1. Introduction
Proteomics holds great promise in personalized medicine for cancer in the post-genomic era. In the past decade, clinical proteomics has significantly evolved in terms of technology development, optimization, and standardization, as well as in advanced bioinformatics data integration and analysis. Great strides have been made for characterizing a large number of proteins qualitatively and quantitatively in a proteome, including the use of sample fractionation, protein microarrays, and mass spectrometry (MS). It is believed that differential proteomic analysis of high-quality clinical biospecimen (tissue and biofluids) can potentially reveal protein/peptide biomarkers responsible for cancer by means of their altered levels of expression and/or post-translational modifications (PTMs). Multiple Reaction Monitoring (MRM-MS), a multiplexed platform using stable isotope dilution mass spectrometry (SID-MS) with sensitivity and reproducibility approaching that of traditional enzyme-linked immunosorbent assays (ELISAs) commonly used in the clinical setting, has emerged as a potentially promising technique for next-generation high-throughput protein biomarker measurements for diagnostics and therapeutics.
2. The role of protein biomarkers in medicine
According to the Food and Drug Administration (FDA), a biomarker is a characteristic that is objectively measured and evaluated as an indicator of normal biologic or pathogenic processes or pharmacological responses to a therapeutic intervention. [1–2]. From a biochemical point of view, a biomarker is often a protein, or a panel of proteins and their PTMs, the presence or quantitative characteristics of which are measured commonly using methods based on antibodies. To have a protein classified as an ideal biomarker, several criteria have to be met in order to enable unbiased diagnosis, particularly in patients without specific symptoms. First, it has to be highly specific towards the given disease (i. e., low false positives) and highly sensitive (i. e., low false negatives). Second, the assay for the biomarker should be easily performed and standardized by trained healthcare professionals. Last, readability of the test results should be transparent and clear for clinicians. All of these factors will affect the biomarker’s performance in the clinical settings. Unfortunately, many of these requirements are not being fulfilled by most approved and currently-used biomarkers [3]. Theoretically, every disease may be uncovered and characterized by its unique diagnostic biomarkers, which can be viewed as a specific protein undergoing changes or a panel of up- and down-regulated proteins and/or proteins with altered PTMs due to disease [4–5]. Prognostic and predictive biomarkers are additional types of biomarkers. The distinction between prognostic and predictive biomarkers lies in the fact that a prognostic marker is a single molecular trait or signature of traits that separate different patient populations with respect to the risk of a disease in the absence of treatment (e. g., risk for cancer recurrence), whereas a predictive biomarker is the one that distinguishes patient populations with respect to the risk of a disease in response to a particular (targeted) treatment (e. g., Gefitinib for non-small cell lung cancer).
The protein biomarker panels for cancer diagnostics and therapeutics are currently of considerable interest in biomedicine [6–8]. The discovery of proteins and peptides “leaked” by tumors into clinically-accessible body fluids such as blood has led to the possibility of diagnosing cancer at an early stage, predicting cancer progression or monitoring response to therapy by testing for the presence of cancer-relevant biomarkers in these biofluids via non-invasive procedures. However, clinical proteomic studies without proper study design and implementation of robust analytical techniques would significantly hamper the efforts and success in discovering new biomarkers for diagnostic and therapeutic use. To fully realize the potential of biomarkers in medicine, a well-designed biomarker development pipeline needs to take into account all aspects involved in such a complex project, ranging from pre-analytical quality biospecimens and analytical platforms, to the proper design of clinical trials. Recently, issues related to biospecimen quality in clinical proteomics have been raised, including the lack of accessibility and type of high quality biological matrix under study, unbiased sample collection, and consensus quality control (QC) protocols for sample processing and storage. Blood as a source of cancer biomarkers, has advantages over other bodily fluids (e. g., nipple aspirate fluids) because: (1) it is easily accessible; (2) its collection is minimally invasive, low risk and economical; and (3) there are routine practices for processing crude blood to plasma in clinical laboratories. However, blood for biomarker discovery poses a great analytical challenge due to its having the widest dynamic range (approximately 1012) of cellular protein species in the body, requiring many additional proteomic approaches such as fractionation. Furthermore, analytical technologies and platforms currently being developed must demonstrate their robustness and reproducibility within and across laboratories in order to identify and verify “true” molecular signatures due to the disease rather than experimental fluctuations arising from instruments, operators and environmental factors. Lastly, clinical qualification of a biomarker or a panel of biomarkers as defined by the FDA and demonstrated in a clinical trial must satisfy all analytical requirements for its intended use [9].
3. The need to reconstruct the current biomarker development pipeline
During the last 10 to 15 years, proteomic technology development has developed to accommodate the growing demand for biomarker research using biospecimens collected from tissue, blood and other biofluids [10–17]. Innovative approaches including protein microarrays, aptamer arrays, bead-based flow cytometry and MS to identify and sequence proteins in a high-throughput and quantitative manner have furthered our understanding in molecular mechanisms involved in diseases. Furthermore, depletion of high-abundance proteins from plasma and/or multi-dimensional chromatographic fractionation coupled with MS has expanded the dynamic range of detection for low abundance proteins in serum and plasma, two major sources for biomarkers [12].
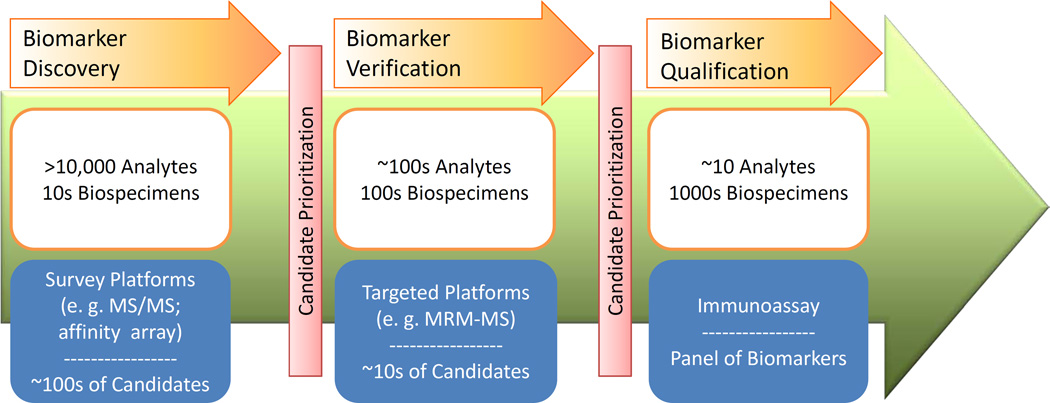
In the context of this review, biomarker qualification refers to the process and acceptance of a biomarker for marketing by the FDA [9], while analytical validation refers to the process of validating a device or platform for precise and accurate measurement of target analytes. The term “verification” (Figure 1), strongly advocated by The Office of Cancer Clinical Proteomics Research at the National Cancer Institute (NCI) for biomarker research, is defined as an intermediary step between discovery and qualification for a protein biomarker (see additional information in Section 4.2). Currently, we do not suffer from a lack of protein biomarker candidates. To date, there are over 1,000 cancer-relevant protein biomarker candidates described in the scientific literature [18]. However, the rate of introduction of new protein analytes approved by the FDA for marketing has remained relatively flat over the past 15 years, averaging 1.5 new biomarkers per year (median of 1 per year) [19]. This discrepancy points to a gap between protein biomarker discovery and qualification, which are commonly performed by MS and immunoassays, respectively, in the current biomarker development pipeline. Several factors at every stage of the pipeline contribute to this discrepancy: (1) pre-analytical (biospecimen collection, processing and storage) variability; (2) analytical variability or the lack of analytical validation of platforms; (3) lack of standardization of technologies and data analysis format for method and result comparison; (4) the lack of a statistically-powered number of patient samples to represent the entire patient population; and (5) the lack of knowledge in the proteomics community on the evaluation criteria required for these distinct processes in the biomarker development pipeline. Furthermore, the high cost and length of time associated with the development of an ELISA have prevented some or most of these published biomarker candidates from moving beyond the discovery phase. These factors have hampered meaningful interpretation of real biological differences associated with diseases [20–22]. Recognizing these principal obstacles in biomarker research, numerous efforts have been made by the proteomics community to alleviate the bottleneck in translational research and biomarker development.
