Abstract
Entosis, a cell-in-cell process, has been implicated in the formation of aneuploidy associated with an aberrant cell division control. Microtubule plus-end-tracking protein TIP150 facilitates the loading of MCAK onto the microtubule plus ends and orchestrates microtubule plus-end dynamics during cell division. Here we show that TIP150 cooperates with MCAK to govern entosis via a regulatory circuitry that involves Aurora A-mediated phosphorylation of MCAK. Our biochemical analyses show that MCAK forms an intra-molecular association, which is essential for TIP150 binding. Interestingly, Aurora A-mediated phosphorylation of MCAK modulates its intra-molecular association, which perturbs the MCAK–TIP150 interaction in vitro and inhibits entosis in vivo. To probe if MCAK–TIP150 interaction regulates microtubule plasticity to affect the mechanical properties of cells during entosis, we used an optical trap to measure the mechanical rigidity of live MCF7 cells. We find that the MCAK cooperates with TIP150 to promote microtubule dynamics and modulate the mechanical rigidity of the cells during entosis. Our results show that a dynamic interaction of MCAK–TIP150 orchestrated by Aurora A-mediated phosphorylation governs entosis via regulating microtubule plus-end dynamics and cell rigidity. These data reveal a previously unknown mechanism of Aurora A regulation in the control of microtubule plasticity during cell-in-cell processes.
Keywords: Aurora A, TIP150, MCAK, entosis, microtubule plus-end, kinesin
Introduction
Entosis or the process of forming homotypic, cell-in-cell structures (Overholtzer and Brugge, 2008), has been observed in many types of tumors, including carcinomas (Abodief et al., 2006), sarcomas (Hong, 1981), angiomyolipomas (Yokoo et al., 2000), and melanomas (Fais, 2007). Several lines of evidence demonstrate the role of cell-in-cell invasion in tumor progression (Lu and Kang, 2009). Different from the well-characterized phagocytosis and autophagy, which normally target dead, dying, or pathogenic cells and are driven by phagocytic hosts (Florey and Overholtzer, 2012; Underhill and Goodridge, 2012), entosis is considered to occur when target cells invade non-phagocytic hosts. These cells are usually viable after the internalization (Overholtzer and Brugge, 2008).
Although the observation of entosis has been documented for many years, the underlying molecular mechanisms are still poorly understood. Previous studies demonstrated that entosis depends on the calcium-dependent cell adhesion protein, E-cadherin (Overholtzer et al., 2007), which transduces a contractile force between the actomyosin cytoskeleton and the plasma membrane (Chu et al., 2004; Borghi et al., 2012). Myosin is activated by Rho GTPase through its effector kinases, ROCK1 and ROCK2 (Matsumura, 2005; Parsons et al., 2010; Rath and Olson, 2012). Entosis is an interaction of paired cells, whereas the Rho-ROCK-myosin pathway is required specifically by ingested cells (Overholtzer et al., 2007), indicating that these cells invade their hosts by using contractile force. Our early study indicates the importance of ezrin in regulating dynamic cytoskeleton-membrane interactions during entosis (Xia et al., 2008; Wang et al., 2009). Recent studies show that entosis is also regulated by a Par3-Lgl antagonism via the Rho-ROCK-myosin pathway (Wan et al., 2012). These studies primarily focused on the regulation of entosis by actin cytoskeleton dynamics. However, the role of the microtubule-based cytoskeleton in entosis has yet to be defined.
It has been shown that perturbation of the contractile ring formation by entosis led to generation of binucleate cells and production of aneuploid cell lineages (Krajcovic et al., 2011; Krajcovic and Overholtzer, 2012). Moreover, chromosomal instability and aneuploidy have been considered as hallmarks of cancer, even though the cause and consequence of aneuploidy have remained elusive (Gordon et al., 2012; Pfau and Amon, 2012).
Mounting evidence suggests that BTAK/STK15/Aurora A kinase contributes to aneuploidy (Giet et al., 2005; Vader and Lens, 2008). In fact, Aurora A is amplified in many types of solid tumor (Zhou et al., 1998; Fraizer et al., 2004; Lassus et al., 2011). Overexpression of Aurora A promotes abnormal centrosome amplification and potentiates H-Ras-mediated oncogenic transformation (Zhou et al., 1998; Tatsuka et al., 2005). Conditional overexpression of Aurora A in the mammary epithelium of transgenic mice resulted in formation of binucleated cells (Zhang et al., 2004). Recent study showed that entosis in anaphase resulted in binucleation of breast cancer cells (Krajcovic et al., 2011). However, whether Aurora A involves in regulation of entosis has not been probed.
We have recently identified a unique microtubule (MT) plus-end-tracking protein, TIP150, which interacts with and targets mitotic centromere-associated kinesin (MCAK) at the plus-ends (Jiang et al., 2009), by which TIP150 regulates kinetochore microtubule plus-end dynamics in cell division (Ward et al., 2013). In the present report, we show that TIP150 and MCAK-dependent MT dynamics are required for entosis and that the interaction of TIP150 and MCAK is modulated by Aurora A phosphorylation. Using dual-color labeled entosis and optical trap measurements, we demonstrate that the orchestration of entosis by TIP150, MCAK, and Aurora A is, at least partially dependent on their regulation of cell rigidity. Our results define a previously unknown mechanism of Aurora A regulation of microtubule dynamics in entosis.
Results
TIP150-dependent MT dynamics is required for entosis
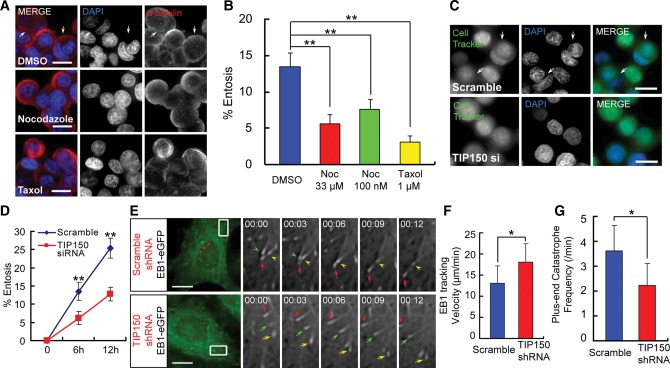
Our early study revealed the role of actin-based cytoskeleton dynamics in cell-in-cell process (Wang et al., 2009). Microtubule, as a major cytoskeleton network, is a fibrous polymer that determines cellular plasticity and dynamics such as cell division and migration. To determine if perturbation of MT dynamics interrupts the process of entosis, MCF7 cells were treated with DMSO (control), taxol (to stabilize MTs), or nocodazole (to depolymerize MTs). As shown in Figure 1A and B, the rate of entosis decreased in cells treated with either taxol or nocodazole, validating that entosis is disrupted by perturbation of MT dynamics. Given our recent discovery of functional interaction of MCAK–TIP150 in governing microtubule plus-end dynamics (Jiang et al., 2009; Ward et al., 2013), we sought to examine the roles and potential regulation of TIP150 in cell-in-cell process using siRNA-mediated knockdown. As shown in Figure 1C, MCF7 cells transfected with scramble siRNA were effectively internalized, which is consistent with previous observation (Overholtzer and Brugge, 2008). Typically, siRNA-based knockdown results in an 8-fold reduction of TIP150 protein level in MCF7 cells judged by western blotting analyses (Supplementary Figure S1A), which yields an ∼50% inhibition in cell-in-cell process (Figure 1C and D).
Figure 1.
Microtubule plus-end-tracking protein TIP150 is required for cell-in-cell process. (A) MCF7 cells were fixed and stained with DAPI and α-tubulin antibody after 6 h of suspension culture in the presence of DMSO (control), 33 μM or 100 nM nocodazole, or 1 μM taxol. Arrows indicate internalized cells. The scale bars are 15 μm. (B) Quantification of internalizing cells after 6 h suspension culture treated with DMSO, nocodazole, or taxol. The rate of entosis is decreased in cells treated with either nocodazole or taxol. Error bars indicate SD for three independent experiments in which at least 600 cells were quantified for each experiment. **P < 0.01. (C) Images of MCF7 cells transfected with scramble or TIP150 siRNA. Cells were labeled with CellTracker Green and fixed after 6 h of suspension culture. Arrows indicate internalized cells. The scale bars are 15 μm. (D) Quantification of internalizing cells at 6 and 12 h. Knockdown of TIP150 reduces the rate of entosis of MCF7 cells. Data are means ± SD. Each point represents at least 600 cells analyzed over three independent experiments. **P < 0.01. (E) Time-lapse imaging of EB1-GFP-stable MCF7 cells transiently transfected with scramble or TIP150 shRNA. Locations of the shRNAs are indicated by co-expression with CENP-B-mCherry. The scale bars are 15 μm. Enlarged insets are shown in grayscale. Time in seconds is shown in the top left corner. Growing and pausing/shrinking MT plus-ends are indicated with arrows and arrowheads, respectively. The frequency of plus-ends catastrophe is decreased by TIP150 knockdown. (F and G) Histograms of MT plus-ends tracking velocity and catastrophe frequency in scramble or TIP150 shRNA-expressing cells (for each condition, at least 900 microtubule plus-ends in 30 cells from three independent experiments were analyzed). Error bars indicate SD. *P < 0.05.
Since TIP150 is an MT plus-end-tracking protein (Jiang et al., 2009; Xia et al., 2012; Ward et al., 2013), perturbation of entosis in TIP150-deficient cells suggests a critical role of MT dynamics during cell-in-cell process. To examine this possibility, an MCF7 cell line stably expressing EB1-GFP was generated to determine if MT dynamics was affected by TIP150 knockdown as EB1-GFP marks the polymerizing plus-ends of microtubules (Piehl et al., 2004). A TIP150 shRNA construct tagged with CENP-B1–167-mCherry allowed visualization of live cells positively transfected (Supplementary Figure S2A). In cells positively transfected with TIP150 shRNA, TIP150 protein was barely detected, indicating an efficient suppression of target protein (Supplementary Figure S2B). MT shrinkage and catastrophe were frequently observed in scramble-control cells but not in TIP150-deficient cells (Figure 1E). Quantitative analyses of 500 MT plus-ends in 40 cells demonstrate that suppression of TIP150 promotes the tracking velocity of EB1 comets but decreases the frequency of plus-end catastrophes (Figure 1F and G), confirming the essential role of TIP150 in regulating MT dynamics. Thus, we conclude that TIP150 is required for governing microtubule dynamics during cell-in-cell process.
