Abstract
The abundance and ancient origins of transposable elements (TEs) in eukaryotic genomes has spawned research into the potential symbiotic relationship between these elements and their hosts. In this review, we introduce the diversity of TEs, discuss how distinct classes are uniquely regulated in development, and describe how they appear to have been coopted for the purposes of gene regulation and the orchestration of a number of processes during early embryonic development. Although young, active TEs play an important role in somatic tissues and evolution, we focus mostly on the contributions of the older, fixed elements in mammalian genomes. We also discuss major challenges inherent in the study of TEs and contemplate future experimental approaches to further investigate how they coordinate developmental processes.
Keywords: transposon, endogenous retrovirus, evolution, development, placentation, gene regulation
Introduction
TEs comprise a substantial fraction of eukaryotic genomes; they account for over forty times the nucleotide content as protein-coding exons [1], equating to nearly half of the human genome [2]. Recent data suggest that the percentage may even be closer to two-thirds [3]. This fact re-raises a long-standing question: do TEs serve any functional role for their host? Although there is a considerable literature on this topic [4–7], there remain few direct data demonstrating a requirement for TEs during embryo development. This review discusses research focusing on recent evidence that TEs function in numerous early embryonic developmental processes. We start with a brief review of the various classes of eukaryotic TEs and the general types of processes that have lead to the genomes we see today.
Snapshot of mammalian genomic TEs
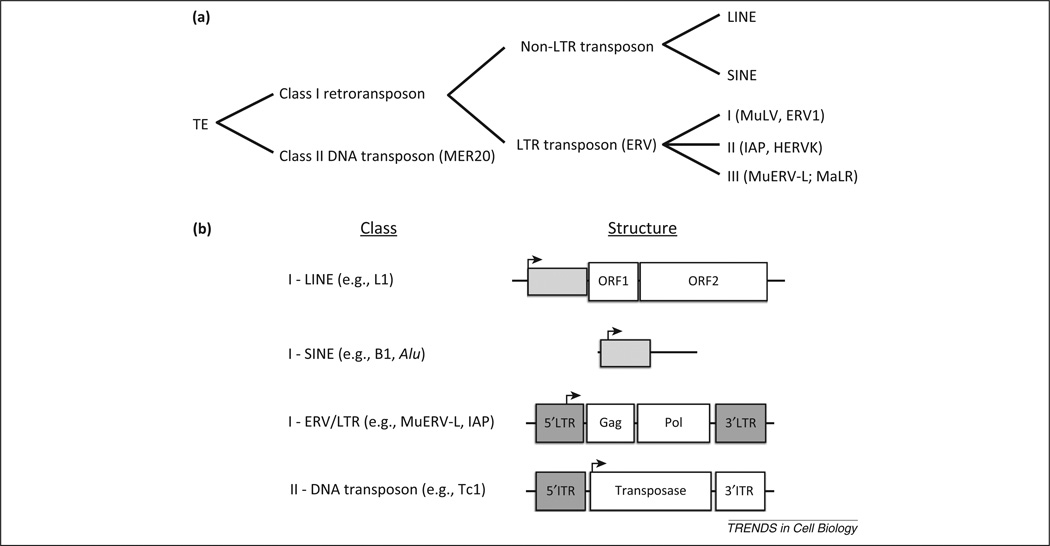
TEs are broadly characterized as either Class I retrotransposons or Class II DNA transposons (Figure 1a). Class II DNA transposons (Tc1/mariner) encode a transposase enzyme that excises the parental sequence and mediates reintegration in another location (Figure 1b). Hence, they use a ‘cut and paste’ mechanism and do not readily accumulate in copy number. We refer the reader to an insightful review on DNA transposons for further information [8]. By contrast, Class I retrotransposons are transcribed into an RNA intermediate that may then be reverse transcribed into DNA and reintegrated as an additional copy elsewhere in the genome. This ‘copy and paste’ mechanism explains the abundance of these elements.
Figure 1.
Classification of mammalian transposable elements (TEs). (a) TE are broadly categorized as DNA transposons or retrotransposons. Retrotransposons are further defined based on the presence of a long terminal repeat (LTR) that contains the element’s regulatory information. Non-LTR retrotransposons comprise long interspersed nuclear elements (LINEs) and short interspersed nuclear elements (SINEs), whereas LTR retrotransposons (endogenous retroviruses [ERVs]) are further divided among three subclasses: I, II, and III. Examples of each subclass follow in parentheses. (b) The generic structures of major classes of TE are shown. LINE elements harbor a promoter sequence that is retained in the final transcript, shown in light grey, and two ORFs that mediate reverse transcription and transposition. SINE elements do not encode protein and rely on machinery encoded by LINE elements for their transposition. Most endogenous viral elements (EVEs) in mammalian genomes are endogenous retroviruses, some of which encode functional Gag and Pol proteins. More rarely, envelope proteins may also be present (not shown). These sequences are flanked by a direct LTR. Hence, these are named LTR-class retrotransposons. Most DNA transposons encode a transposase that cleaves and reinserts the parental element in a new location. The transposase is flanked by inverted terminal repeats (ITR).
Mammalian class I retrotransposons are further divided into two subdivisions defined by the presence or absence of flanking long terminal repeats (LTRs) (Figure 1b). Non-LTR retrotransposons include long interspersed nuclear elements (LINEs) and short interspersed nuclear elements (SINEs). Both of these elements have transpositionally active subfamilies in human (LINE-1, Alu) and mouse (LINE-1, SINE-B1, SINE-B2), although SINEs require protein encoded by LINEs for retrotransposition [9–12]. LTR retrotransposons [including endogenous retroviruses (ERVs)] derive from infectious retroviruses that integrated in the germline [13,14]. They are thus present clonally in all cells of their host. Most ERVs do not encode a functional envelope protein and are incapable of horizontal transmission. ERVs are further categorized into three classes – I, II, or III – depending on the exogenous retrovirus genus they most closely resemble [15]. Numerous subtypes of ERVs are active transposons in mice, including intracisternal A-particles (IAPs) [16] and MuERV-L [17] (discussed below). At least one human subtype, HERV-K, is capable of producing intact viral particles in humans [18]. ERVs, however, are not the only endogenous viral elements (EVEs) embedded in animal genomes [19]. At least ten non-retroviral families have endogenous counterparts, some of which are transcribed and harbor open reading frames [20]. With this in mind, we expect to see future studies implicating non-retroviral EVEs in an array of important biological phenomena for their host.