Figure 1. The Envisioned NCI-CPTC Biomarker Development Pipeline from Biomarker Discovery to Biomarker Qualification.
4. Leading efforts to standardize proteomic technologies
4.1. The Association for Biomolecular Research Facilities (ABRF) and the Human Proteome Organization (HUPO)
ABRF and HUPO have taken initial steps to assess experimental variability in proteomic research and attempted to set standards for the community. In 2002, ABRF initiated Proteomics Research Group (PRG) studies (www.abrf.org/prg) on a yearly basis, where the PRG sends out samples processed at a centralized location to voluntary study participants and compiles data collected using a wide variety of techniques. The study topics ranged from the identification of components in a protein mixture, phosphorylation site mapping of a protein, de novo peptide sequencing to advanced quantitative proteomics and relative protein quantification in a clinical matrix. ABRF has published the results of several studies presenting the challenges that the proteomics community faces in terms of comparability of data collected using different methods/platforms and reproducibility of results [23–25]. Additionally, in 2009, HUPO (www.hupo.org) conducted an inter-laboratory study to assess the common problems encountered in proteomics research by distributing a test sample comprising of 20 recombinant human proteins at equivalent molarities to 27 study participants [26]. Centralized data analysis demonstrated that a major contributing factor to erroneous data reporting was attributable to databases and search engines.
4.2. NCI-Clinical Proteomic Technologies for Cancer (NCI-CPTC)
The NCI-CPTC initiative was created in 2006 (http://proteomics.cancer.gov) to systematically address irreproducibility issues due to the lack of standardization in common proteomics practices. During the past 4 years, it has made significant contributions essential for setting the standards for proteomic research. First, “verification” has been incorporated into the CPTC biomarker development pipeline (Figure 1). In this pipeline, biomarker discovery is the initial step for comprehensively analyzing the protein content (including splice variants, PTMs, single nucleotide polymorphisms [SNPs], etc.) in biospecimens, and to select and prioritize disease-related proteins for verification. Verification, the bridge between discovery and qualification, is the process of credentialing prioritized biomarker candidates using analytically robust, reproducible and quantitative assays on statistically-powered number of samples with clinical relevance. Credentialed proteins successfully passing this stage are considered verified biomarkers, which potentially are of high value for translating into clinical qualification studies. Although verification stage is not limited to a particular analytical platform, in the CPTC network, it currently centers on the application of multiplexed MRM-MS proteomic platform. Second, efforts to reduce pre-analytical and analytical variability on different instrument platforms within and across laboratories using metrics, Standard Operating Procedures (SOPs) and reference materials have produced promising results [11, 27]. Additionally, high quality reagents, including well-characterized antibodies, for the cancer community have been generated and are publicly accessible. Finally, collaborations with other organizations have resulted in successful partnerships. These include the interaction with the FDA to understand the regulatory science involved in diagnostic assay clearance and approval, with the National Institute on Standards and Technology on the development of Standard Reference Materials, and with the American Association of Clinical Chemistry for quality control and assay validation in clinical labs. The interaction with the FDA has already resulted in the publication of two mock 510(k) pre-submission documents based on multiplexed immunoaffinity MRM-MS assay (PepCa10) for the quantification of 10 peptides corresponding to 5 proteins and for multiplexed immunoaffinity arrays for the quantification of multiple glycoprotein isoforms, both used for the detection of breast cancer [28–29] (for FDA’s classification of In Vitro Diagnostic Medical Devices and the regulatory routes [premarket notification or 510(k), and/or premarket approval mechanisms] for device marketing, please visit http://www.fda.gov/MedicalDevices/DeviceRegulationandGuidance/IVDRegulatoryAssistance/ucm123682.htm). These detailed review documents represent an important step forward in applying quantitative proteomic technologies to cancer biomarker research to in order to deliver a clinically-useful product which will meet the review criteria of the Office of In Vitro Diagnostics of the FDA, and which will ultimately have a positive impact on patient outcome.
5. Development of MRM-MS proteomic platform for biomarker verification
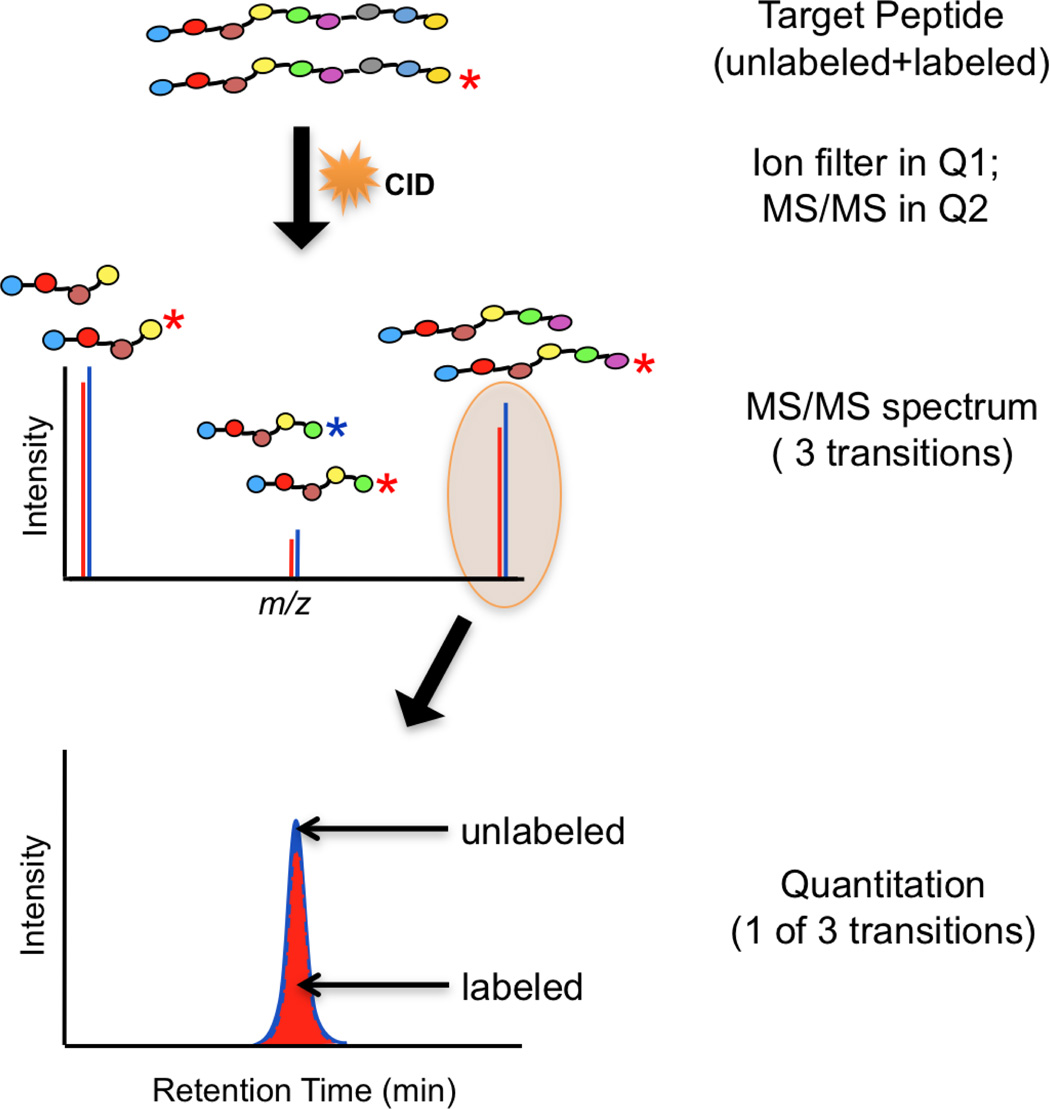
The biomarker verification stage of current NCI-CPTC pipeline currently utilizes a recently-renewed technology called Selected Reaction Monitoring (SRM) or MRM-MS on triple quadrupole mass spectrometers (QQQ-MS) that generates unique fragment ions associated with their corresponding precursor ions which can be quantified even in a very complicated matrix. Specifically, the MRM technique relies on selecting specific precursor-product ion pairs, or transitions. The first quadrupole (Q1) is set to transmit only a particular precursor peptide ion into the second quadrupole (Q2) where collisionally-induced dissociation (CID) yields fragment ions. A signature fragment ion of particular mass-to-charge ratio (m/z) value or several fragment ions is then allowed into the third quadrupole (Q3) and subsequently measured by the detector. The quantitation of peptides is achieved by measuring the intensity of the fragment ions. The general components of MRM-MS-based approaches for targeted peptide quantitation is shown in Figure 2A, where a few of the most abundant fragment ions detected in Q3, as a result of CID in Q2, are used as signature transitions to quantify their corresponding precursor ion intensities.
