Abstract
Ku80 facilitates DNA repair and therefore should suppress cancer. However, ku80−/− mice exhibit reduced cancer, although they age prematurely and have a shortened life span. We tested the hypothesis that Ku80 deletion suppresses cancer by enhancing cellular tumor suppressive responses to inefficiently repaired DNA damage. In support of this hypothesis, Ku80 deletion ameliorated tumor burden in APCMIN mice, and increased a p53-mediated DNA damage response, DNA lesions, and chromosomal rearrangements. Thus, contrary to its assumed role as a caretaker tumor suppressor, Ku80 facilitates tumor growth most likely by dampening baseline cellular DNA damage responses.
Introduction
DNA damage drives malignant tumorigenesis, and processes that ameliorate this damage and its sequelae can be categorized as either gatekeeper or caretaker tumor suppressors, depending on their mode of action (1). Gatekeepers control checkpoints that determine the fate of damaged cells whereas caretakers prevent DNA damage or facilitate its repair (2). DNA damage checkpoint machineries monitor the genome for problems that adversely affect DNA replication or mitosis, and halt cell cycle progression to allow time for repair. If the damage is severe or irreparable, these machineries engage either cell death (apoptosis) or permanent cell cycle arrest (cellular senescence) pathways. These cellular damage responses are crucial anti-cancer mechanisms, and require the activity of potent tumor suppressor proteins, such as p53, which are frequently mutated in both heritable and spontaneous cancers (3). Caretakers are also considered to be tumor suppressors because genomic instability drives cancer progression (4) and some heritable forms of cancer are due to mutations in DNA repair genes (5). Unlike gatekeepers, defects in caretakers are not commonly seen in spontaneous tumors. Thus, gatekeepers appear to be more important than caretakers for suppressing spontaneous tumors.
Nonhomologous end joining (NHEJ) repairs DNA double-strand breaks (DSBs) by joining ends without using a homologous template strand, and has been described as a caretaker (6, 7). Known components of mammalian NHEJ include Ku70, Ku80, DNA-PKCS, Artemis, Xrcc4, DNA ligase IV and Xrcc4-like factor. Ku70 and Ku80 form a heterodimer (termed Ku) that complexes with the 460 kD DNA-PKCS to form a holoenzyme referred to as DNA-PK (DNA dependent-protein kinase). Artemis and DNA-PKCS form a complex that opens hairpins and processes overhangs. These DNA ends are then ligated by Xrcc4-DNA ligase IV in a complex with Xrcc4-like factor. By rapidly processing and ligating broken DNA, NHEJ suppresses general genomic instability.
Ku80-deletion causes an increase in the incidence of DNA DSBs (8) and chromosomal aberrations (9) and also increases the incidence of cancer in mice that carry mutations in additional genes like p53 (10, 11). Based on these observations, Ku80 has been termed a tumor suppressor in the caretaker category (6, 7). But this interpretation should be viewed with caution since p53-defective mice are rare or nonexistent in nature; thus, Ku80 may not have been selected for a caretaker function. Interestingly, ku80−/− mice have a reduced, rather than an increased, cancer incidence, although they age prematurely and have a shortened life span (9, 12, 13). We proposed that the low cancer incidence in ku80−/− mice is due to the Ku80 deletion, and is not an indirect consequence of the shortened life span. We further hypothesized that ku80−/− cells exhibit persistent gatekeeper responses, owing to inefficiently repaired DNA breaks, and that these responses ultimately impede oncogenesis.
To test this hypothesis, we analyzed ku80−/− mice in cancer-prone backgrounds that have either defective or intact gatekeeper responses, predicting that Ku80 deletion will exacerbate oncogenesis in gatekeeper-defective mice but ameliorate oncogenesis in gatekeeper-intact mice. In support of this idea, we previously showed that Ku80 deletion enhances p53-dependent cellular senescence in cultured fibroblasts (10) and exacerbates the development of pro-B cell lymphomas (10) and medulloblastomas (11) in p53-deficient mice. Thus, p53 impacts the Ku80-mutant phenotype to enhance cellular senescence and suppress at least two forms of cancer. However, neither pro-B cell lymphoma nor medulloblastoma are common spontaneous tumors in mice, leaving open the question of whether gatekeeper responses are indeed responsible for the low levels of spontaneous cancer in ku80−/− mice.
Here, we evaluate the impact of Ku80 deletion on spontaneous oncogenesis, and investigate the possibility that Ku80 deficiency induces p53-mediated DNA damage responses to poorly repaired DNA lesions. To study spontaneous oncogenesis, we analyzed APCMIN mice deleted for Ku80 because they develop spontaneous intestinal tumors at high incidence within the life span of ku80−/− mice. Importantly, APCMIN mice retain functional DNA damage checkpoints (14). We show that Ku80 deletion significantly reduces tumor burden in APCMIN mice. In both MEFs and small intestine, we show that Ku80 deletion induces a p53-mediated DNA damage response that elevates p21 expression, causes persistent DNA damage, and increases chromosomal rearrangements. These findings indicate that Ku80 deletion reduces spontaneous oncogenesis, most likely by inducing a p53-mediated DNA damage response to persistent DNA lesions.
Materials and Methods
Phenotypic observations/mouse husbandry
We crossed Ku80+/− mice (C57BL/6J X 129SvEv cross) (15) to APCMIN mice (Jackson Laboratories, C57BL/6J-ApcMin/J) to generate all genotypes presented in figure 1. Thus all mice are controlled for genetic background and environment since they are brothers, sisters and cousins raised under identical conditions (the same room, cages, bedding and food). Mice were housed in a specific pathogen-free environment in microisolator cages and observed 5–6 times per week for their entire life spans. Moribund mice (significant loss of weight and responsiveness) were monitored multiple times per day and euthanatized when incapable of reaching the water source. Morbidities were scored by Kaplan-Meier analysis and statistical significance was determined by the log-rank test. Euthanatized mice were examined by necropsy and organs were removed and fixed for histology. Genotyping has been described for Ku80 (9) and APCMIN (http://jaxmice.jax.org/strain/002020.html).
