Abstract
Mitochondrial reactive oxygen species (ROS) are widely implicated in physiological and pathological pathways. We propose that it is critical to understand the specific sites of mitochondrial ROS production and their mechanisms of action. Mitochondria possess at least eight distinct sites of ROS production in the electron transport chain and matrix compartment. In this chapter, we describe the nature of the mitochondrial ROS-producing machinery and the relative capacities of each site. We provide detailed methods for the measurement of H2O2 release and the conditions under which maximal rates from each site can be achieved in intact skeletal muscle mitochondria.
1. INTRODUCTION
The best-described role for mitochondria is as the primary supplier of cellular ATP. However, during this energy-transducing process, electrons can escape the electron transport chain and partially reduce oxygen to give rise to reactive oxygen species (ROS). There are at least eight sites capable of producing considerable amounts of ROS in the mitochondria (Brand, 2010). Most mitochondrial sites catalyze a monovalent reduction of oxygen to generate superoxide, but some sites are capable of direct formation of hydrogen peroxide (H2O2) from divalent reduction of oxygen. However, since we directly measure H2O2 with our detection system (after dis-mutation of superoxide to H2O2), and in many cases we cannot distinguish whether the originally formed species was superoxide or H2O2, we will use the general term ROS to mean superoxide/H2O2.
The mitochondrial electron transport chain ROS producers with the greatest capacity, in order of magnitude, are complex III (at the site of quinol oxidation: site IIIQo), and complex I (at the ubiquinone-binding site: site IQ) and complex II (at the flavin: site IIF) (Quinlan, Gerencser, Treberg, & Brand, 2011; Quinlan, Orr, et al., 2012; Treberg, Quinlan, & Brand, 2011), with each site generating greater than 1 nmol H2O2 min –1 mg protein –1 under Vmax conditions in skeletal muscle mitochondria. Of course, the rates of mitochondrial ROS production from a given site are tissue specific. For example, the rates of H2O2 measured from mitochondrial glycerol 3-phosphate dehydrogenase (mGPDH) are significantly lower from heart mitochondria than brown fat mitochondria due to the higher expression level of mGPDH in brown fat, whereas both brown fat and heart mitochondria are capable of very high rates of ROS production from site IQ (Orr, Quinlan, Perevoshchikova, & Brand, 2012). Other mitochondrial ROS-producing sites include the flavin of complex I (site IF) (Kussmaul & Hirst, 2006), electron-transferring flavoprotein ubiquinone oxidoreductase (ETFQOR) (Perevoshchikova, Quinlan, Orr, & Brand, 2013), the pyruvate dehydrogenase complex (PDH), and oxoglutarate dehydrogenase (OGDH) complex (Bunik & Sievers, 2002), as well as less well-defined sites such as proline dehydrogenase and dihydroorotate dehydrogenase (Brand, 2010).
Due to their central role in cell metabolism and their ability to produce ROS, mitochondria are thought to be involved in many different physiological and pathological events, including cell differentiation, insulin secretion, cancer, and neurological disorders (Boden et al., 2012; Ralph & Neuzil, 2009; Sundaresan, Yu, Ferrans, Irani, & Finkel, 1995; Witte, Geurts, de Vries, van der Valk, & van Horssen, 2010). The free radical theory of aging (Harman, 1956) states that mitochondrial ROS are detrimental and ultimately causative of the aging process. In favor of this hypothesis, oxidative damage to mitochondrial DNA is observed in aged individuals (Golden & Melov, 2001). However, it is now appreciated that the role of ROS in aging is probably more nuanced than is allowed by the free radical theory. Increasing evidence indicates that a transient increase in ROS formation is a positive adaptive event that is associated with increased stress resistance and life span extension (Ristow & Schmeisser, 2011; Schulz et al., 2007; Zarse et al., 2012). In this sense, low levels of ROS seem to be positive, while high levels of ROS are detrimental (hormesis).
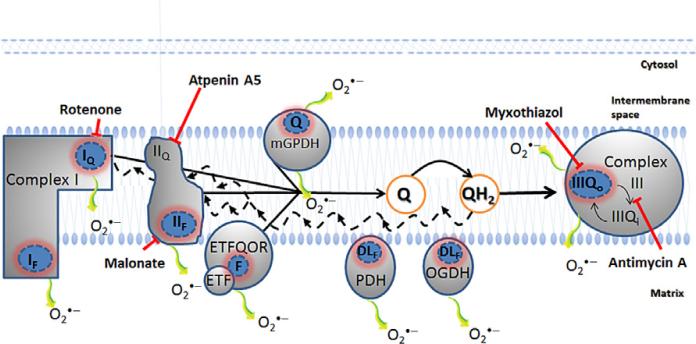
The apparently critical role of mitochondrial ROS in cellular processes makes characterization and quantification of the specific sites of ROS production of particular interest. In this chapter, we describe the experimental protocols that we use to measure site-specific ROS production from isolated mitochondria. We present mechanistic information about the sites and how we manipulate the conditions of our assays to maximize the rate from each site. This information provides a framework from which to interpret less contrived experiments, that is, experiments under “native” or non-Vmax conditions. Figure 12.1 illustrates the eight known mitochondrial sites that will be discussed in this context.
Figure 12.1.
Eight sites of mitochondrial superoxide/H2O2 production are identified. Complex I oxidizes NADH from the tricarboxylic acid cycle and produces superoxide at the flavin site (IF) and ubiquinone-binding site (IQ), both sites release superoxide to the matrix side. Complex II, a tricarboxylic acid cycle enzyme bound to the mitochondrial inner membrane, oxidizes succinate to fumarate and generates superoxide/H2O2 both in the reverse and forward reactions through the flavin site (IIF). Glycerol 3-phosphate dehydrogenase (mGPDH) is part of an important shuttle for oxidation of cytosolic NADH and releases superoxide to both matrix and cytosol. The electron-transferring flavoprotein (ETF) and electron-transferring flavoprotein ubiquinone oxidoreductase (ETFQOR) represent the final steps for β-oxidation; it is not known which of the two sites is responsible for superoxide/H2O2 production during β-oxidation. The IIIQo site of complex III is also a source of superoxide to both cytosol and matrix. In addition to the electron transport chain sites, pyruvate dehydrogenase (PDH) and oxoglutarate dehydrogenase (OGDH) both contain the dihydrolipoamide (DL) dehydrogenase subunit that can generate super-oxide/H2O2, possibly through the DL or flavin moieties. The black solid line shows complex I, complex II, mGPDH, and ETFQOR feeding electrons into the ubiquinone (Q)-pool. With a very reduced Q-pool (QH2), the electrons can run in reverse through complex I and complex II, promoting ROS generation at sites IQ, IF, and IIF (black dashed lines). The blunted arrows indicate the sites where the inhibitors rotenone, malonate, atpenin A5, myxothiazol, and antimycin A bind. The use of such inhibitors is necessary to pharmacologically isolate the sites of ROS production from each other.
2. H2O2 PRODUCTION MEASUREMENTS IN ISOLATED MITOCHONDRIA
Since the discovery of superoxide dismutase in mammalian cells and the resultant evidence for in situ ROS production (Boveris, Oshino, & Chance, 1972; McCord & Fridovich, 1969), there has been great interest in assessing cellular ROS levels and rates of production. As will be detailed in this chapter, there are multiple sites within the mitochondrion that are capable of reducing oxygen to superoxide. After its formation, superoxide is dismutated to hydrogen peroxide (H2O2) either spontaneously or by the matrix located manganese-superoxide dismutase (Mn-SOD) or the cytosolic copper/zinc-superoxide dismutase. H2O2 possesses a longer half-life than superoxide and is considered to be less reactive. Furthermore, in contrast to superoxide, H2O2 is membrane permeant which enables its release from the mitochondrial matrix to the intermembrane space (Nohl & Jordan, 1980). Its stability and the membrane permeability make H2O2 the chosen species for detection of mitochondrial ROS production.
2.1. Techniques for the measurement of mitochondrial H2O2 release
The most commonly used and sensitive techniques for hydrogen peroxide detection are the enzyme-linked fluorescent techniques where horseradish peroxidase (HRP) reacts with hydrogen peroxide generating a compound which readily reacts with a probe (AH2 in Eq. 12.1) that either becomes nonfluorescent or generates a fluorescent compound (A in Eq. 12.1) (Meng, High, Antonello, Washabaugh, & Zhao, 2005).
| [12.1] |
Over the years, several H2O2 probes have been used, including scopoletin (Boveris, Martino, & Stoppani, 1977), p-hydroxyphenylacetate (Hyslop & Sklar, 1984), and homovanillic acid (Ruch, Cooper, & Baggiolini, 1983). Presently, however, Amplex Red (N-acetyl-3,7-dihydroxyphenoxazine) is the preferred probe by most groups when investigating ROS production from mitochondria. Amplex Red is not fluorescent itself but produces the fluorescent compound resurofin (7-hydroxy-3H-phenoxazin-3-one) when it reacts with HRP–H2O2 (Mohanty, Jaffe, Schulman, & Raible, 1997; Zhou, Diwu, Panchuk-Voloshina, & Haugland, 1997). Since resurofin is stable and accumulates in the extramitochondrial environment, the fluorescent signal will increase when H2O2 is released from the mitochondria. It is possible to detect resurofin by monitoring its absorbance at 557–620 nm, but the most sensitive detection is with spectrofluorimetric techniques (Staniek & Nohl, 1999).
Studies have shown that HRP/Amplex Red is selective for H2O2 (Mohanty et al., 1997) and highly sensitive, with the capacity to detect H2O2 amounts as low as 5 pmol (Zhou et al., 1997). In contrast to other well-known probes like scopoletin or homovallinic acid, the wavelengths used in the Amplex Red assay are relatively high (ex/em 560/590), making the assay less prone to interference from autofluorescent changes in biological samples (Zhou et al., 1997). Membranes are not permeable to HRP/ Amplex Red, and therefore the assay detects only extramitochondrial H2O2 (Mohanty et al., 1997). Since the mitochondrial ROS-producing sites generate predominantly superoxide, the Amplex Red technique relies heavily on the endogenous Mn-SOD to convert the superoxide to H2O2. We also routinely add exogenous SOD to ensure detection of the superoxide that is released to extramitochondrial space (e.g., superoxide from mGPDH is released to both sides of the mitochondrial membrane). This also provides a useful tool when characterizing a specific ROS-producing site: withdrawal or addition of exogenous SOD makes it possible to distinguish superoxide produced to the matrix from that produced to the intermembrane space (St-Pierre, Buckingham, Roebuck, & Brand, 2002).