Figure 2. Selected/Multiple Reaction Monitoring Mass Spectrometry (SRM/MRM-MS).
(A). A schematic of a triple quadrupole mass spectrometer (TQMS) commonly used for MRM/SRM-MS analysis. Q1 and Q3 represent two quadrupoles as mass filters in a TQMS. CID is collisionally-induced dissociation during MS/MS in Q2. RF refers to radiofrequency; (B). MRM plots for a target peptide (9 amino acids, no asterisk) and its stable isotope-labeled internal standard (labeled with a red asterisk) using a single transition to yield quantitative information (ratio of the peak areas of both in red and blue, depicted at the bottom of the figure).
Although the use of SRM/MRM-MS to quantify biomolecules (e. g., drugs and metabolites) was widely adopted in research laboratories and pharmaceutical industry (hormones, drugs, metabolites) many years ago, it has only been recently applied, in combination with SID-MS, to quantify peptides and proteins. SID-MRM-MS for protein assays is predicated on the measurement of signature proteotypic tryptic peptides that uniquely and stoichiometrically represent the protein candidates of interest. MRM-based assay development usually starts with a selection of 3–5 peptides per protein [16] to improve specificity of the assay for targeted analytes (i. e., proteins in this case). In addition, synthetic-stable isotope-labeled versions of each peptide (or heavy peptides), chemically identical to their endogenous, analyte peptide counterparts with the exception of their masses (typically 6–10 Da more, depending on the label used), are used as internal standards. Specific fragment ion signals derived from the endogenous unlabeled species are measured and compared to those from the exogenously labeled peptides. The ratio of these values provides a precise measure of the concentration of the corresponding protein. As illustrated in Figure 2B, the MRM plots for a target peptide composed of 9 amino acids and its stable isotope-labeled internal standard (*) using a single transition yield quantitative information based on the ratio of the peak areas of both fragment ions labeled in blue (unlabeled) and red (labeled). The specificity of such quantitative measurements increases as the number of monitored peptides corresponding to a protein and the number of transitions for each peptide selected for monitoring increase [20]. This technique is ideal for sensitive and specific quantitation in a multiplex fashion (i. e., a single assay measures multiple analytes simultaneously in a single liquid chromatography-MS/MS [LC-MS/MS] run).
The typical steps involved in developing an MRM-MS protein assay are as follows: (1) selection of surrogate or signature peptides diagnostic for each protein; (2) protein extraction from biological matrices; (3) proteolytic digestion of proteins usually with trypsin; (4) iterative testing of synthetic peptides and transitions by liquid chromatography LC-MRM-MS (including the heavy isotopically labeled internal standard peptides); (5) assay validation on biological samples; and (6) assay testing SOP/method documentation. Since MRM-MS sensitivity on QQQ-MS/MS is critically dependent on ionization conditions and tuning of instrument parameters, such as collision energy and cone voltage, for the generation of maximal product ion signal, step (4) is an important part of the assay process. It would be very helpful to build spectral fragmentation libraries of proteotypic peptides for the proteomics community to aid in future MRM-based assay development. Additionally, scheduling MRM-MS scans on target analytes based on their different chromatographic retention time can certainly aid in ensuring quantification on desired ions when background interferences are significant (e. g., matrix effects from complex biological samples such as blood and urine). It allows precursor/product ion pairs to be monitored in a single analysis, thus increasing throughput. However, this requires superb system stability of LC pumps and columns, thus putting high demands on the quality control aspects of LC systems and factors which affect LC performance such as ambient temperature. Another component to this analysis is bioinformatics which allows user-friendly, customized planning of analysis on target analytes. One such software suite called Skyline developed in MacCoss’s group is currently downloadable from https://brendanx-uw1.gs.washington.edu/labkey/project/home/software/Skyline/begin.view [30]. Once the ratio of peak area under the monitored fragment ions of light (native) to that of heavy (its internal standard) peptides is measured, the concentration of that protein can be determined [16].
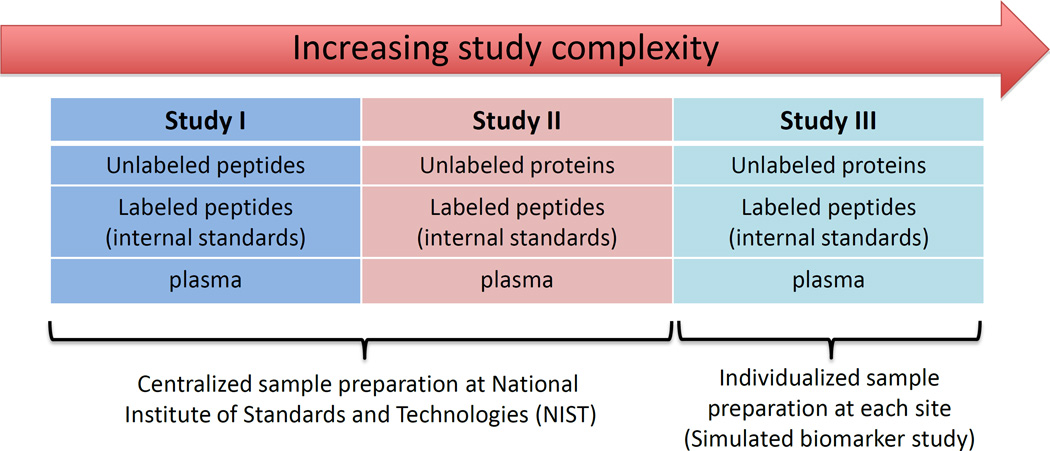
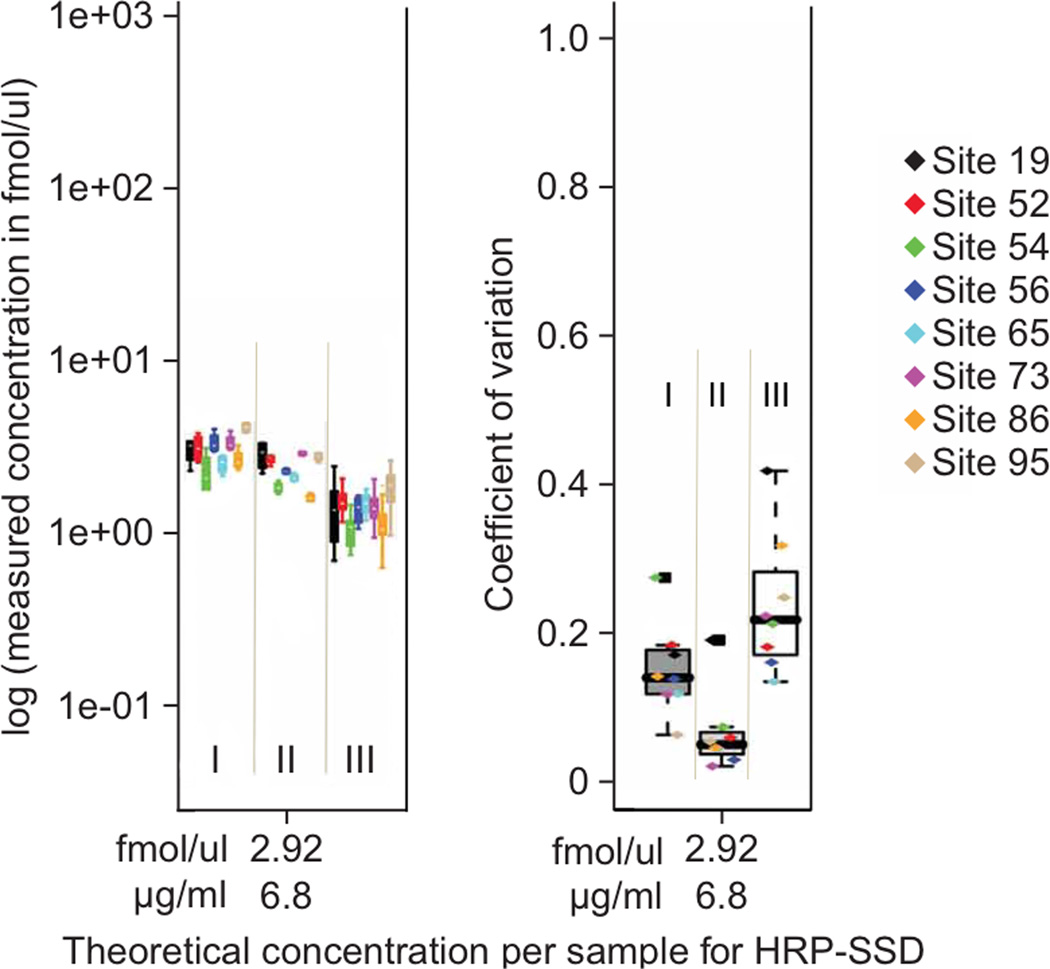
For this technology to be suitable for use in pre-clinical studies where large numbers of candidate protein biomarkers (i. e., hundreds of patient samples) must be rapidly screened (verification), it must be demonstrated that protein quantification can be achieved reproducibly within and across laboratories on different instrument platforms. In 2009, the NCI-CPTC network spearheaded a first-of-its-kind inter-laboratory study composed of 3 sub-studies (Figure 3A) designed to increase the level of difficulty in sample preparation (i. e., more sources of variability) at 8 individual sites [11]. Intra-laboratory variability and reproducibility in all 3 sub-studies were evaluated by comparing the measured concentrations of 7 target proteins to the actual concentrations across the range of spiked-in analytes (9 concentration points down to the limit of quantitation [LOQ] at 2.92 nM), and by determining the coefficients of variation (CVs) for these quantitative measurements (Figure 3B). The results showed that the reproducibility and