The balance of selective forces acting on TEs is complex. Selection for active transposition has kept many subfamilies active for millions of years after their initial infection, whereas deleterious transposition events are rapidly purged from the genome [21]. Suspected TE-induced tumorigenic events have been reported [22], providing further evidence that new integration events are sometimes harmful to the host. One mechanism by which an ERV can be largely eliminated is through non-allelic homologous recombination between its two adjacent LTRs in cis, thereby excising the intervening sequence and leaving a single ‘solo’ LTR in its place. Solo LTRs, which outnumber full-length ERVs within mammalian genomes [23], can also be generated when two non-allelic LTRs recombine in trans. This process simultaneously leads to segmental duplications on the alternate chromosome. Some elements remain subject to neutral forces and slowly drift from their parental sequence and eventually lose the ability to transpose [21], whereas others remain intact for longer periods but are epigenetically silenced by various mechanisms. One such mechanism is DNA methylation, which accelerates the process of TE mutation through deamination of 5-methylcytosine residues to thymine [24–26]. The cumulative effects of these factors explain the genomes we see today: a heterogeneous mixture of elements – full length and truncated, active and broken, modern and ancient – distributed throughout the genome. Many of the resultant TE sequences are under strong purifying selection, despite being transcriptionally or transpositionally inactive, suggesting they retain function [27]. With this backdrop in mind, we will first highlight the mechanisms utilized by the host to keep TEs transcriptionally silent and then turn to the functional role that TEs play in their hosts.
Regulation of ERVs in the early mouse embryo
One area under active investigation is how host cells control the activity of TEs to prevent widespread retrotransposition. Recent evidence demonstrates that mouse cells utilize histone modification machinery to keep ERV subfamilies silenced in early embryos and embryonic stem (ES) cells, and that this silencing is independent of the DNA methylation machinery that is required for their silencing in somatic cells [15,28]. One such subfamily is the class III ERV MuERV-L, which belongs to an ancient endogenous retrovirus family termed ERV-L [29]. These ERV-L elements are likely to predate the placental mammal radiation (70 Mya) and are present in at least ten approximately full-length copies in all placentals examined to date, but generally do not encode intact viral proteins and, like most ERVs, do not harbor an env gene [17,29]. The exception to this includes ERV-L elements from mouse (MuERV-L), all extant simians (including humans [HERV-L]), and uranotherians (including elephants), which retain coding (mouse) or near-coding capacity (humans and uranotherians) for gag and pol genes [17,29–31]. The increased coding capacity is linked with the expansion of these elements in copy number in each of these lineages. In the case of the mouse, this expansion occurred after the mouse–rat divergence, because rats have ancestral levels of ERV-L [29] whereas mice contain roughly 350 fully coding elements per haploid genome [32]. The recent amplification and activity of these elements in the mouse lineage has necessitated tight control over transcription to prevent mutagenic events.
Many factors participate in the silencing of MuERV-L elements in ES cells and blastocyst-stage embryos, including the histone demethylase LSD1/KDM1A, the corepressor KAP1/Trim28, and histone deacetylases (HDACs) [33,34] (Figure 2). However, MuERV-L is normally expressed at the onset of zygote genome activation but is silenced by the blastocyst stage in wild type embryos; by contrast, Kdm1a mutant blastocysts continue to express MuERV-L and subsequently fail to complete gastrulation [32,33,35] (Figure 2). Chromatin IP experiments in Kdm1a mutant ES cells demonstrate that the de-repressed MuERV-L elements have increased levels of acetylated histones and methylation of histone H3 lysine 4 (H3K4), consistent with their activated status and KDM1A’s known ability to demethylate histone H3K4. MuERV-L elements also appear to be silenced in ES cells by the histone methyltransferase G9A, the pluripotency marker REX1/ZFP42, and the polycomb-associated protein RYBP, because genetic removal or knockdown of these factors in ES cells also leads to de-repression of MuERV-L elements [36–38]. Current evidence is consistent with these repressors acting directly on MuERV-L sequences, including evidence from chromatin immunoprecipitation (ChIP) assays in the case of ZFP42 [36] and RYBP [37] and changes in histone modification associated with loss of KDM1A [33], but indirect mechanisms cannot be excluded. Indeed, de-repression of MuERV-L elements in Kdm1a mutants is concomitant with decreases in histone H3 lysine 9 (H3K9) methylation, which cannot be accounted for directly by loss of KDM1A. Furthermore, although ZFP42 and KDM1A are both linked with repression of MuERV-L and to each other [36], KDM1A is required to silence the viral promoter whereas Rex1 associates with the viral Gag and dUTPase genes [33,36]. Thus, numerous questions regarding the silencing mechanism of MuERV-L elements remain to be solved.
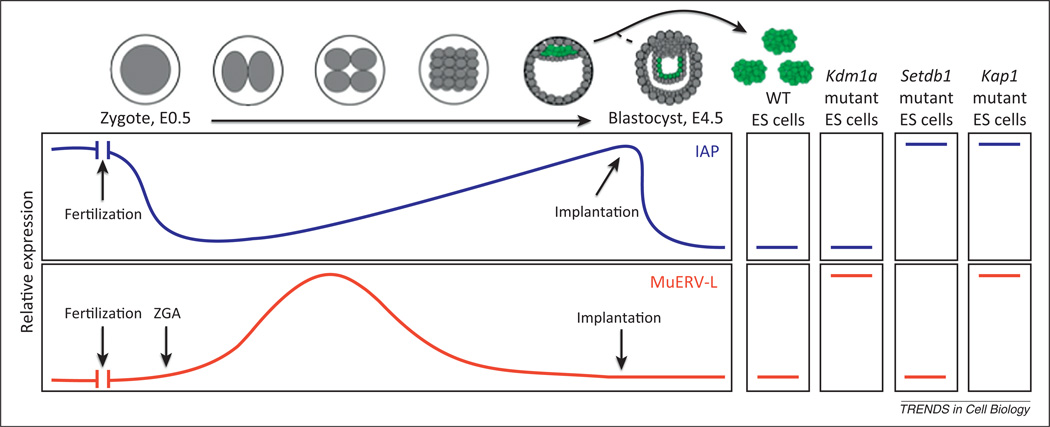
Figure 2.
Endogenous retrovirus (ERV) expression in early development and embryonic stem (ES) cells. Intracisternal A-particles (IAPs) and MuERV-L elements show divergent expression patterns during early mouse development. IAPs are expressed in oocytes and transcripts are carried over into the fertilized embryo. Early embryonic expression remains low but increases until the blastocyst stage. On implantation, IAPs are again silenced. By contrast, MuERV-L is not expressed in oocytes but is expressed at the onset of zygotic genome activation (ZGA) shortly after fertilization. MuERV-L is silenced by the late morula stage. Wild type (WT) ES cells (green) are generally derived from the inner cell mass from E3.5 embryos, but some characteristics of ES cells are more in common with post-implantation epiblast from E4.5 embryos (such as IAP expression). In wild type ES cells, both IAPs and MuERV-Ls are silenced. MuERV-L silencing is compromised in Kdm1a and Kap1 mutants but not Setdb1 mutants, whereas IAPs are activated in Setdb1 and Kap1 mutants but not Kdm1a mutants.
In contrast to the MuERV-L elements, the class II IAP elements are expressed at increasingly higher levels up to the blastocyst stage, after which they are silenced (Figure 2) [15,39–41]. Within ES cells and peri-implantation embryos, the histone methyltransferase SETDB1/ESET and the corepressor KAP1/TRIM28 mediate the silencing of IAP elements via trimethylation of H3K9 (H3K9Me3) and histone H4 lysine 20 (H4K20Me3) [34,42]. Importantly, however, SETDB1-mediated H3K9me3 is not found at MuERV-L elements and MuERV-Ls are not affected by SETDB1 deletion, although MuERV-Ls are activated in KAP1 mutants. Thus, distinct KAP1 complexes must be present that distinguish MuERV-L elements and IAP elements.