Amplex Red reacts with HRP-H2O2 in a 1:1 stoichiometry. However, a linear relationship exists between H2O2 and resurofin fluorescence only when Amplex Red is fivefold in excess above the H2O2 concentration. This is because resurofin is prone to oxidation and generation of a nonfluorescent compound when it reacts directly with H2O2 at high concentrations (Mohanty et al., 1997). However, high concentrations of resurofin (above 5 μM) can also cause an “inner filter” effect resulting in an underestimation of the signal and thus a nonlinear relationship with H2O2 (Brotea & Thibert, 1988). In our experiments, we use 50 μM Amplex Red and our range of experimental detection is up to ~2 nmol H2O2, after which we start to see nonlinearity in the H2O2:resorufin fluorescence relationship.
2.2. Background correction and calibration curve
As with many fluorescent probes, Amplex Red is light sensitive and prone to oxidation and should therefore be protected from light during both experiments and storage (Zhou et al., 1997). During a standard assay, there is some spontaneous oxidation of Amplex Red/UltraRed causing a small background signal, which in our hands corresponds to 8–10 pmol H2O2 min –1 mg protein –1 (typical assay conditions described below). Assuming that this rate is constant during the experiment, this background can be subtracted from all rates determined in the same sample/cuvette (this should, of course, be checked by running a background sample for the time duration of the experiment).
A benefit of the 1:1 stoichiometry provided by Amplex Red is that resorufin fluorescence is easily calibrated to molar units of H2O2. An essential feature of this calculation is established by a calibration curve between the fluorescent signal and a known concentration of H2O2. Several points are important when establishing this calibration curve:
The calibration curve should be generated in the exact experimental medium and in the presence of the isolated mitochondria. Previous studies have shown that mitochondria will scavenge some of the H2O2 during calibration (St-Pierre et al., 2002).
Many of the compounds used in different protocols have the potential to affect the fluorescent signal. For example, bovine serum albumin (BSA) has a moderate quenching effect (Muller et al., 2008), and therefore calibrations should be done in its presence. The same is true for many of the pharmacological compounds and even some substrates commonly used, which may quench or have fluorescent properties. Thus, it is essential to generate the calibration curves under conditions that mimic the experimental conditions.
Since H2O2 will decay when in dilute solution, calibration curves should always be generated using freshly diluted H2O2. Typically, we make a working stock solution of 0.25 μM H2O2 and titrate from 0.25 to 1.5 nmol H2O2.
2.2.1 A brief note on Amplex UltraRed
We have recently switched from Amplex Red to Amplex UltraRed because it improves upon the performance of Amplex Red by providing brighter fluorescence and enhanced sensitivity in peroxidase-coupled assays. One of the disadvantages of many HRP-H2O2-related probes is their fluorescence instability with changes in pH. Amplex UltraRed exhibits much less sensitivity to pH changes and is more stable in the presence of H2O2 and thiols than its predecessor. Amplex UltraRed is stable for hours in the presence of mitochondria (unpublished observation) suggesting lower reactivity than its predecessor and is therefore preferred in our experiments. The structural differences between the compounds are not reported by the vendor (Invitrogen).
2.3. H2O2 detection: Platereader- versus cuvette-based assays
The rates of H2O2 generation by isolated mitochondria can be evaluated using a standard spectrofluorimeter equipped with one or more cuvette holders, or in a platereader capable of the appropriate excitation/emission. In our laboratory, the cuvette-based assays are performed in a Varian Cary Eclipse spectrofluorimeter or a Shimadzu RF5301-PC spectrofluorimeter. We typically use the excitation/emission wavelength pair 560/590 nm, respectively, in a total volume of 2 mL. For assays using 96-well microplates, we use a BMG Labtech microplate reader with filters suitable for excitation at 540 nm and emission at 590 nm, in a total volume of 0.2 mL.
The choice to use a platereader- or cuvette-based assay is the experimenter's prerogative, but there are constraints that might indicate the use of one technique as more sensible than the other. The principal advantage of cuvette-based assays is accuracy. In general, the experimenter has more control over the conditions in the cuvette and is able to monitor the conditions in real time. The mixing and temperature control are usually more accurate in the cuvette than in the platereader, and the maintenance of a well-mixed suspension at the appropriate temperature is critical for precisely measuring rates of H2O2 generation. Another benefit to the cuvette-based assay is that different additions can be performed in series while the run is monitored, giving more flexibility to the assay. However, due to the limited number of cuvettes, the total assay time can be longer. The volume of the assay also necessitates the use of large amounts of protein (0.2–0.3 mg protein mL –1).
Our assays in the cuvette are performed in a total volume of 2 mL. Each run is started with mitochondria (0.3 mg protein mL –1), 50 μM Amplex UltraRed (10 μM stock solution in DMSO), 5 units mL –1 HRP (1000 units mL –1 stock solution in H2O), and 25 units mL –1 SOD (5000 units mL –1 stock solution in H2O). A typical buffer is 120 μM KCl, 5 μM HEPES, 5 μM KH2PO4, 2.5 μM MgCl2, 1 μM EGTA, and 0.3% (w/v) BSA, pH 7.0, 37 °C. Typically, we allow the mitochondria to oxidize endogenous substrates for 3–5 min before we add any inhibitors. After this initial phase, two protocols are possible: one with the inhibitors present before the substrate and the other with the substrate present before the inhibitors. In the first scenario, we collect a background rate in the presence of the inhibitors for several more minutes before we add the substrate of interest. In the second scenario, we add a substrate in the absence of inhibitors, collect a rate for several minutes and then add inhibitors in series. Typically, we perform experiments in both ways to dissect out the specific behavior of a given site or sites.
When working with a limited amount of protein (e.g., mitochondria isolated from cell culture or Drosophila), it is recommended to measure the rates of H2O2 production using the platereader since only 0.015 mg of mitochondrial protein are required per well. It is also preferred to create each test condition in parallel rather than to make additions in series, to minimize the number of times the platereader is opened, and to avoid fluctuations in the assay temperature. Typically, in our protocols, we establish a number of different experimental conditions in parallel in different wells and perform one addition to the wells mid run. For example, at time zero, the assay starts with the Amplex UltraRed/HRP detection system, mitochondria, and the desired inhibitors. After the establishment of a baseline, the run is paused to add the substrate and the signal is monitored for another 5 min.
Our assay in the platereader is set to a final volume of 0.2 mL in a buffer similar to the one exemplified above. The run starts with a volume of 180 μL, where 60 μL is from the mitochondrial stock (0.25 mg mL –1), 60 μL from HRP stock (16.66 units mL –1), 40 μL of the Amplex UltraRed stock (0.25 mM), and 20 μL of the inhibitor stocks (the concentration varies according to the drug). All the components are diluted in the same assay buffer (e.g., the original 10 mM Amplex UltraRed stock in DMSO is diluted 40 × into assay buffer before use in the platereader assay). After several minutes to establish the background rate, 20 μL of substrate is added. We have observed that in the platereader the background rate, in the presence of Amplex UltraRed, HRP, and mitochondria, is in general higher than when using a standard spectrofluorimeter. This is likely due to the high intensity of incident light and broader spectrum of emitted light from the platereader. It is important to subtract the appropriate background for each condition. For this reason, a control parallel well with the H2O2-consuming enzyme catalase should be run to get a measure of the non-H2O2-specific probe oxidation. The H2O2 calibration curve is typically performed in multiple wells, with each well-containing mitochondria, Amplex UltraRed/ HRP, the relevant inhibitors, and one concentration of H2O2 (typically, we titrate from 0.25 to 1.25 nmol H2O2).
2.4. Accounting for the antioxidant system
There are significant antioxidant systems within mitochondria, especially for the decomposition of H2O2 by glutathione (GSH) peroxidase (Schafer & Buettner, 2001). The fact that much of the mitochondrial ROS is generated within the matrix compartment means that any matrix antioxidant system has preferential access to H2O2 prior to its diffusion out into the medium where the detection system is present. Thus, the use of H2O2 production by mitochondria as a quantitative measure of the superoxide formed must assume zero or minimal interference from the antioxidant system. As the antioxidant system always functions to some extent, this assumption is unlikely to be correct and the measured rates of mitochondrial H2O2 production will always underestimate the true rates of production.
Matrix GSH peroxidases can decompose H2O2 into H2O, forming GSSG (oxidized glutathione disulfide), which is reduced back to GSH as a result of NADPH oxidation by glutathione reductase. Since GSH is central to this peroxidase system, GSH-depleting agents are able to compromise the mitochondrial capacity to decompose H2O2. The observed mitochondrial H2O2 production can be increased by pretreatment with 1-chloro-2,4-dinitrobenzene (CDNB), which acts as a GSH-depleting agent through the irreversible conjugation of CDNB and GSH by glutathione-S transferase (Han, Canali, Rettori, & Kaplowitz, 2003; Treberg, Quinlan, & Brand, 2010) We have found that careful pretreatment of mitochondria with CDNB will give an increased ROS signal without significant damage to mitochondrial components. However, care should be taken when treating with CDNB because excess or acute CDNB treatment will result in increased ROS from complex I through a non-GSH pool-dependent mechanism (Liu, Fiskum, & Schubert, 2002; Treberg et al., 2010).