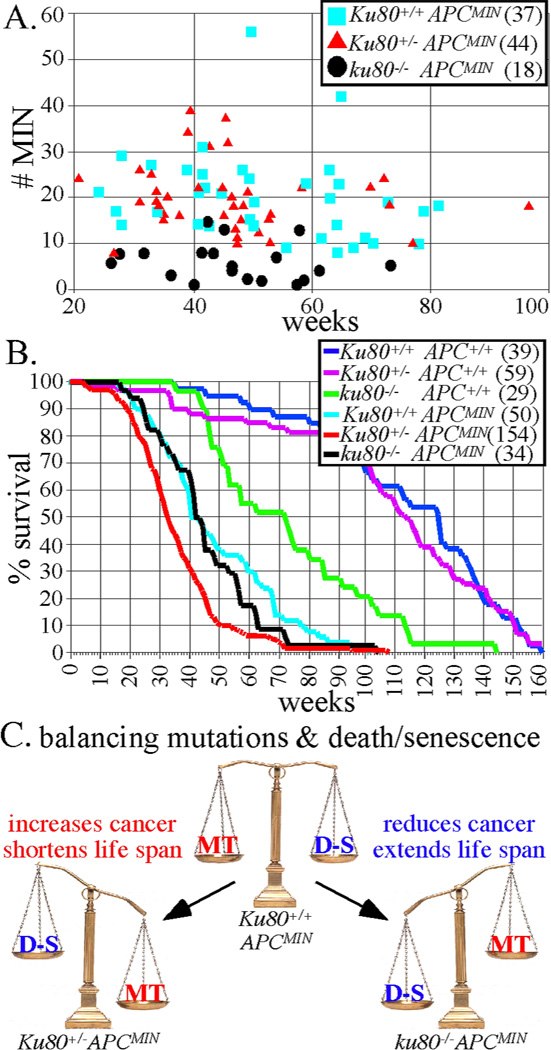
Figure 1.
Ku80-deletion reduces intestinal tumors in APCMIN mice. A, Number of MIN tumors for APCMIN mice with +/+. +/− and −/− Ku80 genotypes. Number of mice observed in parentheses. B, Life spans for all cohorts of mice. Number of mice observed in parentheses. C, Balance between DNA mutations (MT) that exacerbate cancer and cell death or senescence (D-S) that ameliorate cancer.
For mutation analysis, Ku80+/− mice on a C57/BL/6J background were crossed with C57BL/6J pUR288-(lacZ)-transgenic mice line 60—(integration sites on chromosomes 3 and 4) and bred to generate ku80−/− animals hemizygous for pUR288-lacZ. Ku80+/+ littermate animals served as wild type (wt) controls. The animals were maintained in the animal facilities of the University of Texas Health Science Center at San Antonio on a 12-hour light/12-hour dark cycle at a standard temperature of 23 °C. Standard lab chow (Harlan Teklad, Madison WI) and water were supplied ad libitum. Animals were sacrificed by CO2 inhalation followed by cervical dislocation at 4–5 months of age.
MEF Isolation
To generate MEFs, 13.5 day old embryos were separated from the placenta and surrounding membranes, rinsed in 70% ethanol and placed into 1.5 ml eppendorf tubes containing phosphate buffered saline (PBS). Repeated pipetting with a transfer pipette finely minced the embryos. Trypsin-EDTA was added to the tubes, they were incubated at 37°C for 15min and the contents were seeded onto 15 cm plates in M10 medium (Minimum Essential Medium with 10% fetal bovine serum, 2 mM glutamine, 30 mg penicillin/ml, 50 mg streptomycin/ml). This culture was considered passage zero. All cultures were maintained in a 3% O2 and 5% CO2 atmosphere.
Protein extraction and Western blot
Whole cell lysates were prepared from MEFs or mouse small intestine. Tissues were collected, washed with PBS, and snap-frozen in liquid N2. Frozen tissues were ground into a powder using a mortar and pestle and suspended in 150 mM NaCl, 10 mM Tris-HCl pH 8.0, 1 mM EDTA pH 8.0, 1% NP-40, and 1% sodium deoxycholate containing protease inhibitors (Complete, Mini protease inhibitor tablets; Roche)(lysis buffer). After extensive homogenization and sonication, extracts were clarified by centrifugation (10 min at 10,000 × g). MEF were grown to confluency on 15 cm plates, washed with PBS, trypsinized, collected by centrifugation, and lysed in lysis buffer. Extracts were clarified by centrifugation for 10 min at 10,000 × g. Cell or tissue extracts (80 µg) were resolved by 10% SDS-PAGE and proteins were detected by Western blot using mouse monoclonal antibodies to p53 (Ab-1, Clone PAb 240, LabVision; Fremont, CA), phospho-p53 (Ser15, #9284, Cell Signaling Technology; Danvers, MA), p21 (sc-397, Santa Cruz Biotechnology; Santa Cruz, CA), p16 (sc-1207, Santa Cruz Biotechnology; Santa Cruz, CA), or β-actin (ab6276, Abcam; Cambridge, MA) according to the manufacturer’s instructions.
Irradiation
Mice were treated with a single dose of whole body 137Cs gamma irradiation (Mark1 gamma radiation source, Shepard and Associates) of 8 Gy at an exposure rate of 2.44 Gy/min and sacrificed 4 h later to collect tissues. MEF for figure 2 were treated with a single dose of 5 Gy at an exposure rate of 2.44 Gy/min and lysed in lysis buffer 2 h later. Control mice and MEF were placed in the radiation chamber for the same amount of time, but were not irradiated. MEF for figure 4 were cultured on glass slides and treated with a single dose of 10 Gy X-irradiation at rates equal to or above 0.75 Gy/min using a Pantak X-ray generator (320 kV/10 mA with 0.5 mm copper filtration), and lysed in lysis buffer 2 h later.