precision of these quantitative measurements for 9 of 10 peptides tested across 8 laboratories ranged from 4 to 14%, 4 to 13% and 10 to 23% inter-laboratory CVs at or near the estimated LOQ of 2.92 nM for studies I, II and III, respectively when SOPs were adopted. Intra-laboratory CVs were usually <15% and <25% at the identical concentration for studies I/II and III. The progressive increases in CVs from studies I to III clearly indicate that sample handling contributes more to assay variability than instrumental variability, further highlighting the high data quality obtainable from MRM-MS. Although the current MRM assay performance under real biomarker conditions (study III) is below that generally stated for clinical assays using ELISAs (CVs typically <10 to 15%), the performance achieved here is sufficient for the pre-clinical verification stage (bridge) of candidate biomarkers present at more than ∼2 to 6 µg/mL in plasma with a linear dynamic range spanning three orders of magnitude. Furthermore, inter-laboratory and intra-laboratory CVs improved with increasing analyte concentration in all cases, whether by spiking in more analytes or by enrichment techniques.
Figure 3. Robustness of an MRM-MS Platform, an NCI-CPTC inter-laboratory study11.
(A). The design of three MRM-MS experiments (I, II, III) with increasing complexity of sample preparation performed by NCI-CPTC network teams in 2009; (B). Reproducibility of MRM-MS measurements of peptide HRP-SSD (SSDLVALSGGHTFGK) from horse radish peroxidase. Intralaboratory assay coefficients of variation (CVs) for studies I-III shown in the left panel illustrates experimentally determined log concentration (y axis) versus theoretical (spike-in) concentration (x axis) at the limit of quantitation (LOQ) in plasma. Interlaboratory CVs (y-axis) for peptide HRP-SSD versus theoretical (spike-in) concentration (x-axis) at LOQ for studies I-III are shown in the right panel (adopted from Ref. 11: Addona et. al. 2009). Actual intralaboratory CV values for individual laboratories are shown with color-coded markers within each box plot. The CV values are calculated based on the single best performing transition (lowest combined CV) for all studies.
Many biomarkers of current clinical importance, such as prostate-specific antigen (PSA), carcinoembryonic antigen (CEA), and the troponins (Tns), are present in the mid-pg/mL to low ng/mL range in plasma [31]. To use MRM assays for protein biomarkers clinically, this technology still has to improve its sensitivity for detection and quantitation. Several approaches have been developed in this direction. One such approach has demonstrated that a combination of abundant protein depletion with minimal fractionation of tryptic peptides by strong cation exchange (SCX) prior to SID-MRM-MS provides LOQ signal-to-noise ratios of >10 in the 1–20 ng/mL range with CVs of 10%-20% at the LOQs for proteins in plasma [32]. This includes the development of assays for a number of known markers of cardiovascular disease [16], providing additional proof of the power of MRM approaches for configuring assays for proteins for which antibodies are unavailable. An additional approach called SISCAPA (Stable Isotope Standards and Capture by Anti-Peptide Antibodies) can significantly increase the sensitivity of detection and quantitation of proteins in plasma by target peptide enrichment [33] and has been deployed in clinical setting [34]. Using several anti-peptide antibodies against protein targets to enrich for the peptides subsequently subjected to MRM-MS analysis, >1000-fold increase in sensitivity has been achieved. QQQ-MS provide greater sensitivity, wider dynamic range quantitation and detailed, sequence-based characterization of multiple peptides digested from several proteins (unlike the lower-specificity optical or electrochemical signals generated as surrogates for the analytes in conventional immunoassay) [16, 29]. Moreover, the capability for simultaneous measurement and characterization of targeted analytes allows three important advances in protein assays, on which the mock 510(k) PepCa10 test is based [29]: (1) it permits facile multiplexed measurement of many molecular species without significant interference; (2) it facilitates the use of analyte-identical internal standards of same structure with different m/z to control all aspects of the assay workflow; (3) it results in reduced sample handling, high sensitivity and wide dynamic range; and (4) it allows site-specific quantification of post-translationally modified peptides (phosphopeptides [35], glycopeptides [36], etc.) as important biomarker candidates. These advantages constitute a step forward in assay quality control, potentially shifting some of the performance and reliability burden from technical standardization of reagents and instruments to real-time observation and evaluation of the analytes themselves. Future SISCAPA inter-laboratory studies within the NCI-CPTC network will be underway shortly, using metrics, SOPs, high quality reagents and reference materials. While SISCAPA is usually done with elution, followed by LC/MRM-MS detection, elution followed by MALDI/MRM-MS is also possible. A similar technique, without elution of the analytes, is immunoMALDI (iMALDI), where beads are placed directly on a MALDI target, with the affinity-bound peptides still attached [37–38]. The MALDI matrix solvent elutes the peptides from the beads. The presence of the peptide, and its peak height or peak area, is then determined from an MS spectrum for quantitation in the MS mode, while peptide identities are confirmed with MS/MS. In principle, iMALDI can be performed with only a MALDI-MS instrument, but it can also be used “in the “MRM mode on a MALDI-MS/MS instrument”.
