Abstract
Cancer is an inflammatory disease of tissue that is largely influenced by the interactions between multiple cell types, secreted factors, and signal transduction pathways. While single-cell sequencing continues to refine our understanding of the clonotypic heterogeneity within tumors, the complex interplay between genetic variations and non-genetic factors ultimately affects therapeutic outcome. Much has been learned through bulk studies of secreted factors in the tumor microenvironment, but the secretory behavior of single cells has been largely uncharacterized. Here we directly profiled the secretions of ELR+ CXC chemokines from thousands of single colorectal tumor and stromal cells, using an array of subnanoliter wells and a technique called microengraving to characterize both the rates of secretion of several factors at once and the numbers of cells secreting each chemokine. The ELR+ CXC chemokines are highly redundant, pro-angiogenic cytokines that signal via either or both of the CXCR1 and CXCR2 receptors, exerting profound impacts on tumor growth and progression. We find that human primary colorectal tumor and stromal cells exhibit polyfunctional heterogeneity in the combinations and magnitudes of secretions for these chemokines. In cell lines, we observe similar variance: phenotypes observed in bulk can be largely absent among the majority of single cells, and discordances exist between secretory states measured and gene expression for these chemokines among single cells. Together, these measures suggest secretory states among tumor cells are complex and can evolve dynamically. Most importantly, this study reveals new insight into the intratumoral phenotypic heterogeneity of human primary tumors.
Introduction
Tumors comprise a complex, heterogeneous population of cells. Genomic characterization of tumors has revealed the clonotypic variations evident from mutations, amplifications, rearrangements, and translocations of oncogenes, among other genetic aspects that promote tumor growth and survival.1 This genetic variability strongly influences the observed phenotypic heterogeneity in cancer. Nonetheless, non-genetic factors such as epigenetics and stochastic variations in the tumor microenvironment also affect the states of cells present in the tumor.2 For example, dysregulated signaling and inflammation in the microenvironment can suppress anti-tumor immunity; recruit and reprogram supportive stromal cells; and promote tumor growth, invasion, angiogenesis, metastasis, and resistance to treatment.3-6 Both genetic and non-genetic sources of phenotypic heterogeneity can limit the efficacy of treatments and enable the emergence of resistance.7-9 While genomic sequencing has begun to refine the clonotypic heterogeneity within tumors,10-13 further understanding of how functional variations manifest within the tumor microenvironment is needed to inform new strategies for disrupting the intercellular networks involved in maintenance of the tumor.
Chemokines and their respective receptors play an important role in mediating intercellular communication within the tumor microenvironment.14 These factors have been implicated in the maintenance and robustness of tumor-host interactions, as well as chemotaxis.3-6 Their effects on the growth and progression of the tumor can be both indirect, through recruitment of pro-inflammatory leukocytes, and direct, through autocrine or paracrine signaling. The proangiogenic CXC chemokines that bind CXCR1 and CXCR2 have a common tri-peptide motif—Glu-Leu-Arg (ELR)—preceding a conserved motif of two non-adjacent cysteines (CXC).15-17 These ELR+ CXC chemokines and their receptors are often found to be upregulated in tumor relative to normal tissue, and are potential therapeutic targets.18-25 Both CXCL1 and CXCL5 have been observed to increase in tumors with progression of colorectal cancer by immunohistochemistry, qPCR, and ELISA using tissue lysates.26 CXCL8 is also more highly expressed in human colorectal carcinomas than normal tissue.27 The increased levels of these chemokines suggest they may have a role in the initiation and transformation of colorectal cancers, but relatively little is known about how these factors are released into the microenvironment at the single-cell level.
Here, we examined the heterogeneity among the secretory states of cells from colorectal tumors for CXCL1, CXCL5, and CXCL8. To make these measurements, we used a method called quantitative microengraving, a technique that assesses the secretion of multiple proteins from thousands of single cells in parallel.28-33 Characterizing cells from human colorectal tumor, stroma, adjacent normal tissue, and a lung metastasis, we find that single cells exhibit a range of secretory phenotypes for these three chemokines, with various magnitudes of chemokines released. These varied states of secretion are also evident among cell lines derived from colorectal tumors. Furthermore, we demonstrate that the polyfunctional phenotypes inferred from bulk measures obscure the heterogeneity in responses by single cells, and that the secretory states of individual cells can be discordant with the expression of transcripts encoding these chemokines. Together, these results indicate that the secretion of ELR+ CXC chemokines by cells resident in colorectal tumors is highly heterogeneous and plastic. In view of their known redundancies in binding to a common receptor (CXCR2), these data suggest that the heterogeneous release of these chemokines by individual cells promotes a robust, evolving, yet noisy, signaling network within the tumor microenvironment.
Methods
Clinical samples
Human primary colorectal adenocarcinoma and adjacent normal epithelium were obtained as discarded samples from Massachusetts General Hospital, as part of the Institutional Board Review approved protocol 201-P-000632/16.df. Patients were males and females between the ages of 46 and 79 years old with at least stage III colorectal adenocarcinoma. Normal epithelium was obtained as a discarded sample from an 18 year-old patient with carcinoma in situ. A lung metastasis was also obtained as a discarded sample, from a 55 year-old male with stage IV rectal cancer, from the Lahey Clinic.
