Abstract
Divergent selection based on aquatic larval ecology is a likely factor in the recent isolation of two broadly sympatric and morphologically identical African mosquito species, the malaria vectors Anopheles gambiae and An. coluzzii. Population-based genome scans have revealed numerous candidate regions of recent positive selection, but have provided few clues as to the genetic mechanisms underlying behavioral and physiological divergence between the two species, phenotypes which themselves remain obscure. To uncover possible genetic mechanisms, we compared global transcriptional profiles of natural and experimental populations using gene-based microarrays. Larvae were sampled as second and fourth instars from natural populations in and around the city of Yaoundé, capital of Cameroon, where the two species segregate along a gradient of urbanization. Functional enrichment analysis of differentially expressed genes revealed that An. coluzzii—the species that breeds in more stable, biotically complex and potentially polluted urban water bodies—over-expresses genes implicated in detoxification and immunity relative to An. gambiae, which breeds in more ephemeral and relatively depauperate pools and puddles in suburbs and rural areas. Moreover, our data suggest that such over-expression by An. coluzzii is not a transient result of induction by xenobiotics in the larval habitat, but an inherent and presumably adaptive response to repeatedly encountered environmental stressors. Finally, we find no significant overlap between the differentially expressed loci and previously identified genomic regions of recent positive selection, suggesting that transcriptome divergence is regulated by trans-acting factors rather than cis-acting elements.
Keywords: comparative transcriptomics, cryptic species, ecological and environmental genomics, M and S molecular forms, microarrays, transcriptional variation
Introduction
Cryptic species—those difficult or impossible to distinguish morphologically at any life stage—are widespread both geographically and across the tree of life (Bickford et al. 2007; Pfenninger & Schwenk 2007). Due to their morphological similarity, cryptic species are likely to be very similar ecologically (Leibold & McPeek 2006). Thus, for recently diverged cryptic species to coexist stably, competition is presumably limited by physiological and behavioral adaptations to alternative local habitats or resources, which confer some degree of specialization and niche partitioning (Chesson 2000; Montero-Pau et al. 2011). Identifying the genetic basis of such adaptive phenotypic divergence can offer insight into molecular mechanisms underlying ecological isolation, a grand challenge of ecological genomics.
If physiological or behavioral traits distinguishing cryptic species are known in advance and can be scored in controlled laboratory crosses, quantitative trait locus mapping is the most direct way to relate phenotype with an underlying genotype. However, where this approach is unfeasible, comparative genomic approaches involving genome scans of nucleotide divergence along chromosomes (Li et al. 2008; Nosil et al. 2009; Storz 2005) or gene expression profiling (Romero et al. 2012; Whitehead & Crawford 2006b; Zheng et al. 2011) allow biological discovery and hypothesis generation in the absence of a priori information about the nature of ecological divergence between species. There have long been good theoretical grounds for the expectation that transcriptional rewiring underlies many phenotypic changes between species (Britten & Davidson 1969, 1971). More recently, growing empirical evidence supports the important role of transcriptional variation in the evolution of physiological and behavioral differences between species (Borneman et al. 2007; Matzkin 2012; Nowick et al. 2009; Whitehead & Crawford 2006a; Wittkopp et al. 2008). Thus, whole-transcriptome profiling of sympatric and closely related cryptic species can be a powerful approach to discover the functional basis of their ecological isolation (Whitehead 2012).
Two of the principal malaria vector mosquitoes in tropical Africa belong to the Anopheles gambiae sibling species complex. Formerly designated as An. gambiae M and S molecular forms (della Torre et al. 2001), and recently named An. coluzzii and An. gambiae (Coetzee et al. 2013), these sister species are very recently diverged and strictly sympatric in many parts of West and Central Africa (della Torre et al. 2005). Despite the absence of intrinsic genetic barriers to gene exchange in hybrids (Diabate et al. 2007; Hahn et al. 2012), assortative mating (Dabire et al. 2013; Diabate et al. 2006; Diabate et al. 2009), and ecologically based reproductive isolation (Diabate et al. 2008; Diabate et al. 2005) limit the amount of localized interspecific gene flow and introgression, such that both species appear to be evolving collectively on independent trajectories across much of their shared range (Reidenbach et al. 2012).
As adults, An. coluzzii and An. gambiae can be found resting in the same houses and blood feeding on the same human hosts. However, in the savannas of West Africa, the aquatic immature stages are associated with alternative breeding sites (Gimonneau et al. 2012a). Presumed to be the ancestral species, An. gambiae breeds in temporary rain dependent pools and puddles which are largely free of predators. In such sites, An. gambiae outcompetes An. coluzzii (Diabate et al. 2008; Lehmann & Diabate 2008). An. coluzzii is found in more stable sites such as rice fields and reservoirs, and is behaviorally more adept at avoiding the predators that abound in such habitats (Diabate et al. 2008; Gimonneau et al. 2010; Gimonneau et al. 2012b). In the rainforests of Central Africa, larval habitat partitioning also occurs, but here it appears to be associated with gradients of urbanization rather than rice agriculture (Kamdem et al. 2012). In the Cameroon capital Yaoundé, An. gambiae larvae are found in temporary rain dependent pools in the rural suburbs, but An. coluzzii breeds in more stable collections of water within the city center, some of which serve as dumping grounds, polluted by the decay of organic material into ammonia and other nitrogenous breakdown products (Kamdem et al. 2012) (Fig. 1). As predicted, dose-mortality tests of larvae collected in and around Yaoundé confirmed that An. coluzzii is more resistant to ammonia than An. gambiae (Tene Fossog et al. 2013a). Similarly, dose-mortality tests of larvae sampled along the coast of the Gulf of Guinea also have shown that An. coluzzii resists salinity better than An. gambiae (B. Tene Fossog, D. Ayala, P. Acevedo, P. Kengne, P. Awono-Ambene, F. Njiokou, C. Antonio-Nkondjio, M. Pombi, N. Besansky, C. Costantini, unpublished data). Taken together with evidence from ecological niche modeling (Costantini et al. 2009; Simard et al. 2009), the habitat segregation of these species is consistent with an ecological speciation process involving niche expansion by An. coluzzii into larval habitats of marginal quality.
Figure 1.

Representative larval breeding habitats of An. gambiae (rural; left panel) and An. coluzzii (urban; right panel) sampled during this study in Yaoundé, Cameroon.
Genome scans of divergence between An. gambiae and An. coluzzii have revealed numerous candidate regions involved in adaptive natural selection (Lawniczak et al. 2010; Neafsey et al. 2010; Reidenbach et al. 2012; White et al. 2011), but few clues as to the functional basis of their divergence. We reasoned that differences in gene regulation between these sister taxa may help uncover the molecular basis for species-specific traits. Previously, we compared transcriptional profiles between laboratory-adapted colonies of An. gambiae and An. coluzzii, and focused mainly on adult females (Cassone et al. 2008). Here, we sample co-occurring natural populations of both taxa, and examine mixed sexes at the larval stage. Given that larvae of An. coluzzii appear better able to withstand a variety of environmental stressors—both biotic and abiotic—we predicted that larval samples of this species might express stress-responsive genes (e.g. immune genes, detoxification genes) at higher levels than their An. gambiae counterparts. To explore this question, we performed whole transcriptome profiling of An. gambiae and An. coluzzii larvae at two developmental stages (2nd and 4th instar), sampled from rural and urban habitats in and around Yaoundé, Cameroon that are exclusive to An. gambiae (rural sites) or An. coluzzii (urban sites) (Kamdem et al. 2012). Regardless of ontogenic stage, An. gambiae and An. coluzzii RNA samples showed differential gene expression in accord with our hypothesis—namely, elevation of transcripts implicated in immunity and detoxification in An. coluzzii relative to An. gambiae. Our results suggest that the successful exploitation of more stable but marginal larval breeding habitats by An. coluzzii may have been facilitated by adaptive transcriptional responses involving the upregulation of stress-responsive genes.
Materials and methods
Mosquito sampling
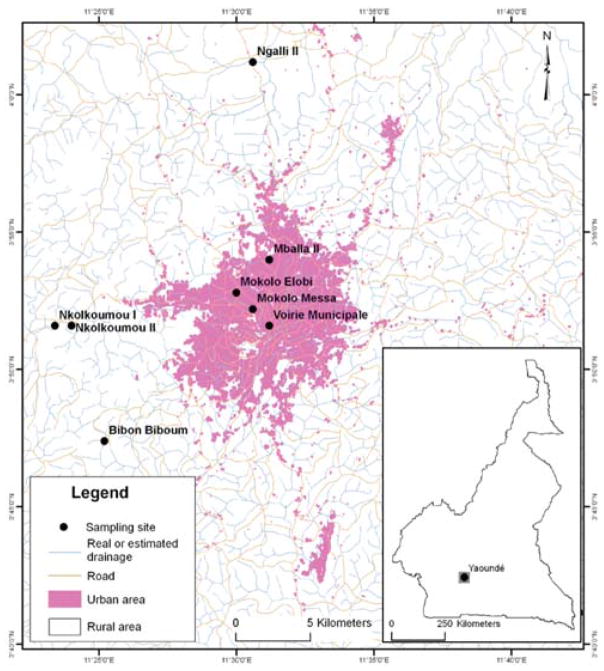
Collections were made in May 2009 from eight localities within a 50 × 50 km area encompassing Yaoundé, Cameroon (Figure 2; Table 1). In this area, intensive longitudinal larval and adult surveys of species composition conducted during one week monthly over the course of one year (May 2008 to April 2009) have revealed spatial segregation of An. coluzzii and An. gambiae along a gradient of urbanization (Kamdem et al. 2012). At opposite ends of the gradient, larval habitats were consistently exclusive to An. coluzzii (urban endpoint) or An. gambiae (rural endpoint). Based on these prior surveys, localities sampled during the present study (May 2009) at opposite ends of the urbanization gradient were expected to contain only one or the other species. The expected species composition at each locality was confirmed by molecular identifications performed individually on DNA extracted from a subset of 15 larvae, using an rDNA-based PCR assay (Santolamazza et al. 2004); all identifications agreed with expectation (Kamdem et al. 2012). All larval collections (and larval sacrifices for RNA sampling) were carried out at the same diel time to control for gene expression differences associated with photoperiod (Rund et al. 2011). At each locality, larvae were collected by dipper from multiple larval habitats.
Figure 2.
Sampling localities in and around Yaoundé, Cameroon. Four urban localities (pink shading) and four rural localities contain habitats exclusive to An. gambiae and An. coluzzii, respectively. Areas were shaded pink based on an index of urbanization (reflecting the proportion of built-up surface per spatial sampling unit), extracted from remotely sensed data (Kamdem et al. 2012).
Table 1.
