Abstract
What mechanisms underlie the transitions responsible for the diverse shapes observed in the living world? While bacteria display a myriad of morphologies1, the mechanisms responsible for the evolution of bacterial cell shape are not understood. We investigated morphological diversity in a group of bacteria that synthesize an appendage-like extension of the cell envelope called the stalk2,3. The location and number of stalks varies among species, as exemplified by three distinct sub-cellular positions of stalks within a rod-shaped cell body: polar in the Caulobacter genus, and sub-polar or bi-lateral in the Asticcacaulis genus4. Here we show that a developmental regulator of Caulobacter crescentus, SpmX5, was co-opted in the Asticcacaulis genus to specify stalk synthesis at either the sub-polar or bi-lateral positions. We show that stepwise evolution of a specific region of SpmX led to the gain of a new function and localization of this protein, which drove the sequential transition in stalk positioning. Our results indicate that evolution of protein function, co-option, and modularity are key elements in the evolution of bacterial morphology. Therefore, similar evolutionary principles of morphological transitions apply to both single-celled prokaryotes and multicellular eukaryotes.
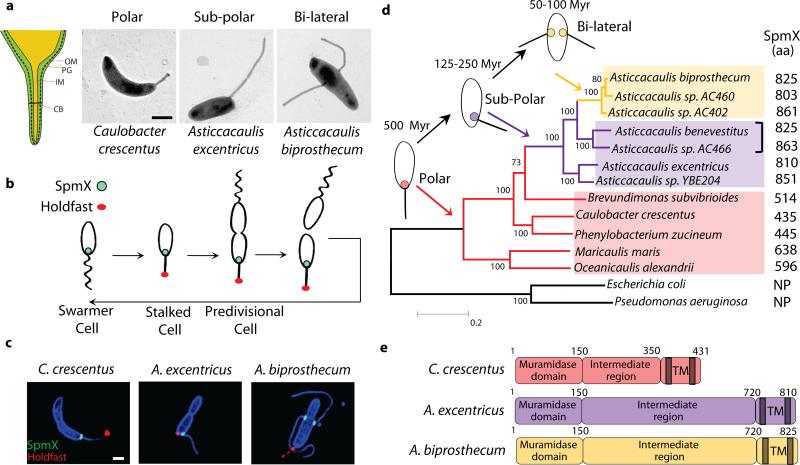
Stalks are a common feature in aquatic bacterial species living in oligotrophic environments3,6. When these species are subjected to nutrient limitation, stalks elongate to increase the effective length and surface area of the cells7, thereby increasing the rate of nutrient uptake2,8. The thin cylindrical stalk is composed of inner and outer membranes separated by peptidoglycan6, and compartmentalized by proteinaceous structures called “cross-bands”9,10 (Fig. 1a). In the Caulobacteraceae family, stalk synthesis occurs at a specific stage of a dimorphic life cycle in which a non-replicating motile swarmer cell differentiates into a sessile stalked cell11 (Fig. 1b). In C. crescentus, the stalk is positioned at a single cell pole; in Asticcacaulis excentricus, the stalk is synthesized at a sub-polar position off-center of a cell pole; and in Asticcacaulis biprosthecum, two stalks are positioned bi-laterally on the cell body4 (Fig. 1a).
Figure 1.
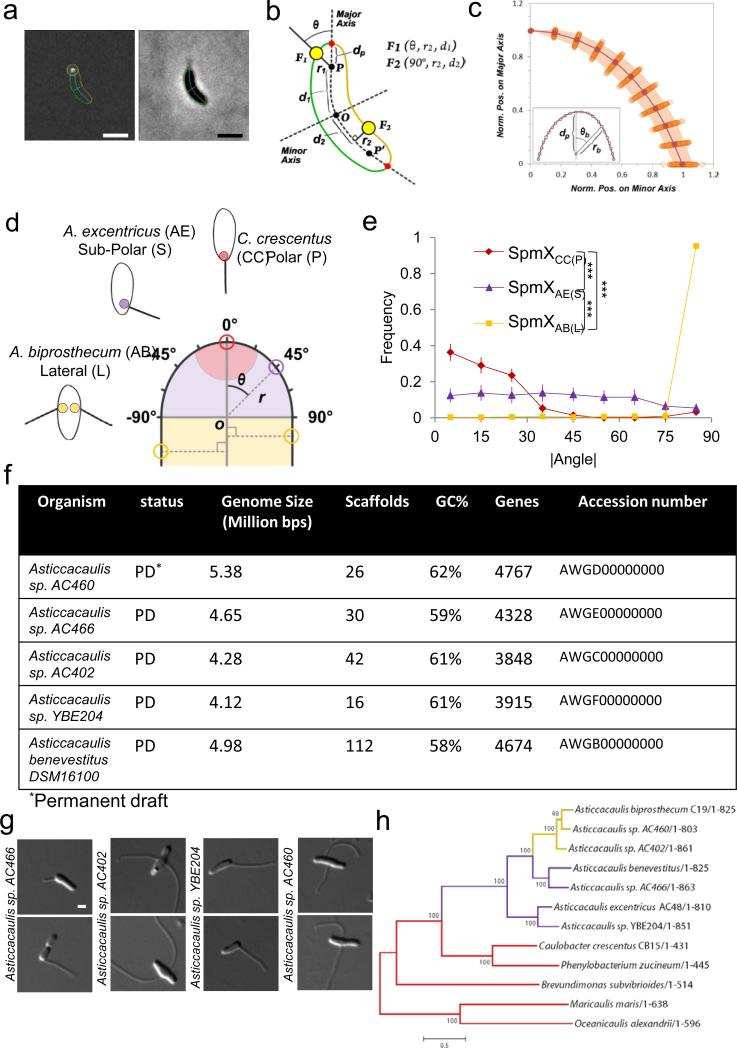
Natural variation and evolution of stalk positioning correlates with SpmX localization. (a) Schematic of the stalk (left). OM, outer membrane; PG, peptidoglycan; IM, inner membrane; CB, cross-band. Transmission electron micrographs of representative species (right). (b) Dimorphic life cycle of Caulobacter crescentus. SpmX (green) and holdfast (red). (c) Structured illumination microscopy images of cells with outer membrane protein stain Pacific Blue SE (blue), SpmX-EGFP (green), and fluorescent lectin bound holdfast (red). Data are representative of three biological repetitions. Scale bars, 1 μm. (d) Phylogenetic tree and inferred evolutionary trajectory of stalk positioning with the predicted origin of morphology calibrated by 16S rRNA identity26 (Myr, million years). Colors of shading, branches, and SpmX (filled circles) denote the polar (red), sub-polar (purple), and bi-lateral (yellow) stalk positioning, respectively. The size of SpmX is indicated in amino acids (aa). NP, orthologs not present. Bracket indicates sub-polar stalked species that share the last common ancestor with the A. biprosthecum clade. Scale bar, number of substitutions per site. (e) Domain organization of SpmX, transmembrane domains (TM) are shown as grey bars.
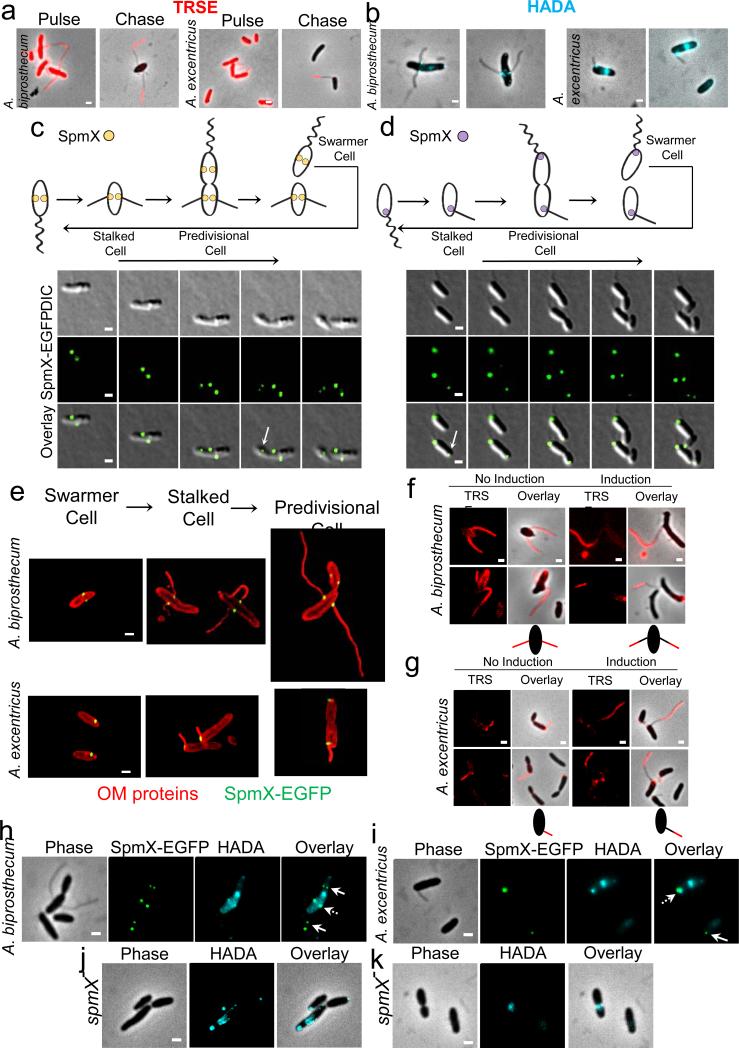
The natural variation in stalk location provides an opportunity to study the mechanisms underlying the precise targeting of cell envelope growth zones to generate different morphologies. Stalks in C. crescentus are synthesized from their base12 by insertion of peptidoglycan within a small area of the cell body13,14. To test whether this mechanism is conserved in the Asticcacaulis genus, we used pulse-chase labeling with Texas Red Succinimidyl Ester (TRSE)15,16 to study cell envelope growth and a fluorescent D-amino acid (FDAA) to label regions of peptidoglycan synthesis13. The stalks of A. excentricus and A. biprosthecum are also synthesized by insertion of peptidoglycan at their base (Extended Data Fig. 1a and b), suggesting that all three species share the same stalk synthesis mechanism.
