Abstract
Background
Chronic alcohol exposure results in liver injury that is driven in partby inflammatory cytokines such as tumor necrosis factor-α (TNF). Hepatocytes are normally resistant to the cytotoxic effects of TNF, but they become sensitized to TNF by chronic alcohol exposure. Recently we reported that the decrease in the ratio of S-adenosylmethionine (SAM) to S-adenosylhomocysteine (SAH) that occurs with alcoholic liver injury renders hepatocytes sensitive to TNF cytotoxicity. The purpose of the present study was to determine whether inhibition of the transcription factor NFκB contributed to TNF-induced cell death in hepatocytes with high levels of SAH.
Methods
Primary human hepatocytes or HepG2 cells were pre-incubated with a combination of adenosine plus homocysteine to increase SAH levels. Following exposure toTNF, viability was determined by the MTT assay, and activation of the NFκB pathway was assessed by measuring degradation of cytosolic IκB-α, phosphorylation and translocation of NFκB to the nucleus, and expression of NFκB-dependent genes.TNF-induced apoptotic signaling pathways were assessed by monitoring levels of the anti-apoptotic protein, A20, and cleavage products of the caspase-8 substrate, RIP1.
Results
NFκB-mediated gene expression was inhibited in cells with high SAH, despite the fact that TNF-induced degradation of the cytoplasmic inhibitor IkB-α and accumulation of NF-κB in the nucleus persisted for much longer. In contrast to control cells, the NF-κB that accumulated in the nucleus of cells with high SAH levels was not phosphorylated at serine 536, a modification associated with activation of the transactivation potential of this transcription factor. The inhibition of transactivation by NF-κB resulted in lower mRNA and protein levels of the anti-apoptotic protein A20 and increased cleavage of RIP1.
Conclusions
High SAH levels inhibitedNFκB-mediatedgene expression and sensitized primary hepatocytes and HepG2 cells to the cytotoxic effects of TNF. It is likely that cross-talk with other transcription factors is perturbed under these conditions, resulting in still other changes in gene expression.
Keywords: Tumor necrosis factor, S-adenosylhomocysteine, NF-kappaB, alcoholic liver disease, sensitization
INTRODUCTION
The inflammatory cytokine tumor necrosis factor-α (TNF) plays a critical role in the development of alcoholic liver disease. Blocking the production of TNF or its interaction with TNF receptors protects hepatocytes from cell death in animal models of alcoholic liver disease (Koop et al., 1997, Iimuro et al., 1997). However, TNF alone cannot induce cell death in hepatocytes; they must be rendered sensitive to this effect. In alcoholic liver disease, sensitization of hepatocytes to TNF has been linked to alcohol-induced alterations in methionine metabolism (McClain et al., 2002, Mato et al., 2008). Upon chronic alcohol exposure, there is a decrease in S-adenosylmethionine (SAM) and an increase in S-adenosylhomocysteine (SAH), resulting in an inhibition of SAM-dependent transmethylation reactions (Halsted et al., 1996, Lieber et al., 1990). We have shown that an increase in SAH relative to SAM is sufficient to sensitize the liver and hepatocytes to TNF cytotoxicity (Song et al., 2004, Chawla et al., 1998, Song et al., 2007).
The purpose of the present study was to determine the mechanism by which increased SAH levels leads to sensitization of hepatocytes to TNF. Agents that block new RNA or protein synthesis, such asgalactosamine, actinomycin D and cycloheximide, are often used experimentally to sensitize hepatocytes to the cytotoxicity of TNF (Galanos et al., 1979, Nagaki and Moriwaki, 2008), suggesting that up-regulation of protective genes is an important aspect of resistance to TNF. The transcription factor NFκB has been shown to be a critical mediator of resistance to TNF cytotoxicity in a number of cell types (Wang et al., 1996, Beg and Baltimore, 1996). Cells that are resistant to TNF cytotoxicity expressNFκB-dependent anti-apoptotic genes such asA20 (Arvelo et al., 2002, Daniel et al., 2004, Opipari et al., 1992, Krikos et al., 1992), and TNF-resistant cells can be made sensitive to TNF cytotoxicity by inhibiting NFκB activity (Van Antwerp et al., 1996).
HepG2 cells have been a useful model in which to study the cytotoxicity of TNF in hepatocytes. As in primary hepatocytes and liver tissue, TNF alone is insufficient to induce death in this hepatocellular carcinoma cell line (Hill et al., 1995).In the current study, HepG2 cells were exposed to a combination of adenosine and homocysteine to increase SAH levels and sensitize them to TNF cytotoxicity. The results showed that NFκB activity is indeed inhibited under these conditions. Interestingly, early steps in the activation of NFκB, including degradation of the cytoplasmic inhibitor IκB-α and translocation of NFκB to nucleus, were not completely blocked in cells with high SAH levels, but the expression of NFκB-dependent genes was no longer inducible upon TNF exposure. We conclude that SAH inhibits NFκB-mediated gene expression by blocking the formation of active transcriptional complexes at the promoters of genes involved in protection against TNF cytotoxicity.
METHODS
Cell culture
HepG2 cells were purchased from ATCC (Manassas, VA) and cultured in DMEM containing L-glutamine (Hyclone Laboratories, Inc., Logan, UT), supplemented with 10% heat-inactivated fetal bovine serum (Hyclone Laboratories, Inc., Logan, UT), 100 U/ml penicillin and 100 µg/ml streptomycin (Hyclone Laboratories, Inc., Logan, UT) in a humidified atmosphere of 5% CO2.Cryoplateable primary human hepatocytes were obtained from BioreclamationIVT (Baltimore, MD). Hepatocytes were thawed and plated according to supplier’s protocols.
TNF cytotoxicity assay
Cells were plated in 96-well plates at a density of 12,500 cells/well. Media was replaced 24 hours later with DMEM supplemented with 1% FBS and containing adenosine (Ado; Sigma-Aldrich, St. Louis, MO), homocysteine (Hcy; Santa Cruz Biotechnology, Inc., Santa Cruz, CA) or the combination, as indicated in figure legends. After 2 hour pre-incubation, TNF (R & D Systems, Inc. Minneapolis, MN) was added to the medium to achieve indicated concentrations. Cell viability was assessed 24 hours after TNF addition by the MTT assay.
MTT assay
MTT (Sigma-Aldrich, St. Louis, MO) was added to the cultures at a final concentration of 0.2 mg/ml, and cells were incubated for an additional 30 to 60 minutes. MTT solution was removed, formazan crystals were dissolved in DMSO and absorbance was measured at 540 nm.
Measurement of SAH
SAH was extracted in 5% metaphosphoric acid and measured by HPLC as described previously (Watson et al., 2011).
NFκB-luciferase reporter assay
NFκB transcriptional activity was measured as in our previous studies (Heilman et al., 2011). The NFκB-luciferase reporter (Stratagene/Agilent Technologies, Santa Clara, CA) was co-transfected with pCMV-β-galactosidase (a kind gift from K. Cameron Falkner) usingLipofectamine (Invitrogen/Life Technologies, Grand Island, NY). Twenty-four hours post-transfection, cells were pre-treated for 2 hours with or without Ado+Hcy, and then stimulated with 50 ng/ml TNF. Cell lysates were prepared in Reporter Lysis Buffer (Promega, Madison, WI), luciferase activity was measured with the Luciferase Assay System (Promega, Madison, WI), andβ-galactosidase activity was measured by incubation with chlorophenol red β-galactopyranoside (Roche, Indianapolis, IN) for 1 hour at 37°C (Falkner et al., 1998). Results are expressed as the ratio of luciferase to β-galactosidase activities.