Geographic coordinates of sampling localities in Yaoundé, Cameroon
| Description | Locality Name | Longitude; Latitude |
|---|---|---|
| Rural (An. gambiae) | Ngalli II | 11°30′36″E; 4°01′12″N |
| Nkolkoumou I | 11°23′38″E; 3°51′39″N | |
| Nkolkoumou II | 11°24′09″E; 3°51′44″N | |
| Bibon Biboum | 11°25′12″E; 3°47′24″N | |
| Urban (An. coluzzii) | Mballa II | 11°31′12″E; 3°54′00″N |
| Mokolo Elobi | 11°30′22″E; 3°52′49″N | |
| Mokolo Messa | 11°30′04″E; 3°52′21″N | |
| Voirie Municipale | 11°31′12″E; 3°51′36″N |
RNA sampling and processing
Individual larvae were killed at the intended sampling times by submersion in tubes containing 1.5 ml of RNAlater RNA stabilization reagent (QIAGEN), to preserve their RNA. All such tubes were transferred from Yaoundé to the University of Notre Dame, where they were stored at −20C prior to RNA extraction. Samples for RNA extraction consisted of 20–40 pooled larvae from the same locality, ontogenic stage, and treatment. Samples from the four rural (or urban) sampling locations (Figure 2; Table 1) were considered as four biological replicates. Total RNA was extracted using the RNeasy Mini Kit (QIAGEN). RNA integrity was assessed by electrophoresis of 1 μl aliquots on 1.5% agarose gels. Total RNA was treated with DNase I (Invitrogen) to remove any contaminating DNA, and concentrated. RNA was quantified using RiboGreen Assays (Molecular Probes/Invitrogen) and the SpectraMAX M2 microplate reader (Molecular Devices). RNA was amplified and converted into double-stranded cDNA using the TransPlex Whole Transcriptome Amplification Kit (Sigma-Aldrich). Quality and quantity of cDNA were assessed using the 2100 Bioanalyzer (Agilent).
Nimblegen microarray processing
Labeling and amplification of 1 μg cDNA using validated Cy3 dye randomers (TriLink) followed the standard sample labeling protocol (Roche Nimblegen). Using the Nimblegen Hybridization Kit (Roche Nimblegen), 6 μg of labeled cDNA was hybridized to 12 × 135 custom Nimblegen arrays designed from genebuild AgamP3.5 (Cassone et al. 2011). Each array interrogated 13,254 predicted AgamP3.5 genes, using at least five duplicated 60-mer probes per gene. Probes were chosen to interrogate gene regions in which there were no nucleotide differences between the species, based on their reference genome assemblies (Lawniczak et al. 2010). Arrays were washed using the Nimblegen Wash Buffer Kit and scanned in the UND Genomics Core Facility using the MS 200 Microarray Scanner (Roche NimbleGen). Array image data quality was assessed and raw fluorescence intensity values generated using NimbleScan v2.5 software (Roche NimbleGen). A total of four 12-plex chips, totaling 48 arrays (2 species × 2 ontogenic stages × 3 treatments × 4 replicates) were run (see Experimental design, below). Labeled cDNA samples from the different treatments and instar stages were evenly distributed across the 12-plex chips, and no batch effects were detected. Data from 3 arrays were omitted prior to analysis due to poor data quality (Table 2). The data have been deposited with Array Express under accession #E-MEXP-3147.
Table 2.
Experimental design for microarray-based transcriptional profiling of An. coluzzii and An. gambiae larvae from Yaoundé, Cameroon
| Species | Treatment | Instar | Replicates hybridized (analyzed) |
|---|---|---|---|
| An. coluzzii | None | 2nd | 4 |
| 4th | 4 (3) | ||
| 24 h auto-translocation | 2nd | 4 (3) | |
| 4th | 4 | ||
| 24 h allo-translocation | 2nd | 4 | |
| 4th | 4 | ||
| An. gambiae | None | 2nd | 4 |
| 4th | 4 | ||
| 24 h auto-translocation | 2nd | 4 (3) | |
| 4 | 4 | ||
| 24 h allo-translocation | 2nd | 4 | |
| 4th | 4 |
XYS files containing the raw expression intensity values were imported into Bioconductor (Gentleman et al. 2004). Using custom R scripts (Supplemetary File S1), a filter was applied to omit oligos that were misannotated or affected by physical imperfections, as previously described (Cassone et al. 2011). The oligo sets were further filtered to contain only those interrogating expressed genes, defined as having an average oligo set intensity value greater than 1500 in any two replicates for at least one condition (species x ontogenic stage x treatment). The final set of filtered expressed oligos was background adjusted, normalized, and summarized using the RMA function. Normalized oligo sets were converted to their respective An. gambiae gene ID based on the AgamP3.5 gene build, with oligo sets interrogating the same gene collapsed into a single value based on mean oligo intensity.
Experimental design and statistical analysis
Larval RNA samples—each with four biological replicates—were derived from two species (An. gambiae and An. coluzzii), two ontogenic stages (2nd and 4th instar), and three larval treatments (Table 2). Treatments refer to larval environmental conditions prior to sacrifice for RNA extraction. Larvae that were sacrificed immediately upon collection in the field were considered to be “untreated.” The other two treatments involve larvae that were transferred to the OCEAC insectary in Yaoundé, where they were cultured for 24 h under standard insectary conditions (27±2°C, 85±5% RH, and 12 h light/dark). The water in which they were cultured for 24 h was derived either from the larval habitats from which they were collected (a treatment referred to as “auto-translocation”) or was derived from larval habitats exclusive to the other species (a treatment referred to as “allo-translocation”).
Bayes-moderated t-tests in the limma package of R were used to examine gene expression differences between untreated 2nd- or 4th-instar An. coluzzii and An. gambiae larvae. Ontogenic stages were considered separately given profound transcriptomic changes during development (Koutsos et al. 2007). The significance threshold was defined at P<0.05 (FDR<0.25)(Benjamini & Hochberg 1995), as our aim was to explore functional enrichment rather than an assessment of individual genes. In addition, we set a minimum threshold log2-fold difference of 1.8 between species.
To assess whether short-term exposure to alternative water sources significantly impacted gene expression, we evaluated the 2nd and 4th instar larval samples cultured for 24 h in an “auto” or “allo” water source (excluding the untreated samples). For each instar stage, we conducted Bayes-moderated t-tests to assay treatment-specific differences. We ran two t-tests per larval stage that employed slightly different definitions of treatment. In one, treatment was defined as a 24 h exposure to water from breeding sites of the same or opposite species; the hypothesis was that exposure of either species to the opposite water source would perturb gene expression profiles relative to samples maintained in water from their normal breeding sites. In the other, treatment was defined as a 24 h exposure to water from An. coluzzii or An. gambiae larval habitats; here, the hypothesis was that exposure of either species to potentially polluted water in which An. coluzzii was breeding would perturb expression profiles relative those from samples exposed to water from relatively pristine An. gambiae habitats.
Functional enrichment of GO and other annotation terms in candidate gene lists was investigated for 2nd- and 4th-instar larvae using the DAVID functional annotation tool (Huang et al. 2009). The DAVID clustering module combines functionally related gene groups into annotation clusters. The clusters are assigned an enrichment score which represents the minus log-transformed geometric mean of the modified Fisher Exact (EASE) Scores within the cluster (Hosack et al. 2003; Huang et al. 2007). Significantly enriched annotation clusters were defined as those containing at least five genes and a minimum enrichment score of 1.3 (P<0.05).
Quantitative RT-PCR validation
Total RNA from untreated 2nd instar larvae of both species was also used for quantitative real-time reverse transcription PCR (qRT-PCR) to validate differential expression detected by microarray. For this purpose, single-stranded cDNA was synthesized from DNAse-treated total RNA using TaqMan Reverse Transcription Reagents (Applied Biosystems, Foster City, CA) with random hexamers instead of oligo d(T)16. A subset of four genes were tested that by microarray were up-regulated in An. coluzzii with a log2-fold difference of at least 2.5—(AGAP0)09728, -00424, -04433 and -09380 (Supplementary Table S1). Primers to these targets were designed using Primer Express 3.0 (Applied Biosystems, Foster City, CA) (Supplementary Table S1). Ribosomal protein S7 (AGAP010592) was the internal control reference gene for relative quantification, amplified in a separate tube using forward and reverse primers as given in Dong et al. (2006). After optimization, qRT-PCR reactions (15 μl) were performed in triplicate using SYBR Green PCR Master Mix (Applied Biosystems, Foster City, CA), 300 nm of each primer, and an AB7500 Real Time PCR System. After an initial step at 50C for 2 min, cycling conditions were 95C for 15 s, 60C for 1 min, and 95C for 15 s, for 40 cycles. Relative expression was calculated using the comparative CT method, after evaluating the kinetic real-time PCR efficiency of target and reference amplifications to ensure they were approximately equal (ABI user bulletin, http://docs.appliedbiosystems.com/pebiodocs/04303859.pdf). The qRT-PCR results confirmed the microarray data (Supplementary Table S1). The qRT-PCR data are available as Supplementary Table S2.
Reanalysis of Affymetrix array data
For reanalysis of the gene expression data of Cassone et al. (2008) based on the Affymetrix Anopheles/Plasmodium GeneChip, we restricted our consideration to the 4th instar larval samples, and performed two An. coluzzii-An. gambiae contrasts. In the first, we compared the An. coluzzii colony from Cameroon (An. coluzzii CAM, a derivative of the parental Yaoundé colony established from a district of Yaoundé) to its nearest available geographic counterpart, the An. gambiae Pimperena colony (established from Pimperena, Mali). In the second, two colonies from Mali were compared: the same An. gambiae Pimperena colony and An. coluzzii Mali-NIH. Using the BioMart tool from VectorBase (Megy et al. 2012), probes from the Affymetrix array were mapped onto the current AgamP3.7 annotation. Starting with the filtered, background adjusted, normalized and summarized 4th instar data, genes with a minimum threshold log2-fold difference of 1.8 between species were subject to Bayes-moderated t-tests to identify significant differential expression (P<0.05) for each contrast.
Comparison to differentiated regions of the genome
To ask whether differentially expressed genes also showed signals of nucleotide differentiation, we collected data on such genes from two previous studies: Neafsey et al. (2010) and Reidenbach et al. (2012). Significantly differentiated genes from Neafsey et al. were taken from the supplementary information of that paper, and significantly differentiated genes from Reidenbach et al. were supplied by the author. In order to determine whether there was a significant overlap between the genes found to be differentially expressed and those found to be differentiated at the nucleotide level, we used a binomial expectation where the probability of success was defined as the proportion of genes found to be differentially expressed with P<0.05. This expectation was used to determine the fraction of all genes with significant nucleotide differentiation that are also expected to show significant differential expression. P-values were calculated based on the observed overlap and the probability mass function of a binomial distribution with the given probability of success, calculated separately for each set of differentially expressed genes considered (e.g. 2nd instar, 4th instar, etc.).
Results
Of the 48 microarrays that were hybridized, 45 passed quality control filters (Table 2). Each microarray interrogated 13,254 genes (based on the AgamP3.5 gene build). Of these, 692 genes were eliminated from further consideration due to missing data (blank intensity values for one or more samples), and an additional 2,579 genes were omitted because they were not detected as expressed (see Methods). Expression patterns from the remaining 9,983 genes were compared between An. coluzzii and An. gambiae larvae sampled from natural breeding sites in and around Yaoundé, Cameroon.
