Abstract
Adenosine monophosphate-activated protein kinase (AMPK) acts as a master mediator of metabolic homeostasis. It is considered as a significant millstone to treat metabolic syndromes including obesity, diabetes, and fatty liver. It can sense cellular energy or nutrient status by switching on the catabolic pathways. Investigation of AMPK has new findings recently. AMPK can inhibit cell growth by the way of autophagy. Thus AMPK has become a hot target for small molecular drug design of tumor inhibition. Activation of AMPK must undergo certain extent change of the structure. Through the methods of structure-based virtual screening and molecular dynamics simulation, we attempted to find out appropriate small compounds from the world's largest TCM Database@Taiwan that had the ability to activate the function of AMPK. Finally, we found that two TCM compounds, eugenyl_beta-D-glucopyranoside and 6-O-cinnamoyl-D-glucopyranose, had the qualification to be AMPK agonist.
1. Introduction
One study has found that the cell “starvation” signal transduction pathway reveals the process of cell “starvation” signal transduction mechanisms. This finding is considered as a significant millstone to treat metabolic syndromes including obesity, diabetes, and fatty liver [1, 2]. Adenosine monophosphate-activated protein kinase (AMPK) was always a hot issue in the recent 10 years because it plays an essential role in sensing cellular energy or nutrient status [3, 4]. AMPK exists in several organs including skeletal muscles, brain, and liver [5]. For example, when skeletal muscles have exercised for a period of time, activated AMPK helps the cells adapt to energy challenge by increasing glucose uptake [6]. AMPK acts as a master mediator of metabolic homeostasis [7]. The effect of AMPK activation is switching on the catabolic pathways consisting of cellular glucose uptake, glycolysis, fatty acid oxidation, inhibition of triglyceride, cholesterol, and protein synthesis [8, 9]. The upstream signal molecule, such as adiponectin, stimulates AMPK to utilize glucose and fatty acid [10]. The activated AMPK in the hypothalamus increases the appetite or desire for food intake [11, 12]. Adiponectin stimulates AMPK to protect the heart against myocardial ischemia [13, 14]. Another upstream signal molecule, leptin, activates AMPK to stimulate fatty acid oxidation [15]. AMPK in the hypothalamus mediates thyroid hormone to regulate energy balance, too [16].
AMPK consists of three subunits, α, β, and γ [17]. Each of the subunits has a specific structure and function [18, 19]. The following mechanism explains how the three subunits work. When the cells need energy, adenosine triphosphate (ATP) converts to adenosine diphosphate (ADP) and AMP by releasing energy simultaneously. At the normal condition, excess AMP binds to the γ subunit of AMPK [20]. After binding of γ subunit exposes the active site, Thr172, on the catalytic α subunit of AMPK [21]. For all of the three subunits, α subunit is the most important. Activation of AMPK follows conformational change of α subunit and phosphorylation at Thr172 [22]. The phosphorylation is induced by the upstream signal molecule such as liver kinase B1 (LKB1) [23]. The β subunit is located between α and γ subunits and is associated with the function of glycogen sensor [24]. ADP can also bind to γ subunit and protect AMPK from dephosphorylation but cannot cause conformational change [25].
Physiological study has approved that blood glucose is partly regulated by AMPK [26]. Activating AMPK by its agonist decreases insulin resistance induced by obesity [27]. Drug design for AMPK activator provides the hope for treating type 2 diabetes mellitus [28]. AMPK has become the drug target for managing diabetes and metabolic syndrome [29]. Metformin is the classic antidiabetes medicine involved in activation of AMPK [30, 31]. Investigation of AMPK has new findings recently. Activated AMPK has the function of anti-inflammation by a mimic state of pseudostarvation [32]. AMPK can regulate cell growth via signal integration [33]. The LKB1-AMPK pathway can mediate cellular autophagy or apoptosis by phosphorylating p27kip1 [34]. Thus the LKB1-AMPK pathway has been approved in the field of tumor suppression [35]. AMPK inducing phosphorylation of Unc-51-like kinase 1 (ULK1) can cause autophagy in an energetic exhaustion status. ULK1 is a serine/threonine-protein kinase [36, 37].
Due to progress of modern technology, the binding phenomena of protein dynamics motion and structure changing can be analyzed by computational simulation [38, 39]. In silico investigation of biology or computational systems biology helps us to explore the protein-protein or protein-molecule interaction [40, 41]. These methods make it possible to participate in computer-aided drug design (CADD) [42, 43]. CADD techniques save our time to select appropriate drug compound rapidly compared with traditional one-by-one biochemistry [44, 45]. CADD procedures consist of virtual screening, validations, and analysis [46, 47]. Virtual screening and analysis employ docking and molecular dynamics (MD) simulation [48–50]. MD can predict how long the compound needs to form stable complex structure with target protein [51]. A series of statistics or score systems facilitate docking and MD accuracy [52]. Best candidates from virtual screening and MD can be selected as potential therapeutic drugs [53].
With progress in medical technology, many diseases can be resolved nowadays [54–56]. When we know how the diseases happen, we can handle them in smart ways [57–59]. Because of coordinating cell growth and autophagy, AMPK has become an emerging target for drug design of tumor inhibition [60]. Design of small molecular AMPK agonist is possible [61]. Small molecular drug design by CADD has been applied in systems biology extensively [62–64]. Traditional Chinese medicine (TCM) has therapeutic effect on many diseases [65–68]. In this study, we attempted to find out appropriate small compounds from the world's largest TCM Database@Taiwan that had the ability to activate the function of AMPK [69].
2. Materials and Methods
2.1. Compound Database
To identify potential AMPK activators from TCM, we downloaded all small molecules from TCM Database@Taiwan (http://tcm.cmu.edu.tw/) to carry on AMPK agonist screening [69].
2.2. Homology Modeling
The AMPK protein sequence was obtained from the Uniprot Knowledgebase (Q13131, human). The 3D structure of rat AMPK was obtained from Protein Data Bank (PDB ID: 2Y94). The sequence of human AMPK (Q13131) and homologous crystal structure of rat AMPK (2Y94) were aligned by the modeler mode in Accelrys Discovery Studio (DS) 2.5. The identity and similarity can be calculated according to the result of sequence alignment. Homology modeling of AMPK was established by the Build Homology Models protocol in DS 2.5. The reasonable AMPK model was further validated by Ramachandran plot with Rampage protocol and Verify score with Profiles-3D protocol in DS 2.5.
2.3. Structure-Based Virtual Screening
The ligands from TCM Database@Taiwan and the control ligand, adenosine monophosphate (AMP), were prepared for specified techniques. Ligand docking was performed by the LigandFit module in DS 2.5. The force field of Chemistry at HARvard Molecular Mechanics (CHARMm) was utilized to minimize all docking poses [70]. Absorption, distribution, metabolism, excretion, and toxicity (ADMET) were applied for all TCM compounds [71]. The scoring systems such as Dock score, piecewise linear potentials (PLP), and potential of mean force (PMF) were calculated by the LigandFit module in DS 2.5 [72, 73].
2.4. Disorder Prediction
We drew disorder disposition to exclude disordered residues by the program of PONDR-FIT in the DisProt website [74, 75].
2.5. Molecular Dynamics (MD) Simulation
The package of GROMACS (GROningen MAchine for Chemical Simulations) was used for MD simulation. We employed SwissParam to determine topology and parameters of small compounds for GROMACS simulation. The cytoplasmic condition was set with transferable intermolecular potential 3P (TIP3P) water at 0.9% sodium chloride concentration. After docking, selected protein-ligand complexes were conducted under the following phases: minimization, heating, equilibration, and production. The minimization protocol included 500 steps of steepest descent and 500 steps of conjugated gradient. The heating time was 50 ps from 50 K to 310 K. The equilibration time was 150 ps at 310 K. The production time was 5000 ps with constant temperature dynamics method. The time for temperature decay was 0.4 ps. We utilized the trajectory analysis to illustrate root mean square deviation (RMSD), Gyrate, solvent accessible surface (SAS), root mean square fluctuation (RMSF), total energy, database of secondary structure assignment (DSSP), matrices of smallest distance of residues for individual ligands, and protein during MD. We calculated the formation and distance of hydrogen bond (H-bond), too. Best distance of H-bond was set at 0.3 nm.
2.6. Ligand Pathway
To analyze the ligand pathway, we employed the software of LigandPath module to illustrate the possible pathway of each ligand. A surface probe and minimum clearance were set at 6 Å and 3 Å, respectively.
3. Results
3.1. Homology Modeling
According to sequence alignment between Q13131_human and template (2Y94), the overall identity was 89.4% and similarity was 89.6% (Figure 1). Ramachandran plot of AMPK-modeled structure showed that 91.7% of residues were in the favored area, 5.9% were in the allowed area, and only 2.4% were in the disallowed area (Figure 2). The Verify score showed that most residues were positive values except residues from 290 to 360 (Figure 3).
Figure 1.
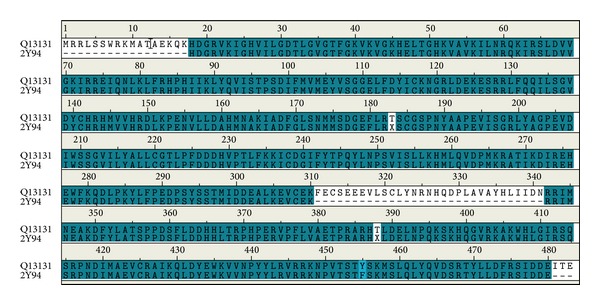
Sequence alignment between Q13131 human and template (2Y94). The identity is 89.4% and the similarity is 89.6%.
Figure 2.