TIP150 and MCAK cooperate to control MT dynamics and entosis
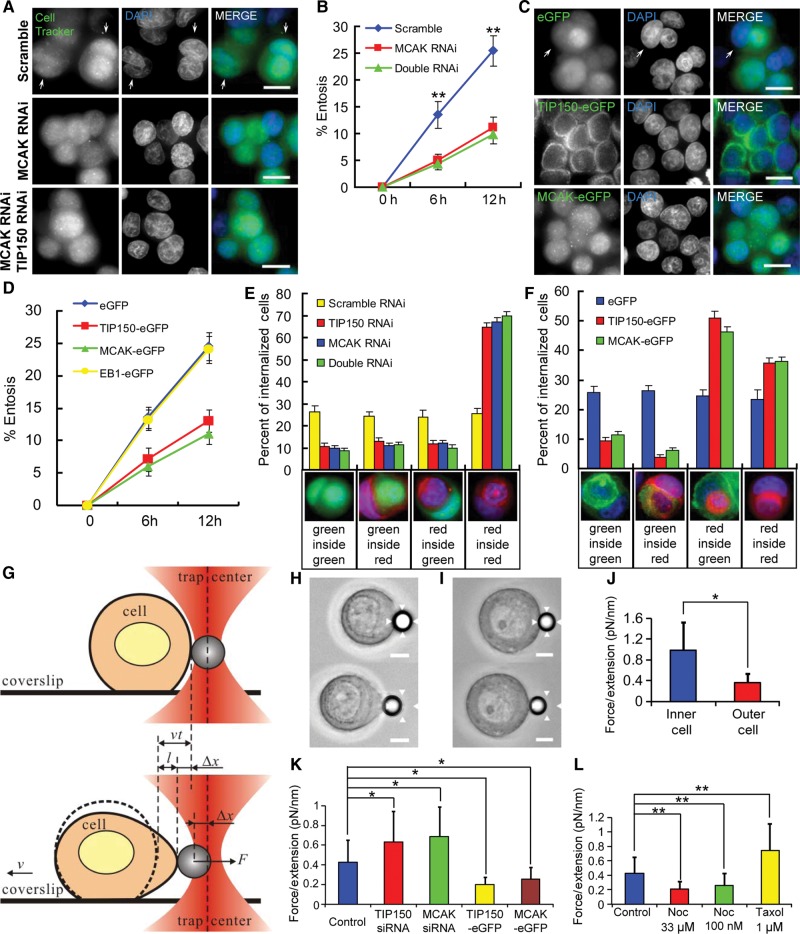
Our previous study demonstrated that TIP150 specifies the plus-end tracking of MCAK (Jiang et al., 2009), and MCAK is essential for MT depolymerization (Desai et al., 1999; Maney et al., 2001; Ohi et al., 2003). To test if MCAK participates in entosis, we carried out RNA interference to suppress the level of MCAK. As shown in Supplementary Figure S1A, the RNA interference markedly suppressed the MCAK protein level judged by western blotting analyses. For cells with depleted MCAK by RNA interference, the rate of entosis decreased by more than one half compared with that of control cells (Figure 2A and B), which exhibited a rate similar to that of TIP150-depleted cells (Figure 1B). Suppression of both TIP150 and MCAK exhibited no additive effect on the inhibition of entosis (Figure 2A and B), suggesting that these two proteins function in the same pathway.
Figure 2.
Interaction of TIP150 with MCAK is essential for entosis. (A) MCF7 cells transfected with scramble, MCAK, or MCAK and TIP150 siRNAs were labeled with CellTracker Green and fixed after 6 h of suspension culture. Arrows indicate internalized cells. The scale bars are 15 μm. (B) Quantification of internalizing cells transfected with scramble, MCAK, or MCAK and TIP150 siRNAs at 6 and 12 h. MCAK RNA interference has a similar but not a synergistic effect with TIP150 knockdown. Data are means ± SD. Each point represents at least 400 cells analyzed over three independent experiments. **P < 0.01. (C) Images of GFP, TIP150-GFP, or MCAK-GFP stable cells after 6 h of suspension culture. Arrows indicate internalized cells. The scale bars are 15 μm. (D) Quantification of cell-in-cell process as a function of time. Overexpression of TIP150/MCAK but not EB1 reduces the entosis rate. Data are means ± SD. Each point represents at least 400 cells analyzed over three independent experiments. **P < 0.01. (E) Percent of dual-color labeled internalizing cells after 6 h of suspension culture. Red-labeled control cells were mixed 1:1 with green-labeled cells transfected with scramble, MCAK, or MCAK and TIP150 siRNAs. Below the x-axis are examples of different color combinations. Data are means ± SD for three independent experiments in which at least 600 cells were quantified for each experiment. (F) Percent of dual-color labeled internalizing cells after 6 h of suspension culture. Red-labeled control cells were mixed 1:1 with cells stably expressing GFP, TIP150-GFP, or MCAK-GFP. Below the x-axis are examples of different color combinations. TIP150/MCAK overexpressing cells tend to be invaded by control cells. Data are means ± SD for three independent experiments in which at least 600 cells were quantified for each experiment. (G) Sketch of stretching a cell by optical tweezers. A handle bead adhering to a cell is trapped by the optical tweezer. When the coverslip is moving with a velocity v, the cell adhering to the coverslip will be stretched by trapping force F, which is induced by the bead's deviation Δx from the trap center. In a given time interval t, the cell (dash curve) moves a distance vt. The length of l = vt−Δx denotes the stretching extension under F. The dependence of F to l can denote cell rigidity. (H and I) Example of optical trap experiments shows a cell with low rigidity (H) deformed and a cell with high rigidity (I) difficult to be deformed. The bead adhered on the cell is trapped by the optical tweezer. The range of the optical tweezer was indicated with arrowhead. The scale bars are 5 μm. (J) Analysis of the dependence of F (trapping force) to l (stretching extension) shows inner cell has higher cell rigidity than outer cell in paired entosis cells. Data are means ± SD. *P < 0.05. See also Supplementary Figure S4A. (K) Analysis of the dependence of F (trapping force) to l (stretching extension) shows cells transfected with TIP150/MCAK siRNA have increased cell rigidity, while cells stably expressing TIP150/MCAK-eGFP have decreased cell rigidity. Data are means ± SD. *P < 0.05. See also Supplementary Figure S4B–E. (L) Analysis of the dependence of F (trapping force) to l (stretching extension) shows cells treated with paclitaxel have increased cell rigidity, while cells treated with nocodazole exhibit decreased cell rigidity. Data are means ± SD. **P < 0.01. See also Supplementary Figure S4F–H.
Since knockdown of TIP150 and MCAK perturbed entosis, we sought to examine if overexpression of TIP150 and MCAK exhibits any adverse effects on MCF7 cells. As shown in Figure 2C and D, up-regulation of either TIP150 or MCAK but not EB1 decreased the rate of entosis, consistent with the rate in TIP150 or MCAK knockdown cells, respectively. As entosis is an interaction and engagement of two cells, one of which is being internalized, dual-color labeled entosis experiments were performed to determine if there were differences between TIP150/MCAK up-regulated and down-regulated cells. Cells transfected with scramble siRNA, TIP150 siRNA, MCAK siRNA, or both TIP150 and MCAK siRNAs were labeled with CellTracker Green, and mixed 1:1 with untreated CellTracker Red-labeled control cells. Cells were fixed and analyzed after 6 h of suspension culture. The results showed that the internalization was equally distributed between cells transfected with green-labeled scramble siRNA and red-labeled control cells, whereas TIP150, MCAK, or double down-regulated cells were rarely internalized into or enwrapped by control cells (Figure 2E). Cells stably overexpressing TIP150- or MCAK-GFP were also rarely internalized into control red cells; these cells tended to be outer cells (Figure 2F). These interesting outcomes led us to speculate that entosis is regulated by MT dynamics and cell rigidity (Supplementary Figure S3A), the latter was demonstrated to be regulated by microtubule cytoskeleton (Pelling et al., 2007). Since MTs are hyper-stabilized in cells depleted TIP150 and MCAK, these cells have reduced MT dynamics but a high rigidity. Thus, they can neither invade nor be invaded by control cells (illustrated in Supplementary Figure S3B and C). In contrast, cells overexpressing TIP150 or MCAK had decreased MT dynamics, a low rigidity (Supplementary Figure S3D and E), and appeared to be more likely to be invaded by control cells.
To test this hypothesis, we measured cell rigidity with optical trap experiment. The principal of this experiment is illustrated as a model (Figure 2G). When a cell adhering to coverslip moves at a fixed velocity v, the movement of an optical tweezer trapped beads adhering to the cell will be hindered by a trapping force F induced by a displacement (Δx) from trap center. In a given time interval t, the cell moves a distance of vt, but the beads deviates only Δx from trap center. The stretching length can be denoted by l = vt−Δx. The cell rigidity is defined by the dependence relation of F to l (Figure 2G). MCF7 cells behave different rigidity could be accurately differentiated with our optical trap system (Figure 2H and I). To examine the rigidity of the paired entosis cells, we selected cells in ongoing but not yet completely entosis to prevent inaccurate measurement. As shown in Figure 2J and Supplementary Figure S4A, our measurement demonstrated that inner cells exhibit greater rigidity than that of outer cells. We also noticed cells transfected with TIP150 or MCAK siRNA had stronger rigidity, whereas cells stably expressing TIP150-eGFP or MCAK-eGFP had weaker rigidity (Figure 2K and Supplementary Figure S4B–E). These results support our hypothesis of cell rigidity regulation by TIP150 and MCAK. Furthermore, MCF7 cell rigidity decreased with nocodazole treatment while increased with paclitaxel treatment, indicating microtubule plasticity is a function of cell rigidity (Figure 2L and Supplementary Figure S4F–H).
In line with the optical trap experiments, MCF7 cells were transiently transfected to express GFP-tagged TIP150, MCAK, or other plus-end-tracking proteins, and were fixed and stained to visualize MT depolymerization activity. To our surprise, MT depolymerization occurred in cells overexpressing TIP150 or MCAK, but not EB1, CLASP2, or CLIP170 (Supplementary Figure S5A and B). Cells co-transfected with TIP150-GFP and MCAK siRNA exhibited regular MT stability evident by IF staining, indicating that MT depolymerization requires cooperation of TIP150 and MCAK.
TIP150 promotes the MT plus-end retention of MCAK which is negatively regulated by Aurora A
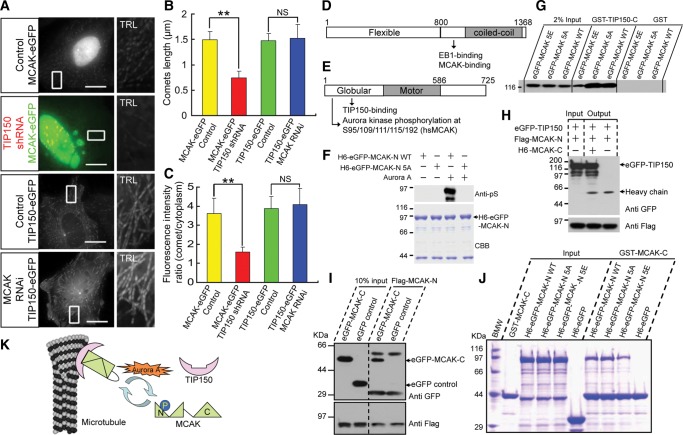
The molecular mechanism underlying the interaction between TIP150 and MCAK was then further examined. Down-regulation of TIP150 liberates the plus-end accumulation of MCAK, but depletion of MCAK did not perturb the plus-end-tracking property of TIP150 (Figure 3A). Both the plus-end comet length and the fluorescence intensity ratio of MCAK-GFP were decreased in TIP150-deficient cells (Figure 3B and C). Thus, we conclude that loading of MCAK to the MT plus-ends requires TIP150.