Disaggregation of human primary tissue
Roughly 1 cm3 of human primary tissue, transported on ice, was washed with 50 mL of PBS then diced with No. 10 scalpels (BD Parker) into 1 mm-thick pieces. The slices were submerged in 1 mL of RPMI media (Cellgro) containing collagenase A (Roche) at 1 mg/mL and DNAse1 (Roche) at 105 units/mL for 30 min at 37 °C. Then, the solution was put through a 40 μm cell strainer (BD Parker), centrifuged ×300g for 5 min, and resuspended in F medium containing the rho-kinase inhibitor Y-27632.34 Cells were loaded into the nanowells immediately following disaggregation and cultured for 2 days at 37 °C in F medium with Y-27632, without feeder cells, to promote recovery prior to assaying for protein secretion. The entire process of disaggregating and loading cells into the nanowells was completed within roughly 1 h.
Cell culture
SW1222, LIM1215, Colo205, HT29, HCT116, HCT15, LS180, and LS147T colorectal tumor cell lines were obtained from the American Type Culture Collection (ATCC). The HCT15 colorectal tumor cell line was the kind gift of Michael S. Goldberg (Dana Farber). SW1222, LIM1215, Colo205 and HCT15 were maintained in RPMI1640 media (Cellgro) supplemented with 10% v/v HI-FBS, 100 units/mL penicillin and 100 μg/mL streptomycin (Cellgro). HT-29, HCT116, LS180, and LS147T were maintained in DMEM media (Cellgro) supplemented with 10% v/v HI-FBS, 100 units/mL penicillin and 100 μg/mL streptomycin.
Quantitative microengraving
Quantitative microengraving was performed, using poly-l-lysine slides coated with capture antibodies, as previously described.28 Prepared poly-l-lysine slides were coated with an 80 μL mixture of capture antibodies at 25 μg/mL each in a buffer comprising 50 mM sodium borate, 8 mM sucrose and 50 mM NaCl (pH 9.0), under a lifter slip in a humidity chamber overnight at 4 °C. The slides were blocked with non-fat milk in PBST (PBS with 0.05% (v/v) Tween 20) for 30 min and washed 3x with PBST and PBS prior to use. Monoclonal capture (CXCL1, clone 20326; CXCL5, clone 33170; CXCL8, clone 6217) and polyclonal detection antibodies were purchased from R&D Systems.
Replica molding with a custom-built mold was used to form a 1 mm-thick polydimethylsiloxane (PDMS, Dow Corning) array of 72 × 24 blocks, each containing a 7 × 7 grid of subnanoliter wells (50 μm × 50 μm × 50 μm). Following oxygen plasma treatment for 2 min, the array was loaded with a cell suspension, washed with growth media, and sealed with a capture slide in a hybridization chamber (Agilent Technologies, G2534A) for a set interval of time under normal cell culture conditions. Then, the capture slide was removed under a bath of media, blocked with 3% non-fat milk in PBST for 30 min, incubated with a mixture of 1 μg/mL fluorescent detection antibodies for 45 min, washed with PBST and PBS, and dried with N2 compressed gas.
The slide was imaged on a Genepix 4200AL microarray scanner and analyzed using the accompanying software (GenePix 7, MDS Corp). This software was used to quantify fluorescence intensities and score positive events as having median fluorescence intensities greater than the median plus two standard deviations of wells with negative signals. Features with saturated pixels (> 1%) or low signal-to-noise ratio (< 1) were removed for quality control. Standard curves were formed by spotting detection antibodies diluted in water onto poly-l-lysine slides (1 μL per spot) in a dilution series, drying under vacuum, and scanning on the Genepix 4200AL (Fig. S1). These standard curves were used to convert the fluorescence intensities corresponding to specific numbers of molecules of detection antibody present per 50 × 50 μm feature, assuming a 1:1 stoichiometric ratio between detection antibody and analyte in the dilute limit for the quantity of analyte.28 Reference curves for microengraving were prepared with each lot of reagents, and verified for consistency with each measurement performed. The lower limit of detection for quantitative microengraving was assessed from the presence of a signal greater than the background plus two standard deviations, and related to the number of molecules from our standard curves. GraphPad PRISM software was used to perform the Mann-Whitney test on measured secretion rates and the Chi-Square and unpaired t-tests on frequencies of secretions. A schematic of the workflow is shown in Fig. S2.
Image cytometry
Cells on the PDMS array were stained with 4 μM calcein violet (Invitrogen), 1 μM SYTOX green nucleic acid stain (Invitrogen), one test of anti-human CD326 PerCp-eFluor710 (eBioscience), and one test of anti-human CD45 AlexaFluor 647 (Biolegend), in 250 μL of growth media under a lifter slip for 1 h. Imaging of the array was performed on an automated epifluorescence microscope (Zeiss) and cells in the nanowells were enumerated using a custom software program (Enumerator; available for academic use upon request). Immunophenotyping was performed after secretion measurements were obtained, so any cells that were lost from the nanowells after microengraving would have been neglected from the analysis. Only wells with one cell were included in the analysis.
qRT-PCR
qRT-PCR was performed using TaqMan One-Step RT-PCR Master Mix (Invitrogen) and TaqMan gene expression assays for CXCL1, 5, 8, CXCR1, 2, ActB, and GAPDH (Invitrogen), on a Roche LightCycler 480II. Ct values were obtained using the second derivative method, and data were normalized to GAPDH using the delta-delta-Ct method.