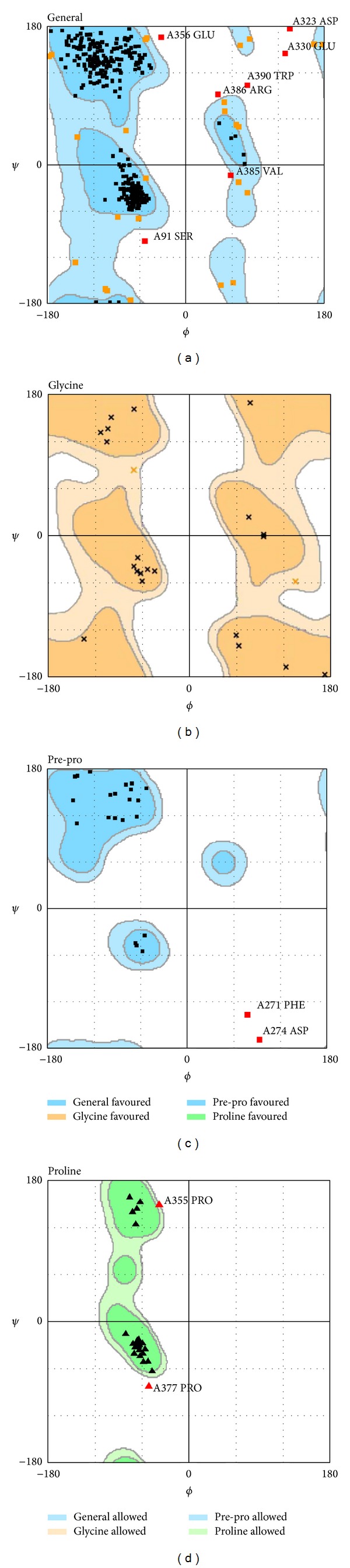
Ramachandran plot of AMPK-modeled structure. Number of residues in favored region (~98.0% expected) is 422 (91.7%). Number of residues in allowed region (~2.0% expected) is 27 (5.9%). Number of residues in disallowed region is 11 (2.4%).
Figure 3.
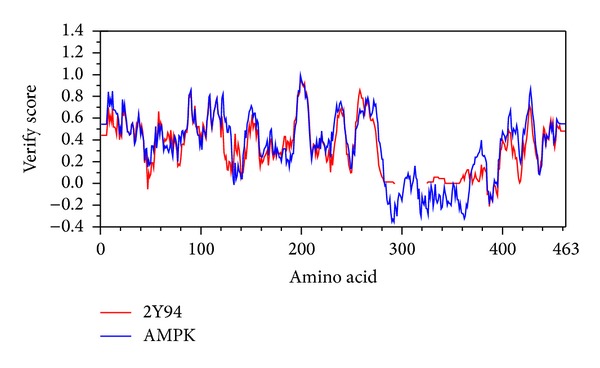
Verify score of AMPK-modeled structure. Most residues are positive values except residues from 290 to 360.
3.2. Structure-Based Virtual Screening
Table 1 showed absorption level of ADMET, Dock score, PLP1, PLP2, and PMF of the top 7 TCM compounds ranked by absorption level of ADMET. By the integration of these data, we selected the first 2 compounds, eugenyl_beta-D-glucopyranoside and 6-O-cinnamoyl-D-glucopyranose, as candidates for further survey (Figure 4). Docking poses of eugenyl_beta-D-glucopyranoside, 6-O-cinnamoyl-D-glucopyranose, and the control (AMP) with AMPK were illustrated in Figure 5. Eugenyl_beta-D-glucopyranoside formed H-bond with Glu94, Asn138, and Asp151 (Figure 5(a)). 6-O-Cinnamoyl-D-glucopyranose formed H-bond with Val18, Asp133, Lys135, and Asp151 (Figure 5(b)). AMP formed H-bond with Gly19, Val90, Ser91, and Glu94 (Figure 5(c)).
Table 1.
Top 7 candidates of scoring function from TCM Database@Taiwan screening. (ADMET_absorption level 0 to 3: good to very low absorption.)
| Name | ADMET_Absorption_Level | Dock score | PLP1 | PLP2 | PMF |
|---|---|---|---|---|---|
| Eugenyl_beta-D-glucopyranoside | 0 | 58.932 | 73.12 | 78.35 | 66.59 |
| 6-O-Cinnamoyl-D-glucopyranose | 0 | 60.305 | 72.41 | 70.61 | 66.65 |
| Loroglossin | 1 | 69.903 | 74.94 | 71.77 | 72.83 |
| Eleutheroside B | 1 | 66.207 | 92.33 | 93.2 | 72.04 |
| Quercetin-3-O-glucopyranoside | 1 | 62.949 | 85.39 | 87.14 | 66.12 |
| Synapic aldehyde 4-O-bata-D-glucopyranoside | 1 | 62.262 | 94.7 | 94.75 | 73.44 |
| (−)-9alpha-Hydroxysophocarpine | 1 | 59.13 | 80.89 | 74.13 | 63.9 |
|
| |||||
| AMP | 3 | 57.025 | 72.1 | 69.07 | 62.96 |
Figure 4.
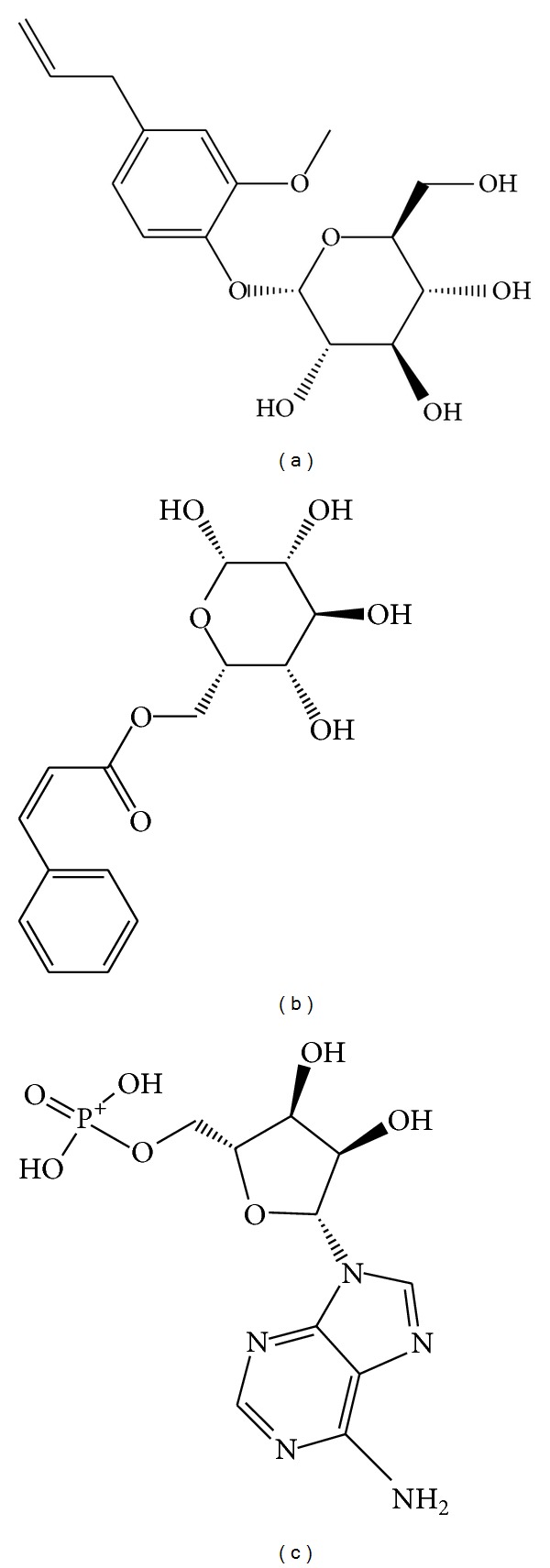
Scaffold of top 2 candidate compounds: (a) eugenyl_beta-D-glucopyranoside, (b) 6-O-cinnamoyl-D-glucopyranose, and the control (c) AMP.
Figure 5.
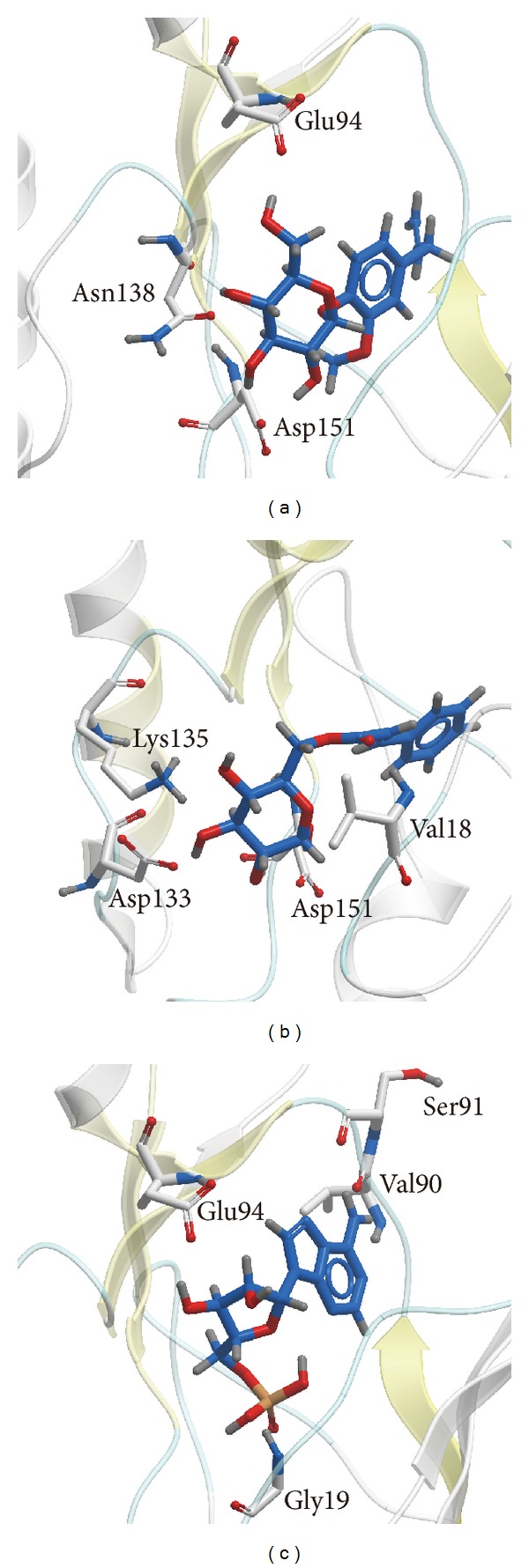
Docking poses of (a) eugenyl_beta-D-glucopyranoside, (b) 6-O-cinnamoyl-D-glucopyranose, and (c) AMP. The common residue is ASP151.
