Abstract
Setting
The country of Georgia has a high burden of multidrug and extensively drug-resistant tuberculosis (M/XDR-TB).
Objective
To assess the performance of the Genotype MTBDRsl assay in the detection of resistance to Kanamycin (KAN), Capreomycin (CAP), Ofloxacin (OFX), and XDR.
Design
Consecutive AFB smear positive sputum specimens identified as MDR by MTBDRplus testing were evaluated with the MTBDRsl assay and conventional second-line drug susceptibility testing (DST).
Results
Among 159 specimens, amplification was adequate in 154 (97%), including 9 of 9 culture negative and 2 of 3 contaminated specimens. Second-line DST revealed 17 (12%) M. tuberculosis isolates were XDR. Compared to DST, the MTBDRsl had 41% sensitivity and 98% specificity in detecting XDR and an 81% sensitivity and 99% specificity in detecting OFX resistance. Sensitivity was low in detecting resistance to KAN (29%) and CAP (57%) while specificity was 99% and 94%, respectively. Median times from sputum collection to second-line DST and MTBDRsl results were 70–104 versus 10 days.
Conclusion
The MTBDRsl assay had a rapid turn around time; however detection of second-line drug-resistance was poor compared to DST. Further genetic mutations associated with resistance to second-line drugs should be included in the assay to improve test performance and clinical utility.
Keywords: Line Probe Assays, tuberculosis, drug-resistance
INTRODUCTION
A major threat to tuberculosis (TB) control efforts is the increasing global burden of drug-resistant TB. Inappropriate treatment regimens and poor adherence to therapy are the most common causes of drug-resistant TB and in large part have led to the development and transmission of multi-drug resistant (MDR)-TB (resistance to isoniazid and rifampicin) and extensively-drug resistant (XDR)-TB (resistance to isoniazid, rifampicin, fluoroquinolones, and any injectable agent). The World Health Organization (WHO) has estimated a worldwide prevalence of 660,000 cases of MDR-TB and 150,000 MDR-TB related deaths annually.1 Especially worrisome is the increasing prevalence of difficult to treat XDR-TB, which has been found in 84 countries and is estimated to be present in 9% of patients with MDR-TB.1 The emergence of XDR-TB has led to the development of virtually untreatable TB in many settings.2, 3
The highest rates of drug-resistant TB occur in former Soviet republics including the country of Georgia, which is one of twenty-seven high burden MDR-TB countries.1 Georgian National TB Program (NTP) data from 2011 found the prevalence of MDR-TB among newly diagnosed patients to be 10.8% and 31.4% among previously treated patients; 6.4% of those with MDR had XDR-TB. With the support of the Global Fund, the Georgian NTP has achieved universal access to diagnosis and treatment of MDR- and XDR-TB and more recently validated and implemented the commercially available MTBDRplus assay into clinical practice.4
The development of commercially available molecular diagnostics tests to detect drug-resistant TB, including the Xpert TB/RIF and MTBDRplus assays, have been hailed as significant achievements and provide clinicians accurate tests to use for the rapid detection of rifampicin resistant and MDR-TB. In 2009, Hain Lifescience introduced a new line probe assay (LPA), the MTBDRsl, for the rapid detection of mutations associated with resistance to fluoroquinolones, aminoglycosides, cyclic peptides, and ethambutol.5 Investigations on the utility of the MTBDRsl assay are limited and WHO recommendations are based on low quality evidence.6 In addition, study results have varied by geographic location and few have been performed using clinical specimens. MTBDRsl implementation projects will help inform current guidelines and set an agenda for future research efforts. Our primary objective was to assess the performance of the MTBDRsl assay compared to conventional culture and DST methods when implemented into the workflow of a high volume National TB Reference Laboratory (NRL).
METHODS
Setting
The study took place at the NRL of the Georgian National TB Program (NTP) in Tbilisi, Georgia, which processed ~18,000 sputum specimens in 2011. AFB smear positive sputum specimens from TB suspects throughout Georgia from November 2011 through April 2012 were collected. While data on HIV status was not available, prior research has demonstrated a low HIV prevalence (~1%) among tuberculosis patients in Georgia.7 Approval for this study was received from the Georgian National Center for Tuberculosis and Lung Disease and Emory University Institutional Review Boards (IRBs).