Single-cell retrieval and qRT-PCR
Single cells were classified by secretory phenotype using microengraving. The arrays containing the cells were treated for 15 min with 0.25% trypsin-EDTA (Invitrogen), and then retrieved from the nanowells by robotic micromanipulation (ALS Aviso CellCelector).33 Lysis and reverse transcription of single cells was performed using the Ambion Single Cell-to-CT Kit (Invitrogen). TaqMan Gene Expression Assays (Invitrogen) were used with 40 cycles of PCR on the Fluidigm Biomark with TaqMan GTXpress Master Mix (Invitrogen). Data are presented in terms of threshold expression (Et), defined by 40 - Ct.35
Protein microarrays
A fluorescent-based, sandwich-style assay was used to estimate the quantity of secreted chemokines from different human colorectal tumor cell lines. Briefly, monoclonal antibodies for capturing secreted human CXCL1, 5, and 8 were diluted in a buffer comprising 50 mM sodium borate, 8 mM sucrose and 50 mM NaCl (pH 9.0) to a final concentration of 250 μg/mL and spotted by hand on the surface of poly-L-lysine slides (1 μL/spot) for at least 1 h at 25 °C. Slides were then placed in non-fat milk (3% w/v in PBS, pH 7.4) for 30 min at 25 °C and washed two times with PBST, dipped in water, and spun dry. The slides were each placed in 24-well Microplate Microarray Hardware (ArrayIt, MMH96). A series of each recombinant analyte CXCL1, CXCL5, and CXCL8 (R&D Systems), serially diluted in DMEM media from 0.01 to 100 ng/mL, were added to the wells (100 μL of solution per well) in order to generate standard curves (Fig. S1). In parallel, 100 μL of bulk supernatants from cell cultures were added to other wells of these partitioned slides. After incubating for 1 hr at 37 °C, the excess media containing unbound protein was removed, and each well was washed twice with PBST (2×250 μL). Then, the slide was placed on a Tecan HS 400 Pro Hybridization Station at 25 °C for incubation with 1 μg/mL of the following polyclonal detection antibodies in PBST: anti-CXCL1 labeled with Alexa488, anti-CXCL5 labeled with Alexa647, and anti-CXCL8 labeled with Alexa594. The labeled microarray slides were then scanned using a Genepix 4200AL microarray scanner (Molecular Devices) in order to quantify fluorescence intensities, generate standard curves for analyte standards, and calculate concentrations of the bulk culture supernatant.
Results
Single cells from human tumor and normal tissue secrete ELR+ CXC chemokines heterogeneously
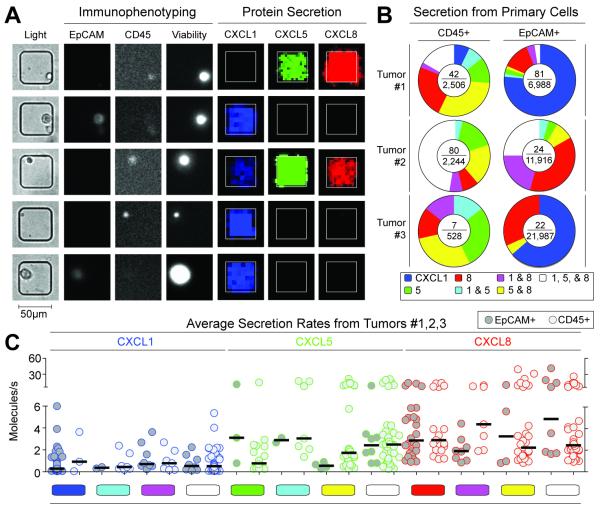
To measure directly the secretory behavior of single cells from human tumor and normal tissue, we performed quantitative microengraving for CXCL1, 5, and 8, then immunophenotyped the same cells via on-chip cytometry for CD45 and EpCAM (Fig. 1a). Cells found to be secreting versus non-secreting exhibited detectable versus undetectable levels of secretion, respectively, within the limits of detection of our assay. Small percentages of both individual tumor cells (EpCAM+; 0.1-1.2%) and leukocytes (CD45+; 1.3-3.6%) secreted quantities of CXCL1, 5, and 8 over 4 h that were greater than the limits of detection for each analyte measured (approximately 0.1 molecules/s) (Fig. 1b, Fig. S1). The combinations of secreted chemokines measured for these cells varied from single analytes (CXCL1, 5, or 8 only) to all other discrete combinations. There was no consistent difference between the percentages of populations of tumor cells and leukocytes isolated from the tumors that secreted detectable quantities. We repeated the measurements four days later and different distributions of secretory phenotypes were observed (Fig. S3), suggesting that the secretory states of tumor cells are not fixed and are sensitive to their environmental conditions.
Fig. 1. Immunophenotyping and secretion of chemokines by human primary colorectal tumor cells.
(A) Micrographs comprising transmitted light and epifluoresence images of single cells from a disaggregated tumor measured by microscopy and their secretory state measured by microengraving. (B) Pie charts of the distribution of secretion states from three human primary colorectal tumor samples (#1-3) measured two days post surgery. The center of each pie chart shows the number of observed single-cell secretion events over the total number of live CD45+ or EpCAM+ single cells, respectively. (C) Scatter plots of rates of secretion for chemokines released by single, live CD45+ leukocytes and EpCAM+ tumor cells classified by observed secretory state. Limits of detection for 4 h assays were ~0.1 molecules/s. Color-coded bars indicate the same combinations noted in Fig. 1b. The fraction of secreting leukocytes detected was higher than the tumor cells in tumor #2 (p<0.0001) and in tumor #3 (p<0.0001), but no statistically-significant differences were observed for tumor #1.