3.3. Disorder Prediction
Main residues (Val18 to Asp151) of AMPK-modeled structure for the 2 candidates and the control were not in the disordered area, so there was no influence to the shape of the binding site (Figure 6).
Figure 6.
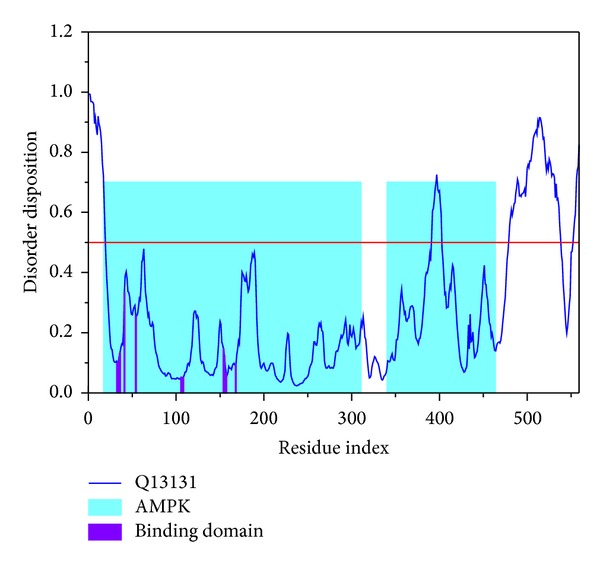
Disorder disposition of AMPK-modeled structure. Main residues (Val18 to ASP151) are in the nondisordered region (below the red line).
3.4. Molecular Dynamics (MD) Simulation
MD trajectories generated by GROMACS were illustrated. We utilized root mean square deviation (RMSD) to show the deviation degree of each ligand or protein from the beginning to the end of MD. When the ligand formed a complex with the protein (AMPK), eugenyl_beta-D-glucopyranoside, 6-O-cinnamoyl-D-glucopyranose, and the control (AMP) had first larger deviations at 1600 ps, 1450 ps, and 2000 ps, respectively. AMP had a larger average deviation than the candidates (Figure 7(a)). We utilized Gyrate to measure the average distance of the atoms to the center of each mass. In other words, Gyrate showed the compact degree of each ligand or protein. 6-O-Cinnamoyl-D-glucopyranose had the largest ligand Gyrate values, but eugenyl_beta-D-glucopyranoside had the largest protein Gyrate values. All the proteins had a trend to compact status at the end of MD (Figure 7(b)). We utilized solvent accessible surface (SAS) to measure the surface area of each ligand or protein in contact with the water. 6-O-Cinnamoyl-D-glucopyranose had the largest ligand SAS values, but eugenyl_beta-D-glucopyranoside had the largest protein SAS values. All the proteins had a trend to decreased SAS value at the end of MD (Figure 7(c)).
Figure 7.
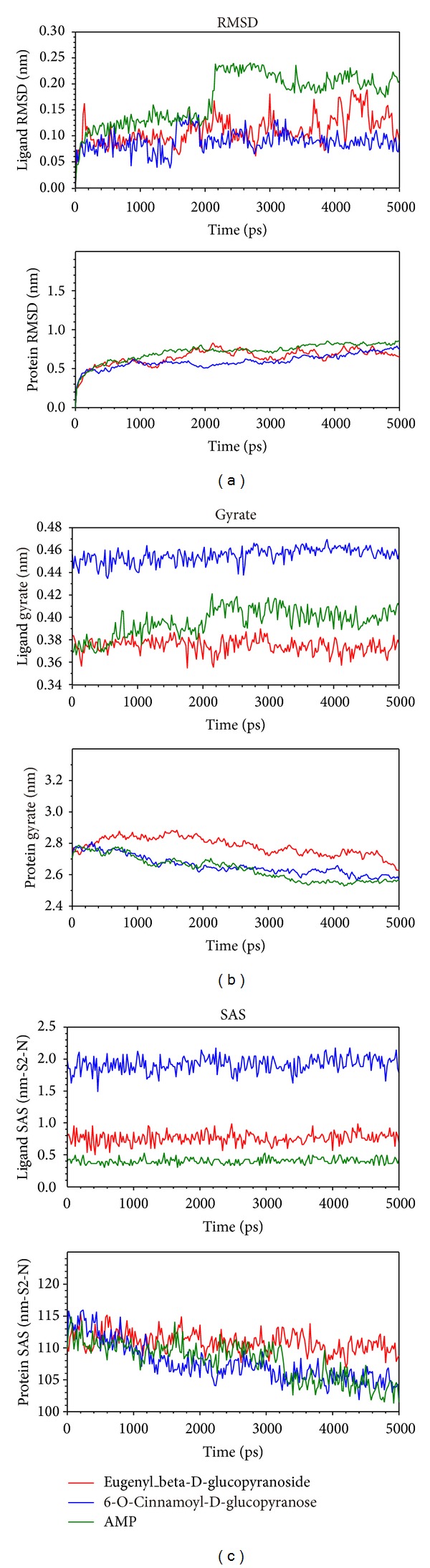
Ligand and protein (a) root mean square deviation (RMSD), (b) Gyrate, and (c) solvent accessible surface (SAS).
Root mean square fluctuation (RMSF) showed the deviation of individual residue of the protein during MD. All the compounds had similar graphs with larger fluctuations at the range between residue 320 and residue 400 (Figure 8). AMP had a lower average total energy (−1923000 kJ/mol) than the 2 candidates (−1920000 and −1921000 kJ/mol) (Figure 9). Database of secondary structure assignment (DSSP) and secondary structural feature ratio variations of the protein was used to discuss the change of structural component during MD. All the candidates and the control had similar findings. Among the components of important second degree structure, the ratio of α-helix was higher than β-sheet, and β-sheet was higher than bend. The ratio of α-helix increased slightly, but the ratio of bend decreased comparatively (Figure 10). We utilized matrices of the smallest distance to find any variation of residue distance when the protein complexed with the ligand. There was not any significant difference compared to the candidates with the control (Figure 11).
Figure 8.
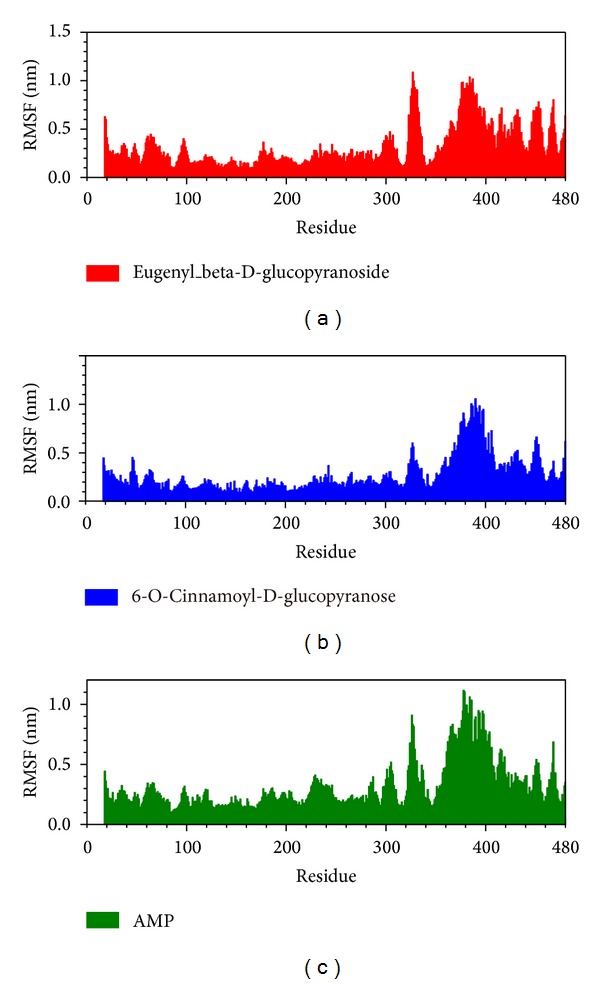
Root mean square fluctuation (RMSF) for (a) eugenyl_beta-D-glucopyranoside, (b) 6-O-cinnamoyl-D-glucopyranose, and (c) AMP.
Figure 9.