Culture and Drug Susceptibility Testing (DST)
Two routine sputum specimens were obtained from each patient and direct smears with Ziehl-Neelsen staining were examined by light microscopy at local microscopy centers in Georgia. One AFB smear positive sample was sent to the NRL where it was processed using standard methodologies (decontaminated in a BSL 2+ area with N-acetyl-L-cysteine-sodium hydroxide, centrifuged, and the sediment was then suspended in 1.5 ml of phosphate buffer).8 The processed specimen was inoculated on to both Löwenstein-Jensen (LJ) based solid medium and the BACTEC MGIT 960 broth culture system. Positive cultures by either method were confirmed to be M. tuberculosis complex using the MTBDRplus assay.9 DST for first-line drugs was done using conventional methods as previously described.4, 10 DST to second-line drugs (SLDs) was performed using the proportion method on LJ medium with the following drug concentrations: ethionamide-40.0 µg/ml; ofloxacin-2.0 µg/ml; para-aminosalicylic acid-0.5 µg/ml, capreomycin-40.0 µg/ml and KM-30.0 µg/ml.11 The Georgian NRL has undergone external quality assessment by the Antwerp WHO Supranational TB Reference Laboratory (SNRL) annually since 2005. In 2012, SNRL quality assurance certification was given for DST of isoniazid, rifampicin, kanamycin (KAN), capreomycin (CAP) and ofloxacin (OFX).
Molecular Testing
All molecular testing was performed using a portion of the same sputum specimen used for culture. A 500-µl portion of decontaminated sample was used to perform the MTBDRplus assay according to manufacturer’s instructions. A portion of extracted DNA was kept refrigerated (+4C) until receiving MTBDRplus assay results. If both rifampicin and isoniazid resistance were detected, the MTBDRsl assay was performed. The saved DNA pellet was centrifugated at 13,000 G for 5 minutes and 5 µl of supernatant was removed. The DNA was added to 45 µl amplification mix and amplified using 42 PCR cycles based on manufacturers recommendation for clinical specimens, further followed by hybridization and test readout steps. Negative controls where used for quality assurance with each run of MTBDRsl assay.
Definitions
New TB cases were patients who had received ≤ 30 days of anti-TB drug therapy; retreatment cases were those with a prior history of receiving TB treatment for >30 days. A completely interpretable MTBDRsl result was defined as a test strip with all control markers positive.
Data Analysis
All data were entered into an online REDCap database12 and analyzed using SAS 9.3. The sensitivity, specificity, positive predictive value (PPV), and negative predictive value (NPV) of the MTBDRsl assay in detecting resistance to OFX, CAP, and KAN were calculated using conventional DST results as the reference standard. Turnaround time was calculated as time between the date of sputum collection and date of culture, DST, and MTBDRsl results. The degree of agreement between test results was assessed using the kappa (κ) statistic. A p-value of <0.05 was considered statistically significant.