One important factor to consider using enrichment strategies is the percentage of analytical recovery of peptides in biological matrices such as human serum or plasma. To address this, a calibration curve made in an appropriate matrix and included with each batch of samples to compensate for this loss would permit the signal of each peptide in each sample to reflect an actual concentration in a multiplexed fashion. Additionally, incomplete digestion due to the high concentration of matrix proteins likely plays a large role in the reduced yield of peptides. Proteolytic digestion variability in different samples also complicates the measurements, and PTMs, single nucleotide polymorphisms (SNP), and other protein modifications can potentially affect the quantitation by shifting the peptide masses. Interferences from other proteins can also affect the final results [39]. In both cases, spiking in an exogenous protein, preferably the properly-folded, stable isotope-labeled version of the native protein of interest, to each sample becomes very important in order to gauge and normalize digestion efficiencies. However, the high cost and extensive efforts associated with the development of high quality reagents, including heavy isotope versions of target proteins and internal standard peptides, as well as anti-protein and anti-peptide antibodies, could limit the broad use of these internal standards for quality control purposes.
6. Future Perspectives
With recent advances in protein-based technologies, it is expected that clinical proteomics, in the near future, will focus on developing highly multiplexed and automated technologies for more accurate quantification of proteins and their isoforms, as well as differences in PTMs between normal and diseased states in a statistically robust manner (i. e., a sufficient number of biospecimen), in order to understand the disease at the molecular level, incorporating known genomic information whenever available.
The first step in biomarker research that needs to be considered is the development and improvement of biospecimen science. Careful studies on the stability of proteins in biological matrices using different methods of collection, processing, storage and distribution need to be carried out. Standardized technologies along with protocols aimed at preserving sample integrity and minimizing pre-analytical variation will need to improve rapidly in order to meet the increasing demand for proteomics-based screening for cancer, especially with the vast amount of knowledge on genetic aberrations identified in cancer that continues to be accumulated.
Currently, proteomic technologies for protein and PTM identification and quantification using MS are still dominated by bottom-up proteomics where peptides are analyzed after proteolysis and pierced together like a jigsaw puzzle. Relative quantification using label-free or isotopic labeling techniques including iTRAQ, 18O, and SILAC for comparative analyses between control and diseased states is still prevalent in biomarker research [40–44]. Although useful and semi-quantitative, these techniques still belong to the biomarker discovery stage because they are non-targeted and do not quantitatively measure the absolute concentrations of specific protein biomarkers present in a biological matrix, which are commonly used by clinicians for diagnosis, prognosis and selection of treatment options. MRM-MS/SISCAPA/iMALDI platforms solve this problem by introducing known amounts of chemically identical, isotopically-labeled heavy peptide counterparts to their target analytes. For quantitation, however, lack of reproducibility and efficiency in proteolysis to generate these native peptides creates uncertainties in the accurate measurements of proteins in complex biological matrices as previously discussed. In this aspect, top-down mass spectrometric approaches especially with sequencing information in addition to molecular weight information to study intact proteins and their PTMs would seem to have the advantage of requiring less sample processing, possibly shortening assay time and yielding more accurate quantification of proteins in biological matrices such as plasma and tissue over bottom-up proteomics. Realistically, however, it is not trivial to characterize intact proteins especially those with high molecular weight (∼>100 kDa), membrane and/or low abundance proteins using MS due to the inherent difficulties associated with the analysis. Such difficulties include, but are not limited to, poor ionization efficiency of proteins lacking basic amino acids; insolubility of membrane proteins; the lack of high quality anti-protein or anti-PTM antibodies for affinity enrichment; difficulty and high cost associated with generating pure proteins and their PTMs as internal standards with high incorporation rate of heavy isotopes. Top-down mass spectrometric approaches at its current stage still need to be developed and optimized for quantitative measurements in a high-throughput manner, ideally with automated workflows to minimize sample handling variation when used in pre-clinical and clinical settings.
Current immunological approaches (e. g., sandwich ELISAs) for intact protein quantification certainly are more commonly used than mass spectrometric methods in clinical laboratories, especially when coupled with automation and multiplexing capability. However, ELISAs have certain inherent problems: including lack of concordance across platforms, antibody availability, and the Hook effect [39]. MS has the unique ability to measure m/z, which are characteristic of each protein and its proteolytic peptides, its isoforms and modifications, potentially providing high specificity for these measurements (retention time, precursor ion m/z and fragment ion m/z values), overcoming many of the problems inherent with ELISA-based methods, and holding the potential to be routinely used in a clinical setting. With the emergence of new technologies such as nano-pores and nano-wires [45–47], it is conceivable that the sensitivity of detection and quantitation of proteins, as well as sample throughput capability will increase tremendously. It is anticipated that proteomics will continue to evolve into a more reliable, quantitative science by developing, improving and standardizing new and current multiplexing technologies/platforms, minimizing sample handling, and decreasing the complexity of use and dependency on expert operators through automation.
In order for proteomics research to successfully translate to real clinical utility, several developments need to occur. These include addressing the current roadblocks in biomarker research, utilization of rigorously-assessed and standardized technologies for quantitative protein research, and capitalizing on deep genomic sequencing efforts by the medical research community. A summary of these roadblocks is given below:
6.1. Addressing the roadblocks in biomarker research
-
Lack of standardization and uniformity in proteomic research at the analytical level
The proteomics community including ABRF, HUPO and NCI-CPTC has been making strides in standardizing analytical platforms to generate reproducible proteomics data.
The adoption of NCI-CPTC program achievements including the SOPs, metrics and reference materials by the community will take time.
-
Lack of proper biospecimen quality assessment (QA) and QC
-
The NCI Office of Biorepositories and Biospecimen Research (OBBR) was established in 2005 in recognition of the critical role that biospecimens play in cancer research. OBBR is responsible for developing a common biorepository infrastructure that promotes resource sharing and team science in order to facilitate multi-institutional, high-throughput genomic and proteomic studies.
There is a considerable lack of scientific data for assessing the effects of specimen-handling variables on molecular testing of human tissues. New research in Biospecimen Science will define the precise relationships between biospecimen handling and the quality and reproducibility of data for cancer research. The NCI Biospecimen Research Network was initiated to systematically address the impact of specific variables in individual specimen types on molecular data from given analysis platforms.
-
Lack of proper study design encompassing a meaningful biological/clinical question in mind (why and how)
Lack of statistically robust number of high quality biospecimen for biomarker “verification”
Lack of high-quality reagents and standards at a reasonable cost
Lack of low-cost automated, easy-to-use instruments/platforms
6.2. Realizing the great potential of clinical proteomics in cancer biomarker research
High-throughput, large-scale genomic studies to characterize genomic aberration typical of cancer are ongoing, and represent unique opportunities for the cancer community to investigate protein expression profiles of wild-type and aberrant proteins and PTMs which are associated with this disease at a functional level.
Currently-available proteomic technologies such as sample fractionation coupled with shotgun analyses, multiplexed MRM-MS combined with affinity enrichment, and immunoaffinity arrays, can be optimized and standardized in the cancer proteomics community for biomarker development.