Extended Data Figure 1.
SpmX localization precedes and is required for stalk synthesis in Asticcacaulis. (a) Stalk synthesis occurs at the junction between the cell body and stalk in A. biprosthecum and A. excentricus. Cell surface proteins were pulse-labeled with TRSE, excess TRSE was removed and cells were allowed to grow for 7-8 doublings. The cell body and cell body proximal stalk label was diluted by cell elongation, whereas the cell body distal part of the stalk remained stained, indicating that new material is incorporated at the base of the stalk in both A. biprosthecum (left) and A. excentricus (right). (b) Fluorescent D-amino acids track stalk peptidoglycan synthesis to its base. Cells were stained with the fluorescent D-amino acid (FDAA) 7-hydroxycoumarin-amino-D-alanine (HADA) (blue), which labels regions of peptidoglycan synthesis, in A. biprosthecum (left) and A. excentricus (right). (c-d) SpmX localization precedes stalk synthesis in Asticcacaulis. Time lapse microscopy tracks the dynamics of native SpmX-EGFP localization in A. biprosthecum (c) and in A. excentricus (d), images taken every 50 (c) and 30 (d) minutes, respectively. Arrows indicate the early localization of SpmX-EGFP in the swarmer compartment of pre-divisional cells. Schematics of dynamic localization patterns are shown on top. (e) Structured illumination micrographs of the localization of SpmX-EGFP at different stages of the life cycle in A. biprosthecum (top) and in A. excentricus (bottom). Cells were stained with the red outer membrane protein stain TRSE and SpmX-EGFP was expressed from its native chromosomal locus. SpmX-EGFP localizes at the future position of stalk synthesis in the daughter swarmer cell compartment of the pre-divisional cell. (f-g) SpmX is required for stalk elongation in A. biprosthecum (f) and A. excentricus (g). Cells expressing a xylose-inducible spmX allele were depleted of SpmX and the cell body and stalks were stained with the red outer membrane protein stain TRSE. Cells were then grown in the presence or absence of the xylose inducer. Schematics of the pulse-chase results are shown below each group. (h-i) SpmX is required for the initiation of stalk peptidoglycan synthesis. Cells were stained for 5 min with HADA. The blue HADA staining correlates with SpmX-EGFP at the base of the stalks in A. biprosthecum (h) and A. excentricus (i). Dashed arrows indicate where peptidoglycan synthesis co-localizes with the SpmX-EGFP foci at the base of the stalks. Arrows indicate the localization of SpmX-EGFP in the swarmer cell or the swarmer cell compartment. (j-k) HADA staining of the spmX− mutant in A. biprosthecum (j) and A. excentricus (k). HADA staining no longer produces the foci seen at the base of stalks in wild-type strains. All data are representative of at least two biological repetitions analyzing at least 100 cells each. Scale bars, 1 μm.
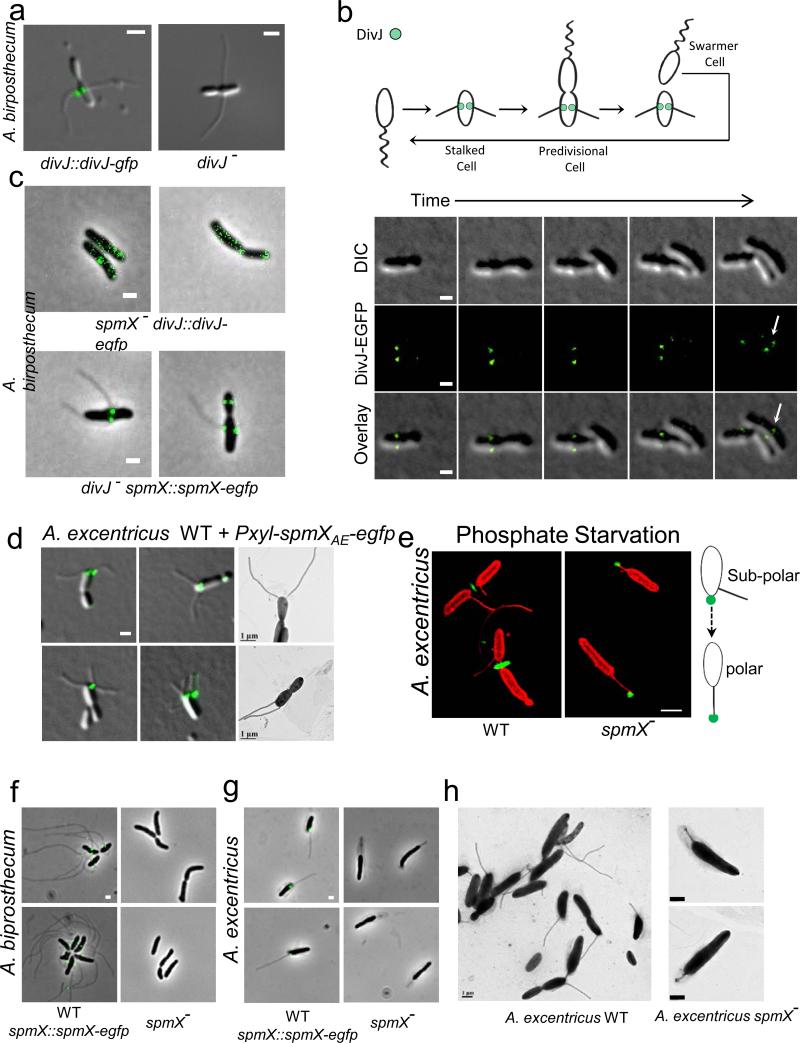
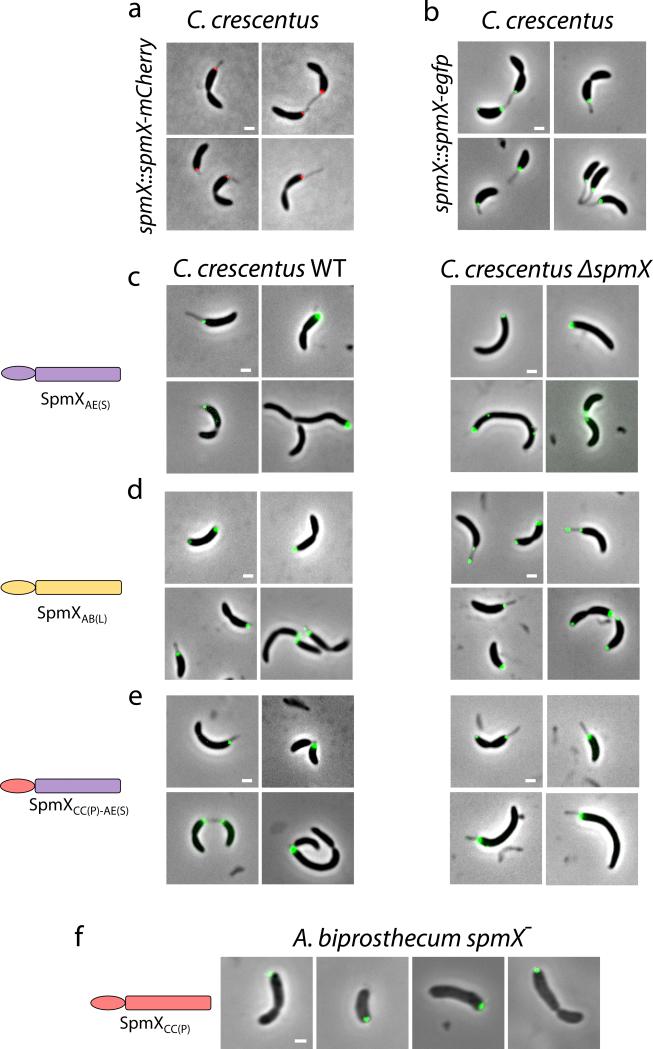
In light of the above results, we hypothesized that if a conserved stalk morphogen exists, it must localize to the base of stalks. Since many proteins localize at the pole in C. crescentus17, we took advantage of the non-polar localization of the stalks in the Asticcacaulis genus to identify stalk morphogen candidates. We constructed fluorescent protein fusions to orthologs of the pole-localized proteins from C. crescentus DivJ, PleC, PopZ, and SpmX and analyzed their localization in A. biprosthecum. Strikingly, only the regulatory histidine kinase DivJ18 (Extended Data Fig. 2a) and its localization and activation factor SpmX5 (Fig. 1c) localized at the base of the stalks in A. biprosthecum. During the cell cycle, A. biprosthecum DivJ-EGFP localized at the base of stalks only after cytokinesis, during swarmer to stalked cell differentiation (Extended Data Fig. 2b). In stark contrast, SpmX-EGFP localized to bilateral positions in the incipient swarmer half of the predivisional cell prior to cytokinesis and subsequent stalk synthesis (Extended Data figure 1c and e). Therefore SpmX localization precedes both DivJ localization and stalk synthesis, potentially marking the future site of stalk synthesis.