RESULTS
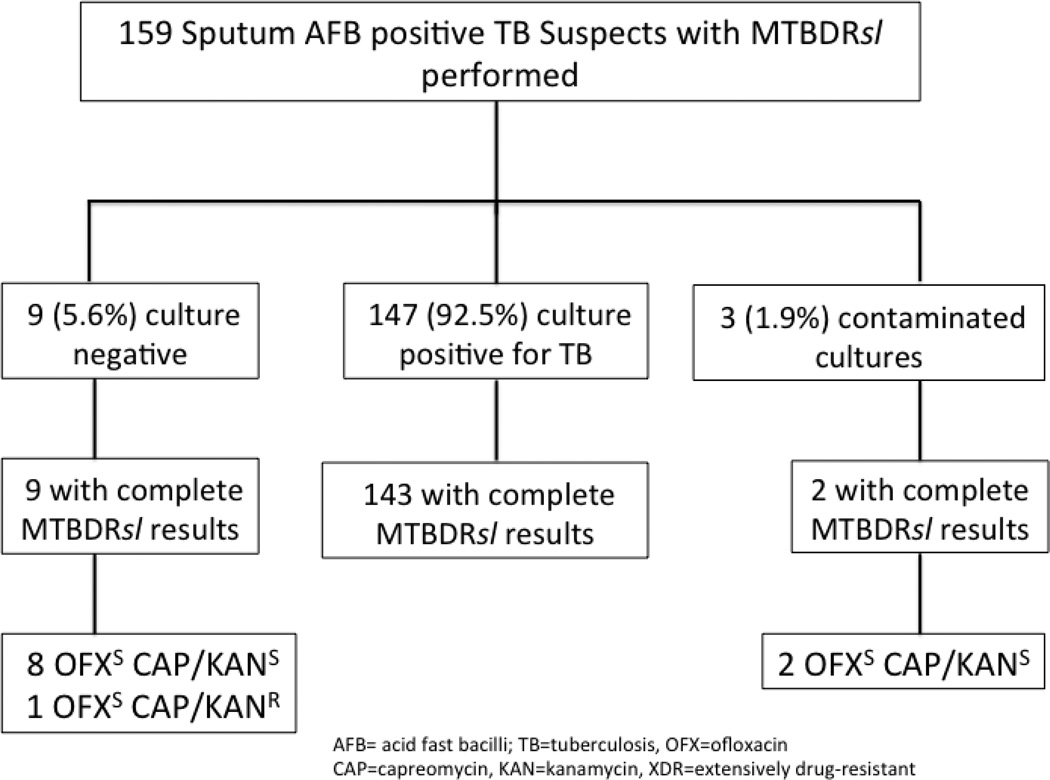
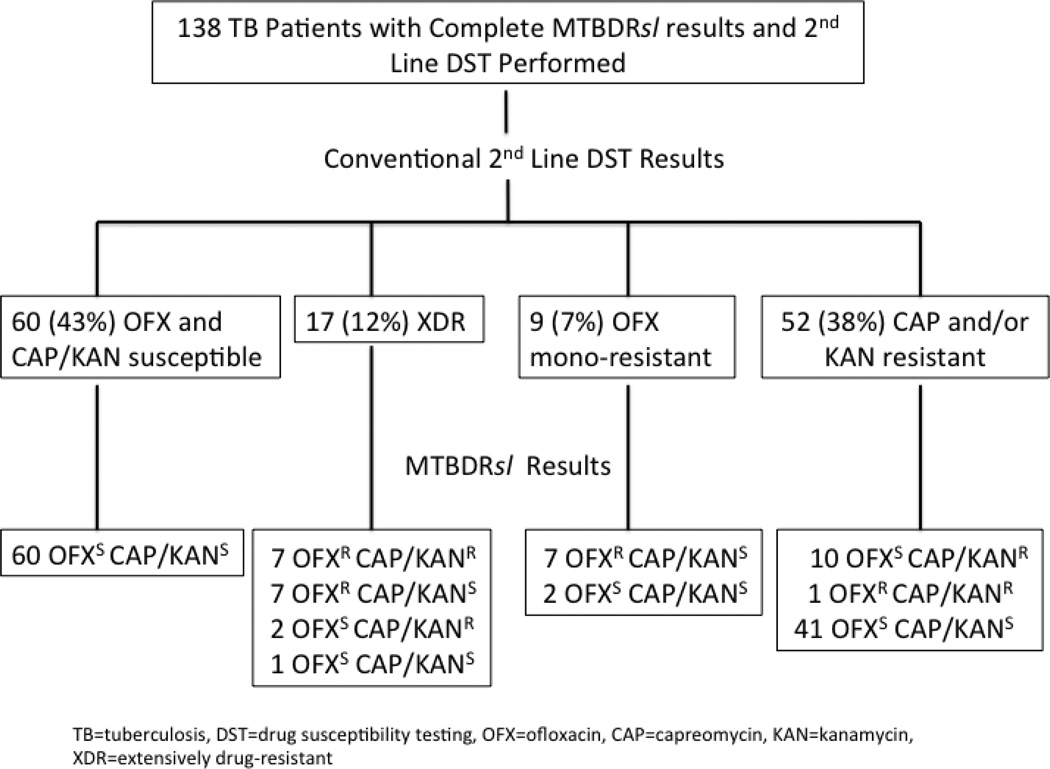
A total of 159 patients with a smear positive AFB sputum specimen and MTBDRplus result indicating resistance to rifampin and isoniazid were enrolled. Among these patients, 69 (43%) were new TB cases, and 90 (57%) were retreatment TB cases. Overall, 147 (92.5%) samples were culture positive for M. tuberculosis by either solid or liquid culture, 9 were culture negative, and 3 had contaminated cultures (Figure 1). Of 147 culture positive patients, 142 (97%) had complete first and second-line DST results. First-line DST of M. tuberculosis was performed in liquid media in 57 (40%) or solid media 85 (60%); all second-line DST was performed using solid media. Excluding four patients with non-interpretable MTBDRsl results, conventional second-line DST revealed 60 (43%) M. tuberculosis isolates with no OFX, CAP, or KAN resistance, 17 (12%) with XDR, 9 (7%) with OFX resistance alone, and 52 (38%) isolates resistant to CAP and/or KAN without OFX resistance (Figure 2).
Figure 1.
Sputum culture results for all AFB smear positive tuberculosis suspects and corresponding complete MTBDRsl assay results
Figure 2.
Distribution of MTBDRsl assay results according to phenotypic drug-resistance patterns using conventional drug susceptibility testing
MTBDRsl Assay
Among the 147 sputum samples with a positive culture for M. tuberculosis, 143 (97%) had a completely interpretable MTBDRsl assay. The 4 non-interpretable MTBDRsl assay results were due to inadequate amplification. The MTBDRsl assay gave interpretable results for most (11 of 12, 92%) specimens with negative or contaminated sputum cultures (Figure 1). Comparison of resistance patterns generated by the MTBDRsl assay and conventional methods among M. tuberculosis isolates recovered from the 138 patients with an interpretable MTBDRsl assay and second-line DST is shown in Figure 2. Performance parameters of the MTBDRsl assay as compared to conventional second-line DST are displayed in Table 1. Sensitivity of the MTBDRsl assay in the detection of OFX resistance (80.8%, 95% CI 65.6–95.9%) was moderate, and it was poor for the detection of CAP (56.5%, 95% CI 36.3–76.8%) and KAN (28.8%, 95% CI 17.9–39.7%) resistance as well as detection of XDR (41.2%, 95% CI 17.8–64.6%). Specificity of the MTBDRsl assay in the detection of OFX (99.1%, 95% CI 97.4–100), CAP (93.9%, 95% CI 89.5–98.3%), and KAN (98.6%. 95% CI 95.9–100%) resistance and XDR (98.3%, 95% CI 96.0–100%) was high. There was good agreement between the MTBDRsl assay and DST in detection of OFX resistance (κ=0.85, 95% CI 0.73–0.96), and poor agreement in the detection of CAP (κ=0.53, 95% CI 0.34–0.73) and KAN (κ=0.28,95% CI 0.16–0.40) resistance and XDR (κ=0.49, 95% CI 0.25–0.74).