Our CDNB pretreatment protocol (Treberg et al., 2010) is based on Han et al. (2003). The final skeletal muscle mitochondria pellet is resuspended to a concentration of 5 mg protein mL –1 in our standard mitochondrial isolation buffer (100 mM KCl, 50 mM Tris, 2 mM EGTA, pH 7.1 at 25 °C). A 50 mM stock solution of CDNB is prepared in ethanol. The mitochondria are treated with either 35 μM CDNB or ethanol control for 5 min at room temperature. Following the 5 min incubation, the suspensions are diluted with an equal volume of ice-cold isolation buffer and centrifuged for 5 min at 15,000 ×g (at 2–4 °C). The pellets are then washed twice more by resuspending in ice-cold buffer and centrifuging as above to remove any residual CDNB/ethanol. The final pellets are resuspended to approximately 20 mg protein mL –1.
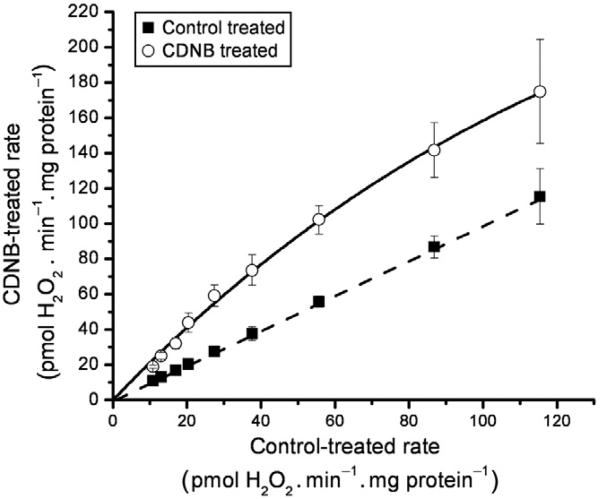
Following CDNB treatment, a calibration curve can be built for a given set of conditions. For example, a titration with malate in the presence rote-none will generate matrix-directed superoxide. When this condition is compared between the CDNB- and control-treated mitochondria, we observe an increase in the rate of H2O2 production from the CDNB-treated mitochondria. We can then plot the CDNB rate against the control rate (as in Quinlan, Treberg, Perevoshchikova, Orr, & Brand, 2012; Treberg et al., 2010). Figure 12.2 is an example of a CDNB calibration curve (see legend for details). This CDNB calibration curve can be used to get a closer approximation of the production rates before interference by the antioxidant system. As mentioned above, this treatment is an approximation and is very particular to the conditions, so it should be determined robustly for every experimental system.
Figure 12.2.
CDNB calibration curve. We generated this relationship by titrating malate (0.01–5 mM) into CDNB- and ethanol control-treated mitochondria. The rates of H2O2 production were measured after the addition of 4 μM rotenone and assumed to be purely matrix directed. Points were fitted to give hyperbolic parameter values. The dashed line indicates a 1:1. Adapted from Quinlan, Treberg, et al. (2012).
3. MAXIMUM PRODUCTION RATES FROM SPECIFIC SITES: THE NATURE AND CAPACITY OF THE MACHINERY
We are particularly interested in the nature of the mitochondrial ROS-producing machinery, and we have invested considerable effort in recent years to determine the specific behavior and capacity of several mitochondrial ROS-producing sites (Lambert & Brand, 2004b; Orr et al., 2012; Perevoshchikova et al., 2013; Quinlan et al., 2011; Quinlan, Orr, et al., 2012; Quinlan, Treberg, et al., 2012; Treberg et al., 2011). In this section, we give an overview of the mechanisms and conditions needed to achieve maximum rates of ROS production from complex I, complex II, complex III, mGPDH, electron-transferring flavoprotein–ETF ubiquinone oxidoreductase (ETF-ETFQOR), and OGDH. The experiments described here were performed in simple KCl buffers using isolated mitochondria. Rat skeletal muscle mitochondria were isolated at 4 °C in Chappell-Perry buffer (CP1; 100 mM KCl, 50 mM Tris, 2 mM EGTA, pH 7.1 at 25 °C) by standard procedures (for a detailed protocol, see Affourtit, Quinlan, & Brand, 2012). Mitochondria were diluted to a final concentration of 0.3 mg protein mL –1 in KHE buffer (120 mM KCl, 5 mM HEPES, 1 mM EGTA, in the presence or absence of 5 mM phosphate and 2.5 mM magnesium, and supplemented with 0.3% (w/v) BSA, pH 7.0 at 37 °C). The measurements were performed in a Varian Cary spectrofluorimeter at the wavelength couple ex=560 nm and em=590 nm in the presence of 50 μM Amplex UltraRed, 5 units mL –1 HRP, and 25 units mL –1 SOD. The samples were added to a four-sided clear cuvette, under constant stirring and temperature control. The inhibitors were added to the concentrations described in each section below.
3.1. Complex I, NADH-ubiquinone oxidoreductase
Complex I (NADH-ubiquinone oxidoreductase) oxidizes NADH to NAD+ and reduces ubiquinone (Q) to quinol (QH2). During this process two electrons are transferred and four protons are pumped from the matrix to intermembrane space (Brown & Brand, 1988; Treberg & Brand, 2011). The overall reaction can be presented as:
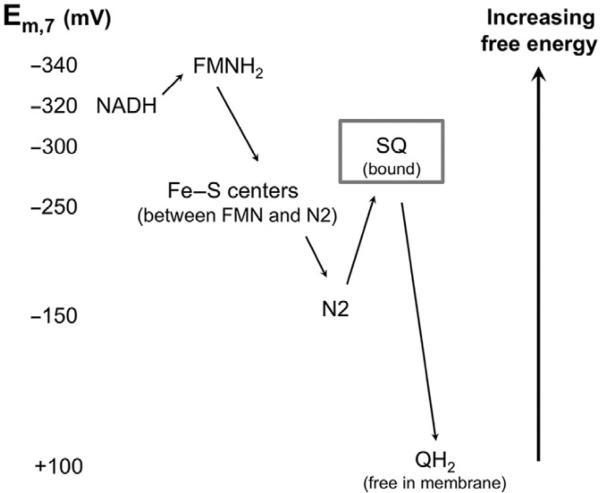
The electron transfer through the complex occurs at several steps using intermediate redox centers with increasing value of their redox potentials (Fig. 12.3).
Figure 12.3.
Reduction potentials of complex I redox centers. Electrons are moved from NADH to QH2 through redox centers of increasing potentials. Center N2 is the terminal and highest mid-point potential [4Fe–4S] cluster. SQ is the proposed semiquinone ROS producer. Adapted from Treberg and Brand (2011).
As well as transducing energy, NADH-ubiquinone oxidoreductase is known to produce superoxide at two different sites within the complex (Treberg & Brand, 2011). The existence of a second site of superoxide production by complex I has been disputed (Kussmaul & Hirst, 2006; Pryde & Hirst, 2011), but we and others think that the experimental evidence supports two different sites of superoxide production rather than a single site (Ohnishi, Shinzawa-Itoh, Ohta, Yoshikawa, & Ohnishi, 2010; Treberg et al., 2011; Vinogradov & Grivennikova, 2005).
The first superoxide-producing candidate is the NADH-oxidizing site or flavin site (site IF). Superoxide production by this site is thought to occur by the leak of electrons from the reduced flavin (FMNH and/or FMNH2) to molecular oxygen (Kussmaul & Hirst, 2006; Pryde & Hirst, 2011). For purposes of simplicity and consistency, we will refer to the fully reduced flavin, FMNH2, as the electron donor to oxygen (Kussmaul & Hirst, 2006). The oxidation of NADH at complex I reduces FMN. Because electrons can be transferred to oxygen from the flavin only when it is reduced, the steady-state concentration of reduced FMNH2 will determine the rate of superoxide production. In turn, the higher the NADH/NAD ratio, the higher the rates of superoxide production from site IF (Hansford, Hogue, & Mildaziene, 1997; Kushnareva, Murphy, & Andreyev, 2002; Starkov & Fiskum, 2003).
Any protocol that fully reduces the NADH pool would be predicted to maximize ROS production from site IF, but depending on the source of electrons that are used to accomplish full NADH reduction, recruitment of other matrix ROS producers (e.g., OGDH) may or may not be predicted. It is necessary to distinguish site IF from other matrix located enzymes that also respond to the reduced NADH pool. As an example, a protocol with 5 mM glutamate plus 5 mM malate and 4 μM rotenone will fully reduce the NADH pool and provide a measurable ROS signal. However, this signal does not necessarily all arise from site IF but could be partially assigned to OGDH. With respect to this, matrix sites have not routinely been distinguished from each other in intact mitochondria, and quoted rates of site IF ROS production usually include ROS production from OGDH.
The second site of superoxide production within complex I is the ubiquinone-reducing site, site IQ. The leak of electrons to molecular oxygen is proposed to occur from the semiquinone (QH·–) moiety formed as an intermediate during electron transfer from center N2 to oxidized ubiqui-none (Treberg & Brand, 2011). Superoxide production at site IQ can be instigated by two different conditions. The first condition, commonly referred to as “reverse electron transfer,” is achieved in the presence of a high proton motive force and oxidized NAD pool. This involves a full reversal of complex I in which QH2 is oxidized and NAD+ is reduced. It is proposed that a semiquinone superoxide producer is stabilized in the complex in the presence of high protonmotive force. In this case, reduction of the ubiquinone pool (Q-pool) and generation of a high protonmotive force can be achieved through several pathways. For example, maximum rates of superoxide production from site IQ can be approached during oxidation of succinate or glycerol 3-phosphate (electrons enter the Q-pool through complex II or glycerol 3-phosphate dehydrogenase, respectively), which both reduce the Q-pool and generate a sufficiently high protonmotive force. High protonmotive force was found to be an important component to trigger reverse electron flow into site IQ (Adam-Vizi & Chinopoulos, 2006; Korshunov, Skulachev, & Starkov, 1997; Lambert & Brand, 2004b; Miwa, St-Pierre, Partridge, & Brand, 2003; Votyakova & Reynolds, 2001). However, between the two components of protonmotive force (protonmotive force=Δψm – 60ΔpH, where Δψm is the membrane potential across the mitochondrial inner membrane), ΔpH has the strongest influence (Lambert & Brand, 2004b). The rate of IQ superoxide production can be suppressed by collapsing the ΔpH with the K+/H+ ionophore nigericin or inorganic Pi, without abolishing Δψm (Lambert & Brand, 2004b). The rate of superoxide production in this case is sensitive to the IQ site inhibitor rotenone, which blocks the oxidation of QH2 and limits the formation of the superoxide-generating semiquinone. In the presence of rotenone, the Q site likely stays fully reduced as QH2 and does not contribute to super-oxide production.