Transcriptional divergence between An. coluzzii and An. gambiae larvae from natural populations
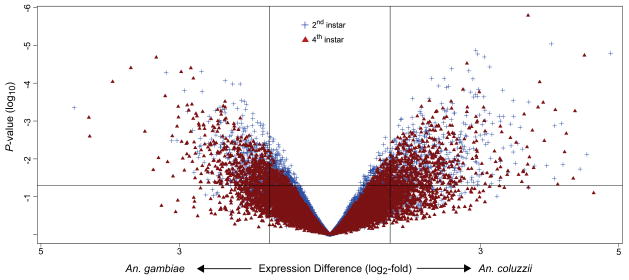
Initially, we examined differential gene expression between An. coluzzii and An. gambiae larvae that were sacrificed immediately upon sampling from their natural larval habitats. The results are summarized in a volcano plot (Figure 3). Of the 9,983 expressed genes, 8.9% (n = 890) differed significantly between species at the 2nd instar stage and displayed log2-fold-changes between 1.8 and 4.7 (Supplementary File S2). A comparable number of genes (957; 9.6%) were significantly differentially expressed between 4th instar larvae of An. coluzzii and An. gambiae with log2-fold-change between 1.8 and 4.4 (Supplementary File S2). The pattern of differential gene expression reflected a bias toward upregulation in An. coluzzii at both ontogenic stages, particularly in the younger (2nd instar) age class (Figure 3). At the log2-fold-change threshold of 1.8, the trend was marginal; approximately 56% (n = 499) and 52% (n = 501) of genes were more highly expressed in An. coluzzii larvae at the 2nd and 4th instar stage, respectively. However, this trend was more pronounced at higher log2-fold-change thresholds. For example, above a fold-change threshold of 2.0, 67% (n = 383) and 55% (n = 372) of genes in 2nd and 4th instar larvae were overexpressed in An. coluzzii. Increasing the threshold to 3.0, 96% (n = 48) and 75% (n = 45) of genes were more highly expressed in An. coluzzii.
Figure 3.
Volcano plot displaying gene expression differences (log2-fold change) between untreated larval samples of An. coluzzii and An. gambiae. Each symbol represents one gene that had detectable expression in either species, in samples of 2nd instar larvae (blue cross) or 4th instar larvae (red triangle). Relative differences in signal intensity along the X-axis reflect up-regulation in An. coluzzii when positive, and up-regulation in An. gambiae when negative; vertical lines indicate the log2-fold-change cut-off of +/−1.8. The Y-axis displays log10-transformed P-values associated with Bayes-moderated t-tests of differential gene expression; the horizontal line indicates the 0.05 threshold level for significance.
The DAVID data mining tool (Huang et al. 2009; Huang et al. 2007) was used to classify the genes differentially expressed between An. coluzzii and An. gambiae into functionally related groups (annotation clusters). For both ontogenic stages we partitioned candidate genes into two lists (up-regulated in An. coluzzii or An. gambiae) and mined each for annotation clusters with enrichment scores greater than 1.3 containing at least 5 genes (Figure 4).
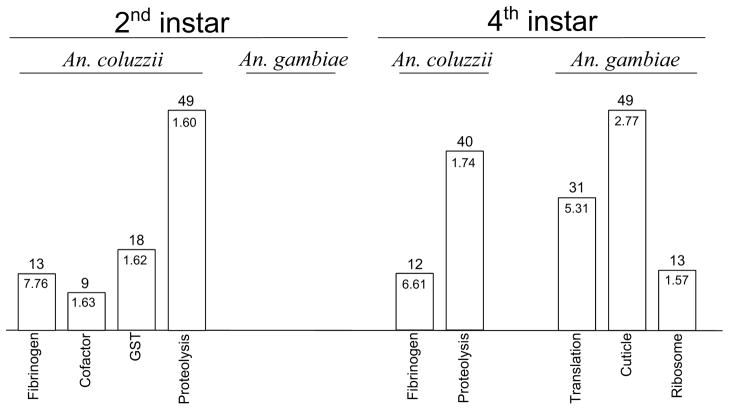
Figure 4.
Functional enrichment of genes that are over-expressed in field-collected samples of 2nd or 4th instar larvae of An. coluzzii and An. gambiae. The numbers of significant genes for each DAVID annotation cluster are indicated above each bar. Enrichment scores are displayed within each bar. GST, glutathione S transferase.
In An. coluzzii larvae at the 2nd instar, four significantly enriched annotation clusters were identified among the 499 genes significantly upregulated at this stage. All four clusters contained genes that potentially function in stress responses. In the cofactor metabolism cluster were nine genes, some of which are predicted to encode iron-sulfur or metallo-sulfur proteins with potential roles in an oxidative stress response. Also in this cluster were genes encoding enzymes from the citric acid cycle (TCA), including the pace-making enzyme citrate synthase, as well as aconitase and succinate dehydrogenase (AGAP0012048, -07852, and -00618, respectively). The glutathione S-transferase (GST) cluster contained 18 genes, among them GSTs whose products are considered vital for the detoxification of xenobiotic compounds (Board & Menon 2013). In addition, there were genes encoding a molecular chaperone and proteins responsible for protein turnover and removal of damaged and abnormal proteins, via the ubiquitin-proteasome system (a ubiquitin ligase, and components of the proteasome complex). The large proteolysis cluster also contained some genes that may function in protein turnover and removal. These include ubiquitin carriers, ubiquitin ligase, and ubiquitin hydrolases. Additionally, nine of the 49 genes in this annotation cluster are immune-related, including clip domain serine proteases (AGAP003250, -03251, -04719, -11789, -11791), serine protease inhibitors (AGAP001375, -03139, -09670) and an autophagy gene (AGAP004023). A connection to immune response is also hinted by the fibrinogen annotation cluster, which had an enrichment score of 7.76—far higher than any other clusters identified in the gene list from 2nd instar An. coluzzii larvae. Fibrinogen domain-containing proteins are required for coagulation in vertebrates, but in invertebrates they appear to play a role in defense against pathogens (Hanington & Zhang 2011). In this regard, it is intriguing that two eupolytin genes were overexpressed in 2nd instar An. coluzzii larvae (and three at the 4th instar stage). In the ground beetle Eupolyphaga sinensis, the eupolytin-1 gene encodes a protease that has been shown to hydrolyze fibrinogen and activate plasminogen (Yang et al. 2011); in An. coluzzii, cleavage of fibrinogen by eupolytin may function in the activation of these defense molecules. One other annotation cluster had a significant enrichment score of 1.33 (P<0.05), although it did not pass our filter requiring membership of at least five genes, and thus is not shown in Figure 4. The three genes in this cluster, identified by DAVID through their relationship to arginine and glutamine metabolism, encode enzymes that catalyze the last three steps in the urea cycle, responsible for the disposal of ammonia in many animal species. These are argininosuccinate synthase, argininosuccinate lyase, and arginase (AGAP003015, -08141, and -08783, respectively). It is noteworthy that in older (4th instar) An. coluzzii larvae, the only two significantly enriched annotation clusters identified matched two of the clusters from the 2nd instar stage (“proteolysis” and “fibrinogen”) and had similar enrichment scores, suggesting that these functions may be upregulated throughout larval development.
In contrast to An. coluzzii, functional annotation clustering of candidate genes upregulated in An. gambiae identified no significantly enriched functions in 2nd instar larvae, among the 391 upregulated genes. The functions enriched in 4th instar larvae among 456 upregulated genes—“translation”, “cuticle”, and “ribosome”—may relate to growth and development. Overall, there was no overlap of significantly enriched functions between An. gambiae and An. coluzzii.
The fact that two of the annotation clusters identified among upregulated genes in An. coluzzii at both ontogenic stages contain immune-related genes with potential roles in immune (or environmental stress) response prompted us to manually inspect the candidate gene lists for all genes assigned as immune-related in the reference genome assembly (Bartholomay et al. 2010; Waterhouse et al. 2007; Waterhouse et al. 2010), whether or not they were assigned to clusters of enriched annotation terms by DAVID. Of the 419 immune-related genes in the PEST (AgamP3.7) assembly (and updates to AgamP3.7 by R. Waterhouse, personal communication), 340 were detected as expressed in our experiments. Comparing 2nd instar larvae of An. coluzzii and An. gambiae, 46 were significantly differentially expressed, of which 41 (89%) were upregulated in An. coluzzii (Table 3). Although the pattern was not as pronounced in 4th instar larvae, of the 42 immune-related genes that were significantly differentially expressed between species, most (30; 71%) were upregulated in An. coluzzii (Table 3). The overrepresentation of upregulated immune genes in An. coluzzii is significant at both ontogenic stages [2nd instar, χ2 = 35.08 (1), P≪0.001; 4th instar, χ2 = 10.16 (1), P=0.001)].
Table 3.
Immune-related genes1 (AGAP last five digits) upregulated in field-collected An. coluzzii or An. gambiae larvae
| Immune Gene Family1 | An. coluzzii | An. gambiae | ||
|---|---|---|---|---|
| 2nd instar | 4th instar | 2nd instar | 4th instar | |
| Anti-Microbial Peptides | 06722* | |||
| Autophagy Genes | 04023 | 00933 | ||
| Glucan Binding Proteins | 04455 | 02798 | ||
| Caspases | 12544* | 12544* | ||
| Clip Domain Serine Proteases | 03250*; 03251; 04719; 11789; 11791 | 03250; 04719 | 09214* | 02270; 09214; 13184 |
| C-Type Lectins | 00871; 07409* | 00940; 02911 | 10193* | 07411; 10193*; 10196; |
| Fibrinogen-Related Proteins | 02005; 04916; 04917; 04918; 04996*; 04997*; 09184; 09728*; 10761; 10763*; 11197; 11223; 11225; 11239 | 04916; 04917; 04918; 05848*; 06914; 09728*; 10759; 10762*; 10811*; 11225; 11231; 11239 | 10772* | 10772*; 11277 |
| IMD Pathway | 06473 | 06473 | ||
| JAKSTAT Signal Transduction | 08354; 010423 | |||
| Leucine-Rich Repeat Proteins | 07033*; 07036* | 07039* | ||
| Lysozymes | 07344* | |||
| MD2-Like Receptors | 12352 | 01956; 02852; 07415 | ||
| Nimrod | 12386 | |||
| Peptidoglycan Recognition Proteins | 05552 | |||
| Peroxidases | 03714; 13327 | 10734 | ||
| Prophenoloxidases | 04975; 04981 | 04975* | ||
| Scavenger Receptors | 01375*; 03139; 04847; 09670 | |||
| Serine Protease Inhibitors | 01377 | |||
| Small RNA Regulatory Pathway | 11717* | 11537; 12523 | 09781 | |
| Thioester-Containing Proteins | 10815 | 10814 | ||
| Toll-Like Receptors | 06974* | 06974 | ||
Gene assignments based on (Bartholomay et al. 2010; Waterhouse et al. 2007; Waterhouse et al. 2010). Asterisks indicate genes upregulated 24 h after transfer to laboratory culture.