Table 1.
Performance parameters of MTBDRsl in detecting any resistance to OFX, CAP, KAN and XDR compared to conventional Drug Susceptibility Testing (reference standard)#,^ [n=138]
| OFX | CAP | KAN | XDR | |
|---|---|---|---|---|
| True Susceptible | 111 | 108 | 71 | 119 |
| True Resistant | 21 | 13 | 19 | 7 |
| False Susceptible | 5 | 10 | 47 | 10 |
| False Resistant | 1 | 7 | 1 | 2 |
| Sensitivity# | 80.8 (65.6–95.9) | 56.5 (36.3–76.8) | 28.8 (17.9–39.7) | 41.2 (17.8–64.6) |
| Specificity# | 99.1 (97.4–100) | 93.9 (89.5–98.3) | 98.6 (95.9–100) | 98.3 (96.0–100) |
| PPV*,# | 95.5 (86.8–100) | 65.0 (44.1–85.9) | 95.0 (85.5–100) | 77.8 (50.1–100) |
| NPV*,# | 95.7 (92.0–99.4) | 91.5 (86.4–96.5) | 59.8 (50.1–68.7) | 92.2 (87.5–96.8) |
| Kappa | 0.85 (0.73–0.96) | 0.53 (0.34–0.73) | 0.28 (0.16–0.40) | 0.49 (0.25–0.74) |
Values are percentages with 95% confidence interval in parentheses
PPV=positive predictive value, NPV=negative predictive value
OFX= ofloxacin, CAP= capreomycin, KAN= kanamycin, XDR=extensive drug-resistance
Time to Results
Time to detection of drug resistance to OFX, CAP, and KAN was significantly shorter for the MTBDRsl assay as compared to conventional culture methods and DST (Table 2). The median time for detection of resistance by the MTBDRsl assays was 10 days as compared to conventional methods in which first-line DST was performed on liquid media and second-line DST on solid media (70 days) and those who had first and second-line DST performed on solid media (104 days).
Table 2.
Median time to results in days for detection of TB and associated drug resistance (N=138)*
| Solid Media 1st Line DST |
Liquid Media 1st Line DST |
MTBDRplus assay |
Liquid 1st Line DST/ Solid 2nd Line DST |
Solid Media 1st and 2nd Line DST |
MTBDRsl assay |
|
|---|---|---|---|---|---|---|
| All Cases | 71 (64–81) | 21 (18–27) | 5 (3–7) | 70 (65–76) | 104 (97–112) | 10 (7–12) |
| XDR TB | 71 (64–76) | 23 (21–28) | 6 (4–8) | 71 (64–71) | 102 (96–107) | 12 (8–19) |
Values are median number of days with 25th–75th percentile values in parentheses
DST= drug susceptibility testing
Genetic Mutations
The distributions of genetic mutations of drug-resistant M. tuberculosis isolates with an interpretable MTBDRsl assay (n=138) are shown in Tables 3 and 4. The most common resistance mutation for any OFX resistance was D94G (48%) followed by A90V (29%). Additionally, a similar percentage had lack of binding to the gyrA WT2 (48%) and WT3 (29%) probes. The majority of isolates lacked binding to a WT probe and had a drug resistance mutation (13/21, 62%). Almost all CAP and KAN phenotypic drug-resistant M. tuberculosis isolates had an A1401G mutation (100% and 84%, respectively), and lacked binding to the WT1 probe (92% and 90%, respectively). There were 6 M. tuberculosis isolates (2 to both OFX and KAN and/or CAP and 4 only to OFX) that had a drug resistant mutation without lack of binding to the corresponding WT probe (Table 3 and 4). The one false resistance fluoroquinolone isolate lacked binding to the WT2 probe. Among the 7 false resistant CAP isolates, 5 had drug resistance mutations and all 7 (100%) were phenotypically resistant to KAN. The one false resistant KAN isolate had an A1401G mutation and was phenotypically resistant to CAP (data not shown).
Table 3.