The second condition known to stimulate IQ superoxide production can be reached during forward electron flow (oxidation of reduced NADH) in the presence of Q site inhibitors (rotenone) and protonmotive force generated by ATP hydrolysis (Lambert & Brand, 2004a; Treberg et al., 2011). Under this condition, both sites (IF and IQ) contribute to the total observed rate, but IQ shows the higher rate of superoxide production and a strong ΔpH sensitivity (Lambert & Brand, 2004a). As in the first condition, the IQ site produces superoxide by the leak of electrons from the semiquinone moiety. However, when electrons are fluxing in the forward direction, the formation of ΔpH (generated by ATP hydrolysis) reverses the proton pumps and drives electrons from QH2 to center N2 (Treberg et al., 2011). Therefore, semiquinone can be formed even in the presence of rotenone. Here, center N2 is reduced by forward electron flow as a result of the highly reduced NADH pool. This condition stabilizes the semiquinone in site IQ and results in superoxide production.
Experimentally, maximum rates from site IQ are best achieved from reverse electron transport. Conditions favoring Vmax are found in a buffer lacking phosphate so that a large pH gradient is maintained. Although site IQ still generates ROS with phosphate present and a minimized ΔpH, the rates are lower. The chosen substrate should be one that will reduce the Q-pool and generate a high protonmotive force. In most tissues, succinate oxidation will accomplish this. However, any substrate that donates electrons to the Q-pool and generates a protonmotive force will suffice (e.g., glycerol 3-phosphate). Rotenone (4 μM) should be added to dissect the rate that is specific to site IQ versus the rate arising from other sites (e.g., sites IF and IIIQo both likely contribute to the total observed rates under conditions of a high protonmotive force and reduced NAD and Q-pools).
3.2. Complex II, succinate:ubiquinone oxidoreductase
Succinate dehydrogenase (SDH), succinate:ubiquinone oxidoreductase, or complex II are the common names for the only tricarboxylic acid cycle enzyme that is also part of the mitochondrial electron transport chain; it couples the two-electron oxidation of succinate to the reduction of the Q-pool. When succinate is oxidized to fumarate, electrons are moved from the flavin moiety through three iron–sulfur clusters to the membrane-embedded subunits that contain the cytochrome b heme moiety and the hydrophobic subunits that provide the binding site for ubiquinone (Sun et al., 2005; Yankovskaya et al., 2003).
There are several lines of evidence that indicate that complex II is capable of ROS production. The enzyme isolated from Escherichia coli or purified from bovine heart has been reported to generate superoxide under some conditions (Messner & Imlay, 2002; Zhang, Yu, & Yu, 1998). Recently, we found in isolated skeletal muscle mitochondria that a large SDH-specific ROS signal can be observed when the succinate concentration is low and electron efflux out of the complex is limited (details below) (Quinlan, Orr, et al., 2012). From this study, we drew the conclusion that SDH is capable of generating ROS at high rates both in the forward (from succinate to the Q-pool) and reverse (from the reduced Q-pool backward to the flavin site) reactions (Quinlan, Orr, et al., 2012). It has been suggested that both the flavin site (site IIF) and the ubiquinone-binding site (site IIQ) are capable of donating electrons directly to O2 and producing ROS (Paranagama et al., 2010). However, our experimental data support the hypothesis that ROS formation occurs only at site IIF (Quinlan, Orr, et al., 2012).
In isolated mitochondria, succinate is commonly used to stimulate high rates of oxygen consumption and ROS generation. Most of the ROS signal observed during the oxidation of 5 mM succinate is rotenone sensitive and therefore attributable to the ubiquinone-binding site of complex I (site IQ) due to the backflow of electrons from the reduced Q-pool (as described in Section 3.1). After addition of rotenone, the remaining rate can be further suppressed by addition of the complex III Qo site inhibitor, myxothiazol. Under this condition, when sites IQ and IIIQo are inhibited (and site IF generates little ROS because the NAD pool remains mostly oxidized), it is still possible to measure significant rates of H2O2 production. We found that in the presence of rotenone and myxothiazol, peak H2O2 production was triggered at low succinate concentrations (e.g., 400 μM). In fact, we observed a bell-shaped dependence of the ROS production rates on succinate concentration, with the left end of the curve presumably limited by substrate availability and the right end limited by succinate itself. We observed that the ROS production from complex II was exquisitely sensitive to its substrate and substrate analogues (i.e., fumarate, oxaloacetate, malonate) and also partially sensitive to inhibitors of the Q-binding site (carboxins and atpenin A5).
To assess peak ROS production by complex II in the forward direction, the mitochondria should be incubated with electron transport chain inhibitors before the substrate. The inhibitors are: 4 μM rotenone (to block reverse electron transfer from the reduced QH2 pool at site IQ) and 2 μM myxothiazol (to inhibit any contribution of site IIIQo to ROS formation). The rationale of adding the inhibitors before succinate is to limit the accumulation of the products of succinate oxidation that will reduce NADH (this condition favors malate exchange on the dicarboxylate carrier rather than malate oxidation and NADH formation by malate dehydrogenase). Therefore, the ROS signal is cleaner without concerns of contributions from those ROS producers that are in communication with the NADH pool (e.g., site IF and OGDH). From here, a titration of succinate should be performed to look for the peak rate under this condition. There is a “sweet spot” that is likely to be different in different mitochondrial preparations. In skeletal muscle mitochondria, we found that peak ROS production by complex II was stimulated by 400 μM succinate (generating ~1.1 nmol H2O2 min – mg protein –1) (Quinlan, Orr, et al., 2012). In this condition, the ROS rates are predictably sensitive to the addition of the competitive inhibitor malonate (0.5 mM). However, a 50% reduction in the rate is also observed after the addition of the Q-binding site inhibitor atpenin A5 (1 μM). It is important to note in this condition that inhibitors of the Q-binding site have direct effects on substrate oxidation (i.e., they behave noncompetitively and lower the Vmax of the enzyme). Indeed, upon analysis of the enzyme activation state in the presence of atpenin A5, we drew the conclusion that the inhibition in ROS rates observed after addition of atpenin A5 could be ascribed to inhibitory effects at site IIF, not inhibition of ROS production at IIQ (Quinlan, Orr, et al., 2012).
To assess maximal ROS production by complex II in the reverse direction, mitochondria are again incubated in the presence of the inhibitors (4 μM rotenone and 2 μM myxothiazol) before the addition of substrate. For an easily interpretable ROS signal, we have worked with glycerol 3-phosphate as the reductant of the Q-pool (20 mM in skeletal muscle mitochondria, plus 200 nM free Ca2+ to increase the affinity of mGPDH for its substrate—see Section 3.4). Glycerol 3-phosphate reduces Q directly through mGPDH without affecting tricarboxylic acid cycle intermediates. Under this condition, the SDH contribution to the total signal can be assessed by its sensitivity to either 0.5 mM malonate or 1 mM atpenin A5. It is also of note that the equivalent response to two inhibitors that bind at different sites is strong evidence that the ROS arise from a single site (IIF). ROS production driven by reverse electron transport from QH2 into complex II during oxidation of glycerol 3-phosphate can also be observed during state 4 respiration, in the absence of complex III inhibitors, but it is a smaller proportion of the total in this condition (Quinlan, Orr, et al., 2012).
3.3. Complex III, cytochrome bc1 complex
Complex III is an important site of energy transduction in the electron transport chain; it transfers reducing equivalents from ubiquinol in the lipid phase to the higher potential acceptor in the aqueous phase (cytochrome c) while performing a critical role in energy conservation. Complex III operates by a Q-cycle mechanism (Crofts, 2004; Mitchell, 1975; Trumpower, 1990). The enzyme's kinetic mechanism proceeds through a bifurcated electron transfer at the quinol oxidation site (Qo site) that results in the first electron moving down the high potential chain (Rieske Fe–S to cytochrome c1 to cyto-chrome c) as the second electron is sent down the low potential chain (cyto-chrome b566 to cytochrome b562) where it ultimately reduces a ubiquinone (in the Qi site) to regenerate quinol as part of the Q-cycle. In this way, two turns of the Q-cycle result in the oxidation of two quinols in the Qo site and the reduction of one ubiquinone in the Qi site. Energy conservation is achieved by the net movement of two electrons from quinol to cyto-chrome c, which drives the loss of two protons from the matrix and the appearance of four protons in the intermembrane space (ΔpH), and the transfer of two negative charges inward to the matrix (Δψm).
Complex III is suspected to be a physiologically relevant source of mitochondrial ROS (Bell et al., 2007; Klimova & Chandel, 2008). It is well established that the complex generates significant amounts of superoxide in the presence of antimycin A, an inhibitor of the quinol reduction site (Qi site) (Boveris & Cadenas, 1975; Muller, Roberts, Bowman, & Kramer, 2003). These high rates arise because antimycin blocks electron flux out of the b cytochromes in the low potential chain and increases formation of the superoxide-precursor semiquinone in the Qo site (Van den Berg et al., 1979). As the precursor to superoxide is the semiquinone (Muller, Crofts, & Kramer, 2002), one would predict that the highest rates of superoxide production would be observed when site IIIQo is saturated with substrate (fully reduced Q-pool). However, somewhat surprisingly, it has been observed that conditions that slightly oxidize the Q-pool result in maximal rates of superoxide production from complex III (Drose & Brandt, 2008; Erecinska, Wilson, & Miyata, 1976; Quinlan et al., 2011). For example, mitochondria oxidizing 5 mM succinate in the presence of antimycin will generate superoxide at high rates (~1.2 nmol H2O2 min –1 mg protein –1), but these rates are only ~50% of the maximal rates that are achieved when the condition is 5 mM succinate plus 5 mM malonate (Quinlan et al., 2011). In a recent study, we correlated the maximal rates of production from site IIIQo with an intermediate reduction state of cytochrome b566 (70–80% reduced). We modeled this behavior and found that the phenomenon could be explained by a decrease in the rate constant for quinol oxidation (semiquinone formation) when antimycin is present and an even greater decrease when cytochrome b566 is maximally reduced. We also found that the reduction state of cytochrome b562 was correlated to the stability of the semiquinone in the Qo site (Quinlan et al., 2011).