Assays which combine MRM-MS analysis with affinity enrichment are sensitive and specific with high-throughput capability, which provides analyte verification in a large number of samples prior to costly and time-consuming clinical qualification.
Technology standardization efforts, as demonstrated by the NCI-CPTC initiative and others to break down analytical barriers in proteomics, have laid the foundation for future endeavors into understanding the molecular basis of cancer development and progression.
6.3. Focusing on key points for future proteomic biomarker research
Develop and improve biospecimen science by proteomics-QA and QC.
Emphasize technology/platform optimization and standardization for data reproducibility/transportability for analytical validation.
Continue new technology development in order to improve protein analysis and to increase sample throughput (i. e., lower analysis cost per sample), improved sensitivity and dynamic range.
Advocate and support multi-disciplinary approaches to biomarker development.
-
Integrate with cancer genomics and other scientific disciplines to create a “systems biology” approach to elucidate pathways/functions implicated in cancer etiology:
During the past decade, several groundbreaking discoveries in life science were made including the successful completion of the human genome sequencing project, which represents a true milestone in biomedicine [48]. This has provided an important knowledge base, enabling rapid development in life science-oriented research in areas such as diagnostics, gene therapy, new drug targets discovery and development of personalized therapies [49–51]. The completion of the human genome project now presents an even more challenging task for scientists: the characterization of the human proteome. Large-scale multi-disciplinary, team science-based initiatives in characterizing disease genomes are already underway in order to understand diseases at a molecular level. The Cancer Genome Atlas (TCGA) [52–54] and The International Cancer Genome Consortium (ICGC) [55] are characterizing cancer genomes in order to understand different cancers at the genetic level. To date, these initiatives have characterized several different tumor types along with their subtypes using high-throughput sequencing technologies and statistically-meaningful sets of clinical samples carefully collected using SOPs. As a result, genetic alterations associated with cancer including copy number aberration, mutation, microdeletion and others have been discovered through the use of multi-dimensional datasets and high level integrative analysis. The next logical step is to characterize, to the best of our abilities, proteins, their altered forms and PTMs, as well as protein-protein interaction/networks. This endeavor will, for the first time, either corroborate or complement the data on genetic aberrations detected in these tumors, providing a deeper understanding of cancer in the context of biology and clinical utility.
A step towards realizing this goal is by incorporating cancer genomics and cancer biology-based concepts from the research community using metrics-driven technologies/platforms to identify and quantify proteins from tissues and biofluids, followed by large-scale, high-throughput proteomic analyses at two clearly distinct stages: (1) biomarker discovery; and (2) biomarker verification; both of which require a statistically-powered number of clinical samples with clinical relevance prior to the costly and time-consuming biomarker qualification stage.
-
It is anticipated that these combined approaches will aid in ultimately translating proteomics into the clinic by:
developing “verified” cancer biomarkers that can be translated into products by other initiatives and activities involved in clinical qualification studies;
creating human cancer protein maps from large-scale, high-quality data and biospecimens corroborating, or complementing genomic findings, if available; and creating protein pathways/networks involved in oncogenesis, metastasis, etc.;
producing a high-quality, publically-accessible database to store and organize all datasets;
developing analytically robust, multiplexed, quantitative assays for peptide/PTM targets with all reagents being publicly available.
7. Concluding Remarks
A foundation for successful biomarker research through careful experimental design and technology standardization is starting to be established. Progress has been made in making the scientific community aware of the need for standardization of technologies/platforms and data analysis.
MRM-MS (+/− affinity enrichment) as a multiplexed, sensitive, reproducible and quantitative “verification” platform to rigorously test target analytes in pre-clinical studies holds great promise. It has the potential to produce comparable quantitative results to current clinical tests with optimization/standardization, but additional technology optimization is still needed for routine use.
Novel technologies still need to be developed in order to improve protein analysis, sample throughput, sensitivity and dynamic range.
Looking into the future, a systems biology approach has great potential for integrating knowledge on cancer biology.
Acknowledgements
The authors alone are responsible for the content and writing of the paper. Certain commercial equipment, instruments, or materials identified in this paper to specify adequately the experimental procedure does not imply recommendation or endorsement by the National Cancer Institute, National Institutes of Health, nor does it imply that the materials or equipment identified are necessarily the best available for the purpose.
Abbreviations
- QA
quality assessment
- QC
quality control
- PTM
post-translational modification
- MRM-MS
multiple reaction monitoring mass spectrometry
- SISCAPA
stable isotope standards and capture by anti-peptide antibodies
- ELISA
enzyme-linked immunosorbent assay
- iMALDI
(immunoMALDI)
- QQQ-MS
(triple quadrupole mass spectrometry)
- SID-MS
(stable-isotope dilution-MS)
- NCI
(National Cancer Institute)
- CPTC
(Clinical Proteomic Technologies for Cancer initiative)
Footnotes
Superscript in Figure 3 denotes reference number. Figure 3B is adopted from Addona et. al., 2009 from reference 11.
Financial disclosure
The authors report no conflicts of interest.
8. References
- 1.Atkinson AJ, Colburn WA, Degruttola VG. Biomarkers and surrogate endpoints: preferred definitions and conceptual framework. Clin Pharmacol Ther. 2001;69:89–95. doi: 10.1067/mcp.2001.113989. [DOI] [PubMed] [Google Scholar]
- 2.Zhang X, Li L, Wei D, Yap Y, Chen F. Moving cancer diagnostics from bench to bedside. Trends Biotechnol. 2007;25:166–173. doi: 10.1016/j.tibtech.2007.02.006. [DOI] [PubMed] [Google Scholar]
- 3.Anderson L. Candidate-based proteomics in the search for biomarkers of cardiovascular disease. J Physiol. 2005;563:23–60. doi: 10.1113/jphysiol.2004.080473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Etzioni R, Urban N, Ramsey S, McIntosh M, Schwartz S, Reid B, Radich J, Anderson G, Hartwell L. The case for early detection. Nat Rev Cancer. 2003;3:243–252. doi: 10.1038/nrc1041. [DOI] [PubMed] [Google Scholar]
- 5.Rifai N, Gillette MA, Carr SA. Protein biomarker discovery and validation: the long and uncertain path to clinical utility. Nat Biotechnol. 2006;24:971–983. doi: 10.1038/nbt1235. [DOI] [PubMed] [Google Scholar]