Pattern of genetic mutations in Mycobacterium tuberculosis isolates with phenotypic ofloxacin drug-resistance and molecular fluoroquinolone drug resistance using the Genotype MTBDRsl assay (N=21)
| Gene | Band | Gene Region | OFX Mono R* N=7 |
XDR* N=14 |
Any OFX R* N=21 |
|---|---|---|---|---|---|
| gyrA | ΔWT1 only | 85–90 | 1 (14) | 1 (7) | 2 (10) |
| ΔWT2 only | 89–93 | - | 1 (7) | 1 (5) | |
| ΔWT2 + MUT1 | 89–93, A90V | 2 (29) | 2 (14) | 4 (19) | |
| ΔWT2 + MUT2 | 89–93, S91P | - | 1 (7) | 1 (5) | |
| ΔWT2 + MUT1+MUT3D | 89–93, A90V, D94H | 1 (14) | - | 1 (5) | |
| ΔWT2 + ΔWT3 + MUT2 | 89–93, 92–97, S91P | - | 1 (7) | 1 (5) | |
| ΔWT3 + MUT3A | 92–97, D94A | - | 1 (7) | 1 (5) | |
| ΔWT3 + MUT3C | 92–97, D94G | 1 (14) | 4 (29) | 5 (24) | |
| MUT1 +MUT3B/3C | A90V, D94N/Y, D94G | - | 1 (7) | 1 (5) | |
| MUT3C only | D94G | 2 (29) | 2 (14) | 4 (19) |
Δ, indicated lack of wild type band; OFX=ofloxacin; XDR=extensively drug-resistant; R=resistance;
Values are numbers, with percentages in parentheses
Table 4.
Pattern of genetic mutations in Mycobacterium tuberculosis isolates with phenotypic capreomycin or kanamycin drug-resistance and molecular injectable agent resistance using the Genotype MTBDRsl assay
| Gene | Band | Gene Region | Any CAP R* N=13 |
Any KAN R* N=19 |
CAP S/KAN R* N=7 |
CAP and KAN R* N=12 |
XDR* N=9 |
|---|---|---|---|---|---|---|---|
| rrs | ΔWT1 only | 1401-2 | 2 (11) | 2 (29) | |||
| ΔWT1 + MUT1 | 1401-2, A1401G | 12 (92) | 15 (79) | 4 (57) | 11 (92) | 7 (78) | |
| ΔWT2 | 1484 | ||||||
| MUT1 only | A1401G | 1 (8) | 1 (5) | 1 (8) | 1 (11) | ||
| MUT2 only | G1484T | 1 (5) | 1 (14) | 1 (11) |
Δ, indicated lack of wild type band; CAP=capreomycin; KAN=kanamycin; XDR=extensively drug-resistant; R=resistance; S=sensitive;
Values are numbers, with percentages in parentheses
DISCUSSION
In a country with a high burden of drug-resistant TB, we demonstrated that the MTBDRsl assay can be successfully implemented into the routine workflow of a high volume National Reference Laboratory and that results can be provided in a timely fashion; however, the performance of the assay was suboptimal. We found a moderate sensitivity for OFX (81%) but poor sensitivity for CAP (57%), KAN (29%) and XDR (41%) detection as compared to conventional methods including cultures plus DST. Specificity was much higher (≥93%) for all categories. Our study is only the third published report evaluating the MTBDRsl assay under routine diagnostic conditions and our results are in agreement with recent meta-analyses finding an overall poor performance of the MTBDRsl assay.6, 13, 14 Improvements of the MTBDRsl assay particularly in regards to detecting KAN and CAP resistance and/or newer technologies are needed for the rapid and accurate detection of second-line anti-tuberculosis drug resistance.
Our study results provide critical information on the performance of the MTBDRsl assay when implemented into normal workflow using clinical specimens. Five studies have been published evaluating MTBDRsl performance using clinical specimens and only two of these used non-frozen clinical specimens and was done under routine diagnostic conditions.14–18 One of the studies was carried out in the Western Cape Province, South Africa and found disparate results as compared to our findings. Among 516 patients, they found high sensitivity of the MTBDRsl in detecting OFX (90.7%), AMK (100%), and XDR (92.3%) and high specificity for all categories (≥98%).18 The excellent performance of the MTBDRsl assay in their setting may have been related to distinct MDR and XDR M. tuberculosis strains circulating in the Western Cape Province and also that they tested for AMK and not KAN phenotypic resistance.19 Mutations in the rrs have been found more commonly among AMK resistance versus KAN resistant strains.20 Another of the prior studies was conducted in Russia and their findings were more similar to our results including low sensitivity for KAN (9.4%) and XDR (13.6%).14 Additionally, our results are in line with those found in recent meta-analyses including a comprehensive review of published and unpublished by a WHO expert group.6 Among published reports, sensitivity of the MTBDRsl assay in detecting resistance to fluoroquinolones (87%), CAP (82%), and KAN (44%) was poor. The WHO report further found the sensitivity of the MTBDRsl in detecting XDR to vary greatly among studies (22.6–100%) but did find overall high specificity (91.8–100%). Our findings provide further evidence supporting WHO recommendations declaring the MTBDRsl assay unfit to replace conventional phenotypic DST or to design individualized MDR or XDR treatment regimens. Given its high specificity, the MTBDRsl may have a role ”ruling in” XDR disease among high-risk patients.
