Abstract
Objective
Elevated levels of plasma trimethyl amine N-oxide (TMAO), the product of gut microbiome and hepatic-mediated metabolism of dietary choline and L-carnitine, have recently been identified as a novel risk factor for the development of atherosclerosis in mice and humans. The goals of this study were to identify the genetic factors associated with plasma TMAO levels.
Approach and Results
We used comparative genome-wide association study (GWAS) approaches to discover loci for plasma TMAO levels in mice and humans. A GWAS in the Hybrid Mouse Diversity Panel identified a locus for TMAO levels on chromosome 3 (p=2.37×10−6) that co-localized with a highly significant (p=1.07×10−20) cis expression quantitative trait locus for solute carrier family 30 member 7 (Slc30a7). This zinc transporter could thus represent at least one positional candidate gene responsible for the association signal at this locus in mice. A GWAS for plasma TMAO levels in 1973 humans identified two loci with suggestive evidence of association (p~3.0×10−7) on chromosomes 1q23.3 and 2p12. However, genotyping of the lead variants at these loci in 1892 additional subjects failed to replicate their association with plasma TMAO levels.
Conclusions
The results of these limited observational studies indicate that, at least in humans, genes play a marginal role in determining TMAO levels and that any genetics effects are relatively weak and/or complex. Variation in diet or the repertoire of gut microbiota may be more important determinants of plasma TMAO levels in mice and humans, which should be investigated in future studies.
Keywords: trimethyl amine N-oxide, genetics, atherosclerosis, mouse, human
Introduction
Choline is a key nutrient with various metabolic roles in lipid metabolism and cell membrane structure, and it serves as a precursor for synthesis of the neurotransmitter acetylcholine 1–3. Dietary choline is also an important source of methyl groups that are required for proper metabolism of certain amino acids, such as homocysteine and methionine 3. A variety of animal studies have shown that choline deficiency adversely affects brain and cognitive development during fetal and neonatal life 1,4–6, which has led to specific nutritional guidelines recommending adequate intake of choline for infants and pregnant or lactating women 7,8.
One route for the initial catabolism of dietary choline (in the form of phosphotidylcholine) is mediated by intestinal microbes and leads to the formation of trimethyl amine (TMA). TMA is efficiently absorbed from the gastrointestinal tract and subsequently oxidized by the liver to form trimethyl amine N-oxide (TMAO). This latter reaction is catalyzed by one or more of the flavin monooxygenase (FMO) enzymes, of which there are at least six gene family members in higher mammals 9. Interestingly, mutations of FMO3 that result in deficiency of this enzyme are the cause of trimethylaminuria, otherwise known as fish malodor syndrome 10. This relatively rare recessive disorder is characterized by the near absence of plasma TMAO levels and highly elevated TMA levels, depending on the functional severity of the mutation in FMO3. The pungent odor of rotting fish that characterizes trimethylaminuria is due to the release of the volatile gas TMA through the breath, skin, and urine.
Recently, we uncovered a novel mechanism through which gut microbiota and hepatic-mediated metabolism of dietary choline promotes atherosclerosis and increases risk of coronary artery disease (CAD) 11,12. These studies demonstrated that plasma TMAO levels in humans were positively associated with the presence of multiple CAD phenotypes, including atherosclerotic plaque burden and future risk of myocardial infarction (MI), stroke, or death in a dose-dependent fashion. A similar relationship was observed between plasma TMAO levels and aortic lesion development among various inbred mouse strains 13. More recently, we also demonstrated that L-carnitine, a trimethylamine abundant in red meat, is also metabolized by intestinal microbiota to produce TMAO in mice and humans and that L-carnitine supplementation accelerated atherosclerosis in mice 14. Notably, short-term administration of broad spectrum antibiotics eliminated the production of TMAO in both mice and humans and decreased atherosclerosis in mice. Moreover, TMAO supplementation in mice, or dietary supplementation of either choline or L-carnitine, in the presence of intact gut microbiota led to alterations in cholesterol and sterol metabolism in multiple distinct compartments, including reduction in reverse cholesterol transport, providing a mechanistic rational for the association between TMAO levels and atherosclerotic cardiovascular phenotypes 14. Taken together, these studies provide evidence consistent with the pro-atherogenic role of TMAO in mammals and support the notion that gut microbiota play an obligatory role in the formation TMAO from dietary choline and L-carnitine.
It is reasonable to assume that variation in plasma TMAO levels could also be affected by intrinsic genetic factors of the host. However, with the exception of FMO3, the genes that control plasma TMAO levels are not known. Therefore, the aim of the present study was to use comparative genome-wide association study (GWAS) approaches in mice and humans to identify novel genetic determinants associated with plasma TMAO levels.