Real time RT-PCR
Total RNA was harvested from cells using TRIzol reagent (Life Technologies, Grand Island, NY). The amount and purity of RNA was quantified with the NanoDrop 1000 system (Thermo Fisher Scientific, Waltham, MA), and 1ug total RNA was converted to cDNA using qScriptcDNASuperMix (Quanta Biosciences, Inc., Gaithersburg, MD) following the manufacturer’s protocol. Real-time RT-PCR reactions were assembled using PerfeCta SYBR Green FastMix, Low ROX (Quanta Biosciences, Inc., Gaithersburg, MD) following the manufacturer’s protocol, and quantitative PCR was performed on an Applied Biosystems 7500 Real-Time PCR System (Life Technologies, Grand Island, NY). CT values were normalized to those of TATA binding protein, and expressed relative to the normalized values from untreated controls. Fold-induction was calculated as 2−ΔΔCT.
Subcellular fractionation and western blotting. Nuclear and cytosolic fractions were prepared as described previously (Hill et al., 1999), with the exception that 10 µMNaF and 1 mMNa3VO4 were added to the buffers to inhibit phosphatase activity. Whole cell lysates were prepared in 25 mMHepes, pH7.2, 150 mM potassium acetate, 2 mM EDTA, 1% NP-40, 1 mM DTT, 10 µMNaF, 1 mM Na3VO4, and a protease inhibitor cocktail (Sigma-Aldrich).Total protein was quantified usingthe Bio-Rad DC protein assay with γ-globulin as the standard (Bio-Rad Laboratories, Hercules, CA). Twenty micrograms total protein were separated by SDS-PAGE electrophoresis and transferred to nitrocellulose using standard protocols. Antibodies were purchased from Santa Cruz Biotechnology, Inc. (IκB-α (C-21), NF-κB p65 (C-20), A20 (A-12), HRP-conjugated anti-mouse, anti-goat, anti-rabbit); Cell Signaling Technology (phospho-IκB- (Ser32/36) (5A5), phospho-NF-κB p65 (Ser536), Boston, MA); BD Biosciences (RIP1); and Sigma-Aldrich (β-actin, clone AC-15, St. Louis, MO). HRP-conjugated secondary antibodies were detected with ECL2 western blotting substrate (Pierce/Thermo Scientific, Rockford, IL). Band intensities were quantified with ImageJ software.
RESULTS
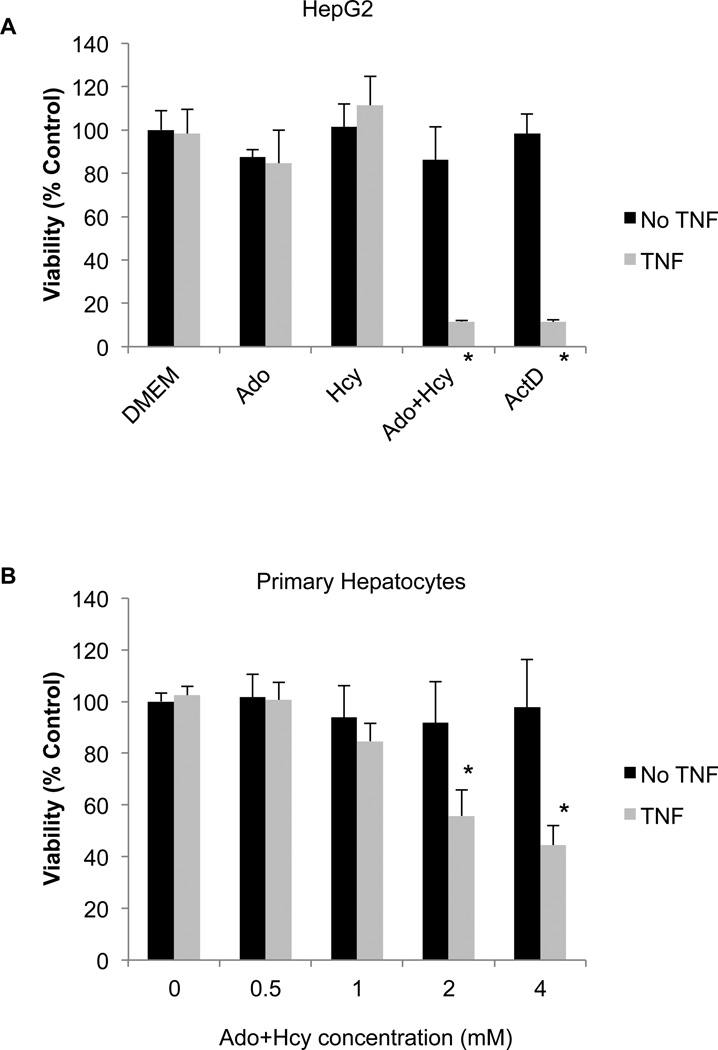
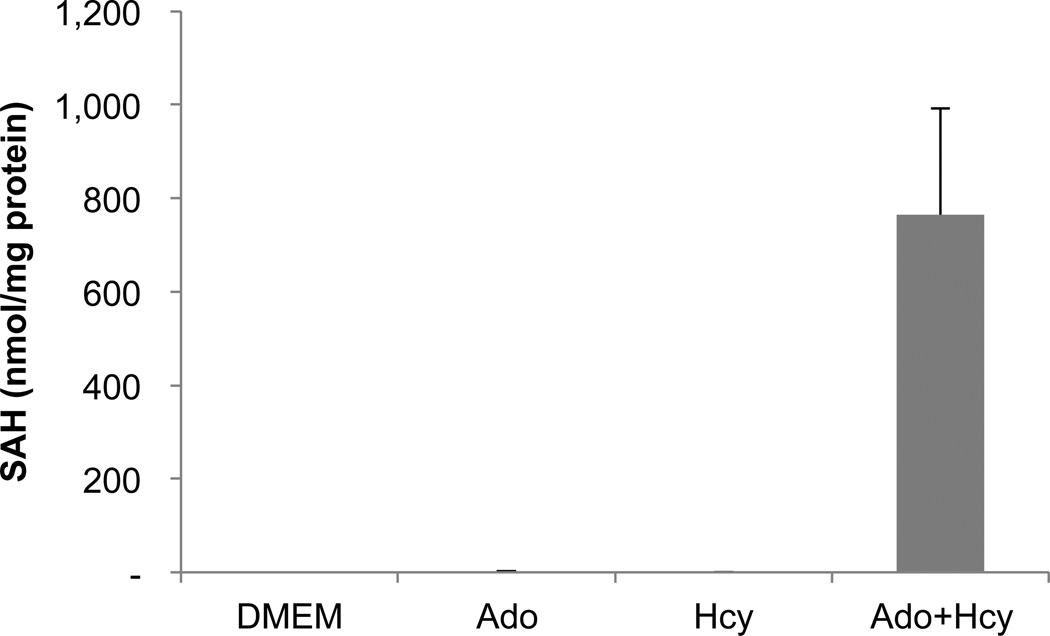
To model the increase in hepatic SAH that is observed in alcoholic liver disease, the HepG2 hepatocyte cell line was incubated with a combination of adenosine (Ado) and homocysteine (Hcy), as in our previous studies (Song et al., 2004, Song et al., 2007). Ado and Hcy serve as co-substrates for the enzyme SAH hydrolase. Under normal conditions, SAH hydrolase catalyzes the breakdown of SAH into Ado and Hcy, but when Ado and Hcy levels are high the reverse reaction is favored, resulting in the synthesis of SAH.Indeed, addition of Ado+Hcy to the culture medium resulted in an increase inintracellular SAH levels.Under these conditions, the cells became sensitive to the cytotoxicity of TNF (Fig. 2). The extent of cell killing was similar to that seen with the classical TNF sensitizer actinomycin D (Fig. 2). Ado or Hcy alone did not sensitize cells to TNF cytotoxicity (Fig. 2), nor did they increase SAH levels to the same extent as seen with the combination of the two (Fig. 1). Similar results were seen with primary human hepatocytes. Although higher concentrations of Ado+Hcy were required, there was a clear concentration-dependent sensitization to the cytotoxic effects of TNF (Fig. 2B). Identical results were obtained with hepatocytes from 2 different donors.