The relationship between the reduction states of cytochrome b566 and b562 is critical to the mechanism of superoxide production at complex III. We observed that in the presence of antimycin and an applied membrane potential (generated through ATP hydrolysis), the peak superoxide production at intermediate b566 reduction was lost. This phenomenon can be explained by the electron distribution between the two cytochromes. Due to its more positive mid-point potential, b562 is the preferred electron acceptor when there is no membrane potential (West, Mitchell, & Rich, 1988). However, in the presence of an electrical potential across the membrane, the distribution of electrons between cytochrome b566 and cytochrome b562 is expected to be equalized. This is an important observation for the physiological mechanism of superoxide production from site IIIQo: under normal physiological conditions, as cytochrome b566 becomes more reduced, superoxide production from this site will increase exponentially. Indeed, this is what we and others have found (Quinlan et al., 2011; Quinlan, Treberg, et al., 2012; Rottenberg, Covian, & Trumpower, 2009).
To design an experiment to observe maximum rates from site IIIQo, mitochondria should be incubated with 2 μM antimycin A and sub-saturating substrate. This condition can be met as described above by titrating succinate with malonate until an intermediate Q-pool reduction state is achieved, or another substrate combination (e.g., glutamate plus malate) can be titrated down to subsaturating concentrations. These titrations will exhibit a bell-shaped curve for superoxide production from site IIIQo, with the highest rates associated with 70–80% reduction of cyto-chrome b566. However, in the presence of a membrane potential (i.e., in the absence of antimycin A), one would predict the superoxide rates from site IIIQo to increase as the reduction of cytochrome b566 increases, without a peak observed at an intermediate reduction state (Quinlan et al., 2011; Quinlan, Treberg, et al., 2012). The rate that is assigned as specific to IIIQo should be determined by its sensitivity to Qo site inhibitors myxothiazol or stigmatellin.
3.4. Glycerol 3-phosphate dehydrogenase
mGPDH is located at the outer leaflet of the mitochondrial inner membrane (Klingenberg, 1970) and comprises part of the glycerolphosphate shuttle. mGPDH oxidizes glycerol 3-phosphate to dihydroxyacetone phosphate, and in turn, dihydroxyacetone phosphate is reduced back to glycerol 3-phosphate by the cytosolic form of GPDH. In this way, reducing equivalents in the cytosol (from NADH oxidation) are transported to the mitochondria (through QH2 formation) where they can contribute to mitochondrial generation of ATP (Klingenberg, 1970).
The cytosol-facing orientation of mGPDH means that its substrate, glycerol 3-phosphate, does not require transport into the mitochondria. mGPDH is an FAD-linked ubiquinone oxidoreductase that donates its electrons directly to the Q-pool. Oxidation of glycerol 3-phosphate is known to stimulate mitochondrial ROS production (Drahota et al., 2002; Miwa et al., 2003; Tretter, Takacs, Hegedus, & Adam-Vizi, 2007). By virtue of its point of electron entry (a direct reductant of the Q-pool), mGPDH is capable of generating ROS from many sites in the mitochondria (i.e., sites IIIQo, IF, IQ, and IIF may all be reduced when electrons enter through mGPDH). However, mGPDH is also known to generate ROS (likely superoxide) itself, which are released to both the matrix and the intermembrane space (Orr et al., 2012).
ROS production from mGPDH is affected by several factors. The enzyme has a calcium-binding domain, which results in an increased affinity for its substrate (decreased KM) when calcium concentrations are increased (Wernette, Ochs, & Lardy, 1981). Furthermore, since electrons from the enzyme are donated to the Q-pool, the redox poise of the Q-pool also affects ROS production from mGPDH (Orr et al., 2012). In a recent study, we showed that in mitochondria the important variables controlling ROS production from mGPDH were expression of the enzyme (dependent on tissue type), substrate concentration, calcium concentration, and reduction state of the Q-pool (Orr et al., 2012).
When designing an experiment to assess mGPDH ROS production, there are several important factors to take into consideration. First, the secondary effects of glycerol 3-phosphate itself need to be assessed and mitigated. Progressive inhibition of H2O2 production, respiration, and membrane potential has been observed at higher glycerol 3-phosphate concentrations (Orr et al., 2012). We found that this response was usually observed at concentrations above 20 mM and could be attributed to the type and amount of counter ion in the glycerol 3-phosphate preparation. To minimize artifacts caused by excess glycerol 3-phosphate, we perform experiments in the presence of 250 nM free Ca2+ to decrease the enzyme's KM for glycerol 3-phosphate (Orr et al., 2012). We also use only the di-sodium salt of glycerol 3-phosphate and keep the final concentration in the experiment below 20 mM. The total concentration of glycerol phosphate can also be decreased by employing only the active isomer (sn-glycerol 3-phosphate) as substrate. However, even with these precautions, glycerol 3-phosphate has effects on the Amplex UltraRed detection system and those should be taken into account by performing the H2O2 calibration in its presence. Once these factors have been taken into account, to assess superoxide production from mGPDH without contribution from other sites, the experiment should be performed in the presence of 4 μM rotenone, 0.5 mM malonate, and 2 μM myxothiazol. This will block electron flux through complexes I, II, and III, respectively and should provide a relatively pure mGPDH signal as long as NAD remains oxidized.
3.5. Electron-transferring flavoprotein–ETF ubiquinone oxidoreductase
ETF–ETFQOR transfers electrons from β-oxidation and the branched-chain aminoacidmetabolicpathwaystotheQ-pool.Bythispathway,energyfromfat and amino acid oxidation can be used to generate protonmotive force and contributetoATPsynthesis.ETFisasolubleFAD-containingproteininthematrix that accepts electrons from nine primary flavoprotein dehydrogenases, including the four straight-chain-specific fatty acyl-CoA dehydrogenases (medium-, short-,long-,andvery-long-chaindehydrogenases),aswellasseveraldehydrogenases involved in the catabolism of amino acids (isovaleryl-CoA dehydrogenase, Ikeda & Tanaka, 1983b; glutaryl-CoA dehydrogenase, Lenich & Goodman, 1986; and short-branched-chain-CoA dehydrogenase, Ikeda & Tanaka, 1983a) and choline (sarcosine and dimethylglycine dehydrogenases, Frisell & Mackenzie, 1962). ETFQOR is an inner membrane-bound protein that contains two main redox centers, FAD and Fe–S, and has a ubiquinone-binding site (Watmough & Frerman, 2010; Zhang, Frerman, & Kim, 2006). Its main function is to oxidize reduced ETF and transfer electrons to the Q-pool. Flavoprotein dehydrogenases transfer a total of two electrons to ETF with the formation of semireduced ETF as an intermediate (Ramsay, Steenkamp, & Husain, 1987; Ruzicka & Beinert, 1977). The rate constant of the first electron transfer reaction is several fold faster than that of the second electron transfer reaction and therefore the FAD moiety in ETF is thought to be stabilized as a semiflavin (Hall & Lambeth, 1980; Ramsay et al., 1987). This makes ETF a good candidate for superoxide or H2O2 production and this has been shown at the level of the isolated enzyme (Rodrigues & Gomes, 2012). Electron transfer events in ETFQOR are also thought to form the semireduced state of the FAD moiety and likely semiquinone as an intermediate. However, ETFQOR is proposed to possess a low reactivity toward oxygen based on the crystal structure of the protein and, to the best of our knowledge, there are no data supporting or opposing this suggestion (Zhang et al., 2006). Therefore, H2O2 production detected upon reduction of the ETF-ETFQOR system is attributed to two proteins (ETF and ETFQOR) without clear distinction between them at this point.
When designing an experiment to assess ROS production during fatty acid oxidation, there are several important factors to take into account. The choice of substrate is important; we prefer palmitoylcarnitine to palmitoyl-CoA because palmitoyl-CoA requires modification to palmitoylcarnitine to cross the mitochondrial inner membrane. This reaction is catalyzed by carnitinepalmitoyl transferase-1, which can have strong rate limitation in this system (Eaton, 2002). Palmitate cannot be used as a substrate for β-oxidation in isolated mitochondria without addition of ATP, CoA, and the cytoplasmic enzyme acyl-CoA synthetase. Therefore, when investigating ROS production from long-chain fatty acid oxidation, we use 15 μM palmitoylcarnitine. We prefer the l-isomer, rather than the dl mix because the mixture of isomers seems to have a stronger uncoupling effect at increasing concentrations, possibly due to higher contamination with free palmitate, which acts as a detergent. For our stock solutions, we prepare l-palmitoylcarnitine to 10 mM in our assay buffer in the absence of BSA and phosphate. The l-palmitoylcarnitine is incubated at 37 °C to ensure it is fully dissolved, and then it is aliquoted and frozen. Before use, it is warmed again to 37 °C to ensure full dissolution in the buffer. In the assay, we supplement the l-palmitoylcarnitine with 2 mM free carnitine. Carnitine severs two purposes here: one, it is required to maximize accumulation of palmitoylcarnitine inside the mitochondria; and two, it supports high flux through β-oxidation by forming acetylcarnitine from the excess acetyl-CoA that accumulates as an end product (Perevoshchikova et al., 2013). Instead of carnitine, supplementation of l-palmitoylcarnitine with 5 mM malate can stimulate the oxidation of acetyl-CoA in the TCA cycle, but it is less desirable as a substrate because it will recruit other enzymes into the pathway and make dissection of the ROS sites more complicated.