Among the three primary tumors characterized, the median rates of secretion for a given chemokine did not vary significantly as a function of the specific functional phenotype observed (CXCL1, 0.3 to 0.9 molecules/s; CXCL5, 0.6 to 3.1 molecules/s; CXCL8, 1.9 to 4.8 molecules/s) (Fig. 1c). This indifference of individual cells to their own secretory state suggests that there are little, if any, additive autocrine effects among these three chemokines. We note that the ranges of rates are also similar to those previously reported for cytokines secreted from human primary cells so it is also possible that their release is constrained by the specific secretory capacity of a given cell.28 Further supporting this possible mechanism, the overall median rates for all three chemokines did not differ significantly between tumor cells and leukocytes. Together, these results suggest that single tumor cells can exhibit diverse and independent secretory profiles from one another and that these profiles can evolve in a dynamic manner.
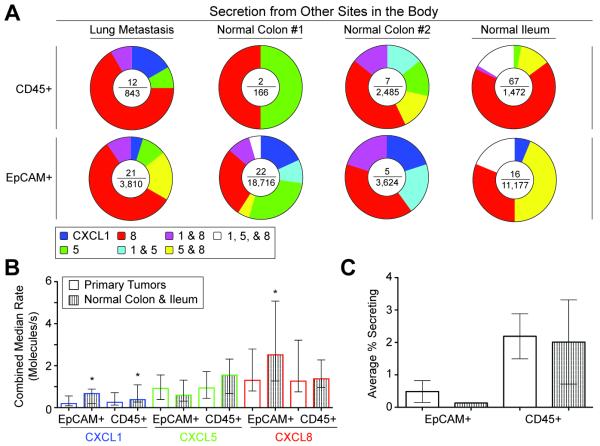
We then compared the secretory phenotypes of single cells isolated from other tissues, including a lung metastasis, two normal colon samples, and one normal ileum epithelium. Individual cells from these samples also exhibited diverse secretory profiles for CXCL1, 5 and 8 (Fig. 2a). Similar to cells from primary tumors, there were few epithelial cells or leukocytes secreting detectable quantities over 4 h. For two of the three normal samples, a higher percentage of leukocytes secreted chemokines than EpCAM+ epithelial cells (normal colon #1, p<0.005; and normal ileum, p<0.0001). Median secretion rates of CXCL1, 5, and 8 among secreting cells were also of similar magnitude between cells from both tumor and normal samples (Fig. 2b) with statistically-significant differences in CXCL1 for EpCAM+ and CD45+ cells, and in CXCL8 for EpCAM+ cells. The percentage of EpCAM+ cells isolated from tumors secreting detectable quantities of chemokines (0.49%) was slightly greater than those cells (0.13%) from normal tissue; the percentage of secreting CD45+ cells from the primary tumors (2.2%) was comparable to that from normal tissue (2.0%) (Fig. 2c). Neither of these differences was statistically significant. Previous reports have demonstrated that CXC chemokines are overexpressed in tumor microenvironments in bulk relative to normal tissues when measured by qPCR or ELISA from tissue lysates or intracellular staining by immunohistochemistry.26,27 Our data suggest that the detectable secretion of these chemokines may be regulated and that cells from the tumor may not have substantially enhanced capacities for secretion.
Fig. 2. Secretion of chemokines from other sites of the body.
(A) Pie charts of the distributions of secretory states observed from single cells of a lung metastasis, two normal colon samples, and normal ileum epithelium, measured two days post surgery. The center of each pie chart depicts the number of single-cell secretion events over the total number of live single cells. (B) Bargraph of the combined median rates of secretions of CXCL1, 5, and 8 for EpCAM+ and CD45+ cells from both normal colon and ileum and human primary colorectal tumors. EpCAM+ cells of normal tissue had higher median secretion rates of CXCL1 and CXCL8 than those of tumor tissue (p<0.05). CD45+ cells of normal tissue secreted higher median levels of CXCL1 than those of tumor tissue (p<0.05). Error bars indicate interquartile range. (C) Bargraph of percentages of EpCAM+ and CD45+ cells with measured secretions of CXCL1, 5, and 8. Error bars indicate s.e.m.
Colon tumor cell lines express mRNA for ELR+ CXC chemokines and receptors and secrete these chemokines in bulk
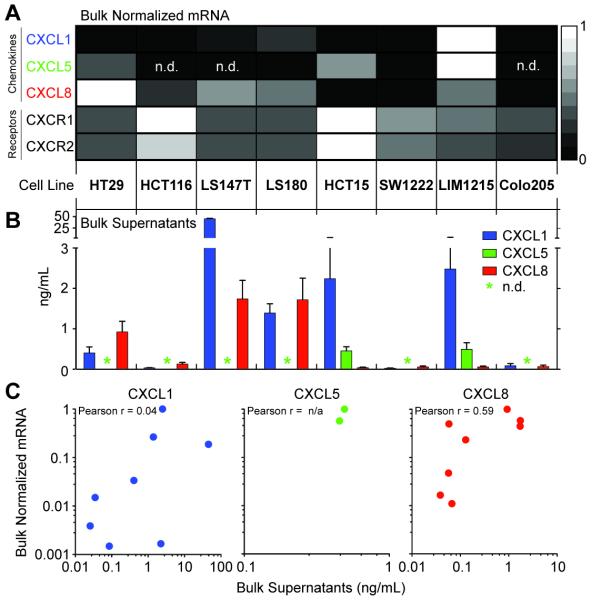
The polyfunctional diversity in secretory states observed among the heterogeneous populations of cells isolated from primary tumors led us to further characterize whether colon cancer cell lines might also exhibit a breadth of secretory states for ELR+ CXC chemokines. CXCL1 and CXCL5 signal exclusively via CXCR2, but CXCL8 signals via both CXCR1 and CXCR2. We therefore performed gene expression analysis for CXCL1, CXCL5, and CXCL8, and the chemokine receptors, CXCR1 and CXCR2 for each of eight cell lines (Fig. 3a). Five of the eight cell lines expressed transcripts for the three chemokines; HCT116, LS147T, and Colo205 did not express CXCL5. The fractional quantities of each gene varied among cell lines, however—for example, HT29, LS180, and SW1222 were near the limit of detection for CXCL5. All of the cell lines expressed similar quantities of transcripts for both chemokine receptors. These data together show that five of the eight cell lines considered here can express the three chemokines, and that they all have the capacity to produce receptors for them.