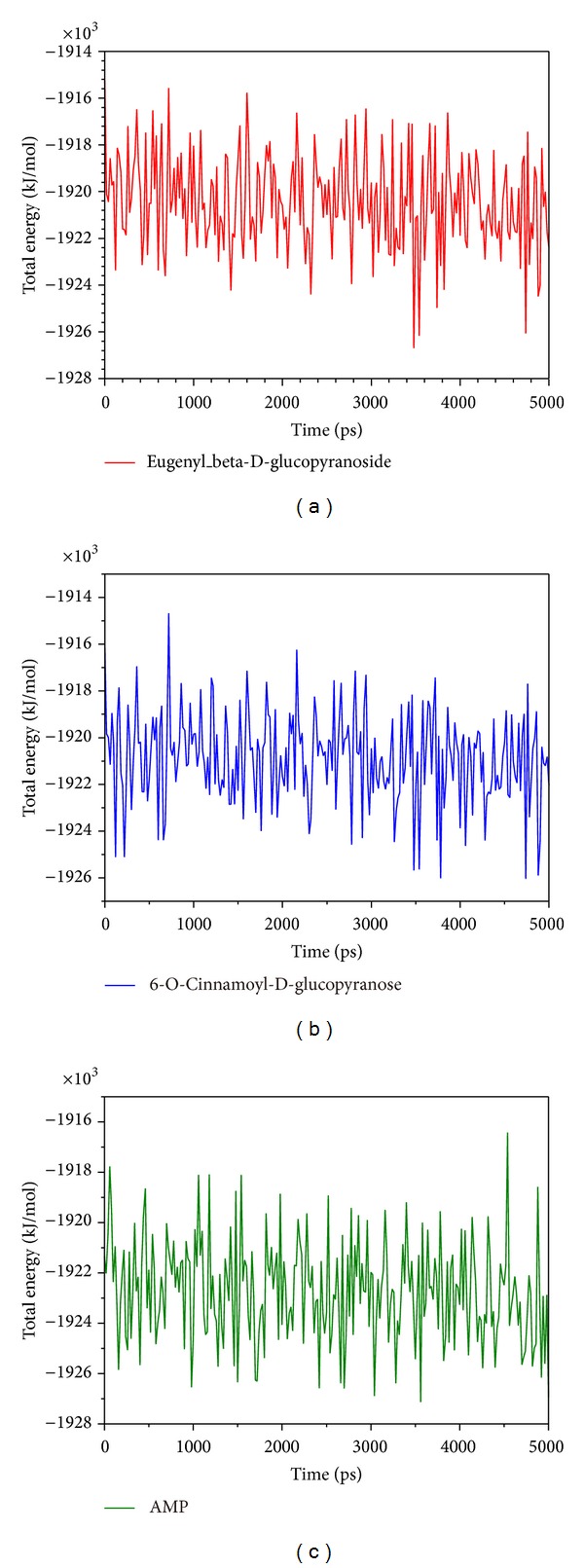
Total energy for (a) eugenyl_beta-D-glucopyranoside, (b) 6-O-cinnamoyl-D-glucopyranose, and (c) AMP.
Figure 10.
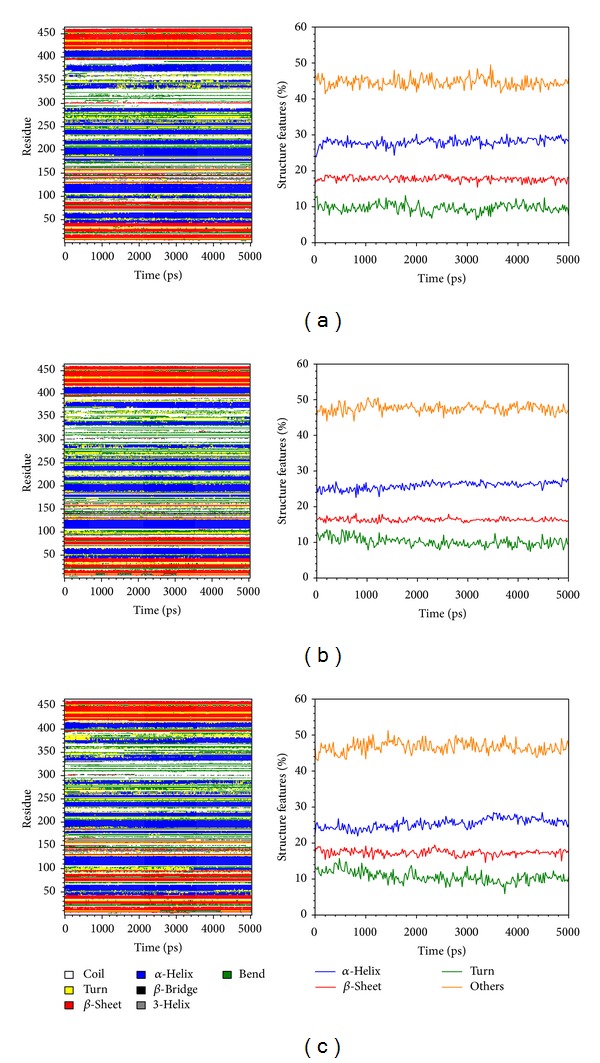
Database of secondary structure assignment (DSSP) and secondary structural component for (a) eugenyl_beta-D-glucopyranoside, (b) 6-O-cinnamoyl-D-glucopyranose, and (c) AMP.
Figure 11.
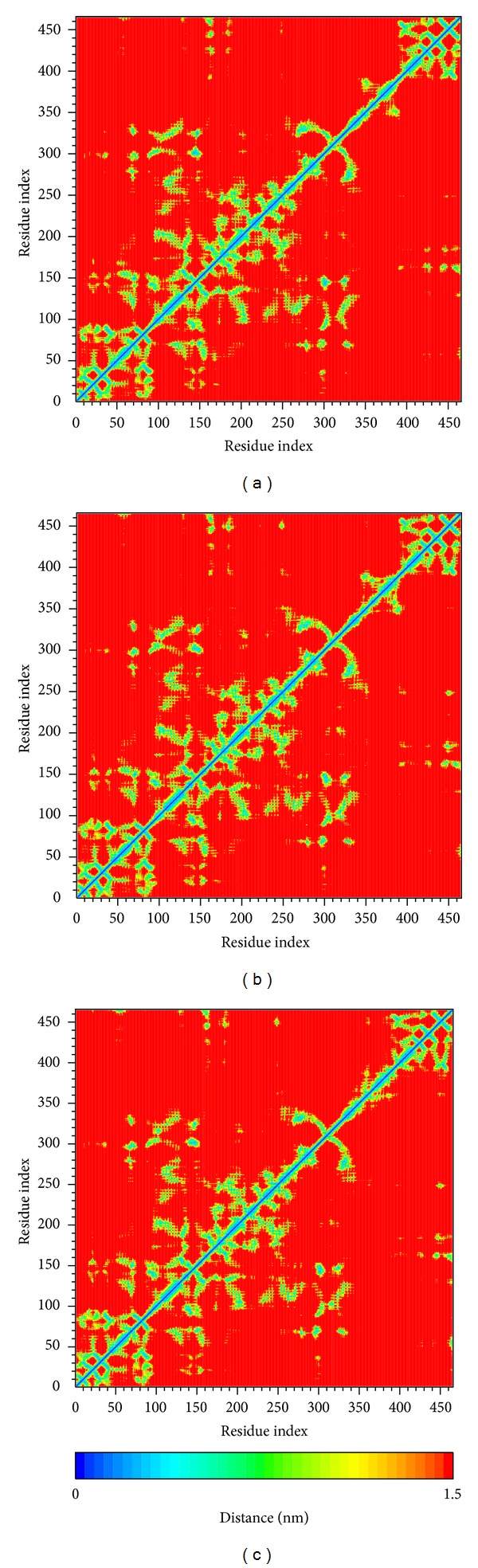
Matrices of the smallest distance of residues for (a) eugenyl_beta-D-glucopyranoside, (b) 6-O-cinnamoyl-D-glucopyranose, and (c) AMP.
Table 2 showed occupancy of H-bond between eugenyl_beta-D-glucopyranoside, 6-O-cinnamoyl-D-glucopyranose, and AMP with AMPK protein during MD. The formation of H-bond between eugenyl_beta-D-glucopyranoside with Asn138 and Asp151, 6-O-cinnamoyl-D-glucopyranose with Val18, Asp133, Lys135, and Asp151, and AMP with Gly19, Val90, and Glu94 was consistent with the results of docking. AMP also formed H-bond with Asp151, so we could conclude that Asp151 was key residue for top 2 TCM candidates and the control bound with AMPK.
Table 2.
Occupancy of H-bond between eugenyl_beta-D-glucopyranoside, 6-O-cinnamoyl-D-glucopyranose, and AMP with AMPK protein.
| Ligand | H-bond | Ligand | Amino acid | Occupancy (%) |
|---|---|---|---|---|
| Atom | ||||
| Eugenyl_beta-D-glucopyranoside | 1 | O15 | THR20:HN | 23.51% |
| 2 | H38 | THR20:OG1 | 14.34% | |
| 3 | O15 | PHE21:HN | 13.55% | |
| 4 | H34 | GLY22:O | 20.32% | |
| 5 | O13 | LYS39:HZ3 | 19.92% | |
| 6 | H36 | ASN138:OD1 | 1.59% | |
| 7 | H38 | ASN138:OD1 | 1.99% | |
| 8 | H34 | ASP151:OD1 | 14.34% | |
| 9 | H36 | ASP151:OD1 | 20.32% | |
| 10 | H38 | ASP151:OD1 | 7.17% | |
| 11 | H36 | ASP151:OD2 | 21.51% | |
|
| ||||
| 6-O-Cinnamoyl-D-glucopyranose | 1 | O22 | VAL18:HN | 16.33% |
| 2 | O22 | GLY19:HN | 21.51% | |
| 3 | H26 | ASP133:OD1 | 43.43% | |
| 4 | H28 | ASP133:OD1 | 30.28% | |
| 5 | H26 | ASP133:OD2 | 37.85% | |
| 6 | H28 | ASP133:OD2 | 41.43% | |
| 7 | O6 | LYS135:HZ3 | 15.54% | |
| 8 | H30 | GLU137:OE1 | 27.49% | |
| 9 | H30 | GLU137:OE2 | 33.07% | |
| 10 | H23 | ASP151:OD1 | 24.70% | |
| 11 | H23 | ASP151:OD2 | 64.94% | |
|
| ||||
| AMP | 1 | O4 | GLY19:HN | 13.94% |
| 2 | O4 | VAL90:HN | 28.29% | |
| 3 | O6 | VAL90:HN | 11.55% | |
| 4 | N14 | VAL90:HN | 40.24% | |
| 5 | H30 | VAL90:O | 0.80% | |
| 6 | O6 | GLU94:HN | 10.76% | |
| 7 | H28 | GLU94:OE1 | 6.77% | |
| 8 | H30 | GLU94:OE1 | 22.31% | |
| 9 | H28 | GLU94:OE2 | 4.38% | |
| 10 | H30 | GLU94:OE2 | 19.92% | |
| 11 | H36 | ASP151:OD1 | 77.29% | |
| 12 | H36 | ASP151:OD2 | 47.81% | |
| 13 | O20 | PHE152:HN | 64.54% | |
We showed the distance of H-bonds between eugenyl_beta-D-glucopyranoside and essential amino acids of AMPK. The O13 of eugenyl_beta-D-glucopyranoside formed H-bond with Lys39 at early stage of MD, and H34 formed H-bond with Gly22 at late stage of MD. H36 and H38 formed H-bonds with Asn138 only at initial stage of MD. H36 formed H-bond with Asn151 at early stage of MD (Figure 12).