Extended Data Figure 2.
DivJ localizes to the base of the stalk and the effects of SpmX overexpression and phosphate starvation on stalk synthesis. (a) DivJ localizes to the base of the stalk (left) and the divJ− mutant still synthesizes stalks (right) in A. biprosthecum. (b) Localization dynamics of DivJ-EGFP tracked by time-lapse microscopy, images taken every 80 minutes. Note that DivJ-EGFP only localizes in the daughter cell after cytokinesis (arrow). (c) SpmX is required to localize DivJ (top) but DivJ is not required to localize SpmX (bottom) in A. biprosthecum. (d) Micrographs of wild-type A. excentricus cells overexpressing SpmX-EGFP. (e) Structured illumination micrographs of wild-type (left) and spmX− (right) A. excentricus cells growing under low phosphate conditions. Cells (red) are stained with TRSE and the polar holdfast (green) is stained with a fluorescent WGA lectin. Phosphate starvation induces polar stalk synthesis in the A. excentricus spmX− mutant, pushing the holdfast to its tip as shown on the right schematic. (f-g) Micrographs of wild-type (left, with SpmX-EGFP) and spmX− (right) A. biprosthecum (f) or A. excentricus (g) grown under phosphate starvation. (h) Transmission electron micrographs of A. excentricus grown under phosphate starvation. The diffuse structure around the cell body and the stalk is a sheath whose synthesis is induced in response to environmental stress39. All data are representative of at least two biological repetitions analyzing at least 100 cells each. Scale bars, 1 μm.
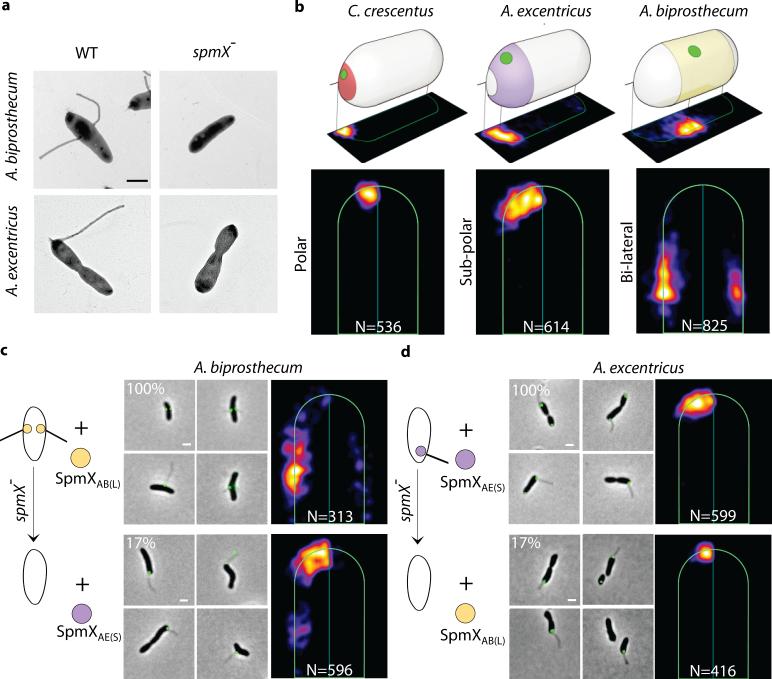
Interestingly, while the A. biprosthecum divJ− mutant still synthesized bi-lateral stalks (Extended Data Fig. 2a), the A. biprosthecum spmX− mutant was stalkless (Fig. 2a). Moreover, while newly synthesized peptidoglycan material co-localized with SpmX-GFP in wild-type cells, no bi-lateral foci of FDAA staining were observed in the absence of SpmX (Extended Data Fig. 1h and j), demonstrating that SpmX is required for stalk peptidoglycan synthesis in A. biprosthecum. Finally, stalk elongation only occurred when SpmX was expressed (Extended Data Fig. 1f), suggesting that SpmX is required both for the initiation and the elongation of stalk synthesis in A. biprosthecum. Similar results were obtained for A. excentricus (Fig. 1c, middle; Extended Data Fig. 1d, e, g, i, and k), suggesting that the role of SpmX is conserved in both Asticcacaulis species. Notably, SpmX is not required for stalk synthesis in C. crescentus5. Since the Caulobacter genus diverged earlier than the Asticcacaulis genus (Fig. 1d), we conclude that SpmX has been co-opted for stalk synthesis in the Asticcacaulis genus. However, despite its newly acquired role in stalk synthesis, the ancestral function of SpmX in DivJ localization has been retained in A. biprosthecum (Extended Data Fig. 2c).
Figure 2.