- 6.García-Foncillas J, Bandrés E, Zárate R, Remírez N. Proteomic analysis in cancer research: potential application in clinical use. Clin Transl Oncol. 2006;8(4):250–261. doi: 10.1007/BF02664935. [DOI] [PubMed] [Google Scholar]
- 7.Kelleher MT, Fruhwirth G, Patel G, Ofo E, Festy F, Barber PR, Ameer-Beg SM, Vojnovic B, Gillett C, Coolen A, Kéri G, Ellis PA, Ng T. The potential of optical proteomic technologies to individualize prognosis and guide rational treatment for cancer patients. Target Oncol. 2009;4(3):235–252. doi: 10.1007/s11523-009-0116-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang P, Whiteaker JR, Paulovich AG. The evolving role of mass spectrometry in cancer biomarker discovery. Cancer Biol Ther. 2009;8(12):1083–1094. doi: 10.4161/cbt.8.12.8634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goodsaid FM, Frueh FW, Mattes W. Strategic paths for biomarker qualification. Toxicology. 2008;245:219–223. doi: 10.1016/j.tox.2007.12.023. [DOI] [PubMed] [Google Scholar]
- 10.Findeisen P, Neumaier M. Mass spectrometry based proteomics profiling as diagnostic tool in oncology: current status and future perspective. Clin Chem Lab Med. 2009;47(6):666–684. doi: 10.1515/CCLM.2009.159. [DOI] [PubMed] [Google Scholar]
- 11.Addona TA, Abbatiello SE, Schilling B, Skates SJ, Mani DR, Bunk DM, Spiegelman CH, Zimmerman LJ, Ham AJ, Keshishian H, Hall SC, Allen S, Blackman RK, Borchers CH, Buck C, Cardasis HL, Cusack MP, Dodder NG, Gibson BW, Held JM, Hiltke T, Jackson A, Johansen EB, Kinsinger CR, Li J, Mesri M, Neubert TA, Niles RK, Pulsipher TC, Ransohoff D, Rodriguez H, Rudnick PA, Smith D, Tabb DL, Tegeler TJ, Variyath AM, Vega-Montoto LJ, Wahlander A, Waldemarson S, Wang M, Whiteaker JR, Zhao L, Anderson NL, Fisher SJ, Liebler DC, Paulovich AG, Regnier FE, Tempst P, Carr SA. Multi-site assessment of the precision and reproducibility of multiple reaction monitoring-based measurements of proteins in plasma. Nat Biotechnol. 2009;27(7):633–641. doi: 10.1038/nbt.1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moritz RL, Clippingdale AB, Kapp EA, Eddes JS, Gilbert HS, Connolly LM, Simpson RJ. Application of 2-D free-flow electrophoresis/RP-HPLC for proteomic analysis of human plasma depleted of multi high-abundance proteins. Proteomics. 2005;5:3402–3413. doi: 10.1002/pmic.200500096. [DOI] [PubMed] [Google Scholar]
- 13.Li Y, Lee HJ, Corn RM. Detection of protein biomarkers using RNA aptamer microarrays and enzymatically amplified surface plasmon resonance imaging. Anal. Chem. 2007;79:1082–1088. doi: 10.1021/ac061849m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pierobon M, Calvert V, Belluco C, Garaci E, Deng J, Lise M, Nitti D, Mammano E, De Marchi F, Liotta L, Petricoin E. Multiplexed cell signaling analysis of metastatic and nonmetastatic colorectal cancer reveals COX2-EGFR signaling activation as a potential prognostic pathway biomarker. Clin Colorectal Cancer. 2009;8(2):110–117. doi: 10.3816/CCC.2009.n.018. [DOI] [PubMed] [Google Scholar]
- 15.Ramachandran N, Raphael JV, Hainsworth E, Demirkan G, Fuentes MG, Rolfs A, Hu Y, LaBaer J. Next-generation high-density self-assembling functional protein arrays. Nat Methods. 2008;5(6):535–538. doi: 10.1038/nmeth.1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kuhn E, Addona T, Keshishian H, Burgess M, Mani DR, Lee RT, Sabatine MS, Gerszten RE, Carr SA. Developing multiplexed assays for troponin I and interleukin-33 in plasma by peptide immunoaffinity enrichment and targeted mass spectrometry. Clin Chem. 2009;55(6):1108–1117. doi: 10.1373/clinchem.2009.123935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Beirne P, Pantelidis P, Charles P, Wells AU, Abraham DJ, Denton CP, Welsh KI, Shah PL, du Bois RM, Kelleher P. Multiplex immune serum biomarker profiling in sarcoidosis and systemic sclerosis. Eur Respir J. 2009;34(6):1376–1382. doi: 10.1183/09031936.00028209. [DOI] [PubMed] [Google Scholar]
- 18.Polanski M, Anderson L. A list of candidate cancer biomarkers for targeted proteomics. Biomark Insights. 2007;1:1–48. [PMC free article] [PubMed] [Google Scholar]
- 19.Anderson NL. The clinical plasma proteome: a survey of clinical assays for proteins in plasma and serum. Clin Chem. 2010;56(2):177–185. doi: 10.1373/clinchem.2009.126706. [DOI] [PubMed] [Google Scholar]
- 20.Anderson L, Hunter CL. Quantitative mass spectrometric multiple reaction monitoring assays for major plasma proteins. Mol Cell Proteomics. 2006;5(4):573–588. doi: 10.1074/mcp.M500331-MCP200. [DOI] [PubMed] [Google Scholar]
- 21.Paulovich AG, Whiteaker JR, Hoofnagle AN, Wang P. The interface between biomarker discovery and clinical validation: The tar pit of the protein biomarker pipeline. Proteomics-Clinical Applications. 2008;2(10–11):1386–1402. doi: 10.1002/prca.200780174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Makawita S, Diamandis EP. The bottleneck in the cancer biomarker pipeline and protein quantification through mass spectrometry-based approaches: current strategies for candidate verification. Clin Chem. 2010;56(2):212–222. doi: 10.1373/clinchem.2009.127019. [DOI] [PubMed] [Google Scholar]
- 23.Turck CW, Falick AM, Kowalak JA, Lane WS, Lilley KS, Phinney BS, Weintraub ST, Witkowska HE, Yates NA. The Association of Biomolecular Resource Facilities Proteomics Research Group 2006 study: relative protein quantitation. Mol Cell Proteomics. 2007;6(8):1291–1298. doi: 10.1074/mcp.M700165-MCP200. [DOI] [PubMed] [Google Scholar]
- 24.Arnott D, Gawinowicz MA, Kowalak JA, Lane WS, Speicher KD, Turck CW, West KA, Neubert TA. ABRF-PRG04: differentiation of protein isoforms. J Biomol Tech. 2007;18(2):124–134. [PMC free article] [PubMed] [Google Scholar]
- 25.Falick AM, Kowalak JA, Lane WS, Phinney BS, Turck CW, Weintraub ST, West KA, Neubert TA. ABRF-PRG05: de novo peptide sequence determination. J Biomol Tech. 2008;19(4):251–257. [PMC free article] [PubMed] [Google Scholar]
- 26.Bell AW, Deutsch EW, Au CE, Kearney RE, Beavis R, Sechi S, Nilsson T, Bergeron JJ HUPO Test Sample Working Group. A HUPO test sample study reveals common problems in mass spectrometry-based proteomics. Nat Methods. 2009;6(6):423–430. doi: 10.1038/nmeth.1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Paulovich AG, Billheimer D, Ham AJ, Vega-Montoto L, Rudnick PA, Tabb DL, Wang P, Blackman RK, Bunk DM, Cardasis HL, Clauser KR, Kinsinger CR, Schilling B, Tegeler TJ, Variyath AM, Wang M, Whiteaker JR, Zimmerman LJ, Fenyo D, Carr SA, Fisher SJ, Gibson BW, Mesri M, Neubert TA, Regnier FE, Rodriguez H, Spiegelman C, Stein SE, Tempst P, Liebler DC. Interlaboratory study characterizing a yeast performance standard for benchmarking LC-MS platform performance. Mol Cell Proteomics. 2010;9(2):242–254. doi: 10.1074/mcp.M900222-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rodriguez H, Tezak Z, Mesri M, Carr SA, Liebler DC, Fisher SJ, Tempst P, Hiltke T, Kessler LG, Kinsinger CR, Philip R, Ransohoff DF, Skates SJ, Regnier FE, Anderson NL, Mansfield E on behalf of the Workshop Participants. Analytical Validation of Protein-Based Multiplex Assays: A Workshop Report by the NCI-FDA Interagency Oncology Task Force on Molecular Diagnostics. Clin. Chem. 2010;56(2):237–243. doi: 10.1373/clinchem.2009.136416. [DOI] [PubMed] [Google Scholar]