Figure 12.
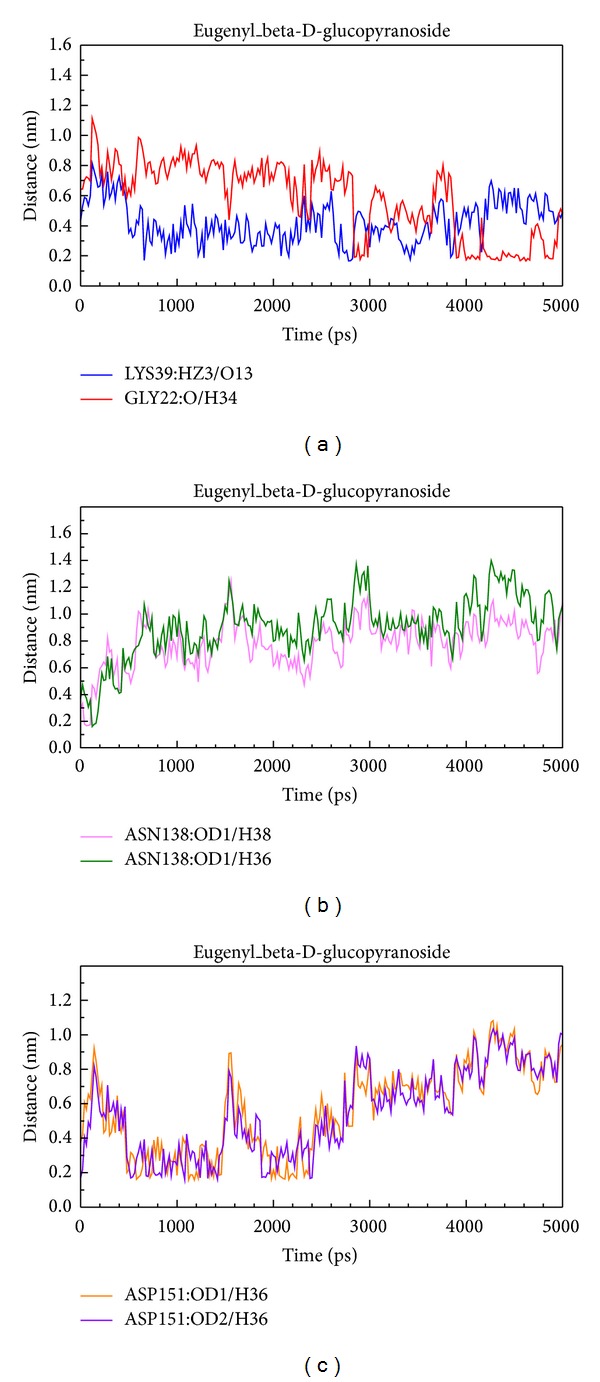
Distance of hydrogen bonds between eugenyl_beta-D-glucopyranoside and essential amino acids of AMPK.
We showed distance of H-bonds between 6-O-cinnamoyl-D-glucopyranose and essential amino acids of AMPK. The H30 of 6-O-cinnamoyl-D-glucopyranose formed H-bond with Glu137 at late stage of MD. H26 formed H-bond with OD2 of Asp133 at early stage of MD and formed H-bond with OD1 of Asp133 at late stage of MD. H23 formed H-bond with OD2 of Asp151 at early and late stage of MD and formed H-bond with OD1 at middle stage of MD (Figure 13).
Figure 13.
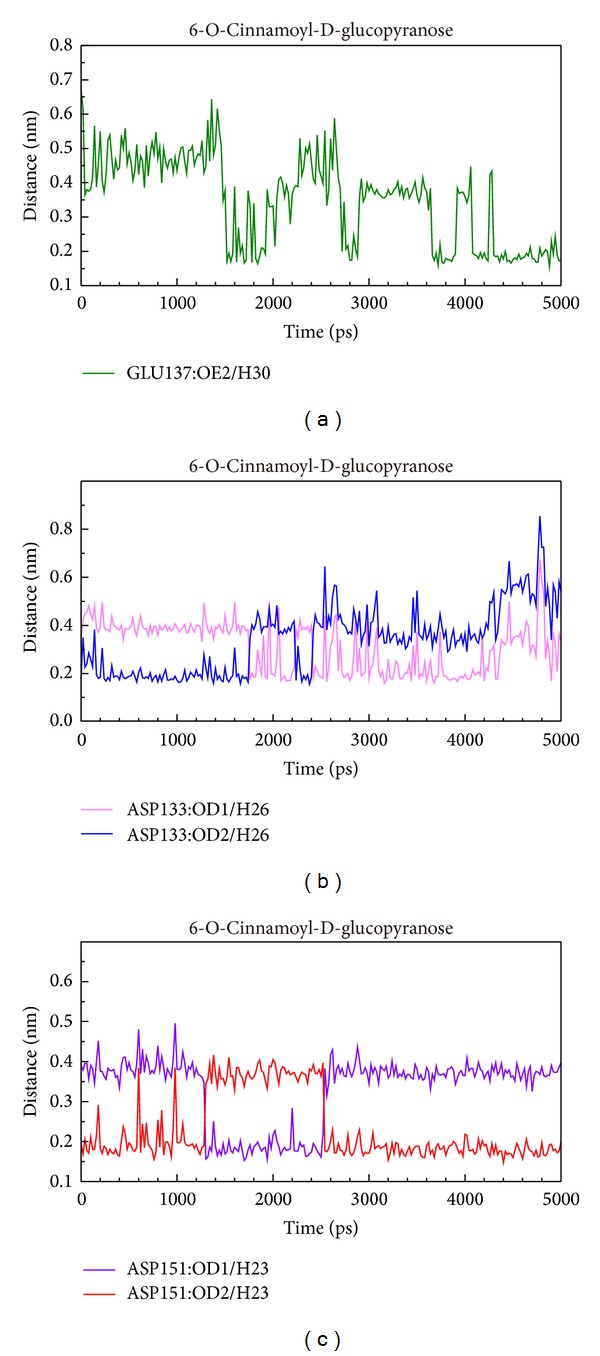
Distance of hydrogen bonds between 6-O-cinnamoyl-D-glucopyranose and essential amino acids of AMPK.
We showed distance of hydrogen bonds between AMP and essential amino acids of AMPK. The O4 of AMP formed H-bond with Gly19 at early stage of MD, and O20 formed H-bond with Phe152 at all stages of MD. H30 formed H-bond with Glu94 at early stage of MD. O4 formed H-bond with Val90 at early stage of MD, and N14 formed H-bond with Val90 at late stage of MD. H36 formed H-bond with OD1 of Asp151 at early and late stage of MD and formed H-bond with OD2 of Asp151 at middle stage of MD (Figure 14).
Figure 14.
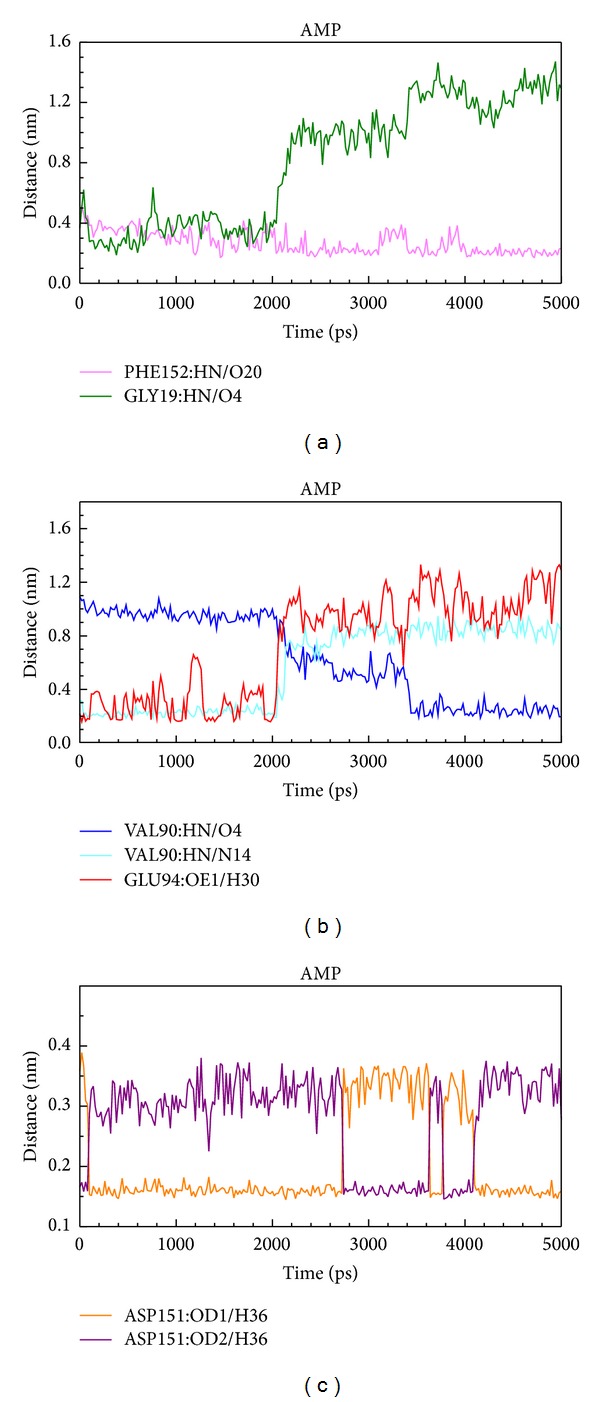
Distance of hydrogen bonds between AMP and essential amino acids of AMPK.