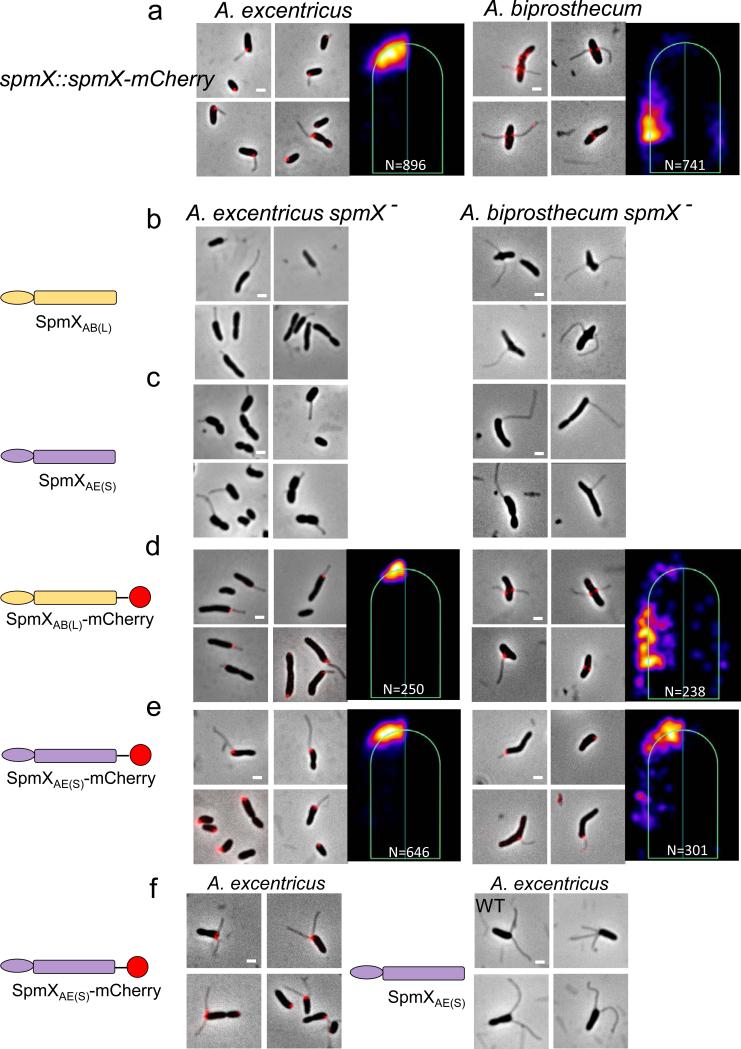
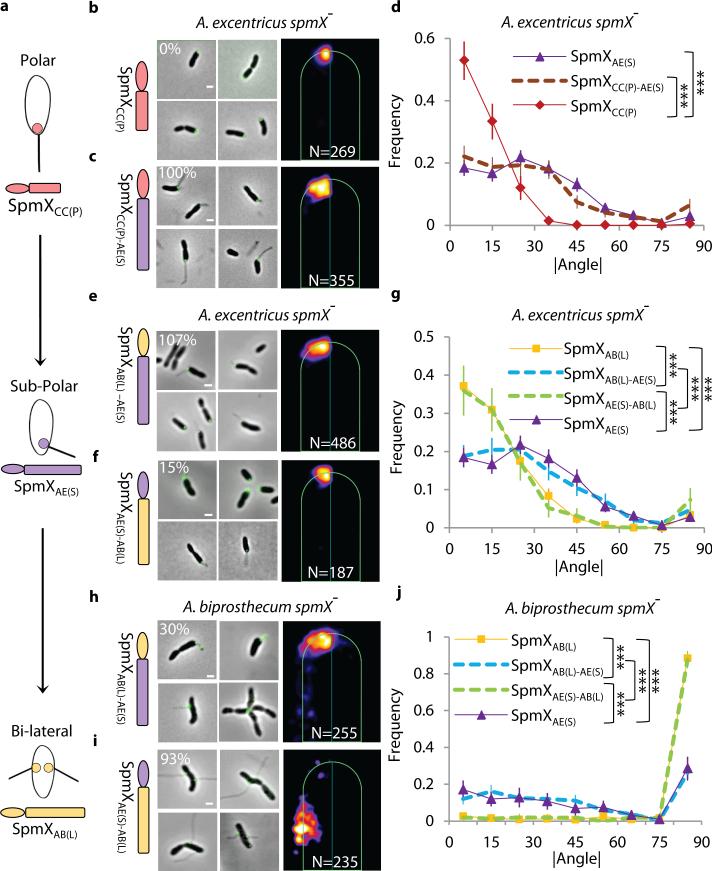
SpmX specifies the location of stalk synthesis in Asticcacaulis. (a) SpmX is required for stalk synthesis in Asticcacaulis. Transmission electron microscopy images of Asticcacaulis species and their respective spmX− mutants. Data are representative of five biological repetitions. (b) Heat maps of the localization patterns of SpmX-EGFP in three species with differentially positioned stalks. The number (N) of foci quantified is shown at the bottom of each map. For simplicity throughout the paper, we use SpmXCC(P), SpmXAE(S) and SpmXAB(L) to denote SpmX from C. crescentus (CC), A. excentricus (AE), and A. biprosthecum (AB), with the subscripts P, S, and L indicating their native polar, sub-polar, and lateral positioning, respectively. (c-d) Microscopy images and heat maps of the A. biprosthecum and A. excentricus spmX− mutant expressing SpmXAB(L)-EGFP or SpmXAE(S)-EGFP. The percentage represents the stalk synthesis ability for each strain compared to the control spmX− mutant expressing native SpmX-EGFP.
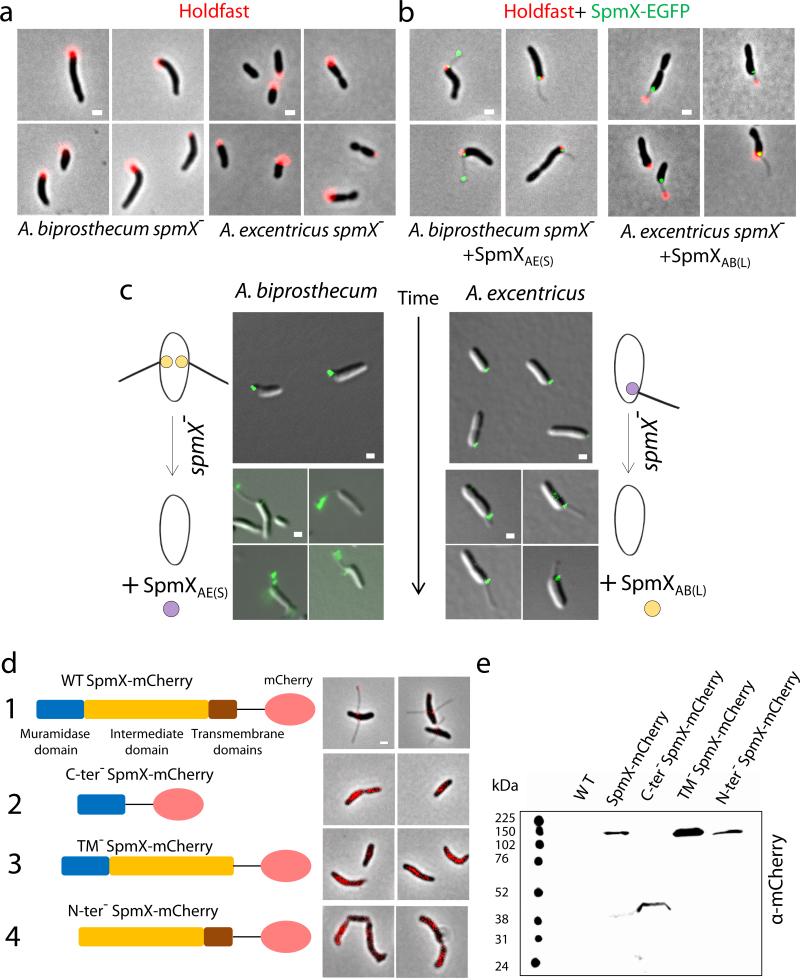
To test the hypothesis that SpmX could play a pivotal role in the evolutionary transitions in stalk positioning, we carried out cross-complementation experiments by expressing heterologous SpmXs and SpmX fusions in wild-type or spmX mutant strains of the two Asticcacaulis species and quantitatively analyzed SpmX localization. (Fig. 2 and Extended Data Fig. 3-6). When we cross-complemented SpmX-EGFP in either the homologous or heterologous wild-type backgrounds, SpmX both localized and drove stalk synthesis at its host-specific location, suggesting that the endogenous SpmX may be able to recruit the heterologous SpmX (Extended Data Fig. 4b, c, h, and i). To test this possibility, we expressed heterologous SpmX in absence of the native spmX gene. Strikingly, when SpmX from the sub-polar stalked species A. excentricus (SpmXAE(S)-EGFP) was expressed in the bi-lateral stalked species A. biprosthecum spmX− mutant, it localized to and drove stalk synthesis at a sub-polar position (Fig. 2c; Extended Data Fig. 7). Therefore, A. excentricus SpmX can recruit the heterologous stalk synthesis machinery of A. biprosthecum to synthesize a stalk at an ectopic sub-polar position. In contrast, when SpmX from the bi-lateral stalked species A. biprosthecum (SpmXAB(L)-EGFP) was expressed in the sub-polar stalked species A. excentricus spmX− mutant, it localized mostly to poles where it induced stalk synthesis (Fig. 2d; Extended Data Fig. 7). These results indicate that while the sub-polar positional information exists in A. biprosthecum and can be recognized by SpmXAE(S), the specific bi-lateral positional information present in A. biprosthecum is absent or not recognizable in A. excentricus.
Extended Data Figure 3.