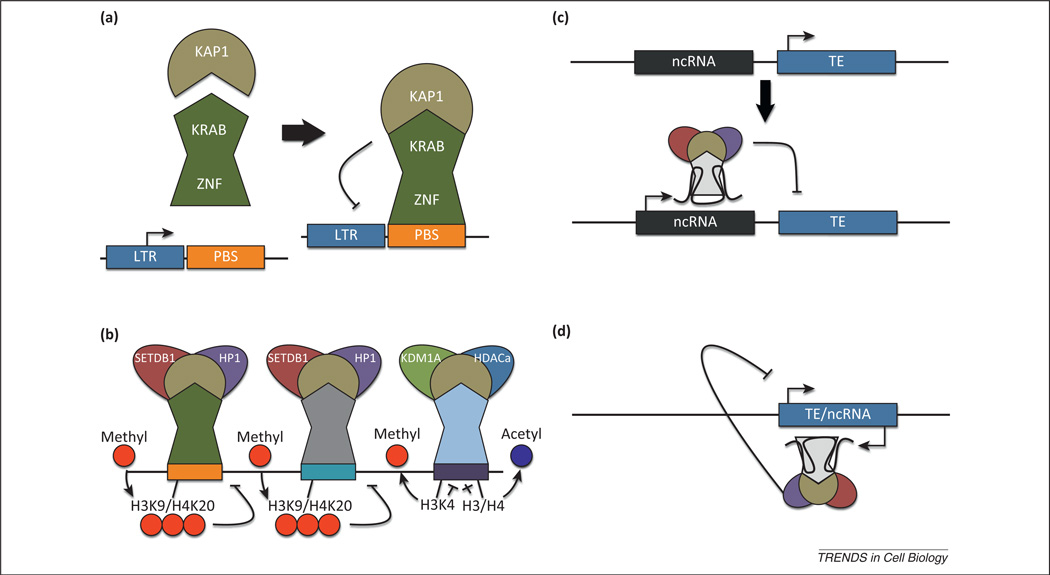
So how exactly are ERVs and other transposable elements recognized by distinct activating and silencing machinery? In the case of MuERV-L elements, the LTR promoter and primer-binding site (PBS), which is essential for tRNA-mediated priming of reverse transcription, contain all of the regulatory information required for their expression and silencing, because reporter genes cloned downstream of these elements perfectly mark MuERV-L-expressing cells within embryos as well as within ES cells [33,38]. A 500-bp region encompassing the 5′ untranslated region (UTR) of IAPs also recruits KAP1 and SETDB1 repression complexes, although it is unclear what factors are critical for this site-specific recognition [34,43,44]. One possibility is that the repressive machinery is directly recruited via Kruppel-associated box domain–zinc finger proteins (KRAB–ZFPs) specific for each ERV (Figure 3a,b) [15,45]. This idea is supported by the striking correlation between the number of LTR retroelements and the number of zinc finger (ZF) domains found in vertebrates and by the strong positive selection observed for the ZF domains [46]. One member of this family, ZFP809, recognizes the PBS within murine leukemia virus (MuLV), which in turn recruits the KAP1 corepressor via its KRAB domain to silence the MuLV [47]. Thus, a large pool of ZFPs may have competed in an evolutionary ‘arms race’ with retroelements to recognize and silence their promoters via recruitment of KAP1 repression complexes [46].
Figure 3.
Hypothetical mechanisms for transposable element (TE) recognition and silencing. (a) Kruppel-associated box domain–zinc finger proteins (KRAB-ZFPs) harbor an N-terminal KRAB domain that mediates interactions with the corepressor KAP1 and a C-terminal zinc finger domain that mediates DNA binding with sequence specificity. The zinc finger domains may bind to the primer binding site (PBS) in some ERVs, whereas it may bind to the LTR, gag, or pol domain in others. (b) The zinc finger domains across hundreds of KRAB–ZFPs are under positive selection, suggesting they may be coevolving with TEs to silence their expression. Individual KRAB–ZFPs recognize unique classes of TE and recruit distinct KAP1 complexes. SETDB1-containing complexes catalyze the addition of methyl groups to histone H3 lysine 9 (H3K9) and histone H4K20, whereas KDM1A/HDAC complexes catalyze removal of methyl groups from H3K4 and acetyl groups from multiple residues on histones H3 and H4. (c) Alternatively, noncoding RNAs (ncRNAs), capable of forming complex secondary structures, may silence nearby TEs by recruiting corepressor complexes through RNA–protein interactions. (d) Antisense ncRNAs transcribed from a TE may recruit corepressor complexes to silence its own expression. Such long ncRNAs have been shown to recruit KDM1A complexes to the hox locus.
An alternative possibility is that the silencing machinery is recruited and tethered to ERVs by non-coding RNAs in cis (Figure 3c,d). Forward- and reverse-strand transcripts of IAP and MuERV-L elements have been identified in mouse embryos [48]. In addition, corepressor complexes containing KDM1A, which participates in silencing MuERV-L elements, are recruited to other genomic loci by long non-coding RNAs in cis, including the Hox locus [49]. Thus it is possible that non-coding RNAs initiated upstream or within ERVs themselves are important for mediating ERV silencing in cis.
Despite the many factors utilized by the host to silence ERVs and other transposons, there are periods in development when ERVs are able to evade silencing (Figure 2). These periods appear to correlate with major waves of epigenomic reprogramming. During preimplantation development, for example, when most of the genome is demethylated to reset the genome to pluripotency, IAP elements are partially demethylated and transcribed [15]. Additionally, MuERV-L elements are transcribed at the one- and two-cell stage at a time when the paternal genome is stripped of its protamine packaging and actively demethylated in a mechanism dependent on the Tet3 dioxygenase [50]. Thus, ERVs and other retroelements may take advantage of developmental epigenomic reprogramming windows to evade silencing by chromatinmodifying enzymes [28].
Retroviruses contribute genes important to placenta function
Much progress has been made in understanding the regulation of TEs in mice. A similar burst in evidence supporting a functional role for TEs is also emerging. The best example comes from the syncytin proteins, which have been captured from distinct retroviral envelope (env) genes independently in multiple mammalian lineages [51] (Figure 4a). Consistent with the function of envelope proteins in mediating fusion of viruses with cell membranes, the SyncytinA gene in mice is required for placental development by mediating cell–cell fusion in the trophoblast [52]. Envelope proteins are also known to be immunosuppressive and it has been speculated that infection by a retrovirus and domestication of an env gene was a prerequisite for the evolution of placental mammals, because mammals have highly adaptive immune systems yet fail to recognize their own allogeneic embryos [53]. Gag proteins from retroviruses have also been domesticated for the purpose of blocking other virus infections. Most notably, the Fv1 restriction gene in mice is derived from the MuERV-L Gag protein and this protein mediates restriction from infection with MuLVs [17,54,55]. Thus, ERVs have provided useful gene products domesticated for specialized functions in placental mammals.