Fig. 2.
HepG2 cells and primary human hepatocytes are sensitized to TNF cytotoxicity by pre-treatment with the combination of adenosine and homocysteine (Ado+Hcy). (A) HepG2 cells were pre-treated for 2 hours with medium alone (DMEM), 500 µM adenosine (Ado), 500 µMhomocysteine (Hcy) or the combination (Ado+Hcy; 500 µM each), then exposed to 10 ng/ml TNF (light bars) or vehicle (DMEM; dark bars) for an additional 24 hours. MTT was added during the final hour of exposure to assess cell viability. Actinomycin D (ActD; 0.4 µg/ml) was included as a positive control for sensitization to TNF cytotoxicity. (B) Primary human hepatocytes were pre-treated with the indicated concentrations of Ado+Hcy, then exposed to 100 ng/ml TNF (light bars) or vehicle (DMEM; dark bars) for an additional 24 hours. MTT was added during the final hour of exposure to assess cell viability. The experiments were performed in triplicate using hepatocytes from 2 different donors, and the data from both donors was pooled for statistical analyses. Data show mean ± s.d. of the pooled data. *- Significantly different (p<0.05) compared to no TNF control.
Fig. 1.
SAH levels are increased in cells exposed to the combination of adenosine and homocysteine. HepG2 cells were incubated for 2 hours in medium alone (DMEM) or in medium supplemented with 500 µM adenosine (Ado), 500 µMhomocysteine (Hcy), or the combination (Ado+Hcy). Intracellular SAH was measured by HPLC as described in Materials and Methods. Data represent the means ± sd of 3 independent measurements.
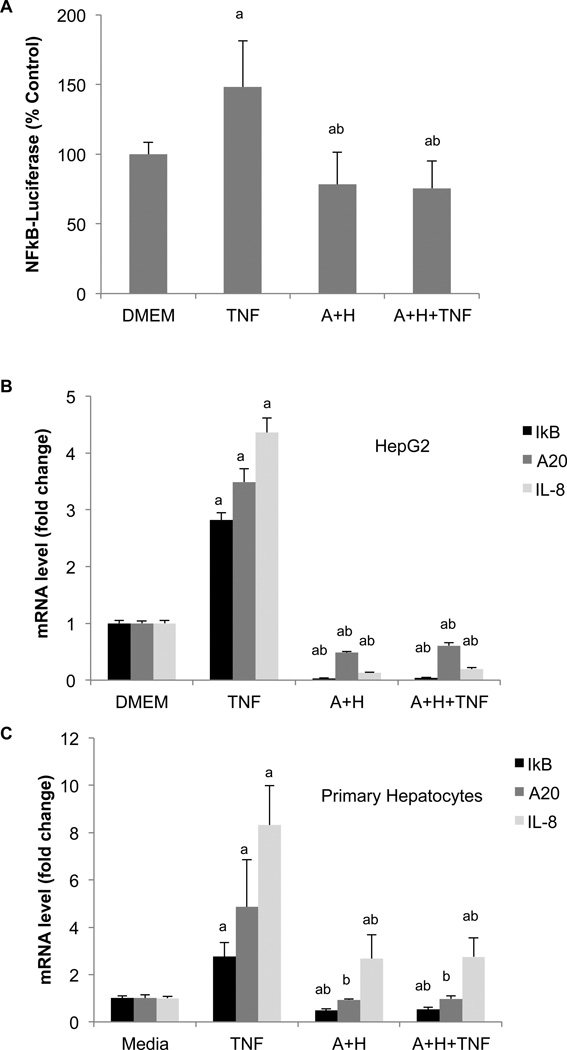
Because actinomycin D inhibits new RNA synthesis, it is believed that actinomycin D sensitizes cells to TNF by blocking the expression of protective genes (Hill et al., 1995, Nagaki and Moriwaki, 2008). Previous studies have shown that the transcription factor NFκB controls the expression of these protective genes (Van Antwerp et al., 1996), and inhibition of NFκB can also render cells sensitive to TNF (Wang et al., 1996). To determine whether Ado+Hcy treatment inhibitedNFκB-mediated gene expression, NFκB activity was measured with an NFκB-luciferase reporter. The results showed that NFκB activity was activated in resistant control cells, but that this activation was blocked in cells treated with Ado+Hcy (Fig. 3A).
Fig. 3.
NFκB-dependent gene expression is inhibited by SAH. (A) Expression of an NFκB-luciferase reporter construct in inhibited by SAH. HepG2 cells were co-transfected with plasmids encoding an NFκB-luciferase reporter and β-galactosidase under the control of the CMV promoter. 24 hours after transfection, the cells were pre-treated for 2 hours with either medium (DMEM) or 500 µM adenosine and homocysteine (A+H), then either stimulated with 10 ng/ml TNF or left unstimulated for 3 hours. Luciferase activity was normalized to β-galactosidase activity and expressed as a percent of the value in DMEM (control) cells. (B) HepG2 cells were pre-treated with medium alone (DMEM) or 500 µMAdo+Hcy (A+H), then treated with 10 ng/ml TNF were indicated for 3 hours. RNA was isolated and analyzed by real time RT-PCR as described in the Methods. (C) Primary human hepatocytes were pre-treated with 4 mMAdo+Hcy (A+H) or medium alone, then treated with 100 ng/ml for 3 hours. RNA was isolated and analyzed as in panel (B). a- Significantly different (p<0.05) compared to medium alone. b- Significantly different (p<0.05) compared to TNF alone.
In unstimulated cells, NFκB is normally maintained in an inactive form in a cytoplasmic complex with IκB-α. Upon stimulation, IκB-α is degraded, allowing NFκB to move into the nucleus and bind to specific sequences within the promoters of its target genes. One of these targets is IκB-α, which is re-synthesized and thereby limits the duration of NFκB activity (Sun et al., 1993). Expression of IκB-α was induced upon stimulation with TNF in control cells, but not in cells treated with Ado+Hcy (Fig. 3B). In fact, IκB-α expression was inhibited by Ado+Hcy even in the absence of TNF stimulation, suggesting that basal NFκB activity was inhibited. Similarly, basal and TNF-induced expression of two other endogenous NFκB targets, IL-8 and A20, were inhibited in HepG2 cells with high SAH levels (Fig. 3B). NFκB-mediated gene expression was also inhibited in primary human hepatocytes treated with Ado+Hcy. As in HepG2 cells, TNF-induced expression of IκB-α, A20 and IL-8 was inhibited in hepatocytes pre-treated with Ado+Hcy (Fig. 3C). Basal IκB-α mRNA levels were also lower in Ado+Hcy-treated cells. In contrast to HepG2 cells, however, basal A20 levels were unaffected, and basal IL-8 levels were increased by Ado+Hcy in primary hepatoctyes (Fig. 3C).