- 29.Regnier FE, Skates SJ, Mesri M, Rodriguez H, Tezak Z, Kondratovich MV, Alterman MA, Levin JD, Roscoe D, Reilly E, Callaghan J, Kelm K, Brown D, Philip R, Carr SA, Liebler DC, Fisher SJ, Tempst P, Hiltke T, Kessler LG, Kinsinger CR, Ransohoff DF, Mansfield E, Anderson NL. Protein-Based Multiplex Assays: Mock Presubmissions to the US Food and Drug Administration. Clin. Chem. 2010;56(2):165–171. doi: 10.1373/clinchem.2009.140087. [DOI] [PubMed] [Google Scholar]
- 30.MacLean B, Tomazela DM, Shulman N, Chambers M, Finney GL, Frewen B, Kern R, Tabb DL, Liebler DC, MacCoss MJ. Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics. 2010;26(7):966–968. doi: 10.1093/bioinformatics/btq054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gerszten RE, Carr SA, Sabatine M. Integration of Proteomic-Based Tools for Improved Biomarkers of Myocardial Injury. Clin Chem. 2010;56:194–201. doi: 10.1373/clinchem.2009.127878. [DOI] [PubMed] [Google Scholar]
- 32.Keshishian H, Addona T, Burgess M, Kuhn E, Carr SA. Quantitative, multiplexed assays for low abundance proteins in plasma by targeted mass spectrometry and stable isotope dilution. Mol Cell Proteomics. 2007;6:2212–2229. doi: 10.1074/mcp.M700354-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Anderson NL, Jackson A, Smith D, Hardie D, Borchers C, Pearson TW. SISCAPA peptide enrichment on magnetic beads using an in-line bead trap device. Mol Cell Proteomics. 2009;8(5):995–1005. doi: 10.1074/mcp.M800446-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ciccimaro E, Hanks SK, Yu KH, Blair IA. Absolute quantification of phosphorylation on the kinase activation loop of cellular focal adhesion kinase by stable isotope dilution liquid chromatography/mass spectrometry. Anal Chem. 2009;81(9):3304–3313. doi: 10.1021/ac900204f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hoofnagle AN, Becker JO, Wener MH, Heinecke JW. Quantification of thyroglobulin, a low-abundance serum protein, by immunoaffinity peptide enrichment and tandem mass spectrometry. Clin Chem. 2008;54(11):1796–1804. doi: 10.1373/clinchem.2008.109652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Drake PM, Cho W, Li B, Prakobphol A, Johansen E, Anderson NL, Regnier FE, Gibson BW, Fisher SJ. Sweetening the pot: adding glycosylation to the biomarker discovery equation. Clin Chem. 2010;56(2):223–236. doi: 10.1373/clinchem.2009.136333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reid JD, Holmes DT, Mason DR, Shah B, Borchers CH. Towards the Development of an Immuno MALDI (iMALDI) Mass Spectrometry Assay for the Diagnosis of Hypertension. J Am Soc Mass Spectrom. 2010 Feb 1; doi: 10.1016/j.jasms.2010.01.024. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 38.Jiang J, Parker CE, Fuller JR, Kawula TH, Borchers CH. An immunoaffinity tandem mass spectrometry (iMALDI) assay for detection of Francisella tularensis. Anal Chim Acta. 2007;605(1):70–79. doi: 10.1016/j.aca.2007.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hoofnagle AN, Wener MH. The fundamental flaws of immunoassays and potential solutions using tandem mass spectrometry. J Immunol Methods. 2009;347(1–2):3–11. doi: 10.1016/j.jim.2009.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhao L, Lee BY, Brown DA, Molloy MP, Marx GM, Pavlakis N, Boyer MJ, Stockler MR, Kaplan W, Breit SN, Sutherland RL, Henshall SM, Horvath LG. Identification of candidate biomarkers of therapeutic response to docetaxel by proteomic profiling. Cancer Res. 2009;69(19):7696–7703. doi: 10.1158/0008-5472.CAN-08-4901. [DOI] [PubMed] [Google Scholar]
- 41.Kristiansen TZ, Harsha HC, Grønborg M, Maitra A, Pandey A. Differential membrane proteomics using 18O-labeling to identify biomarkers for cholangiocarcinoma. J Proteome Res. 2008;7(11):4670–4677. doi: 10.1021/pr800215n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pan C, Olsen JV, Daub H, Mann M. Global effects of kinase inhibitors on signaling networks revealed by quantitative phosphoproteomics. Mol Cell Proteomics. 2009;8(12):2796–2808. doi: 10.1074/mcp.M900285-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zamò A, Cecconi D. Proteomic analysis of lymphoid and haematopoietic neoplasms: there's more than biomarker discovery. J Proteomics. 2010;73(3):508–520. doi: 10.1016/j.jprot.2009.08.012. [DOI] [PubMed] [Google Scholar]
- 44.Piersma SR, Fiedler U, Span S, Lingnau A, Pham TV, Hoffmann S, Kubbutat MH, Jimeénez CR. Workflow comparison for label-free, quantitative secretome proteomics for cancer biomarker discovery: method evaluation, differential analysis, and verification in serum. J Proteome Res. 2010 Feb 12; doi: 10.1021/pr901072h. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 45.Zheng G, Patolsky F, Cui Y, Wang WU, Lieber CM. Multiplexed electrical detection of cancer markers with nanowire sensor arrays. Nat. Biotechnol. 2005;23:1294–1301. doi: 10.1038/nbt1138. [DOI] [PubMed] [Google Scholar]
- 46.Archakov AI, Ivanov YD. Analytical nanobiotechnology for medicine diagnostics. Mol Biosyst. 2007;3(5):336–342. doi: 10.1039/b618285b. [DOI] [PubMed] [Google Scholar]
- 47.Sakamoto JH, van de Ven AL, Godin B, Blanco E, Serda RE, Grattoni A, Ziemys A, Bouamrani A, Hu T, Ranganathan SI, De Rosa E, Martinez JO, Smid CA, Buchanan RM, Lee SY, Srinivasan S, Landry M, Meyn A, Tasciotti E, Liu X, Decuzzi P, Ferrari M. Enabling individualized therapy through nanotechnology. Pharmacol Res. 2010 Jan 5; doi: 10.1016/j.phrs.2009.12.011. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hamburg MA, Collins FS. The Path to Personalized Medicine. N Engl J Med. 2010 Jun 15; doi: 10.1056/NEJMp1006304. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 49.Workman P. Strategies for treating cancers caused by multiple genome abnormalities: from concepts to cures? Curr Opin Investig Drugs. 2003;4(12):1410–1415. [PubMed] [Google Scholar]
- 50.Nibbe RK, Chance MR. Approaches to biomarkers in human colorectal cancer: looking back, to go forward. Biomark Med. 2009;3(4):385–396. doi: 10.2217/BMM.09.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jolly Graham A, Potti A. Translating genomics into clinical practice: applications in lung cancer. Curr Oncol Rep. 2009;11(4):263–268. doi: 10.1007/s11912-009-0037-z. [DOI] [PubMed] [Google Scholar]
- 52.Verhaak RG, Hoadley KA, Purdom E, Wang V, Qi Y, Wilkerson MD, Miller CR, Ding L, Golub T, Mesirov JP, Alexe G, Lawrence M, O'Kelly M, Tamayo P, Weir BA, Gabriel S, Winckler W, Gupta S, Jakkula L, Feiler HS, Hodgson JG, James CD, Sarkaria JN, Brennan C, Kahn A, Spellman PT, Wilson RK, Speed TP, Gray JW, Meyerson M, Getz G, Perou CM, Hayes DN Cancer Genome Atlas Research Network. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17(1):98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cancer Genome Atlas Research Network. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455(7216):1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hodgson JG, Yeh RF, Ray A, Wang NJ, Smirnov I, Yu M, Hariono S, Silber J, Feiler HS, Gray JW, Spellman PT, Vandenberg SR, Berger MS, James CD. Comparative analyses of gene copy number and mRNA expression in glioblastoma multiforme tumors and xenografts. Neuro Oncol. 2009;11(5):477–487. doi: 10.1215/15228517-2008-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.International Cancer Genome Consortium. International network of cancer genome projects. Nature. 2010;464(7291):993–998. doi: 10.1038/nature08987. [DOI] [PMC free article] [PubMed] [Google Scholar]