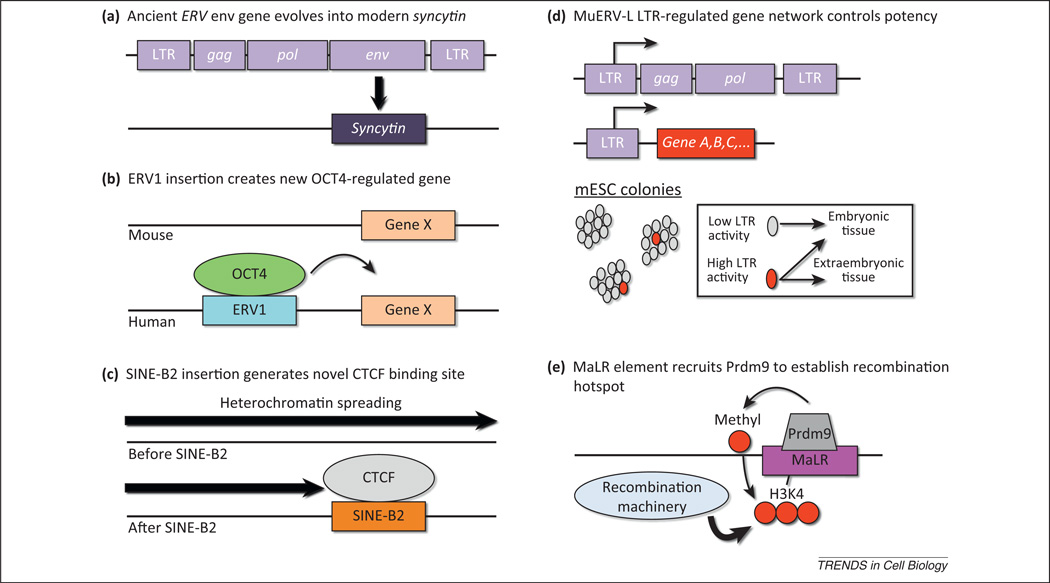
Figure 4.
Strategies by which transposable elements (TEs) have been coopted for development. Host organisms have evolved various means to take advantage of TE sequences incorporated in their genome, some of which are illustrated here. (a) Ancient endogenous retrovirus (ERV) env genes have evolved into present-day syncytins, important for syncytiotrophoblast formation in placental mammals. (b) ERV1 and other transposons carry transcription factor-binding sites for OCT4, which can regulate genes near insertion sites, leading to novel regulatory patterns in evolving mammals. (c) Short interspersed nuclear element (SINE)-B2 elements carry sequence motifs recognized by the insulator-binding protein CTCF and thereby regulate chromatin architecture. In the illustration, heterochromatin spreading is blocked by a newly inserted SINE-B2 element. (d) MuERV-L elements and their remnant ‘solo’ long terminal repeats (LTRs) have been coopted to participate in gene-regulatory networks, by serving as primary or alternative promoters of nearby genes. A subset of mouse embryonic stem cells expresses MuERV-L LTR-driven chimeric transcripts, which correlates with increased potency. (e) Certain classes of TE (Mammalian apparent LTR retrotransposons [MaLRs]) may recruit PR domain containing 9 (Prdm9), a histone H3 lysine 4 (H3K4) methyltransferase. Trimethylated H3K4 (H3K4me3) enhances recruitment of machinery important for initiating meiotic recombination, thus leading to the formation of species-specific recombination hotspots.
TEs rewire gene-regulatory networks
In addition to providing proteins that are critical for embryo development, TEs have exerted equally important effects on gene-regulatory networks. When TEs integrate into new sites within the genome, they can disrupt or alter patterns of gene expression because TE sequences also carry potential regulatory information. We present supporting evidence below that transposon-mediated rewiring of gene-expression networks is likely to have directly contributed to the evolution and diversification of mammals.
There are several striking examples of TEs that contributed to novel gene-regulatory networks in mammals by carrying transcription factor-binding sites. This is perhaps best exemplified by the genome-wide binding profiles of the transcription factors OCT4 and NANOG and the insulator-binding protein CTCF in different mammalian species. Using ChIP sequencing (ChIP-Seq) in mouse ES cells (mESCs) and human ES cells (hESCs), it was demonstrated that only about 5% of OCT4 and NANOG transcription factor-binding sites are orthologously occupied in both species and that up to 25% of the sites bound by these factors were contributed by TEs, most notably the ERV1 repeat family [56]. Importantly, many of the genes near these OCT4-binding sites were also functional targets of OCT4, because they were affected by depletion of OCT4 by RNAi (Figure 4b).
By contrast, CTCF-binding sites are more highly conserved between mammalian species (estimated 15–30% conservation between human and mouse) [56,57]. Despite this conservation, novel CTCF-binding sites also appear to be expanding due to the activity of SINE-B2 elements, which harbor CTCF-binding sites [57]. These novel transposon-derived CTCF-binding sites are also likely to be functional, because they are associated with histone marks typical of CTCF-mediated boundary elements that separate active and repressed domains (Figure 4c). In addition, tandem genes that are split by novel SINE-mediated CTCF-binding sites have greater transcriptional divergence than equivalent tandem genes in species that lack the CTCF-binding site, suggesting that the novel sites are functional barrier elements.
A third example of transposon-mediated rewiring of a gene-expression network appears to have been particularly important for the evolution of placental mammals. Using comparative gene expression data from endometrial tissue from mammals, approximately 1500 genes were shown to be recruited into endometrial tissue in placental mammals [58]. Almost 13% of these genes were within 200 kb of the eutherian-specific TE MER20. Importantly, the MER20 elements harbored binding sites for numerous transcription factors, had epigenetic signatures of enhancers, silencers, and boundary elements, and functioned as cAMP- and progesterone-dependent repressors in reporter-gene assays, supporting the hypothesis that insertions of MER20 elements contributed to placentation in mammals.
In addition to serving as enhancers and boundary elements, TEs can serve as primary or alternative promoters of genes or sometimes entire networks of genes (Figure 4d). LTR-driven transcripts are found in numerous mammalian species, including human and mouse, and are developmentally regulated [59–61]. By screening cDNA libraries, numerous transcripts during zygote genome activation were found to be chimeric transcripts of MuERV-L-type retrovirus LTRs and non-retroviral genes [62]. More recently, it was demonstrated by deep RNA sequencing of two-cell-stage mouse embryos that there are over 100 chimeric genes initiating from within MuERV-L LTRs [38]. IAP elements also drive expression of chimeric transcripts in ES cells lacking IAP-repressive machinery, suggesting that LTRs of multiple families are capable of serving as alternative promoters [63].