3.5. Ligand Pathway
3D simulation of ligand pathway bound with AMPK helped us understand the process of combination. Eugenyl_beta-D-glucopyranoside, 6-O-cinnamoyl-D-glucopyranose, and AMP had different pathways bound with AMPK protein (Figure 15).
Figure 15.
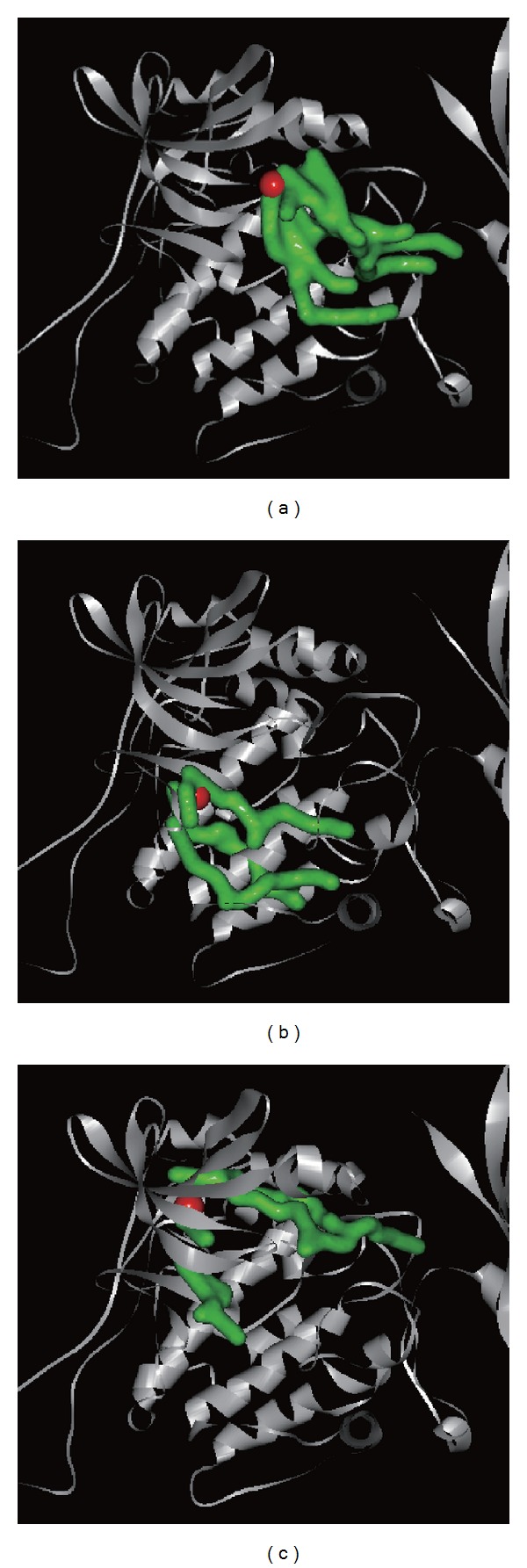
3D simulation of ligand pathway for (a) eugenyl_beta-D-glucopyranoside, (b) 6-O-cinnamoyl-D-glucopyranose, and (c) AMP bound with AMPK protein.
4. Discussion
4.1. Compound Database
The TCM small compounds for this study were from the world's largest TCM Database@Taiwan (http://tcm.cmu.edu.tw/). The TCM Database contained 453 kinds of herb plants, animals, and minerals. They consist of more than 20000 pure compounds. The TCM Database is a useful and precious treasury for exploring the mystery of traditional Chinese medicine.
4.2. Homology Modeling
Because the catalytic α subunit is the most important for all the three subunits of AMPK, we selected human AMPK (Q13131, α subunit) and rat templates (2Y94, α subunit) for homology modeling. The high percentage of identity and similarity of sequence alignment between Q13131 and 2Y94 indicated that the sequence alignment was reliable. The high percentage of residues in the favored and allowed area indicated that the AMPK-modeled structure was reasonable. The Verify scores of most residues were positive which indicated that the AMPK-modeled structure was reliable. We estimated that the negative values from residue 290 to residue 360 might be attributed to loss of reasonable structure from residue 311 to residue 341 illustrated in Figure 1.
4.3. Structure-Based Virtual Screening
Whether absorption level of ADMET, Dock scores, PLP1, PLP2 or PMF, all the top 7 TCM compounds were better than the control (AMP). Absorption level of ADMET from 0 to 3 means good to very low absorption. We selected the first 2 compounds, eugenyl_beta-D-glucopyranoside and 6-O-cinnamoyl-D-glucopyranose, as candidates due to their good absorption level. Both eugenyl_beta-D-glucopyranoside and 6-O-cinnamoyl-D-glucopyranose formed H-bond with Asp151. Both eugenyl_beta-D-glucopyranoside and AMP formed H-bond with Glu94. The main residues bound for the candidates and the control were located from Val18 to Asp151. The results mean that loss of reasonable structure from residue 311 to residue 341 did not disturb us for adopting the AMPK-modeled structure.
4.4. Molecular Dynamics (MD) Simulation
By integrating the figures of RMSD, Gyrate, and SAS, individual structure of the ligand and the protein underwent certain extent change during MD. This finding was essential for explanation of AMPK activation. Activation of AMPK followed its conformational change. Further analysis of RMSD, eugenyl_beta-D-glucopyranoside, 6-O-cinnamoyl-D-glucopyranose, and the control (AMP) had first larger deviation at 1600 ps, 1450 ps, and 2000 ps, which means that all the compounds could induce conformational change of AMPK. By further analysis of Gyrate and SAS, 6-O-cinnamoyl-D-glucopyranose had the largest ligand Gyrate and SAS values, but eugenyl_beta-D-glucopyranoside had the largest protein Gyrate and SAS values. We speculated that both the candidates could induce different kinds of conformational change of AMPK protein.
All the compounds had larger fluctuations at the range between residue 320 and residue 400 in RMSF. The larger fluctuations were not reliable due to loss of reasonable structure from residue 311 to residue 341. Based on the similar graph in RMSF and total energy, we could speculate that eugenyl_beta-D-glucopyranoside, 6-O-cinnamoyl-D-glucopyranose, and the control (AMP) had the same ability to activate AMPK. Our hypothesis was further approved by DSSP and smallest distance matrices. The structure of AMPK had similar change when it complexed with the 2 candidates and the control. Eugenyl_beta-D-glucopyranoside, 6-O-cinnamoyl-D-glucopyranose, and the control had the ability to induce AMPK conformational change.
According to occupancy of H-bond between eugenyl_beta-D-glucopyranoside, 6-O-cinnamoyl-D-glucopyranose, and AMP with AMPK protein, we could conclude that Asp151 was key residue for top 2 TCM candidates and the control bound with AMPK. By further analysis of distance of H-bonds between eugenyl_beta-D-glucopyranoside and essential amino acids of AMPK, the compound formed H-bond with Asp151 only at early stage of MD. At late stage of MD, the compound formed H-bond with Gly22 instead. By further analysis of distance of H-bonds between 6-O-cinnamoyl-D-glucopyranose and essential amino acids of AMPK, the compound formed H-bond with Asp151 at all stages of MD. By further analysis of distance of H-bonds between AMP and essential amino acids of AMPK, the compound formed H-bond with Asp151 at all stages of MD. Interestingly, H36 of eugenyl_beta-D-glucopyranoside formed H-bond with OD1 and OD2 of ASP151; H36 of AMP also formed H-bond with OD1 and OD2 of ASP151, but their figure patterns were quite different. However, H23 of 6-O-cinnamoyl-D-glucopyranose formed H-bond with OD1 and OD2 of ASP151 and the figure pattern was similar to that of AMP. We could confirm that all the candidates and the control formed stable complexes with AMPK. In addition, all the candidates and the control induced conformational change of AMPK due to changing position of H-bonds.
5. Conclusion
AMPK was a hot issue in the past decade because it plays an important role in sensing cellular energy or nutrient status. AMPK can regularize cell growth by the way of pseudostarvation leading to autophagy. Thus AMPK has become a great target for drug design of tumor inhibition. Activation of AMPK followed its conformational change. We tried to find potential compounds that could bind to AMPK by virtual screening of the world's largest TCM Database (http://tcm.cmu.edu.tw/) and had the ability to activate the function of AMPK. We selected eugenyl_beta-D-glucopyranoside and 6-O-cinnamoyl-D-glucopyranose as candidates for further investigation. Through the methods of MD simulation consisting of RMSD, Gyrate, SAS, RMSF, total energy, DSSP, smallest distance matrices, occupancy and distance of H-bonds, and ligand pathway, we could conclude that the candidates and the control (AMP) formed stable complexes with AMPK. Asp151 was key residue for the candidates and the control bound with AMPK. These compounds also had the ability to induce AMPK conformational change. Thus eugenyl_beta-D-glucopyranoside and 6-O-cinnamoyl-D-glucopyranose had the qualification to be AMPK agonist.
