 10.1016/j.cgh.2008.05.006. [DOI] [PubMed] [Google Scholar]
- 2.Fujita H, Ohuchida K, Mizumoto K, et al. α-Smooth muscle actin expressing stroma promotes an aggressive tumor biology in pancreatic ductal adenocarcinoma. Pancreas. 2010;39:1254–1262. doi: 10.1097/MPA.0b013e3181dbf647. [DOI] [PubMed] [Google Scholar]
- 3.Wang K, Ma W, Wang J, et al. Tumor:stroma ratio is an independent predictor for survival in esophageal squamous cell carcinoma. J Thorac Oncol. 2012;7:1457–1461. doi: 10.1097/JTO.0b013e318260dfe8. [DOI] [PubMed] [Google Scholar]
- 4.Woenne EC, Lederle W, Zwick S, et al. MMP inhibition blocks fibroblast-dependent skin cancer invasion, reduces vascularization and alters VEGF-A and PDGF-BB expression. Anticancer Res. 2012;30:703–711. [PubMed] [Google Scholar]
- 5.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 6.De Wever O, Demetter P, Mareel M, et al. Stromal myofibroblasts are drivers of invasive cancer growth. Int J Cancer. 2008;123:2229–2238. doi: 10.1002/ijc.23925. [DOI] [PubMed] [Google Scholar]
- 7.Cukierman E, Bassi DE. Physico-mechanical aspects of extracellular matrix influences on tumorigenic behaviors. Semin Cancer Biol. 2010;20:139–145. doi: 10.1016/j.semcancer.2010.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Erkan M, Adler G, Apte MV, et al. StellaTUM: current consensus and discussion on pancreatic stellate cell research. Gut. 2012;61:172–178. doi: 10.1136/gutjnl-2011-301220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hwang RF, Moore T, Arumugam T, et al. Cancer-associated stromal fibroblasts promote pancreatic tumor progression. Cancer Res. 2008;68:918–926. doi: 10.1158/0008-5472.CAN-07-5714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Froeling FE, Feig C, Chelala C, et al. Retinoic acid-induced pancreatic stellate cell quiescence reduces paracrine Wnt-β-catenin signaling to slow tumor progression. Gastroenterology. 2011;141:1486–1497. doi: 10.1053/j.gastro.2011.06.047. [DOI] [PubMed] [Google Scholar]
- 11.Olive KP, Jacobetz MA, Davidson CJ, et al. Inhibition of Hedgehog signaling enhances delivery of chemotherapy in a mouse model of pancreatic cancer. Science. 2009;324:1457–1461. doi: 10.1126/science.1171362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Von Hoff DD, Ramanathan RK, Borad MJ, et al. Gemcitabine plus nab-paclitaxel is an active regimen in patients with advanced pancreatic cancer: a phase I/II trial. J Clin Oncol. 2011;29:4548–4554. doi: 10.1200/JCO.2011.36.5742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Froeling FE, Mirza TA, Feakins RM, et al. Organotypic culture model of pancreatic cancer demonstrates that stromal cells modulate E-cadherin, β-catenin, and Ezrin expression in tumor cells. Am J Pathol. 2009;175:636–648. doi: 10.2353/ajpath.2009.090131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Froeling FE, Marshall JF, Kocher HM. Pancreatic cancer organotypic cultures. J Biotechnol. 2010;148:16–23. doi: 10.1016/j.jbiotec.2010.01.008. [DOI] [PubMed] [Google Scholar]
- 15.Gaggioli C, Hooper S, Hidalgo-Carcedo C, et al. Fibroblast-led collective invasion of carcinoma cells with differing roles for RhoGTPases in leading and following cells. Nat Cell Biol. 2007;9:1392–1400. doi: 10.1038/ncb1658. [DOI] [PubMed] [Google Scholar]
- 16.Oyama K, Okawa T, Nakagawa H, et al. AKT induces senescence in primary esophageal epithelial cells but is permissive for differentiation as revealed in organotypic culture. Oncogene. 2007;26:2353–2364. doi: 10.1038/sj.onc.1210025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li NF, Kocher HM, Salako MA, et al. A novel function of colony-stimulating factor 1 receptor in hTERT immortalization of human epithelial cells. Oncogene. 2009;28:773–780. doi: 10.1038/onc.2008.412. [DOI] [PubMed] [Google Scholar]
- 18.Bachem MG, Schneider E, Gross H, et al. Identification, culture, and characterization of pancreatic stellate cells in rats and humans. Gastroenterology. 1998;115:421–432. doi: 10.1016/s0016-5085(98)70209-4. [DOI] [PubMed] [Google Scholar]
- 19.Gentleman RC, Carey VJ, Bates DM, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.The R project of statistical computing. www.r-project.org.