Fig. 3. Bulk mRNA expression and production of chemokines by tumor cell lines.
(A) Relative gene expression of bulk mRNA for CXCL1, 5, 8, and their receptors, CXCR1 and 2. Data were measured in triplicate and are normalized across the rows via the delta-delta CT method using GAPDH as the housekeeping gene. (B) Bargraph of the concentrations of CXCL1, 5, and 8, measured by quantitative protein microarrays in triplicate, from the supernatants from cultures of 100,000 cells of each cell line. (C) Scatterplots of normalized mRNA versus secreted protein for CXCL1, 5, and 8. Calculated Pearson correlation coefficients are shown.
We then measured the bulk quantities of CXCL1, 5, and 8 present in the supernatants collected from the same cell lines by quantitative protein microarrays (Fig. 3b). The amounts of CXCL8 correlated well with the amounts of expressed mRNA, while CXCL1 was not correlated with the gene expression across the eight lines (Fig. 3c). Only the two lines with highest amounts of CXCL5 mRNA—HCT15 and LIM1215—had detectable quantities of CXCL5. In sum, these data indicate that each of the eight cell lines is capable of producing at least two of the three chemokines.
Single cells from colon tumor cell lines also exhibit heterogeneous secretions of ELR+ CXC chemokines
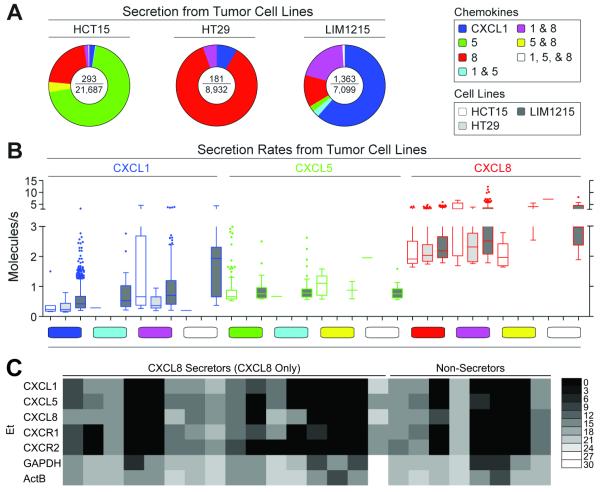
We then chose three cell lines—HCT15, HT29, and LIM1215—that each exhibited bulk production of two or three chemokines to study more extensively, at the single-cell level. Secretions of CXCL1, 5, and 8 from individual cells from each cell line were characterized by quantitative microengraving (Fig. 4a). Interestingly, most secreting cells did not exhibit the functional phenotypes inferred from the bulk measures for each line. For example, both CXCL1 and CXCL8 were evident in the supernatants from the HT29 cell line, but only 5.5% of HT29s secreted detectable quantities of both CXCL1 and CXCL8 simultaneously. The majority of HT29s secreting detectable quantities of chemokines released CXCL8 only (86.2%), while the remaining secreted CXCL1 only (8.3%). Similarly, HCT15s and LIM1215s had measureable amounts of all three chemokines in bulk, but individual cells secreting detectable quantities of all three chemokines concurrently were rare among the HCT15s (0.3%) and LIM1215s (0.9%). There was also variation among the measured rates of secretion for single cells in the three tumor cell lines (Fig. 4b). These data suggest that the breadth of secretory states for chemokines can vary at the single-cell level, even among cell lines, and that the pro-inflammatory milieu in a tumor microenvironment may develop from a combination of discrete single-cell behaviors reflecting complex phenotypes not evident from bulk measures.
Fig. 4. Single-cell production of chemokines by tumor cell lines.
(A) Pie charts of the distribution of secretory states observed from single cells of three colon cancer cell lines assayed for 2 h by microengraving. The center of each pie chart depicts the number of single-cell secretion events over the total number of live single cells. (B) Scatter plots of the rates of secretion of chemokines measured for live single cells, and classified according to secretory state, from the three colon cancer cell lines. Limits of detection for 2 h assays were approximately 0.2 molecules/s. (C) Single-cell gene expression measured by qRT-PCR for HT29 cells selected based on secretory state following microengraving for 2 h for CXCL8. Ct values were converted to Et values as described in Methods.35
Given that the bulk gene expression data for these lines showed all three genes expressed in these lines but the individual cells showed variances in their dominant secretions, we then evaluated the mRNA expression of CXCL1, 5, 8, and CXCR1 and 2, in both single HT29 cells secreting detectable quantities of CXCL8 only and those not secreting detectable amounts of any of the three chemokines (CXCL1, 5, or 8) (Fig. 4c). Both phenotypes showed varying levels of CXCL1, 5, 8, and CXCR1 and 2 mRNA, with many cells expressing transcripts for analytes not detected. Though the measures of single-cell secretion are constrained by the lower limit of detection of the assays here, these data suggest that there is discordance between the gene expression of these chemokines and the dominant secretory state of the cells over a given interval in time. This discordance also further supports a model wherein secretory states may evolve dynamically in their local environments.