Short exposure to alternative aqueous environments has no significant impact on the larval transcriptome
Different transcriptional patterns between untreated larvae (i.e. sacrificed immediately upon sampling from native habitats) of An. coluzzii and An. gambiae may have been the result of a plastic response to distinctive abiotic (or biotic) features of their native larval habitats. In particular, the stress responsive genes overexpressed in An. coluzzii (e.g. immune-related genes, GSTs) may have been induced by the presence of xenobiotic chemicals or higher concentrations of organic waste such as ammonia in water from its larval habitat, pollutants that should be largely absent from the An. gambiae breeding sites that we sampled. None of the four sampling localities where An. coluzzii was breeding were in close proximity to areas used for intense urban farming (in contrast to, e.g., Nkolondom; Tene Fossog et al. 2013b), nor were the larval habitats created by the practice of agriculture (e.g. furrows), and thus the urban breeding sites were unlikely to be contaminated by insecticides commonly applied by farmers. However, urban larval breeding sites are frequently associated with domestic waste and organic pollutants (Antonio-Nkondjio et al. 2011). As an initial assessment of whether the observed transcriptional differences may have resulted from a plastic response by An. coluzzii to pollutant exposure, we performed a short-term semi-field translocation experiment.
Upon sampling from their native habitats, An. coluzzii and An. gambiae larvae at the 2nd and 4th instar stages were transported to the OCEAC insectary and reared for 24 h, either in water taken from their native habitats (auto-translocation), or in water taken from the habitat of the other species (allo-translocation), before being sacrificed and subject to global transcriptome analysis. We tested for significantly different patterns of gene expression by Bayes-moderated t-tests, repeating the test to cover two possible definitions of “treatment”: water source “auto” (native) versus “allo” (foreign), or water source from “coluzzii” versus “gambiae” larval habitat (see Methods). Strikingly, treatment—regardless of its definition and the ontogenic stage—showed no significant differential expression, with all genes having an FDR = 1. Our results suggest that, at least on the basis of the short (24 h) exposure measured in this experiment, transcriptional differences between species are not adequately explained by a plastic response to acute pollutant exposure in An. coluzzii larval habitats.
Transcriptome divergence between An. coluzzii and An. gambiae larvae is similar between field and laboratory samples
Previously, we measured transcriptional profiles between laboratory colonies of An. coluzzii and An. gambiae, including 4th instar larvae (Cassone et al. 2008). Comparison between those results and the present study is difficult, not only due to the genetic effects of colonization and relaxed selection under artificial rearing conditions, but also due to diverse geographic origins of the mosquitoes and technical differences between microarray platforms. The 2008 study was based on the Anopheles/Plasmodium Affymetrix GeneChip, which aside from inherent differences (such as probe length) relative to Nimblegen chips, was designed from the first An. gambiae genome assembly and annotation, and thus fails to interrogate numerous genes predicted in more recent annotations. Despite these important caveats, we reasoned that if basic trends in transcriptomic profiles are indeed inherent to species differences, and not merely a reflection of recent local adaptation or transcriptional plasticity, we should see these same general trends in contrasts between colonies as well as natural populations.
We began by comparing 4th instar transcriptomes of the An. coluzzii colony from Yaoundé, Cameroon (CAM) to its geographically nearest available An. gambiae counterpart, from Mali (Pimperena). Of the 529 genes significantly differentially expressed between 4th instar larvae of An. coluzzii CAM and An. gambiae Pimperena colonies above a log2-fold-change of 1.8 (Supplementary File S2), 291 (55%) were up-regulated in An. coluzzii CAM. As seen with field-collected larvae, increasing the fold-change threshold increased the proportion of genes that were upregulated in this species relative to An. gambiae Pimperena.
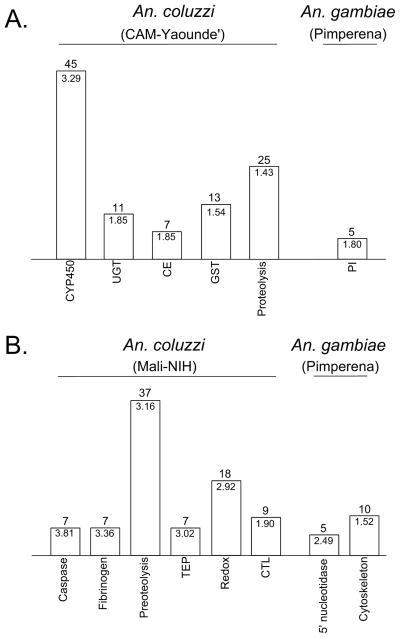
A functional enrichment analysis of the genes upregulated in An. coluzzii CAM larvae revealed six significant annotation clusters pointing to detoxification and proteolysis functions (Figure 5A; Supplementary File S2). Although genes clustered by these shared annotation terms do not necessarily all function in proteolysis or detoxification, the common enrichment of these themes in field and colony An. coluzzii larvae is striking. Of the six annotation clusters, four are associated with detoxification processes: an ion/metal-binding cluster containing numerous cytochrome P450s (CYP450s), superoxide dismutase and alkaline phosphatase (45 total genes, enrichment score 3.299), a transferase cluster containing five UDP-glucuronosyl transferases (UGTs) (11 genes, score 1.85), a carboxylesterase cluster (7 genes, score 1.85), and finally, a cluster featuring five GSTs and a chaperone involved in protein turnover (13 genes, score 1.54). The large proteolysis cluster included immune-related genes such as caspases, clip-domain serine proteases, and a serine protease inhibitor (25 genes, score 1.43). Indeed, the serine protease inhibitor gene SRPN2 was the most highly over-expressed gene detected in An. coluzzii CAM larvae (log2-fold change 7.88). As observed for field-collected An. coluzzii larvae, the CAM proteolysis cluster also contained eupolytin genes that may function in activation of fibrinogen. Notable for its absence was any detectable enrichment of fibrinogen genes among those significantly upregulated in 4th instar larvae from An. coluzzii CAM, although four fibrinogen genes were among those upregulated, showing large fold-changes ranging from 2.6 to 4.1 relative to An. gambiae Pimperena. Inspection of the probe sets present on the Affymetrix array revealed that 12 of 43 fibrinogen genes annotated in the current An. gambiae reference assembly are not interrogated by any probe on this outdated array, suggesting that the absence of enrichment for fibrinogen is inconclusive. Of note, an annotation cluster of C-type lectins that—like fibrinogens—mediate several invertebrate immune responses just missed our significance threshold (8 genes, score 1.2). Moreover, additional genes with potential roles in innate immunity were also over-expressed in An. coluzzii, although not clustered by DAVID: two thioester-containing proteins (TEP1, TEP9), gambicin (GAM1), defensin (DEF3), a glucan binding protein (GNBP2), a scavenger receptor (SCRBQ1), and an inhibitor of apoptosis (IAP7).
Figure 5.
Functional enrichment of genes that are overexpressed in 4th instar larvae from laboratory colonies of An. coluzzii and An. gambiae. A, An. coluzzii CAM from Yaoundé, Cameroon compared to An. gambiae Pimperena from Mali. B, Comparison of two colonies from Mali: An. coluzzii Mali-NIH and An. gambiae Pimperena. The numbers of significant genes for each DAVID annotation cluster are indicated above each bar. Enrichment scores are displayed within each bar. CYP450, cytochrome P450; UGT, UDP-glucuronosyltransferase; CE, carboxylesterase; GST, glutathione S transferase; PI, peptidase inhibitor; TEP, thioester-containing protein; Redox, oxidoreductase; CTL, C-type lectin.
In contrast to the strong enrichment of detoxification and proteolysis annotation terms among 291 genes upregulated in An. coluzzii CAM larvae, only one small annotation cluster involving peptidase inhibition was detected among 244 genes upregulated in An. gambiae Pimperena larvae (5 genes, enrichment score 1.8). Taken together, the transcriptional profiles of 4th instar larvae from CAM and Pimperena colonies are broadly similar to those measured from field-collected larvae: An. coluzzii larvae seem to be characterized by inherently higher levels of expression from classes of genes that may play a role in helping this species overcome the stress of more marginal habitats. This result is striking not merely because this trend seems to have persisted through the colonization process, but also because it has persisted for so long. The An. coluzzii CAM colony was derived from a parental Yaoundé colony established in 1988 (Tchuinkam et al. 1993), and in the 25 years since then (>250 generations) has been reared under benign laboratory conditions.
Population structure of An. gambiae across Africa appears to be quite shallow (Lehmann et al. 2003), but An. coluzzii populations are notably structured between the forest ecozone of Cameroon and West African savanna populations (Lee et al. 2009; Pinto et al. 2013; Slotman et al. 2007). Despite this, 4th instar larvae from the An. coluzzii Mali-NIH colony also overexpress immune defense genes—including fibrinogens—relative to their An. gambiae Pimperena counterparts (Figure 5B, Supplementary File S2), in keeping with our findings from natural populations in Cameroon. In addition to several fibrinogens, immune-related genes over-expressed in An. coluzzii Mali-NIH larvae included six caspases, several clip-domain serine proteases, C-type lectins, and five thioester-containing proteins (including TEP1). Among the genes in the enriched “Proteolysis” annotation cluster were four CYP450s. By contrast, and consistent with our results from other contrasts between colonies and population samples from this pair of species, only two annotation clusters were enriched in the set of genes overexpressed in An. gambiae Pimperena, and these had no obvious relationship to stress response, neither immune defense nor detoxification.
Discussion
The results of ecological niche modeling and distribution mapping of An. gambiae and An. coluzzii in both West and Central Africa are consistent with a process of ecological speciation, in which the more recently derived taxon, An. coluzzii, has undergone niche expansion into anthropogenic larval habitats of marginal quality, promoted by larval competition in the original, optimal habitat (Costantini et al. 2009; Simard et al. 2009). In the savanna, An. coluzzii habitats are irrigated rice fields, which would have been a notable landscape feature only after the domestication of African rice from a wild ancestor within the last 2,000–3,000 years (Li et al. 2011; Porteres 1976). Unlike the temporary pools and puddles exploited by An. gambiae, rice fields are relatively stable, biotically complex and rich in a notable environmental stressor: predators. The ecological variables associated with larval habitat segregation in the rainforests of Cameroon differ, but relatively recent niche expansion by An. coluzzii into a more stable larval habitat of lower (marginal) quality appears to be a common denominator. In the equatorial forest region, An. coluzzii is associated with densely populated urban centers such as Douala and Yaoundé (Antonio-Nkondjio et al. 2011; Kamdem et al. 2012), whose explosive growth—characterized by inferior housing and poor urban planning, sanitation and drainage of surface water (Antonio-Nkondjio et al. 2011)—is a recent phenomenon in the decades since the end of World War II and independence (Franqueville 1984). In these cities, An. coluzzii breeds in both temporary and permanent water bodies that, while largely free from top predators (larvivorous fish), may contain variable amounts of other biotic and abiotic environmental stressors, such as competition from Culex mosquito larvae and exposure to pollutants from decaying organic matter, household and industrial waste, or insecticide run-off from urban agriculture (Antonio-Nkondjio et al. 2011). Urban breeding sites vary greatly in water quality and other physicochemical characteristics, but on average they contain 2- to 100-fold higher concentrations of ammonia, nitrates, phosphate, sulfate, and other chemicals, have higher turbidity, conductivity and total hardness, and tend to be more alkaline than rural breeding sites (Antonio-Nkondjio et al. 2011; Tene Fossog et al. 2013a). Particularly in the dry season when suitable temporary and less polluted larval habitats become scarce, An. coluzzii can occupy large organically polluted breeding sites in association with competing larvae of Culex species, mainly Cx. quinquefasciatus (Antonio-Nkondjio et al. 2011; Kamdem et al. 2012).