It is clear that ERVs are utilized as alternative promoters for numerous genes, but are the LTR-initiated transcripts functional? Several lines of evidence support this hypothesis. First, most of the LTR-initiated transcripts derived from MuERV-L LTRs have open reading frames. Second, several of the genes that utilize LTRs as alternative promoters are critical for the specification of early embryonic lineages, including the transcription factors GATA4 and TEAD4. Third, cells within mESC cultures that express the LTR-initiated transcripts have functional properties distinct from those of cells that do not express these transcripts; namely, the ability to contribute to extra-embryonic lineages of the placenta in mouse chimera assays (Figure 4d). Fourth, removal of cells within mESC cultures that express LTR-initiated transcripts affects the potency of the cultures [38]. Thus, it is likely that TE insertions are constantly creating novel regulatory patterns that can become fixed in a population and contribute to the evolution of new structures such as the placenta.
The presence of MuERV-L and its associated LTR-initiated transcripts in early mouse embryos begs the question of whether a similar phenomenon is present in human embryos, which harbor hundreds of almost full-length HERV-L elements [31]. We speculate that the answer is yes, based on the fact that HERV-L elements are transcribed in placenta [31] and hESCs and chimeric transcripts of HERV-L LTRs and non-retroviral genes have been detected in hESCs (T.S. Macfarlan, unpublished). Thus, virus-derived promoters are also likely to play a role in regulating human gene expression during embryo development.
TEs and epigenetic inheritance
ERV-derived alternative promoters also play important roles in epigenetic inheritance. At the agouti locus in viable yellow (A(vy)/a) mice, transcription initiates from an IAP element inserted upstream of the agouti gene and causes ectopic expression of the agouti protein, leading to yellow fur and obesity [64]. (A(vy)/a) mice, however, display variable expressivity, ranging from yellow to full agouti, based on the frequency of cells that initiate expression of the agouti gene from within the IAP and, fascinatingly, the phenotype is related to the phenotype of the dam, in that yellow females transmit the yellow phenotype and agouti females transmit the agouti phenotype at higher frequencies. Interestingly, the inheritance of the agouti epi-allele is sensitive to methyl donor supplementation in the maternal diet [65]. Whether such epigenetically sensitive alleles exist in humans is unclear, but searching for genes that utilize ERVs as alternative promoters may provide a useful strategy for finding them. In an intriguing recent study, many genes in mESCs were found to be under the negative control of IAP elements (via KAP1-mediated silencing), suggesting that the epigenetic effects of ERVs on neighboring genes may be significantly more widespread [66].
ERVs have also contributed to other crucial epigenetic phenomena in mammals: X-inactivation and genomic imprinting. The DXPAS34 element found at the X-inactivation center is required for both random and imprinted X inactivation and was suggested to have derived from ancestral ERV-L elements in mammals. [67]. Additionally, the KRAB-ZFP and KAP1 repression system discussed previously as a system for silencing TEs (Figure 3a,b) also maintains the silencing of imprinted alleles during preimplantation development [68–70]. It has even been speculated that ERVs themselves may be mono-allelically expressed from paternally inherited alleles in the trophoblast, because of relaxed selection in paternal germlines to epigenetically silence ERVs, relative to the maternal germline, which has strong selective pressure to block horizontal transmission of ERVs in utero [13]. Thus, epigenetic inheritance patterns including X inactivation and genomic imprinting may have evolved as a byproduct of attempts by the host to silence invading retroviral sequences and their endogenous counterparts.
TEs and meiotic recombination ‘hotspots’
We speculate that TEs also play an important role during gametogenesis by providing nucleotide substrates for meiotic recombination. A series of papers have cumulatively suggested a mechanism whereby the histone methyltransferase ZF protein PR domain containing 9 (Prdm9) trimethylates H3K4 (H3K4me3) on TE sequences, which then recruits recombination machinery to mediate recombination at sites referred to as hotspots (Figure 4e). Several lines of evidence support this hypothesis. First, mouse recombination hotspots were recently mapped genome wide and both Prdm9 binding motifs and several classes of TE were enriched at these hotspots, particularly Mammalian apparent LTR retrotransposons (MaLRs) and certain subclasses of SINE [71]. Second, Prdm9 is highly polymorphic and allelic variants are associated with altered hotspot usage [72]. Third, Prdm9 is specifically expressed in germ cells in meiotic prophase in fetal female gonads and postnatal testis [73]. Finally, Prdm9 was shown to generate H3K4me3 residues specifically at hotspots in mouse [74]. It should be noted that Prdm9 knockout mice still display meiotic recombination, but the hotspots are instead found at H3K4me3-marked promoters that were not affected by the Prdm9 deletion [75]. Hence, the H3K4me3 modification plays an important role in recombination machinery recruitment, but the Prdm9-deposited H3K4me3 normally outcompetes the H3K4me3-marked promoters, most likely due to other, concomitant epigenetic features also present at these loci. One potential advantage of this process is that Prdm9 spares functional promoter elements from potential errors introduced during recombination by enhancing recombination at distal TEs [75]. Although the advantages of meiotic recombination are apparent, it is unclear what drove the selection for Prdm9 allelic diversity. The fact that TEs are enriched at many of these hotspots suggests a coevolutionary process between TEs and Prdm9, which is reminiscent of the TE/KRAB–ZFP arms race hypothesis discussed above (Figure 3a,b).
Challenges and experimental approaches to the study of TEs in development
Attempts to link TEs to developmental processes are met with some major challenges. One is that it is difficult to distinguish among highly similar TEs based on their nucleotide sequence. This may be important if one is to establish a relationship between expression of a given TE with expression of an adjacent gene, the epigenetic regulation and chromatin state of a particular element, or the activity of particular elements over evolutionary time. Several interrelated factors contribute to this. First, TEs are present at very high copy numbers and many share high sequence homology, especially among younger and more active families. Second, published reference genomes currently do not reflect the immense individual sequence variability expected in TEs at a defined locus. Third, current high-throughput sequencing technology, an important experimental approach used by many in this field, typically generates short (< 100 bp) sequencing reads. The combination of these factors precludes assignment of a given sequence to a particular locus with high confidence in many cases. Indeed, reads that align to more than one locus with high confidence are often eliminated from further analysis.
A corollary to this problem is that transposition events during development generate new alleles. If this occurs in the germline, these may be passed to the next generation and the effects of the event therefore need to be investigated specifically in those harboring this allele. Again, this requires richer reference genome datasets. Without knowledge of such allelic variation, it is difficult to make predictions or test hypotheses about the effects of a particular transposition event or the presence of a TE at a specific locus and how that perturbs development.
Retrotransposition can also occur in somatic tissues, leading to somatic mosaicism. One interesting example is the demonstration that L1 elements retrotranspose in mouse, rat, and human neural precursors [76,77]. Instances of L1 reintegration events take place near neuronally expressed genes and, accordingly, it has been hypothesized that this plays a role in generating functional neuronal diversity [77]. This suggests an important developmental role for TEs that is not directly inherited and could help to explain how selective forces may still favor the retention of active elements. Rigorous testing of such hypotheses is difficult, however, because studying the resulting mosaicism necessitates assays with single-cell resolution. This was accomplished in a more recent study where single-cell genome sequencing of 300 human cortical and caudate neurons found the number of retrotransposition events to be between 0.04 and 0.6 per neuron [78], which is much less than previous estimates in human brain assayed by quantitative PCR [76] and has called the importance of the phenomenon into question. It is possible that this discrepancy is due to differences in retrotransposition rates between the neuroanatomic regions tested, but a full understanding awaits further study. We ask whether a more important feature of L1 activation in neural precursors is to drive expression of neighboring genes through the formation of chimeric transcripts or local chromatin effects. This, to our knowledge, has not been tested to date.