The poor sensitivity of the MTBDRsl assay reflects our limited knowledge of drug-resistance mechanisms and mutations. A recent review of genetic mutations causing resistance to injectable second-line agents evaluated over 1500 M. tuberculosis isolates and found that the A1401G mutation could explain only 76% of CAP and 56% of KAN resistance and furthermore that it was present in 7% of CAP susceptible isolates.20 Our results were worse with mutations in the 1401 region of the rrs gene present in 57% and 29% of CAP and KAN phenotypically resistant isolates, respectively, and in 9% of CAP susceptible isolates. Ongoing work has found that additional mutations in the rrs, eis promoter, tlyA, and gidB genes may be associated with injectable drug resistance and might explain the poor sensitivity of the MTBDRsl assay. In regard to fluoroquinolones, we detected mutations in the 90, 91 and 94 codons of the gyrA gene in 81% of OFX R cases, which is similar to other reports.13 Mutations in the gyrB gene or in genes encoding the MfpA protein may also cause fluoroquinolone resistance and could explain some cases with a normal gyrA gene and phenotypic fluoroquinolone resistance. Mutations in the eis gene could be responsible for the poor sensitivity of the MTBDRsl in detecting KAN resistance. Utilizing both the MTBDRsl and DNA sequencing Huang and colleagues found the sensitivity of the MTBDRsl for detection of drug resistance to KAN could be increased by approximately 27% by adding new alleles of the eis promoter into molecular analysis.21 Additional genomic studies among Beijing strains from Russia, which also has high rates of KAN resistance, found that a significant number of CAP and AMK sensitive but KAN resistant strains harbored mutations in the eis gene.22, 23 This line of evidence may in part explain the poor performance of the MTBDRsl assay in Georgia, as the Beijing strain is the most common genotype in the country.24
An important finding of our study is the feasibility of implementing an “add on” rapid molecular test for XDR detection into a busy NRL that already performs a LPA for MDR detection. The majority of MTBDRsl assays had sufficient amplification and interpretable results (97%); a rate higher than the percentage of cultures positive for M. tuberculosis (92%). We also found that incorporated into normal workflow MTBDRsl assay results were available in less than two weeks as compared to 70–104 days using conventional DST methods. If the sensitivity of the MTBDRsl or other future LPA’s can be improved, this rapid turnaround time could help ensure earlier treatment with effective regimens, which could result in improved treatment outcomes, decreased development of amplified drug resistance, and disease transmission prevention.25
Genetic sequencing of M. tuberculosis isolates with discordant MTBDRsl and DST results would have helped identify non-assay mutations responsible for second-line drug resistance. As a consequence of only testing the initial sputum specimen we may have had false negative MTBDRsl results and hence lower MTBDRsl sensitivity due to heteroresistant bacilli populations. Heteroresistance is the phenomenon of simultaneous occurrence of drug resistant and drug sensitive organisms in the same sample. A recent study found 5% and 8% of M. tuberculosis isolates with phenotypic drug resistance to OFL and AMK, respectively, without molecular markers of drug resistance.26 Subsequent DNA sequencing of single colonies selected in the presence of OFL and AMK revealed underlying mutations in 78% and 100% of the isolates, thus demonstrating heteroresistance. A further study found that the MTBDRplus assay had a poor sensitivity in detecting INH resistance in a sample of 1% resistant bacteria.27 These findings demonstrate the challenges posed by heteroresistance bacilli in regards to genetics based drug resistance testing. The performance of the MTBDRsl was compared to WHO recommended methods for phenotypic second-line DST; however, these methods have not been fully standardized and there are limited studies evaluating the reproducibility of DST for second-line anti-tuberculosis drugs.28, 29
CONCLUSION
In conclusion, we have demonstrated the feasibility of implementing the MTBDRsl assay into a “real world” setting in a high-burdened drug-resistant TB country, but also found the assay lacks sufficient accuracy to be recommended for clinical use in this setting. For the MTBDRsl or other new assays to have a clinical impact on the treatment and transmission of XDR-TB they need to include additional genetic mutations responsible for second-line drug-resistance especially.
ACKNOWLEDGEMENTS
This work was supported in part by the NIH Fogarty International Center [D43TW007124 and D43TW007124-06S1], and the Emory Global Health Institute.
Footnotes
AUTHOR CONTRIBUTIONS
N.T. and R.R.K. conceived the idea for this study in collaboration with N.B., R.A. and H.M.B. N.B. performed the laboratory work and data entry. R.R.K. performed the data analyses. N.T. and R.R.K. drafted the manuscript. N.B., R.A., and H.M.B. reviewed and edited the manuscript. All authors reviewed and approved the final version of the manuscript.