Materials and Methods
Materials and Methods are available in the online-only Data Supplement
Results
Association of the FMO Cluster with FMO3 Gene Expression, plasma TMAO levels, and CAD in Humans
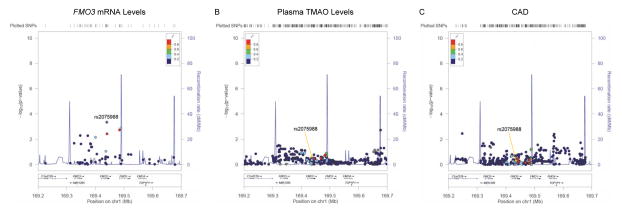
In previous studies, we reported that Fmo3 expression varied significantly among inbred strains from the Hybrid Mouse Diversity Panel (HMPD) and that a major locus regulating its expression mapped directly over Fmo3, suggesting cis-acting regulation in mice 13. Furthermore, Fmo3 expression was positively correlated with both plasma TMAO levels and atherosclerosis in mice. Based on these observations, we first used a targeted approach to evaluate whether genetic associations could specifically be observed with the human FMO locus on chromosome 1q24.3. To evaluate association of the FMO cluster with hepatic FMO3 mRNA levels, we used a previously published liver gene expression dataset 15. These analyses were carried out in a subset of 151 Caucasian subjects for whom complete gene expression and genotype data were publicly available. Fifty seven single nucleotide polymorphisms (SNPs) were available for analysis in a specified ~451kb region containing FMO3, FMO6P, FMO2, FMO1, and FMO4, including 200kb of flanking sequence (100kb from each end). As shown in Figure 1A, one SNP (rs2075988) yielded age- and sex-adjusted association with FMO3 mRNA levels (p=4.5×10−4) that remained significant after correction for multiple testing (0.05/57; Bonferroni-corrected p=8.8×10−4). Cis eQTL were not observed for any other members of the FMO gene family at this locus (data not shown).
Figure 1. Association of the FMO locus with FMO3 mRNA levels, plasma TMAO levels, and risk of CAD in humans.
Using a publicly available eQTL liver dataset, 57 SNPs were tested for association with hepatic FMO3 mRNA levels, one of which (rs2075988) yielded a significant p-value (4.5×10−4) after Bonferroni correction for multiple testing (A). In the GeneBank cohort, none of the 471 SNPs tested in the FMO locus yielded significant association with plasma TMAO levels (B). Evaluation of the FMO locus with risk of CAD using 388 SNPs available from the results of the CARDIoGRAM consortium did not reveal any significant associations (C). The same genomic interval spanning ~451kb across the FMO cluster on chromosome 1q24.3 is shown for all three plots and the variant most strongly associated with FMO3 mRNA levels is given as the reference SNP (rs2075988).
We next determined whether variation at the FMO cluster influenced plasma TMAO levels using the GWAS results from the GeneBank sdtudy, a cohort of patients undergoing elective cardiac evaluation at the Cleveland Clinic. Table 1 describes the clinical characteristics of the 3865 individuals used in the present study. As expected for a patient population undergoing coronary angiography as part of their clinical evaluation, the majority of these subjects were male, had prevalent CAD, and were taking lipid-lowering medication (Table 1). In this analysis, 471 SNPs were available but none were significantly associated with plasma TMAO levels (Figure 1B). Lastly, we evaluated whether the FMO locus was associated with risk of CAD in the CARDIoGRAM consortium, which represents a meta-analysis of GWAS data from a discovery set of ~22,000 CAD cases and ~65,000 controls 16. In CARDIoGRAM, 388 SNPs were available for analyses, of which 21 yielded p-values < 0.05 for association with CAD (Figure 1C). However, none of these associations were significant at the Bonferroni-corrected significance threshold (p=1.3×10−4; 0.05/388). Furthermore, the SNP that exhibited the strongest association with FMO3 mRNA levels (rs2075988) did not demonstrate evidence for association with either plasma TMAO levels or risk of CAD (Figure 1).
Table 1.
Clinical Characteristics of the Study Population.
| Trait | N = 3865 |
|---|---|
| Age (years) | 64 ± 11 |
| Male/Female | 6372/2789 |
| Number with CAD at baseline (%) | 6776 (76) |
| CAD Severity | |
| 0 vessels (%) | 2766 (30) |
| 1 or 2 vessels (%) | 3392 (37) |
| ≥ 3 vessels (%) | 3003 (33) |
| Number of MACE (%) | 1285 (14) |
| BMI (kg/m2) | 29.6 ± 6.2 |
| Total Cholesterol (mg/dl) | 170 ± 41.1 |
| HDL Cholesterol (mg/dl) | 40.0 ± 13.5 |
| LDL Cholesterol (mg/dl) | 99.0 ± 33.5 |
| Triglycerides (mg/dl) | 151.5 ± 110.1 |
| TMAO (μM) | 6.2 ± 13.0 |
| Taking lipid lowering medication (%) | 5751 (63) |
Data is shown as mean ± SD or numbers of individuals (%).