Figure 3.
Aurora A phosphorylation perturbs MCAK–TIP150 interaction. (A) Images of MCAK-GFP or TIP150-GFP stable cells transfected with or without TIP150 shRNA or MCAK siRNA. The scale bars are 15 μm. Enlarged insets are trails (TRL) of 31 frames with 3-sec intervals. (B and C) Histograms of MT plus-ends comet lengths, or fluorescence intensity ratios at the growing MT tips and in surrounding cytoplasm of indicated cells (for each condition, at least 900 microtubule plus-ends in 30 cells from three independent experiments were analyzed); error bars indicate SD, **P < 0.01; NS, P > 0.05. (D) Schematic diagram of TIP150 domains. Full-length TIP150 is divided into TIP150-N (1–800 aa) and TIP150-C (801–1368 aa). (E) Schematic diagram of MCAK domains. Full-length MCAK is divided into MCAK-N (1–586 aa) and MCAK-C (587–725 aa). The phosphorylation sites (S95/109/111/115/192, 5S for short) of Aurora kinase are located at the globular domain of MCAK. (F) Validation of MCAK 5S phosphorylation by Aurora A. Purified 6×His (H6)-GFP-MCAK-N and 5S mutant 5A (1 μg) were incubated with 50 μM ATP, with or without recombinant Aurora A kinase (50 ng). Anti-phosphoserine antibody blotting (upper); CBB staining (lower). (G) GST-TIP150-C and GST proteins were used as affinity matrices to absorb GFP-tagged wild-type MCAK and mutants. Binding activity was examined by anti-GFP blotting. TIP150-C bound to MCAK wild-type and the 5A mutant but less to the 5E mutant. (H) Beads with bound Flag-MCAK-N were used as affinity matrices to absorb GFP-TIP150 proteins from soluble 293 T cell lysates with or without purified H6-MCAK-C. Binding activity was analyzed by anti-GFP blotting (upper). Flag-MCAK-N was analyzed by anti-Flag blotting (lower). (I) Beads with bound Flag-MCAK-N were used as affinity matrices to absorb GFP-MCAK-C or GFP proteins from soluble 293T cell lysates. Binding activity was analyzed by anti-GFP blotting (upper). Flag-MCAK-N was analyzed by anti-Flag blotting (lower). (J) Binding activity assay for GST-MCAK-C to purified H6-GFP-MCAK-N WT/5A/5E. The input and bound proteins were analyzed by SDS–PAGE and CBB staining. Quantitatively, MCAK-C binds tighter to MCAK-N WT and 5A than to the 5E mutant (see also Supplementary Figure S6). (K) Molecular mechanism accounting for the MCAK–TIP150 interaction and MT plus-end tracking.
In frogs and in hamsters, MCAK is phosphorylated on its N-terminus by Aurora B kinase (Andrews et al., 2004; Lan et al., 2004; Ohi et al., 2004), but in interphase cells Aurora B is predominantly located in the nucleus (Mackay et al., 2010), making it less likely to be involved in MT regulation in entosis. In contrast, another Aurora kinase family member, Aurora A, which shares 70% identity with Aurora B in the catalytic domain is found to localize to the cytoplasm at interphase (Carmena et al., 2009), and is reported to phosphorylate MCAK to control ran-dependent spindle bipolarity at mitosis (Zhang et al., 2008). Thus, we sought to test if Aurora A regulates MCAK–TIP150 interaction through phosphorylation of MCAK.
To facilitate efficient recombinant protein production and purification, TIP150 and MCAK were truncated into TIP150-N (1–800 aa), TIP150-C (801–1368 aa), MCAK-N (1–586 aa), and MCAK-C (587–725 aa) with appropriate affinity tags (Figure 3D and E). Our previous work demonstrated that the TIP150-C directly binds to MCAK-N (Jiang et al., 2009). Early analyses suggest that the MCAK-N contains putative Aurora A substrates (Zhang et al., 2008). Indeed, western blotting analyses using an anti-phosphoserine antibody to probe in vitro phosphorylated samples demonstrated that Aurora A phosphorylates wild-type MCAK, but not MCAK5A (Figure 3F), confirming that the five serine residues (5S) of MCAK-N are substrates of Aurora A (Figure 3E). To determine if phosphorylation of MCAK-5S modulates the MCAK–TIP150 interaction, HEK293 T cell lysates expressing GFP-tagged wild-type MCAK and its mutants were used for binding to GST-TIP150-C and GST (negative control). TIP150 bound more tightly to MCAKWT and MCAK5A than to MCAK5E (Figure 3G), suggesting the possibility that Aurora A phosphorylation negatively regulates MCAK binding with TIP150.
Experiments were then performed to explore the mechanisms underlying regulation of MCAK–TIP150 interaction. As shown in Figure 3H, MCAK-N and TIP150 were co-immunoprecipitated only in the presence of MCAK-C. Thus, the N–C interaction of MCAK is necessary for its binding to TIP150. Consistent with this, GFP-MCAK-C specifically bound to Flag-MCAK-N (Figure 3I). If phosphorylation of MCAK-5S abolishes the MCAK–TIP150 interaction by regulating intra-molecular association of MCAK, MCAK N-C folding should be minimized by a mutation mimicking phosphorylation. Consistent with this notion, MCAKWT/5A, but not the MCAK5E N-terminus, bound efficiently to GST-MCAK-C (Figure 3J and Supplementary Figure S6), confirming that MCAK N-C association was regulated by Aurora A phosphorylation. Thus, we conclude that TIP150 binds and recruits MCAK to MT plus-ends when MCAK is in an N-C folded state; this binding is negatively regulated by Aurora A phosphorylation (Figure 3K).
Phosphorylation of MCAK by Aurora A reduces its plus-end tracking and MT depolymerizing activity
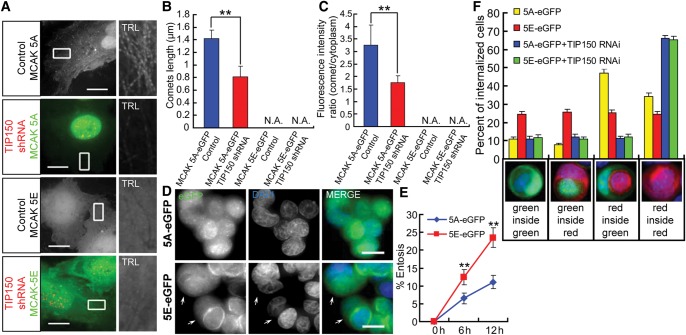
Since MCAK-5S phosphorylation disrupts the MCAK–TIP150 interaction in vitro, we speculate that MCAK-5S phosphorylation may perturb MCAK plus-end tracking in vivo. Consistent with this notion, MCAK5A-GFP tracks MT plus-ends similar to wild-type MCAK (Figures 4A and 3A), whereas MCAK5E-GFP rarely located to the plus-ends (Figure 4A), which agrees with previous reports (Andrews et al., 2004; Lan et al., 2004; Ohi et al., 2004). Similar to MCAKWT, the plus-end tracking of MCAK5A was diminished by TIP150 RNA interference, validating the role of TIP150 in localizing MCAK to the plus-ends of microtubules. In fact, both plus-end comet length and the fluorescence intensity ratio of MCAK5A were decreased in TIP150-deficient cells. Apparently, MCAK5E exhibited no comet-like localization regardless of TIP150 expression (Figure 4B and C). MCF7 cells were transiently transfected with GFP-tagged MCAK5A or MCAK5E mutants to determine the effect of MCAK-5S phosphorylation on the MT depolymerizing activity of MCAK. Overexpression of MCAK5A, but not MCAK5E, depolymerized MTs (Supplementary Figure S7A and B). These results indicate that Aurora A kinase plays a role in inhibiting the plus-end-tracking property and the MT depolymerizing activity of MCAK.
Figure 4.
Aurora A-elicited phosphorylation-mimicking mutants of MCAK promote entosis. (A) Live cell images of cells stably expressing the eGFP-tagged MCAK-5A or -5E mutant transiently transfected with or without TIP150 shRNA. Locations of the shRNAs were indicated by co-expressing with CENP-B-mCherry. Enlarged insets are trails (TRL) of 31 frames with 3-sec intervals and shown in grayscale. Knockdown of TIP150 weakens plus-end tracking of MCAK-5A, but MCAK-5E has no MT tracking activity whether expression of TIP150 is depressed or not. The scale bars are 15 μm. (B) Histograms of MT plus-end comet lengths in indicated cells (for each condition, at least 900 microtubule plus-ends in 30 cells from three independent experiments were analyzed); error bars indicate SD. **P < 0.01; N.A., not available. (C) Ratio of fluorescence intensities at the growing MT tips and in surrounding cytoplasm of indicated cells (for each condition, at least 900 microtubule plus-ends in 30 cells from three independent experiments were analyzed); error bars indicate SD. **P < 0.01; N.A., not available. (D) Images of MCAK-5A/5E-GFP stable cells after 6 h of suspension culture. Arrows indicate internalized cells. The scale bars are 15 μm. (E) Quantification of internalizing cells expressing MCAK-5A/5E-GFP at 6 and 12 h. Overexpression of MCAK-5A but not MCAK-5E results in decreased entosis, suggesting that Aurora A-elicited phosphorylation of MCAK promotes entosis. Data expressed as means ± SD. Each point represents at least 600 cells analyzed over three independent experiments. **P < 0.01. (F) Percent of dual-color labeled internalizing cells after 6 h of suspension culture. Red-labeled control cells were mixed 1:1 with MCAK-5A/5E-GFP stable cells. Below the x-axis are examples of different color combinations. Note that cells overexpressing MCAK-5A but not -5E tend to be invaded by control cells. But after being transiently transfected with TIP150 siRNA, both MCAK-5A and -5E cells essentially did not invade and were not invaded by control cells. Data expressed as means ± SD for three independent experiments in which at least 600 cells were quantified for each experiment.