The authors report no conflicts of interest.
References
- 1.WHO. Geneva: World Health Organization; 2012. Global tuberculosis control: WHO report 2012. WHO/HTM/20126. 2012. [Google Scholar]
- 2.Abubakar I, Zignol M, Falzon D, Raviglione M, Ditiu L, Masham S, et al. Drug-resistant tuberculosis: time for visionary political leadership. Lancet Infect Dis. 2013 doi: 10.1016/S1473-3099(13)70030-6. Epub 2013/03/28. [DOI] [PubMed] [Google Scholar]
- 3.Raviglione M. XDR-TB: entering the post-antibiotic era? Int J Tuberc Lung Dis. 2006;10(11):1185–1187. Epub 2006/11/30. [PubMed] [Google Scholar]
- 4.Tukvadze N, Kempker RR, Kalandadze I, Kurbatova E, Leonard MK, Apsindzelashvili R, et al. Use of a molecular diagnostic test in AFB smear positive tuberculosis suspects greatly reduces time to detection of multidrug resistant tuberculosis. PLoS One. 2012;7(2):e31563. doi: 10.1371/journal.pone.0031563. Epub 2012/02/22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nehren, Germany: Hain Lifescience; [accessed April 15, 2013]. Genotype MTBDRsl, product page. http://www.hain-lifescience.de.en/products/microbiology/mycobacteria/genotype-mtbdrsl.html. [Google Scholar]
- 6.WHO. Geneva: World Health Organization; 2013. The use of Molecular Line Probe Assay for the detection of resistance to second-line anti-tuberculosis drugs: Expert group meeting report, February 2013.. WHO/HTM/TB/201301. 2013. [Google Scholar]
- 7.Richards DC, Mikiashvili T, Parris JJ, Kourbatova EV, Wilson JC, Shubladze N, et al. High prevalence of hepatitis C virus but not HIV co-infection among patients with tuberculosis in Georgia. Int J Tuberc Lung Dis. 2006;10(4):396–401. Epub 2006/04/11. [PubMed] [Google Scholar]
- 8.Parsons LM, Somoskovi A, Gutierrez C, Lee E, Paramasivan CN, Abimiku A, et al. Laboratory diagnosis of tuberculosis in resource-poor countries: challenges and opportunities. Clin Microbiol Rev. 2011;24(2):314–350. doi: 10.1128/CMR.00059-10. Epub 2011/04/13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nehren, Germany: Hain Lifescience; [accessed August 115, 2011]. Genotype MTBDRplus, product page. http://www.hain-lifescience.de.en/products/microbiology/mycobacteria/genotype-mtbdrpus.hrm. [Google Scholar]
- 10.WHO. Geneva: World Health Organization; 2009. Guidelines for surveillance of drug resistance in tuberculosis-4th ed.. WHO/HTM/TB/2009422. 2009. [Google Scholar]
- 11.WHO. Geneva: World Health Organization; 2008. Policy guidance on drug susceptibility testing (DST) of second-line antituberculosis drugs. WHO/HTM/TB/2008392. 2008. [PubMed] [Google Scholar]
- 12.Harris PA, Taylor R, Thielke R, Payne J, Gonzalez N, Conde JG. Research electronic data capture (REDCap)--a metadata-driven methodology and workflow process for providing translational research informatics support. Journal of biomedical informatics. 2009;42(2):377–381. doi: 10.1016/j.jbi.2008.08.010. Epub 2008/10/22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Feng Y, Liu S, Wang Q, Wang L, Tang S, Wang J, et al. Rapid diagnosis of drug resistance to fluoroquinolones, amikacin, capreomycin, kanamycin and ethambutol using genotype MTBDRsl assay: a meta-analysis. PLoS One. 2013;8(2):e55292. doi: 10.1371/journal.pone.0055292. Epub 2013/02/06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kontsevaya I, Ignatyeva O, Nikolayevskyy V, Balabanova Y, Kovalyov A, Kritsky A, et al. Diagnostic accuracy of the genotype MTBDRsl assay for rapid diagnosis of extensively drug-resistant tuberculosis in HIV-coinfected patients. J Clin Microbiol. 2013;51(1):243–248. doi: 10.1128/JCM.02513-12. Epub 2012/11/16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lacoma A, Garcia-Sierra N, Prat C, Maldonado J, Ruiz-Manzano J, Haba L, et al. GenoType MTBDRsl for molecular detection of second-line-drug and ethambutol resistance in Mycobacterium tuberculosis strains and clinical samples. J Clin Microbiol. 2012;50(1):30–36. doi: 10.1128/JCM.05274-11. Epub 2011/11/15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hillemann D, Rusch-Gerdes S, Richter E. Feasibility of the GenoType MTBDRsl assay for fluoroquinolone, amikacin-capreomycin, and ethambutol resistance testing of Mycobacterium tuberculosis strains and clinical specimens. J Clin Microbiol. 2009;47(6):1767–1772. doi: 10.1128/JCM.00081-09. Epub 2009/04/24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miotto P, Cabibbe AM, Mantegani P, Borroni E, Fattorini L, Tortoli E, et al. GenoType MTBDRsl performance on clinical samples with diverse genetic background. Eur Respir J. 2012;40(3):690–698. doi: 10.1183/09031936.00164111. Epub 2012/01/24. [DOI] [PubMed] [Google Scholar]