We have found (Perevoshchikova et al., 2013) that the maximum rate of H2O2 production by the ETF–ETFQOR system is achieved during oxidation of 15 μM palmitoylcarnitine plus 2 mM free carnitine and 0.5 mM malonate (to inhibit ROS production arising from complex II). Additionally, the Q-pool needs to be highly reduced (accomplished in the presence of 2 μM myxothiazol), and the antioxidant defense system compromised (in the presence of 4 μM of the uncoupler FCCP). In this case, uncoupling of mitochondria may compromise the antioxidant defense system by decreasing NAD(P)H formation through the energy-dependent transhydrogenase and thereby limiting reduction of the GSH pool (Rydstrom, 2006). With the addition of substrate, the upstream redox centers (NAD+, ETFQOR, ETF) are reduced; and we propose it is this condition that results in a stabilized semireduced ETF and the maximum (but still low) rates from this system (Perevoshchikova et al., 2013).
3.6. Dihydrolipoamide-containing enzyme complexes
PDH and OGDH complexes are both mitochondrial matrix enzyme complexes that contain an FAD-linked dihydrolipoamide (DL) dehydrogenase component. Flavins can make radicals, including both superoxide and H2O2, and the isolated PDH and OGDH complexes have been shown to generate ROS (Bunik & Sievers, 2002; Starkov et al., 2004). However, it has not been definitively shown that the flavin is the ROS-producing site in these enzymes. The second product of the reaction catalyzed by these enzymes, the thiyl radical of the complex-bound DL, may be a significant source of ROS production as well (Bunik, 2003).
We have limited experience with ROS production by PDH. However, we have some experience with OGDH. In skeletal muscle mitochondria, we have observed significant rates of production from OGDH in the presence of phosphate and ADP, which lower the Kd of the enzyme. An assay designed to observe a pure ROS signal from OGDH in situ is not possible without correction for site IF, which will always contribute to the observed signal in intact mitochondria. However, with 5 mM oxoglutarate as substrate, 2 mM ADP and 5 mM phosphate present as activators, and 0.5 mM malonate present to inhibit complex II, the addition of rotenone (4 μM) will generate a substantial rate from OGDH+IF. We surmise that most of the rate observed under this condition can be assigned to OGDH (unpublished observation).
4. CONCLUDING REMARKS
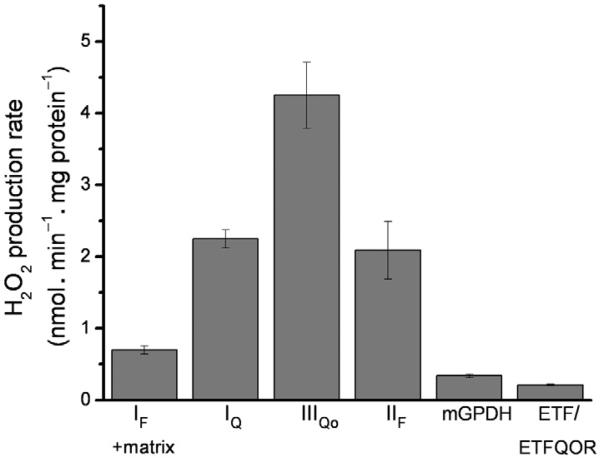
The information given here is intended to provide the reader with a mechanistic understanding of the major mitochondrial ROS producers. We have presented it in the context of the maximum attainable rates, but this informationisreadilytransferabletolesscontrivedsystems.Insummary,Fig.12.4isa compilation of the maximum rates of H2O2 production from each site in skeletal muscle mitochondria. In concert with a mechanistic understanding of the ROS producers in mitochondria, we are developing a system that allows us to predict the contributions from different sites under complex and near-physiological conditions. We recently outlined the principles of using “endogenous reporters” topredictthe ROS contributionsfromspecific sites(Quinlan, Treberg, et al., 2012). In particular, we have calibrated the rate of ROS production from site IF to the reduction state of the endogenous NAD(P)H pool, and the rate of ROS production from site IIIQo to the reduction state of cyto-chrome b566. The endogenous reporter technique, combined with the above information, allows us to assess ROS production frommitochondria-oxidizing complex substrate mixes and predict which sites are the dominant ROS producers under near-physiological conditions. It is important to recognize, when interpreting an undifferentiated ROS signal, that any given scenario may foster a single or multiple sites of mitochondrial ROS production. Understanding the conditions that foster one site over another allows rational interpretation of a global ROS signal and enhanced understanding of physiological and pathological ROS pathways.
Figure 12.4.
Maximum rates of superoxide/H2O2 production from different mitochondrial sites. Data were corrected for H2O2 consumption by matrix peroxidases using the CDNB protocol described in Section 2.4. In all cases, measurements were made under the conditions that we have found to maximize superoxide production rates from these sites, as described in Section 3. The rate from the complex I flavin site includes any other matrix sites that may respond to NADH reduction and is therefore referred to as IF+matrix. Data for this graph are means±SEM (n≥3) and were compiled from Orr et al. (2012), Perevoshchikova et al. (2013), Quinlan et al. (2011), Quinlan, Orr, et al. (2012), and Treberg et al. (2010).
ACKNOWLEDGMENTS
This work was supported by National Institutes of Health, grants P01 AG025901, PL1 AG032118, and R01 AG03354 (to M. D. B.); TL1 AG032116 (to C. L. Q.); and The Ellison Medical Foundation, grant AG-SS-2288-09 (to M. D. B.). Fellowship support was from The Glenn Foundation (to I. V. P.), the Brazilian Government through the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (to R. L. S. G.), and The Carlsberg Foundation (to M. H.-M.).
REFERENCES
- Adam-Vizi V, Chinopoulos C. Bioenergetics and the formation of mitochondrial reactive oxygen species. Trends in Pharmacological Sciences. 2006;27:639–645. doi: 10.1016/j.tips.2006.10.005. [DOI] [PubMed] [Google Scholar]
- Affourtit CL, Quinlan CL, Brand MD. Measurement of proton leak and electron leak in isolated mitochondria. Methods in Molecular Biology. 2012;810:165–182. doi: 10.1007/978-1-61779-382-0_11. [DOI] [PubMed] [Google Scholar]
- Bell EL, Klimova TA, Eisenbart J, Moraes CT, Murphy MP, Budinger GRS, et al. The Qo site of the mitochondrial complex III is required for the transduction of hypoxic signaling via reactive oxygen species production. The Journal of Cell Biology. 2007;177:1029–1036. doi: 10.1083/jcb.200609074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boden MJ, Brandon AE, Tid-Ang JD, Preston E, Wilks D, Stuart E, et al. Overexpression of manganese superoxide dismutase ameliorates high-fat diet-induced insulin resistance in rat skeletal muscle. American Journal of Physiology. Endocrinology and Metabolism. 2012;303:E798–E805. doi: 10.1152/ajpendo.00577.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boveris A, Cadenas E. Mitochondrial production of superoxide anions and its relationship to the antimycin insensitive respiration. FEBS Letters. 1975;54:311–314. doi: 10.1016/0014-5793(75)80928-8. [DOI] [PubMed] [Google Scholar]
- Boveris A, Martino E, Stoppani AOM. Evaluation of the horseradish peroxidase-scopoletin method for the measurement of hydrogen peroxide formation in biological systems. Analytical Biochemistry. 1977;80:145–158. doi: 10.1016/0003-2697(77)90634-0. [DOI] [PubMed] [Google Scholar]
- Boveris A, Oshino N, Chance B. The cellular production of hydrogen peroxide. The Biochemical Journal. 1972;128:617–630. doi: 10.1042/bj1280617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand MD. The sites and topology of mitochondrial superoxide production. Experimental Gerontology. 2010;45:466–472. doi: 10.1016/j.exger.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brotea GP, Thibert RJ. Fluorometric determination of hydrogen peroxide using resorufin and peroxidase. Microchemical Journal. 1988;37:368–376. [Google Scholar]
- Brown GC, Brand MD. Proton/electron stoichiometry of mitochondrial complex I estimated from the equilibrium thermodynamic force ratio. The Biochemical Journal. 1988;252:473–479. doi: 10.1042/bj2520473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunik VI. 2-Oxo acid dehydrogenase complexes in redox regulation. European Journal of Biochemistry. 2003;270:1036–1042. doi: 10.1046/j.1432-1033.2003.03470.x. [DOI] [PubMed] [Google Scholar]
- Bunik VI, Sievers C. Inactivation of the 2-oxo acid dehydrogenase complexes upon generation of intrinsic radical species. European Journal of Biochemistry. 2002;269:5004–5015. doi: 10.1046/j.1432-1033.2002.03204.x. [DOI] [PubMed] [Google Scholar]
- Crofts AR. The cytochrome bc1 complex: Function in the context of structure. Annual Reviews of Physiology. 2004;66:689–733. doi: 10.1146/annurev.physiol.66.032102.150251. [DOI] [PubMed] [Google Scholar]
- Drahota Z, Chowdhury SK, Floryk D, Mracek T, Wilhelm J, Rauchova H, et al. Glycerophosphate-dependent hydrogen peroxide production by brown adipose tissue mitochondria and its activation by ferricyanide. Journal of Bioenergetics and Biomembranes. 2002;34:105–113. doi: 10.1023/a:1015123908918. [DOI] [PubMed] [Google Scholar]
- Drose S, Brandt U. The mechanism of mitochondrial superoxide production by the cytochrome bc1 complex. The Journal of Biological Chemistry. 2008;283:21649–21654. doi: 10.1074/jbc.M803236200. [DOI] [PubMed] [Google Scholar]
- Eaton S. Control of mitochondrial beta-oxidation flux. Progress in Lipid Research. 2002;41:197–239. doi: 10.1016/s0163-7827(01)00024-8. [DOI] [PubMed] [Google Scholar]