- 21.Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3 doi: 10.2202/1544-6115.1027. Article 3. [DOI] [PubMed] [Google Scholar]
- 22.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Statist Soc. 1995;57:289–300. [Google Scholar]
- 23.Girke T. Intersect and Venn diagram functions. http://faculty.ucr.edu/∼tgirke/Documents/R_BioCond/My_R_Scripts/overLapper.R.
- 24.Gene Expression Omnibus. http://www.ncbi.nlm.nih.gov/geo/
- 25.Primer Blast. http://www.ncbi.nlm.nih.gov/tools/primer-blast/
- 26.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 27.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nature Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Royston P, Altman DG. Regression using fractional polynomials of continuous covariates: Parsimonious parametric modelling. Appl Statist. 1994;43:429–467. [Google Scholar]
- 29.Cuzick J. A Wilcoxon-type test for trend. Statist Med. 1985;4:87–90. doi: 10.1002/sim.4780040112. [DOI] [PubMed] [Google Scholar]
- 30.Kalabis J, Wong GS, Vega ME, et al. Isolation and characterization of mouse and human esophageal epithelial cells in 3D organotypic culture. Nat Protoc. 2012;7:235–246. doi: 10.1038/nprot.2011.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Provenzano PP, Cuevas C, Chang AE, et al. Enzymatic targeting of the stroma ablates physical barriers to treatment of pancreatic ductal adenocarcinoma. Cancer Cell. 2012;21:418–429. doi: 10.1016/j.ccr.2012.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Giampieri S, Manning C, Hooper S, et al. Localized and reversible TGFβ signalling switches breast cancer cells from cohesive to single cell motility. Nat Cell Biol. 2009;11:1287–1296. doi: 10.1038/ncb1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Heuberger J, Birchmeier W. Interplay of cadherin-mediated cell adhesion and canonical Wnt signaling. Cold Spring Harb Perspect Biol. 2010;2:a002915. doi: 10.1101/cshperspect.a002915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ai J, Tang Q, Wu Y, et al. The role of polymeric immunoglobulin receptor in inflammation-induced tumor metastasis of human hepatocellular carcinoma. J Natl Cancer Inst. 2012;103:1696–1712. doi: 10.1093/jnci/djr360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Andl CD, McCowan KM, Allison GL, et al. Cathepsin B is the driving force of esophageal cell invasion in a fibroblast-dependent manner. Neoplasia. 2011;12:485–498. doi: 10.1593/neo.10216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Andl CD, Fargnoli BB, Okawa T, et al. Coordinated functions of E-cadherin and transforming growth factor-β receptor II in vitro and in vivo. Cancer Res. 2006;66:9878–9885. doi: 10.1158/0008-5472.CAN-05-4157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Koenig A, Mueller C, Hasel C, et al. Collagen type I induces disruption of E-cadherin-mediated cell-cell contacts and promotes proliferation of pancreatic carcinoma cells. Cancer Res. 2006;66:4662–4671. doi: 10.1158/0008-5472.CAN-05-2804. [DOI] [PubMed] [Google Scholar]
- 38.Vermeulen L, De Sousa EMF, van der Heijden M, et al. Wnt activity defines colon cancer stem cells and is regulated by the microenvironment. Nat Cell Biol. 2010;12:468–476. doi: 10.1038/ncb2048. [DOI] [PubMed] [Google Scholar]