Persistent phosphorylation of MCAK attenuates its inhibitory effect on entosis
Modulation of MCAK–TIP150 interaction by Aurora A phosphorylation prompted us to examine the effect of MCAK phosphorylation on entosis. Stable overexpression of MCAK5A-GFP inhibited entosis (Figure 4D) to the same extent as MCAKWT (Figures 4E and 2D). In contrast, MCAK5E-eGFP stable cells exhibited entosis at a rate similar to that of the GFP control (Figures 4E and 2D). As determined with experiments involving dual-color labeled cells in entosis, MCAK5A-GFP stable cells, like MCAKWT, were rarely internalized into control red cells and were more likely to function as host cells. However, MCAK5E-GFP resulted in distribution of outer versus inner cell functions equal to that of the red-labeled control cells (Figure 4F). When TIP150 was down-regulated, cells overexpressing both MCAK5A and MCAK5E rarely invaded or were invaded by control cells. Therefore, by modulating the interaction between MCAK and TIP150, phosphorylation overrides the perturbation of MCAK overexpression on entosis.
Aurora A kinase orchestrates entosis in a TIP150/MCAK-dependent manner
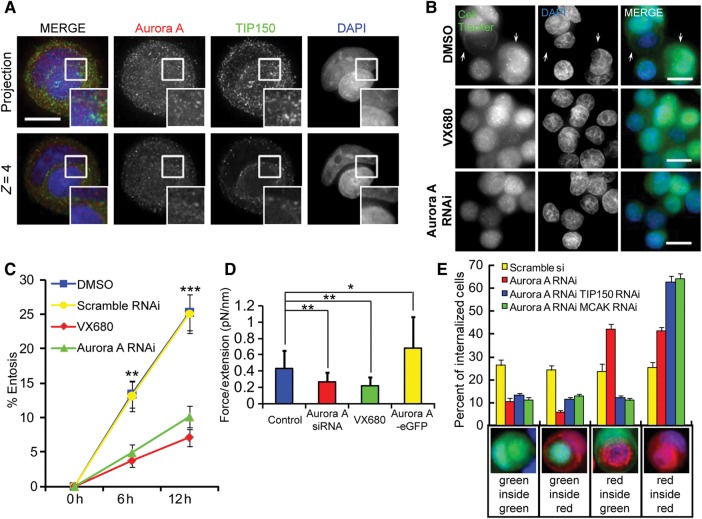
These results demonstrated that perturbation of MT dynamics and entosis could be induced by either up- or down-regulation of the MCAK–TIP150 interaction, which was regulated by Aurora A kinase activity. Thus, the reversible post-translational modification by Aurora A may act as a regulator for the interaction of TIP150 and MCAK, and, consequently, MT dynamics and entosis. To test this hypothesis, we sought to determine the localization of Aurora A relative to TIP150 and DNA. As shown in Figure 5A, TIP150 was recruited to the microtubule plus-ends in the cytoplasm, and Aurora A was predominantly spread in the cytoplasm (Supplementary Figure S8). Merged images showed that distribution of Aurora A was partially overlapped that of TIP150, supporting its role in regulating the MCAK–TIP150 interaction (Figure 5A).
Figure 5.
Dynamic regulation of Aurora A kinase activity is necessary for entosis to proceed. (A) Immunofluorescence staining of TIP150 and Aurora A in entotic MCF7cells. Up-panel, images are projections of 12 Z-stack frames with 1-μm intervals (see Supplementary Figure S8 for single-frame tomography); down-panel, the fifth single plan (4 μm from the bottom) of the same cell in up-panel. Inframe shows partial co-localization of TIP150 and Aurora A. The scale bar is 10 μm. (B) MCF7 cells treated with DMSO (control) or 1 μM Aurora kinase inhibitor VX680 or transfected with Aurora A siRNA were labeled with CellTracker Green and fixed after 6 h of suspension culture. Arrows indicate internalized cells. The scale bars are 15 μm. (C) Quantification of internalizing cells treated as in D at 6 and 12 h. Inhibition of Aurora A reduces the rate of entosis. Data are means ± SD. Each point represents at least 600 cells analyzed over three independent experiments. **P < 0.01. (D) Analysis of the dependence of F (trapping force) to l (stretching extension) shows cells transfected with Aurora A siRNA or treated with VX680 have decreased cell rigidity, while cells stably expressing Aurora A-eGFP have increased cell rigidity. Data are means ± SD. *P < 0.05, **P < 0.01. See also Supplementary Figure S10A–C. (E) Percent of dual-color labeled internalizing cells after 6 h culture in suspension culture. Red-labeled control cells were mixed 1:1 with green-labeled cells transfected with Scramble or Aurora A siRNA together with or without TIP150/MCAK siRNA. Below the x-axis are examples of different color combinations. Cells with individual knockdown of Aurora A tended to be invaded by control cells. But with double knockdown of Aurora A and MCAK or TIP150, cells did not invade and were not invaded by control cells. Data are means ± SD for three independent experiments in which at least 600 cells were quantified for each experiment.
If Aurora A kinase activity is necessary for MT dynamics to drive entosis, the rate of entosis should be decreased by inhibition of Aurora A kinase. To test this hypothesis, we compared plus-ends tracking of MCAK-GFP before and after treatment of VX680, a chemical inhibitor of Aurora A. After VX680 treatment, the number of the plus-end comets was significantly reduced whereas comet length and the fluorescence intensity of comets were largely unaffected (Supplementary Figure S9A, C and D). Suppression of Aurora A by siRNA exhibits similar effect as VX680 treatment (Supplementary Figure S9B–D). These results indicate that inhibition of Aurora A promotes depolymerase activity of MCAK. In line with these, entosis was perturbed in MCF7 cells treated either with the Aurora kinase inhibitor, VX680, or transfected with Aurora A siRNA (Figure 5B and C). To test if cell rigidity is regulated by Aurora A, we carried out optical trap analyses. As shown in Figure 5D and Supplementary Figure S10, optical tweezer measurements demonstrated that either suppression of Aurora A by siRNA or inhibition of its kinase activity by VX680 decreases cell rigidity while overexpressing Aurora A increases cell rigidity. To determine if cells deficient in Aurora A were more vulnerable to invasion by control cells, similar to cells overexpressing MCAKWT or MCAK5A, dual-color analysis of entosis was performed. As shown in Supplementary Figure S1B, the knockdown efficiency was confirmed by western blotting. Consistent with the hypothesis, suppression of Aurora A alone or in combination with TIP150 or MCAK modulates the cell-in-cell process (Figure 5E). In the dual-color experiment, suppression of Aurora A alone by siRNA increases the tendency of cells to be invaded by control cells while suppression of both Aurora A and TIP150/MCAK results in a dramatic inhibition of entosis. In those double knockdown cells, both invading and engulfing processed are inhibited. Thus, we conclude that Aurora A kinase activity is necessary for entosis to proceed via a pathway dependent upon the interaction between TIP150/MCAK.
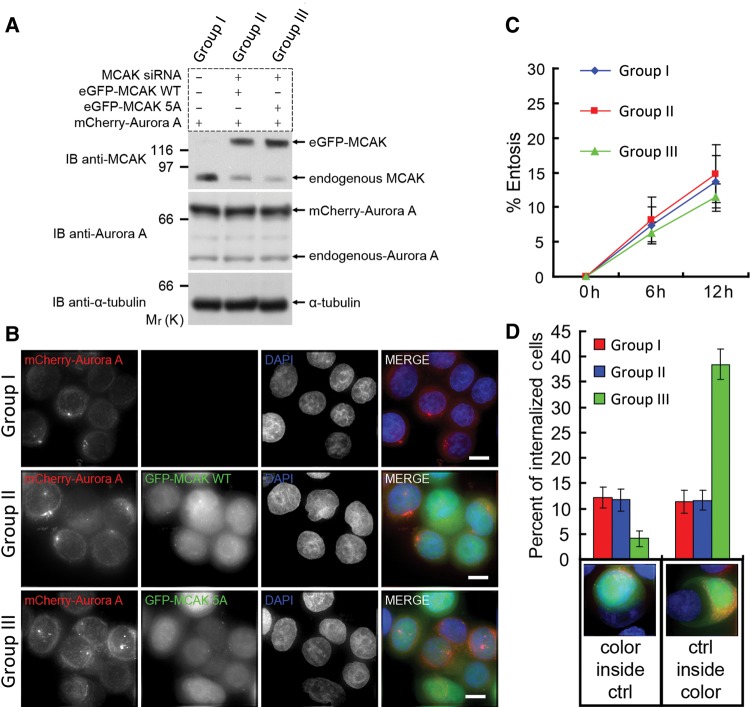
We then sought to examine the effect of overexpressing Aurora A on plus-ends tracking of MCAK. Overexpression of mCherry-Aurora A, but not mCherry tag alone, decreased both comet length and fluorescence intensity (Supplementary Figure S11), suggesting that increasing of Aurora A activity negatively regulates the plus-ends tracking affinity of MCAK. To validate whether a dynamically regulated MCAK–TIP150 interaction by Aurora A-elicited phosphorylation is essential for entosis processing, we sought to overexpress Aurora A in the presence of wild type and non-phosphorylatable MCAK mutant. As shown in Figure 6A, forced overexpression of mCherry-Aurora A yields a typical 4.3 ± 0.5 fold increase of exogenous Aurora A protein level in MCF7 cells. To examine the cooperation of MCAK with Aurora A in cell-in-cell process, aliquots of MCF7 cells were transiently transfected to update MCAK siRNA targeted to 3′-UTR and GFP-tagged MCAK. As shown in Figure 6A (top panel), the siRNA gave a typical 75%–85% suppression of endogenous MCAK level. The expression of exogenous MCAK (wild type and S5A mutant) exhibits similar level of endogenous MCAK judged by western blotting (Figure 6A). As shown in Figure 6B, expression of Aurora A alone or in the presence of wild-type MCAK and non-phosphorylatable MCAK decreased the efficiency of homotypic cell-in-cell process to the same extent (comparing Figure 6C to 2B).
Figure 6.
Aurora A orchestrates entosis in a MCAK-dependent manner. (A) Assessment of exogenous protein expression levels. GROUP I is MCF7 cell transient transfected with mCherry-Aurora A; GROUP II is eGFP-MCAK-WT stable MCF7 cell line transfected with mCherry-Aurora A and MCAK siRNA targeting 3′-UTR; GROUP III is eGFP-MCAK-5A stable cell line transfected with mCherry-Aurora A and MCAK siRNA. Protein levels of exogenously expressed GFP-MCAK, mCherry-Aurora A, and endogenous MCAK and Aurora A in the three groups were judged by immunoblotting with anti-MCAK and anti-Aurora A antibodies, respectively. (B) Aliquots of cells from the aforementioned three groups were suspended and incubated for 6 h to induce homotypic cell-in-cell process before centrifugation and formaldehyde fixation. The scale bar is 10 μm. (C) Quantification of entosis cells treated as in B at 6 and 12 h. All three groups exhibit reduced rates of cell-in-cell processes. Data are means ± SD. Each point represents at least 400 cells analyzed over three independent experiments. Differences between the three groups are not significant. (D) Aliquots of the aforementioned three groups of cells expressing various exogenously marked proteins were individually mixed 1:1 with un-transfected MCF7 (control) cells and incubated suspended to allow the process of cell-in-cell for 6 h. Treated cells were then fixed and stained with DAPI to mark nucleus for annotation of entosis. Below the x-axis are examples of different color combinations. Overexpression of Aurora A in the presence of wild type inhibits target cells invading or being invaded by control cells, but the non-phosphorylatable MCAK mutant can override the effect of Aurora A overexpression, resulting in increasing of target cells invaded by control cells. GROUP III cells tended to be invaded by control cells, but GROUP I and II cells did not. Data are means ± SD for three independent experiments in which at least 600 cells were quantified for each experiment.