- Erecinska M, Wilson DF, Miyata Y. Mitochondrial cytochrome b-c complex: Its oxidation-reduction components and their stoichiometry. Archives of Biochemistry and Biophysics. 1976;177:133–143. doi: 10.1016/0003-9861(76)90423-9. [DOI] [PubMed] [Google Scholar]
- Frisell WR, Mackenzie CG. Separation and purification of sarcosine dehydrogenase and dimethylglycine dehydrogenase. The Journal of Biological Chemistry. 1962;237:94–98. [PubMed] [Google Scholar]
- Golden T, Melov S. Mitochondrial DNA mutations, oxidative stress, and aging. Mechanisms of Ageing and Development. 2001;122:1577–1589. doi: 10.1016/s0047-6374(01)00288-3. [DOI] [PubMed] [Google Scholar]
- Hall CL, Lambeth JD. Studies on electron transfer from general acyl-CoA dehydrogenase to electron transfer flavoprotein. The Journal of Biological Chemistry. 1980;255:3591–3595. [PubMed] [Google Scholar]
- Han D, Canali R, Rettori D, Kaplowitz N. Effect of glutathione depletion on sites and topology of superoxide and hydrogen peroxide production in mitochondria. Molecular Pharmacology. 2003;64:1136–1144. doi: 10.1124/mol.64.5.1136. [DOI] [PubMed] [Google Scholar]
- Hansford RG, Hogue BA, Mildaziene V. Dependence of H2O2 formation by rat heart mitochondria on substrate availability and donor age. Journal of Bioenergetics and Biomembranes. 1997;29:89–95. doi: 10.1023/a:1022420007908. [DOI] [PubMed] [Google Scholar]
- Harman D. Aging: A theory based on free radical and radiation chemistry. Journal of Gerontology. 1956;11:298–300. doi: 10.1093/geronj/11.3.298. [DOI] [PubMed] [Google Scholar]
- Hyslop PA, Sklar LA. A quantitative fluorimetric assay for the determination of oxidant production by polymorphonuclear leukocytes: Its use in the simultaneous fluorimetric assay of cellular activation processes. Analytical Biochemistry. 1984;141:280–286. doi: 10.1016/0003-2697(84)90457-3. [DOI] [PubMed] [Google Scholar]
- Ikeda Y, Tanaka K. Purification and characterization of 2-methyl-branched chain acyl coenzyme A dehydrogenase, an enzyme involved in the isoleucine and valine metabolism, from rat liver mitochondria. The Journal of Biological Chemistry. 1983a;258:9477–9487. [PubMed] [Google Scholar]
- Ikeda Y, Tanaka K. Purification and characterization of isovaleryl coenzyme A dehydrogenase from rat liver mitochondria. The Journal of Biological Chemistry. 1983b;258:1077–1085. [PubMed] [Google Scholar]
- Klimova T, Chandel NS. Mitochondrial complex III regulates hypoxic activation of HIF. Cell Death and Differentiation. 2008;15:660–666. doi: 10.1038/sj.cdd.4402307. [DOI] [PubMed] [Google Scholar]
- Klingenberg M. Localization of the glycerol-phosphate dehydrogenase in the outer phase of the mitochondrial inner membrane. European Journal of Biochemistry. 1970;13:247–252. doi: 10.1111/j.1432-1033.1970.tb00924.x. [DOI] [PubMed] [Google Scholar]
- Korshunov SS, Skulachev VP, Starkov AA. High protonic potential actuates a mechanism of production of reactive oxygen species in mitochondria. FEBS Letters. 1997;416:15–18. doi: 10.1016/s0014-5793(97)01159-9. [DOI] [PubMed] [Google Scholar]
- Kushnareva Y, Murphy AN, Andreyev A. Complex I-mediated reactive oxygen species generation: Modulation by cytochrome c and NAD(P)+ oxidation-reduction state. The Biochemical Journal. 2002;368:545–553. doi: 10.1042/BJ20021121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kussmaul L, Hirst J. The mechanism of superoxide production by NADH: Ubiquinone oxidoreductase (complex I) from bovine heart mitochondria. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:7607–7612. doi: 10.1073/pnas.0510977103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert AJ, Brand MD. Inhibitors of the quinone-binding site allow rapid superoxide production from mitochondrial NADH:ubiquinone oxidoreductase (complex I). The Journal of Biological Chemistry. 2004a;279:39414–39420. doi: 10.1074/jbc.M406576200. [DOI] [PubMed] [Google Scholar]
- Lambert AJ, Brand MD. Superoxide production by NADH:ubiquinone oxidoreductase (complex I) depends on the pH gradient across the mitochondrial inner membrane. The Biochemical Journal. 2004b;382:511–517. doi: 10.1042/BJ20040485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenich AC, Goodman SI. The purification and characterization of glutarylcoenzyme A dehydrogenase from porcine and human liver. The Journal of Biological Chemistry. 1986;261:4090–4096. [PubMed] [Google Scholar]
- Liu Y, Fiskum G, Schubert D. Generation of reactive oxygen species by the mitochondrial electron transport chain. Journal of Neurochemistry. 2002;80:780–787. doi: 10.1046/j.0022-3042.2002.00744.x. [DOI] [PubMed] [Google Scholar]
- McCord JM, Fridovich I. Superoxide dismutase. An enzymic function for erythrocuprein (hemocuprein). The Journal of Biological Chemistry. 1969;244:6049–6055. [PubMed] [Google Scholar]
- Meng Y, High K, Antonello J, Washabaugh MW, Zhao Q. Enhanced sensitivity and precision in an enzyme-linked immunosorbent assay with fluorogenic substrates compared with commonly used chromogenic substrates. Analytical Biochemistry. 2005;345:227–236. doi: 10.1016/j.ab.2005.07.026. [DOI] [PubMed] [Google Scholar]
- Messner KR, Imlay JA. Mechanism of superoxide and hydrogen peroxide formation by fumarate reductase, succinate dehydrogenase, and aspartate oxidase. The Journal of Biological Chemistry. 2002;277:42563–42571. doi: 10.1074/jbc.M204958200. [DOI] [PubMed] [Google Scholar]
- Mitchell P. The protonmotive Q cycle: A general formulation. FEBS Letters. 1975;59:137–139. doi: 10.1016/0014-5793(75)80359-0. [DOI] [PubMed] [Google Scholar]
- Miwa S, St-Pierre J, Partridge L, Brand MD. Superoxide and hydrogen peroxide production by Drosophila mitochondria. Free Radical Biology & Medicine. 2003;35:938–948. doi: 10.1016/s0891-5849(03)00464-7. [DOI] [PubMed] [Google Scholar]
- Mohanty JG, Jaffe JS, Schulman ES, Raible DG. A highly sensitive fluorescent micro-assay of H2O2 release from activated human leukocytes using a dihydroxyphenoxazine derivative. Journal of Immunological Methods. 1997;202:133–141. doi: 10.1016/s0022-1759(96)00244-x. [DOI] [PubMed] [Google Scholar]
- Muller F, Crofts AR, Kramer DM. Multiple Q-cycle bypass reactions at the Qo site of the cytochrome bc1 complex. Biochemistry. 2002;41:7866–7874. doi: 10.1021/bi025581e. [DOI] [PubMed] [Google Scholar]
- Muller FL, Liu Y, Abdul-Ghani MA, Lustgarten MS, Bhattacharya A, Jang YC, et al. High rates of superoxide production in skeletal-muscle mitochondria respiring on both complex I- and complex II-linked substrates. The Biochemical Journal. 2008;409:491–499. doi: 10.1042/BJ20071162. [DOI] [PubMed] [Google Scholar]
- Muller FL, Roberts AG, Bowman MK, Kramer DM. Architecture of the Qo site of the cytochrome bc1 complex probed by superoxide production. Biochemistry. 2003;42:6493–6499. doi: 10.1021/bi0342160. [DOI] [PubMed] [Google Scholar]
- Nohl H, Jordan W. The metabolic fate of mitochondrial hydrogen peroxide. European Journal of Biochemistry. 1980;111:203–210. doi: 10.1111/j.1432-1033.1980.tb06094.x. [DOI] [PubMed] [Google Scholar]
- Ohnishi ST, Shinzawa-Itoh K, Ohta K, Yoshikawa S, Ohnishi T. New insights into the superoxide generation sites in bovine heart NADH-ubiquinone oxidoreductase (Complex I): The significance of protein-associated ubiquinone and the dynamic shifting of generation sites between semiflavin and semiquinone radicals. Biochimica et Biophysica Acta. 2010;1797:1901–1909. doi: 10.1016/j.bbabio.2010.05.012. [DOI] [PubMed] [Google Scholar]
- Orr AL, Quinlan CL, Perevoshchikova IV, Brand MD. A refined analysis of superoxide production by mitochondrial sn-glycerol 3-phosphate dehydrogenase. The Journal of Biological Chemistry. 2012;287:42921–42935. doi: 10.1074/jbc.M112.397828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paranagama MP, Sakamoto K, Amino H, Awano M, Miyoshi H, Kita K. Contribution of the FAD and quinone binding sites to the production of reactive oxygen species from Ascaris suum mitochondrial complex II. Mitochondrion. 2010;10:158–165. doi: 10.1016/j.mito.2009.12.145. [DOI] [PubMed] [Google Scholar]
- Perevoshchikova IV, Quinlan CL, Orr AL, Brand MD. Sites of superoxide and hydrogen peroxide production during fatty acid oxidation in rat skeletal muscle. Free Radical Biology & Medicine. 2013 doi: 10.1016/j.freeradbiomed.2013.04.006. In press. http://dx.doi.org/10.1016/j.freeradbiomed.2013.04.006. [DOI] [PMC free article] [PubMed]