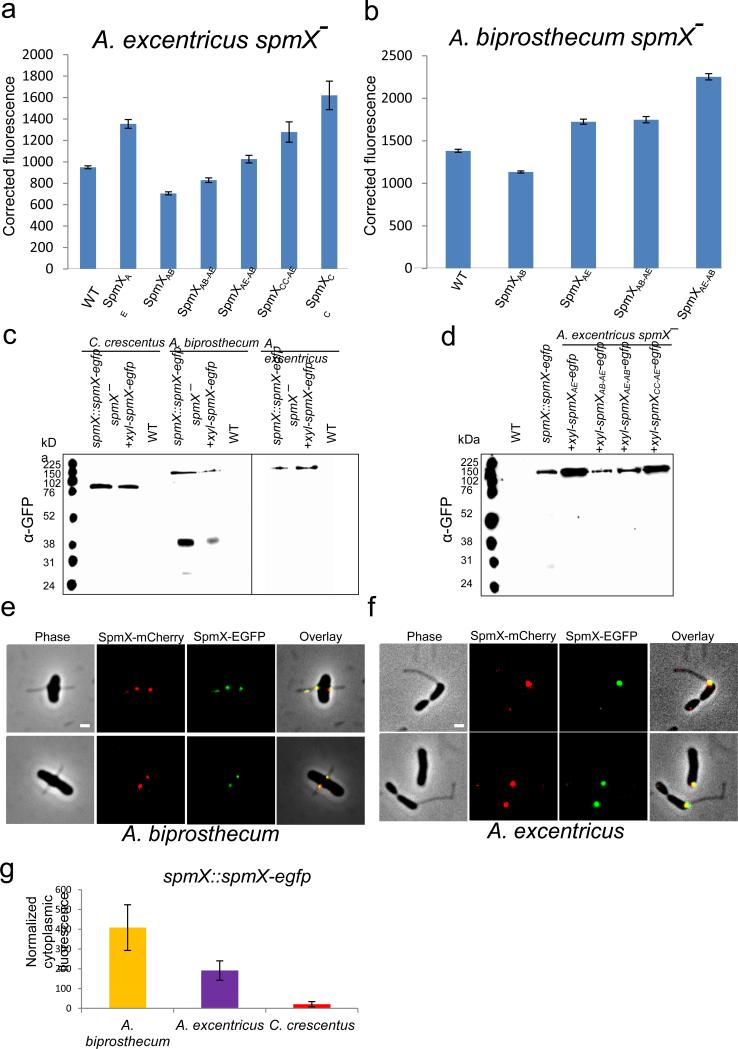
Expression and integrity of various SpmX-EGFP fusions. (a-b) Expression level of SpmX-EGFP in strains used in this study. All fusion proteins were expressed from a xylose-inducible promoter on a replicating plasmid in the presence of 0.05% (w/v) xylose. SpmX-EGFP expression level was measured by quantifying the fluorescence intensity of the fusion proteins. Corrected fluorescence is calculated as (integrated fluorescence-integrated background)/area. Measurements for each strain were done in duplicate and at least 500 cells were quantified in each case. Error bars denote standard error of the mean. (c) Western blot of SpmX-EGFP fusion proteins expressed from either the chromosomal locus or a replicating plasmid in different species probed with anti-GFP antibody. Wild-type strains were used as controls. Note that only SpmXAB-EGFP appears to have a clipping/degradation pattern. Data are representative of three biological repetitions. (d) Western blot of various chimeric/heterologous SpmX-EGFP fusions when expressed in the A. excentricus spmX− mutant, probed by anti-GFP antibody. Note that the relative levels of expression correlate with the results from quantitative fluorescence analysis (a). The size of SpmX-EGFP fusions ranges from 76 kDa to 120 kDa and free EGFP is expected to be around 31 kDa. Data are representative of three biological repetitions. (e-f) SpmX-mCherry and SpmX-EGFP share the same localization pattern in A. biprosthecum (left, Pearson r=0.79±0.1) and A. excentricus (right, Pearson r=0.81±0.08) cells. Both strains are expressing SpmX-EGFP fusions from the chromosomal locus and the SpmX-mCherry fusions from a replicating plasmid. Scale bars, 1 μm. Data are representative of two biological repetitions. (g) The elevated cytoplasmic fluorescence in A. biprosthecum may correlate with the clipping/degradation pattern of SpmXAB-EGFP. Data are representative of two biological repetitions analyzing at least 100 cells each. Error bars, standard deviation.
Extended Data Figure 6.
SpmX fusion proteins are functionally equivalent to the native SpmXs. (a) Microscopic images and heatmaps of A. excentricus (left) or A. biprosthecum (right) expressing SpmX-mCherry fusions from the chromosomal locus. Note that the localization as well as the function in stalk synthesis is identical to that of the respective spmX::spmX-egfp and wild-type strains (Fig. 1 and 2). (b-c) Microscopic images of the A. excentricus spmX− (left) and A. biprosthecum spmX− (right) strains expressing non-tagged SpmXs. Note that SpmXAB(L) induces mostly polar stalks in the A. excentricus spmX− mutant (b) and SpmXAE(S) induces mostly sub-polar stalks in the A. biprosthecum spmX− mutant (c). Both SpmXs were able to complement the stalkless phenotype. (d-e) Microscopic images and heatmaps of expression of SpmX-mCherry fusions in the A. excentricus spmX− (left) and A. biprosthecum spmX− (right) mutants. The phenotypes are identical to that of the non-tagged SpmXs (as well as SpmX-EGFPs in Fig. 3). (f) Expression of SpmXAE(S)-mCherry (left) and SpmXAE(S) (right) in wild-type A. excentricus both induce multiple stalk synthesis, as is the case for SpmX-EGFP (Extended Data Fig. 2). All data are representative of three biological repetitions analyzing at least 100 cells each. Scale bars, 1 μm.
Extended Data Figure 4.
Localization of SpmX-EGFP fusions in various strain backgrounds. Full length or chimeric SpmX-EGFPs were expressed in wild-type A. biprosthecum and A. excentricus as indicated on the left. Representative pictures are shown. All data are representative of three biological repetitions analyzing at least 100 cells each. Scale bars, 1 μm.
Extended Data Figure 7.
SpmX is required for stalk positioning in Asticcacaulis and the integrity of SpmX is critical for its function in A. biprosthecum. (a) Holdfast (red) localizes to the pole in both A. biprosthecum (left) and A. excentricus (right) spmX− mutants. (b) Dual-labeling images of SpmX-EGFP (green) and holdfast (red) in the A. biprosthecum spmX− mutant complemented by SpmXAE(S) (left) and the A. excentricus spmX− mutant complemented by SpmXAB(L) (right). (c) SpmXAE(S) localizes and initiates stalk synthesis at the sub-polar position in the A. biprosthecum spmX− mutant (left). SpmXAB(L) localizes to polar or sub-polar locations before triggering either polar and sub-polar stalk synthesis in the A. excentricus spmX− mutant (right). We noticed that SpmX-EGFP sometimes localizes in the stalk, likely due to stalk outgrowth from its base since it should not be able to diffuse to positions in already synthesized stalks, where diffusion is constrained by cross-bands9 (Fig. 1a). (d) From top to bottom, 1. SpmX-mCherry (825aa+mCherry, 121 kDa) localizes to the base of stalks. This fusion protein was expressed from its native chromosomal locus (spmX::spmX-mcherry). 2-4. Different truncated alleles of SpmX fail to complement the stalkless phenotype of the A. biprosthecum spmX− mutant. Both C-ter− SpmX-mCherry (1-150aa+mCherry, 45 kDa) and TM− SpmX-mCherry (1-750aa+mCherry, 111 kDa) were expressed from the native spmX chromosomal locus replacing the natural allele. N-ter− SpmX-mCherry (150-825aa+mCherry, 104 kDa) was expressed from a replicating plasmid (YB7129) in the A. biprosthecum spmX− mutant. (e) Western blot of truncated SpmX-mCherry fusions detected with anti-mCherry polyclonal antibodies. Data are representative of two biological repitions. Micrographs are representative of three biological repetitions analyzing at least 100 cells each. Scale bars, 1 μm.
Remarkably, these observations also suggest that A. excentricus possesses the ability to synthesize polar stalks in the absence of its endogenous SpmX. Indeed, phosphate starvation, which stimulates stalk synthesis in wild-type strains of all three species (Extended Data Fig. 2f and g), rescued stalk synthesis in A. excentricus spmX− cells, but stalks were located at the pole (Extended Data Fig. 2e, g, and h). Using holdfast polysaccharide adhesin as a polar marker (Fig. 1c), we found that stalks from phosphate starved A. excentricus spmX− cells were tipped by a holdfast (Extended Data Fig. 2e), confirming that they were synthesized polarly.We infer that A. excentricus possesses an alternative polar stalk synthesis mechanism that is normally masked by the endogenous SpmX-driven sub-polar stalk synthesis mechanism (as detailed in Supplementary Information). In contrast, the A. biprosthecum spmX− mutant remained stalkless when starved for phosphate (Extended Data Fig. 2f), indicating that the spmX− independent pathway for stalk biosynthesis has been lost in A. biprosthecum, or is no longer regulated by phosphate starvation. We conclude that both Asticcacaulis species possess the ability to synthesize stalks at exogenous positions, which is masked by the effects of the endogenous SpmX in wild-type cells. We hypothesized that the exogenous positions of stalk synthesis are phylogenetically ancestral, and we next sought to infer the evolutionary trajectory of stalk positioning.