To examine if overexpression of Aurora A in the presence of MCAK (wild type and mutant) alters the cell-in-cell process between those transfected cells and un-transfected cells, aliquots of transfected MCF7 cells (GROUP I, II and III) were incubated with un-transfected MCF cells in 1:1 ratio. As shown in Figure 6D, overexpression of Aurora A in the presence of endogenous MCAK (Group I) or exogenously expressed GFP-MCAK (Group II) dramatically decreased the efficiency of transfected cells to uptake untransfected cells compared with that of cells overexpressing Aurora A and non-phosphorylatable MCAK (Group III). Interestingly, cells expressing non-phosphorylatable MCAK exhibited a reduced capability in entering untransfected MCF7 cells, suggesting that microtubule plus-end dynamics orchestrated by MCAK–TIP150 interaction is essential for cell-in-cell process. Interestingly, persistent interaction of MCAK-5A mutant with TIP150 inhibits the entry of those cells into untransfected MCF7 cells, suggesting dynamic growth of microtubule plus-ends provides an input for cell-in-cell process. Therefore, we conclude that Aurora A cooperates with MCAK in regulating microtubule plus-end dynamics which is essential for entosis.
Discussion
The MT plus-end-tracking proteins establish a complex structure platform that serves as a molecular machine to regulate MT dynamics (Akhmanova and Steinmetz, 2008) and thereby orchestrates cellular dynamic events such as cell division and migration (Tirnauer et al., 2002; Wen et al., 2004; Xia et al., 2012). In this report, we extended our early study on the role of actin-based cytoskeleton in NK cell internalization (Wang et al., 2009) into essential function of microtubule plus-end dynamics in cell-in-cell process, and revealed a critical role of MCAK–TIP150 interaction in cell entry into target cell. Surprisingly, both knockdown and overexpression of TIP150/MCAK perturbed cell-in-cell process but not initial cell–cell contacts, suggesting microtubule plus-end dynamics is essential for entosis. In addition, our study revealed a critical role of Aurora A-mediated phosphorylation of MCAK in entosis via regulating MCAK–TIP150 interaction. These findings offer a mechanistic link between the Aurora A-mediated phosphorylation of MCAK in cell-in-cell process.
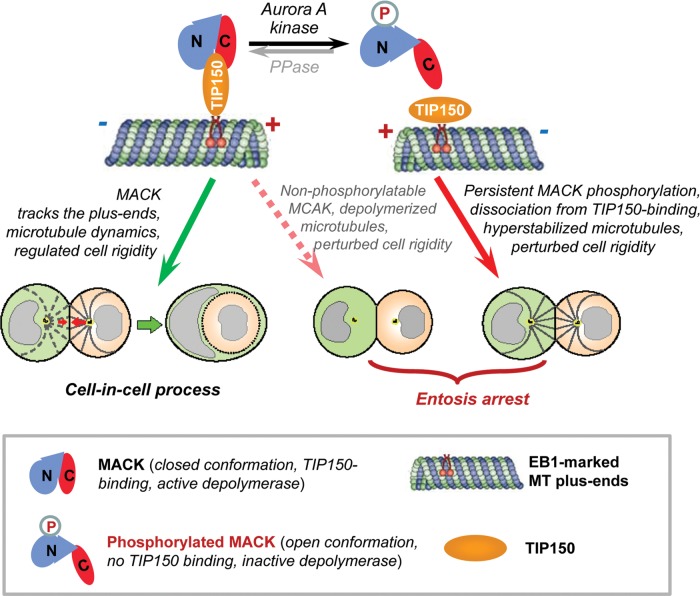
Phosphorylation of MCAK has been functionally linked to microtubule dynamics and plasticity. Our previous study demonstrated that Aurora A-induced phosphorylation of MCAK perturbs its association with TIP150 (Jiang et al., 2009). However, the functional relevance of this phospho-regulation in interphase cells was never addressed despite its role in orchestration spindle bipolarity (Zhang et al., 2008). The present results identify reversible phosphorylation of MCAK as a potential molecular mechanism to regulate its functional activity in cell-in-cell process, which is necessary for MCAK to bind to TIP150 (Figure 7).
Figure 7.
Schematic model accounting for Aurora A–MCAK–TIP150 axis during entosis. Microtubule depolymerase MCAK binds to TIP150 by which it tracks microtubule plus-end for regulating microtubule plus-end dynamics and cell rigidity during entosis. Aurora A interacts with and phosphorylates the N-terminal MCAK to release its intra-molecular N-C association by which MCAK–TIP150 interaction is abolished. In this case, Aurora A-phosphorylated MCAK is no longer associated with microtubule plus-end via TIP150 to exerts its depolymerase activity by which microtubule plus-ends become static and cell rigidity regulation is perturbed. It should be noted that MCAK phosphorylation is dynamically regulated either by phosphorylation/dephosphorylation cycle or by spatial separation of enzyme-substrate contact. The identity of the phosphatase underlying entosis is unclear. On the other hand, non-phosphorylatable MCAK exhibits greater microtubule depolymerase activity which results in perturbed cell rigidity. The perturbation of cell rigidity by either hyperstabilization of microtubules or destabilization of microtubules harnesses the progression of entosis. Thus, dynamic interaction of MCAK–TIP150 orchestrated by Aurora A phosphorylation is essential for entosis.
MCAK belongs to the Kin I family of ATPases (Tournebize et al., 2000). Unlike conventional kinesins that transport cargo in cells, Kin I kinesins lack motility but stimulate microtubule depolymerization by promoting catastrophe (Ogawa et al., 2004). The kinesin-13 family, consisting of Kif2a, Kif2b, and Kif2c/MCAK, has a conserved motor domain in the middle of the protein and is essential for the control of microtubule length in mitosis and of microtubule dynamics in interphase (Zhang et al., 2011). MCAK/kif2c, the best characterized member of the Kin I subfamily, localizes dynamically at various mitotic structures, such as inner centromeres, outer kinetochores, centrosomes, spindle microtubule plus ends, and the spindle midzone (Walczak et al., 1996). MCAK promotes catastrophe of both stable and dynamic MTs via depolymerizing MTs at both ends in vitro (Desai et al., 1999). The structural-functional analysis indicated that the C-terminal domain of MCAK is required for its MT depolymerization activity in vitro (Ems-McClung et al., 2007), which may attribute to its activity to remove tubulin subunits (Cooper et al., 2010). Our recent study showed that PLK1 phosphorylates MCAK and promotes its depolymerase activity during mitosis (Zhang et al., 2011). In this study, we surprisingly found that MCAK exhibits a greater binding activity to TIP150 in a close conformation which promotes its localization to the plus-end and depolymerase activity. We speculate that MCAK is a potential convergent point for regulating microtubule dynamics to exhibit context-dependent functional specificity. It would be of great interest and significant to examine whether and how site-specific phosphorylation regulates MCAK molecular conformation at single molecule level given the molecular heterogeneity.
Our early work demonstrated that actin-based cytoskeleton and regulator ezrin participate in stabilization of adherent complexes and subsequent remodeling of actin-based cytoskeleton upon cell–cell contact (Wang et al., 2009). With the plug-in of optical trap experiments, our present study shows that microtubule-based cytoskeleton participates in entosis by regulating cell rigidity. Although several previous reports link interactions between the microtubule and actin cytoskeleton and perturbation of MT dynamics with nocodazole treatment alters the distribution of E-cadherin and the activation of myosin (Stehbens et al., 2006), we found overexpressing of Aurora A, EB1, MCAK, or TIP150 had no apparent effect on the distribution of either general or phospho-myosin light chain II (Supplementary Figure S12A and B). In addition, staining of E-cadherin also had no obvious change unless when extremely overexpressing MCAK or Aurora A, which is rarely observed in our experiments (Supplementary Figure S12C and D). We also observed when expressing or depressing MCAK/TIP150/Aurora A or being treated with nocodazole or paclitaxel, cells were still adhered to each other while their entosis rate strongly decreased (Figures 1A, C, 2A, C, 5B and 6B). These results indicate disruption of microtubule dynamics dependent cell rigidity should be a main mechanism of entosis perturbation in cells with Aurora A–MCAK–TIP150 axis. Our study shows that inhibition of Aurora A kinase activity leads to persistent retention of MCAK to MT plus-ends by TIP150, which promotes MT depolymerization and reduces cell rigidity. In contrast, disruption of the MCAK–TIP150 interaction excessively stabilizes MTs and decreases MT dynamics. Interestingly, either suppression of the cell rigidity or a decrease in MT dynamics prevents the process of entosis (Figure 7). Our study presented here aims to provide an outline so that further mechanistic studies of phospho-regulation of MCAK in the cell internalization process can be pursued in greater detail.
Aurora-A is considered as a candidate oncogene as it is overexpressed in many tumors (Ke et al., 2003). It has been shown that overexpression of Aurora-A in diploid human breast epithelial cells causes aberrant centrosome replication and promotes aneuploidy, indicating that there are intrinsic links between aberrant regulation of Aurora-A activity and pathogeneses of aneuploidy and chromosomal instability (Ke et al., 2003). Since entosis promotes aneuploidy (Krajcovic et al., 2011), it would be of great clinic relevance and importance to investigate whether entosis is promoted in Aurora A-overexpressing human samples and whether dynamic MCAK–TIP150 interaction was perturbed in these cancers.
Taken together, we have characterized the role of MCAK–TIP150 interaction in cell internalization into target cell and revealed that Aurora A phosphorylation governs entosis via regulating a dynamic interaction between MCAK and TIP150. Our finding of phospho-regulation of MCAK by Aurora A in cell-in-cell process demonstrates a critical role of MCAK–TIP150 in regulating microtubule plus-end dynamics related to the cell-in-cell process. The fact that suppression of Aurora A phosphorylation attenuates entosis and hyper-activation of Aurora A is seen in advanced solid tumors points to the importance of Aurora A–MCAK–TIP150 axis in cellular plasticity.