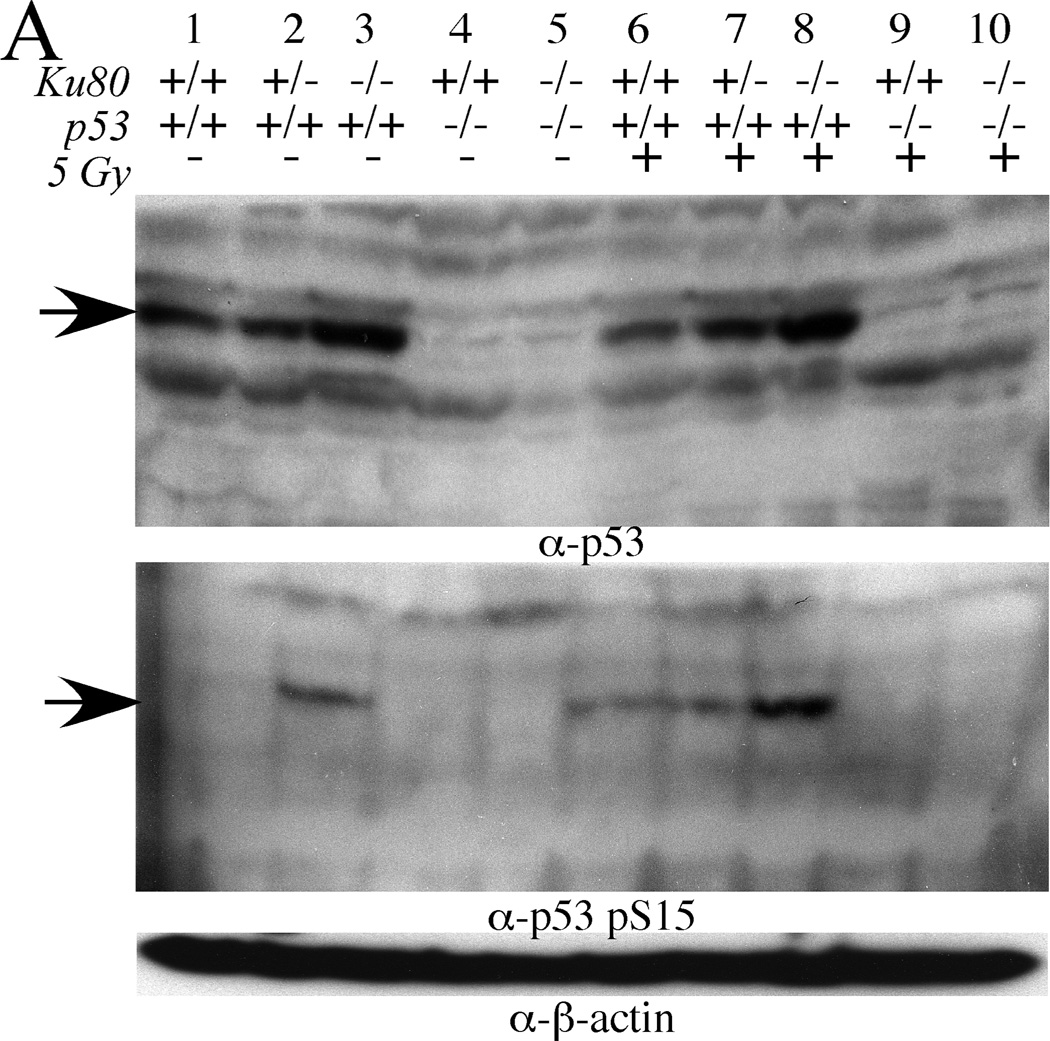
Figure 2.
Ku80-deletion increases p53-mediated DNA damage response in MEFs. (A, B) Western blots of passage 3 MEFs. A, Total p53 (top panel, arrow), p53 pS15 (middle panel, arrow) and β-actin (bottom panel). B, Total p21 (top panel), p16 (middle panel) and β-actin (bottom panel).
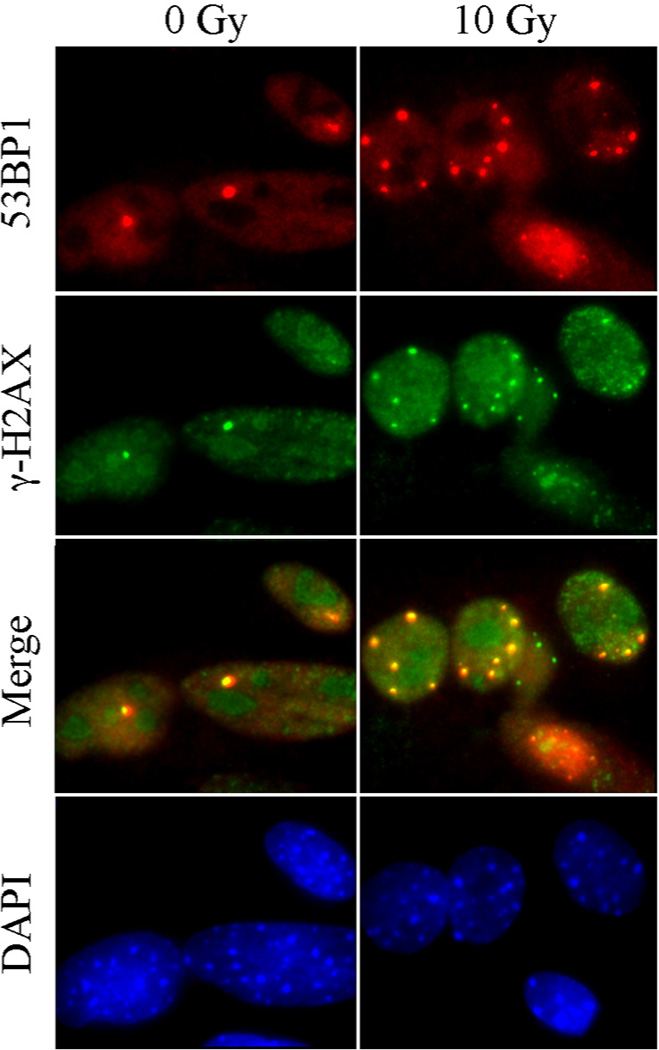
Figure 4.
53BP1 and γ-H2AX colocalize in persistent or spontaneous DNA damage foci in MEFs. MEFs grown on glass slides were mock-irradiated or irradiated with 10 Gy X-rays and let recover for 48 h before fixation and staining with the indicated antibodies. Note that the yellow foci in the merge panel demonstrate co-localization between 53BP1 (red) and γ-H2AX (green).
RNA extraction and cDNA synthesis
Total RNA was prepared by grinding frozen small intestine into a powder using a mortar and pestle. The powdered tissue was homogenized in Trizol Reagent (Invitrogen, Carlsbad, CA) according to manufacturer’s instructions and reverse transcribed using oligo dT (SuperscriptII, Invitrogen, Carlsbad, CA).