GWAS for Plasma TMAO Levels in Mice
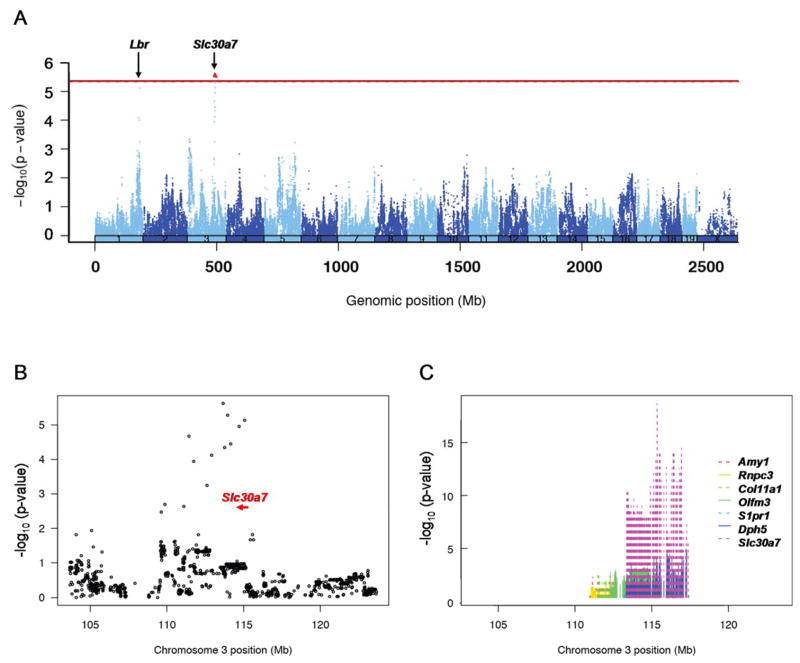
To identify novel genetic factors associated with plasma TMAO levels in mammals, we next used the HMDP to carry out an unbiased genome-wide association study (GWAS) in mice. This newly developed genetics platform consists of ~100 classic inbred and recombinant inbred mouse strains that are maximally informative for association analysis and have been used to carry out GWAS for other quantitative traits relevant to human diseases, including atherosclerosis, metabolites, and hepatic mRNA levels 17–20. For the present study, we carried out a GWAS for plasma TMAO levels in male mice on a chow diet and identified one locus on mouse chromosome 3 between 110–115Mb that exceeded the genome-wide significance threshold for association in the HMDP (p= 2.37×10−6; Figure 2A and B). The 10Mb region centered around the lead SNP on chromosome 3 contains several genes and exhibited a highly significant cis eQTL (p=1.07×10−20) for the gene encoding solute carrier family 30 member 7 (Slc30a7; Figure 2C). The co-localization of QTLs for plasma TMAO and Slc30a7 mRNA levels suggest that this zinc transporter could represent at least one positional candidate gene responsible for the association signal at this locus. Suggestive evidence for association of plasma TMAO levels (p=7.62×10−6) was also observed with a region on mouse chromosome 1 at 184Mb (Figure 2A), although this locus did not achieve genome-wide significance. The lead SNP on chromosome 1 maps to within 40kb of the Lamin beta receptor (Lbr) gene but ~20Mb distal from the Fmo gene cluster (162–163Mb).
Figure 2. Manhattan plot for GWAS of plasma TMAO levels in mice.
A GWAS for plasma TMAO levels in the HMDP identifies a significant locus over the Slc30a7 gene (red dot) at 110–115Mb on chromosome 3 and a suggestive locus on chromosome 1 ~40kb away from the Lbr gene (A). A regional plot for chromosome 3 shows the location and transcriptional orientation of Slc30a7 (indicated by red arrow) in relation to the peak SNPs in this region (B). Of the genes in this locus, a highly significant (p=1.07×10−20) cis-acting eQTL is observed for Slc30a7 (C). The red line indicates the genome-wide threshold for significance in the HMDP (p=4.1×10−6). Plasma TMAO and hepatic mRNA levels were quantitated in male mice from ~100 HMDP strains (n=3–8 mice per strain) and analyzed for association with ~107,000 SNPs, after correcting for population structure using the EMMA algorithm.