In order to improve the phylogenetic resolution of stalk positioning, we sequenced the genomes of several additional Asticcacaulis strains (Extended Data Fig. 5f and g) and inferred their phylogeny (see materials and methods; Fig. 1d). Based on parsimony, the emergence of the polar stalk morphology occurred before the divergence between the Caulobacteraceae and the Hyphomonadaceae family (Maricaulis maris and Oceanicaulis alexandrii) (Fig. 1d). No known Asticcacaulis isolates synthesize polar stalks, implying that the transition in stalk positioning from polar to sub-polar occurred very early. In addition, two sub-polar stalked strains, Asticcacaulis benevestitus and Asticcacaulis sp. AC466 (Fig. 1d, bracket), diverged from the same ancestor that led to the sub-clade containing A. biprosthecum, indicating that sub-polar stalk synthesis is ancestral to bi-lateral stalk synthesis. In conclusion, stalk positioning evolved from an ancestral single polar stalk to a single sub-polar stalk, and subsequently to bi-lateral stalks.
Extended Data Figure 5.
Overview of the sub-cellular localization quantification method and characteristics of newly sequenced genomes. (a) Output images provided by the customized ImageJ-based software package. Fluorescent (left) and corresponding phase contrast (right) images are shown. The cell boundary is represented in green or orange according to the side of the cell. The major and the minor axes of the cell are represented in cyan. In each cell, fluorescent foci of SpmX-GFP (white spot, left panel) were detected and outlined by a yellow circle centered to their respective sub-pixel resolution positions. The dark blue line links the focus coordinates to its relative position on the major axes. (b) Schematic representation of the polar coordinate system in which each point is defined by three coordinates: the distance from the pole or from the major axis (r1, r2), the angle relative to the major axis (θ1, θ2) and the distance from the mid-cell (d1, d2). A null angular coordinate means that the focus is localized at the tip of the cell pole, whereas 90° means that the focus is localized on cell sides. dP represents the distance between the pole (P or P’) and the cell boundary (red dot). F1 and F2 represent sub-polar and lateral localized objects with their respective coordinates. (c) Variation of the position of discrete points on the cell boundary relative to the pole (P or P’) as determined by the polar coordinates (n=100). Each point represents the normalized position on the major (rb/dp × cos(θb)) and the minor (rb/dp × sin(θb)) axis of discrete positions along the cell boundary in the pole region. The shaded region indicates the interquartile range (IQR) as a measure of the dispersion between the upper and lower quartiles of values observed for discrete values of θb. (d) Schematic showing how localization of SpmX-EGFP is quantified by measuring the angle (θ) and the radial distance (r) of each focus using the geometric center of the pole as the origin of a polar coordinate system within a normalized cell body (see a-c). Red, purple, and yellow colored circles represent SpmX in C. crescentus (CC), A. excentricus (AE), and A. biprosthecum (AB), respectively. P, S, and L in parentheses are shorthand to denote the native polar, sub-polar, and lateral stalk positioning of their respective SpmX proteins. (e) Density plot of the absolute values of measured angles (|θ|) of heatmaps in Figure 2b (same N). Measurements are binned for every 10 degrees. Error bars in the angle profiles denote standard deviation of the sample evaluated by the Jackknifing method (see Methods). (f) The general characteristics of five Asticcacaulis genomes. PD denotes the permanent draft status of the genome. (g) DIC micrographs of the sequenced strains in (f) except for A. benevestitus, which was previously published39. Scale bars, 1 μm. (h) Phylogenetic tree inferred from SpmX sequences of different species. The alignment from Extended Data Figure 8 was used to infer this tree based on the maximum likelihood method. Note that since E. coli and P. aeruginosa do not have spmX orthologs, species from the Hyphomonadaceae family serve as the outgroup (Maricaulis maris and Oceanicaulis alexandrii). We estimated the statistical support for each node by performing 500 bootstrap repetitions. The overall SpmX protein tree topology matches that of the species tree (Fig. 1d). The only exception is that Brevundimonas subvibriodes is placed differently, due to the difficulty of resolving its phylogeny40. Scale, number of substitutions per position.
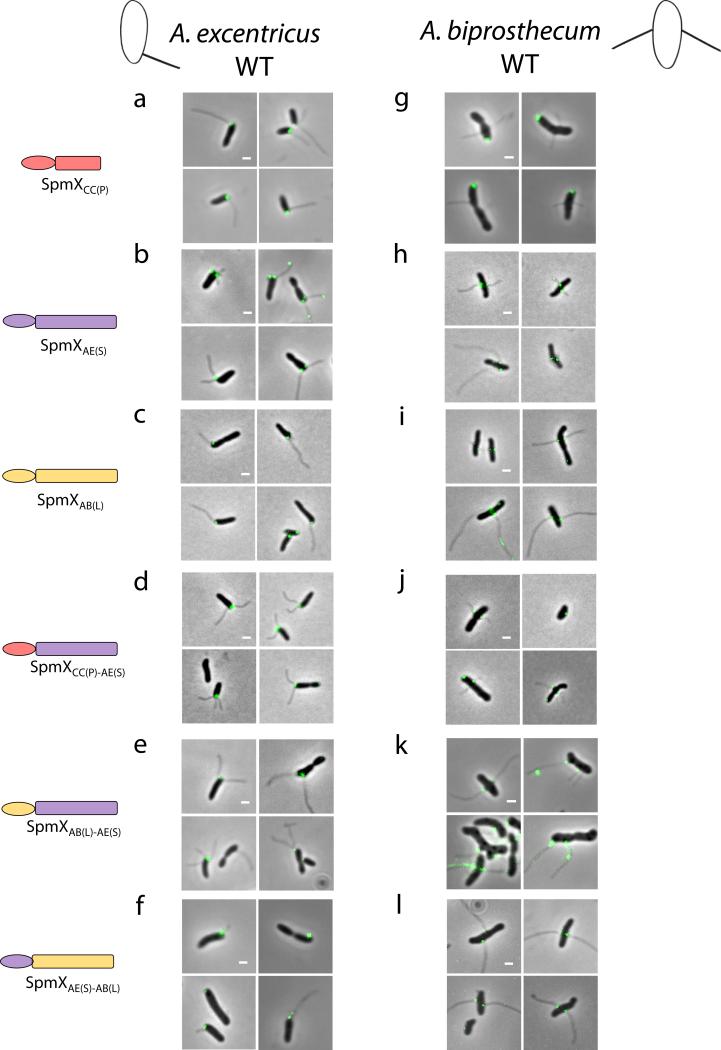
We next sought to understand how SpmX has evolved at the protein level by testing the requirement of its major domains for localization and stalk synthesis (Fig. 1e and Extended Data Fig. 8). We constructed a set of truncated alleles removing various domains of A. biprosthecum SpmX, which failed to localize or rescue the stalkless phenotype of the A. biprosthecum spmX− mutant (Extended Data Fig. 7d and e). The muramidase domain and the C-terminal region (intermediate region and transmembrane domains) of SpmX are indispensable for its localization and function. To determine what region of SpmX evolved to specify the location of stalk synthesis, we constructed chimeric SpmX proteins by mixing and matching the muramidase and the C-terminal regions of different SpmX proteins. In each case, the phenotype of the spmX− mutants expressing the various chimeras correlated with the source of their C-terminal region (Fig. 3, Extended Data Fig. 4, and Supplementary Information). We conclude that mutations in the SpmX C-terminal region are responsible for the evolution of SpmX’s ability to drive stalk synthesis from polar to sub-polar to bi-lateral positions.
Extended Data Figure 8.
Alignment of SpmX sequences from species/strains used in this study (as shown in Fig. 1d). Sequences were aligned in Jalview (http://www.jalview.org/). Colored residues denote conservation across species. Vertical dashed lines denote the boundaries of the N-terminal muramidase domain (black) and the two transmembrane domains (red), respectively. Chimeras were constructed using the end of the muramidase domains as the boundary between N-terminal and C-terminal moieties (Fig. 3). Red arrows indicate the three major species used in this study: A. biprosthecum, A. excentricus, and C. crescentus, respectively. From top to bottom, the order of the sequences reflects the tree topology as deduced from Figure 1d. The colored vertical lines next to the names of the species/strains denote their respective stalk positioning as shown in Fig. 1d (yellow, bi-lateral; purple, sub-polar; red, polar). The numbers next to the names of the species/strains indicate the length of their respective SpmX proteins.
Figure 3.