- 39.Goentoro L, Kirschner MW. Evidence that fold-change, and not absolute level, of β-catenin dictates Wnt signaling. Mol Cell. 2009;36:872–884. doi: 10.1016/j.molcel.2009.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sphyris N, Mani SA. pIgR: frenemy of inflammation, EMT, and HCC progression. J Natl Cancer Inst. 2012;103:1644–1645. doi: 10.1093/jnci/djr421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kong B, Michalski CW, Hong X, et al. AZGP1 is a tumor suppressor in pancreatic cancer inducing mesenchymal-to-epithelial transdifferentiation by inhibiting TGFβ-mediated ERK signaling. Oncogene. 2010;29:5146–5158. doi: 10.1038/onc.2010.258. [DOI] [PubMed] [Google Scholar]
- 42.Dumartin L, Whiteman HJ, Weeks ME, et al. AGR2 is a novel surface antigen that promotes the dissemination of pancreatic cancer cells through regulation of cathepsins B and D. Cancer Res. 2011;71:7091–7102. doi: 10.1158/0008-5472.CAN-11-1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sheikh AA, Vimalachandran D, Thompson CC, et al. The expression of S100A8 in pancreatic cancer-associated monocytes is associated with the Smad4 status of pancreatic cancer cells. Proteomics. 2007;7:1929–1940. doi: 10.1002/pmic.200700072. [DOI] [PubMed] [Google Scholar]
- 44.Hummerich L, Muller R, Hess J, et al. Identification of novel tumour-associated genes differentially expressed in the process of squamous cell cancer development. Oncogene. 2006;25:111–121. doi: 10.1038/sj.onc.1209016. [DOI] [PubMed] [Google Scholar]
- 45.Stairs DB, Bayne LJ, Rhoades B, et al. Deletion of p120-catenin results in a tumor microenvironment with inflammation and cancer that establishes it as a tumor suppressor gene. Cancer Cell. 2012;19:470–483. doi: 10.1016/j.ccr.2011.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46*.Yu M, Ting DT, Stott SL, et al. RNA sequencing of pancreatic circulating tumor cells implicates WNT signaling in metastasis. Nature. 2012;487:510–513. doi: 10.1038/nature11217. This reference is cited only in the Supplementary material. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S11. PIGR and E-cadherin expression in human PDACs.
Figure S1. Construction of organotypics and the influence of stellate cells on ECM matrix contraction and increasing cancer cell number.
Figure S2. Stellate cells have pro-survival effect on cancer cells.
Figure S3. Stellate cells enhance cancer cell invasion.
Figure S4. Primary pancreatic stellate cells in mini-organotypic 3D co-culture caused ECM gel contraction and an increase in cancer cell number.
Figure S5. Primary stellate cells have a pro-survival, pro-invasive effect on cancer cells.
Figure S6. Activated stellate cells have a pro-migratory effect on cancer cells.
Figure S7. Stellate cells alter the expression of key molecules.
Figure S8. Retrieval of mRNA from organotypic gels and gene-expression microarray analysis.
Figure S9. Gene-expression microarray analysis.
Figure S10. PIGR and E-cadherin expression in human PDACs.
Table S1. STR profile of cell lines used for the experiments.
Table S2. Primer sequences used for qRT–PCR.
Table S3. Antibodies used for immunostaining.
Table S4. Statistical summary analysis for individual outcomes in each experiment and corresponding n values.
Table S5. 146 probes demonstrating differentially expressed genes (n = 126).
Table S6. List of genes in specific subsets shown in the correlation plot in Figure 3e.