GWAS for Plasma TMAO Levels in Humans
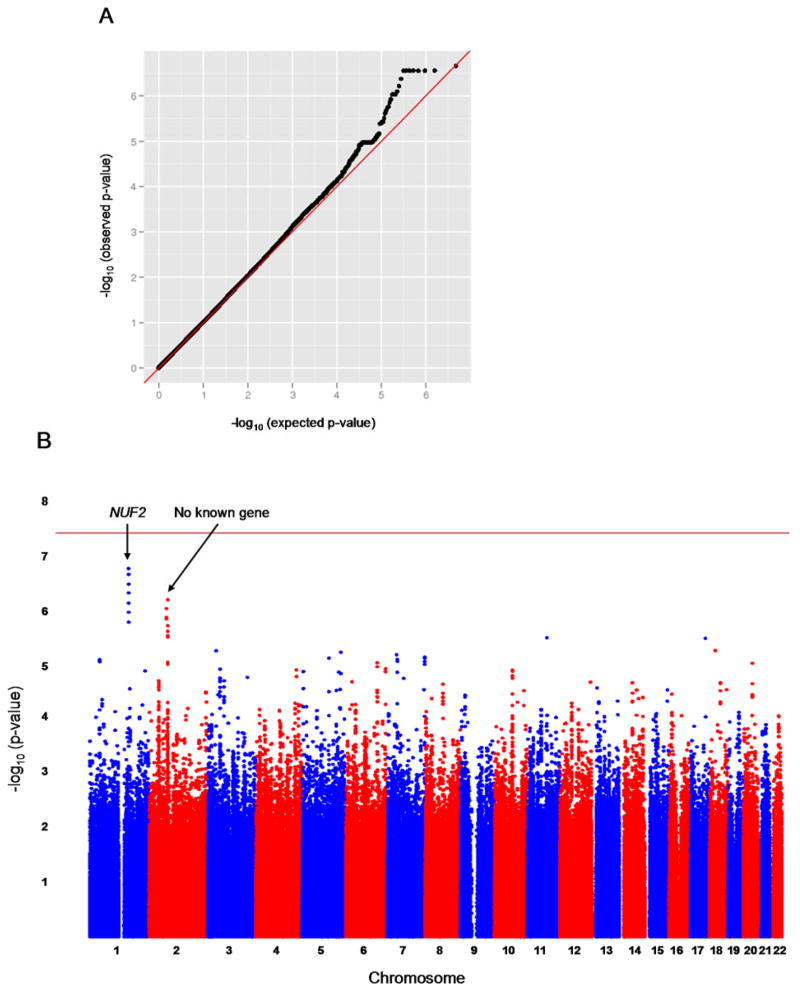
To complement the mouse studies, we carried out a two-stage GWAS in GeneBank. In the first stage, ~2.4 million genotyped and imputed autosomal SNPs were evaluated for association with plasma TMAO levels in 1973 subjects with adjustment for age and sex. The quantile-quantile (Q-Q) plot for these analyses is shown in Figure 3A and the observed genomic inflation factor (λ) was 1.007, indicating that the GWAS results are not confounded by underlying population stratification. As shown by the Manhattan plot in Figure 3B, two loci with suggestive evidence of association were identified on chromosomes 1q23.3 and 2p12. The lead SNP at the chromosome 1 locus (rs17359359; p=2.8 × 10−7) is located ~47kb telomeric of NUF2, which is a component of the kinetochore complex that is required for chromosome segregation, but, to our knowledge, has no known relationship to TMAO metabolism. This locus is also located ~8Mb telomeric to the FMO gene cluster and is clearly distinct since there is no apparent long-range linkage disequilibrium (LD) between these two loci. By comparison, the lead SNP at the chromosome 2p12 locus (rs885187; p=2.8 × 10−7) does not map near any known gene. Based on previously observed sex differences in plasma TMAO levels, we also carried out a GWAS in males and females separately. However, these analyses did not reveal sex-specific effects on chromosomes 1q23.3 and 2p12 or identify other loci (Supplemental Figure I).
Figure 3. Results of a GWAS for plasma TMAO levels in humans.
The Q-Q plot of the GWAS results for plasma TMAO levels in humans (n=1973) shows slight deviation of the observed p-values from the expected distribution under the null hypothesis of no association (A). The observed genomic control factor in these analyses was 1.007, indicating that the results are not confounded by underlying population stratification. A GWAS analysis in humans identifies two loci on chromosomes 1 and 2 exhibiting suggestive evidence of association with plasma TMAO levels but no locus that exceeds the genome-wide threshold for significance (indicated by the horizontal red line) (B).
In stage 2, we evaluated the chromosome 1 locus further by genotyping rs17359359 in 1892 additional GeneBank subjects for whom plasma TMAO levels were available. These analyses failed to replicate the association of rs17359359 with plasma TMAO levels in stage 2 (p=0.85), and a combined analysis with all subjects attenuated the overall association (p=1.8 × 10−4; Table 2). Based on the chromosome 3 locus identified in the HMDP (Figure 2A and B), we used synteny mapping and evaluated the association of plasma TMAO levels with SNPs in the 1Mb genomic region centered on the human SLC30A7 ortholog located on chromosome 1p21.2. In the GWAS dataset (n=1973), one SNP located ~225kb telomeric to SLC30A7 (rs12402441) demonstrated nominal association (p=0.006) with plasma TMAO levels (Table 2). However, the association of rs12402441 with plasma TMAO levels did not replicate in the stage 2 samples, and a combined analysis with all subjects was not significant (Table 2). In the combined dataset, there was also no evidence for an interaction between sex and either SNP on plasma TMAO levels (rs17359359, pint=0.33; rs12402441, pint=0.11). The sex-specific effects of rs17359359 and rs12402441 when males and females were analyzed separately are shown in Supplemental Table I.
Table 2.