- 18.Barnard M, Warren R, Van Pittius NG, van Helden P, Bosman M, Streicher E, et al. Genotype MTBDRsl line probe assay shortens time to diagnosis of extensively drugresistant tuberculosis in a high-throughput diagnostic laboratory. Am J Respir Crit Care Med. 2012;186(12):1298–1305. doi: 10.1164/rccm.201205-0960OC. Epub 2012/10/23. [DOI] [PubMed] [Google Scholar]
- 19.Chihota VN, Muller B, Mlambo CK, Pillay M, Tait M, Streicher EM, et al. Population structure of multi- and extensively drug-resistant Mycobacterium tuberculosis strains in South Africa. J Clin Microbiol. 2012;50(3):995–1002. doi: 10.1128/JCM.05832-11. Epub 2011/12/16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Georghiou SB, Magana M, Garfein RS, Catanzaro DG, Catanzaro A, Rodwell TC. Evaluation of genetic mutations associated with Mycobacterium tuberculosis resistance to amikacin, kanamycin and capreomycin: a systematic review. PLoS One. 2012;7(3):e33275. doi: 10.1371/journal.pone.0033275. Epub 2012/04/06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang WL, Chi TL, Wu MH, Jou R. Performance assessment of the GenoType MTBDRsl test and DNA sequencing for detection of second-line and ethambutol drug resistance among patients infected with multidrug-resistant Mycobacterium tuberculosis. J Clin Microbiol. 2011;49(7):2502–2508. doi: 10.1128/JCM.00197-11. Epub 2011/05/13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Casali N, Nikolayevskyy V, Balabanova Y, Ignatyeva O, Kontsevaya I, Harris SR, et al. Microevolution of extensively drug-resistant tuberculosis in Russia. Genome research. 2012;22(4):735–745. doi: 10.1101/gr.128678.111. Epub 2012/02/02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gikalo MB, Nosova EY, Krylova LY, Moroz AM. The role of eis mutations in the development of kanamycin resistance in Mycobacterium tuberculosis isolates from the Moscow region. J Antimicrob Chemother. 2012;67(9):2107–2109. doi: 10.1093/jac/dks178. Epub 2012/05/18. [DOI] [PubMed] [Google Scholar]
- 24.Niemann S, Diel R, Khechinashvili G, Gegia M, Mdivani N, Tang YW. Mycobacterium tuberculosis Beijing lineage favors the spread of multidrug-resistant tuberculosis in the Republic of Georgia. J Clin Microbiol. 2010;48(10):3544–3550. doi: 10.1128/JCM.00715-10. Epub 2010/08/13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wells WA, Boehme CC, Cobelens FG, Daniels C, Dowdy D, Gardiner E, et al. Alignment of new tuberculosis drug regimens and drug susceptibility testing: a framework for action. Lancet Infect Dis. 2013 doi: 10.1016/S1473-3099(13)70025-2. Epub 2013/03/28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Streicher EM, Bergval I, Dheda K, Bottger EC, Gey van Pittius NC, Bosman M, et al. Mycobacterium tuberculosis population structure determines the outcome of geneticsbased second-line drug resistance testing. Antimicrob Agents Chemother. 2012;56(5):2420–2427. doi: 10.1128/AAC.05905-11. Epub 2012/02/15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Folkvardsen DB, Svensson E, Thomsen VO, Rasmussen EM, Bang D, Werngren J, et al. Can molecular methods detect 1% isoniazid resistance in Mycobacterium tuberculosis? J Clin Microbiol. 2013;51(5):1596–1599. doi: 10.1128/JCM.00472-13. Epub 2013/03/01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Horne DJ, Pinto LM, Arentz M, Lin SY, Desmond E, Flores LL, et al. Diagnostic accuracy and reproducibility of WHO-endorsed phenotypic drug susceptibility testing methods for first-line and second-line antituberculosis drugs. J Clin Microbiol. 2013;51(2):393–401. doi: 10.1128/JCM.02724-12. Epub 2012/11/16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim SJ, Espinal MA, Abe C, Bai GH, Boulahbal F, Fattorin L, et al. Is second-line anti-tuberculosis drug susceptibility testing reliable? Int J Tuberc Lung Dis. 2004;8(9):1157–1158. Epub 2004/10/01. [PubMed] [Google Scholar]