Abstract
Chemokines critically regulate chemotaxis in normal and pathologic states, but there is limited understanding of how multicellular interactions generate gradients needed for cell migration. Previous studies of chemotaxis of CXCR4+ cells toward chemokine CXCL12 suggest the requirement of cells expressing scavenger receptor CXCR7 in a source-sink system. We leveraged an established microfluidic device to discover that chemotaxis of CXCR4 cells toward distinct isoforms of CXCL12 required CXCR7 scavenging only under conditions with higher than optimal levels of CXCL12. Chemotaxis toward CXCL12-β and -γ isoforms, which have greater binding to extracellular molecules and have been largely overlooked, was less dependent on CXCR7 than the more commonly studied CXCL12-α. Chemotaxis of CXCR4+ cells toward even low levels of CXCL12-γ and CXCL12-β still occurred during treatment with a FDA-approved inhibitor of CXCR4. We also detected CXCL12-γ only in breast cancers from patients with advanced disease. Physiological gradient formation within the device facilitated interrogation of key differences in chemotaxis among CXCL12 isoforms and suggests CXCL12-γ as a biomarker for metastatic cancer.
Introduction
Chemotaxis of cells along a concentration gradient is essential for normal development, tissue homeostasis, and pathogenesis of diseases including metastatic cancer, atherosclerosis, and multiple sclerosis1. Chemotaxis controls trafficking of normal stem cells, and there are ongoing efforts to enhance homing of stem cells to injured tissues for regenerative medicine2. The source-sink model of chemotaxis is one common process to generate gradients and drive cell migration in vitro and in vivo3 The balance between chemotactic molecule secretion (source) and degradation (sink) critically determines gradient profiles and responsiveness of migrating cells3b, c, 4. Recent studies also demonstrate that gradients of chemokine bound to the extracellular matrix, rather than soluble molecules, drive chemotaxis5 by increasing local concentrations of chemokine, limiting degradation, and enhancing presentation to receptors6. Therapeutic targeting of source-sink chemotaxis as an emergent phenotype of multiple cells, receptors, and microenvironmental factors rather than a singular molecular event provides flexibility in drug targets but requires evaluation of the entire integrated system.
Chemokine CXCL12-α and its receptors CXCR4 and CXCR7 are a prominent example of source-sink chemotaxis in normal physiology and pathologic conditions7,8. CXCR7 functions as a scavenger receptor, controlling availability of CXCL12 by removing it from the extracellular space and degrading it3b, c, 4. Two recent studies highlight that CXCL12 secretion and CXCR7 scavenging are obligate partners in generating sustained, local gradients of CXCL12-α in vivo, allowing cells with CXCR4 to migrate toward CXCL12 source cells9. Loss of CXCR7 in both zebrafish and an in vitro device we developed prevented normal migration of CXCR4+ cells due to loss of chemokine gradients and/or desensitization of CXCR4 from elevated levels of CXCL12-α3b, c. While prior studies show that CXCR7 is required for CXCR4-dependent migration toward CXCL12-α, these studies overlook the importance of variable interactions of CXCL12 isoforms with receptors and the extracellular space. Studies of CXCL12 isoforms in chemotaxis have been particularly challenging because only the α-isoform efficiently stimulates chemotaxis in conventional transwell assays, while other isoforms require supraphysiologic concentrations to drive cell migration10.
To investigate interrelationships between a source-sink model and binding of chemotactic molecules to extracellular surfaces, we used our established microfluidic source-sink model of CXCL12, CXCR4, and CXCR7 (Fig. 1). We tested three of the six naturally expressed CXCL12-isoforms (α, β, and γ, common to humans, mice, and rats) that span low-to-high affinities for receptors CXCR4, CXCR7, and the extracellular environment11. Secreted forms of these CXCL12 isoforms share a common N-terminal 68 amino acid core that comprises the entirety of CXCL12-α. CXCL12-β and -γ have four and 30 additional amino acids at the C-termini, respectively. C-termini of CXCL12-β and -γ are enriched with basic amino acids that enhance interactions with negatively-charged extracellular molecules and surfaces10, 11b, 12. In particular, CXCL12-γ binds to major components of the extracellular matrix, such as the glycosaminoglycan heparan sulfate, with more than two orders of magnitude greater affinity than the most commonly studied isoform, CXCL12-α. However, CXCL12-γ binds with lower affinity to receptor CXCR4, and scavenging by CXCR7 is also less efficient. Association of chemotactic molecules with extracellular components also may enhance chemotaxis by increasing local concentrations of chemokine, favoring oligomerization that may be necessary for chemokine activity, limiting proteolytic degradation, and enhancing presentation to receptors 6. These opposing interactions between CXCL12-isoforms and extracellular surfaces or receptors produce marked disparities in bound versus soluble concentrations of each isoform10, 11b. Effects of different isoforms of CXCL12 on gradient formation and chemotaxis within physiological source-sink environments are unknown.
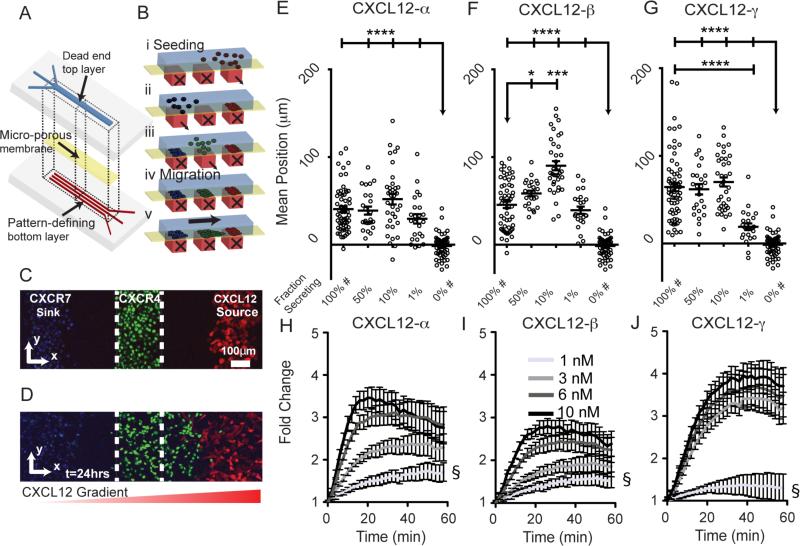
Figure 1. Microfluidic source-sink-migration device.
(A, B) Schematic multilayered microfluidic device fabrication and representation of patterned cell seeding. Controlled flow through bottom channels results in 200 µm wide cell patterns with 200 µm gaps. Parts i, ii, and iii depict stepwise addition of CXCL12-secreting cells, CXCR7+ cells, and CXCR4+ cells, respectively. Parts iv and v represent conditions before and after migration. (C, D) Representative confocal images of patterned cells in the device at t=0 (C corresponds to B iv) and after 24 hours (D corresponds to B v). The CXCL12 secreting cells co-express FP650 (red); CXCR4+ cells express NLS-AcGFP (green); and CXCR7+ cells are stained with Hoescht 33342 (blue). The dashed white line denotes channel boundaries that define the starting position. The graded red triangle below (D) denotes the gradient direction. (E-G) Average position of CXCR4+ cells after 24 hours of migration toward CXCL12-α, -β, or -γ. Each point represents the mean position of~300±50 cells from 1 of 6 view fields from 10-11 devices. Fraction of secreting cells denotes the relative dilution of CXCL12-isoform secreting cells patterned with non-secreting cells. Data are shown as mean values ± S.E.M. (n=6 view fields for 4-11 devices per condition). The bars represent statistical comparison between pairs of conditions. The arrow denotes multiple paired comparisons to the same condition (*p<0.05, ***p<0.005, ****p<0.0001). Data for 100% and 0% secreting cells are marked (#) to designate the same data plotted for comparison in multiple figures. Matched conditions were performed in parallel. (H-J) Cells expressing a luciferase complementation reporter for association of CXCR4 and β-arrestin 2 were incubated with increasing equimolar concentrations of synthetic CXCL12-α, β, or γ. Data were graphed as mean values ± S.E.M. (n=4 measurements) from one of two representative experiments. Fold change in bioluminescence is relative to untreated cells at corresponding time points. The symbol § demarcates statistical differences by Tukey post hoc test between concentrations for the final time point. For CXCL12-α, 1nM is different from 6nM (p<0.01) and 10 nM (p<0.01). For CXCL12-β, 1nM is different from 6nM (p<0.05) and 10 nM (p<0.01). For CXCL12-γ, 1nM is different from all other concentrations (p<0.0001). Comparisons between isoforms are in supplemental information (Fig. S6).