Effect of SNPs Identified Through GWAS in Humans and Mice on Plasma TMAO Levels in the GeneBank Cohort.
| rs17359359 | rs12402441 | |||||||
|---|---|---|---|---|---|---|---|---|
| Stage | GG | AG | AA | a p-value | AA | AG | GG | a p-value |
| GWAS | 5.3 ± 8.0 (n=1727) | 8.2 ± 20.3 (n=238) | 9.9 ± 8.1 (n=8) | 2.8 × 10−7 | 5.8 ± 10.8 (n=1773) | 4.5 ± 4.7 (n=186) | 4.5 ± 3.7 (n=14) | 0.017 |
| Replication | 6.6 ± 14.5 (n=1495) | 8.3 ± 23.2 (n=186) | 4.0 ± 2.9 (n=9) | 0.71 | 6.6 ± 13.3 (n=1598) | 8.8 ± 31.2 (n=158) | 4.5 ± 1.5 (n=10) | 0.68 |
| Combined | 5.9 ± 11.5 (n=3222) | 8.3 ± 21.6 (n=424) | 6.8 ± 6.5 (n=17) | 1.1 × 10−4 | 6.2 ± 12.1 (n=3371) | 6.5 ± 21.5 (n=344) | 4.5 ± 2.9 (n=24) | 0.14 |
Mean (± SD) plasma TMAO levels (μM) are shown as a function of genotype.
p-values obtained using linear regression with natural log transformed values after adjustment for age and sex.
Discussion
Using a combined mouse-human GWAS approach, we sought to identify the genetic determinants of plasma TMAO levels in mammals. Several factors served as the motivation for these studies, including recent studies demonstrating that TMAO can be generated from gut microbiota-mediated metabolism of either dietary choline or L-carnitine and that elevated plasma levels are strongly pro-atherogenic in both mice and humans 11,12,14. Subsequent reports further showed that plasma TMAO levels in mice are regulated by both sex hormones, which could account in part for the observed dimorphism between male and female mice, and increased Fmo3 gene expression via bile acid-mediated activation of the farnesoid X receptor (FXR) 13. Of note, in humans, no differences in plasma TMAO levels were observed between males and females 12. The collective results of these comprehensive, albeit limited, observational studies indicate that genes play a marginal role in determining TMAO levels and that any genetics effects are either complex or relatively weak. This is particularly true in humans and raises the possibility that variation in dietary composition or the repertoire of gut microbiota may be more important determinants of plasma TMAO levels.
Using the HMDP, we identified one locus on chromosome 3 containing Slc30a7 that was associated with plasma TMAO levels in male mice at the genome-wide significance threshold. This locus also exhibited evidence for cis gene regulation of Slc30a7, which is a subfamily member of the cation diffusion facilitator family of transporters and has essential functions in dietary zinc absorption 21. Although a biological mechanism for how Slc30a7 would regulate plasma TMAO levels is not directly evident, it has been reported that the zinc finger protein, YY1, regulates the expression both rabbit and human FMO1 22. Interestingly, the activity of certain bacterial monoooxygenases have also been shown to use zinc as a co-factor 23. However, more detailed functional studies will be required to determine whether Slc30a7 could impact TMAO levels by influencing zinc-mediated activity of one or more of the FMOs in mice. We also note that while this locus on mouse chromosome 3 also yielded a highly significant cis eQTL for Slc30a7, we cannot exclude the possibility that another gene in this interval harboring functional coding variation is the causal genetic factor for plasma TMAO levels. Since our GWAS with the HMDP was only with male mice, it is also possible that inclusion of females would provide additional support for association of the Slc30a7 locus as well as identify other genomic regions controlling plasma TMAO levels that are potentially specific to females. For example, we previously reported that plasma TMAO levels are several-fold higher in female mice compared to males, a portion of which is attributable to differences in sex hormones 13.
As a comparative analysis to our studies with the HMDP, we also carried out a GWAS for plasma TMAO levels in the GeneBank cohort. This analysis identified two suggestive loci in the discovery phase but our attempt to replicate the NUF2 locus on chromosome 1 was unsuccessful. Based on the Slc30a7 locus identified in the mouse GWAS, we also evaluated the syntenic region on human chromosome 1p21.2 for association with plasma TMAO levels. While one SNP in this region yielded nominal association with plasma TMAO levels in humans, this signal also did not replicate in the stage 2 samples. Given the high concordance rate (>98.8%) for genotypes of the same DNA samples used in stages 1 and 2, we do not believe technical variability to have been a factor for the lack of replication in stage 2 and conclude that these loci likely represent false positive signals. However, despite the lack of genetic variation around the human SLC30A7 ortholog being associated with plasma TMAO levels, it is still possible that this transporter still plays a biological role in regulating TMAO levels in both species. Furthermore, we did not obtain any evidence for sex-specific effects at these loci or identify any others when the GWAS was carried out in males and females separately. Taken together, these results suggest that variation in plasma TMAO levels in humans may be due to weak genetic effects and that larger sample sizes will be required to identify the underlying regulatory factors.