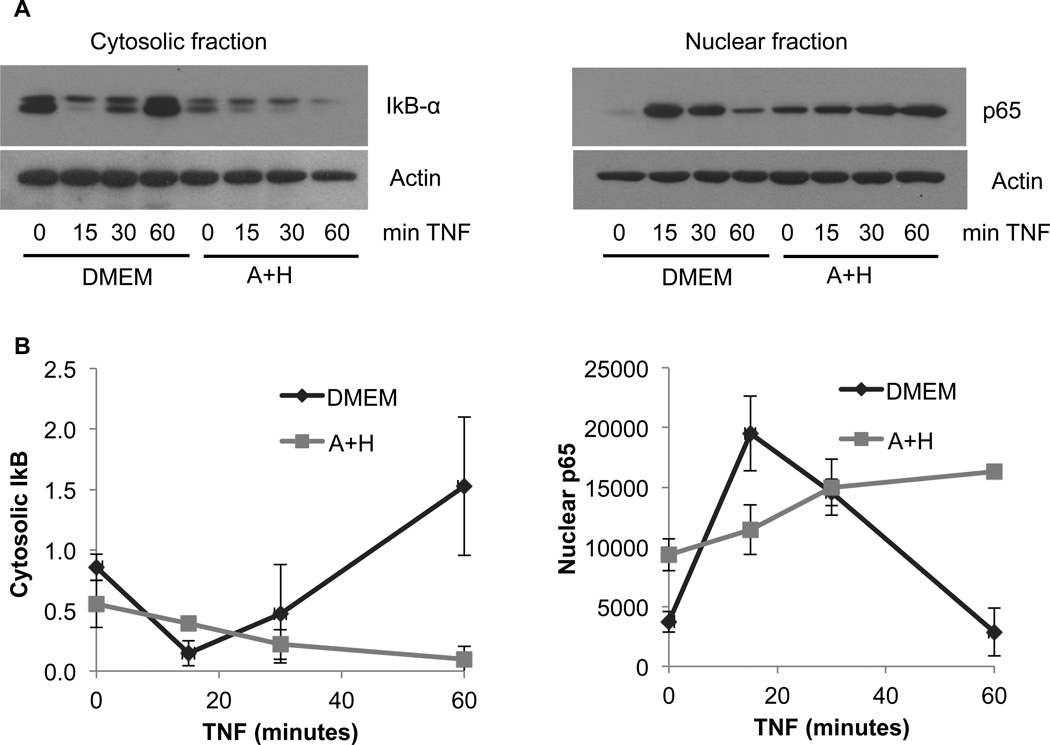
To determine the mechanism by which Ado+Hcy inhibitedNFκB-mediated gene expression, activation of the pathway by degradation of IκB-α and nuclear translocation of NFκB was examined by western blotting of cytosolic and nuclear fractions (Fig. 4). In control cells, TNF stimulation resulted in transient degradation of cytosolic IκB-α within 15 minutes, followed by the reappearance of IκB-α and a return to baseline levels within 60 minutes. The kinetics of IκB-α degradation and re-synthesis were mirrored by the movement of the NFκB subunit p65 into, and then out of, the nucleus over the same time period. These results are typical of activation of the canonical NFκB signaling pathway by TNF. In contrast to the results in control cells, the Ado+Hcy pre-treated cells responded much differently. Baseline levels (i.e., in cells not stimulated with TNF) of cytosolic IκB-α and nuclear p65 were lower and higher, respectively, than in control cells. Stimulation with TNF resulted in more persistentstimulation of the NFκB signaling pathway, as evidenced by continuing degradation of IκB-α and increasingly high levels of p65 in the nuclear fractions over the time course of the exposure (Fig. 4). The prolonged nuclear localization of NFκB is most likely the result of inhibition of IκB-α re-synthesis; IκB-α expression is normally induced by NFκB (see Fig.3), providing a mechanism to replenish IκB-α and limit the duration of NFκB activity. A second NFκB target gene, A20, also functions as a negative feedback inhibitor of NFκB activation by deubiquitinating RIP1 and disrupting the recruitment of IKK-γ to the TNFR1 signaling complex (Shembade and Harhaj, 2012). Because SAH inhibits these negative feedback arms of the canonical pathway, NFκB levels remain high in the nucleus. However, this NFκB is not transcriptionally active, as evidenced by the low levels of IκB-α, A20 and IL-8 mRNA (Figs. 3B and 3C) and lack of expression of the NFκB-dependent luciferase reporter (Fig. 3A).
Fig. 4.
TNF-induced degradation of IκB-α and nuclear translocation of NFκB is not inhibited by SAH. HepG2 cells were pre-treated for 2 hours with either medium alone (DMEM) or the combination of 500 µM adenosine and 500 µMhomocysteine (A+H). The cells were then stimulated with 10 ng/ml TNF for the indicated times, and cytosolic and nuclear fractions were prepared as described in Materials and Methods. (A) Representative western blot showing degradation of IκB-α in cytosolic fraction and translocation of the NFκB subunit p65 to the nucleus in response to TNF. (B) Densitometric analyses of western blots from 3 independent experiments performed as in (A).
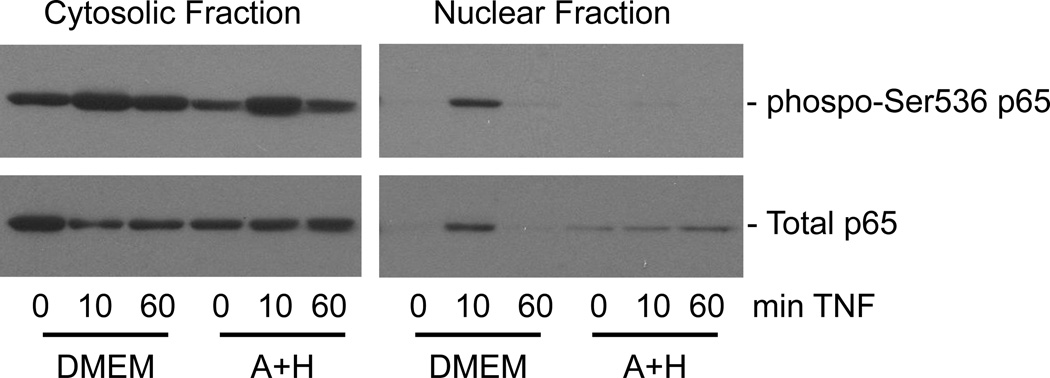
Nuclear accumulation alone is not sufficient for the activation of NFκB-mediated gene expression; other proteins must be recruited for the assembly of active transcriptional complexes at the promoters of target genes. One way that this is regulated is through posttranslational modifications of NFκB, such as phosphorylation of Ser536 within the transactivation domain of the p65 subunit (Neumann and Naumann, 2007). TNF stimulation results in rapid and transient phosphorylation of Ser536 (Fig. 5). Most of the NFκB that accumulates in the nucleus of control cells is in the phosphorylated form. In contrast, phosphorylation of Ser536 in the cytosolic pool of p65 is somewhat inhibited in cells with high SAH levels, and it is the unphosphorylated form of p65 that builds up in the nucleus of these cells over time (Fig. 5).
Fig. 5.
The phosphorylated form of p65 does not accumulate in the nucleus of Ado+Hcy-treated cells. Cells were pre-treated for 2 hours with medium alone (DMEM) or 500 µM adenosine and homocysteine (A+H), then stimulated with 10 ng/ml TNF for the indicated times. Cytosolic and nuclear fractions were prepared, and equal amounts of total protein (15 µg) were analyzed by western blotting.