Using unique capabilities of our microfluidic device, we discover that levels of secreted CXCL12 isoforms dictate the requirement for CXCR7-dependent scavenging in chemotaxis of CXCR4+ cells. CXCR7-scavenging is necessary for chemotaxis of CXCR4 cells under conditions with higher levels of CXCL12, while reducing amounts of CXCL12 partially rescues chemotaxis without functional scavenging by CXCR7. Even at concentrations 10 to 20-fold lower, we also show for the first time in vitro that CXCL12-γ, an isoform with highest binding to the extracellular environment, drives chemotaxis of CXCR4 cells to an extent similar to CXCL12-β and greater than CXCL12-α. Exploiting capabilities of this device for drug testing, we demonstrate that AMD3100, the only FDA-approved inhibitor of CXCR4, fails to entirely block migration of CXCR4+ cells toward CXCL12-β or -γ. Moreover, we show for the first time expression of CXCL12-γ in primary human breast cancers and suggest that this isoform is associated with metastatic disease. These results demonstrate that intrinsic biophysical and biochemical differences among chemokine isoforms regulate cell migration and emphasize the need for drugs that more effectively target CXCL12-β and -γ.
Experimental Methods
Plasmid construction
The CXCL12 fusions to Gaussia luciferase (GLuc) were generated by PCR or gene synthesis (supplied in pIDTSMARTKan blunt, Integrated DNA Technologies) as indicated in Supplemental Table S1, products 1,3-5. These were constructed in pEGFP-N1 digested with XhoI and NotI to remove the EGFP open reading frame. A Gly/Ser linker and EcoRI site were included between the CXCL12 and GL open reading frames. The CXCL12-GL fusions were amplified by PCR with appropriate primers shown in Table S1, products 9,10, and inserted into the PacI sites of FU650W (constructed from FUGW as described13) for product 9 or the XbaI and NotI sites of pLVX-EF1α-IRES-mCherry (Clontech) for product 10. Unfused versions of CXCL12 isoforms and secreted Gaussia luciferase were amplified with primers as indicated in Table S1, products 2, 6-8 and cloned into the XbaI and NotI sites of pLVX-EF1α-IRES-mCherry (note that NheI and XbaI have compatible cohesive ends). Vector FUAcGFP-nucW was generated by amplifying nuclear-targeted AcGFP from pAcGFP1-Nuc (Clontech) as indicated in Table S1, product 11, and cloned into the PacI site of FUGW.
Stable cell lines
We transduced MDA-MB-231 cells with lentiviral vectors described above to generate populations of cells expressing fusions of CXCL12-α, β, or γ to GL with co-expressed fluorescent protein FP65014. Expression of FP650 and the IRES-linked mCherry enabled flow cytometry sorting and identification of cells within the microfluidic device. We previously described MDA-MB-231 cells stably transduced with CXCR4-GFP (231-CXCR4)15. To facilitate imaging and image analysis, we transduced 231 CXCR4 cells with FUAcGFP-nucW. We previously reported MDA-MB-231 cells stably transduced with CXCR7-GFP (231-CXCR7) or CXCR7-Δ322-GFP (231- CXCR7-Δ322)4e. We used MDA-MB-231 cells stably expressing click beetle green and red luciferase complementation reporters for association of β-arrestin 2 and CXCR7 or CXCR4, respectively (Coggins, submitted). These reporter cell lines are comparable to the firefly luciferase complementation systems we previously have described for these protein interactions16.
Murine tumor models
We implanted 2 x 105 E0771 mouse breast cancer cells stably expressing firefly luciferase and GFP along with 1 x 105 immortalized mouse mammary fibroblasts (gift of Harold Moses, Vanderbilt) orthotopically into 4th inguinal mammary fat pads of syngeneic C57Bl/6 mice (Taconic). We harvested 8-10 mm diameter tumors and extracted RNA using Trizol (Life Technologies) according the manufacturer's directions. We also generated human breast cancer xenografts by implanting 5 x 105 231-CXCR4 cells co-expressing firefly luciferase into 4th inguinal mammary fat pads of NSG mice4d. We extracted RNA from these tumors as described above.
Western blot analysis
We measured phosphorylation of AKT as we have previously described17. Synthetic CXCL12 isoforms for this assay were purchased from R&D Systems.
Secreted CXCL12 ELISA analysis
We contracted the University of Michigan Cancer Center Immunology Core to perform CXCL12 ELISA per specifications for R&D Systems CXCL12-α ELISA. We previously demonstrated complementary ELISA- and bioluminescence-based CXCL12 measurement17.
Quantitative RT-PCR for CXCL12-isoforms
We measured mRNA levels of CXCL12-α, β, or γ fusions to Gaussia luciferase in stably transduced MDA-MB-231 cells by qRT-PCR using PCR primers common to all isoforms and SYBR Green detection: 5’ tgcccttcagattgttgcacg 3’ and 5’ ctccaggtactcttggatccac 3’, based on our prior protocol13. To analyze expression of CXCL12 isoforms in mouse tumors and bone marrow, we extracted RNA with Trizol reagent (Life Technologies) and further purified RNA with a column-based kit and on-column treatment with DNaseI (Qiagen). We performed qRT-PCR as described above with isoform-specific primers: common forward primer 5’-tgcccttcagattgttgcacg-3’, α-reverse primer 5’-ggctgttgtgcttacttgtttaaagc-3’, mouse β-reverse primer 5’-ctgactcacacctctcacatcttg-3’, human β-reverse primer 5’-ggcgtctgaccctctcacatcttg-3’, and γ-reverse primer 5’-gaactagtttttccttttctgggcagcc-3’. The primers are the same for human and mouse except for the reverse primer for CXCL12-β. We used a cDNA array of normal human breast tissue and breast cancers (Origene, Breast Cancer cDNA Array II). Human primers were used for xenograft tumors and the human cDNA array. For both cells and tissues, we amplified GADPH as a control and quantified data as ΔCt values. We defined tissues as positive for an isoform of CXCL12 based on amplification at <40 qRT-PCR cycles and appropriate size PCR product identified by gel electrophoresis.
Microfluidic device fabrication and preparation
We fabricated the microfluidic migration device as described previously3c. We used a top channel with 100μm depth instead of 200 μm. We patterned cells in three 200μm wide strips spaced 200 μm apart. For time course confocal imaging and for imaging of receptor localization within the device, we fabricated the microfluidic device with the total top layer thickness of ~2mm to allow imaging.
Microfluidic device experimental setup
We seeded cells as described previously3c. For treatment with AMD3100, we supplemented both the final batch of cells and parallel medium with 1 μM AMD3100 (Tocris) in DMEM with 10% FBS. Within an hour of seeding the final batch of cells (CXCR4-GFP), we imaged each device to match corresponding phase contrast and epifluorescence images. We matched parallel device conditions among isoforms, dilutions of secreting cells, and treatments for each run.
Bioluminescence imaging
We imaged fusions of CXCL12-isoforms to Gaussia luciferase and luciferase complementation between CXCR4 or CXCR7 and β-arrestin 2 as described previously16-17.
Image acquisition and analysis
For time point analyses of cells in the microfluidic device, we acquired phase contrast and fluorescence images at 4 and 6 locations along the length of the device channel for t=0 and 24 hours, respectively (n=4-11 device setups per condition). We captured images using an inverted Olympus IX70 microscope with a 10X objective. We developed a semi-automated NIH ImageJ (64bit) script to measure location of NLS-AcGFP tagged CXCR4+ cells relative to the channel boundaries (Supplemental Info). The script returned coordinates of each CXCR4+ cell based on NLS-GFP (fluorescence) relative to the matched channel boundaries (phase contrast). We measured the average position for each view field in the direction of the source cells. For time-lapse microscopy, we used a custom CO2 and temperature controlled stage for live-cell imaging with a 10X, 0.3 NA objective and upright confocal microscope (Olympus MPE Twin). For each CXCL12-isoform, we imaged a single view field of three separate device setups at 15-minute increments for 20 hours total. Using a modified ImageJ script, we measured position of cells over time. We imaged CXCR4-GFP localization in the microfluidic channel by confocal microscopy using a 60X, 1.0NA objective. We captured z-stack images (2.5μm increments) at 5 positions along the channel length and displayed representative z-compressions that were adjusted identically in parallel for demonstration (n = 2 devices per condition).
Transwell migration experiments
We performed transwell migration assays as reported previously18. Briefly, we incubated 231-cells overexpressing CXCR4+ in transwells with equal levels of CXCL12-isoforms, based on Gaussia luciferase activity. We observed migrated cells based on crystal violet staining.
Statistical analysis
We made all plots and statistical comparisons using GraphPad Prism. We plotted time point data as mean ± standard error of the mean (S.E.M.). For time course data, we plotted the mean ± standard deviation (S.D.) of percentile lines, which we created in Microsoft Excel. For all statistical comparisons of migration data we performed paired, two-sided statistical comparisons using the Mann-Whitney U-test (non-parametric) with exact p-values. For analysis of CXCL12-uptake we performed simple two-sided t-test. For analyses of bioluminescence complementation (Figs. 1, 3, S6, and S7) and CXCR4-GFP internalization over time (Fig. 4E) we performed two-way ANOVA and post hoc Tukey multiple comparisons test within rows. For two-way ANOVA we used time as the row effect versus CXCL12-isoform or concentration for the column effect.