To date, FMO3 is the only genetic factor known to affect plasma TMAO levels in humans. FMO3 is comprised of 10 exons spanning 26.9 kb on chromosome 1q24.3 and encodes a 532-residue enzyme. At the amino acid level, FMO3 shares ≥79% homology with the mouse Fmo3 protein and other members of the human FMO family. Interestingly, we previously demonstrated that FMO1, FMO2, and FMO3 were able to generate TMAO from TMA, but that FMO3 was by far the most active family member 13. Since the Mendelian disease trimethylaminuria is caused by rare mutations that lead to FMO3 deficiency, we leveraged our own data in GeneBank and those from public sources to evaluate whether common variants at the FMO locus were associated with FMO3 gene expression, plasma TMAO levels, and risk of CAD. However, these analyses in humans did not reveal any strong associations with SNPs surrounding FMO3. It is possible that the imputed genotypes from the GWAS data we used did not provide sufficient coverage of the variation around FMO3 (or the entire FMO locus). Based on data for subjects of European ancestry from the 1000 Genomes Project, 59 tagging SNPs with minor allele frequencies ≥ 1% would cover FMO3 at an r2 ≥ 0.8. However, only 15 tagging SNPs across FMO3 were present in our analyses of plasma TMAO levels in GeneBank. In addition, rare variants in FMO3 that could influence gene expression, TMAO production, and risk of CAD would also not necessarily be represented by our imputed GWAS data. By comparison, our previous studies in mice revealed a relatively strong cis eQTL for Fmo3 expression over the Fmo locus. However, the present analyses for plasma TMAO levels in the HMDP did not yield association with the Fmo locus at the genome-wide level (data not shown). These observations suggest that the relationship between FMO3 gene expression and plasma TMAO levels in both mice and humans is complex and that other regulatory mechanisms, including post-transcriptional and/or post-translational modifications, may exist.
The discordance between rare mutations in FMO3 that dramatically reduce plasma TMAO levels and the lack of common genetic determinants associated with this metabolite implies that variation in TMAO levels in humans and mice may be influenced by other factors, such as gut microbial and/or dietary composition. For example, we previously defined the relative abundances of bacteria at each taxonomic level in relation to the production of TMAO through pyrosequencing of 16S rRNA genes in both mice and humans. One notable difference in these analyses was the source of gut bacteria since the contents of the caecum were used for mice whereas stool samples were used for the human analyses. This may explain, at least in part, why a direct comparison of bacterial taxa associated with plasma TMAO concentrations did not identify any genre common to both species. This observation is consistent with prior reports indicating that microbes identified from the distal gut of the mouse do not necessarily represent those typically detected in humans 24,25. Thus, while sharing many taxa, the microbial composition observed in mice is architecturally and globally different than in humans. In spite of these differences, we were still able to demonstrate associations between dietary patterns (e.g. vegan/vegetarian vs. omnivore or normal chow vs. choline/carnitine supplemented) and both plasma TMAO levels and proportions of specific taxa of fecal microbes in humans and cecal microbes in mice 14. These observations suggest that high dietary intake of L-carnitine or choline would lead to increased plasma TMAO levels, particularly if specific bacterial taxa that metabolize these nutrients to TMA are present in the gut. It is possible that the effects of host genetic factors would also manifest under such dietary conditions. However, compared to mice housed under standardized environmental conditions, the diet in free ranging humans is far more heterogeneous, which would add further complexity and diversity to any potential interactions with the gut microbiome.
Despite our comprehensive efforts to identify loci associated with plasma TMAO levels, we also note several potential limitations of our study. First, we used GWAS approaches in mice and humans that mostly tests association with common genetic variation, which would not necessarily detect the effects of rare variants on plasma TMAO levels. Second, our human GWAS was carried out in subjects of European ancestry and it is possible that genetic variants that are either specific to or present at higher frequency in other ethnicities could influence TMAO levels. Third, while including ~100 inbred strains, it is still possible that the HMDP does provide sufficient genetic variation to capture all of the effects on plasma TMAO levels in mice, compared to the substantially greater genetic diversity present in outbred human populations. Additionally, the pathway(s) leading to variation in TMAO levels in mice and humans may not be entirely similar. Lastly, as discussed above, variability in dietary composition, particularly in humans, and the gut microbiome clearly factor into plasma TMAO levels, and are thus likely to be strong confounding variables that our study did not take into consideration.
In conclusion, our results indicate that Slc30a7 may represent a novel gene for TMAO levels in mice but that the contribution of genetic factors in humans is more complex. These observations suggest that the inter-relationships between dietary choline and L-carnitine levels with the composition of gut microbes are perhaps more likely determinants of variation in plasma TMAO levels. Exploring such interactions as part of future studies may help to identify the intrinsic genetic factors that influence plasma TMAO levels and their influence on the development of atherosclerosis.