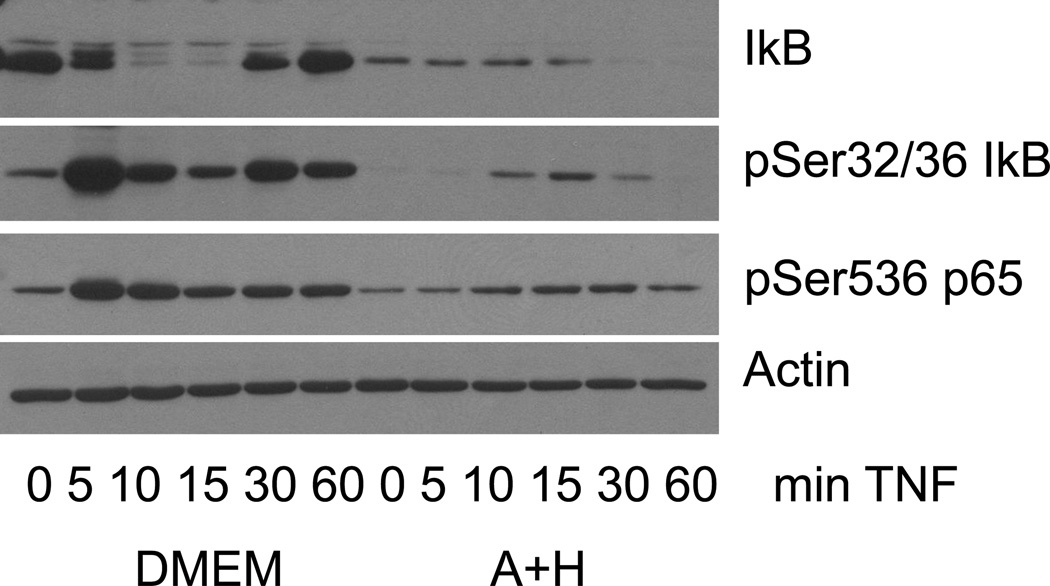
Phosphorylation of p65 at Ser536 can be mediated by at least five different enzymes, including multiple components of the IKK complex (Neumann and Naumann, 2007). If Ado+Hcy acts at the level of the IKK complex, which phosphorylatesIκB-αin response to TNF stimulation, then phosphorylation of IκB-α could also be affected. To examine this possibility, the time course of TNF-induced p65 phosphorylation was compared to the time course of IκB-α phosphorylation (Fig. 6). Maximal phosphorylation of both p65 and IκB-α occurred 5 minutes after TNF stimulation in control cells, whereas peak phosphorylation was delayed in Ado+Hcy-treated cells. In addition, the degree of phosphorylation of p65 and IκB-α was much less in Ado+Hcy-treated cells than in control cells. The phosphorylation of p65 paralleled the phosphorylation of IκB-α in control cells and in Ado+Hcy-treated cells, but the onset and extent of phosphorylation was inhibited in Ado+Hcy-treated cells. These results suggest that the activity of at least one of the components of the IKK complex is altered in cells with high levels of SAH.
Fig. 6.
Phosphorylation of both IκB-α and p65 is delayed and inhibited in cells with high SAH levels. Whole cell lysates were prepared from cells pre-treated with either medium alone (DMEM) or medium supplemented with 500 µM adenosine and homocysteine (A+H) and then stimulated with 10 ng/ml TNF for the indicated times. Western blotting was performed on 15 µg total protein using antibodies specific for the indicated proteins.
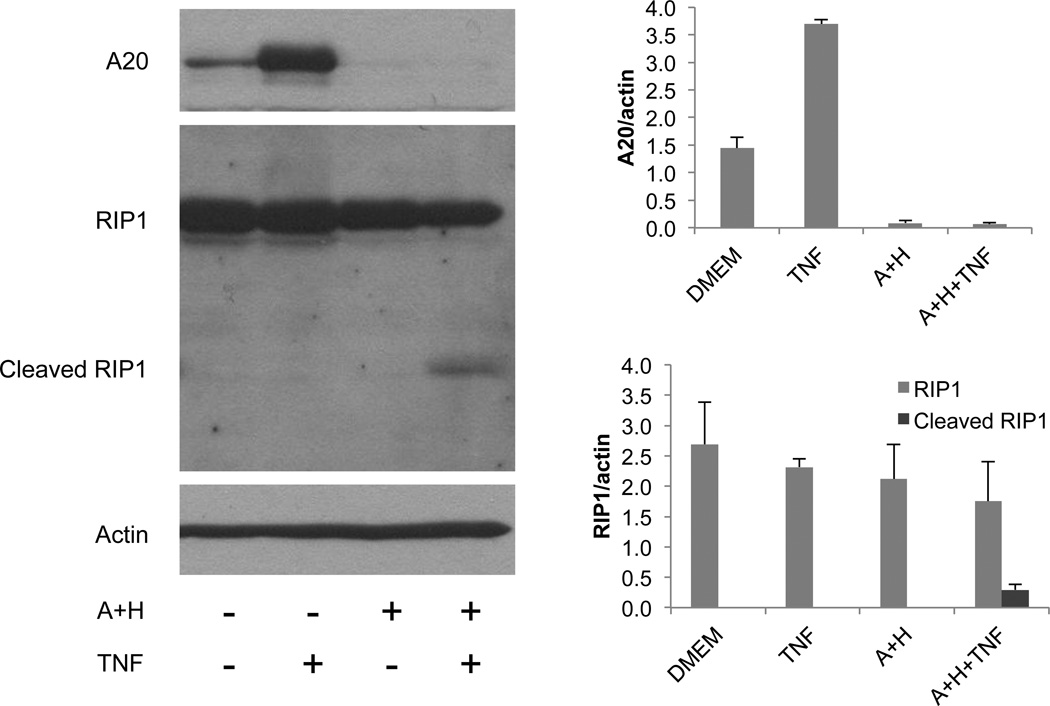
The data in Fig. 3 show that A20 mRNA levels are low in cells with high SAH. Fig. 7 shows that A20 protein levels correlate very well with mRNA levels, where both basal and TNF-stimulated A20 levels are inhibited by Ado+Hcy. In addition to being a feedback inhibitor of NFκB activation, A20 also functions as an anti-apoptotic protein that protects against TNF cytotoxicity (Lee et al., 2000).When TNF binds to its receptor TNFR1, multiple proteins are recruited to form what is known as TNFR1 complex I. Complex I signals NFκB activation and resistance to cell death. In cells lacking A20, formation of TNFR1 complex II is favored, allowing activation of caspase-8 and cell death. RIP1 is a component of both complexes, but becomes cleaved by caspase-8 upon the transition from complex I to complex II (Vandenabeele et al., 2010). RIP1 cleavage is seen in TNF-sensitive (Bellail et al., 2012) and TRAIL-sensitive cells (Dong et al., 2012) undergoing death, but not in resistant cells. To determine whether loss of A20 expression in cells with high SAH levels was associated with RIP1 cleavage, a western blot was performed. The data show that the RIP1 cleavage product is seen in response to TNF only in cells that were pre-treated with Ado+Hcy (Fig. 7).
Fig. 7.
Decreased A20 protein levels are associated with increased apoptotic signaling in TNF-stimulated cells with high SAH. Basal and TNF-induced A20 protein levels (upper panel) are decreased in whole cell lysates from cells treated with 500 µM adenosine and homocysteine (A+H). Cleavage of RIP1 is induced by TNF in cells with low A20 and high SAH levels. Actin is shown as a loading control. The graph at the bottom represents the results of densitometric analysis (mean +/− SD) of western blotting of 3 separate samples for each condition.
DISCUSSION
A number of cell types can be made sensitive to TNF cytotoxicity by genetic or pharmacologic inhibition of NFκB activity (Xu et al., 1998, Beg and Baltimore, 1996, Van Antwerp et al., 1996, Wang et al., 1996). The studies presented here demonstrate that intracellular accumulation of the endogenous methionine metabolite SAH can also inhibit NFκB and convert TNF-resistant HepG2 cells to TNF-sensitive cells. This sensitization was accompanied by an inhibition of both basal and TNF-induced expression of NFκB-dependent genes, including the TNFAIP3 gene encoding the anti-apoptotic A20 protein. In TNF-stimulated cells, loss of A20 expression was associated with the transition of the anti-apoptotic TNF receptor complex I to the pro-apoptotic complex II, as evidenced by RIP-1 cleavage. Therefore, both basal and TNF-induced NFκB-dependent A20 expression contribute to resistance to the cytotoxic effects of TNF in HepG2 cells and in primary hepatocytes by favoring anti-apoptotic signaling pathways, and the balance is shifted in favor of pro-apoptotic pathways in cells with high SAH levels.