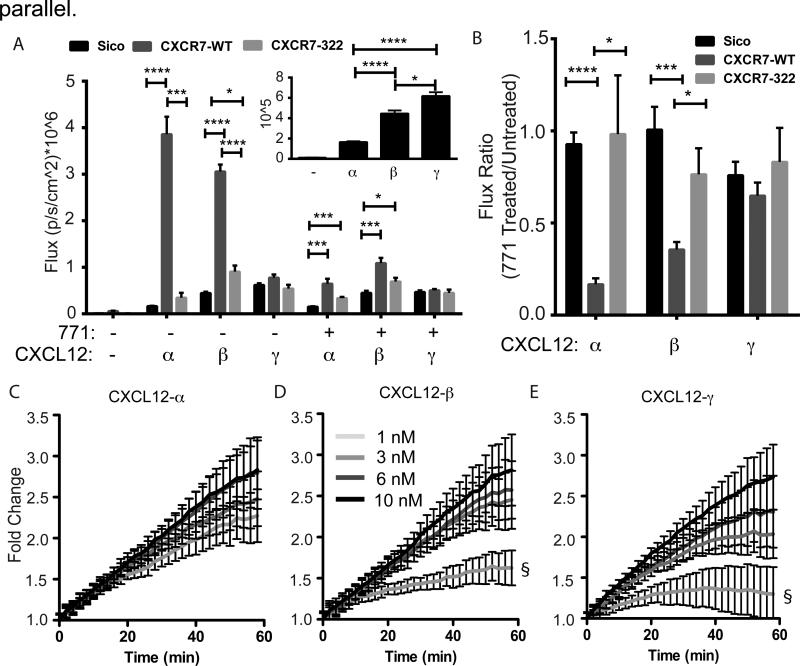
Figure 3. CXCR7-dependent scavenging of CXCL12-isoforms.
(A) 231 cells expressing CXCR7-GFP-WT, CXCR7-Δ322-GFP or no CXCR7 were incubated for 1 hour with equal levels (based on Gaussia luciferase activity) of cell-secreted CXCL12- α, β, or γ. Following incubation and acid wash to remove extracellular CXCL12, we measured internal Gaussia luciferase activity to quantify internalization of CXCL12. Photon flux values are reported as mean ± S.E.M. (n=4 measurements) from one of three representative experiments. The inset highlights only CXCL12-isoforms binding to CXCR7-negative 231-Sico cells. (B) Ratio of internalized bioluminescence signal (A) between cells incubated with inhibitor of CXCL12 binding to CXCR7 (771) relative to untreated cells. Statistical demarcations compare data between bars (* p< 0.05, ***p<0.005, **** p<0.0001). (C-E) Cells expressing a luciferase complementation reporter for association of CXCR7 and β-arrestin 2 were incubated with increasing equimolar concentrations of synthetic CXCL12-α, β, or γ. Data were graphed as mean values ± S.E.M. (n=4 measurements) from one of two representative experiments. Gray-scale code for concentrations indicated in panel D is the same for all isoforms. Fold change in bioluminescence is relative to untreated cells at corresponding time points. The symbol § demarcates statistical differences by Tukey post hoc test between concentrations for the final time point. There are no statistical differences between concentrations of CXCL12-α (C). For CXCL12-β, 1nM is different from 3nM (p<0.01), 6nM (p<0.01), and 10 nM (p<0.0001). For CXCL12-γ, 1nM is different from 3nM (p<0.05), 6nM (p<0.01), and 10 nM (p<0.0001). Comparisons between isoforms are in supplemental information (Fig. S5).
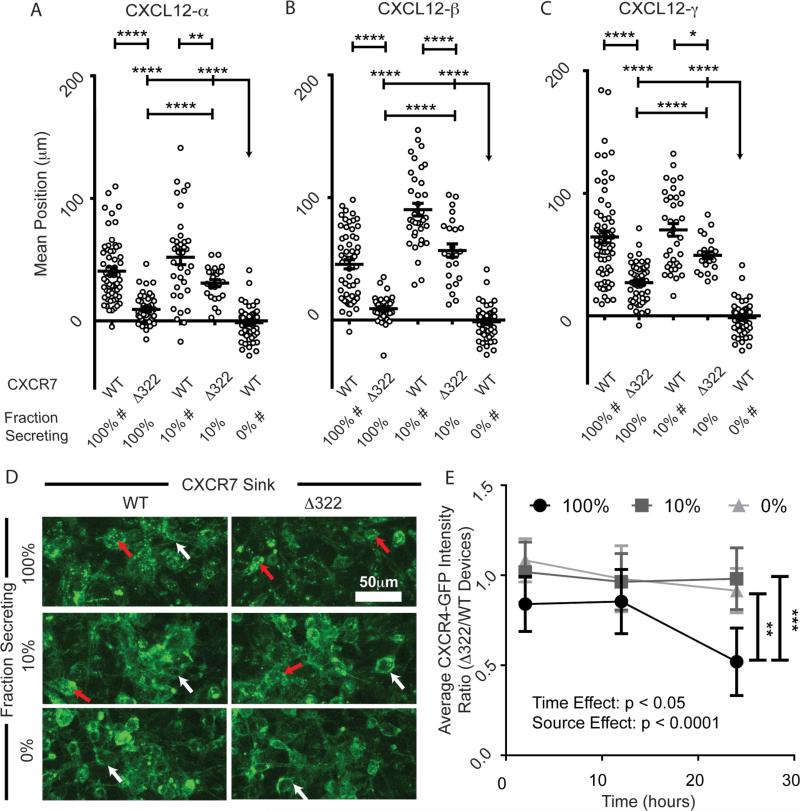
Figure 4. CXCR7 scavenging is necessary for chemotaxis of CXCR4+ cells in response to higher levels of CXCL12.
(A-C) Migration of CXCR4+ cells toward various fractions of cells secreting different isoforms of CXCL12 in the presence of cells expressing either CXCR7-WT or a mutant lacking the carboxy terminus of the receptor (CXCR7-Δ322). Data are graphed as average position ± S.E.M. of migrating CXCR4+ cells after 24 hours (n=6 view fields each for 4-11 devices per condition, similar to previous figures). Fraction of secreting cells denotes the relative dilution of CXCL12-isoform secreting cells. The bar represents the statistical comparison between pairs of conditions. The arrow denotes multiple paired comparisons to the same condition (*p<0.05, **p<0.01, ****p<0.0001). Data for 100%, 10%, and 0% secreting cells are marked (#) to designate the same data plotted for comparison in multiple figures. Matched conditions were performed in parallel. (D) Representative Z-stack compressions of confocal images of CXCR4-GFP+ cells after 24 hours patterned in the context of dilutions of CXCL12-β (0, 10, and 100%) and with WT and CXCR7-Δ322 cells. Red arrows highlight intracellular CXCR4-GFP vesicles. White arrows denote cell membrane CXCR4-GFP. (E) Time course quantification of CXCR4-GFP intensity in devices patterned in the context of dilutions of CXCL12-β source cells (0, 10, and 100%). The plot depicts the mean ± SEM of the ratio between average CXCR4-GFP fluorescence intensity for devices with Δ322-CXCR7 relative to WT-CXCR7 (n=5 images from one of two representative experiments). Two-way ANOVA reveals significant time and source effects without significant interactions. The bars represent statistical significance by the Tukey post hoc test only for the 100% source at the 24 hour time point (** p<0.01, ***, p<0.005).
Results
Microfluidic source-sink-migration system
Using a multi-layered device (Fig. 1A), we patterned cells that secrete individual CXCL12 isoforms (source) in a geometrically defined location relative to MDA-MB-231 breast cancer cells expressing CXCR7 (CXCR7+, sink) or CXCR4 (CXCR4+, migrating cells) (Fig. 1B-D). In this device, intercellular interactions between source and sink cells generate chemotactic gradients in situ in the context of serum-containing medium and other molecules secreted by cells. We also emphasize that the only difference among source cells is the isoform of secreted CXCL12, so differences in chemotaxis of CXCR4 cells arise from distinct biologic effects of each isoform. The device allows us to quantify changes in position of all CXCR4+ cells through time lapse imaging (Fig. S1) or endpoint analyses to give chemotaxis data as a pooled frequency distribution or mean position per view field (see Fig S2 A-D or 1E-G, respectively, as examples). By changing the percentage of secreting cells mixed in with non-secreting cells, the system also tests the effect of changing the amount of source CXCL12 produced. We also analyzed the role of the sink cells by either eliminating CXCR7 cells or by patterning cells expressing non-functional forms of CXCR7. CXCL12 conditions did not cause differences in growth in these settings based on the number of 231-CXCR4 cells uniformly doubling over 24 hours (data not shown).