Materials and methods
Cell culture
MCF7 and HEK293 T cells, obtained from the American Type Culture Collection, were maintained as subconfluent monolayers in Advanced DMEM (Invitrogen) with 10% fetal bovine serum (FBS, HyClone), 100 U/ml of penicillin, and 100 μg/ml of streptomycin (Invitrogen). Stable MCF7 clones expressing GFP, EB1-GFP, TIP150-GFP, MCAK-GFP, MCAK5A-GFP, or MCAK5E-GFP were selected in the presence of 0.5–1 µg/ml of the antibiotic, G418 (Calbiochem).
siRNA transfection
MCF7 cells, grown on 24-well plates, were transfected with 40 nM TIP150 siRNA (5′-CAUACAGCAAAGUGCGAGA-3′, described previously (Jiang et al., 2009) (synthesized by Dhamacon), MCAK siRNA (targeting 3′-UTR, synthesized by Qiagen), or Aurora A siRNA (5′-UUCUUAGACUGUAUGGUUA-3′, synthesized by GenePharma), using Lipofectamine 2000 (Invitrogen).
Inhibitors and reagents
MCF7 cells were exposed to 33 μM nocodazole (Sigma), 1 μM taxol (Cytoskeleton) or 1 μM VX680 (Selleckchem) for not <6 h before fixation. For staining, cells were exposed to CellTracker Green CMFDA and CellTracker Red CMTPX (2 μM, Invitrogen) for 20 min at 37°C in PBS buffer before re-suspension and mixing for entosis.
Antibodies
The following antibodies were used: anti-α-tubulin (Sigma; #F1804, 1:2000 for IF and 1:5000 for western blots); anti-TIP150 mAb (Jiang et al., 2009;1:400 for IF and 1:1000 for western blots); anti-GFP mAb (Sigma; #G6539, 1:2000); anti-FLAG mAb (Sigma, #F1804, 1:2000); anti-Aurora A rabbit mAb (Cell Signaling, #4718S; 1:500 for IF and 1:2000 for western blots); anti-MCAK Rabbit Ab (Jiang et al., 2009; 1:1000); anti-phosphoserine mAb (Sigma, #P3430, 1:1000); anti-E-cadherin rabbit mAb (Cell Signaling, #3195S; 1:400 for IF); anti-Myosin Light Chain 2 pAb (Cell Signaling, #3672; 1:400 for IF); and anti-phospho-Myosin Light Chain 2 (Ser19) mAb (Cell Signaling, #3675; 1:400 for IF). Secondary antibodies were purchased from Jackson ImmunoResearch and used at concentrations of 1:400 for IF and 1:2000 for western blotting.
Plasmid construction
TIP150-eGFP, MCAK-eGFP, MCAK5A-eGFP, MCAK5E-eGFP, eGFP-MCAK-C (MCAK587–725), EB1-eGFP, CLASP2-eGFP, Clip170-eGFP were described previously (Jiang et al., 2009; Liu et al., 2009; Zhang et al., 2011). Aurora A cDNA was cloned to a pcDNA 3.1 myc-his B vector containing mCherry to generate mCherry-Aurora A.
For phosphorylation and pull-down experiments, TIP150801–1368 and MCAK587–725 were cloned by PCR from human TIP150 and MCAK cDNA to the pGEX-6P-1 vector (GE Healthcare) to generate GST-TIP150-C and GST-MCAK-C, respectively. MCAK587–725 was further subcloned to the pET-28a(+) vector (Novagen) by PCR to generate H6-MCAK-C. To construct mammalian expression Flag-MCAK-N, MCAK1–586 was amplified by PCR and inserted into the p3Xflag-myc-CMV-24 vector (Sigma). To generate bacterial expression H6-eGFP-MCAKWT/5A/5E-N plasmids, cDNAs encoding 1–586 aa of MCAKWT/5A/5E were amplified by PCR and inserted into the pET-22b-GFP vector (Xia et al., 2012).
The shRNA and the CENP-B1–167-mCherry co-expression plasmids were generated by reconstruction of the PLKO.1 cloning vector (Addgene). The method was described previously (Xia et al., 2012). Briefly, synthesized TIP150/scramble shRNA sequences were inserted after the U6 promoter of the vector. CENP-B1–167-mCherry was then cloned to displace the puromycin resistance marker after the hPGK promoter on the PLKO.1 TIP150/scramble shRNA plasmids. All of the plasmids with the desired insertions were sequenced at Beijing Genomics Institute (Shanghai).
Recombinant protein preparation
The plasmids of GST-fusion or 6×His (H6)-tagged proteins were transformed into Escherichia coli strain BL21 or Rosetta (DE3), and expression was induced with 0.5 mM isopropyl β-D-thiogalactopyranoside, with shaking overnight at 16°C. Bacteria expressing GST, GST-TIP150-C, or GST-MCAK-C were suspended and lysed by sonication in PBS buffer containing 1 μg/ml phenylmethylsulfonyl fluoride (PMSF), followed by purification with glutathione-Sepharose 4B. Bacteria expressing H6-eGFP-MCAKWT/5A/5E-N or H6-MCAK-C were resuspended and lysed by sonication in lysis buffer (50 mM NaH2PO4, pH 8.0; 300 mM NaCl; 10 mM imidazole) with 1 μg/ml PMSF. H6-tagged proteins were bound to Ni-NTA resin (Qiagen) and eluted with elution buffer (50 mM NaH2PO4, pH 8.0; 300 mM NaCl; 200 mM imidazole). All purified proteins were analyzed and confirmed by SDS/PAGE.
Pull-down assays
GST or GST-tagged proteins immobilized on agarose beads were incubated with cell lysates or soluble proteins in PBS buffer containing 0.2% Triton X-100 at 4°C for 4 h. The resins were washed twice with PBS buffer, followed by boiling in SDS/PAGE buffer. The samples were subjected to SDS/PAGE, and proteins were detected by western blots.
Immunoprecipitation
For immunoprecipitation (IP) of protein complexes, HEK293 T cells expressing the indicated proteins were lysed in lysis buffer (PBS, pH 7.4; 0.2% Triton X-100; 1 mM EDTA; 1 mM dithiothreitol, DTT containing 10 U/ml of DNase I and protease inhibitors (Sigma)). The lysates were clarified by centrifugation at 14000 rpm for 10 min at 4°C. The FLAG-tagged proteins were precipitated with anti-FLAG-M2 beads (Sigma). Beads were washed three times with lysis buffer and then boiled and loaded onto 8% SDS–PAGE for CBB staining or western blotting.
Phosphorylation in vitro
Recombinant Aurora A kinase (50 ng, Millipore) was incubated with 1 μg of purified 6×His-tagged substrates in 50 μl of reaction buffer containing 50 μM ATP, 25 mM potassium-HEPES (pH 7.4), 5 mM MgSO4, 0.1 M KCl, 2 mM MgCl2, 0.1 mM CaCl2, 5 mM EGTA, and 1 mM DTT for 1 h at 30°C. Kinase reactions were terminated with Laemmli sample buffer, boiled for 5 min, and subjected to SDS–PAGE for western blotting analysis with anti-phosphoserine mAb. Input was detected with Coomassie brilliant blue staining (CBB).
Western blots
Samples were subjected to SDS/PAGE and transferred onto nitrocellulose membrane. Proteins were probed by appropriate primary and secondary antibodies and detected by enhanced chemiluminescence.
Entosis assays
Monolayer MCF7 cells were treated with indicated inhibitors for 2 h before being stained with 2 μM CellTracker Green for 20 min at 37°C in serum-free PBS buffer. The buffer was then replaced with fresh, pre-warmed culture medium, and the cells were incubated at 37°C for 20 min to remove redundant dye. (All these staining and wash processes were in the presence of the indicated inhibitors. For cells expressing GFP-tagged proteins, these processes were skipped.) The stained cells were trypsinized to a single cell suspension, mixed at 100000 cells per milliliter, and incubated in growth medium (containing the indicated inhibitors) in 24-well plates. To prevent cell adherence, the plates were pre-coated with 6 mg/ml poly-HEMA (Sigma-Aldrich) in ethanol, and incubated at 37°C until dry (described previously by Overholtzer et al., 2007). At the indicated times, cells were cytospun at 1000 rpm for 5 min onto coverglasses in a Hettich Universal 32 Tabletop Centrifuge (DJB Labcare Ltd, UK). Cells were then fixed in 3.7% paraformaldehyde at room temperature (rt) for 15 min. The fixed cells were permeablized and stained with PBS containing 0.1% Triton X-100 supplemented with 4′,6-diamidino-2-phenylindole (DAPI, Sigma, 1:10000). After washing with PBS three times, cells were mounted onto slides with Vectashield Mounting Medium (#H-1000, Vector Laboratories, Inc.)
Since the comet-like localization of plus-end-tracking proteins can be visualized after cold ethanol fixation but not after formaldehyde fixation, in entosis experiments, the plus-end-tracking proteins have no obvious comet-like staining. The percentages of entotic cells were quantified with an Olympus 40 × 1.35 NA objective on an Olympus IX71 microscope with softWORX software (Applied Precision, Inc.). To validate the quantification, images were acquired every 1 μm at the z-axis to generate 3D image stacks. Percentages were calculated from 500 cells in at least three independent experiments. Cells enwrapped at least half-way by a neighboring cell were considered to be entotic or internalizing. Cell pairs participating in entosis were counted as one. Because internalization at time zero was low, this rate of entosis was graphed as zero. The rate of entosis after 12 h could not be determined due to higher level of cell aggregation.
For dual-color labeled entosis experiments, control cells were labeled with CellTracker Red and mixed 1:1 with treated cells labeled with CellTracker Green. Cells were analyzed as described above after 6 h of suspension culture.
Immunofluorescence microscopy
For immunofluorescence staining of entosis cells with TIP150 and Aurora A, cells were first trypsinized and plated on polyhema-coated plates for 6 h as described in entosis assays. After suspension culture, cells were piped out and plated on coverglass to facilitate adherence. Two hours later the adherent cells were fixed in methanol for 10 min at −20°C. Fixed samples were washed in PBS, blocked in 2% BSA in PBST (PBS buffer supplemented with 0.5% Tween-20) for 30 min at rt. Primary and secondary antibodies were incubated at rt for 1 h and 45 min, respectively. DNA was stained with DAPI. Images were acquired every 1 μM at z-axis to generate 3D image stacks using Zeiss LSM710 Confocal Microscope with a Zeiss Plan-Apochromat 63×/1.40NA Oil DIC objective. The 3D image stacks were projected with ZEN software (Carl Zeiss Microscopy, LLC), and mounted in figures with Photoshop and Illustrator (Adobe).
For microtubule depolymerization assay, cells plating on coverglasses were first transfected with eGFP-tagged plus-end-tracking proteins and siRNA for 48 h, then fixed with pre-warmed 3.7% Paraformaldehyde at 37°C for 15 min to avoid microtubule depolymerization during experimental procedure. Cells were fixed, blocked and stained with antibodies as above. This method has been well established previously (Andrews et al., 2004; Ogawa et al., 2004). Images were acquired every 0.5 μM at z-axis to generate 3D image stacks using Olympus IX71 microscope with an Olympus 60×/1.42 Plan APO N objective. The 3D image sections were projected with SoftWORX software, and mounted in figures with Photoshop and Illustrator. As above mentioned, the classic comet-like localization of the eGFP-tagged plus-end-tracking proteins could not be clearly visualized due to paraformaldehyde fixation.