In addition to its role as an inhibitor of apoptosis, A20 also functions as an inhibitor of NFκB activation. Induction of A20, therefore, serves to limit the duration of NFκB activity following a stimulus such as TNF. In this respect, A20 is analogous to IκB-α: both genes are activated by NFκB, and both encode proteins that terminate NFκB signaling (Sun et al., 1993, Lee et al., 2000). A20 does so by de-ubiquitinating RIP1 and disrupting its interactions IKK-γ and the subsequent activation of the IKK complex. IκB-α terminates NFκB activity by replacing the IκB-α that was degraded and re-forming inhibitory complexes with NFκB. Because A20 and IκB-α inhibit signaling events leading to NFκB activation, overexpression of either can inhibit the extent to which NFκB can be activated. However, Ado+Hcy does not induce overexpression of these proteins; rather, they are both inhibited. Therefore, this does not appear to be the mechanism by which SAH inhibits NFκB. The decreased basal expression of the NFκB target genes A20, IκB-α and IL-8 most likely reflects the inhibition of transactivation potential of nuclear NFκB by SAH. Our data show that phosphorylation of Ser536 within the transactivation domain of p65 is decreased by SAH, even under basal conditions. Phosphorylation at this site promotes the recruitment of co-activators necessary for expression of target genes, and loss of this activating posttranslational modification may account for the inability of NFκB to mediate gene expression.
The mechanism by which SAH inhibitedNFκB was somewhat unusual in that the cytoplasmic signaling pathways were only partially inhibited, allowing NFκB to translocate to the nucleus in response to TNF stimulation. However, this nuclear NFκB was not active, due, at least in part, to a lack of phosphorylation of the transactivation domain. Because the expression of the feedback inhibitors A20 and IκB-α were not activated, NFκB remained in the nucleus for much longer. It is unclear, however, to what extent the presence of transcriptionally inactive NFκB in the nucleus interferes with the activities of other transcription factors.
Other agents that affect methionine metabolism have been shown to have similar effects on NFκB activity in other cell types. Treatment of RAW 264.7 cells with SAM, the immediate upstream metabolite of SAH, decreased LPS-induced NFκB transcriptional activity without affecting IκB-α degradation or nuclear translocation of NFκB (Gobejishvili et al., 2011). In another study using LPS-stimulated RAW 264.7 cells, Jeong et al, showed that deazaadenosine (DZA), an inhibitor of SAH hydrolase, inhibited the transcriptional activity of NFκB while at the same time inducing the degradation of IκB-α (Jeong et al., 1999). Interestingly, another SAH hydrolase inhibitor, DZAari, had no effect on NFκB in that study (Jeong et al., 1999). Additional studies are needed to differentiate among the effects of SAM, SAH and enzymes that interact with these metabolites in the regulation of NFκB in response to different stimuli and in different cell types.
Inhibition of two other enzymes that are not directly related to methionine metabolism also had effects on NFκB activity that were similar to those reported here. Inhibition of PDE4B by rolipram increased cAMP levels and inhibited LPS-induced NFκB transcriptional activity without affecting IκB-α degradation or NFκB nuclear translocation (Gobejishvili et al., 2008). Similarly, inhibitors of thioredoxinreductase, an enzyme involved in antioxidant defenses, blocked the transcriptional activity of NFκB, but had no effect on the upstream steps activated by TNF or LPS (Heilman et al., 2011). The phosphorylation state of Ser536 was not measured in the nuclear pool of p65 in those earlier studies, but at least one of the inhibitors (curcumin) blocked phosphorylation of total p65 in response to TNF (Heilman et al., 2011).
SAH is produced in multiple subcellular compartments by SAM-dependent methyltransferases within those compartments. Cytoplasmic and mitochondrial pools of SAH are increased in hepatocytes of mice chronically exposed to alcohol (Song et al., 2007), and this may be influenced by both altered methionine metabolism and the effects on transport of SAM and SAH across the mitochondrial membrane (Fernandez et al., 2009). Much less is known about what happens to SAH in the nuclear compartment. DNA methyltransferases and histone methyltransferases, which use SAM and generate SAH, are present in the nucleus and have a tremendous impact on gene expression through effects on chromatin remodeling. These methylransferases are subject to product inhibition by SAH (Mull et al., 2006) and, therefore, chromatin structure could be altered by high concentrations of SAH in the nucleus. Either nuclear or cytoplasmic SAH levels may be responsible for the observation that NFκB was not phosphorylated in the nuclei of cells exposed to Ado+Hcy. Phosphorylation of p65 at Ser536 occurs within the cytoplasm, as shown by our data showing an increase in phosphorylated p65 within that compartment (Fig. 6) as well as the reported localization of the kinases responsible for this modification (Neumann and Naumann, 2007). While SAH does not appear to completely block phosphorylation in the cytoplasm, it may block the translocation of the phosphorylated form of NFκB from the cytoplasm to the nucleus, possibly by interfering with its interaction with importin α3 or α4 (Fagerlund et al., 2005). Alternatively, SAH could exert its effects within the nucleus by facilitating dephosphorylation within the nucleusor by increasing the rate of export from the nucleus. Therefore, it is possible that SAH acts at multiple levels may to modulate transcription independent of, or in concert with, cytoplasmic signaling cascades.
The data presented hereconfirm and extend our previous finding that elevated SAH levels sensitize hepatocytes to TNF cytotoxicity (Song et al., 2007, Song et al., 2004). The combination of Ado+Hcy increased intracellular SAH levels to 760 nmol/mg protein. This value was slightly higher than the value of 250 nmol/mg protein that we reported previously (Song et al., 2004). The difference could have been due to our use of medium containing 1% fetal bovine serum in the current study and 10% fetal bovine serum in the earlier study. Others have shown that the combination of Ado+Hcy sensitized other cell types to TNF (Ratter et al., 1999, Bergmann et al., 1994). Bergmann et al., reported that L929 cells were sensitized to TNF by the combination of Ado+Hcy, but that NFκB activation was not altered (Bergmann et al., 1994). This latter conclusion was based on the observation that Ado+Hcy did not inhibit TNF-induced nuclear translocation of NFκB as measured by electrophoretic mobility shift assay.Transcriptional activity was not measured in that study, but in view of the present results it is likely that the NFκB seen in the nuclei of Ado+Hcy-treated L929 cells was transcriptionally inactive. Factors other than NFκB inhibition may increase the extent to which hepatocytes become sensitive to TNF (Fredriksson et al., 2011, Osawa et al., 2001), and chronic alcohol exposure has effects other than perturbation of SAM and SAH metabolism (McClain et al., 2002). For example, TNF-induced hepatocyte apoptosis has been observed even in the presence of activated NFκB when mitochondrial glutathione was depleted (Mari et al., 2008), a condition associated with chronic alcohol exposure (Hirano et al., 1992). Nevertheless, the results presented here demonstrate NFκB signaling is impaired in cells with high SAH levels, and that this leads to increased sensitivity to the cytotoxic effects of TNF.