Characterization of secretedGaussialuciferase CXCL12-isoform-fusions
For the CXCL12-isoform secreting (source) cells we stably transduced MDA-MB-231 breast cancer cells with an isoform of CXCL12 fused to Gaussia luciferase (GLuc). We previously described that fusion of CXCL12-α to GLuc provides a sensitive, quantitative measure of chemokine levels without affecting ligand activity17. We focused on CXCL12-α, -β, and -γ, the most common human isoforms and those shared with mice and rats11a. While we emphasized CXCL12 secretion by cancer cells, previous studies show that both malignant and stromal cells may secrete this chemokine19. We expressed CXCL12-isoforms using two different vectors (Fig. S3), allowing us to create GLuc-fused and -unfused chemokines to demonstrate that fusion to GLuc retains biological activity of each isoform (Supplemental Results and Fig. S3-5). We note that, despite only modest differences in mRNA levels, amounts of secreted CXCL12-γ were 10-fold lower than CXCL12-α and -β by GLuc activity and ELISA reactivity (Fig. S2E-F). Although the polyclonal ELISA antibodies were developed against CXCL12-α, the assay detects the N-terminal core, which is common to all isoforms. Relatively lower amounts of secreted CXCL12-γ protein as compared with mRNA may reflect previously reported differences in intracellular trafficking of CXCL12-γ20, and we have observed this effect with multiple vectors and cell types. GLuc-fused CXCL12-isoforms also signalled via AKT at comparable levels as synthetic isoforms based on our ELISA and GLuc concentration estimates (Fig. S5). Overall, these data allow us to quantitatively compare levels of CXCL12 isoforms secreted by source cells.
CXCL12-β and -γ have surprisingly high chemotaxis potency in anin vitro source-sink model
Chemotaxis of CXCR4+ cells toward CXCL12-γ was slightly more than CXCL12-β (p<0.05) and significantly more than CXCL12-α (p = 0.0001) after 24 hours (Fig 1E-G, comparison among 100% secreting conditions for each isoform with statistics not marked on plots). These results differ notably from transwell assays in which CXCR4+ cells show higher sensitivity migration toward cell-secreted or recombinant CXCL12-α (Fig. S2G)10, 21. Rueda and colleagues previously showed a ten-fold higher concentration of CXCL12-γ was required to drive chemotaxis of CXCR4 cells to the same extent as CXCL12-α in transwells. However, their study required 100nM CXCL12-γ to promote chemotaxis, substantially higher than amounts expected in vivo or produced in our system. Using time-lapse microscopy we also show that CXCL12-γ induced more immediate and rapid migration of both leading and trailing edges, followed by continued migration comparable to sustained effects of CXCL12-α and -β (Fig. S1B-D).
Chemotactic responses of CXCR4+ cells toward CXCL12 can show a bell-shaped curve with reduced effects at both high and low concentrations of chemokine10. To evaluate such effects, we progressively reduced concentrations of CXCL12 by patterning lower percentages of CXCL12-secreting cells mixed with non-secreting parental cells while keeping the total number of cells in the source region constant (p<0.0001; Fig 2A-C). Chemotaxis toward CXCL12-α showed no significant peak before dropping off at 1% relative concentration. CXCL12-β elicited a clear bell-shaped response with peak chemotaxis at 50% and 10% relative concentrations (p<0.05 and p<0.0001, respectively) (Fig 2B), indicating that 100% source cells produce a higher than optimal concentration of chemokine. Chemotaxis towards CXCL12-γ also showed no significant peak before dropping off at 1% (p<0.0001). To investigate mechanisms underlying greater responsiveness of CXCR4+ cells toward CXCL12-γ, we quantified activation of CXCR4 by recruitment of the cytosolic adapter protein β-arrestin 2, which is implicated in chemotaxis22. We treated cells expressing a luciferase complementation reporter for association of CXCR4 and β-arrestin 2 with increasing concentrations of recombinant CXCL12-α, -β, or -γ16a, 23. CXCL12-γ stimulated greater recruitment of β-arrestin 2 to CXCR4 than the other isoforms, potentially contributing to enhanced chemotaxis toward the γ-isoform (Fig 1H-J, S6)22.
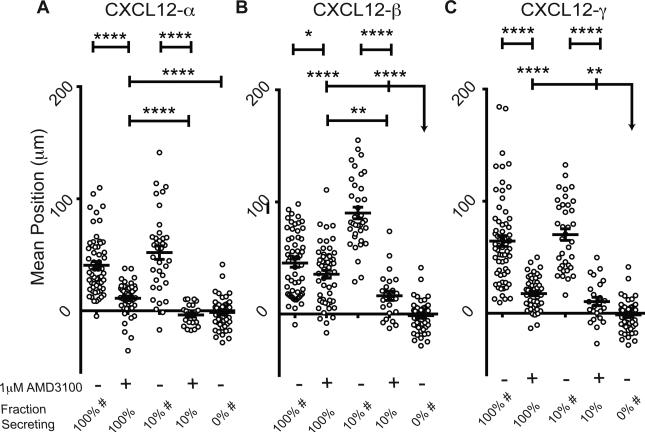
Figure 2. AMD3100 limits migration of CXCR4+ cells toward CXCL12 isoforms.
(A-C) Positions of CXCR4+ cells within migration devices were determined after 24 hours of migration in the absence or presence of 1 µM AMD3100. Data are plotted as average positions ± S.E.M. (n=6 view fields each for 4-11 devices per condition, similar to previous figures). Fraction of secreting cells denotes the relative percent of CXCL12-isoform secreting cells relative to control cells patterned in the source position. The bar represents the statistical comparison between pairs of conditions. The arrow denotes multiple paired comparisons to the same condition (*p<0.05, **p<0.01, ****p<0.0001). Data for 100%, 10%, and 0% secreting cells are marked (#) to designate the same data plotted for comparison in multiple figures. Matched conditions were performed in
Inhibiting CXCL12-CXCR4 dependent chemotaxis
AMD3100 is a small-molecule competitive inhibitor of CXCL12-CXCR4 binding used clinically to mobilize hematopoietic stem cells21b, 24. Little is known about effects of AMD3100 against different isoforms of CXCL12. Using the microfluidic source-sink model, we found AMD3100 to be less effective than may be expected from transwell studies against just the α-isoform (Fig. 2A-C). While AMD3100 effectively eliminated chemotaxis in transwell assays with CXCL12-α (Fig S2G), in the source-sink device CXCL12-β or -γ secreting cells still drove migration in the presence of AMD3100 (p<0.0001 and p<0.01, respectively for β- and γ-isoforms). These data indicate that even very low, physiologic levels of CXCL12-β or -γ may continue to drive detectable chemotaxis in the presence of a validated CXCR4 inhibitor.
Limited CXCR7-dependent scavenging of CXCL12-γ
Cells expressing wild-type CXCR7 internalized more CXCL12-α and -β than cells with either CXCR7-Δ322 (p<0.005 and p<0.0001 for α and β, respectively) or no CXCR7 (p<0.0001 for both α and β) (Fig. 3A). In cells with wild-type (WT) CXCR7, intracellular accumulation of CXCL12-α was highest and followed by -β, with only minimal CXCR7-dependent accumulation of CXCL12-γ relative to CXCR7-Δ322 or control cells without CXCR7 (Fig. 3A). This order was reversed for cells without CXCR7, suggesting that higher binding affinity of CXCL12-γ to glycosaminoglycans on cell membranes confers CXCR7-independent accumulation (Fig 3A inset). Likewise, a small molecule inhibitor of CXCL12 binding to CXCR7 (CCX771) decreased CXCR7-dependent internalization for CXCL12-α and -β, but not -γ (Fig. 3B). Accordingly, lower amounts of CXCL12-α and -β than -γ induced CXCR7-dependent recruitment of β-arrestin 2, which is involved in CXCR7 internalization, as determined by luciferase complementation (Fig. 3C-E, S7)4e, 16b. These data show inefficient scavenging of CXCL12-γ by CXCR7.
Levels of CXCL12 determine requirement for CXCR7 scavenging in chemotaxis
We previously demonstrated that CXCR7 functions as a sink to determine the magnitude and shape of a CXCL12-α gradient in our microfluidic device3c. To investigate to what extent CXCR7 scavenging is required for chemotaxis of CXCR4 cells, we used sink cells that does not internalize CXCL124e. When using high numbers of CXCL12 source cells, chemotaxis of CXCR4+ cells toward all CXCL12-isoforms decreased when CXCR7-Δ322 sink cells replaced WT-CXCR7 cells (p<0.0001) (Fig 4A-C). However, differences in chemotaxis between CXCR7-Δ322 and CXCR7-WT were substantially less with only 10% of the source cells producing CXCL12. For devices with CXCR7-Δ322 sink cells, we observed greater chemotaxis when the relative concentration was 10% rather than 100% (p<0.0001). These data emphasize the dynamic balance between levels of CXCL12 secretion and CXCR7-dependent scavenging in CXCR4-dependent chemotaxis. CXCL12 scavenging by CXCR7 is required for chemotaxis of CXCR4+ cells at higher levels of chemokine, while functioning CXCR7 is less important for chemotaxis with lower levels of CXCL12.