Live cell imaging
Time-lapse imaging of cultured cells was accomplished with a 40× or 60× oil-immersion 1.4 NA objective on an Olympus IX71 microscope. Images were recorded at 37°C in Lab-Tek chambered 1.0 borosilicate coverglasses (Nunc) in CO2-independent medium (Gibco) containing 10% (vol/vol) FBS, 100 U/ml penicillin, and 100 μg/ml streptomycin. For plus-end-tracking experiments, cells expressing eGFP-tagged plus-end-tracking proteins were imaged with 500-msec exposures at a temporal resolution of 3 sec for a total of 1.5 min. The trails (TRL) were generated by projection of the frames with SoftWORX software and mounted by Photoshop and Illustrator. Analyses of comet lengths, fluorescence intensity ratios, velocities, and catastrophe frequencies were performed with ImageJ (NIH) as previously described (Komarova et al., 2002). The fluorescence intensity ratio was an average between the fluorescence intensities of plus-end comets and the following MT lattice. The EB1 tracking velocity was an average of instantaneous plus-end-tracking velocities. Catastrophe frequency was the average of reciprocals of insistent growing time of MTs. Data analyses were performed by use of OriginPro 7.0 or Microsoft Office Excel.
Optical trap measurements
The optical tweezers system was described previously (Zhong et al., 2013). This optical tweezers were built on an inverted microscope (Olympus IX71, Japan) using a fiber laser (AFL-1064-40-R-CL, Amonics Limited, Hong Kong) with a wavelength of 1064 nm and a nominal out power of 10 W (Watt). The laser beam was expanded to a diameter of 7 mm to overfill the back-aperture of a water-immersion objective with a high NA of 1.2 (UPLSAPO, 60×, Olympus, Japan). The tightly focused beam can trap dielectric beads and cells steadily in a chamber. When the chamber was moved by a piezoelectric stage (P-545.3R7, PI, Germany), the trapped bead was fixed at an initial location in our field of view. A CCD camera (Photometrics CoolSNAP HQ2, America) was used to monitor our manipulations.
For adhesion of beads to cells, experiments were performed at rt (25°C–26°C). The beads with a diameter of 5 μm (4.987 ± 0.040 µm, 4205A, Duke Scientific) were incubated with lectin (sigma, 0.5 mg/ml lectin in PBS buffer) for 90 min, followed by three washes with PBS, and finally diluted 1:10 in cell culture buffer.
Flow chambers prepared from coverslips sealed with parafilm were pre-absorbed with poly-l-lysine solution (0.01%, Sigma) for 30 min at rt, washed with cell culture buffer, incubated with MCF7 cells for 20 min. After removing of the unbound cells, Lectin-coated beads were introduced into the chamber.
Through moving piezoelectric stage, the free beads were trapped by optical tweezers and manipulated to contact cells for 3 min. Since coated lectin on bead's surface combined with glycoproteins from cell membrane (Kaji et al., 2003), the bead adhered to cell tightly and was moved as a handle to exert a force on the cell.
When a cell adhering to coverslip moves at a fixed velocity v, the movement of a trapped bead adhering to the cell will be hindered by a trapping force F induced by a displacement from trap center, as shown in Figure 2G. As a result, the force exerted on the cell membrane will induce the cell to deform. In a given time interval t, the cell moves a distance of vt, but the bead deviates only Δx from trap center. The stretching length can be denoted by l = vt−Δx. Meanwhile, the force meets a linear relationship of F = −κΔx, where κ is the stiffness of optical trap. When the deviation of bead is smaller than 0.6rbead (rbead, radius of bead), the stiffness can be regarded as a constant for large particles including ellipsoid (Zhou et al., 2012). In our measurements, the piezoelectric stage moved at v = 100 nm/sec, and all movements of stretching were recorded by the CCD camera. The deviation Δx can be tracked positions of bead in image sequences with an image analysis technique of cross-correlation calculation (Gelles et al., 1988). When the deviation was beyond 0.6rbead (1.5 µm), the subsequent positions were discarded. According to tracked deviation of the bead and calibrated stiffness, both tension force (F) and corresponding stretching length (l) can be determined. The dependence relation of F to l can denote cell rigidity.
When a trapped bead moved freely from trap center by shutting optical trap, its trajectory can be traced by image analysis. According to the relationship of <Δx2(τ) ≥ 2Dτ (τ, time interval, D, diffusion coefficient) (Lin et al., 2000), the viscosity of assay buffer was about 8.6 × 10−4 Ns/m2 under laser power of 1.78 W at objective entrance aperture. With calibration using viscous drag method (Buosciolo et al., 2004), the stiffness for 5-μm bead trapped in assay buffer was 405.7 pN/μm at a depth of 20 μm from coverslip. For an optical trap consisted of the water-immersion objective, the stiffness will vary slightly at different depths (Vermeulen et al., 2006). Thus, the stiffness can be regarded as a constant for stretching different cells at varying depths.
Statistics
Data analysis and statistics were performed with OriginPro 7.0 or Microsoft Office Excel.
Supplementary material
Supplementary material is available at Journal of Molecular Cell Biology online.
Funding
This work is supported by Chinese 973 Project Grants (2010|CB912103, 2012|CB917200, 2013CB911203, and 2012CB713704), Chinese Academy of Science Grant (KSCX2-YW-H-10), Anhui Province Key Project Grant (08040102005), International Collaboration Grant (MOST2009DFA31010), Chinese Natural Science Foundation Grants (31320103904, 91313303, 90913016, and 31100992), MOE20113402130010, the Fundamental Research Funds for Central Universities (WK2060190018 and WK2340000021), MOE Innovative Team IRT13038, and National Institutes of Health Grants (DK56292, CA164133, and G12RR03034). X.Y. is a Cheung Kong Scholar.
Conflict of interest: none declared.
Supplementary Material
Acknowledgements
We thank Dr Donald Hill (University of Alabama–Birmingham, USA) for critical reading and members of our groups for insightful discussion.
References
- Abodief W.T., Dey P., Al-Hattab O. Cell cannibalism in ductal carcinoma of breast. Cytopathology. 2006;17:304–305. doi: 10.1111/j.1365-2303.2006.00326.x. [DOI] [PubMed] [Google Scholar]
- Akhmanova A., Steinmetz M.O. Tracking the ends: a dynamic protein network controls the fate of microtubule tips. Nat. Rev. Mol. Cell Biol. 2008;9:309–322. doi: 10.1038/nrm2369. [DOI] [PubMed] [Google Scholar]
- Andrews P.D., Ovechkina Y., Morrice N., et al. Aurora B regulates MCAK at the mitotic centromere. Dev. Cell. 2004;6:253–268. doi: 10.1016/s1534-5807(04)00025-5. [DOI] [PubMed] [Google Scholar]
- Borghi N., Sorokina M., Shcherbakova O.G., et al. E-cadherin is under constitutive actomyosin-generated tension that is increased at cell-cell contacts upon externally applied stretch. Proc. Natl Acad. Sci. USA. 2012;109:12568–12573. doi: 10.1073/pnas.1204390109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buosciolo A., Pesce G., Sasso A. New calibration method for position detector for simultaneous measurements of force constants and local viscosity in optical tweezers. Opt. Commun. 2004;230:357–368. [Google Scholar]
- Carmena M., Ruchaud S., Earnshaw W.C. Making the Auroras glow: regulation of Aurora A and B kinase function by interacting proteins. Curr. Opin. Cell Biol. 2009;21:796–805. doi: 10.1016/j.ceb.2009.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu Y.S., Thomas W.A., Eder O., et al. Force measurements in E-cadherin-mediated cell doublets reveal rapid adhesion strengthened by actin cytoskeleton remodeling through Rac and Cdc42. J. Cell Biol. 2004;167:1183–1194. doi: 10.1083/jcb.200403043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper J.R., Wagenbach M., Asbury C.L., et al. Catalysis of the microtubule on-rate is the major parameter regulating the depolymerase activity of MCAK. Nat. Struct. Mol. Biol. 2010;17:77–82. doi: 10.1038/nsmb.1728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai A., Verma S., Mitchison T.J., et al. Kin I kinesins are microtubule-destabilizing enzymes. Cell. 1999;96:69–78. doi: 10.1016/s0092-8674(00)80960-5. [DOI] [PubMed] [Google Scholar]
- Ems-McClung S.C., Hertzer K.M., Zhang X., et al. The interplay of the N- and C-terminal domains of MCAK control microtubule depolymerization activity and spindle assembly. Mol. Biol. Cell. 2007;18:282–294. doi: 10.1091/mbc.E06-08-0724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fais S. Cannibalism: a way to feed on metastatic tumors. Cancer Lett. 2007;258:155–164. doi: 10.1016/j.canlet.2007.09.014. [DOI] [PubMed] [Google Scholar]
- Florey O., Overholtzer M. Autophagy proteins in macroendocytic engulfment. Trends Cell Biol. 2012;22:374–380. doi: 10.1016/j.tcb.2012.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraizer G.C., Diaz M.F., Lee I.L., et al. Aurora-A/STK15/BTAK enhances chromosomal instability in bladder cancer cells. Int. J. Oncol. 2004;25:1631–1639. [PubMed] [Google Scholar]
- Gelles J., Schnapp B.J., Sheetz M.P. Tracking kinesin-driven movements with nanometre-scale precision. Nature. 1988;331:450–453. doi: 10.1038/331450a0. [DOI] [PubMed] [Google Scholar]
- Giet R., Petretti C., Prigent C. Aurora kinases, aneuploidy and cancer, a coincidence or a real link? Trends Cell Biol. 2005;15:241–250. doi: 10.1016/j.tcb.2005.03.004. [DOI] [PubMed] [Google Scholar]
- Gordon D.J., Resio B., Pellman D. Causes and consequences of aneuploidy in cancer. Nat. Rev. Genet. 2012;13:189–203. doi: 10.1038/nrg3123. [DOI] [PubMed] [Google Scholar]
- Hong I.S. The exfoliative cytology of endometrial stromal sarcoma in peritoneal fluid. Acta Cytol. 1981;25:277–281. [PubMed] [Google Scholar]
- Jiang K., Wang J., Liu J., et al. TIP150 interacts with and targets MCAK at the microtubule plus ends. EMBO Rep. 2009;10:857–865. doi: 10.1038/embor.2009.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaji H., Saito H., Yamauchi Y., et al. Lectin affinity capture, isotope-coded tagging and mass spectrometry to identify N-linked glycoproteins. Nat. Biotechnol. 2003;21:667–672. doi: 10.1038/nbt829. [DOI] [PubMed] [Google Scholar]