Other secreted factors in the tumor microenvironment are heterogeneous
The diversity among the functional phenotypes observed for single cells producing chemokines suggested that such heterogeneity might also apply to other secreted factors commonly measured from tumors or the constituent cells. To expand our study of the tumor microenvironment, we developed a panel of antibodies to detect 26 different secreted factors previously reported from bulk analyses of primary tumors and cell lines (Fig. S4). These analytes included chemokines (CCL2, CCL5, CXCL8, CXCL12), interleukins (IL-2, IL-4, IL-5, IL-6, IL-7, IL-10, IL-13, IL-17A), monokines (IL-1β, TNF), a matrix metalloprotease (MMP9), and the following growth factors: transforming (TGF-β1, BMP-2, BMP-6), hematopoietic cell (M-CSF, GM-CSF), epidermal (EGF), hepatocyte (HGF), platelet-derived (PDGF-BB), nerve (BDNF, beta-NGF), and vascular endothelial (VEGF). We found that the viable cells isolated from six human primary colorectal tumors exhibited a broad diversity in their individual secretory signatures (Fig. S5). Interestingly, single-cell secretions of CXCL8 and MMP9 were the two most common factors observed among these samples (CXCL1 and CXCL5 were not screened in this panel). MMP9 is a matrix metalloprotease that degrades extracellular matrix and facilitates tumor cell invasion. MMP9 is reportedly overexpressed in tumor relative to normal tissue and expression has been associated with poorer prognosis in colorectal cancer.36 These data further emphasize the dynamic complexity of the tumor microenvironment as evidenced by the diverse operational states exhibited by individual cells comprising the tumor.
Discussion
The tumor microenvironment comprises a complex network of cells and proteins that facilitate the maintenance and survival of the tumor. In this paper, we directly profiled the secretory behavior of individual tumor, stromal, and adjacent epithelial cells, using quantitative microengraving over short time scales (2-4 h) with a resolution not common in cancer research. Characterization of tumors using bulk, integrated measures such as ELISA and qPCR have identified many secreted factors and genes for secreted factors that are constitutively present in the microenvironment or arise from perturbations (e.g., treatment with drugs) to the microenvironment. Our single-cell analysis here reveals, however, that individual cells can contribute to this inflammatory milieu using a complex set of functional phenotypes. We find that the phenotypic state suggested by a bulk measure does not accurately reflect the states of the individual cells contributing to the overall milieu (Fig. S6). This insight emphasizes that bulk measures can reflect average states discordant from those at the single-cell level and obscure rare operational behaviors of single cells by their intrinsic reduction of spatiotemporal dimensionality.7
We demonstrate that single cells isolated from human primary tumors and tumor cell lines can exhibit a variety of secretory states at any given instance, and show that these phenotypic states of secretion can evolve. Indeed, only a small percentage of cells secrete detectable quantities of chemokines over short time intervals (2-4 h). These single-cell behaviors suggest that small fractions of highly active cells, acting with different functional phenotypes, could contribute to alter the tumor microenvironment dynamically (Fig. 5). In this model, the robustness and redundancy of intercellular signaling via the ELR+ CXC chemokines can be appreciated—especially since CXCL1, 5, and 8 all have similar EC50’s for CXCR2.37 Interestingly, similar secretory behaviors were also present among single cells of normal tissue. Therefore, an increased frequency of highly active secreting cells within the tumor, driven by autocrine and paracrine signaling—rather than the increased magnitude of production by any individual cell—could promote elevated levels and may support tumor progression via positive feedback.
Fig. 5. Proposed model for ELR+ CXC chemokine signaling in the tumor microenvironment.
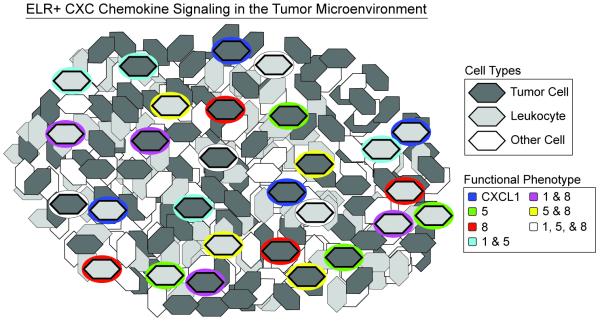
A small fraction of leukocytes and tumor cells secrete robustly at any given instance in time with discrete functional phenotypes that can change with time. The measurements in this study cannot infer the spatial positioning of the cells in the tumor and positioning shown is schematic.
We examined here the heterogeneity and polyfunctionality in secretions of three ELR+ CXC chemokines, but there are four others (CXCL2, 3, 6, and 7) not included in this study. These additional factors likely compound the phenotypic heterogeneity observed here further. While there are opportunities to therapeutically modulate these networks, our data suggest that such targets are complex and may require approaches that target all possible phenotypic states within the tumor microenvironment. It also suggests that therapeutics that target common receptors such as CXCR1 or CXCR2 may be more effective than those targeting one of the pleotropic chemokines (e.g., CXCL8). For instance, the knockdown of CXCR2 in ovarian cancer cells leads to arrested cell cycle in G0-G1, decreased angiogenesis, reduced secretion of ELR+ CXC chemokines, and increased apoptosis.25
The heterogeneous secretory states observed for isolated single cells from tumors represent the potential landscape of phenotypes available to both epithelial cells and leukocytes. The measurements on individual cells here do not account for how they may behave when in contact with different cells within the tissue. One opportunity for further characterization of this perturbation on the states of individual cells is to examine the synergistic or antagonistic effects of cell-cell interactions on the secretory states or phenotypic behaviors.38 Such measures should further reveal the dynamic intercellular networks governing communication within the tumor microenvironment. Also, one implication of the observations here is that in vitro models that rely on static production of chemokines or growth factors to maintain gradients may imperfectly reflect the temporal dynamics of such factors as modulated by stochastic bursts in secretion by cells. From our measurements, a new understanding of ELR+ CXC chemokine signaling in the tumor microenvironment begins to emerge, involving temporal and discrete fluctuations among small subsets of highly active secreting cells.