Upon prolonged stimulation with CXCL12, CXCR4 internalizes and is degraded4d, 21b. In our experiments, localization and fluorescence intensity of a CXCR4-GFP fusion protein in migrating cells could be used as metrics of CXCR4 signaling and desensitization (Fig 4D-E). Using CXCL12-β secreting cells as the source, we found significantly higher CXCR4-GFP internalization when the WT CXCR7 scavenging cells were replaced with the scavenging-deficient CXCR7-Δ322 cells (p < 0.05). Such CXCR4 internalization effects, however, were significantly mitigated when using a lower 10% relative concentration of CXCL12-β cells independent of WT or mutant CXCR7 sink cells (Fig. 4E). These results show that CXCR7-scavenging prevents degradation of CXCR4 and loss of chemotaxis under conditions with relatively higher levels of CXCL12. Conversely, by lowering the amount of CXCL12, migrating cells retain a signaling pool of CXCR4-GFP at the cell membrane and overall levels of receptor sufficient for chemotaxis even with CXCR7-Δ322 cells.
CXCL12-γ in primary breast tumors
To link our data to breast cancer biology in vivo, we analyzed expression of CXCL12-α, -β, and -γ in orthotopic breast tumor implants from syngeneic and human xenograft mouse models and a cDNA array from bulk tissues derived from normal breast and primary human breast cancers. We detected CXCL12-α, -β, and -γ in mouse tumors and with rank order of frequency of expression being α > β > γ (Fig. S8), based on the number of samples amplifying at <40 qRTPCR cycles. Despite quantitatively lower levels of CXCL12-β and γ-isoforms, nearly all samples had detectable signal. In human tissues, we detected transcripts for CXCL12-α and -β more consistently in human tissues (normal and tumor) as compared with the γ-isoform. CXCL12-β was expressed slightly more frequently in cancers. Remarkably, we detected CXCL12-γ only in primary tumors from patients with advanced disease (Table I). We note that these data do not assign the cell type(s) in tumors that produce CXCL12 isoforms. While our microfluidic device emphasized secretion of CXCL12 by cancer cells, stromal cells in tumors also are sources of this chemokine4d, 19a, 25.
Table I.
CXCL12 isoforms in human breast cancer.
| CXCL12-Isoforms |
|||
|---|---|---|---|
| Tumor Grade: | α | β | γ |
| Normal | 5/5 | 2/5 | 0/5 |
| Stage I | 11/11 | 7/11 | 0/11 |
| Stage II (A+B) | 13/14 | 7/14 | 0/14 |
| Stage III (A,B,C) | 14/14 | 13/14 | 1/4 |
| Stage IV | 4/4 | 3/4 | 4/4 |
Numbers of samples positive for each isoform of CXCL12 as determined by QRT-PCR and total number of samples for normal breast tissue and various stages of primary human breast cancers. Transcripts amplified below 40 qRT-PCR cycles and confirmed by gel electrophoresis were denoted as positive.
Discussion
CXCL12 is a homeostatic chemokine that drives many finely tuned, dynamic physiologic and pathologic processes, necessitating multiple levels of regulation. Our study focuses on chemotaxis as a dynamic balance among chemokine secretion, endocytosis by sink cells, binding to extracellular surfaces, and regulating chemokine-sensing receptor numbers. These factors must all be included in models for screening therapeutic agents, as changing one has concerted effects on the entire system.
In the simplest conceptual model, secreted ligands freely diffuse from their source and create a concentration-dependent gradient according to Fick's law of diffusion. We highlight mechanisms that modify the shape and magnitude of this gradient in the extracellular space. CXCR7 removes and degrades CXCL12, which decreases local concentrations of chemokine and sharpens the chemokine gradient to facilitate migration3b, 4b-e. The requirement for CXCR7 scavenging in chemotaxis of CXCR4 cells is contingent upon levels of CXCL12. CXCR7 scavenging is particularly critical for chemotaxis of CXCR4 cells under conditions with relatively high levels of CXCL12. However, when source cells secrete only low levels of CXCL12, CXCR4 cells still migrate in the absence of CXCR7 scavenging, albeit at somewhat reduced efficiency. In devices with reduced numbers of source cells, cells retained sufficient CXCR4 at the cell surface to migrate in response to the CXCL12 gradient. These results highlight an essential balance in gradient formation between relative capacities of the source and sink to produce and scavenge ligand, respectively. Our data also, suggest that therapeutic targeting of CXCR7 in chemotaxis will be contingent on relative CXCL12 levels locally.
Although CXCR7 has been highlighted9 as a critical mediator of CXCL12 gradient formation and chemotaxis25,26, we emphasize CXCL12 isoforms as another largely overlooked mechanism for controlling gradient shape, local chemokine levels, and chemotaxis. CXCL12 exists as six human (α, β, γ, δ, ε, and Φ) and three mouse and rat (α, β, and γ) isoforms, which are expressed in time and tissue-specific distributions during development and post-natal life11. CXCL12 isoforms share 68 common amino-terminal amino acids, which comprise all of the most studied CXCL12-α isoform and contain one glycosaminoglycan-binding BBXB domain (B denotes basic and X denotes any amino acid, respectively). Other isoforms differ by addition of 1-41 largely basic amino acids to the carboxy-terminus, accounting for one and four additional BBXB motifs in CXCL12-β and -γ, respectively. Differences in carboxy-termini among isoforms alter biologic and biophysical properties of CXCL12, including binding affinities for receptors and extracellular matrix molecules and activation of downstream signaling10, 11b. In our model, despite adjusting the fraction of secreting cells to produce comparable levels of each isoform, we established for the first time in vitro that CXCL12-γ maintains CXCR4 sensitization and chemotaxis despite low chemokine levels and in the presence of AMD3100. Interestingly, CXCL12-γ binds directly to CXCR4 with lower affinity than CXCL12-α or -β, and yet our results show it functions as a high affinity ligand for CXCR4 in chemotaxis10, 11b. We posit two mechanisms for this observation. One is that that CXCL12-β and -γ form sharper gradients due to relatively increased surface adhesion, both to cell and device surfaces. This notion is supported by the lack of absolute requirement for CXCR7 scavenging cells to elicit chemotaxis. Another mechanism is that high affinity interactions between the more positively charged CXCL12-β and -γ and negatively charged cell surface proteoglycans increase local concentrations of chemokine to bind to CXCR4 and drive migration 26. As a result, these isoforms promote chemotaxis at low total abundance and effectively compete with inhibitor AMD3100 (See Supplemental Discussion and Table S2). By concentrating CXCL12 isoforms on cell surfaces, we propose that heparan sulfates function as stable co-receptors for CXCL12-isoforms and CXCR4, enhancing activation of CXCR4-β-arrestin 2 signaling and chemotaxis relative to other isoforms11b, 22, 27.
While interactions between CXCL12 and glycosaminoglycans are necessary for optimal chemotactic responses to this chemokine, interrelationships among glycosaminoglycan binding, chemokine oligomers, and chemotaxis remain uncertain 26, 28. CXCL12 bound to heparan sulfates may form dimers at physiologic concentrations of chemokine 18, 29. CXCL12 monomers have been reported to preferentially stimulate chemotaxis in some studies, while others have determined that CXCL12 dimers comprise the more active species 18, 28, 30. Disparate results about potency of CXCL12 monomers versus dimers in chemotaxis may be due to variable concentrations and presentations of chemokine and/or glycosaminoglycans in assays. Future studies adding heparan sulfates and other glycosaminoglycans to our device, combined with imaging reporters of dimers, may resolve this uncertainty 18.
With the caveat of small sample size, a provocative observation of our study is detection of CXCL12-α, -β, and -γ in human breast cancer with CXCL12-γ only present in primary tumors from patients with metastases to lymph nodes or other organs. This result underscores previous descriptions of alternative splicing as transformative mechanism in cancer31 and supports further studies of CXCL12-γ as a biomarker for tumor microenvironments that promote metastasis. Similar to CXCL12, alternative splicing of vascular endothelial growth factor produces a functional range of short to long isoforms, with the longest and shortest having the highest and lowest heparan sulfate binding, respectively32. However, in malignant transformation, alternative splicing switches towards expression of the more diffusible, short isoforms of VEGF in lung and colon cancers. Indeed, limited in vivo studies suggest that CXCL12-γ drives chemotaxis to a much greater extent than CXCL12-α or -β, but standard in vitro assays fail to capture this phenotype and instead show CXCL12-γ to be comparatively ineffective10, 33. One study in mice with mutated carboxy-terminal BBXB domains showed exogenous delivery of CXCL12-γ better recruited endothelial progenitor cells and restored vascular repair after acute ischemia as compared with exogenous CXCL12-α33. We further implicate CXCL12-γ beyond post-ischemic repair as a potential marker of advanced human breast cancer.