Real time RT-PCR
Primers used for quantitative RT-PCR were as follows: p21 forward 5'-CCA GGC CAA GAT GGT GTC TT-3' and p21 reverse 5'-TGA GAA AGG ATC AGC CAT TGC-3'; MDM2 forward 5’-TGTCTGTGTCTACCGAGGGTG-3’; MDM2 reverse 5’-TCCAACGGACTTTAACAACTTCA-3’; KILLER/DR5 forward 5’-GAGGGTATTGACTACACCAGCC-3’; KILLER/DR5 reverse 5’-ATGTTGCATCGGGTTTCTACG-3’; GADD45a forward 5’ CTCGGCTGCAGAGCAGAAGA-3’ GADD45a reverse 5’-GGCACAGTACCACGTTATCG-3; BAX forward 5’-GACAGGGGCCTTTTTGCTA - 3’ BAX reverse 5’-TGTCCACGTCAGCAATCATC -3’; glyceraldehyde-3-phosphate dehydrogenase (GAPDH) forward 5'-CCT GTT CGA CAG TCA GCC G-3' and GAPDH reverse 5'-CGA CCA AAT CCG TTG ACT CC-3'. Quantitative RT-PCR was performed using an Applied Biosystems 7900HT Fast Real-Time PCR System (Applied Biosystems, Foster City, CA) and Absolute SYBR Green ROX Mix (ABgene, Epsom, United Kingdom). The ΔΔCt method of relative quantification was employed to calculate fold expression, p21 (MDM2, KILLER/DR5, GADD45a BAX) RNA levels were normalized to GAPDH levels, and all reactions were performed in triplicate.
Immunostaining
MEFs were plated at 5 × 104 cells per well in 4-well chamber slides and cultured at 3% O2 for 5 days, as described (Parrinello, 2003). Cells were washed with PBS, fixed with 4% paraformaldehyde in PBS for 10 min at room temperature, permeabilized with 0.5% Triton X-100 for 10 min, blocked for 30 min and incubated overnight at 4° C with primary antibody. After washing 3 times with PBS, cells were incubated with secondary antibody for 45 min at room temperature, washed once with PBS and again with PBS containing DAPI(1 ug/ml). The cells were photographed using a Nikon Eclipse 800 microscope with a 40X objective and Nikon Digital Camera DXM1200. The primary antibody for 53BP1 was a rabbit polyclonal from Bethyl (BL182) and the secondary antibody was a donkey anti-rabbit conjugated to Alexa fluor 594 from Molecular probes. The primary antibody for γ-H2AX was a mouse monoclonal from Upstate (anti γ-H2AX ser139 antibody clone JBW301) and the secondary antibody was a donkey anti-mouse conjugated to Alexa fluor 488 from Molecular probes.
Plasmid rescue and mutation analysis
DNA was isolated by phenol/chloroform extraction, as described (16). Briefly, 20–30 µg genomic DNA was digested with Hind III (Roche) for 1 h in the presence of magnetic beads (Dynal) pre-coated with lacZ-lacI fusion protein. The beads were washed 3 times to remove excess genomic DNA, and plasmids were eluted from the beads by IPTG. After circularization of the plasmids with T4 DNA ligase, they were ethanol precipitated and used to transform Escherichia coli C (ΔlacZ, galE−) cells. One thousandth of the transformed cells was plated on a titer plate containing X-gal, and the remainder on a selective plate containing p-gal. The plates were incubated for 15 h at 37 °C. Mutant frequencies were determined from the number of colonies on the selective plate divided by the number of colonies on the titer plate, times 1,000. Each mutant frequency is based on at least 300,000 recovered plasmids.
Mutant classification and characterization
Mutant colonies from the selective plates were grown at 37 °C overnight in 96-well round-bottomed plates containing LB medium, kanamycin and ampicillin. One µl was then plated on X-gal to screen for galactose-insensitive host cells and this background was subtracted. One µl was added to a PCR mix and the DNA amplified as described (16). The PCR products were digested with AvaI and size separated on a 1% agarose gel. Mutant plasmids with restriction patterns resembling the wild-type pattern were classed as “no-change” mutations, whereas those deviating from the wild-type restriction pattern were classified as “size-change” mutations. Approximately 48 mutants per condition were analyzed. LacZ genes of selected mutant plasmids were prepared for sequencing by the UC Davis Sequencing facility (Davis, CA). The chromatograms were analyzed with Sequencher (Gene Codes, Ann Arbor, MI). The primers used for the sequence reactions were described (16).
Results and Discussion
Ku80 deletion ameliorates tumor burden for APCMIN mice
We crossed ku80−/− (15) and APCMIN (Jackson laboratories) mice to determine the impact of Ku80 deletion on oncogenesis. The APC gene is mutated in people with familial adenomatous polyposis coli (17, 18). Similar to humans, mice with APC mutations are predisposed to multiple intestinal neoplasias (MIN), both adenomas and adenocarcinomas. APCMIN/MIN embryos die from primitive ectoderm failure before gastrulation (19); therefore, cancer can be observed only in heterozygote mice. APC+/MIN (termed APCMIN) mice die from anemia caused by multiple intestinal neoplasias (20, 21). APC down regulates the Wnt signaling pathway by associating with β-catenin and controls cellular proliferation (22). Importantly, p53 deficiency enhances the multiplicity and invasiveness of intestinal adenomas in APCMIN mice (14). We therefore reasoned that Ku80 deficiency should suppress the MIN phenotype if it activates p53 responses.
Ku80+/− APCMIN breeding pairs generated cohorts for all genotypes. All APCMIN cohorts, independent of Ku80 status, developed intestinal neoplasias. We counted MIN at the end of life and found that complete Ku80 deficiency (ku80−/−) reduced MIN by ~67% (Figure 1A, p<0.0001, non-parametric Wilcoxon rank sum test). On average, Ku80+/+APCMIN and Ku80+/−APCMIN mice developed 20.0 and 18.9 tumors per intestine, whereas ku80−/−APCMIN mice exhibited only 6.2 tumors per intestine. Thus, complete Ku80 deficiency substantially reduced the total number of adenomas and adenocarcinomas at the end of life for APCMIN mice.