The capacity of An. coluzzii to invade and exploit newly available anthropogenic habitats of poorer quality, both in the rural savanna and urbanized equatorial forest, prompted the hypothesis that this taxon has rapidly, and perhaps repeatedly, evolved a tolerance to stressors in the environment, mediated by altered gene expression. Our transcriptome profiling of larvae sampled from the field in Cameroon and from laboratory colonies is consistent with this prediction. Differential gene expression between species generally reflected overexpression in An. coluzzii relative to An. gambiae, and functional annotations enriched among upregulated genes included detoxification and immune defense. Between natural populations from Yaoundé, gene classes overrepresented in An. coluzzii included GSTs, serine proteases, serine protease inhibitors, and a wide spectrum of other immune-related genes, notably fibrinogens. Between laboratory colonies not subject to the same environmental stresses, immune-related and detoxification gene classes were similarly over-expressed. Previous studies have shown that short-term (as little as 24 h) exposure of susceptible mosquito larvae to sub-lethal doses of various xenobiotics (heavy metal, herbicide, or insecticide) induces overproduction of transcripts encoding detoxification enzymes that can confer larval tolerance and potentially cross-tolerance (David et al. 2010; Poupardin et al. 2008). However—although not definitive—our data suggest that the upregulation of detoxification and immune gene expression observed in An. coluzzii was unlikely to have been a transient (plastic) response to xenobiotics present in water from the larval breeding habitat. First, reciprocal translocation for 24 h in water from larval habitats of the opposite species had no impact on gene expression patterns of either species. Physicochemical data were not recorded from the sampled breeding sites during our experiments; however, such data have been collected previously from a variety of larval habitats in and around Yaoundé, and significant differences were few (Antonio-Nkondjio et al. 2011; Tene Fossog et al. 2013a; Tene Fossog et al. 2012). Importantly, none of the larval habitats sampled for this study were near cultivated areas, thus contamination with insecticides used in farming was very unlikely. Moreover, had the presence of other pollutants in An. coluzzii habitats been directly responsible for inducing gene upregulation in this species, our expectation would be that exposure of An. gambiae larvae to water from these habitats should have resulted in upregulation of stress-responsive genes in this species (assuming that 24 h was a sufficiently long exposure), yet this was not observed. Second, the expression profiles of wild and colonized An. coluzzii from Yaoundé were broadly similar, despite laboratory culture of the Yaoundé colony since 1988 (Tchuinkam et al. 1993). Thus, our working hypothesis is that the higher expression of detoxification and immune defense genes in An. coluzzii relative to An. gambiae reflects a constitutive increase in basal expression levels, as observed recently for immune activation genes in Aedes aegypti populations refractory to Dengue virus (Sim et al. 2013). More definitive evidence for this working hypothesis could come from common garden experiments in the laboratory, using the F1 progeny of field-collected females of both species.
To the extent that expression of detoxification and immune defense genes is inherently higher in An. coluzzii relative to An. gambiae, it may be an adaptive response to environmental conditions that have been repeatedly encountered. If this is the case, it is important to stress that there is a dearth of information to support inferences about the ultimate sources of selective pressures acting on An. coluzzii historically, as so little is known of its biogeographic history, ecology, or behavior. One obvious recent (and potentially confounding) factor, certainly the one that has commanded the most attention, is selection by insecticides used to control agricultural pests. Through runoff, insecticides intended to target pests of crops also contaminate the furrows and irrigation ditches associated with agriculture (“cultivated sites”) where mosquitoes can breed. The emergence of insecticide resistance in non-target insects such as mosquitoes has long been recognized (Lines 1988). With respect to the malaria vectors An. gambiae, An. coluzzii, and An. arabiensis (another sibling species), this association has been observed not only with traditional agriculture (Diabate et al. 2002; Muller et al. 2008), but also with the recent and rapidly expanding practice of intensive urban agriculture in African cities (Antonio-Nkondjio et al. 2011; Jones et al. 2012; Yadouleton et al. 2009). Urban An. coluzzii populations are resistant to multiple insecticide classes, and an important mechanism of insecticide tolerance appears to be metabolic, mediated by enzyme-based detoxification (Djouaka et al. 2008; Nwane et al. 2013; Oduola et al. 2012).
Previous microarray analyses of DDT-resistant or permethrin-resistant adult An. coluzzii, collected as larvae from a variety of breeding sites in urban and agricultural areas of southern Benin, Nigeria, and Cameroon, revealed over-expression of CYP450s, GSTs, carboxylesterases, and UGTs (Djouaka et al. 2008; Tene Fossog et al. 2013b). The association of over-expressed detoxification genes with insecticide resistance implicates these genes in metabolic resistance, but as only a handful of individual genes have been shown to metabolize insecticides and many of them may have unrelated functions, caution is merited and firm conclusions await direct experimental validation. Table 4 presents a compilation of the detoxification genes found in the present study to be over-expressed in field-collected or colonized An. coluzzii larvae. It is noteworthy that only one CYP450 (CYP325C2, over-expressed in 4th instar field-collected larvae) has been associated previously with insecticide resistance in An. coluzzii (Tene Fossog et al. 2013b), and of all the detoxification genes over-expressed in our field collections, only one (GSTE2) has been validated to metabolize DDT and permethrin (Daborn et al. 2012; Ranson et al. 2001). The transcriptional profile of the field-collected An. coluzzii samples does not appear to match that expected for a highly resistant strain (in contrast to the An. coluzzii CAM colony, which over-expresses CYP6P3 and CYP6M2, among others). We did not test our larval samples for insecticide resistance, but two recent surveys conducted in Cameroon (in Yaoundé and the coastal city of Douala) reported insecticide resistance bioassays on mosquitoes collected from cultivated sites or polluted/non-polluted larval habitats away from cultivated sites. A panel of insecticides of different classes were applied, either to larvae (Tene Fossog et al. 2012) or to emerged adults (Antonio-Nkondjio et al. 2011). Importantly, both studies identified substantial heterogeneity in levels of resistance to DDT, permethrin, deltamethrin, and bendiocarb, based on the type of larval habitat. In good agreement with a previous survey of permethrin resistance in Benin and Nigeria (Djouaka et al. 2008), the level of insecticide resistance to diagnostic concentrations was much higher in mosquitoes collected from urban agricultural sites versus other urban sites polluted (or not) with organic waste or petroleum products—even if the cultivated versus non-cultivated larval habitats were separated by less than 10 km. Taken together, these data suggest that agricultural insecticides play a very important selective role, affecting all An. coluzzii in Yaoundé to some degree, even those derived from uncultivated breeding sites. However, it is not clear that insecticides are the only selective factor, or more importantly, the initial driver responsible for overexpression of metabolic detoxification genes in An. coluzzii. Alternative environmental factors, biotic and/or abiotic, may have played supporting or even primary roles (Nkya et al. 2013).
Table 4.
Detoxification genes (AGAP last 5 digits) over-expressed in field-collected and colonized An. coluzzii larvae
| Gene class | Field-collected 2nd instar | Field-collected 4th instar | CAM 4th instar | Mali-NIH 4th instar |
|---|---|---|---|---|
| CYP450 | CYP301A1 (06082), CYP9J5 (12296) | CYP325C2 (02205), CYP325B1 (02210), CYP4D16 (13241) | CYP9K1 (00818), CYP6AA1 (02862), CYP6P3 (02865), CYP6P4 (02867). CYP6P1 (02868), CYP6S1 (08204), CYP6M1 (08209), CYP6M2 (08212), CYP6M4 (08214), CYP6Z3 (08217), CYP6Z2 (08218), CYP4H17 (08358), CYP4H27 (08552), CYP9J5 (12296) | CYP6M2 (08212), CYP4C36 (09241), CYP6M4 (08214), CYP12F1 (08022) |
| CE | COEBE4C (05370), COEBE2C (05371), COEBE3C (05372), COEAE3D (05758) | COEAE1D (05756) | COEAE1A (01723), COEAE3D (05758), COEJHE5E (05837), COE13O (11507) | --- |
| GST | GSTD10 (04383), GSTE5 (09192), GSTE2 (09194), GSTE1 (09195), GSTE3 (09197) | GSTE2 (09194), GSTE1 (09195) | GSTT2 (00888), GSTD11 (04378), GSTD6 (04379), GSTD12 (04380), GSTD3 (04382) | GSTT2 (00888), GSTD11 (04378) |
| UGT | UGT5 (02783) | UGT5 (02783) | UGT (06222), UGT (07028), UGT (07029), UGT (11564) | --- |
CYP450, cytochrome P450; CE, carboxylesterase; GST, glutathione S transferase; UGT, UDP-glucuronosyltransferase. Genes underlined and bolded have been validated to metabolize insecticide (Daborn et al. 2012; David et al. 2013; Ranson et al. 2001); those in bold have been associated with insecticide resistance (David et al. 2013; Djouaka et al. 2008; Tene Fossog et al. 2013b)
A second selective factor that may play a role in transcriptional divergence between An. coluzzii and An. gambiae, especially in detoxification genes, could be the toxic effects of ammonia, a byproduct of the breakdown of organic waste present in polluted urban larval habitats. Indeed, in a previous study we measured higher levels of ammonia in such habitats, and higher tolerance of ammonia by An. coluzzii larvae breeding in those habitats in urban Yaoundé, relative to An. gambiae breeding in pristine larval habitats in the rural outskirts of Yaoundé (Tene Fossog et al. 2013a). Disposal of dietary ammonia is a major and universal problem faced by all adult female mosquitoes during digestion of the proteinaceous blood meal, and by larvae that ingest excess protein relative to their growth requirements (Scaraffia et al. 2005; von Dungern & Briegel 2001; Weihrauch et al. 2012). Mosquitoes utilize multiple metabolic routes for the disposal of nitrogenous waste, including the synthesis of urea catalyzed by arginase (Isoe & Scaraffia 2013; Scaraffia et al. 2005; Scaraffia et al. 2008; von Dungern & Briegel 2001), one of three enzymes from the urea (ornithine) cycle found to be significantly overexpressed in An. coluzzii larvae in this study. However, because the typical urea cycle is apparently absent in mosquitoes due to lack of the ornithine carbamoyltransferase gene (Zdobnov et al. 2002), this pathway has been considered non-functional for ammonia disposal (Scaraffia et al. 2005). Instead, environmental ammonia ingested by larvae may be transported and directly excreted from the Malpighian tubules, hindgut, and anal papillae (Donini & O’Donnell 2005; Weihrauch et al. 2012). Neither putative ammonium transporter proteins nor aquaporins were detected as overexpressed in An. coluzzii. However, V-type H+-ATPase (V-ATPase) is known to play a crucial role in transepithelial ammonia transport in many systems (Weihrauch et al. 2012). In both 2nd and 4th instar samples of An. coluzzii, the V-ATPase 21kDa subunit (AGAP009334) was significantly over-expressed by log2-fold change of ~3. Consistent with a role for this V-ATPase in excretion, MozAtlas (Baker et al. 2011) reports statistically significant enrichment of its expression in the adult Malpighian tubules. These observations merit more detailed follow-up study.