We identified interdependence between levels of CXCL12 isoforms and inhibition of chemotaxis with AMD3100. AMD3100 only partially inhibited chemotaxis toward all isoforms with high levels of CXCL12, but the drug more effectively limited migration toward relatively lower amounts of CXCL12. However, even low levels of CXCL12-β or -γ drove CXCR4-dependent chemotaxis in the presence of AMD3100, suggesting that the drug may be less effective against isoforms other than α. Our results are consistent with prior work showing that 10-fold more AMD3100 is needed to block binding of CXCL12-γ to CXCR4 relative to CXCL12-α21b. As CXCL12-isoforms ostensibly confer signaling through the same CXCR4 binding site, AMD3100 should exhibit equal competitive inhibition of all isoforms. This point suggests that higher local concentrations of CXCL12-β and -γ effectively compete with AMD3100 for binding to CXCR4. Although we detected modest amounts of mRNA for CXCL12 isoforms in human cancers, these results suggest relatively few CXCL12-β and -β producing cells are required to drive migration. Shift of alternative splicing programs towards high potency CXCL12-β and -γ may enhance metastasis and drug resistance in advanced disease. Our results indicate the need for more potent agents to block chemotaxis in response to all isoforms of CXCL12.
Our device facilitates gradient formation in a dynamic and cell-autonomous way, unlike systems in which users introduce external gradients. The low, rising gradients in our device are less defined but more physiological than bolus doses in Boyden chambers and linear gradients generated in microfluidic systems. Nonetheless, we used several methods to quantify and perturb chemokine levels to understand their activities relative to reported dissociation constants for CXCL12-isoforms to their receptors. First, we quantified the maximal soluble level of chemokine with a combination of ELISA, Gaussia luciferase assays, and signaling/uptake assays. Second, we varied isoform levels, numbers of cells secreting an isoform, efficiency of CXCR7-dependent scavenging, and specific inhibitor AMD3100. Using these strategies we defined different qualitative functional regimes of the source-sink components. Without improved real-time chemokine measurement techniques and substantial computational modeling of this complex system, it is difficult to dynamically quantitate the soluble and more importantly bound levels of chemokine. Nonetheless, our device offers middle ground between migration assays of varying complexity: standard Boyden chambers, three-dimensional hydrogel systems conditions 34, and in vivo physiology 35.
Our device provides the first in vitro system that recapitulates observations in mouse models of enhanced chemotactic effects of CXCL12-β and -γ relative to α-isoforms10, 36. Importantly, we emphasize that this device detects differences in chemotaxis among CXCL12 isoforms secreted in the context of other molecules secreted by source cells. Our results differ markedly from standard Boyden chamber migration assays in which CXCL12-α is most effective and cells migrate only in response to CXCL12-γ lacking cationic BBXB domains or very high concentrations of wild-type CXCL12-γ10. We propose several differences between our device and Boyden chambers allow us to replicate CXCL12-β and -γ in physiological conditions. Boyden chamber assays expose cells acutely to high levels of CXCL12, which may cause rapid CXCR4 internalization and desensitization21b. We have shown that our device generates low, sustained gradients of CXCL12 that may prevent CXCR4 desensitization3c. Several studies suggest that low levels of CXCL12 “prime” cells for signaling by recruiting additional cell-surface CXCR4, as evidenced by increased HIV infection or β-arrestin 2 recruitment after pre-treatment with low amounts of CXCL1216a, 21a. The highly cationic C-terminus of CXCL12-β and -γ also confers greater non-specific binding to negatively-charged tissue culture plastic, which may remove these chemokines from solution in standard Boyden chamber systems, altering the gradient profile and directionality. Binding of CXCL12 to the migration surface of our microfluidic device may contribute to gradient formation in the direction of CXCR4+ cell migration. Finally, observing migration along a longer distance in our device may be more representative of in vivo processes as compared to having cells squeeze through small pore over a very short distance in Boyden chambers5. These and likely other factors allow our device to accurately reproduce in vivo chemotactic effects of CXCL12 isoforms, providing a facile system to investigate functions and therapeutic targeting of various isoforms in chemotaxis.
On the other end of assay complexity, extension of our cell-based source-sink model into three dimensions would better facilitate incorporation of glycosaminoglycans within a three dimensional hydrogel, and provide more in vivo-like invasion/chemotaxis34. We expect that incorporating heparan sulphate proteoglycan chemistry within a hydrogel matrix will sharpen gradients and limit gradient length scales, particularly for ligands with high matrix affinity. However, our device highlights the cell-autonomous role in gradient formation due to ligand interactions with receptors, cell-surface proteoglycans, and the device surface. In three dimensional hydrogel systems ligand-matrix interactions dominate gradient formation. Nonetheless, understanding how source-sink interactions facilitate gradient and chemotaxis dynamics in the context of three-dimensional matrices and proteoglycans is an important next step to understanding in vivo physiology.
Conclusions
Our study provides new insights into the chemotactic microenvironment and interdependencies between CXCL12-secretion and bioavailability within the extracellular space. The microfluidic device utilized in this work is the first cell culture system that reproduces enhanced chemotactic effects of CXCL12-γ reported in living animals. Interestingly, we found that few CXCL12 secreting cells were required to drive migration and that low levels of CXCL12 may largely bypass the need for CXCR7 to form chemotactic gradients and retain CXCR4 sensitization. These data suggest that even low levels of CXCL12, particularly CXCL12-β and -γ, may be relevant for driving chemotaxis in vivo. This device provides an ideal platform to identify mechanistic differences among CXCL12 isoforms in chemotaxis and identify new inhibitors that are effective against all isoforms of this chemokine. These studies suggest that the collective microenvironment should be considered a biomarker for metastatic cancer, including the distribution of CXCL12-isoforms and relative levels of CXCR4 and CXCR7.
Supplementary Material
Acknowledgements
This work was supported by United States National Institutes of Health grants R01CA136553, R01CA136829, R01CA142750, and P50CA093990. S.P.C. was supported on Advanced Proteome Informatics of Cancer Training Grant # T32 CA140044 and an NSF Predoctoral Fellowship Project Grant # F031543. We thank ChemoCentryx for generously providing CCX771.
Footnotes
Competing financial interests statement
The authors declare no competing financial interests.
References
- 1.a Raman D, Sobolik-Delmaire T, Richmond A. Chemokines in health and disease. Experimental Cell Research. 2011;317(5):575–589. doi: 10.1016/j.yexcr.2011.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Burkhardt AM, Homey B, Zlotnik A. Homeostatic chemokine receptors and organ-specific metastasis. Nature Reviews Immunology. 2011;11(9):597+. doi: 10.1038/nri3049. [DOI] [PubMed] [Google Scholar]; c Balkwill FR. The chemokine system and cancer. The Journal of Pathology. 2012;226(2):148–157. doi: 10.1002/path.3029. [DOI] [PubMed] [Google Scholar]; d Zhang GX, Baker CM, Kolson DL, Rostami AM. Chemokines and chemokine receptors in the pathogenesis of multiple sclerosis. Multiple Sclerosis. 2000;6(1):3–13. doi: 10.1177/135245850000600103. [DOI] [PubMed] [Google Scholar]; e Zernecke A, Shagdarsuren E, Weber C. Chemokines in atherosclerosis: an update. Arterioscler Throm Vasc Biol. 2008;28(11):1897–1908. doi: 10.1161/ATVBAHA.107.161174. [DOI] [PubMed] [Google Scholar]; f Krumbholz M, Theil D, Cepok S, Hemmer B, Kivisäkk P, Ransohoff RM, Hofbauer M, Farina C, Derfuss T, Hartle C, Newcombe J, Hohlfeld R, Meinl E. Chemokines in multiple sclerosis: CXCL12 and CXCL13 up-regulation is differentially linked to CNS immune cell recruitment. Brain. 2006;129(1):200–211. doi: 10.1093/brain/awh680. [DOI] [PubMed] [Google Scholar]