We next determined the effect of this significant reduction in cancer incidence on life span. ku80−/−APCMIN mice survived longer than Ku80+/−APCMIN mice (Figure 1B, p>0.006 log rank test), consistent with Ku80 deficiency suppressing cancer development. However, the Ku80+/+APCMIN mice also lived longer than Ku80+/−APCMIN mice (p>0.001), and survived for about the same length of time as ku80−/−APCMIN mice (p>0.119). Thus, Ku80 haploinsufficiency, but not Ku80-deletion reduced life span in APCMIN mice.
Why did tumor number fail to correlate with life span? It is important to note that tumor number was measured at the end of life. For Ku80+/+APC+/+ and Ku80+/−APC+/+ mice, about 20 tumors per animal were commonly found at the time of death. This finding suggests that cancer onset and/or progression was more severe in the Ku80+/−APC+/+ mice compared to Ku80+/+APC+/+ mice, but total tumor number was the same. However, the total number of tumors at the end of life was much lower for ku80−/−APCMIN mice. About half of these mice exhibited six or fewer tumors and some exhibited as low as one or two tumors (Figure 1A). Therefore, in addition to tumor burden, the early aging phenotype likely contributed to death in ku80−/−APCMIN mice. Many of the ku80−/−APC+/+ mice died from early aging during the life span of the ku80−/−APCMIN mice (Figure 1B) showing that the latter cohort died from a combination of early aging and MIN.
Why does Ku80 gene dosage fail to correlate with life span? The odd response to Ku80 gene dosage suggests a balance between two independent events that either increase or reduce the MIN phenotype. We hypothesized that these events are DNA mutations (MT) and the DNA damage responses that ultimately lead to deleterious cell fates like cell death or senescence (D-S) (Figure 1C). We anticipated that Ku80+/+APCMIN mice would harbor a normal level of spontaneous DNA damage that was quickly repaired, resulting in minimal mutations and minimal cell death or senescence. Ku80+/−APCMIN mice might show a small decrease in damage repair that only modestly increases mutations and deleterious cell fates. However, ku80−/−APCMIN mice would be substantially deficient in repairing spontaneous DNA damage, and the accumulated damage would stimulate a significant DNA damage response resulting in cell death or senescence, which would ultimately reduce cancer development. This possibility is supported by the observation that Ku80 haploinsufficiency modestly increases chromosomal breaks (likely due to a small decrease in DNA repair) but does not affect cell proliferation, whereas total Ku80 deletion greatly increases chromosomal breaks and enhances p53-dependent cellular senescence (8, 10, 13). Thus, we hypothesized that complete Ku80-deletion lowers tumor burden by elevating DNA damage responses, and predicted that ku80−/− cells should harbor a greater level of spontaneous DNA damage that induced constitutive cellular damage responses, compared to either Ku80+/+ or Ku80+/− cells.
Total Ku80 deletion induces p53-mediated DNA damage responses in fibroblasts and small intestine
Total deficiency in NHEJ was shown to induce cellular senescence (10) and neuronal apoptosis (23), both of which depended on p53 activity. We found p53 to be constitutively activated in ku80−/− murine embryonic fibroblasts (MEF, passage 3) without exposure to DNA damaging agents. Western analyses showed elevated levels of total p53 protein in ku80−/−, compared to Ku80+/+ and Ku80+/− fibroblasts (Figure 2A, top panel compare lanes 1 & 2 to lane 3). In addition, ku80−/− cells showed higher levels of p53 phosphorylated at serine 15 (p53 pS15) than control cells with or without radiation exposure (Figure 2A, middle panel, compare lanes 3 & 8 to lanes 1, 2, 6 & 7). p53 serine 15 is phosphorylated by ATM/ATR (ataxia-telangiectasia mutated/ATM and Rad3-related) in response to DNA breaks and is a hallmark of p53 activation (24), supporting the idea shown in figure 1C that complete Ku80 deficiency, but not Ku80 haploinsufficiency, induced cellular responses to inefficiently repaired DNA breaks.
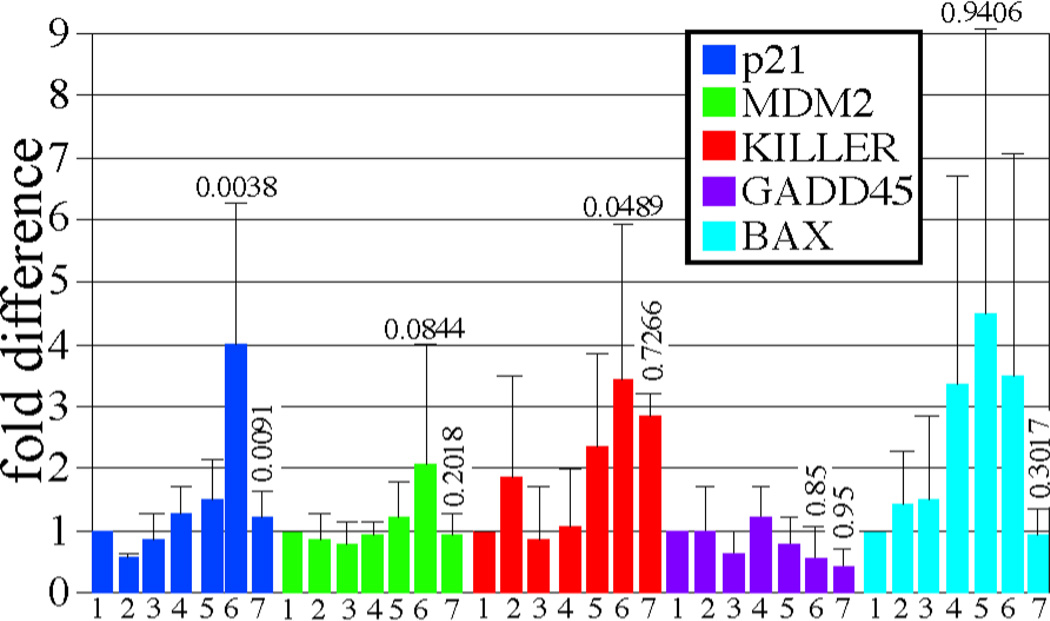
Next, we measured expression levels of p21, a p53 transcriptional target and a cyclin-dependent kinase inhibitor that can induce cellular senescence (2). Complete Ku80 deficiency increased steady state p21 levels, detected by Western blotting (Figure 2B top panel, compare lanes 1 & 2 to lane 3), in a p53-dependent manner (compare lanes 3 & 5). However, Ku80 deficiency did not increase the levels of p16 (Figure 2B middle panel, compare lanes 1 & 2 to lane 3), another cyclin-dependent kinase inhibitor that is important for cellular senescence but is not controlled by p53. Quantitative real time RT-PCR (Q-PCR) showed that complete Ku80 deficiency increased p21 transcription in small intestine 4 h after exposure to 8 Gy γ-radiation (Figure 3, compare blue lanes 4 & 6, p=0.0038, t test) in a p53-dependent manner (Figure 3, compare blue lanes 6 & 7, p=0.0091). By contrast, Ku80 haploinsufficiency did not increase p21 levels (Figure 3 compare blue lanes 4 & 5, p=0.8349). Thus, these observations support our model (Figure 1C) since complete Ku80 deficiency, but not Ku80 haploinsufficiency, enhanced both chronic and acute p53-mediated upregulation of p21 in MEFs and small intestine. Furthermore, these data support our hypothesis that Ku80 deletion inhibits oncogenesis by inducing a p53-mediated DNA damage response.