Evolution of the C-terminal region of SpmX drives the morphological transition in stalk positioning. (a) Schematic of inferred evolutionary trajectory of stalk positioning. (b-g) Microscopy images, heat maps, and angle profile analysis for the A. excentricus spmX− mutant expressing SpmXCC(P) (b), the chimera SpmXCC(P)-AE(S) (c), SpmXAB(L)-AE(S) (e), and SpmXAE(S)-AB(L) (f). (h-j) Microscopy images, heat maps, and angle profile analysis for the A. biprosthecum spmX− mutant expressing the chimera SpmXAB(L)-AE(S) (h) and SpmXAE(S)-AB(L) (i). Percentages indicate stalk synthesis ability compared to the respective controls as in Fig. 2. Error bars in the angle profiles denote standard deviation of the sample evaluated by the Jackknifing method (see Methods). All profiles of absolute angles (|θ|) were generated by measuring the data points in associated heatmaps (same N) and were analyzed by non-parametric statistical methods, as detailed in Supplementary Table 1 and Methods. *** p<0.001. Scale bars, 1μm.
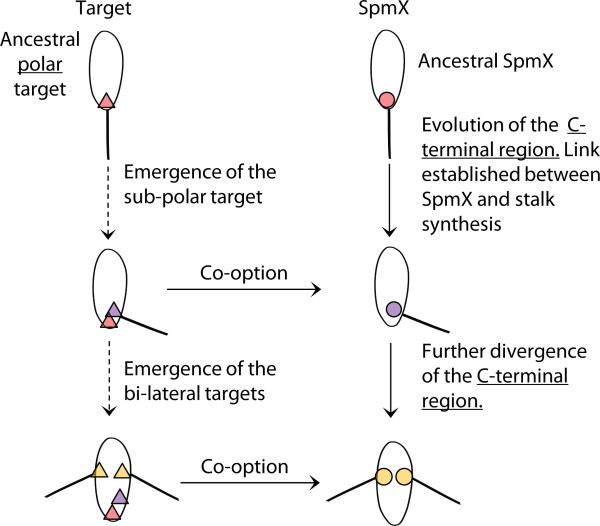
Morphological transitions generate the diversity of biological forms. A few cases have been studied in eukaryotes, highlighting the importance of both changes in regulatory sequences and functional protein evolution19-23. Our study has begun to unravel the elusive mechanisms of morphological transitions in bacteria by showing that evolution of the SpmX morphogen underlies the evolutionary trajectory of stalk positioning in the Caulobacteraceae family. Polar stalk synthesis arose from non-stalked species before the divergence of the Caulobacteraceae and Hyphomonadaceae families, but C. crescentus SpmX is not required for stalk synthesis, likely representing the ancestral state. Through differential protein evolution, changes in the SpmX C-terminal region led to stalk synthesis and positioning functions in the Asticcacaulis clade. Interestingly, the ancient polar targeting mechanism is conserved in Asticcacaulis since SpmXCC(P) can localize to the pole in both A. excentricus and A. biprosthecum (Fig. 3b, Extended Data Fig. 4a, g and Fig. 9f). Inversely, both SpmXAE(S) and SpmXAB(L) can still localize to the polar target in the C. crescentus strains, suggesting the lack of recognizable alternative targets (Extended Data Fig. 9). During the transition from polar to sub-polar stalk positioning, the C-terminal region of SpmX evolved to position and coordinate the synthesis of stalks, coupled with its co-option at the sub-polar target in A. excentricus (Fig. 4). Further divergence of the C-terminal region of SpmX led to its ability to recognize new targets, combined with its co-option at the bi-lateral targets in A. biprosthecum (Fig. 4).
Extended Data Figure 9.
Expression of SpmXAB(L)-EGFP or SpmXAE(S)-EGFP in C. crescentus does not induce ectopic stalk synthesis. (a-b) The localization of SpmXCC(P)-EGFP/SpmXCC(P)-mCherry and the phenotypes of the strains are identical. (c-e) Asticcacaulis SpmX-EGFP variants localize mostly to the pole in wild-type C. crescentus (left) and C. crescentus spmX− (right) strains. No ectopic stalks were observed, indicating that the alternative targets are lacking in C. crescentus. It should also be noted that the expression of variants of SpmXAE-EGFP can cause cell filamentation. (f) SpmXCC-EGFP localizes to the pole in the A. biprosthecum spmX− strain and cannot rescue stalk synthesis. All data are representative of three biological repetitions analyzing at least 100 cells each. Scale bars, 1μm.
Figure 4.
The co-option of SpmX in stalk synthesis leads to sequential morphological transitions. From polar to sub-polar stalk positioning, two events occurred: SpmX underwent an expansion of its C-terminal region and was co-opted for stalk synthesis, and the sub-polar target emerged in A. excentricus. Similarly, during the second transition of sub-polar to bi-lateral positioning, SpmX evolved the ability to recognize the bi-lateral target and the bi-lateral targets emerged in A. biprosthecum. The exact order of events for each transition is unknown. Targets (triangles) and SpmXs (circles) are shown with respective color-coding for the species.
Our results highlight the modular nature of the positioning mechanism that directs the zonal peptidoglycan synthesis responsible for stalk synthesis. This modularity is evident in both Asticcacaulis species, since SpmX always localizes at the base of ectopically synthesized stalks in several genetically engineered strains (Fig. 2 and 3 and Extended Data Fig. 2d). In addition, the fact that changes in the abundance of SpmX alone can alter the number of stalks in A. excentricus (Extended Data Fig. 2d) suggests that simple changes in the regulation of SpmX expression could drive the evolution of a species with multiple sub-polar stalks. Conceptually, to position the stalk around the cell body, the cells only need to evolve to the ability to localize SpmX to a new sub-cellular position, where it recruits the stalk synthesis module. This morphogenetic modularity could be exploited in synthetic biology to generate the optimal cell shape for a given process.
Finally, this study has demonstrated that functional evolution of a regulatory protein into a morphogenetic module made the evolution of stalk positioning possible, which in turn generated distinct cellular morphologies. Therefore, protein evolution, co-option, and modularity can drive morphological transitions in both single-celled prokaryotes and multicellular eukaryotes, contributing to the diversity of Darwin’s “endless forms most beautiful”24 from the microscopic to the macroscopic world.
Methods Summary
Caulobacter crescentus, Asticcacaulis excentricus, and Asticcacaulis biprosthecum strains were used in this study. Strains were grown in liquid PYE medium at 30°C for C. crescentus and 26°C for the Asticcacaulis strains. A detailed list of strains and plasmids and their methods of construction is provided in the Supplementary Information and the Methods section. For the quantitative analysis of fluorescent protein fusion localization, cells were incubated for 18 hours in the presence of inducer, mounted on a 1% (w/v) agarose pad and imaged. Quantitative sub-cellular localization of fluorescent protein fusions was performed at sub-pixel resolution using a specifically developed plug-in for ImageJ25. For bioinformatics analysis, orthologs of SpmX were identified using the BLAST suite hosted by NCBI and Integrated Microbial Genomes (IMG). Phylogenetic trees were generated using the maximum likelihood method, and a concatenation of the products of six housekeeping genes was used to infer the phylogeny of species involved. All methods are detailed in the Methods section.
Supplementary Material
Acknowledgements
We thank members of the Brun lab and Clay Fuqua for critical comments on the manuscript. We thank David Kysela, Velocity Hughes, and Vidhya Silvanose for their help and efforts in environmental sampling and phylogenetic analysis, Luting Zhuo and Chunfeng Huang for their help in statistical analysis, Sidney Shaw for his advice on quantitative image analysis, Michael Lynch and Rudolf Raff for insightful discussions on evolution, and Matthew Hahn and the Center for Genomics and Bioinformatics at Indiana University for their help in sequencing and bioinformatics analysis. We thank the Indiana University Light Microscopy Imaging Center for their help with OMX super resolution microscopy, supported by National Institutes of Health Grant S10RR028697-01, and the Indiana Molecular biology Institute (IMBI) Electron Microscopy facility at Indiana University for their help with electron microscopy. We thank Martin Thanbichler, Jeanne Poindexter, Paul Caccamo and Patrick Viollier for providing us with Caulobacter strain and plasmids, Jeanne Poindexter, Judy Peterson, John Lindquist, and Alvaro Quinones for help in locating the strain collection of the late Jack Pate from which we obtained some of the A. excentricus and A. biprosthecum strains used in this study, and Mark Wortinger, Sally Green, Ellen Quardokus, and Jennifer (Wagner) Herman for early work with Asticcacaulis that helped set the stage for this study. This work was supported by National Institutes of Health Grant GM051986, National Science Foundation Grant MCB0731950, and by a grant from the Indiana University Metabolomics and Cytomics Initiative (METACyt) program, which was funded, in part, by a major endowment from the Lilly Foundation. P.J.B.B. was supported by National Institutes of Health National Research Service Award AI072992.
Footnotes
Author Contributions
C.J., P.J.B.B., and Y.V.B. designed the experiments. C.J. and A.D. performed the research. C.J., P.J.B.B., A.D., and Y.V.B. analyzed and interpreted the data. C.J. and Y.V.B. wrote the paper. C.J., P.J.B.B., A.D., and Y.V.B. edited the paper.