ACKNOWLEDGEMENTS
This work was supported by NIH grants R21 ES021311 (WHW), R01 AA018869 (CJM), P01 AA017103 (CJM), R01 AA018016 (CJM), R37 AA010762 (CJM), P30 AA019360 (CJM), RC2AA019385 (CJM), R01 AA015970 (CJM), R01 DK071765 (CJM) and the Department of Veterans Affairs (CJM). The authors wish to thank David Barker for his assistance with the design of primers for the PCR experiments, and K. Cameron Falkner for his kind gift of the β-galactosidase expression vector.
Sources of Support
This work was supported by NIH grants R21 ES021311 (WHW), R01 AA018869 (CJM), P01 AA017103 (CJM), R01 AA018016 (CJM), R37 AA010762 (CJM), P30 AA019360 (CJM), RC2AA019385 (CJM), R01 AA015970 (CJM), R01 DK071765 (CJM) and the Department of Veterans Affairs (CJM).
REFERENCES
- Arvelo MB, Cooper JT, Longo C, Daniel S, Grey ST, Mahiou J, Czismadia E, Abu-Jawdeh G, Ferran C. A20 protects mice from D-galactosamine/lipopolysaccharide acute toxic lethal hepatitis. Hepatology. 2002;35:535–543. doi: 10.1053/jhep.2002.31309. [DOI] [PubMed] [Google Scholar]
- Beg AA, Baltimore D. An essential role for NF-kappaB in preventing TNF-alpha-induced cell death. Science. 1996;274:782–784. doi: 10.1126/science.274.5288.782. [DOI] [PubMed] [Google Scholar]
- Bellail AC, Olson JJ, Yang X, Chen ZJ, Hao C. A20 ubiquitin ligase-mediated polyubiquitination of RIP1 inhibits caspase-8 cleavage and TRAIL-induced apoptosis in glioblastoma. Cancer discovery. 2012;2:140–155. doi: 10.1158/2159-8290.CD-11-0172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergmann S, Shatrov V, Ratter F, Schiemann S, Schulze-Osthoff K, Lehmann V. Adenosine and homocysteine together enhance TNF-mediated cytotoxicity but do not alter activation of nuclear factor-kappa B in L929 cells. J Immunol. 1994;153:1736–1743. [PubMed] [Google Scholar]
- Chawla RK, Watson WH, Eastin CE, Lee EY, Schmidt J, McClain CJ. S-adenosylmethionine deficiency and TNF-alpha in lipopolysaccharide-induced hepatic injury. Am J Physiol. 1998;275:G125–G129. doi: 10.1152/ajpgi.1998.275.1.G125. [DOI] [PubMed] [Google Scholar]
- Daniel S, Arvelo MB, Patel VI, Longo CR, Shrikhande G, Shukri T, Mahiou J, Sun DW, Mottley C, Grey ST, Ferran C. A20 protects endothelial cells from TNF-, Fas-, and NK-mediated cell death by inhibiting caspase 8 activation. Blood. 2004;104:2376–2384. doi: 10.1182/blood-2003-02-0635. [DOI] [PubMed] [Google Scholar]
- Dong B, Lv G, Wang Q, Wei F, Bellail AC, Hao C, Wang G. Targeting A20 enhances TRAIL-induced apoptosis in hepatocellular carcinoma cells. Biochem Biophys Res Commun. 2012;418:433–438. doi: 10.1016/j.bbrc.2012.01.056. [DOI] [PubMed] [Google Scholar]
- Fagerlund R, Kinnunen L, Kohler M, Julkunen I, Melen K. NF-{kappa}B is transported into the nucleus by importin {alpha}3 and importin {alpha}4. J Biol Chem. 2005;280:15942–15951. doi: 10.1074/jbc.M500814200. [DOI] [PubMed] [Google Scholar]
- Falkner KC, Rushmore TH, Linder MW, Prough RA. Negative regulation of the rat glutathione S-transferase A2 gene by glucocorticoids involves a canonical glucocorticoid consensus sequence. Molecular pharmacology. 1998;53:1016–1026. [PubMed] [Google Scholar]
- Fernandez A, Colell A, Caballero F, Matias N, Garcia-Ruiz C, Fernandez-Checa JC. Mitochondrial S-adenosyl-L-methionine transport is insensitive to alcohol-mediated changes in membrane dynamics. Alcohol Clin Exp Res. 2009;33:1169–1180. doi: 10.1111/j.1530-0277.2009.00940.x. [DOI] [PubMed] [Google Scholar]
- Fredriksson L, Herpers B, Benedetti G, Matadin Q, Puigvert JC, de Bont H, Dragovic S, Vermeulen NP, Commandeur JN, Danen E, de Graauw M, van de Water B. Diclofenac inhibits tumor necrosis factor-alpha-induced nuclear factor-kappaB activation causing synergistic hepatocyte apoptosis. Hepatology. 2011;53:2027–2041. doi: 10.1002/hep.24314. [DOI] [PubMed] [Google Scholar]
- Galanos C, Freudenberg MA, Reutter W. Galactosamine-induced sensitization to the lethal effects of endotoxin. Proc Natl Acad Sci U S A. 1979;76:5939–5943. doi: 10.1073/pnas.76.11.5939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gobejishvili L, Avila DV, Barker DF, Ghare S, Henderson D, Brock GN, Kirpich IA, Joshi-Barve S, Mokshagundam SP, McClain CJ, Barve S. S-Adenosylmethionine Decreases Lipopolysaccharide-Induced Phosphodiesterase 4B2 and Attenuates Tumor Necrosis Factor Expression via cAMP/Protein Kinase A Pathway. J Pharmacol Exp Ther. 2011;337:433–443. doi: 10.1124/jpet.110.174268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gobejishvili L, Barve S, Joshi-Barve S, McClain C. Enhanced PDE4B expression augments LPS-inducible TNF expression in ethanol-primed monocytes: relevance to alcoholic liver disease. Am J Physiol Gastrointest Liver Physiol. 2008;295:G718–G724. doi: 10.1152/ajpgi.90232.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halsted CH, Villanueva J, Chandler CJ, Stabler SP, Allen RH, Muskhelishvili L, James SJ, Poirier L. Ethanol feeding of micropigs alters methionine metabolism and increases hepatocellular apoptosis and proliferation. Hepatology. 1996;23:497–505. doi: 10.1002/hep.510230314. [DOI] [PubMed] [Google Scholar]
- Heilman JM, Burke TJ, McClain CJ, Watson WH. Transactivation of gene expression by NF-kappaB is dependent on thioredoxin reductase activity. Free Radic Biol Med. 2011;51:1533–1542. doi: 10.1016/j.freeradbiomed.2011.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill DB, Devalaraja R, Joshi-Barve S, Barve S, McClain CJ. Antioxidants attenuate nuclear factor-kappa B activation and tumor necrosis factor-alpha production in alcoholic hepatitis patient monocytes and rat Kupffer cells, in vitro. Clinical biochemistry. 1999;32:563–570. doi: 10.1016/s0009-9120(99)00056-9. [DOI] [PubMed] [Google Scholar]
- Hill DB, Schmidt J, Shedlofsky SI, Cohen DA, McClain CJ. In vitro tumor necrosis factor cytotoxicity in Hep G2 liver cells. Hepatology. 1995;21:1114–1119. [PubMed] [Google Scholar]
- Hirano T, Kaplowitz N, Tsukamoto H, Kamimura S, Fernandez-Checa JC. Hepatic mitochondrial glutathione depletion and progression of experimental alcoholic liver disease in rats. Hepatology. 1992;16:1423–1427. doi: 10.1002/hep.1840160619. [DOI] [PubMed] [Google Scholar]