The evidence that some classes of TE play a role in the control of gene networks encourages the development of novel experimental tools that would allow one to precisely target all of the participating TEs for transcriptional activation or repression. This would allow one to perturb the entire network by targeting a single, relatively conserved TE sequence present in high copy number and, thereby, directly test the role that the TE network plays in a particular developmental process. Numerous recent developments provide promising strategies to attempt such experiments. One idea is to build customized transcription factors designed to activate or repress prespecified targets by modifying transcription activator-like effectors (TALEs) for use in mammalian systems [79–85].
Another possibility for studying TEs looms on the horizon. The publication of the first synthetic prokaryotic genome [86] raises the question of whether mammalian chromosomes can be synthesized biochemically. As a starting point, a collaboration is currently underway to synthesize yeast chromosomes (http://syntheticyeast.org). If this approach is successful, many TEs of a particular class could be eliminated from the final sequence to test what their contribution is to gene regulation and development, thus bypassing the need for gene targeting and greatly increasing the extent of genomic modification. We expect some combination of these technologies to greatly aid the study of gene-regulatory networks controlled in part by TEs and their ultimate impact on organismal development.
Concluding remarks
This review has highlighted examples where TEs and their remnants provide specific functions important for host transcriptional control and the regulation of early embryonic processes, including placentation and meiotic recombination. In such contexts, TEs can therefore be viewed as important genomic symbionts. We have also touched on the impact that transpositionally active TEs have on evolution and how they may play a role in tissue function by generating somatic diversity. In this latter context, specific TEs may act as parasites and lead to aberrant function. This leaves us with our current perspective of TEs, not as passive junk sequences nested within larger genomes, but as important players in many biological processes in both health and disease.
Acknowledgments
The authors thank Igor Dawid, Karl Pfeifer, and Carson Miller for critical reading of the manuscript. W.D.G is supported by an ARCS Award, the Rose Hills Foundation, and the Chapman Foundation. S.L.P. is the Benjamin H. Lewis Chair in Neurobiology and an Investigator of the Howard Hughes Medical Institute and his research is supported by NINDS, CIRM, and The Marshall Heritage Foundation. T.S.M is an Earl Stadtman Investigator at the Eunice Kennedy Shriver National Institute of Child Health and Human Development.
References
- 1.Bernstein BE, et al. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lander ES, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 3.de Koning AP, et al. Repetitive elements may comprise over two-thirds of the human genome. PLoS Genet. 2011;7:e1002384. doi: 10.1371/journal.pgen.1002384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Makalowski W. Genomic scrap yard: how genomes utilize all that junk. Gene. 2000;259:61–67. doi: 10.1016/s0378-1119(00)00436-4. [DOI] [PubMed] [Google Scholar]
- 5.Oliver KR, Greene WK. Mobile DNA and the TE-Thrust hypothesis: supporting evidence from the primates. Mobile DNA. 2011;2:8. doi: 10.1186/1759-8753-2-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rebollo R, et al. Transposable elements: an abundant and natural source of regulatory sequences for host genes. Annu. Rev. Genet. 2012;46:21–42. doi: 10.1146/annurev-genet-110711-155621. [DOI] [PubMed] [Google Scholar]
- 7.Feschotte C. Transposable elements and the evolution of regulatory networks. Nat. Rev. Genet. 2008;9:397–405. doi: 10.1038/nrg2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Feschotte C, Pritham EJ. DNA transposons and the evolution of eukaryotic genomes. Annu. Rev. Genet. 2007;41:331–368. doi: 10.1146/annurev.genet.40.110405.090448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boissinot S, Furano AV. The recent evolution of human L1 retrotransposons. Cytogenet. Genome Res. 2005;110:402–406. doi: 10.1159/000084972. [DOI] [PubMed] [Google Scholar]
- 10.Dewannieux M, Heidmann T. LINEs, SINEs and processed pseudogenes: parasitic strategies for genome modeling. Cytogenet. Genome Res. 2005;110:35–48. doi: 10.1159/000084936. [DOI] [PubMed] [Google Scholar]
- 11.Deininger P. Alu elements: know the SINEs. Genome Biol. 2011;12:236. doi: 10.1186/gb-2011-12-12-236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rosser JM, An W. L1 expression and regulation in humans and rodents. Front. Biosci. (Elite Ed.) 2012;4:2203–2225. doi: 10.2741/537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Haig D. Retroviruses and the placenta. Curr. Biol. 2012;22:R609–R613. doi: 10.1016/j.cub.2012.06.002. [DOI] [PubMed] [Google Scholar]
- 14.Stocking C, Kozak CA. Murine endogenous retroviruses. Cell. Mol. Life Sci. 2008;65:3383–3398. doi: 10.1007/s00018-008-8497-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rowe HM, Trono D. Dynamic control of endogenous retroviruses during development. Virology. 2011;411:273–287. doi: 10.1016/j.virol.2010.12.007. [DOI] [PubMed] [Google Scholar]
- 16.Qin C, et al. Intracisternal A particle genes: distribution in the mouse genome, active subtypes, and potential roles as species-specific mediators of susceptibility to cancer. Mol. Carcinog. 2010;49:54–67. doi: 10.1002/mc.20576. [DOI] [PubMed] [Google Scholar]
- 17.Benit L, et al. Cloning of a new murine endogenous retrovirus, MuERV-L, with strong similarity to the human HERV-L element and with a gag coding sequence closely related to the Fv1 restriction gene. J. Virol. 1997;71:5652–5657. doi: 10.1128/jvi.71.7.5652-5657.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Boller K, et al. Human endogenous retrovirus HERV-K113 is capable of producing intact viral particles. J. Gen. Virol. 2008;89:567–572. doi: 10.1099/vir.0.83534-0. [DOI] [PubMed] [Google Scholar]
- 19.Feschotte C, Gilbert C. Endogenous viruses: insights into viral evolution and impact on host biology. Nat. Rev. Genet. 2012;13:283–296. doi: 10.1038/nrg3199. [DOI] [PubMed] [Google Scholar]
- 20.Katzourakis A, Gifford RJ. Endogenous viral elements in animal genomes. PLoS Genet. 2010;6:e1001191. doi: 10.1371/journal.pgen.1001191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nellaker C, et al. The genomic landscape shaped by selection on transposable elements across 18 mouse strains. Genome Biol. 2012;13:R45. doi: 10.1186/gb-2012-13-6-r45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miki Y, et al. Disruption of the APC gene by a retrotransposal insertion of L1 sequence in a colon cancer. Cancer Res. 1992;52:643–645. [PubMed] [Google Scholar]