Accession numbers for new genomic data
Genomic data of strains sequenced in this study are deposited in Genebank under the accession numbers: AWGD00000000 (Asticcacaulis sp. AC460), AWGE00000000 (Asticcacaulis sp. AC466), AWGC00000000 (Asticcacaulis sp. AC402) , AWGF00000000 (Asticcacaulis sp. YBE204), and AWGB00000000 (Asticcacaulis benevestitus DSM16100).
The authors declare no competing financial interests. Readers are welcome to comment on the online version of the paper.
References
- 1.Young KD. The selective value of bacterial shape. Microbiol Mol Biol Rev. 2006;70:660–703. doi: 10.1128/MMBR.00001-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wagner JK, Brun YV. Out on a limb: how the Caulobacter stalk can boost the study of bacterial cell shape. Mol Microbiol. 2007;64:28–33. doi: 10.1111/j.1365-2958.2007.05633.x. [DOI] [PubMed] [Google Scholar]
- 3.Stove-poindexter JL, Cohen-Bazire G. The Fine Structure of Stalked Bacteria Belonging to the Family Caulobacteraceae. J Cell Biol. 1964;23:587–607. doi: 10.1083/jcb.23.3.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pate JL, Ordal EJ. The fine structure of two unusual stalked bacteria. J Cell Biol. 1965;27:133–50. doi: 10.1083/jcb.27.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Radhakrishnan SK, Thanbichler M, Viollier PH. The dynamic interplay between a cell fate determinant and a lysozyme homolog drives the asymmetric division cycle of Caulobacter crescentus. Genes Dev. 2008;22:212–25. doi: 10.1101/gad.1601808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Poindexter JS. Biological Properties and Classification of the Caulobacter Group. Bacteriol Rev. 1964;28:231–95. doi: 10.1128/br.28.3.231-295.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gonin M, Quardokus EM, O'Donnol D, Maddock J, Brun YV. Regulation of stalk elongation by phosphate in Caulobacter crescentus. J Bacteriol. 2000;182:337–47. doi: 10.1128/jb.182.2.337-347.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wagner JK, Setayeshgar S, Sharon LA, Reilly JP, Brun YV. A nutrient uptake role for bacterial cell envelope extensions. Proc Natl Acad Sci U S A. 2006;103:11772–7. doi: 10.1073/pnas.0602047103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schlimpert S, et al. General protein diffusion barriers create compartments within bacterial cells. Cell. 2012;151:1270–82. doi: 10.1016/j.cell.2012.10.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hughes HV, et al. Protein localization and dynamics within a bacterial organelle. Proc Natl Acad Sci U S A. 2010;107:5599–604. doi: 10.1073/pnas.0909119107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brown PJ, Hardy GG, Trimble MJ, Brun YV. Complex regulatory pathways coordinate cell-cycle progression and development in Caulobacter crescentus. Adv Microb Physiol. 2009;54:1–101. doi: 10.1016/S0065-2911(08)00001-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schmidt JM, Stanier RY. The development of cellular stalks in bacteria. J Cell Biol. 1966;28:423–36. doi: 10.1083/jcb.28.3.423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kuru E, et al. In Situ probing of newly synthesized peptidoglycan in live bacteria with fluorescent D-amino acids. Angew Chem Int Ed Engl. 2012;51:12519–23. doi: 10.1002/anie.201206749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aaron M, et al. The tubulin homologue FtsZ contributes to cell elongation by guiding cell wall precursor synthesis in Caulobacter crescentus. Mol Microbiol. 2007;64:938–52. doi: 10.1111/j.1365-2958.2007.05720.x. [DOI] [PubMed] [Google Scholar]
- 15.de Pedro MA, Grunfelder CG, Schwarz H. Restricted Mobility of Cell Surface Proteins in the Polar Regions of Escherichia coli. J Bacteriol. 2004;186:2594–602. doi: 10.1128/JB.186.9.2594-2602.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brown PJ, et al. Polar growth in the Alphaproteobacterial order Rhizobiales. Proc Natl Acad Sci U S A. 2012;109:1697–701. doi: 10.1073/pnas.1114476109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Curtis PD, Brun YV. Getting in the loop: regulation of development in Caulobacter crescentus. Microbiol Mol Biol Rev. 2010;74:13–41. doi: 10.1128/MMBR.00040-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ohta N, Lane T, Ninfa EG, Sommer JM, Newton A. A histidine protein kinase homologue required for regulation of bacterial cell division and differentiation. Proc Natl Acad Sci U S A. 1992;89:10297–301. doi: 10.1073/pnas.89.21.10297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mallarino R, et al. Two developmental modules establish 3D beak-shape variation in Darwin's finches. Proc Natl Acad Sci U S A. 2011;108:4057–62. doi: 10.1073/pnas.1011480108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Loehlin DW, Werren JH. Evolution of shape by multiple regulatory changes to a growth gene. Science. 2012;335:943–7. doi: 10.1126/science.1215193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Werner T, Koshikawa S, Williams TM, Carroll SB. Generation of a novel wing colour pattern by the Wingless morphogen. Nature. 2010;464:1143–8. doi: 10.1038/nature08896. [DOI] [PubMed] [Google Scholar]
- 22.Chan YF, et al. Adaptive evolution of pelvic reduction in sticklebacks by recurrent deletion of a Pitx1 enhancer. Science. 2010;327:302–5. doi: 10.1126/science.1182213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ronshaugen M, McGinnis N, McGinnis W. Hox protein mutation and macroevolution of the insect body plan. Nature. 2002;415:914–7. doi: 10.1038/nature716. [DOI] [PubMed] [Google Scholar]
- 24.Darwin C. On the origin of species by means of natural selection. ix. Vol. 1. J. Murray; London: 1859. p. 502. [Google Scholar]
- 25.Collins TJ. ImageJ for microscopy. Biotechniques. 2007;43:25–30. doi: 10.2144/000112517. [DOI] [PubMed] [Google Scholar]
- 26.Ochman H, Elwyn S, Moran NA. Calibrating bacterial evolution. Proc Natl Acad Sci U S A. 1999;96:12638–43. doi: 10.1073/pnas.96.22.12638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Poindexter JS. Selection for nonbuoyant morphological mutants of Caulobacter crescentus. J Bacteriol. 1978;135:1141–5. doi: 10.1128/jb.135.3.1141-1145.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wan Z, Brown PJ, Elliott EN, Brun YV. The adhesive and cohesive properties of a bacterial polysaccharide adhesin are modulated by a deacetylase. Mol Microbiol. 2013;88:486–500. doi: 10.1111/mmi.12199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Merker RI, Smit J. Characterization of the adhesive holdfast of marine and freshwater caulobacters. Appl Environ Microbiol. 1988;54:2078–85. doi: 10.1128/aem.54.8.2078-2085.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Roy A, Kucukural A, Zhang Y. I-TASSER: a unified platform for automated protein structure and function prediction. Nat Protoc. 2010;5:725–38. doi: 10.1038/nprot.2010.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huang XQ, Miller W. A Time-Efficient, Linear-Space Local Similarity Algorithm. Advances in Applied Mathematics. 1991;12:337–357. [Google Scholar]
- 32.Katoh K, Standley DM. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Molecular biology and evolution. 2013;30:772–80. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ. Jalview Version 2--a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25:1189–91. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tamura K, et al. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–9. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Williams KP, Sobral BW, Dickerman AW. A robust species tree for the alphaproteobacteria. J Bacteriol. 2007;189:4578–86. doi: 10.1128/JB.00269-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.JW T. Bias and confidence in not quite large samples. Ann Math Stats. 1958;614 [Google Scholar]
- 37.Thanbichler M, Iniesta AA, Shapiro L. A comprehensive set of plasmids for vanillateand xylose-inducible gene expression in Caulobacter crescentus. Nucleic Acids Res. 2007;35:e137. doi: 10.1093/nar/gkm818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Horton RM. PCR-mediated recombination and mutagenesis. SOEing together tailor-made genes. Mol Biotechnol. 1995;3:93–9. doi: 10.1007/BF02789105. [DOI] [PubMed] [Google Scholar]
- 39.Vasilyeva LV, et al. Asticcacaulis benevestitus sp. nov., a psychrotolerant, dimorphic, prosthecate bacterium from tundra wetland soil. Int J Syst Evol Microbiol. 2006;56:2083–8. doi: 10.1099/ijs.0.64122-0. [DOI] [PubMed] [Google Scholar]
- 40.Syvanen M. Evolutionary implications of horizontal gene transfer. Annu Rev Genet. 2012;46:341–58. doi: 10.1146/annurev-genet-110711-155529. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.