Acknowledgments
This research was supported by Grants from the National Science Council of Taiwan (nos. NSC102-2325-B039-001 and NSC102-2221-E-468-027-), China Medical University Hospital (DMR-103-001, DMR-103-058, and DMR-103-096), and Asia University (ASIA101-CMU-2, 102-Asia-07). This research was also supported in part by Taiwan Department of Health Cancer Research Center of Excellence (DOH102-TD-C-111-005) and Taiwan Department of Health Clinical Trial and Research Center of Excellence (MOHW103-TD-B-111-03).
Conflict of Interests
The authors declare that they have no conflict of interests regarding the publication of this paper.
References
- 1.Zhang YL, Guo H, Zhang CS, et al. AMP as a low-energy charge signal autonomously initiates assembly of AXIN-AMPK-LKB1 complex for AMPK activation. Cell Metabolism. 2013;18(4):546–555. doi: 10.1016/j.cmet.2013.09.005. [DOI] [PubMed] [Google Scholar]
- 2.Chen L, Xin FJ, Wang J, et al. Conserved regulatory elements in AMPK. Nature. 2013;498(7453):E8–E10. doi: 10.1038/nature12189. [DOI] [PubMed] [Google Scholar]
- 3.Hardie DG, Ross FA, Hawley SA. AMPK: a nutrient and energy sensor that maintains energy homeostasis. Nature Reviews Molecular Cell Biology. 2012;13(4):251–262. doi: 10.1038/nrm3311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cantó C, Gerhart-Hines Z, Feige JN, et al. AMPK regulates energy expenditure by modulating NAD + metabolism and SIRT1 activity. Nature. 2009;458(7241):1056–1060. doi: 10.1038/nature07813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carling D, Mayer FV, Sanders MJ, Gamblin SJ. AMP-activated protein kinase: nature’s energy sensor. Nature Chemical Biology. 2011;7(8):512–518. doi: 10.1038/nchembio.610. [DOI] [PubMed] [Google Scholar]
- 6.Cantó C, Jiang LQ, Deshmukh AS, et al. Interdependence of AMPK and SIRT1 for metabolic adaptation to fasting and exercise in skeletal muscle. Cell Metabolism. 2010;11(3):213–219. doi: 10.1016/j.cmet.2010.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hardie DG. AMP-activated/SNF1 protein kinases: conserved guardians of cellular energy. Nature Reviews Molecular Cell Biology. 2007;8(10):774–785. doi: 10.1038/nrm2249. [DOI] [PubMed] [Google Scholar]
- 8.Bland ML, Birnbaum MJ. Cell biology. ADaPting to energetic stress. Science. 2011;332(6036):1387–1388. doi: 10.1126/science.1208444. [DOI] [PubMed] [Google Scholar]
- 9.Hardie DG. Signal transduction: how cells sense energy. Nature. 2011;472(7342):176–177. doi: 10.1038/472176a. [DOI] [PubMed] [Google Scholar]
- 10.Yamauchi T, Kamon J, Minokoshi Y, et al. Adiponectin stimulates glucose utilization and fatty-acid oxidation by activating AMP-activated protein kinase. Nature Medicine. 2002;8(11):1288–1295. doi: 10.1038/nm788. [DOI] [PubMed] [Google Scholar]
- 11.Kubota N, Yano W, Kubota T, et al. Adiponectin stimulates AMP-activated protein kinase in the hypothalamus and increases food intake. Cell Metabolism. 2007;6(1):55–68. doi: 10.1016/j.cmet.2007.06.003. [DOI] [PubMed] [Google Scholar]
- 12.Minokoshi Y, Alquier T, Furukawa H, et al. AMP-kinase regulates food intake by responding to hormonal and nutrient signals in the hypothalamus. Nature. 2004;428(6982):569–574. doi: 10.1038/nature02440. [DOI] [PubMed] [Google Scholar]
- 13.Shibata R, Sato K, Pimentel DR, et al. Adiponectin protects against myocardial ischemia-reperfusion injury through AMPK- and COX-2-dependent mechanisms. Nature Medicine. 2005;11(10):1096–1103. doi: 10.1038/nm1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miller EJ, Li J, Leng L, et al. Macrophage migration inhibitory factor stimulates AMP-activated protein kinase in the ischaemic heart. Nature. 2008;451(7178):578–582. doi: 10.1038/nature06504. [DOI] [PubMed] [Google Scholar]
- 15.Minokoshi Y, Kim Y-B, Peroni OD, et al. Leptin stimulates fatty-acid oxidation by activating AMP-activated protein kinase. Nature. 2002;415(6869):339–343. doi: 10.1038/415339a. [DOI] [PubMed] [Google Scholar]
- 16.López M, Varela L, Vázquez MJ, et al. Hypothalamic AMPK and fatty acid metabolism mediate thyroid regulation of energy balance. Nature Medicine. 2010;16(9):1001–1008. doi: 10.1038/nm.2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xiao B, Heath R, Saiu P, et al. Structural basis for AMP binding to mammalian AMP-activated protein kinase. Nature. 2007;449(7161):496–500. doi: 10.1038/nature06161. [DOI] [PubMed] [Google Scholar]
- 18.Townley R, Shapiro L. Crystal structures of the adenylate sensor from fission yeast AMP-activated protein kinase. Science. 2007;315(5819):1726–1729. doi: 10.1126/science.1137503. [DOI] [PubMed] [Google Scholar]
- 19.Amodeo GA, Rudolph MJ, Tong L. Crystal structure of the heterotrimer core of Saccharomyces cerevisiae AMPK homologue SNF1. Nature. 2007;449(7161):492–495. doi: 10.1038/nature06127. [DOI] [PubMed] [Google Scholar]
- 20.Oakhill JS, Steel R, Chen Z-P, et al. AMPK is a direct adenylate charge-regulated protein kinase. Science. 2011;332(6036):1433–1435. doi: 10.1126/science.1200094. [DOI] [PubMed] [Google Scholar]
- 21.Hawley SA, Ross FA, Chevtzoff C, et al. Use of cells expressing γ subunit variants to identify diverse mechanisms of AMPK activation. Cell Metabolism. 2010;11(6):554–565. doi: 10.1016/j.cmet.2010.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bungard D, Fuerth BJ, Zeng P-Y, et al. Signaling kinase AMPK activates stress-promoted transcription via histone H2B phosphorylation. Science. 2010;329(5996):1201–1205. doi: 10.1126/science.1191241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Forcet C, Billaud M. Dialogue between LKB1 and AMPK: a hot topic at the cellular pole. Science’s STKE. 2007;2007(404):p. pe51. doi: 10.1126/stke.4042007pe51. [DOI] [PubMed] [Google Scholar]
- 24.McBride A, Ghilagaber S, Nikolaev A, Hardie DG. The glycogen-binding domain on the AMPK beta subunit allows the kinase to act as a glycogen sensor. Cell Metabolism. 2009;9(1):23–34. doi: 10.1016/j.cmet.2008.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xiao B, Sanders MJ, Underwood E, et al. Structure of mammalian AMPK and its regulation by ADP. Nature. 2011;472(7342):230–233. doi: 10.1038/nature09932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Banerjee RR, Rangwala SM, Shapiro JS, et al. Regulation of fasted blood glucose by resistin. Science. 2004;303(5661):1195–1198. doi: 10.1126/science.1092341. [DOI] [PubMed] [Google Scholar]
- 27.Watt MJ, Dzamko N, Thomas WG, et al. CNTF reverses obesity-induced insulin resistance by activating skeletal muscle AMPK. Nature Medicine. 2006;12(5):541–548. doi: 10.1038/nm1383. [DOI] [PubMed] [Google Scholar]
- 28.Cool B, Zinker B, Chiou W, et al. Identification and characterization of a small molecule AMPK activator that treats key components of type 2 diabetes and the metabolic syndrome. Cell Metabolism. 2006;3(6):403–416. doi: 10.1016/j.cmet.2006.05.005. [DOI] [PubMed] [Google Scholar]
- 29.Zhang BB, Zhou G, Li C. AMPK: an emerging drug target for diabetes and the metabolic syndrome. Cell Metabolism. 2009;9(5):407–416. doi: 10.1016/j.cmet.2009.03.012. [DOI] [PubMed] [Google Scholar]
- 30.Shaw RJ, Lamia KA, Vasquez D, et al. Medicine: the kinase LKB1 mediates glucose homeostasis in liver and therapeutic effects of metformin. Science. 2005;310(5754):1642–1646. doi: 10.1126/science.1120781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yee SW, Chen L, Giacomini KM. The role of ATM in response to metformin treatment and activation of AMPK. Nature Genetics. 2012;44(4):359–360. doi: 10.1038/ng.2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.O'Neill LA, Hardie DG. Metabolism of inflammation limited by AMPK and pseudo-starvation. Nature. 2013;493(7432):346–355. doi: 10.1038/nature11862. [DOI] [PubMed] [Google Scholar]
- 33.Inoki K, Ouyang H, Zhu T, et al. TSC2 integrates Wnt and energy signals via a coordinated phosphorylation by AMPK and GSK3 to regulate cell growth. Cell. 2006;126(5):955–968. doi: 10.1016/j.cell.2006.06.055. [DOI] [PubMed] [Google Scholar]
- 34.Liang J, Shao SH, Xu Z-X, et al. The energy sensing LKB1-AMPK pathway regulates p27kip1 phosphorylation mediating the decision to enter autophagy or apoptosis. Nature Cell Biology. 2007;9(2):218–224. doi: 10.1038/ncb1537. [DOI] [PubMed] [Google Scholar]