Supplementary Material
Acknowledgements
The authors thank Alex K. Shalek for his guidance with single-cell gene expression. We also thank the Koch Institute Swanson Biotechnology Center for technical support, specifically the BioMicroCenter. The authors acknowledge Janssen Pharmaceuticals, Inc., for its generous support of this work. This work was also supported in part by the Koch Institute Support (core) Grant P30-CA14051 from the National Cancer Institute. V.A. was supported in part by the National Science Foundation’s Graduate Research Fellowship Program. X.Y. is supported by the National Science Scholarship of the Agency for Science, Technology and Research (A*STAR). A.A. acknowledges financial contribution from the Swiss National Science Foundation (SNSF Fellowships for Advanced Researchers PA00P3 139659). J.C.L. is a Latham Family Career Development Professor and a Camille Dreyfus Teacher-Scholar. The authors dedicate this paper to the memory of Officer Sean Collier, for his caring service to the MIT community and for his sacrifice.
References
- 1.Greaves M, Maley CC. Nature. 2012;481:306–313. doi: 10.1038/nature10762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Marusyk A, Almendro V, Polyak K. Nat Rev Cancer. 2012;12:323–334. doi: 10.1038/nrc3261. [DOI] [PubMed] [Google Scholar]
- 3.Straussman R, Morikawa T, Shee K, Barzily-Rokni M, Qian ZR, Du J, Davis A, Mongare MM, Gould J, Frederick DT, Cooper ZA, Chapman PB, Solit DB, Ribas A, Lo RS, Flaherty KT, Ogino S, Wargo JA, Golub TR. Nature. 2012;487:500–506. doi: 10.1038/nature11183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Balkwill F. Nat Rev Cancer. 2004;4:540–550. doi: 10.1038/nrc1388. [DOI] [PubMed] [Google Scholar]
- 5.Hanahan D, Weinberg RA. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 6.Grivennikov SI, Greten FR, Karin M. Cell. 2010;140:883–899. doi: 10.1016/j.cell.2010.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bendall SC, Nolan GP. Nat Biotechnol. 2012;30:639–647. doi: 10.1038/nbt.2283. [DOI] [PubMed] [Google Scholar]
- 8.Taniguchi K, Okami J, Kodama K, Higashiyama M, Kato K. Cancer Science. 2008;99:929–935. doi: 10.1111/j.1349-7006.2008.00782.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Seol H, Lee HJ, Choi Y, Lee HE, Kim YJ, Kim JH, Kang E, Kim S-W, Park SY. Mod. Pathol. 2012;25:938–948. doi: 10.1038/modpathol.2012.36. [DOI] [PubMed] [Google Scholar]
- 10.Hou Y, Song L, Zhu P, Zhang B, Tao Y, Xu X, Li F, Wu K, Liang J, Shao D, Wu H, Ye X, Ye C, Wu R, Jian M, Chen Y, Xie W, Zhang R, Chen L, Liu X, Yao X, Zheng H, Yu C, Li Q, Gong Z, Mao M, Yang X, Yang L, Li J, Wang W, Lu Z, Gu N, Laurie G, Bolund L, Kristiansen K, Wang J, Yang H, Li Y, Zhang X, Wang J. Cell. 2012;148:873–885. doi: 10.1016/j.cell.2012.02.028. [DOI] [PubMed] [Google Scholar]
- 11.Xu X, Hou Y, Yin X, Bao L, Tang A, Song L, Li F, Tsang S, Wu K, Wu H, He W, Zeng L, Xing M, Wu R, Jiang H, Liu X, Cao D, Guo G, Hu X, Gui Y, Li Z, Xie W, Sun X, Shi M, Cai Z, Wang B, Zhong M, Li J, Lu Z, Gu N, Zhang X, Goodman L, Bolund L, Wang J, Yang H, Kristiansen K, Dean M, Li Y, Wang J. Cell. 2012;148:886–895. doi: 10.1016/j.cell.2012.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gerlinger M, Rowan AJ, Horswell S, Larkin J, Endesfelder D, Gronroos E, Martinez P, Matthews N, Stewart A, Tarpey P, Varela I, Phillimore B, Begum S, McDonald NQ, Butler A, Jones D, Raine K, Latimer C, Santos CR, Nohadani M, Eklund AC, Spencer-Dene B, Clark G, Pickering L, Stamp G, Gore M, Szallasi Z, Downward J, Futreal PA, Swanton C. N. Engl. J. Med. 2012;366:883–892. doi: 10.1056/NEJMoa1113205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Navin N, Kendall J, Troge J, Andrews P, Rodgers L, McIndoo J, Cook K, Stepansky A, Levy D, Esposito D, Muthuswamy L, Krasnitz A, McCombie WR, Hicks J, Wigler M. Nature. 2011;472:90–94. doi: 10.1038/nature09807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Roussos ET, Condeelis JS, Patsialou A. Nat Rev Cancer. 2011;11:573–587. doi: 10.1038/nrc3078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lazennec G, Richmond A. Trends in Molecular Medicine. 2010;16:133–144. doi: 10.1016/j.molmed.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rossi D, Zlotnik A. Annual Review of Immunology. 2000;18:217–242. doi: 10.1146/annurev.immunol.18.1.217. [DOI] [PubMed] [Google Scholar]