- Ke Y.W., Dou Z., Zhang J., et al. Function and regulation of Aurora/Ipl1p kinase family in cell division. Cell Res. 2003;13:69–81. doi: 10.1038/sj.cr.7290152. [DOI] [PubMed] [Google Scholar]
- Komarova Y.A., Akhmanova A.S., Kojima S., et al. Cytoplasmic linker proteins promote microtubule rescue in vivo. J. Cell Biol. 2002;159:589–599. doi: 10.1083/jcb.200208058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krajcovic M., Overholtzer M. Mechanisms of ploidy increase in human cancers: a new role for cell cannibalism. Cancer Res. 2012;72:1596–1601. doi: 10.1158/0008-5472.CAN-11-3127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krajcovic M., Johnson N.B., Sun Q., et al. A non-genetic route to aneuploidy in human cancers. Nat. Cell Biol. 2011;13:324–330. doi: 10.1038/ncb2174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan W., Zhang X., Kline-Smith S.L., et al. Aurora B phosphorylates centromeric MCAK and regulates its localization and microtubule depolymerization activity. Curr. Biol. 2004;14:273–286. doi: 10.1016/j.cub.2004.01.055. [DOI] [PubMed] [Google Scholar]
- Lassus H., Staff S., Leminen A., et al. Aurora-A overexpression and aneuploidy predict poor outcome in serous ovarian carcinoma. Gynecol. Oncol. 2011;120:11–17. doi: 10.1016/j.ygyno.2010.09.003. [DOI] [PubMed] [Google Scholar]
- Lin B., Yu J., Rice S.A. Direct measurements of constrained Brownian motion of an isolated sphere between two walls. Phys. Rev. E. 2000;62:3909–3919. doi: 10.1103/physreve.62.3909. [DOI] [PubMed] [Google Scholar]
- Liu J., Wang Z., Jiang K., et al. PRC1 cooperates with CLASP1 to organize central spindle plasticity in mitosis. J. Biol. Chem. 2009;284:23059–23071. doi: 10.1074/jbc.M109.009670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X., Kang Y. Cell fusion as a hidden force in tumor progression. Cancer Res. 2009;69:8536–8539. doi: 10.1158/0008-5472.CAN-09-2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay D.R., Makise M., Ullman K.S. Defects in nuclear pore assembly lead to activation of an Aurora B-mediated abscission checkpoint. J. Cell Biol. 2010;191:923–931. doi: 10.1083/jcb.201007124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maney T., Wagenbach M., Wordeman L. Molecular dissection of the microtubule depolymerizing activity of mitotic centromere-associated kinesin. J. Biol. Chem. 2001;276:34753–34758. doi: 10.1074/jbc.M106626200. [DOI] [PubMed] [Google Scholar]
- Matsumura F. Regulation of myosin II during cytokinesis in higher eukaryotes. Trends Cell Biol. 2005;15:371–377. doi: 10.1016/j.tcb.2005.05.004. [DOI] [PubMed] [Google Scholar]
- Ogawa T., Nitta R., Okada Y., et al. A common mechanism for microtubule destabilizers-M type kinesins stabilize curling of the protofilament using the class-specific neck and loops. Cell. 2004;116:591–602. doi: 10.1016/s0092-8674(04)00129-1. [DOI] [PubMed] [Google Scholar]
- Ohi R., Coughlin M.L., Lane W.S., et al. An inner centromere protein that stimulates the microtubule depolymerizing activity of a KinI kinesin. Dev. Cell. 2003;5:309–321. doi: 10.1016/s1534-5807(03)00229-6. [DOI] [PubMed] [Google Scholar]
- Ohi R., Sapra T., Howard J., et al. Differentiation of cytoplasmic and meiotic spindle assembly MCAK functions by Aurora B-dependent phosphorylation. Mol. Biol. Cell. 2004;15:2895–2906. doi: 10.1091/mbc.E04-02-0082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overholtzer M., Brugge J.S. The cell biology of cell-in-cell structures. Nat. Rev. Mol. Cell Biol. 2008;9:796–809. doi: 10.1038/nrm2504. [DOI] [PubMed] [Google Scholar]
- Overholtzer M., Mailleux A.A., Mouneimne G., et al. A nonapoptotic cell death process, entosis, that occurs by cell-in-cell invasion. Cell. 2007;131:966–979. doi: 10.1016/j.cell.2007.10.040. [DOI] [PubMed] [Google Scholar]
- Parsons J.T., Horwitz A.R., Schwartz M.A. Cell adhesion: integrating cytoskeletal dynamics and cellular tension. Nat. Rev. Mol. Cell Biol. 2010;11:633–643. doi: 10.1038/nrm2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelling A.E., Dawson D.W., Carreon D.M., et al. Distinct contributions of microtubule subtypes to cell membrane shape and stability. Nanomedicine. 2007;3:43–52. doi: 10.1016/j.nano.2006.11.006. [DOI] [PubMed] [Google Scholar]
- Pfau S.J., Amon A. Chromosomal instability and aneuploidy in cancer: from yeast to man. EMBO Rep. 2012;13:515–527. doi: 10.1038/embor.2012.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piehl M., Tulu U.S., Wadsworth P., et al. Centrosome maturation: measurement of microtubule nucleation throughout the cell cycle by using GFP-tagged EB1. Proc. Natl Acad. Sci. USA. 2004;101:1584–1588. doi: 10.1073/pnas.0308205100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rath N., Olson M.F. Rho-associated kinases in tumorigenesis: re-considering ROCK inhibition for cancer therapy. EMBO Rep. 2012;13:900–908. doi: 10.1038/embor.2012.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stehbens S.J., Paterson A.D., Crampton M.S., et al. Dynamic microtubules regulate the local concentration of E-cadherin at cell-cell contacts. J. Cell Sci. 2006;119:1801–1811. doi: 10.1242/jcs.02903. [DOI] [PubMed] [Google Scholar]
- Tatsuka M., Sato S., Kitajima S., et al. Overexpression of Aurora-A potentiates HRAS-mediated oncogenic transformation and is implicated in oral carcinogenesis. Oncogene. 2005;24:1122–1127. doi: 10.1038/sj.onc.1208293. [DOI] [PubMed] [Google Scholar]
- Tirnauer J.S., Canman J.C., Salmon E.D., et al. EB1 targets to kinetochores with attached, polymerizing microtubules. Mol. Biol. Cell. 2002;13:4308–4316. doi: 10.1091/mbc.E02-04-0236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tournebize R., Popov A., Kinoshita K., et al. Control of microtubule dynamics by the antagonistic activities of XMAP215 and XKCM1 in Xenopus egg extracts. Nat. Cell Biol. 2000;2:13–19. doi: 10.1038/71330. [DOI] [PubMed] [Google Scholar]
- Underhill D.M., Goodridge H.S. Information processing during phagocytosis. Nat. Rev. Immunol. 2012;12:492–502. doi: 10.1038/nri3244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vader G., Lens S.M. The Aurora kinase family in cell division and cancer. Biochim. Biophys. Acta. 2008;1786:60–72. doi: 10.1016/j.bbcan.2008.07.003. [DOI] [PubMed] [Google Scholar]
- Vermeulen K.C., Wuite G.J., Stienen G.J., et al. Optical trap stiffness in the presence and absence of spherical aberrations. Appl. Opt. 2006;45:1812–1819. doi: 10.1364/ao.45.001812. [DOI] [PubMed] [Google Scholar]
- Walczak C.E., Mitchison T.J., Desai A. XKCM1: a Xenopus kinesin-related protein that regulates microtubule dynamics during mitotic spindle assembly. Cell. 1996;84:37–47. doi: 10.1016/s0092-8674(00)80991-5. [DOI] [PubMed] [Google Scholar]
- Wan Q., Liu J., Zheng Z., et al. Regulation of myosin activation during cell-cell contact formation by Par3-Lgl antagonism: entosis without matrix detachment. Mol. Biol. Cell. 2012;23:2076–2091. doi: 10.1091/mbc.E11-11-0940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Guo Z., Xia P., et al. Internalization of NK cells into tumor cells requires ezrin and leads to programmed cell-in-cell death. Cell Res. 2009;19:1350–1362. doi: 10.1038/cr.2009.114. [DOI] [PubMed] [Google Scholar]
- Ward T., Wang M., Liu X., et al. Regulation of a dynamic interaction between two microtubule-binding proteins, EB1 and TIP150, by PCAF orchestrates kinetochore microtubule plasticity and chromosome stability during mitosis. J. Biol. Chem. 2013;288:15771–15785. doi: 10.1074/jbc.M112.448886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen Y., Eng C.H., Schmoranzer J., et al. EB1 and APC bind to mDia to stabilize microtubules downstream of Rho and promote cell migration. Nat. Cell Biol. 2004;6:820–830. doi: 10.1038/ncb1160. [DOI] [PubMed] [Google Scholar]
- Xia P., Wang S., Guo Z., et al. Emperipolesis, entosis and beyond: dance with fate. Cell Res. 2008;18:705–707. doi: 10.1038/cr.2008.64. [DOI] [PubMed] [Google Scholar]
- Xia P., Wang Z., Liu X., et al. EB1 acetylation by P300/CBP-associated factor (PCAF) ensures accurate kinetochore-microtubule interactions in mitosis. Proc. Natl Acad. Sci. USA. 2012;109:16564–16569. doi: 10.1073/pnas.1202639109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoo H., Isoda K., Nakazato Y., et al. Retroperitoneal epithelioid angiomyolipoma leading to fatal outcome. Pathol. Int. 2000;50:649–654. doi: 10.1046/j.1440-1827.2000.01096.x. [DOI] [PubMed] [Google Scholar]
- Zhang D., Hirota T., Marumoto T., et al. Cre-loxP-controlled periodic Aurora-A overexpression induces mitotic abnormalities and hyperplasia in mammary glands of mouse models. Oncogene. 2004;23:8720–8730. doi: 10.1038/sj.onc.1208153. [DOI] [PubMed] [Google Scholar]
- Zhang X., Ems-McClung S.C., Walczak C.E. Aurora A phosphorylates MCAK to control ran-dependent spindle bipolarity. Mol. Biol. Cell. 2008;19:2752–2765. doi: 10.1091/mbc.E08-02-0198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Shao H., Huang Y., et al. PLK1 phosphorylates mitotic centromere-associated kinesin and promotes its depolymerase activity. J. Biol. Chem. 2011;286:3033–3046. doi: 10.1074/jbc.M110.165340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong M.-C., Wei X.-B., Zhou J.-H., et al. Trapping red blood cells in living animals using optical tweezers. Nat. Commun. 2013;4:1768. doi: 10.1038/ncomms2786. [DOI] [PubMed] [Google Scholar]
- Zhou H., Kuang J., Zhong L., et al. Tumour amplified kinase STK15/BTAK induces centrosome amplification, aneuploidy and transformation. Nat. Genet. 1998;20:189–193. doi: 10.1038/2496. [DOI] [PubMed] [Google Scholar]
- Zhou J.H., Zhong M.C., Wang Z.Q., et al. Calculation of optical forces on an ellipsoid using vectorial ray tracing method. Opt. Express. 2012;20:14928–14937. doi: 10.1364/OE.20.014928. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.