A third selective factor potentially contributing to transcriptional divergence between An. coluzzii and An. gambiae may be differential exposure to pathogenic fungi, bacteria, and parasites found in their distinctive larval breeding sites. In a previous study employing genome scans of divergence between natural populations of these species, we uncovered a region of exceptional divergence on chromosome 3L in samples from West Africa, and implicated the TEP1 gene, whose complement C3-like product has antiparasitic and antibacterial activity (White et al. 2011). DNA sequence analysis of these population samples suggested that an allelic variant of TEP1 unique to An. coluzzii had swept to fixation in Mali and Burkina Faso, and was spreading into neighboring Ghana. This allele, closely related but distinct from previously characterized TEP1 resistance alleles, conferred resistance to Plasmodium parasites that invade and replicate within the adult female mosquito (White et al. 2011). However, given the relatively low rates and intensities of Plasmodium infection in nature for both An. coluzzii and An. gambiae adult female populations, we speculated that the most likely source of pathogen-mediated selection for resistance came from the An. coluzzii larval habitat—a longer-lasting and more biotically diverse aquatic milieu than that exploited by An. gambiae, thus presumably harboring distinctive pathogen populations. Although the particular West African resistance allele described in White et al (2011) is apparently absent from Central Africa, the TEP1 gene was over-expressed in our 2nd instar samples of An. coluzzii from Cameroon (log2-fold change 2.4), together with a plethora of other immune-related genes, most notably fibrinogens (Table 3, Supplementary File S2). The TEP1 gene was also the second-ranked over-expressed gene in 4th instar larvae from the CAM colony (compared to An. gambiae Pimperena; log2-fold change 7.1) and the top-ranked over-expressed gene in the Mali-NIH colony (compared to An. gambiae Pimperena; log2-fold change 8.8), closely followed by four other TEP genes (log2-fold changes from 6.1-4.8 in Mali-NIH) and two C-type lectins (log2-fold changes from 6.1-4.0 in Mali-NIH). A non-coincidental relationship between immune and stress responses has recently been highlighted in the model organism Drosophila (Davies et al. 2012). Emerging evidence from the fruitfly suggests that apparently unrelated stresses (e.g. immune, oxidative, osmotic, xenobiotic) are both sensed and countered through overlapping cellular responses mediated by the Malpighian tubule, the insect equivalent of the vertebrate kidney and liver (Davies et al. 2012). If this immune and stress “cross talk” applies more broadly to mosquitoes as expected, it may provide a conceptual framework for understanding the functional enrichment of both immune-related and detoxification genes among those over-expressed in An. coluzzii.
A previous study of transcriptional divergence between An. coluzzii and An. gambiae was conducted on adults that emerged in the OCEAC insectary from larval collections made at two localities 10 km apart in Yaounde (Nkolbisson) and on its outskirts (Nkolondom) (Aguilar et al. 2010). These localities differ from those sampled in the present study in that both, particularly Nkolondom, are sites of urban farming (e.g. cultivated sites where larval habitats are subject to contamination by insecticides). Both An. coluzzii and An. gambiae were collected at each location, although they were present in highly skewed proportions. Based on competitive hybridizations to an Agilent glass slide microarray, the authors reported that transcriptional divergence between the same species across the two localities was ten times greater than transcriptional divergence between different species across the same two localities. This conclusion apparently contradicts our findings in the present study, but the source of the discrepancy is difficult to discern, not least because these previous microarray data are not in a public database. However, the basis of their conclusion seems to rest on the observation that 61 transcripts were in common between different species across sites, while only 6 transcripts were in common between samples of the same species across sites (Aguilar et al. 2010), a finding that may not be robust. Possible factors contributing to the different conclusions may be: probes on the Agilent array that do not interrogate the same genes as our Nimblegen array; technical differences between platforms; differences in statistical methods, analytical approach, and data interpretation; differences in life stage analyzed; and differences in sampling localities (cultivated versus uncultivated). Two common findings between the studies are that in the urban Yaoundé locality, An. coluzzii shows a higher proportion of over-expressed genes, and those genes are enriched for immune functions.
Given our results on the suite of genes that are differentially expressed between An. coluzzii and An. gambiae—genes that are presumably evolving in response to environmental stressors—we asked whether these same genes were identified in previous genome scans for regions under recent positive selection. In our earlier study of gene expression in laboratory colonies (Cassone et al. 2008), we found a slight enrichment for differentially expressed genes in the pericentromeric region of the X chromosome, a large area that had been identified as under selection by some of the first genome scans in An. gambiae (Stump et al. 2005; Turner & Hahn 2007; Turner et al. 2005). However, using two more recent, high-resolution studies of nucleotide differentiation between these two species (Neafsey et al. 2010; Reidenbach et al. 2012), we found no significant enrichment for differentially expressed genes among those genes in windows of high population divergence (P>0.10 for all sets of differentially expressed genes, for comparisons to both datasets). We also repeated these analyses using the genes adjacent to the previously identified targets of natural selection, to account for the possibility that cis-acting changes were not located immediately within the genes they regulated. Again, we found no significant enrichment for differentially expressed genes. Even if differences in gene expression between the two species are driven by natural selection, the mutations responsible for regulating such differences may occur in the coding regions of genes that show no differential expression (i.e. trans-acting regulators). Therefore, we would only expect a significant overlap between these two types of data if most differences are driven by cis-regulation (cf. Fay & Wittkopp 2008). Our results suggest that this is not the case.
Adaptation in the mosquito species considered here may be representative of range expansions in other systems. For instance, Drosophila mojavensis is a cactophilic species of the North American desert, comprised of four recently diverged (<0.5 MYA) subspecies adapted to different cactus hosts with differing sets of allelochemicals (Matzkin 2014, and refs therein). Similar to our findings, transcriptional changes in detoxification genes appear to be associated with cactus host shifts—including those known to confer resistance to DDT (e.g. GSTE2) (Matzkin 2012; Matzkin & Markow 2013). However, as was also true for the D. mojavensis transcriptomic analysis, the fraction of detoxification (and in our case, immune) genes among all differentially expressed genes is small, and it is likely that the adaptive divergence of An. coluzzii and An. gambiae involved selection on many additional genes. Although the designation of An. coluzzii and An. gambiae as separate species may be controversial due to ongoing hybridization and introgression, known differences between them at the larval stage (in preferred larval habitat, ammonia tolerance, development time, and predator avoidance behavior) and at the adult stage (in mate recognition and swarming behaviors) (Dabire et al. 2013; Diabate et al. 2009; Gimonneau et al. 2010; Gimonneau et al. 2012a; Gimonneau et al. 2012b; Kamdem et al. 2012; Lehmann & Diabate 2008; Pennetier et al. 2010; Roux et al. 2013; Tene Fossog et al. 2013a) suggest that their divergence involved multifarious selection on multiple genetic targets that presumably include chemosensory genes and regulators of development. Even for closely related species with few morphological differences, these examples suggest that there can—or must—be multiple adaptive differences between them in order to maintain niche differentiation and ecological specialization.
Supplementary Material
Acknowledgments
We thank R. Waterhouse for providing an updated set of immune-related genes, D. Lawson for assistance with VectorBase, I. Morlais for providing information about OCEAC colonies, and three anonymous reviewers. We are grateful to the OCEAC Malaria Research Laboratory staff for excellent technical and field support. Financial support was provided by NIH grant R01 AI63508 to NJB.
Footnotes
Data accessibility
Microarray data have been deposited with Array Express under accession E-MEXP-3147.
Author contributions
N.J.B., C. Costantini, and M.W.H. designed the study and obtained funding; C.K. and C. Costantini conducted field work; B.J.C. and J.C.T. performed molecular work; B.J.C., C. Cheng, M.W.H. and N.J.B. analyzed data; B.J.C., M.W.H. and N.J.B. wrote the paper.
References
- Aguilar R, Simard F, Kamdem C, et al. Genome-wide analysis of transcriptomic divergence between laboratory colony and field Anopheles gambiae mosquitoes of the M and S molecular forms. Insect Molecular Biology. 2010;19:695–705. doi: 10.1111/j.1365-2583.2010.01031.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonio-Nkondjio C, Fossog BT, Ndo C, et al. Anopheles gambiae distribution and insecticide resistance in the cities of Douala and Yaounde (Cameroon): influence of urban agriculture and pollution. Malar J. 2011;10:154. doi: 10.1186/1475-2875-10-154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker DA, Nolan T, Fischer B, et al. A comprehensive gene expression atlas of sex- and tissue-specificity in the malaria vector, Anopheles gambiae. BMC Genomics. 2011;12:296. doi: 10.1186/1471-2164-12-296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartholomay LC, Waterhouse RM, Mayhew GF, et al. Pathogenomics of Culex quinquefasciatus and meta-analysis of infection responses to diverse pathogens. Science. 2010;330:88–90. doi: 10.1126/science.1193162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate – A practical and powerful approach to multiple testing. J Roy Stat Soc B. 1995;57:289–300. [Google Scholar]
- Bickford D, Lohman DJ, Sodhi NS, et al. Cryptic species as a window on diversity and conservation. Trends in ecology & evolution. 2007;22:148–155. doi: 10.1016/j.tree.2006.11.004. [DOI] [PubMed] [Google Scholar]
- Board PG, Menon D. Glutathione transferases, regulators of cellular metabolism and physiology. Biochimica et Biophysica Acta. 2013;1830:3267–3288. doi: 10.1016/j.bbagen.2012.11.019. [DOI] [PubMed] [Google Scholar]
- Borneman AR, Gianoulis TA, Zhang ZD, et al. Divergence of Transcription Factor Binding Sites Across Related Yeast Species. Science. 2007;317:815–819. doi: 10.1126/science.1140748. [DOI] [PubMed] [Google Scholar]
- Britten RJ, Davidson EH. Gene regulation for higher cells: a theory. Science. 1969;165:349–357. doi: 10.1126/science.165.3891.349. [DOI] [PubMed] [Google Scholar]
- Britten RJ, Davidson EH. Repetitive and non-repetitive DNA sequences and a speculation on the origins of evolutionary novelty. The Quarterly review of biology. 1971;46:111–138. doi: 10.1086/406830. [DOI] [PubMed] [Google Scholar]