- 2.a Wu Y, Zhao R. The Role of Chemokines in Mesenchymal Stem Cell Homing to Myocardium. Stem Cell Reviews and Reports. 2012;8(1):243–250. doi: 10.1007/s12015-011-9293-z. [DOI] [PubMed] [Google Scholar]; b Fukuda S, Pelus LM. Chemokine-mobilized adult stem cells; defining a better hematopoietic graft. Leukemia. 2008;22(3):466+. doi: 10.1038/sj.leu.2405021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.a Yu SR, Burkhardt M, Nowak M, Ries J, Petrasek Z, Scholpp S, Schwille P, Brand M. Fgf8 morphogen gradient forms by a source-sink mechanism with freely diffusing molecules. Nature. 2009;461(7263):533–6. doi: 10.1038/nature08391. doi: 10.1038/nature08391. Epub 2009 Sep 9. [DOI] [PubMed] [Google Scholar]; b Boldajipour B, Mahabaleshwar S, Kardash E, Reichman-Fried M, Blaser H, Minina S, Wilson D, Xu Q, Raz E. Control of chemokine-guided cell migration by ligand sequestration. Cell. 2008;132(3):463–473. doi: 10.1016/j.cell.2007.12.034. [DOI] [PubMed] [Google Scholar]; c Torisawa Y, Mosadegh B, Bersano-Begey T, Steele J, Luker K, Luker G, Takayama S. Microfluidic platform for chemotaxis in gradients formed by CXCL12 source-sink cells. Integr Biol (Camb) 2010;2(11-12):680–686. doi: 10.1039/c0ib00041h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.a Scholpp S, Brand M. Endocytosis controls spreading and effective signaling range of Fgf8 protein. Current biology. 2004;14(20):1834–1841. doi: 10.1016/j.cub.2004.09.084. [DOI] [PubMed] [Google Scholar]; b Naumann U, Cameroni E, Pruenster M, Mahabaleshwar S, Raz E, Zerwes H, Rot A, Thelen M. CXCR7 functions as a scavenger for CXCL12 and CXCL11. PLoS One. 2010;5(2):e9175. doi: 10.1371/journal.pone.0009175. [DOI] [PMC free article] [PubMed] [Google Scholar]; c Luker K, Steele J, Mihalko L, Luker G. Constitutive and chemokine-dependent internalization and recycling of CXCR7 in breast cancer cells to degrade chemokine ligands. Oncogene. 2010;29:4599–4610. doi: 10.1038/onc.2010.212. [DOI] [PMC free article] [PubMed] [Google Scholar]; d Luker K, Lewin S, Mihalko L, Schmidt B, Winkler J, Coggins N, Thomas D, Luker G. Scavenging of CXCL12 by CXCR7 promotes tumor growth and metastasis of CXCR4-positive breast cancer cells. Oncogene. 2012 doi: 10.1038/onc.2011.633. doi: 10.1038/onc.2011.633. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]; e Ray P, Mihalko L, Coggins N, Moudgil P, Ehrlich A, Luker K, Luker G. Carboxy-terminus of CXCR7 regulates receptor localization and function. Int J Biochem Cell Biol. 2012;44(4):669–678. doi: 10.1016/j.biocel.2012.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]; f Fredericks Z, Pitcher J, Lefkowitz R. Identification of the G protein-coupled receptor kinase phosphorylation sites in the human beta2-adrenergic receptor. J Biol Chem. 1996;271(23):13796–13803. doi: 10.1074/jbc.271.23.13796. [DOI] [PubMed] [Google Scholar]; g Sanchez-Alcaniz J, Haege S, Mueller W, Pla R, Mackay F, Schulz S, Lopez-Bendito G, Stumm R, Marin O. Cxcr7 controls neuronal migration by regulating chemokine responsiveness. Neuron. 2011;69(1):77–90. doi: 10.1016/j.neuron.2010.12.006. [DOI] [PubMed] [Google Scholar]; h Wang Y, Li G, Stanco A, Long J, Crawford D, Potter G, Pleasure S, Behrens T, Rubenstein J. CXCR4 and CXCR7 have distinct functions in regulating interneuron migration. Neuron. 2011;69(1):61–76. doi: 10.1016/j.neuron.2010.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Weber M, Hauschild R, Schwarz J, Moussion C, de Vries I, Legler DF, Luther SA, Bollenbach T, Sixt M. Interstitial Dendritic Cell Guidance by Haptotactic Chemokine Gradients. Science. 2013;339(6117):328–332. doi: 10.1126/science.1228456. [DOI] [PubMed] [Google Scholar]
- 6.Proudfoot AE, Handel TM, Johnson Z, Lau EK, LiWang P, Clark-Lewis I, Borlat F, Wells TN, Kosco-Vilbois MH. Glycosaminoglycan binding and oligomerization are essential for the in vivo activity of certain chemokines. Proceedings of the National Academy of Sciences. 2003;100(4):1885–1890. doi: 10.1073/pnas.0334864100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.a Balkwill F. Cancer and the chemokine network. Nat Rev Cancer. 2004;4(7):540–550. doi: 10.1038/nrc1388. [DOI] [PubMed] [Google Scholar]; b Balkwill F. The significance of cancer cell expression of the chemokine receptor CXCR4. Seminars in Cancer Biology. 2004;14(3):171–179. doi: 10.1016/j.semcancer.2003.10.003. [DOI] [PubMed] [Google Scholar]
- 8.Phillips RJ, Burdick MD, Hong K, Lutz MA, Murray LA, Xue YY, Belperio JA, Keane MP, Strieter RM. Circulating fibrocytes traffic to the lungs in response to CXCL12 and mediate fibrosis. Journal of Clinical Investigation. 2004;114(3):438–446. doi: 10.1172/JCI20997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.a Venkiteswaran G, Lewellis SW, Wang J, Reynolds E, Nicholson C, Knaut H. Generation and Dynamics of an Endogenous, Self-Generated Signaling Gradient across a Migrating Tissue. Cell. 2013;155(3):674–687. doi: 10.1016/j.cell.2013.09.046. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Donà E, Barry JD, Valentin G, Quirin C, Khmelinskii A, Kunze A, Durdu S, Newton LR, Fernandez-Minan A, Huber W. Directional tissue migration through a self-generated chemokine gradient. Nature. 2013 doi: 10.1038/nature12635. [DOI] [PubMed] [Google Scholar]
- 10.Rueda P, Balabanian K, Lagane B, Staropoli I, Chow K, Levoye A, Laguri C, Sadir R, Delaunay T, Izquierdo E, Pablos J, Lendinez E, Caruz A, Franco D, Baleux F, Lortat-Jacob H, Arenzana-Seisdedos F. The CXCL12gamma chemokine displays unprecedented structural and functional properties that make it a paradigm of chemoattractant proteins. PLoS One. 2008;3(7):e2543. doi: 10.1371/journal.pone.0002543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.a Yu L, Cecil J, Peng S, Schrementi J, Kovacevic S, Paul D, Su E, Wang J. Identification and expression of novel isoforms of human stromal cell-derived factor 1. Gene. 2006;374:174–179. doi: 10.1016/j.gene.2006.02.001. [DOI] [PubMed] [Google Scholar]; b Laguri C, Sadir R, Rueda P, Baleux F, Gans P, Arenzana-Seisdedos F. The novel CXCL12gamma isoform encodes an unstructured cationic domain which regulates bioactivity and interaction with both glycosaminoglycans and CXCR4. PLoS One. 2007;2:e1110. doi: 10.1371/journal.pone.0001110. [DOI] [PMC free article] [PubMed] [Google Scholar]; c Gleichmann M, Gillen C, Czardybon M, Bosse F, Greiner-Petter R, Auer J, Müller HW. Cloning and characterization of SDF-1γ, a novel SDF-1 chemokine transcript with developmentally regulated expression in the nervous system. European Journal of Neuroscience. 2000;12(6):1857–1866. doi: 10.1046/j.1460-9568.2000.00048.x. [DOI] [PubMed] [Google Scholar]
- 12.Laguri C, Arenzana-Seisdedos F, Lortat-Jacob H. Relationships between glycosaminoglycan and receptor binding sites in chemokines—the CXCL12 example. Carbohydrate Research. 2008;343(12):2018–2023. doi: 10.1016/j.carres.2008.01.047. [DOI] [PubMed] [Google Scholar]
- 13.Luker K, Mihalko L, Schmidt B, Lewin S, Ray P, Shcherbo D, Chudakov D, Luker G. In vivo imaging of ligand receptor binding with Gaussia luciferase complementation. Nat Med. 2012;18(1):172–177. doi: 10.1038/nm.2590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Smith M, Luker K, Garbow J, Prior J, Jackson E, Piwnica-Worms D, Luker G. CXCR4 regulates growth of both primary and metastatic breast cancer. Cancer Res. 2004;64(23):8604–8612. doi: 10.1158/0008-5472.CAN-04-1844. [DOI] [PubMed] [Google Scholar]