- 23.Jern P, Coffin JM. Effects of retroviruses on host genome function. Annu. Rev. Genet. 2008;42:709–732. doi: 10.1146/annurev.genet.42.110807.091501. [DOI] [PubMed] [Google Scholar]
- 24.Lutsenko E, Bhagwat AS. Principal causes of hot spots for cytosine to thymine mutations at sites of cytosine methylation in growing cells. A model, its experimental support and implications. Mutat. Res. 1999;437:11–20. doi: 10.1016/s1383-5742(99)00065-4. [DOI] [PubMed] [Google Scholar]
- 25.Zhang X, Mathews CK. Effect of DNA cytosine methylation upon deamination-induced mutagenesis in a natural target sequence in duplex DNA. J. Biol. Chem. 1994;269:7066–7069. [PubMed] [Google Scholar]
- 26.Holliday R, Grigg GW. DNA methylation and mutation. Mutat. Res. 1993;285:61–67. doi: 10.1016/0027-5107(93)90052-h. [DOI] [PubMed] [Google Scholar]
- 27.Lowe CB, Haussler D. 29 Mammalian genomes reveal novel exaptations of mobile elements for likely regulatory functions in the human genome. PLoS ONE. 2012;7:e43128. doi: 10.1371/journal.pone.0043128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Leung DC, Lorincz MC. Silencing of endogenous retroviruses: when and why do histone marks predominate? Trends Biochem. Sci. 2012;37:127–133. doi: 10.1016/j.tibs.2011.11.006. [DOI] [PubMed] [Google Scholar]
- 29.Benit L, et al. ERV-L elements: a family of endogenous retrovirus-like elements active throughout the evolution of mammals. J. Virol. 1999;73:3301–3308. doi: 10.1128/jvi.73.4.3301-3308.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Greenwood AD, et al. Evolution of endogenous retrovirus-like elements of the woolly mammoth (Mammuthus primigenius) and its relatives. Mol. Biol. Evol. 2001;18:840–847. doi: 10.1093/oxfordjournals.molbev.a003865. [DOI] [PubMed] [Google Scholar]
- 31.Cordonnier A, et al. Isolation of novel human endogenous retrovirus-like elements with foamy virus-related pol sequence. J. Virol. 1995;69:5890–5897. doi: 10.1128/jvi.69.9.5890-5897.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ribet D, et al. Murine endogenous retrovirus MuERV-L is the progenitor of the “orphan” epsilon viruslike particles of the early mouse embryo. J. Virol. 2008;82:1622–1625. doi: 10.1128/JVI.02097-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Macfarlan TS, et al. Endogenous retroviruses and neighboring genes are coordinately repressed by LSD1/KDM1A. Genes Dev. 2011;25:594–607. doi: 10.1101/gad.2008511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rowe HM, et al. KAP1 controls endogenous retroviruses in embryonic stem cells. Nature. 2010;463:237–240. doi: 10.1038/nature08674. [DOI] [PubMed] [Google Scholar]
- 35.Kigami D, et al. MuERV-L is one of the earliest transcribed genes in mouse one-cell embryos. Biol. Reprod. 2003;68:651–654. doi: 10.1095/biolreprod.102.007906. [DOI] [PubMed] [Google Scholar]
- 36.Guallar D, et al. Expression of endogenous retroviruses is negatively regulated by the pluripotency marker Rex1/Zfp42. Nucleic Acids Res. 2012;40:8993–9007. doi: 10.1093/nar/gks686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hisada K, et al. RYBP represses endogenous retroviruses and preimplantation- and germ line-specific genes in mouse embryonic stem cells. Mol. Cell. Biol. 2012;32:1139–1149. doi: 10.1128/MCB.06441-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Macfarlan TS, et al. Embryonic stem cell potency fluctuates with endogenous retrovirus activity. Nature. 2012;487:57–63. doi: 10.1038/nature11244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Biczysko W, et al. Virus particles in early mouse embryos. J. Natl Cancer Inst. 1973;51:1041–1050. doi: 10.1093/jnci/51.3.1041. [DOI] [PubMed] [Google Scholar]
- 40.Calarco PG, Szollosi D. Intracisternal A particles in ova and preimplantation stages of the mouse. Nat. New Biol. 1973;243:91–93. [PubMed] [Google Scholar]
- 41.Chase DG, Piko L. Expression of A- and C-type particles in early mouse embryos. J. Natl. Cancer Inst. 1973;51:1971–1975. doi: 10.1093/jnci/51.6.1971. [DOI] [PubMed] [Google Scholar]
- 42.Matsui T, et al. Proviral silencing in embryonic stem cells requires the histone methyltransferase ESET. Nature. 2010;464:927–931. doi: 10.1038/nature08858. [DOI] [PubMed] [Google Scholar]
- 43.Wolf D, et al. Primer binding site-dependent restriction of murine leukemia virus requires HP1 binding by TRIM28. J. Virol. 2008;82:4675–4679. doi: 10.1128/JVI.02445-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wolf D, Goff SP. TRIM28 mediates primer binding site-targeted silencing of murine leukemia virus in embryonic cells. Cell. 2007;131:46–57. doi: 10.1016/j.cell.2007.07.026. [DOI] [PubMed] [Google Scholar]
- 45.Thomas JH, Schneider S. Coevolution of retroelements and tandem zinc finger genes. Genome Res. 2011;21:1800–1812. doi: 10.1101/gr.121749.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Emerson RO, Thomas JH. Adaptive evolution in zinc finger transcription factors. PLoS Genet. 2009;5:e1000325. doi: 10.1371/journal.pgen.1000325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wolf D, Goff SP. Embryonic stem cells use ZFP809 to silence retroviral DNAs. Nature. 2009;458:1201–1204. doi: 10.1038/nature07844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Svoboda P, et al. RNAi and expression of retrotransposons MuERV-L and IAP in preimplantation mouse embryos. Dev. Biol. 2004;269:276–285. doi: 10.1016/j.ydbio.2004.01.028. [DOI] [PubMed] [Google Scholar]
- 49.Tsai MC, et al. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329:689–693. doi: 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gu TP, et al. The role of Tet3 DNA dioxygenase in epigenetic reprogramming by oocytes. Nature. 2011;477:606–610. doi: 10.1038/nature10443. [DOI] [PubMed] [Google Scholar]
- 51.Dupressoir A, et al. From ancestral infectious retroviruses to bona fide cellular genes: role of the captured syncytins in placentation. Placenta. 2012;33:663–671. doi: 10.1016/j.placenta.2012.05.005. [DOI] [PubMed] [Google Scholar]
- 52.Dupressoir A, et al. Syncytin-A knockout mice demonstrate the critical role in placentation of a fusogenic, endogenous retrovirus-derived, envelope gene. Proc. Natl. Acad. Sci. U.S.A. 2009;106:12127–12132. doi: 10.1073/pnas.0902925106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Heidmann O, et al. Identification of an endogenous retroviral envelope gene with fusogenic activity and placenta-specific expression in the rabbit: a new “syncytin” in a third order of mammals. Retrovirology. 2009;6:107. doi: 10.1186/1742-4690-6-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Best S, et al. Positional cloning of the mouse retrovirus restriction gene Fv1. Nature. 1996;382:826–829. doi: 10.1038/382826a0. [DOI] [PubMed] [Google Scholar]