- Cassone BJ, Molloy MJ, Cheng C, et al. Divergent transcriptional response to thermal stress by Anopheles gambiae larvae carrying alternative arrangements of inversion 2La. Molecular Ecology. 2011;20:2567–2580. doi: 10.1111/j.1365-294X.2011.05114.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassone BJ, Mouline K, Hahn MW, et al. Differential gene expression in incipient species of Anopheles gambiae. Molecular Ecology. 2008;17:2491–2504. doi: 10.1111/j.1365-294X.2008.03774.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chesson P. Mechanisms of maintenance of species diversity. Annual Review of Ecology and Systematics. 2000;31:343–366. [Google Scholar]
- Coetzee M, Hunt RH, Wilkerson R, et al. Anopheles coluzzii and Anopheles amharicus, new members of the Anopheles gambiae complex. Zootaxa. 2013;3619:246–274. [PubMed] [Google Scholar]
- Costantini C, Ayala D, Guelbeogo WM, et al. Living at the edge: biogeographic patterns of habitat segregation conform to speciation by niche expansion in Anopheles gambiae. BMC Ecol. 2009;9:16. doi: 10.1186/1472-6785-9-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dabire KR, Sawadodgo S, Diabate A, et al. Medical and Veterinary Entomology. 2013. Assortative mating in mixed swarms of the mosquito Anopheles gambiae s.s. M and S molecular forms, in Burkina Faso, West Africa. [DOI] [PubMed] [Google Scholar]
- Daborn PJ, Lumb C, Harrop TW, et al. Using Drosophila melanogaster to validate metabolism-based insecticide resistance from insect pests. Insect Biochemistry and Molecular Biology. 2012;42:918–924. doi: 10.1016/j.ibmb.2012.09.003. [DOI] [PubMed] [Google Scholar]
- David JP, Coissac E, Melodelima C, et al. Transcriptome response to pollutants and insecticides in the dengue vector Aedes aegypti using next-generation sequencing technology. BMC Genomics. 2010;11:216. doi: 10.1186/1471-2164-11-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David JP, Ismail HM, Chandor-Proust A, Paine MJ. Role of cytochrome P450s in insecticide resistance: impact on the control of mosquito-borne diseases and use of insecticides on Earth. Philosophical transactions of the Royal Society of London Series B, Biological sciences. 2013;368:20120429. doi: 10.1098/rstb.2012.0429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies SA, Overend G, Sebastian S, et al. Immune and stress response ‘cross-talk’ in the Drosophila Malpighian tubule. Journal of insect physiology. 2012;58:488–497. doi: 10.1016/j.jinsphys.2012.01.008. [DOI] [PubMed] [Google Scholar]
- della Torre A, Fanello C, Akogbeto M, et al. Molecular evidence of incipient speciation within Anopheles gambiae s.s. in West Africa. Insect Molecular Biology. 2001;10:9–18. doi: 10.1046/j.1365-2583.2001.00235.x. [DOI] [PubMed] [Google Scholar]
- della Torre A, Tu ZJ, Petrarca V. On the distribution and genetic differentiation of Anopheles gambiae s.s. molecular forms. Insect Biochemistry and Molecular Biology. 2005;35:755–769. doi: 10.1016/j.ibmb.2005.02.006. [DOI] [PubMed] [Google Scholar]
- Diabate A, Baldet T, Chandre F, et al. The role of agricultural use of insecticides in resistance to pyrethroids in Anopheles gambiae s.l. in Burkina Faso. American Journal of Tropical Medicine and Hygiene. 2002;67:617–622. doi: 10.4269/ajtmh.2002.67.617. [DOI] [PubMed] [Google Scholar]
- Diabate A, Dabire KR, Heidenberger K, et al. Evidence for divergent selection between the molecular forms of Anopheles gambiae: role of predation. BMC Evol Biol. 2008;8:5. doi: 10.1186/1471-2148-8-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diabate A, Dabire RK, Kengne P, et al. Mixed swarms of the molecular M and S forms of Anopheles gambiae (Diptera: Culicidae) in sympatric area from Burkina Faso. Journal of Medical Entomology. 2006;43:480–483. doi: 10.1603/0022-2585(2006)43[480:msotmm]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Diabate A, Dabire RK, Kim EH, et al. Larval development of the molecular forms of Anopheles gambiae (Diptera: Culicidae) in different habitats: a transplantation experiment. Journal of Medical Entomology. 2005;42:548–553. doi: 10.1093/jmedent/42.4.548. [DOI] [PubMed] [Google Scholar]
- Diabate A, Dabire RK, Millogo N, Lehmann T. Evaluating the effect of postmating isolation between molecular forms of Anopheles gambiae (Diptera: Culicidae) Journal of Medical Entomology. 2007;44:60–64. doi: 10.1603/0022-2585(2007)44[60:eteopi]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Diabate A, Dao A, Yaro AS, et al. Spatial swarm segregation and reproductive isolation between the molecular forms of Anopheles gambiae. Proc Biol Sci. 2009;276:4215–4222. doi: 10.1098/rspb.2009.1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djouaka RF, Bakare AA, Coulibaly ON, et al. Expression of the cytochrome P450s, CYP6P3 and CYP6M2 are significantly elevated in multiple pyrethroid resistant populations of Anopheles gambiae s.s. from Southern Benin and Nigeria. BMC Genomics. 2008;9:538. doi: 10.1186/1471-2164-9-538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Y, Aguilar R, Xi Z, et al. Anopheles gambiae immune responses to human and rodent Plasmodium parasite species. PLoS Pathog. 2006;2:e52. doi: 10.1371/journal.ppat.0020052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donini A, O’Donnell MJ. Analysis of Na+, Cl−, K+, H+ and NH4+ concentration gradients adjacent to the surface of anal papillae of the mosquito Aedes aegypti: application of self-referencing ion-selective microelectrodes. The Journal of experimental biology. 2005;208:603–610. doi: 10.1242/jeb.01422. [DOI] [PubMed] [Google Scholar]
- Fay JC, Wittkopp PJ. Evaluating the role of natural selection in the evolution of gene regulation. Heredity. 2008;100:191–199. doi: 10.1038/sj.hdy.6801000. [DOI] [PubMed] [Google Scholar]
- Franqueville A. Collection Mémoires. Vol. 104. ORSTOM; Paris: 1984. Yaoundé, construire une capitale. [Google Scholar]
- Gentleman RC, VJC, DMB, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimonneau G, Bouyer J, Morand S, et al. A behavioral mechanism underlying ecological divergence in the malaria mosquito Anopheles gambiae. Behav Ecol. 2010;21:1087–1092. doi: 10.1093/beheco/arq114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimonneau G, Pombi M, Choisy M, et al. Larval habitat segregation between the molecular forms of the mosquito, Anopheles gambiae in a rice field area of Burkina Faso, West Africa. Medical and Veterinary Entomology. 2012a;26:9–17. doi: 10.1111/j.1365-2915.2011.00957.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimonneau G, Pombi M, Dabire RK, et al. Behavioural responses of Anopheles gambiae sensu stricto M and S molecular form larvae to an aquatic predator in Burkina Faso. Parasit Vectors. 2012b;5:65. doi: 10.1186/1756-3305-5-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn MW, White BJ, Muir CJ, Besansky NJ. No evidence for biased co-transmission of speciation islands in Anopheles gambiae. Phil Trans R Soc B. 2012;367:374–384. doi: 10.1098/rstb.2011.0188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanington PC, Zhang S-M. The primary role of fibrinogen-related proteins in invertebrates is defense, not coagulation. J Innate Immun. 2011;3:17–27. doi: 10.1159/000321882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosack DA, Dennis G, Jr, Sherman BT, Lane HC, Lempicki RA. Identifying biological themes within lists of genes with EASE. Genome Biol. 2003;4:R70. doi: 10.1186/gb-2003-4-10-r70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Tan Q, et al. The DAVID Gene Functional Classification Tool: a novel biological module-centric algorithm to functionally analyze large gene lists. Genome Biol. 2007;8:R183. doi: 10.1186/gb-2007-8-9-r183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isoe J, Scaraffia PY. Urea synthesis and excretion in Aedes aegypti mosquitoes are regulated by a unique cross-talk mechanism. PloS one. 2013;8:e65393. doi: 10.1371/journal.pone.0065393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones CM, Toe HK, Sanou A, et al. Additional selection for insecticide resistance in urban malaria vectors: DDT resistance in Anopheles arabiensis from Bobo-Dioulasso, Burkina Faso. PloS one. 2012;7:e45995. doi: 10.1371/journal.pone.0045995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamdem C, Tene Fossog B, Simard F, et al. Anthropogenic habitat disturbance and ecological divergence between incipient species of the malaria mosquito Anopheles gambiae. PloS one. 2012;7:e39453. doi: 10.1371/journal.pone.0039453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koutsos AC, Blass C, Meister S, et al. Life cycle transcriptome of the malaria mosquito Anopheles gambiae and comparison with the fruitfly Drosophila melanogaster. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:11304–11309. doi: 10.1073/pnas.0703988104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawniczak MK, Emrich SJ, Holloway AK, et al. Widespread divergence between incipient Anopheles gambiae species revealed by whole genome sequences. Science. 2010;330:512–514. doi: 10.1126/science.1195755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, Cornel AJ, Meneses CR, et al. Ecological and genetic relationships of the Forest-M form among chromosomal and molecular forms of the malaria vector Anopheles gambiae sensu stricto. Malar J. 2009;8:75. doi: 10.1186/1475-2875-8-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann T, Diabate A. The molecular forms of Anopheles gambiae: a phenotypic perspective. Infect Genet Evol. 2008;8:737–746. doi: 10.1016/j.meegid.2008.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann T, Licht M, Elissa N, et al. Population Structure of Anopheles gambiae in Africa. Journal of Heredity. 2003;94:133–147. doi: 10.1093/jhered/esg024. [DOI] [PubMed] [Google Scholar]
- Leibold MA, McPeek MA. Coexistence of the niche and neutral perspectives in community ecology. Ecology. 2006;87:1399–1410. doi: 10.1890/0012-9658(2006)87[1399:cotnan]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Li YF, Costello JC, Holloway AK, Hahn MW. “Reverse ecology” and the power of population genomics. Evolution. 2008;62:2984–2994. doi: 10.1111/j.1558-5646.2008.00486.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li ZM, Zheng XM, Ge S. Genetic diversity and domestication history of African rice (Oryza glaberrima) as inferred from multiple gene sequences. Theor Appl Genet. 2011;123:21–31. doi: 10.1007/s00122-011-1563-2. [DOI] [PubMed] [Google Scholar]
- Lines JD. Do agricultural insecticides select for insecticide resistance in mosquitoes? A look at the evidence. Parasitology today. 1988;4:S17–S20. doi: 10.1016/0169-4758(88)90083-x. [DOI] [PubMed] [Google Scholar]
- Matzkin LM. Population transcriptomics of cactus host shifts in Drosophila mojavensis. Molecular Ecology. 2012;21:2428–2439. doi: 10.1111/j.1365-294X.2012.05549.x. [DOI] [PubMed] [Google Scholar]
- Matzkin LM. Ecological Genomics of Host Shifts in Drosophila mojavensis. Advances in Experimental Medicine and Biology. 2014;781:233–247. doi: 10.1007/978-94-007-7347-9_12. [DOI] [PubMed] [Google Scholar]
- Matzkin LM, Markow TA. Transcriptional differentiation across the four cactus host races of Drosophila mojavensis. In: Michalak P, editor. Speciation: Natural Processes, Genetics and Biodiversity. Nova Science Publishers Inc; New York: 2013. pp. 119–135. [Google Scholar]