- 35.Shackelford DB, Shaw RJ. The LKB1-AMPK pathway: metabolism and growth control in tumour suppression. Nature Reviews Cancer. 2009;9(8):563–575. doi: 10.1038/nrc2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Egan DF, Shackelford DB, Mihaylova MM, et al. Phosphorylation of ULK1 (hATG1) by AMP-activated protein kinase connects energy sensing to mitophagy. Science. 2011;331(6016):456–461. doi: 10.1126/science.1196371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim J, Kundu M, Viollet B, Guan K-L. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nature Cell Biology. 2011;13(2):132–141. doi: 10.1038/ncb2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Levitt M, Warshel A. Computer simulation of protein folding. Nature. 1975;253(5494):694–698. doi: 10.1038/253694a0. [DOI] [PubMed] [Google Scholar]
- 39.Sotomayor M, Schulten K. Single-molecule experiments in vitro and in silico. Science. 2007;316(5828):1144–1148. doi: 10.1126/science.1137591. [DOI] [PubMed] [Google Scholar]
- 40.von Mering C, Krause R, Snel B, et al. Comparative assessment of large-scale data sets of protein-protein interactions. Nature. 2002;417(6887):399–403. doi: 10.1038/nature750. [DOI] [PubMed] [Google Scholar]
- 41.Palsson B. The challenges of in silico biology. Nature Biotechnology. 2000;18(11):1147–1150. doi: 10.1038/81125. [DOI] [PubMed] [Google Scholar]
- 42.Kitano H. Computational systems biology. Nature. 2002;420(6912):206–210. doi: 10.1038/nature01254. [DOI] [PubMed] [Google Scholar]
- 43.Palsson B. In silico biology through ‘omics’. Nature Biotechnology. 2002;20(7):649–650. doi: 10.1038/nbt0702-649. [DOI] [PubMed] [Google Scholar]
- 44.van de Waterbeemd H, Gifford E. ADMET in silico modelling: towards prediction paradise? Nature Reviews Drug Discovery. 2003;2(3):192–204. doi: 10.1038/nrd1032. [DOI] [PubMed] [Google Scholar]
- 45.Di Ventura B, Lemerle C, Michalodimitrakis K, Serrano L. From in vivo to in silico biology and back. Nature. 2006;443(7111):527–533. doi: 10.1038/nature05127. [DOI] [PubMed] [Google Scholar]
- 46.Pellecchia M. Fragment-based drug discovery takes a virtual turn. Nature Chemical Biology. 2009;5(5):274–275. doi: 10.1038/nchembio0509-274. [DOI] [PubMed] [Google Scholar]
- 47.Liao G, Wang J, Guo J, et al. In silico genetics: identification of a functional elements regulating HZ-Eα gene expression. Science. 2004;306(5696):690–695. doi: 10.1126/science.1100636. [DOI] [PubMed] [Google Scholar]
- 48.Naylor E, Arredouani A, Vasudevan SR, et al. Identification of a chemical probe for NAADP by virtual screening. Nature Chemical Biology. 2009;5(4):220–226. doi: 10.1038/nchembio.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shoichet BK. Virtual screening of chemical libraries. Nature. 2004;432(7019):862–865. doi: 10.1038/nature03197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bajorath J. Integration of virtual and high-throughput screening. Nature Reviews Drug Discovery. 2002;1(11):882–894. doi: 10.1038/nrd941. [DOI] [PubMed] [Google Scholar]
- 51.Duan Y, Kollman PA. Pathways to a protein folding intermediate observed in a 1-microsecond simulation in aqueous solution. Science. 1998;282(5389):740–744. doi: 10.1126/science.282.5389.740. [DOI] [PubMed] [Google Scholar]
- 52.Karplus M, Petsko GA. Molecular dynamics simulations in biology. Nature. 1990;347(6294):631–639. doi: 10.1038/347631a0. [DOI] [PubMed] [Google Scholar]
- 53.Karplus M, McCammon JA. Molecular dynamics simulations of biomolecules. Nature Structural Biology. 2002;9(9):646–652. doi: 10.1038/nsb0902-646. [DOI] [PubMed] [Google Scholar]
- 54.Liao WL, Tsai FJ. Personalized medicine: a paradigm shift in healthcare. BioMedicine. 2013;3(2):66–72. [Google Scholar]
- 55.Mahamuni SP, Khose RD, Menaa F, Badole SL. Therapeutic approaches to drug targets in hyperlipidemia. BioMedicine. 2012;2(4):137–146. [Google Scholar]
- 56.Ho TY, Lo HY, Li CC, et al. In vitro and in vivo bioluminescent imaging to evaluate anti-Escherichia coli activity of Galla Chinensis . BioMedicine. 2013;3(4):160–166. [Google Scholar]
- 57.Wang C-H, Lin W-D, Tsai F-J. Craniofacial dysmorphism, what is your diagnosis? BioMedicine. 2012;2(2):49–50. [Google Scholar]
- 58.Lam CY, Gritz ER. Incorporating behavioral research to examine the relationship between betel quid chewing and oral cancer in Taiwan. BioMedicine. 2012;2(4):160–166. [Google Scholar]
- 59.Su PY, Liu SJ, Chen YH, et al. Increased IL-8 and IL-1b in the bile of acute cholecystitis patients. BioMedicine. 2013;3(181):p. e185. [Google Scholar]
- 60.Mihaylova MM, Shaw RJ. The AMPK signalling pathway coordinates cell growth, autophagy and metabolism. Nature Cell Biology. 2011;13(9):1016–1023. doi: 10.1038/ncb2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Xiao B, Sanders MJ, Carmena D, et al. Structural basis of AMPK regulation by small molecule activators. Nature Communications. 2013;4:p. 3017. doi: 10.1038/ncomms4017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tsai T-Y, Chang K-W, Chen CY-C. IScreen: world’s first cloud-computing web server for virtual screening and de novo drug design based on TCM database@Taiwan. Journal of Computer-Aided Molecular Design. 2011;25(6):525–531. doi: 10.1007/s10822-011-9438-9. [DOI] [PubMed] [Google Scholar]
- 63.Yang S-C, Chang S-S, Chen H-Y, Chen CY-C. Identification of potent EGFR inhibitors from TCM Database@Taiwan. PLoS Computational Biology. 2011;7(10) doi: 10.1371/journal.pcbi.1002189.e1002189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chen K-C, Sun M-F, Yang S-C, et al. Investigation into Potent Inflammation Inhibitors from Traditional Chinese Medicine. Chemical Biology and Drug Design. 2011;78(4):679–688. doi: 10.1111/j.1747-0285.2011.01202.x. [DOI] [PubMed] [Google Scholar]
- 65.Chang T-T, Sun M-F, Chen H-Y, et al. Screening from the world’s largest TCM database against H1N1 virus. Journal of Biomolecular Structure and Dynamics. 2011;28(5):773–786. doi: 10.1080/07391102.2011.10508605. [DOI] [PubMed] [Google Scholar]
- 66.Chang S-S, Huang H-J, Chen CY-C. Two birds with one stone? Possible dual-targeting H1N1 inhibitors from traditional Chinese medicine. PLoS Computational Biology. 2011;7(12) doi: 10.1371/journal.pcbi.1002315.e1002315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tou WI, Chang SS, Lee CC, Chen CY. Drug design for neuropathic pain regulation from traditional Chinese medicine. Scientific Reports. 2013;3:p. 844. doi: 10.1038/srep00844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chen KC, Chang SS, Huang HJ, et al. Three-in-one agonists for PPAR-alpha, PPAR-gamma, and PPAR-delta from traditional Chinese medicine. Journal of Biomolecular Structure and Dynamics. 2012;30(6):662–683. doi: 10.1080/07391102.2012.689699. [DOI] [PubMed] [Google Scholar]
- 69.Chen CY-C. TCM Database@Taiwan: the world’s largest traditional Chinese medicine database for drug screening In Silico. PLoS ONE. 2011;6(1) doi: 10.1371/journal.pone.0015939.e15939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Brooks BR, Brooks CL, III, Mackerell AD, Jr., et al. CHARMM: the biomolecular simulation program. Journal of Computational Chemistry. 2009;30(10):1545–1614. doi: 10.1002/jcc.21287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Khan MTH. Predictions of the ADMET properties of candidate drug molecules utilizing different QSAR/QSPR modelling approaches. Current Drug Metabolism. 2010;11(4):285–295. doi: 10.2174/138920010791514306. [DOI] [PubMed] [Google Scholar]
- 72.Chen CY. A novel integrated framework and improved methodology of computer-aided drug design. Current Topics in Medicinal Chemistry. 2013;13(9):965–988. doi: 10.2174/1568026611313090002. [DOI] [PubMed] [Google Scholar]
- 73.Chen CY-C. Weighted equation and rules—a novel concept for evaluating protein-ligand interaction. Journal of Biomolecular Structure and Dynamics. 2009;27(3):271–282. doi: 10.1080/07391102.2009.10507315. [DOI] [PubMed] [Google Scholar]
- 74.Chen CY, Tou WI. How to design a drug for the disordered proteins? Drug Discovery Today. 2013;18(19-20):910–915. doi: 10.1016/j.drudis.2013.04.008. [DOI] [PubMed] [Google Scholar]
- 75.Tou WI, Chen CY. May disordered protein cause serious drug side effect? Drug Discovery Today. 2014;19(4):367–372. doi: 10.1016/j.drudis.2013.10.020. [DOI] [PubMed] [Google Scholar]