- 55.Yan Y, et al. Origin, antiviral function and evidence for positive selection of the gammaretrovirus restriction gene Fv1 in the genus Mus. Proc. Natl. Acad. Sci. U.S.A. 2009;106:3259–3263. doi: 10.1073/pnas.0900181106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kunarso G, et al. Transposable elements have rewired the core regulatory network of human embryonic stem cells. Nat. Genet. 2010;42:631–634. doi: 10.1038/ng.600. [DOI] [PubMed] [Google Scholar]
- 57.Schmidt D, et al. Waves of retrotransposon expansion remodel genome organization and CTCF binding in multiple mammalian lineages. Cell. 2012;148:335–348. doi: 10.1016/j.cell.2011.11.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lynch VJ, et al. Transposon-mediated rewiring of gene regulatory networks contributed to the evolution of pregnancy in mammals. Nat. Genet. 2011;43:1154–1159. doi: 10.1038/ng.917. [DOI] [PubMed] [Google Scholar]
- 59.Cohen CJ, et al. Endogenous retroviral LTRs as promoters for human genes: a critical assessment. Gene. 2009;448:105–114. doi: 10.1016/j.gene.2009.06.020. [DOI] [PubMed] [Google Scholar]
- 60.Conley AB, et al. Retroviral promoters in the human genome. Bioinformatics. 2008;24:1563–1567. doi: 10.1093/bioinformatics/btn243. [DOI] [PubMed] [Google Scholar]
- 61.Rebollo R, et al. C-GATE - catalogue of genes affected by transposable elements. Mobile DNA. 2012;3:9. doi: 10.1186/1759-8753-3-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Peaston AE, et al. Retrotransposons regulate host genes in mouse oocytes and preimplantation embryos. Dev. Cell. 2004;7:597–606. doi: 10.1016/j.devcel.2004.09.004. [DOI] [PubMed] [Google Scholar]
- 63.Karimi MM, et al. DNA methylation and SETDB1/H3K9me3 regulate predominantly distinct sets of genes, retroelements, and chimeric transcripts in mESCs. Cell Stem Cell. 2011;8:676–687. doi: 10.1016/j.stem.2011.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Morgan HD, et al. Epigenetic inheritance at the agouti locus in the mouse. Nat. Genet. 1999;23:314–318. doi: 10.1038/15490. [DOI] [PubMed] [Google Scholar]
- 65.Waterland RA, Jirtle RL. Transposable elements: targets for early nutritional effects on epigenetic gene regulation. Mol. Cell. Biol. 2003;23:5293–5300. doi: 10.1128/MCB.23.15.5293-5300.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rowe HM, et al. TRIM28 repression of retrotransposon-based enhancers is necessary to preserve transcriptional dynamics in embryonic stem cells. Genome Res. 2013;23:452–461. doi: 10.1101/gr.147678.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cohen DE, et al. The DXPas34 repeat regulates random and imprinted X inactivation. Dev. Cell. 2007;12:57–71. doi: 10.1016/j.devcel.2006.11.014. [DOI] [PubMed] [Google Scholar]
- 68.Liu Y, et al. An atomic model of Zfp57 recognition of CpG methylation within a specific DNA sequence. Genes Dev. 2012;26:2374–2379. doi: 10.1101/gad.202200.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Quenneville S, et al. In embryonic stem cells, ZFP57/KAP1 recognize a methylated hexanucleotide to affect chromatin and DNA methylation of imprinting control regions. Mol. Cell. 2011;44:361–372. doi: 10.1016/j.molcel.2011.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Li X, et al. A maternal-zygotic effect gene, Zfp57, maintains both maternal and paternal imprints. Dev. Cell. 2008;15:547–557. doi: 10.1016/j.devcel.2008.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Smagulova F, et al. Genome-wide analysis reveals novel molecular features of mouse recombination hotspots. Nature. 2011;472:375–378. doi: 10.1038/nature09869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Baudat F, et al. PRDM9 is a major determinant of meiotic recombination hotspots in humans and mice. Science. 2010;327:836–840. doi: 10.1126/science.1183439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hayashi K, et al. A histone H3 methyltransferase controls epigenetic events required for meiotic prophase. Nature. 2005;438:374–378. doi: 10.1038/nature04112. [DOI] [PubMed] [Google Scholar]
- 74.Parvanov ED, et al. Prdm9 controls activation of mammalian recombination hotspots. Science. 2010;327:835. doi: 10.1126/science.1181495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Brick K, et al. Genetic recombination is directed away from functional genomic elements in mice. Nature. 2012;485:642–645. doi: 10.1038/nature11089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Coufal NG, et al. L1 retrotransposition in human neural progenitor cells. Nature. 2009;460:1127–1131. doi: 10.1038/nature08248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Muotri AR, et al. Somatic mosaicism in neuronal precursor cells mediated by L1 retrotransposition. Nature. 2005;435:903–910. doi: 10.1038/nature03663. [DOI] [PubMed] [Google Scholar]
- 78.Evrony GD, et al. Single-neuron sequencing analysis of l1 retrotransposition and somatic mutation in the human brain. Cell. 2012;151:483–496. doi: 10.1016/j.cell.2012.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Li T, et al. Modularly assembled designer TAL effector nucleases for targeted gene knockout and gene replacement in eukaryotes. Nucleic Acids Res. 2011;39:6315–6325. doi: 10.1093/nar/gkr188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Cermak T, et al. Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucleic Acids Res. 2011;39:e82. doi: 10.1093/nar/gkr218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Cong L, et al. Comprehensive interrogation of natural TALE DNA-binding modules and transcriptional repressor domains. Nat. Commun. 2012;3:968. doi: 10.1038/ncomms1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Moscou MJ, Bogdanove AJ. A simple cipher governs DNA recognition by TAL effectors. Science. 2009;326:1501. doi: 10.1126/science.1178817. [DOI] [PubMed] [Google Scholar]
- 83.Boch J, et al. Breaking the code of DNA binding specificity of TAL-type III effectors. Science. 2009;326:1509–1512. doi: 10.1126/science.1178811. [DOI] [PubMed] [Google Scholar]
- 84.Tremblay JP, et al. Transcription activator-like effector proteins induce the expression of the frataxin gene. Hum. Gene Ther. 2012;23:883–890. doi: 10.1089/hum.2012.034. [DOI] [PubMed] [Google Scholar]
- 85.Zhang F, et al. Efficient construction of sequence-specific TAL effectors for modulating mammalian transcription. Nat. Biotechnol. 2011;29:149–153. doi: 10.1038/nbt.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Gibson DG, et al. Creation of a bacterial cell controlled by a chemically synthesized genome. Science. 2010;329:52–56. doi: 10.1126/science.1190719. [DOI] [PubMed] [Google Scholar]