- Megy K, Emrich SJ, Lawson D, et al. VectorBase: improvements to a bioinformatics resource for invertebrate vector genomics. Nucleic Acids Research. 2012;40:D729–734. doi: 10.1093/nar/gkr1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montero-Pau J, Ramos-Rodriguez E, Serra M, Gomez A. Long-term coexistence of rotifer cryptic species. PloS one. 2011;6:e21530. doi: 10.1371/journal.pone.0021530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller P, Chouaibou M, Pignatelli P, et al. Pyrethroid tolerance is associated with elevated expression of antioxidants and agricultural practice in Anopheles arabiensis sampled from an area of cotton fields in Northern Cameroon. Molecular Ecology. 2008;17:1145–1155. doi: 10.1111/j.1365-294X.2007.03617.x. [DOI] [PubMed] [Google Scholar]
- Neafsey DE, Lawniczak MK, Park DJ, et al. SNP genotyping defines complex gene-flow boundaries among African malaria vector mosquitoes. Science. 2010;330:514–517. doi: 10.1126/science.1193036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nkya TE, Akhouayri I, Kisinza W, David JP. Impact of environment on mosquito response to pyrethroid insecticides: facts, evidences and prospects. Insect Biochemistry and Molecular Biology. 2013;43:407–416. doi: 10.1016/j.ibmb.2012.10.006. [DOI] [PubMed] [Google Scholar]
- Nosil P, Funk DJ, Ortiz-Barrientos D. Divergent selection and heterogeneous genomic divergence. Molecular Ecology. 2009;18:375–402. doi: 10.1111/j.1365-294X.2008.03946.x. [DOI] [PubMed] [Google Scholar]
- Nowick K, Gernat T, Almaas E, Stubbs L. Differences in human and chimpanzee gene expression patterns define an evolving network of transcription factors in brain. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:22358–22363. doi: 10.1073/pnas.0911376106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nwane P, Etang J, Chouasmall yi UM, et al. Multiple insecticide resistance mechanisms in Anopheles gambiae s.l. populations from Cameroon, Central Africa. Parasites & vectors. 2013;6:41. doi: 10.1186/1756-3305-6-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oduola AO, Idowu ET, Oyebola MK, et al. Evidence of carbamate resistance in urban populations of Anopheles gambiae s.s. mosquitoes resistant to DDT and deltamethrin insecticides in Lagos, South-Western Nigeria. Parasites & vectors. 2012;5:116. doi: 10.1186/1756-3305-5-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennetier C, Warren B, Dabire KR, Russell IJ, Gibson G. “Singing on the wing” as a mechanism for species recognition in the malarial mosquito Anopheles gambiae. Current Biology. 2010;20:131–136. doi: 10.1016/j.cub.2009.11.040. [DOI] [PubMed] [Google Scholar]
- Pfenninger M, Schwenk K. Cryptic animal species are homogeneously distributed among taxa and biogeographical regions. BMC evolutionary biology. 2007;7:121. doi: 10.1186/1471-2148-7-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto J, Egyir-Yawson A, Vicente J, et al. Geographic population structure of the African malaria vector Anopheles gambiae suggests a role for the forest-savannah biome transition as a barrier to gene flow. Evolutionary applications. 2013;6:910–924. doi: 10.1111/eva.12075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porteres R. African cereals: Eleusine, fonio, black fonio, teff, Brachiaria, Paspalum, Pennisetum and African rice. In: Harlan JR, de Wet JMJ, Stemler ABL, editors. Origins of African plant domestication. Mouton Publishers; The Hague: 1976. pp. 409–452. [Google Scholar]
- Poupardin R, Reynaud S, Strode C, et al. Cross-induction of detoxification genes by environmental xenobiotics and insecticides in the mosquito Aedes aegypti: impact on larval tolerance to chemical insecticides. Insect Biochemistry and Molecular Biology. 2008;38:540–551. doi: 10.1016/j.ibmb.2008.01.004. [DOI] [PubMed] [Google Scholar]
- Ranson H, Rossiter L, Ortelli F, et al. Identification of a novel class of insect glutathione S-transferases involved in resistance to DDT in the malaria vector Anopheles gambiae. The Biochemical journal. 2001;359:295–304. doi: 10.1042/0264-6021:3590295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reidenbach KR, Neafsey DE, Costantini C, et al. Patterns of genomic differentiation between ecologically differentiated M and S forms of Anopheles gambiae in West and Central Africa. Genome Biology and Evolution. 2012;4:1202–1212. doi: 10.1093/gbe/evs095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero IG, Ruvinsky I, Gilad Y. Comparative studies of gene expression and the evolution of gene regulation. Nature reviews Genetics. 2012;13:505–516. doi: 10.1038/nrg3229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux O, Diabate A, Simard F. Divergence in threat-sensitivity among aquatic larvae of cryptic mosquito species. The Journal of animal ecology. 2013 doi: 10.1111/1365-2656.12163. [DOI] [PubMed] [Google Scholar]
- Rund SS, Hou TY, Ward SM, Collins FH, Duffield GE. Genome-wide profiling of diel and circadian gene expression in the malaria vector Anopheles gambiae. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:E421–430. doi: 10.1073/pnas.1100584108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santolamazza F, Della Torre A, Caccone A. Short report: A new polymerase chain reaction-restriction fragment length polymorphism method to identify Anopheles arabiensis from An. gambiae and its two molecular forms from degraded DNA templates or museum samples. American Journal of Tropical Medicine and Hygiene. 2004;70:604–606. [PubMed] [Google Scholar]
- Scaraffia PY, Isoe J, Murillo A, Wells MA. Ammonia metabolism in Aedes aegypti. Insect Biochemistry and Molecular Biology. 2005;35:491–503. doi: 10.1016/j.ibmb.2005.01.012. [DOI] [PubMed] [Google Scholar]
- Scaraffia PY, Tan G, Isoe J, et al. Discovery of an alternate metabolic pathway for urea synthesis in adult Aedes aegypti mosquitoes. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:518–523. doi: 10.1073/pnas.0708098105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sim S, Jupatanakul N, Ramirez JL, et al. Transcriptomic profiling of diverse Aedes aegypti strains reveals increased basal-level immune activation in dengue virus-refractory populations and identifies novel virus-vector molecular interactions. PLoS neglected tropical diseases. 2013;7:e2295. doi: 10.1371/journal.pntd.0002295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simard F, Ayala D, Kamdem GC, et al. Ecological niche partitioning between the M and S molecular forms of Anopheles gambiae in Cameroon: the ecological side of speciation. BMC Ecol. 2009;9:17. doi: 10.1186/1472-6785-9-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slotman MA, Tripet F, Cornel AJ, et al. Evidence for subdivision within the M molecular form of Anopheles gambiae. Molecular Ecology. 2007;16:639–649. doi: 10.1111/j.1365-294X.2006.03172.x. [DOI] [PubMed] [Google Scholar]
- Storz JF. Using genome scans of DNA polymorphism to infer adaptive population divergence. Molecular Ecology. 2005;14:671–688. doi: 10.1111/j.1365-294X.2005.02437.x. [DOI] [PubMed] [Google Scholar]
- Stump AD, Fitzpatrick MC, Lobo NF, et al. Centromere-proximal differentiation and speciation in Anopheles gambiae. Proc Natl Acad Sci U S A. 2005;102:15930–15935. doi: 10.1073/pnas.0508161102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tchuinkam T, Mulder B, Dechering K, et al. Experimental infections of Anopheles gambiae with Plasmodium falciparum of naturally infected gametocyte carriers in Cameroon: factors influencing the infectivity to mosquitoes. Tropical Medicine and Parasitology. 1993;44:271–276. [PubMed] [Google Scholar]
- Tene Fossog B, Antonio-Nkondjio C, Pierre Kengne P, et al. Physiological correlates of ecological divergence along an urbanization gradient: differential tolerance to ammonia among molecular forms of the malaria mosquito Anopheles gambiae. BMC Ecol. 2013a;13:1. doi: 10.1186/1472-6785-13-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tene Fossog B, Kopya E, Ndo C, et al. Water Quality and Anopheles gambiae Larval Tolerance to Pyrethroids in the Cities of Douala and Yaounde (Cameroon) Journal of tropical medicine. 2012;2012:429817. doi: 10.1155/2012/429817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tene Fossog B, Poupardin R, Costantini C, et al. Resistance to DDT in an urban setting: common mechanisms implicated in both M and S forms of Anopheles gambiae in the city of Yaounde Cameroon. PloS one. 2013b;8:e61408. doi: 10.1371/journal.pone.0061408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner TL, Hahn MW. Locus- and population-specific selection and differentiation between incipient species of Anopheles gambiae. Molecular Biology and Evolution. 2007;24:2132–2138. doi: 10.1093/molbev/msm143. [DOI] [PubMed] [Google Scholar]
- Turner TL, Hahn MW, Nuzhdin SV. Genomic islands of speciation in Anopheles gambiae. PLoS Biol. 2005;3:e285. doi: 10.1371/journal.pbio.0030285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Dungern P, Briegel H. Protein catabolism in mosquitoes: ureotely and uricotely in larval and imaginal Aedes aegypti. Journal of insect physiology. 2001;47:131–141. doi: 10.1016/s0022-1910(00)00096-2. [DOI] [PubMed] [Google Scholar]
- Waterhouse RM, Kriventseva EV, Meister S, et al. Evolutionary dynamics of immune-related genes and pathways in disease-vector mosquitoes. Science. 2007;316:1738–1743. doi: 10.1126/science.1139862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse RM, Povelones M, Christophides GK. Sequence-structure-function relations of the mosquito leucine-rich repeat immune proteins. BMC Genomics. 2010;11:531. doi: 10.1186/1471-2164-11-531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weihrauch D, Donini A, O’Donnell MJ. Ammonia transport by terrestrial and aquatic insects. Journal of insect physiology. 2012;58:473–487. doi: 10.1016/j.jinsphys.2011.11.005. [DOI] [PubMed] [Google Scholar]
- White BJ, Lawniczak MK, Cheng C, et al. Adaptive divergence between incipient species of Anopheles gambiae increases resistance to Plasmodium. Proc Natl Acad Sci U S A. 2011;108:244–249. doi: 10.1073/pnas.1013648108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitehead A. Comparative genomics in ecological physiology: toward a more nuanced understanding of acclimation and adaptation. The Journal of experimental biology. 2012;215:884–891. doi: 10.1242/jeb.058735. [DOI] [PubMed] [Google Scholar]
- Whitehead A, Crawford DL. Neutral and adaptive variation in gene expression. Proceedings of the National Academy of Sciences of the United States of America. 2006a;103:5425–5430. doi: 10.1073/pnas.0507648103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitehead A, Crawford DL. Variation within and among species in gene expression: raw material for evolution. Molecular Ecology. 2006b;15:1197–1211. doi: 10.1111/j.1365-294X.2006.02868.x. [DOI] [PubMed] [Google Scholar]
- Wittkopp PJ, Haerum BK, Clark AG. Regulatory changes underlying expression differences within and between Drosophila species. Nature Genetics. 2008;40:346–350. doi: 10.1038/ng.77. [DOI] [PubMed] [Google Scholar]
- Yadouleton AW, Asidi A, Djouaka RF, et al. Development of vegetable farming: a cause of the emergence of insecticide resistance in populations of Anopheles gambiae in urban areas of Benin. Malaria journal. 2009;8:103. doi: 10.1186/1475-2875-8-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Wang Y, Xiao Y, et al. A bi-functional anti-thrombosis protein containing both direct-acting fibrin(ogen)olytic and plasminogen-activating activities. PloS one. 2011;6:e17519. doi: 10.1371/journal.pone.0017519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zdobnov EM, von Mering C, Letunic I, et al. Comparative genome and proteome analysis of Anopheles gambiae and Drosophila melanogaster. Science. 2002;298:149–159. doi: 10.1126/science.1077061. [DOI] [PubMed] [Google Scholar]
- Zheng W, Gianoulis TA, Karczewski KJ, Zhao H, Snyder M. Regulatory variation within and between species. Annual review of genomics and human genetics. 2011;12:327–346. doi: 10.1146/annurev-genom-082908-150139. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.