Supplementary Material
Significance.
Elevated plasma levels of trimethyl amine N-oxide (TMAO), a metabolite generated from dietary choline and carnitine by intestinal bacteria, has recently been identified as a novel risk factor for coronary artery disease. Notably, elimination of bacteria in the gut through administration of antibiotics reduced TMAO levels in mice and humans and decreased atherosclerosis in mice. However, the genes that control plasma TMAO levels are not well defined. The present study utilizes a comparative genome-wide association study approach in mice and humans to identify the genetic determinants of plasma TMAO levels. In mice, genetic variants near Slc30a7 were significantly associated with plasma TMAO levels whereas no locus was identified in humans. Our findings suggest that, at least in humans, plasma TMAO levels are under complex genetic regulation, that the effects of any underlying genes are relatively weak, and/or that variation in gut bacteria may be more important in determining TMAO levels.
Acknowledgments
Sources of Funding
This study was supported in part by NIH grants K99HL102223, P01HL30568, P01HL28481, R01HL103866, P20HL113452, R01ES021801, a pilot project award the Southern California Clinical and Translational Science Institute (SC CTSI) through NIH grant UL1TR000130, and American Heart Association Scientist Development Grant 12SDG12050473. GeneBank was supported in part by NIH grants P01HL098055, P01HL076491, and R01HL103931. RR has received research funding from Canadian Institutes of Health Research #MOT82810 and Canada Foundation for Innovation # 11966. SLH is also partially supported by a gift from the Leonard Krieger Fund. Mass spectrometry instrumentation used was housed within the Cleveland Clinic Mass Spectrometry Facility with partial support through a Center of Innovation by AB SCIEX.
Abbreviations
- eQTL
cis expression quantitative trait loci
- CAD
coronary artery disease
- CARDIoGRAM
Coronary Artery Disease Genome-wide Replication And Meta-Analysis
- FMO
flavin monooxygenase
- GWAS
genome-wide association study
- HMDP
Hybrid Mouse Diversity Panel
- MACE
major adverse cardiac events
- MI
myocardial infarction
- Q-Q
quantile-quantile
- SNPs
single nucleotide polymorphisms
- Slc30a7
solute carrier family 30 member 7
- TMA
trimethyl amine
- TMAO
trimethyl amine N-oxide
Footnotes
Disclosures
Dr. Hazen (SLH) is named as co-inventor on pending and issued patents held by the Cleveland Clinic relating to cardiovascular diagnostics. Dr. Hazen reports he has been paid as a consultant or speaker for the following companies: Cleveland Heart Lab, Inc., Esperion, Liposciences Inc., Merck & Co., Inc., and Pfizer Inc. Dr. Hazen reports he has received research funds from Abbott, Cleveland Heart Lab, Esperion and Liposciences, Inc. Dr. Hazen has the right to receive royalty payments for inventions or discoveries related to cardiovascular diagnostics from Abbott Laboratories, Cleveland Heart Lab, Inc., Frantz Biomarkers, LLC, and Siemens.
References
- 1.Zeisel SH, Blusztajn JK. Choline and human nutrition. Annu Rev Nutr. 1994;14:269–96. doi: 10.1146/annurev.nu.14.070194.001413. [DOI] [PubMed] [Google Scholar]
- 2.Buchman AL, Ament ME, Sohel M, Dubin M, Jenden DJ, Roch M, Pownall H, Farley W, Awal M, Ahn C. Choline deficiency causes reversible hepatic abnormalities in patients receiving parenteral nutrition: proof of a human choline requirement: a placebo-controlled trial. J Parenter Enteral Nutr. 2001;25:260–8. doi: 10.1177/0148607101025005260. [DOI] [PubMed] [Google Scholar]
- 3.Hollenbeck CB. The importance of being choline. J Am Diet Assoc. 2010;110:1162–5. doi: 10.1016/j.jada.2010.05.012. [DOI] [PubMed] [Google Scholar]
- 4.Meck WH, Williams CL. Metabolic imprinting of choline by its availability during gestation: implications for memory and attentional processing across the lifespan. Neurosci Biobehav Rev. 2003;27:385–99. doi: 10.1016/s0149-7634(03)00069-1. [DOI] [PubMed] [Google Scholar]
- 5.Cermak JM, Holler T, Jackson DA, Blusztajn JK. Prenatal availability of choline modifies development of the hippocampal cholinergic system. Faseb J. 1998;12:349–57. doi: 10.1096/fasebj.12.3.349. [DOI] [PubMed] [Google Scholar]
- 6.Craciunescu CN, Albright CD, Mar MH, Song J, Zeisel SH. Choline availability during embryonic development alters progenitor cell mitosis in developing mouse hippocampus. J Nutr. 2003;133:3614–8. doi: 10.1093/jn/133.11.3614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yates AA, Schlicker SA, Suitor CW. Dietary Reference Intakes: the new basis for recommendations for calcium and related nutrients, B vitamins, and choline. J Am Diet Assoc. 1998;98:699–706. doi: 10.1016/S0002-8223(98)00160-6. [DOI] [PubMed] [Google Scholar]