Figure 3.
Ku80-mutant small intestines exhibit increased p21 levels that are dependent upon p53 in response to 8 Gy γ-radiation as shown by Q-PCR. Lanes 1–3: no IR. Lanes 4–7: 8 Gy. Lanes 1 & 4: Ku80+/+ p53+/+. Lanes 2 & 5: Ku80+/− p53+/+. Lane 3 & 6: ku80−/− p53+/+. Lane 7: Ku80+/+p53−/−. The average of three samples is shown for each cohort. Statistics (student t test) comparing lanes 4 & 6 are above lane 6 and comparing lanes 6 & 7 are above lane 7.
We also tested levels of other p53-induced transcripts in the small intestine with and without exposure to 8Gy γ-radiation. These transcripts include MDM2, KILLER/DR5, GADD45, and BAX (3, 24). Q-PCR suggests a trend that complete Ku80 deficiency increased MDM2 transcription after exposure to 8 Gy γ-radiation (Figure 3, compare green lanes 4 & 6, p>0.0844) in a p53-dependent manner (Figure 3, compare green lanes 6 & 7, p>0.2018). However, this increase is not as dramatic as the increase in p21, and not significant with the numbers tested. The remaining transcripts do not consistently exhibit a pattern that reflects specificity based on Ku80 dosage (GADD45, BAX) and/or p53 regulation (KILLER/DR5, BAX).
Ku80 deficiency increases DNA lesions in fibroblasts and chromosomal rearrangements in small intestine
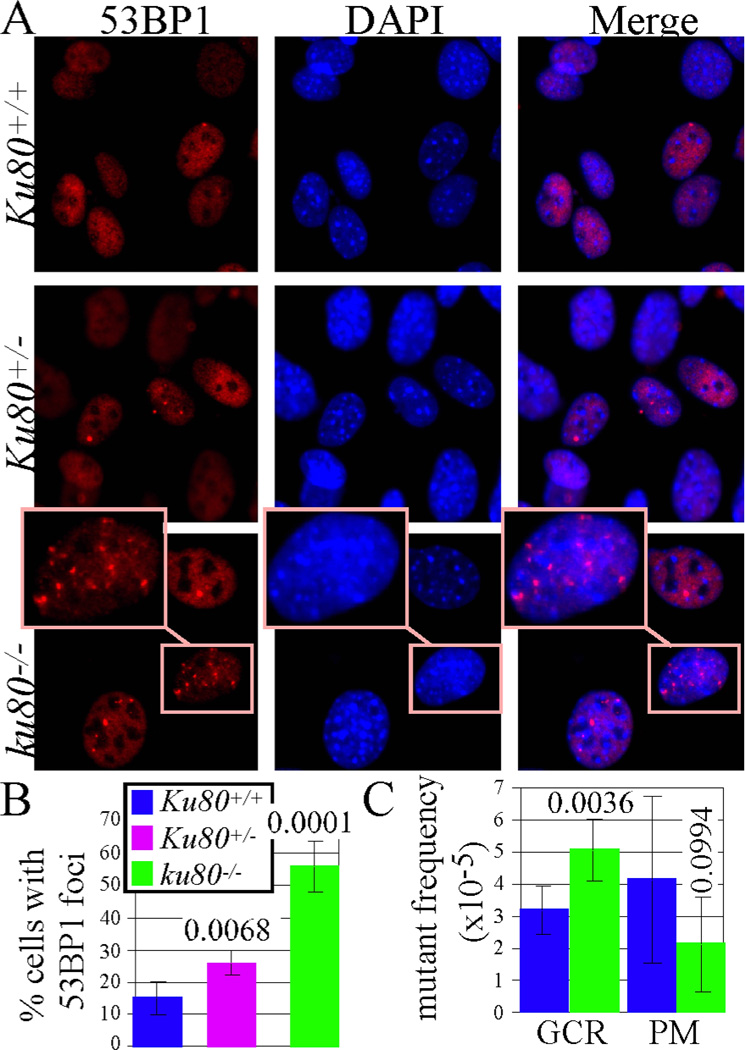
We next asked whether the elevated p21 levels in ku80−/− cells could have resulted from persistent DNA damage. To evaluate DNA lesions, we measured the number of nuclear foci containing 53BP1 (p53 binding protein 1) in MEFs cultured under physiological (3%) O2. We previously showed that ku80−/− fibroblasts are hypersensitive to the oxidative stress of atmospheric (21%) O2 (25). 53BP1 rapidly localizes to DNA DSBs, and is a marker of unrepaired DNA damage and an important component of the DNA damage response (26). Accordingly, we find that 53BP1 and γ-H2AX foci colocalize in control MEFs, as well as in persistent DNA damage foci 48 hours after exposure to 10 Gy X-Rays (Figure 4). Irradiation increased the number of MEF with foci by ~5 fold. Since many γ-H2AX molecules recognize a DNA DSB (27) and since 53BP1 and γ-H2AX foci display perfect co-localization; the spontaneous 53BP1 foci observed in non-irradiated MEFs are therefore likely similar to X-ray induced DSBs. We observed that ~15% of Ku80+/+ MEFs exhibit one or more 53BP1 foci per nucleus, compared to ~25% of Ku80+/− MEFs and ~55% of ku80−/− MEFs (Figure 5A, B). Thus, Ku80 haploinsufficiency caused a modest but significant increase in 53BP1 foci (p=0.0068, sample t test), whereas Ku80 deletion caused a greater increase in 53BP1 foci as compared to both Ku80+/+ MEFs (p>0.0001) and Ku80+/− MEF (p>0.0001). These findings indicate that most of the ku80−/− MEFs harbored persistent unrepaired DNA damage. These data also support our model (Figure 1C) since Ku80 haploinsufficiency increased DNA damage but not p53-mediated DNA damage responses and the levels of DNA lesions were directly dependent on Ku80 gene dosage.