- Iimuro Y, Gallucci RM, Luster MI, Kono H, Thurman RG. Antibodies to tumor necrosis factor alfa attenuate hepatic necrosis and inflammation caused by chronic exposure to ethanol in the rat. Hepatology. 1997;26:1530–1537. doi: 10.1002/hep.510260621. [DOI] [PubMed] [Google Scholar]
- Jeong SY, Ahn SG, Lee JH, Kim HS, Kim JW, Rhim H, Jeong SW, Kim IK. 3-deazaadenosine, a S-adenosylhomocysteine hydrolase inhibitor, has dual effects on NF-kappaB regulation. Inhibition of NF-kappaB transcriptional activity and promotion of IkappaBalpha degradation. J Biol Chem. 1999;274:18981–18988. doi: 10.1074/jbc.274.27.18981. [DOI] [PubMed] [Google Scholar]
- Koop DR, Klopfenstein B, Iimuro Y, Thurman RG. Gadolinium chloride blocks alcohol-dependent liver toxicity in rats treated chronically with intragastric alcohol despite the induction of CYP2E1. Molecular pharmacology. 1997;51:944–950. doi: 10.1124/mol.51.6.944. [DOI] [PubMed] [Google Scholar]
- Krikos A, Laherty CD, Dixit VM. Transcriptional activation of the tumor necrosis factor alpha-inducible zinc finger protein, A20, is mediated by kappa B elements. J Biol Chem. 1992;267:17971–17976. [PubMed] [Google Scholar]
- Lee EG, Boone DL, Chai S, Libby SL, Chien M, Lodolce JP, Ma A. Failure to regulate TNF-induced NF-kappaB and cell death responses in A20-deficient mice. Science. 2000;289:2350–2354. doi: 10.1126/science.289.5488.2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieber CS, Casini A, DeCarli LM, Kim CI, Lowe N, Sasaki R, Leo MA. S-adenosyl-L-methionine attenuates alcohol-induced liver injury in the baboon. Hepatology. 1990;11:165–172. doi: 10.1002/hep.1840110203. [DOI] [PubMed] [Google Scholar]
- Mari M, Colell A, Morales A, Caballero F, Moles A, Fernandez A, Terrones O, Basanez G, Antonsson B, Garcia-Ruiz C, Fernandez-Checa JC. Mechanism of mitochondrial glutathione-dependent hepatocellular susceptibility to TNF despite NF-kappaB activation. Gastroenterology. 2008;134:1507–1520. doi: 10.1053/j.gastro.2008.01.073. [DOI] [PubMed] [Google Scholar]
- Mato JM, Martinez-Chantar ML, Lu SC. Methionine metabolism and liver disease. Annual review of nutrition. 2008;28:273–293. doi: 10.1146/annurev.nutr.28.061807.155438. [DOI] [PubMed] [Google Scholar]
- McClain CJ, Hill DB, Song Z, Chawla R, Watson WH, Chen T, Barve S. S-Adenosylmethionine, cytokines, and alcoholic liver disease. Alcohol. 2002;27:185–192. doi: 10.1016/s0741-8329(02)00224-0. [DOI] [PubMed] [Google Scholar]
- Mull L, Ebbs ML, Bender J. A histone methylation-dependent DNA methylation pathway is uniquely impaired by deficiency in Arabidopsis S-adenosylhomocysteine hydrolase. Genetics. 2006;174:1161–1171. doi: 10.1534/genetics.106.063974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagaki M, Moriwaki H. Implication of cytokines: Roles of tumor necrosis factor-alpha in liver injury. Hepatol Res. 2008;38(Suppl 1):S19–S28. doi: 10.1111/j.1872-034X.2008.00422.x. [DOI] [PubMed] [Google Scholar]
- Neumann M, Naumann M. Beyond IkappaBs: alternative regulation of NF-kappaB activity. The FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 2007;21:2642–2654. doi: 10.1096/fj.06-7615rev. [DOI] [PubMed] [Google Scholar]
- Opipari AW, Jr, Hu HM, Yabkowitz R, Dixit VM. The A20 zinc finger protein protects cells from tumor necrosis factor cytotoxicity. J Biol Chem. 1992;267:12424–12427. [PubMed] [Google Scholar]
- Osawa Y, Banno Y, Nagaki M, Brenner DA, Naiki T, Nozawa Y, Nakashima S, Moriwaki H. TNF-alpha-induced sphingosine 1-phosphate inhibits apoptosis through a phosphatidylinositol 3-kinase/Akt pathway in human hepatocytes. J Immunol. 2001;167:173–180. doi: 10.4049/jimmunol.167.1.173. [DOI] [PubMed] [Google Scholar]
- Ratter F, Gassner C, Shatrov V, Lehmann V. Modulation of tumor necrosis factor-alpha-mediated cytotoxicity by changes of the cellular methylation state: mechanism and in vivo relevance. Int Immunol. 1999;11:519–527. doi: 10.1093/intimm/11.4.519. [DOI] [PubMed] [Google Scholar]
- Shembade N, Harhaj EW. Regulation of NF-kappaB signaling by the A20 deubiquitinase. Cellular & molecular immunology. 2012;9:123–130. doi: 10.1038/cmi.2011.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Z, Zhou Z, Song M, Uriarte S, Chen T, Deaciuc I, McClain CJ. Alcohol-induced S-adenosylhomocysteine accumulation in the liver sensitizes to TNF hepatotoxicity: possible involvement of mitochondrial S-adenosylmethionine transport. Biochem Pharmacol. 2007;74:521–531. doi: 10.1016/j.bcp.2007.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Z, Zhou Z, Uriarte S, Wang L, Kang YJ, Chen T, Barve S, McClain CJ. S-adenosylhomocysteine sensitizes to TNF-alpha hepatotoxicity in mice and liver cells: a possible etiological factor in alcoholic liver disease. Hepatology. 2004;40:989–997. doi: 10.1002/hep.20412. [DOI] [PubMed] [Google Scholar]
- Sun SC, Ganchi PA, Ballard DW, Greene WC. NF-kappa B controls expression of inhibitor I kappa B alpha: evidence for an inducible autoregulatory pathway. Science. 1993;259:1912–1915. doi: 10.1126/science.8096091. [DOI] [PubMed] [Google Scholar]
- Van Antwerp DJ, Martin SJ, Kafri T, Green DR, Verma IM. Suppression of TNF-alpha-induced apoptosis by NF-kappaB. Science. 1996;274:787–789. doi: 10.1126/science.274.5288.787. [DOI] [PubMed] [Google Scholar]
- Vandenabeele P, Galluzzi L, Vanden Berghe T, Kroemer G. Molecular mechanisms of necroptosis: an ordered cellular explosion. Nature reviews. Molecular cell biology. 2010;11:700–714. doi: 10.1038/nrm2970. [DOI] [PubMed] [Google Scholar]
- Wang CY, Mayo MW, Baldwin AS., Jr TNF- and cancer therapy-induced apoptosis: potentiation by inhibition of NF-kappaB. Science. 1996;274:784–787. doi: 10.1126/science.274.5288.784. [DOI] [PubMed] [Google Scholar]
- Watson WH, Song Z, Kirpich IA, Deaciuc IV, Chen T, McClain CJ. Ethanol exposure modulates hepatic S-adenosylmethionine and S-adenosylhomocysteine levels in the isolated perfused rat liver through changes in the redox state of the NADH/NAD(+) system. Biochim Biophys Acta. 2011;1812:613–618. doi: 10.1016/j.bbadis.2011.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Bialik S, Jones BE, Iimuro Y, Kitsis RN, Srinivasan A, Brenner DA, Czaja MJ. NF-kappaB inactivation converts a hepatocyte cell line TNF-alpha response from proliferation to apoptosis. Am J Physiol. 1998;275:C1058–C1066. doi: 10.1152/ajpcell.1998.275.4.C1058. [DOI] [PubMed] [Google Scholar]