- 17.Moser B, Wolf M, Walz A, Loetscher P. Trends Immunol. 2004;25:75–84. doi: 10.1016/j.it.2003.12.005. [DOI] [PubMed] [Google Scholar]
- 18.Ning Y, LaBonte MJ, Zhang W, Bohanes PO, Gerger A, Yang D, Benhaim L, Paez D, Rosenberg DO, Nagulapalli Venkata KC, Louie SG, Petasis NA, Ladner RD, Lenz H-J. Molecular Cancer Therapeutics. 2012;11:1353–1364. doi: 10.1158/1535-7163.MCT-11-0915. [DOI] [PubMed] [Google Scholar]
- 19.Varney ML, Singh S, Li A, Mayer-Ezell R, Bond R. Cancer Letters. 2011;300:180–188. doi: 10.1016/j.canlet.2010.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ginestier C, Liu S, Diebel ME, Korkaya H, Luo M, Brown M, Wicinski J, Cabaud O, Charafe-Jauffret E, Birnbaum D, Guan J-L, Dontu G, Wicha MS. J. Clin. Invest. 2010;120:485–497. doi: 10.1172/JCI39397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ijichi H. Oncoimmunology. 2012;1:569–571. doi: 10.4161/onci.19402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Keane MP, Belperio JA, Xue YY, Burdick MD, Strieter RM. J. Immunol. 2004;172:2853–2860. doi: 10.4049/jimmunol.172.5.2853. [DOI] [PubMed] [Google Scholar]
- 23.Mestas J, Burdick MD, Reckamp K, Pantuck A, Figlin RA, Strieter RM. J. Immunol. 2005;175:5351–5357. doi: 10.4049/jimmunol.175.8.5351. [DOI] [PubMed] [Google Scholar]
- 24.Singh S, Sadanandam A, Nannuru KC, Varney ML, Mayer-Ezell R, Bond R, Singh RK. Clin. Cancer Res. 2009;15:2380–2386. doi: 10.1158/1078-0432.CCR-08-2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang G, Rosen DG, Liu G, Yang F, Guo X, Xiao X, Xue F, Mercado-Uribe I, Huang J, Lin SH, Mills GB, Liu J. Clinical Cancer Research. 2010;16:3875–3886. doi: 10.1158/1078-0432.CCR-10-0483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rubie C, Frick VO, Wagner M, Schuld J, Gräber S. BMC Cancer. 2008;8:178. doi: 10.1186/1471-2407-8-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Baier P, Eggstein S, Wolff-Vorbeck G. Anticancer Research. 2005;25:3581–3584. [PubMed] [Google Scholar]
- 28.Han Q, Bradshaw EM, Nilsson B, Hafler DA, Love JC. Lab Chip. 2010;10:1391–1400. doi: 10.1039/b926849a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Love JC, Ronan JL, Grotenbreg GM, van der Veen AG, Ploegh HL. Nat Biotechnol. 2006;24:703–707. doi: 10.1038/nbt1210. [DOI] [PubMed] [Google Scholar]
- 30.Varadarajan N, Kwon DS, Law KM, Ogunniyi AO, Anahtar MN, Richter JM, Walker BD, Love JC. Proceedings of the National Academy of Sciences. 2012;109:3885–3890. doi: 10.1073/pnas.1111205109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Varadarajan N, Julg B, Yamanaka YJ, Chen H, Ogunniyi AO, McAndrew E, Porter LC, Piechocka-Trocha A, Hill BJ, Douek DC, Pereyra F, Walker BD, Love JC. J. Clin. Invest. 2011;121:4322–4331. doi: 10.1172/JCI58653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Panagiotou V, Love KR, Jiang B, Nett J, Stadheim T, Love JC. Appl. Environ. Microbiol. 2011;77:3154–3156. doi: 10.1128/AEM.00104-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ogunniyi AO, Story CM, Papa E, Guillen E, Love JC. Nat Protoc. 2009;4:767–782. doi: 10.1038/nprot.2009.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu X, Ory V, Chapman S, Yuan H, Albanese C, Kallakury B, Timofeeva OA, Nealon C, Dakic A, Simic V, Haddad BR, Rhim JS, Dritschilo A, Riegel A, McBride A, Schlegel R. Am. J. Pathol. 2012;180:599–607. doi: 10.1016/j.ajpath.2011.10.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dominguez MH, et al. Highly multiplexed quantitation of gene expression on single cells. J. Immunol. Methods. 2013;391:133–145. doi: 10.1016/j.jim.2013.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang Y, Guan X-Y, Dong B, Zhao M, Wu J-H, Tian X-Y, Hao C-Y. J. Cancer Res. Clin. Oncol. 2012;138:2035–2044. doi: 10.1007/s00432-012-1274-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ahuja SK, Murphy PM. J. Biol. Chem. 1996;271(34):20545–20550. doi: 10.1074/jbc.271.34.20545. [DOI] [PubMed] [Google Scholar]
- 38.Yamanaka YJ, Berger CT, Sips M, Cheney PC, Alter G, Love JC. Integrative Biology. 2012;4:1175–1184. doi: 10.1039/c2ib20167d. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.