Figure 5.
Ku80-deletion increases DNA damage and mutations. A, 53BP1 foci in MEFs. Note enlarged inset for one ku80−/− nuclei to better visualize 53BP1 foci. B, Quantitation for 53BP1 nuclear foci after observing 1,239 Ku80+/+, 838 Ku80+/− and 688 ku80−/− nuclei. Statistics (student t test) comparing Ku80+/+ to Ku80+/− nuclei (p=0.0068), Ku80+/+ to Ku80−/− nuclei (p>0.0001) and Ku80+/− to Ku80−/− nuclei (p>0.0001). (C) Gross chromosomal rearrangements (GCR) in small intestine. Statistics comparing GCRs in Ku80+/+ to Ku80−/− mice (p>0.0036) and point mutations in Ku80+/+ to Ku80−/− mice (p>0.0994).
We also determined the number of gross chromosomal rearrangements (GCRs) in the small intestine of Ku80+/+ and ku80−/− mice that harbor the pUR288-lacZ mutation reporter (16). Previously, we showed mutations that change the size of the pUR266-lacZ mutation reporter plasmid were GCRs (large deletions, inversions or translocations) whereas mutations that did not change the size of the mutation reporter were point mutations or small insertions/deletions (28). Complete Ku80 deletion significantly increased GCRs in the small intestine (Figure 4C), supporting the notion that DSB repair is chronically defective in the absence of Ku80 function and that an alternative mutagenic pathway repaired these breaks. In addition, we observed a small but not significant decrease in point mutations. Thus, Ku80 deletion impairs DSB repair such that some breaks ultimately result in rearrangements; however, these rearrangements do not frequently lead to cancer, as long as gatekeeper responses are intact.
Conclusion
DNA repair genes are generally thought to suppress malignant tumorigenesis because they are important for maintaining genomic integrity. Consistent with this view, mutations in some repair genes increase the incidence of cancer in both mice and humans. Ku80 is crucial for the repair of DNA DSBs by the NHEJ pathway, and has been described as a tumor suppressor. However, ku80−/− mice, which age rapidly and have a shortened life span, display a very low cancer incidence, suggesting that Ku80 may not act as a tumor suppressor. Nonetheless, it was not clear whether the low cancer incidence was a secondary consequence of the shortened life span or a primary consequence of Ku80 deletion. Here our results show that complete Ku80 deletion reduced the tumor burden of APCMIN mice, which are prone to intestinal tumors but have intact DNA damage responses. Thus, in chronological time, complete Ku80 deletion reduced tumor burden. Our data further suggest that the reduced tumor burden was due to persistent unrepaired DNA DSBs that elevated a p53-dependent DNA damage response. We conclude that Ku80 is not a tumor suppressor but rather, enables tumor development, likely by dampening gatekeeper responses (29).
Acknowledgments
We thank Ms. Charnae Williams for expert technical assistance and Mr. Gary Chisholm for statistical analysis (The Department of Epidemiology and Biostatistics, The University of Texas Health Science Center at San Antonio). We thank Dr. Philip Leder for p21-mutant MEF, Dr. Norman Sharpless for p16/19-mutant MEFs and Dr. Larry Donehower for assistance with detecting p53 by Western blot. This work was supported by grants R01 CA76317-05A1 and 3P30 CA054174-16S2 to PH, NIH UO1 ES11044 to PH and JV, P01 AG17242 to PH, JC and JV and DOD W81XWH-04-1-0325 to VBH.