- 8.Zeisel SH. Choline: an essential nutrient for humans. Nutrition. 2000;16:669–71. doi: 10.1016/s0899-9007(00)00349-x. [DOI] [PubMed] [Google Scholar]
- 9.Krueger SK, Williams DE. Mammalian flavin-containing monooxygenases: structure/function, genetic polymorphisms and role in drug metabolism. Pharmacol Ther. 2005;106:357–87. doi: 10.1016/j.pharmthera.2005.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Phillips IR, Shephard EA. Flavin-containing monooxygenases: mutations, disease and drug response. Trends Pharmacol Sci. 2008;29:294–301. doi: 10.1016/j.tips.2008.03.004. [DOI] [PubMed] [Google Scholar]
- 11.Wang Z, Klipfell E, Bennett BJ, et al. Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature. 2011;472:57–63. doi: 10.1038/nature09922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tang WH, Wang Z, Levison BS, Koeth RA, Britt EB, Fu X, Wu Y, Hazen SL. Intestinal microbial metabolism of phosphatidylcholine and cardiovascular risk. N Engl J Med. 2013;368:1575–84. doi: 10.1056/NEJMoa1109400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bennett BJ, de Aguiar Vallim TQ, Wang Z, Shih DM, Meng Y, Gregory J, Allayee H, Lee R, Graham M, Crooke R, Edwards PA, Hazen SL, Lusis AJ. Trimethylamine-N-oxide, a metabolite associated with atherosclerosis, exhibits complex genetic and dietary regulation. Cell Metab. 2013;17:49–60. doi: 10.1016/j.cmet.2012.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Koeth RA, Wang Z, Levison BS, et al. Intestinal microbiota metabolism of L-carnitine, a nutrient in red meat, promotes atherosclerosis. Nat Med. 2013;19:576–85. doi: 10.1038/nm.3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schadt EE, Molony C, Chudin E, et al. Mapping the genetic architecture of gene expression in human liver. PLoS Biol. 2008;6:e107. doi: 10.1371/journal.pbio.0060107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schunkert H, Konig IR, Kathiresan S, et al. Large-scale association analysis identifies 13 new susceptibility loci for coronary artery disease. Nat Genet. 2011;43:333–8. doi: 10.1038/ng.784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bennett BJ, Farber CR, Orozco L, et al. A high-resolution association mapping panel for the dissection of complex traits in mice. Genome Res. 2010;20:281–90. doi: 10.1101/gr.099234.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Farber CR, Bennett BJ, Orozco L, et al. Mouse genome-wide association and systems genetics identify Asxl2 as a regulator of bone mineral density and osteoclastogenesis. PLoS Genet. 2011;7:e1002038. doi: 10.1371/journal.pgen.1002038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ghazalpour A, Bennett B, Petyuk VA, et al. Comparative analysis of proteome and transcriptome variation in mouse. PLoS Genet. 2011;7:e1001393. doi: 10.1371/journal.pgen.1001393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Park CC, Gale GD, de Jong S, Ghazalpour A, Bennett BJ, Farber CR, Langfelder P, Lin A, Khan AH, Eskin E, Horvath S, Lusis AJ, Ophoff RA, Smith DJ. Gene networks associated with conditional fear in mice identified using a systems genetics approach. BMC Syst Biol. 2011;5:43. doi: 10.1186/1752-0509-5-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang L, Yu YY, Kirschke CP, Gertz ER, Lloyd KK. Znt7 (Slc30a7)-deficient mice display reduced body zinc status and body fat accumulation. J Biol Chem. 2007;282:37053–63. doi: 10.1074/jbc.M706631200. [DOI] [PubMed] [Google Scholar]
- 22.Luo Z, Hines RN. Regulation of flavin-containing monooxygenase 1 expression by ying yang 1 and hepatic nuclear factors 1 and 4. Mol Pharmacol. 2001;60:1421–30. doi: 10.1124/mol.60.6.1421. [DOI] [PubMed] [Google Scholar]
- 23.Ensign SA, Allen JR. Aliphatic epoxide carboxylation. Annu Rev Biochem. 2003;72:55–76. doi: 10.1146/annurev.biochem.72.121801.161820. [DOI] [PubMed] [Google Scholar]
- 24.Ley RE, Backhed F, Turnbaugh P, Lozupone CA, Knight RD, Gordon JI. Obesity alters gut microbial ecology. Proc Natl Acad Sci U S A. 2005;102:11070–5. doi: 10.1073/pnas.0504978102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ley RE, Hamady M, Lozupone C, Turnbaugh PJ, Ramey RR, Bircher JS, Schlegel ML, Tucker TA, Schrenzel MD, Knight R, Gordon JI. Evolution of mammals and their gut microbes. Science. 2008;320:1647–51. doi: 10.1126/science.1155725. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.