- 15.Song J, Cavnar S, Walker A, Luker K, Gupta M, Tung Y, Luker G, Takayama S. Microfluidic endothelium for studying the intravascular adhesion of metastatic breast cancer cells. PLoS One. 2009;4(6):e5756. doi: 10.1371/journal.pone.0005756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.a Luker K, Gupta M, Luker G. Imaging CXCR4 signaling with firefly luciferase complementation. Anal Chem. 2008;80(14):5565–5573. doi: 10.1021/ac8005457. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Luker K, Gupta M, Steele J, Foerster B, Luker G. Imaging ligand-dependent activation of CXCR7. Neoplasia. 2009;11(10):1022–1035. doi: 10.1593/neo.09724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Luker K, Gupta M, Luker G. Bioluminescent CXCL12 fusion protein for cellular studies of CXCR4 and CXCR7. Biotechniques. 2009;47(1):625–632. doi: 10.2144/000113126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ray P, Lewin S, Mihalko L, SC L-P, Takayama S, Luker K, Luker G. Secreted CXCL12 (SDF-1) forms dimers under physiologic conditions. Biochem J. 2012;442:433–442. doi: 10.1042/BJ20111341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.a Orimo A, Gupta P, Sgroi D, Arenzana-Seisdedos F, Delaunay T, Naeem R, Carey V, Richardson A, Weinberg R. Stromal fibroblasts present in invasive human breast carcinomas promote tumor growth and angiogenesis through elevated SDF-1/CXCL12 secretion. Cell. 2005;121(3):335–348. doi: 10.1016/j.cell.2005.02.034. [DOI] [PubMed] [Google Scholar]; b Boimel P, Smirvova T, Zhou Z, Wyckoff J, Park H, Coniglio S, Patel P, Qian B, Stanley E, Bresnick A, Cox D, Pollard J, Muller W, Condeelis J, Segall J. Contribution of CXCL12 secretion to invasion of breast cancer cells. Breast Cancer Res. 2012;14(1):R23. doi: 10.1186/bcr3108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Torres R, Ramirez JC. A Chemokine Targets the Nucleus: Cxcl12-Gamma Isoform Localizes to the Nucleolus in Adult Mouse Heart. PLoS ONE. 2009;4(10):e7570. doi: 10.1371/journal.pone.0007570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.a Altenburg J, Broxmeyer H, Jin Q, Cooper S, Basu S, Alkhatib G. A naturally occurring splice variant of CXCL12/stromal cell-derived factor 1 is a potent human immunodeficiency virus type 1 inhibitor with weak chemotaxis and cell survival activities. J Virol. 2007;81(15):8140–8148. doi: 10.1128/JVI.00268-07. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Altenburg J, Jin Q, Alkhatib B, Aklhatib G. The potent anti-HIV activity of CXCL12γ correlates with efficient CXCR4 binding and internalization. J Virol. 2010;84(5):2563–2572. doi: 10.1128/JVI.00342-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.a Lagane B, Chow K, Balabanian K, Levoye A, Harriague J, Planchenault T, Baleux F, Gunera-Saad N, Arenzana-Seisdedos F, Bachelerie F. CXCR4 dimerization and {beta}-arrestin-mediated signaling account for the enhanced chemotaxis to CXCL12 in WHIM syndrome. Blood. 2008;112(1):34–44. doi: 10.1182/blood-2007-07-102103. [DOI] [PubMed] [Google Scholar]; b Drury L, Ziarek J, Gravel S, Veldkamp C, Takekoshi T, Hwang S, Heveker N, Volkman B, Dwinell M. Monomeric and dimeric CXCL12 inhibit metastasis through distinct CXCR4 interactions and signaling pathways. Proc Natl Acad Sci U S A. 2011 Oct 11; doi: 10.1073/pnas.1101133108. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Villalobos V, Naik S, Bruinsma M, Dothager R, Pan M-H, Samrakandi M, Moss B, Elhammali A, Piwnica-Worms D. Dualcolor click beetle luciferase heteroprotein fragment complementation assays. Chem Biol. 2011;17(9):1018–1029. doi: 10.1016/j.chembiol.2010.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fricker SP, Anastassov V, Cox J, Darkes MC, Grujic O, Idzan SR, Labrecque J, Lau G, Mosi RM, Nelson KL, Qin L, Santucci Z, Wong RS. Characterization of the molecular pharmacology of AMD3100: a specific antagonist of the G-protein coupled chemokine receptor, CXCR4. Biochem Pharmacol. 2006;72(5):588–96. doi: 10.1016/j.bcp.2006.05.010. [DOI] [PubMed] [Google Scholar]
- 25.Miao Z, Luker K, Summers B, Berahovich R, Bhojani M, Rehemtulla A, Kleer C, Essner J, Nasevicius A, Luker G, Howard M, Schall T. CXCR7 (RDC1) promotes breast and lung tumor growth in vivo and is expressed on tumor-associated vasculature. Proc Natl Acad Sci U S A. 2007;104(40):15735–15740. doi: 10.1073/pnas.0610444104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Netelenbos T, Zuijderduijn S, van den Born J, Kessler F, Zweegman S, Huijgens P, Drager A. Proteoglycans guide SDF-1-induced migration of hematopoietic progenitor cells. J Leukoc Biol. 2002;72:353–362. [PubMed] [Google Scholar]
- 27.a Valenzuela-Fernandez A, Palanche T, Amara A, Magerus A, Altmeyer R, Delaunay T, Virelizier J-L, Baleux F, Galzi J-L, Arenzana-Seisdedos F. Optimal Inhibition of X4 HIV Isolates by the CXC Chemokine Stromal Cell-derived Factor 1alpha Requires Interaction with Cell Surface Heparan Sulfate Proteoglycans. J. Biol. Chem. 2001;276(28):26550–26558. doi: 10.1074/jbc.M100411200. [DOI] [PubMed] [Google Scholar]; b Murphy J, Cho Y, Sachpatzidis A, Fan C, Hodsdon M, Lolis E. Structural and functional basis of CXCL12 (stromal cell-derived factor-1) binding to heparin. J Biol Chem. 2007;282(13):10018–10027. doi: 10.1074/jbc.M608796200. [DOI] [PMC free article] [PubMed] [Google Scholar]; c Sun Y, Cheng Z, Ma L, Pei G. Beta-arrestin2 is critically involved in CXCR4-mediated chemotaxis, and this is mediated by its enhancement of p38 MAPK activation. J Biol Chem. 2002;277(51):49212–49219. doi: 10.1074/jbc.M207294200. [DOI] [PubMed] [Google Scholar]
- 28.Fermas S, Gonnet F, Sutton A, Charnaux N, Mulloy B, Du Y, Baleux F, Daniel R. Sulfated oligosaccharides (heparin and fucoidan) binding and dimerization of stromal cell-derived factor-1 (SDF-1/CXCL 12) are coupled as evidenced by affinity CE-MS analysis. Glycobiology. 2008;18(12):1054–1064. doi: 10.1093/glycob/cwn088. [DOI] [PubMed] [Google Scholar]
- 29.Veldkamp C, Peterson F, Pelzek A, Volkman B. The monomerdimer equilibrium of stromal cell-derived factor-1 (CXCL 12) is altered by pH, phosphate, sulfate, and heparin. Protein Sci. 2005;14(4):1071–1081. doi: 10.1110/ps.041219505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Veldkamp C, Seibert C, Peterson F, De la Cruz N, Haughner JI, Basnet H, Sakmar T, Volkman B. Structural basis of CXCR4 sulfotyrosine recognition by the chemokine SDF-1/CXCL12. Sci Signal. 2008;1:ra4. doi: 10.1126/scisignal.1160755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Venables JP. Aberrant and Alternative Splicing in Cancer. Cancer Research. 2004;64(21):7647–7654. doi: 10.1158/0008-5472.CAN-04-1910. [DOI] [PubMed] [Google Scholar]
- 32.Cheung N, Wong MP, Yuen ST, Leung SY, Chung LP. Tissue-specific expression pattern of vascular endothelial growth factor isoforms in the malignant transformation of lung and colon. Human pathology. 1998;29(9):910–914. doi: 10.1016/s0046-8177(98)90195-2. [DOI] [PubMed] [Google Scholar]
- 33.Rueda P, Richart A, Récalde A, Gasse P, Vilar J, Guérin C, Lortat-Jacob H, Vieira P, Baleux F. i., Chretien F, Arenzana-Seisdedos F, Silvestre J-S. Homeostatic and Tissue Reparation Defaults in Mice Carrying Selective Genetic Invalidation of CXCL12/Proteoglycan Interactions / Clinical Perspective. Circulation. 2012;126(15):1882–1895. doi: 10.1161/CIRCULATIONAHA.112.113290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim BJ, Hannanta-Anan P, Chau M, Kim YS, Swartz MA, Wu M. Cooperative roles of SDF-1α and EGF gradients on tumor cell migration revealed by a robust 3D microfluidic model. PLoS ONE. 2013;8(7):e68422. doi: 10.1371/journal.pone.0068422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.a Condeelis J, Segall J. Intravital imaging of cell movement in tumours. Nature Rev Cancer. 2003;3:921–930. doi: 10.1038/nrc1231. [DOI] [PubMed] [Google Scholar]; b Kedrin D, Gligorijevic B, Wyckoff J, Verkhusha V, Condeelis J, Segall J, van Rheenen J. Intravital imaging of metastatic behavior through a mammary imaging window. Nat Methods. 2008;5(12):1019–1021. doi: 10.1038/nmeth.1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rueda P, Richart A, Récalde A, Gasse P, Vilar J, Guérin C, Lortat-Jacob H, Vieira P, Baleux F. i., Chretien F. Homeostatic and Tissue Reparation Defaults in Mice Carrying Selective Genetic Invalidation of CXCL12/Proteoglycan InteractionsClinical Perspective. Circulation. 2012;126(15):1882–1895. doi: 10.1161/CIRCULATIONAHA.112.113290. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.