References
- 1.Kinzler KW, Vogelstein B. Gatekeepers and caretakers. Nature. 1997;386:761–763. doi: 10.1038/386761a0. [DOI] [PubMed] [Google Scholar]
- 2.Campisi J, d'Adda di Fagagna F. Cellular senescence: when bad things happen to good cells. Nat Rev Mol Cell Biol. 2007;8:729–740. doi: 10.1038/nrm2233. [DOI] [PubMed] [Google Scholar]
- 3.Riley T, Sontag E, Chen P, Levine A. Transcriptional control of human p53-regulated genes. Nat Rev Mol Cell Biol. 2008;9:402–412. doi: 10.1038/nrm2395. [DOI] [PubMed] [Google Scholar]
- 4.Lengauer C, Kinzler KW, Vogelstein B. Genetic instabilities in human cancers. Nature. 1998;396:643–649. doi: 10.1038/25292. [DOI] [PubMed] [Google Scholar]
- 5.Hoeijmakers JH. Genome maintenance mechanisms for preventing cancer. Nature. 2001;411:366–374. doi: 10.1038/35077232. [DOI] [PubMed] [Google Scholar]
- 6.Roth DB, Gellert M. New guardians of the genome. Nature. 2000;404:823–825. doi: 10.1038/35009180. [DOI] [PubMed] [Google Scholar]
- 7.Burma S, Chen BP, Chen DJ. Role of non-homologous end joining (NHEJ) in maintaining genomic integrity. DNA Repair (Amst) 2006;5:1042–1048. doi: 10.1016/j.dnarep.2006.05.026. [DOI] [PubMed] [Google Scholar]
- 8.Karanjawala ZE, Grawunder U, Hsieh CL, Lieber MR. The nonhomologous DNA end joining pathway is important for chromosome stability in primary fibroblasts. Curr Biol. 1999;9:1501–1504. doi: 10.1016/s0960-9822(00)80123-2. [DOI] [PubMed] [Google Scholar]
- 9.Li H, Vogel H, Holcomb VB, Gu Y, Hasty P. Deletion of Ku70, Ku80, or Both Causes Early Aging without Substantially Increased Cancer. Mol Cell Biol. 2007;27:8205–8214. doi: 10.1128/MCB.00785-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lim DS, Vogel H, Willerford DM, Sands AT, Platt KA, Hasty P. Analysis of ku80-mutant mice and cells with deficient levels of p53. Mol Cell Biol. 2000;20:3772–3780. doi: 10.1128/mcb.20.11.3772-3780.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Holcomb VB, Vogel H, Marple T, Kornegay RW, Hasty P. Ku80 and p53 suppress medulloblastoma that arise independent of Rag-1-induced DSBs. Oncogene. 2006;25:7159–7165. doi: 10.1038/sj.onc.1209704. [DOI] [PubMed] [Google Scholar]
- 12.Vogel H, Lim DS, Karsenty G, Finegold M, Hasty P. Deletion of Ku86 causes early onset of senescence in mice. Proc Natl Acad Sci U S A. 1999;96:10770–10775. doi: 10.1073/pnas.96.19.10770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Holcomb VB, Vogel H, Hasty P. Deletion of Ku80 causes early aging independent of chronic inflammation and Rag-1-induced DSBs. Mech Ageing Dev. 2007;128:601–608. doi: 10.1016/j.mad.2007.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Halberg RB, Katzung DS, Hoff PD, et al. Tumorigenesis in the multiple intestinal neoplasia mouse: redundancy of negative regulators and specificity of modifiers. Proc Natl Acad Sci U S A. 2000;97:3461–3466. doi: 10.1073/pnas.050585597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhu C, Bogue MA, Lim DS, Hasty P, Roth DB. Ku86-deficient mice exhibit severe combined immunodeficiency and defective processing of V(D)J recombination intermediates. Cell. 1996;86:379–389. doi: 10.1016/s0092-8674(00)80111-7. [DOI] [PubMed] [Google Scholar]
- 16.Garcia AM, Busuttil RA, Rodriguez A, et al. Detection and analysis of somatic mutations at a lacZ reporter locus in higher organisms: application to Mus musculus and Drosophila melanogaster. Methods Mol Biol. 2007;371:267–287. doi: 10.1007/978-1-59745-361-5_20. [DOI] [PubMed] [Google Scholar]
- 17.Nishisho I, Nakamura Y, Miyoshi Y, et al. Mutations of chromosome 5q21 genes in FAP and colorectal cancer patients. Science. 1991;253:665–669. doi: 10.1126/science.1651563. [DOI] [PubMed] [Google Scholar]
- 18.Groden J, Thliveris A, Samowitz W, et al. Identification and characterization of the familial adenomatous polyposis coli gene. Cell. 1991;66:589–600. doi: 10.1016/0092-8674(81)90021-0. [DOI] [PubMed] [Google Scholar]
- 19.Moser AR, Shoemaker AR, Connelly CS, et al. Homozygosity for the Min allele of Apc results in disruption of mouse development prior to gastrulation. Dev Dyn. 1995;203:422–433. doi: 10.1002/aja.1002030405. [DOI] [PubMed] [Google Scholar]
- 20.Moser AR, Pitot HC, Dove WF. A dominant mutation that predisposes to multiple intestinal neoplasia in the mouse. Science. 1990;247:322–324. doi: 10.1126/science.2296722. [DOI] [PubMed] [Google Scholar]
- 21.Su LK, Kinzler KW, Vogelstein B, et al. Multiple intestinal neoplasia caused by a mutation in the murine homolog of the APC gene. Science. 1992;256:668–670. doi: 10.1126/science.1350108. [DOI] [PubMed] [Google Scholar]
- 22.Senda T, Iizuka-Kogo A, Onouchi T, Shimomura A. Adenomatous polyposis coli (APC) plays multiple roles in the intestinal and colorectal epithelia. Med Mol Morphol. 2007;40:68–81. doi: 10.1007/s00795-006-0352-5. [DOI] [PubMed] [Google Scholar]
- 23.Gao Y, Ferguson DO, Xie W, et al. Interplay of p53 and DNA-repair protein XRCC4 in tumorigenesis, genomic stability and development. Nature. 2000;404:897–900. doi: 10.1038/35009138. [see comments]. [DOI] [PubMed] [Google Scholar]
- 24.Meek DW. The p53 response to DNA damage. DNA Repair (Amst) 2004;3:1049–1056. doi: 10.1016/j.dnarep.2004.03.027. [DOI] [PubMed] [Google Scholar]
- 25.Parrinello S, Samper E, Krtolica A, Goldstein J, Melov S, Campisi J. Oxygen sensitivity severely limits the replicative lifespan of murine fibroblasts. Nat Cell Biol. 2003;5:741–747. doi: 10.1038/ncb1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mochan TA, Venere M, DiTullio RA, Jr, Halazonetis TD. 53BP1, an activator of ATM in response to DNA damage. DNA Repair (Amst) 2004;3:945–952. doi: 10.1016/j.dnarep.2004.03.017. [DOI] [PubMed] [Google Scholar]
- 27.Pilch DR, Sedelnikova OA, Redon C, Celeste A, Nussenzweig A, Bonner WM. Characteristics of gamma-H2AX foci at DNA double-strand breaks sites. Biochem Cell Biol. 2003;81:123–129. doi: 10.1139/o03-042. [DOI] [PubMed] [Google Scholar]
- 28.Dolle ME, Giese H, Hopkins CL, Martus HJ, Hausdorff JM, Vijg J. Rapid accumulation of genome rearrangements in liver but not in brain of old mice. Nat Genet. 1997;17:431–434. doi: 10.1038/ng1297-431. [see comments]. [DOI] [PubMed] [Google Scholar]
- 29.Hasty P. Is NHEJ a tumor suppressor or an aging suppressor? Cell Cycle. 2008;7:1139–1145. doi: 10.4161/cc.7.9.5807. [DOI] [PubMed] [Google Scholar]