- Pryde KR, Hirst J. Superoxide is produced by the reduced flavin in mitochondrial complex I: A single, unified mechanism that applies during both forward and reverse electron transfer. The Journal of Biological Chemistry. 2011;286:18056–18065. doi: 10.1074/jbc.M110.186841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan CL, Gerencser AA, Treberg JR, Brand MD. The mechanism of superoxide production by the antimycin-inhibited mitochondrial Q-cycle. The Journal of Biological Chemistry. 2011;286:31361–31372. doi: 10.1074/jbc.M111.267898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan CL, Orr AL, Perevoshchikova IV, Treberg JR, Ackrell BA, Brand MD. Mitochondrial complex II can generate reactive oxygen species at high rates in both the forward and reverse reactions. The Journal of Biological Chemistry. 2012;287:27255–27264. doi: 10.1074/jbc.M112.374629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan CL, Treberg JR, Perevoshchikova IV, Orr AL, Brand MD. Native rates of superoxide production from multiple sites in isolated mitochondria measured using endogenous reporters. Free Radical Biology & Medicine. 2012;53:1807–1817. doi: 10.1016/j.freeradbiomed.2012.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ralph SJ, Neuzil J. Mitochondria as targets for cancer therapy. Molecular Nutrition & Food Research. 2009;53:9–28. doi: 10.1002/mnfr.200800044. [DOI] [PubMed] [Google Scholar]
- Ramsay RR, Steenkamp DJ, Husain M. Reactions of electron-transfer flavoprotein and electron-transfer flavoprotein: Ubiquinone oxidoreductase. The Biochemical Journal. 1987;241:883–892. doi: 10.1042/bj2410883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ristow M, Schmeisser S. Extending life span by increasing oxidative stress. Free Radical Biology & Medicine. 2011;51:327–336. doi: 10.1016/j.freeradbiomed.2011.05.010. [DOI] [PubMed] [Google Scholar]
- Rodrigues JV, Gomes CM. Mechanism of superoxide and hydrogen peroxide generation by human electron-transfer flavoprotein and pathological variants. Free Radical Biology & Medicine. 2012;53:12–19. doi: 10.1016/j.freeradbiomed.2012.04.016. [DOI] [PubMed] [Google Scholar]
- Rottenberg H, Covian R, Trumpower BL. Membrane potential greatly enhances superoxide generation by the cytochrome bc1 complex reconstituted into phospholipid vesicles. The Journal of Biological Chemistry. 2009;284:19203–19210. doi: 10.1074/jbc.M109.017376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruch W, Cooper PH, Baggiolini M. Assay of H2O2 production by macrophages and neutrophils with homovanillic acid and horse-radish peroxidase. Journal of Immunological Methods. 1983;63:347–357. doi: 10.1016/s0022-1759(83)80008-8. [DOI] [PubMed] [Google Scholar]
- Ruzicka FJ, Beinert H. A new iron-sulfur flavoprotein of the respiratory chain. A component of the fatty acid beta oxidation pathway. The Journal of Biological Chemistry. 1977;252:8440–8445. [PubMed] [Google Scholar]
- Rydstrom J. Mitochondrial NADPH, transhydrogenase and disease. Biochimica et Biophysica Acta. 2006;1757:721–726. doi: 10.1016/j.bbabio.2006.03.010. [DOI] [PubMed] [Google Scholar]
- Schafer FQ, Buettner GR. Redox environment of the cell as viewed through the redox state of the glutathione disulfide/glutathione couple. Free Radical Biology & Medicine. 2001;30:1191–1212. doi: 10.1016/s0891-5849(01)00480-4. [DOI] [PubMed] [Google Scholar]
- Schulz TJ, Zarse K, Voigt A, Urban N, Birringer M, Ristow M. Glucose restriction extends Caenorhabditis elegans life span by inducing mitochondrial respiration and increasing oxidative stress. Cell Metabolism. 2007;6:280–293. doi: 10.1016/j.cmet.2007.08.011. [DOI] [PubMed] [Google Scholar]
- Staniek K, Nohl H. H2O2 detection from intact mitochondria as a measure for one-electron reduction of dioxygen requires a non-invasive assay system. Biochimica et Biophysica Acta: Bioenergetics. 1999;1413:70–80. doi: 10.1016/s0005-2728(99)00083-3. [DOI] [PubMed] [Google Scholar]
- Starkov AA, Fiskum G. Regulation of brain mitochondrial H2O2 production by membrane potential and NAD(P)H redox state. Journal of Neurochemistry. 2003;86:1101–1107. doi: 10.1046/j.1471-4159.2003.01908.x. [DOI] [PubMed] [Google Scholar]
- Starkov AA, Fiskum G, Chinopoulos C, Lorenzo BJ, Browne SE, Patel MS, et al. Mitochondrial alpha-ketoglutarate dehydrogenase complex generates reactive oxygen species. The Journal of Neuroscience. 2004;24:7779–7788. doi: 10.1523/JNEUROSCI.1899-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St-Pierre J, Buckingham JA, Roebuck SJ, Brand MD. Topology of super-oxide production from different sites in the mitochondrial electron transport chain. The Journal of Biological Chemistry. 2002;277:44784–44790. doi: 10.1074/jbc.M207217200. [DOI] [PubMed] [Google Scholar]
- Sun F, Huo X, Zhai Y, Wang A, Xu J, Su D, et al. Crystal structure of mitochondrial respiratory membrane protein complex II. Cell. 2005;121:1043–1057. doi: 10.1016/j.cell.2005.05.025. [DOI] [PubMed] [Google Scholar]
- Sundaresan M, Yu ZX, Ferrans VJ, Irani K, Finkel T. Requirement for generation of H2O2 for platelet-derived growth factor signal transduction. Science. 1995;270:296–299. doi: 10.1126/science.270.5234.296. [DOI] [PubMed] [Google Scholar]
- Treberg JR, Brand MD. A model of the proton translocation mechanism of complex I. The Journal of Biological Chemistry. 2011;286:17579–17584. doi: 10.1074/jbc.M111.227751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treberg JR, Quinlan CL, Brand MD. Hydrogen peroxide efflux from muscle mitochondria underestimates matrix superoxide production—A correction using glutathione depletion. The FEBS Journal. 2010;277:2766–2778. doi: 10.1111/j.1742-4658.2010.07693.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treberg JR, Quinlan CL, Brand MD. Evidence for two sites of superoxide production by mitochondrial NADH-ubiquinone oxidoreductase (complex I). The Journal of Biological Chemistry. 2011;286:27103–27110. doi: 10.1074/jbc.M111.252502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tretter L, Takacs K, Hegedus V, Adam-Vizi V. Characteristics of alpha-glycerophosphate-evoked H2O2 generation in brain mitochondria. Journal of Neurochemistry. 2007;100:650–663. doi: 10.1111/j.1471-4159.2006.04223.x. [DOI] [PubMed] [Google Scholar]
- Trumpower BL. The protonmotive Q cycle. Energy transduction by coupling of proton translocation to electron transfer by the cytochrome bc1 complex. The Journal of Biological Chemistry. 1990;265:11409–11412. [PubMed] [Google Scholar]
- Van den Berg WH, Prince RC, Bashford CL, Takamiya KI, Bonner WD, Jr., Dutton PL. Electron and proton transport in the ubiquinone cytochrome b-c2 oxidoreductase of Rhodopseudomonas sphaeroides. Patterns of binding and inhibition by antimycin. The Journal of Biological Chemistry. 1979;254:8594–8604. [PubMed] [Google Scholar]
- Vinogradov AD, Grivennikova VG. Generation of superoxide-radical by the NADH:ubiquinone oxidoreductase of heart mitochondria. Biochemistry (Moscow) 2005;70:120–127. doi: 10.1007/s10541-005-0090-7. [DOI] [PubMed] [Google Scholar]
- Votyakova TV, Reynolds IJ. DeltaPsi(m)-Dependent and -independent production of reactive oxygen species by rat brain mitochondria. Journal of Neurochemistry. 2001;79:266–277. doi: 10.1046/j.1471-4159.2001.00548.x. [DOI] [PubMed] [Google Scholar]
- Watmough NJ, Frerman FE. The electron transfer flavoprotein: Ubiquinone oxidoreductases. Biochimica et Biophysica Acta. 2010;1797:1910–1916. doi: 10.1016/j.bbabio.2010.10.007. [DOI] [PubMed] [Google Scholar]
- Wernette ME, Ochs RS, Lardy HA. Ca2+ stimulation of rat liver mitochondrial glycerophosphate dehydrogenase. The Journal of Biological Chemistry. 1981;256:12767–12771. [PubMed] [Google Scholar]
- West IC, Mitchell P, Rich PR. Electron conduction between b cytochromes of the mitochondrial respiratory chain in the presence of antimycin plus myxothiazol. Biochimica et Biophysica Acta. 1988;933:35–41. doi: 10.1016/0005-2728(88)90053-9. [DOI] [PubMed] [Google Scholar]
- Witte ME, Geurts JJ, de Vries HE, van der Valk P, van Horssen J. Mitochondrial dysfunction: A potential link between neuroinflammation and neurodegeneration? Mitochondrion. 2010;10:411–418. doi: 10.1016/j.mito.2010.05.014. [DOI] [PubMed] [Google Scholar]
- Yankovskaya V, Horsefield R, Tornroth S, Luna-Chavez C, Miyoshi H, Leger C, et al. Architecture of succinate dehydrogenase and reactive oxygen species generation. Science. 2003;299:700–704. doi: 10.1126/science.1079605. [DOI] [PubMed] [Google Scholar]
- Zarse K, Schmeisser S, Groth M, Priebe S, Beuster G, Kuhlow D, et al. Impaired insulin/IGF1 signaling extends life span by promoting mitochondrial L-proline catabolism to induce a transient ROS signal. Cell Metabolism. 2012;15:451–465. doi: 10.1016/j.cmet.2012.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Frerman FE, Kim JJ. Structure of electron transfer flavoproteinubiquinone oxidoreductase and electron transfer to the mitochondrial ubiquinone pool. Proceedings of the National Academy of Sciences of the United States America. 2006;103:16212–16217. doi: 10.1073/pnas.0604567103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Yu L, Yu CA. Generation of superoxide anion by succinatecytochrome c reductase from bovine heart mitochondria. The Journal of Biological Chemistry. 1998;273:33972–33976. doi: 10.1074/jbc.273.51.33972. [DOI] [PubMed] [Google Scholar]
- Zhou M, Diwu Z, Panchuk-Voloshina N, Haugland RP. A stable non-fluorescent derivative of resorufin for the fluorometric determination of trace hydrogen peroxide: Applications in detecting the activity of phagocyte NADPH oxidase and other oxidases. Analytical Biochemistry. 1997;253:162–168. doi: 10.1006/abio.1997.2391. [DOI] [PubMed] [Google Scholar]