Abstract
S-Nitrosothiols are made by nitric oxide synthases and other metalloproteins. Unlike nitric oxide, S-nitrosothiols are involved in localized, covalent signaling reactions in specific cellular compartments. These reactions are enzymatically regulated. They affect protein interactions involved in virtually every aspect of normal cell biology.
Keywords: Cell signaling, Nitric oxide synthase, Nitrosonium, Protein-protein interaction, S-Nitrosoglutathione, S-Nitrosylation
1. Introduction
Protein S-nitrosylation, the post-translational modification of a cysteine thiol by the attachment of an NO group, is a regulated reaction that is responsible for a broad spectrum of cell signaling effects [1–4]. S-Nitrosylation is analogous to phosphorylation, glutathionylation, palmitoylation, acetylation and other physiological modifications of proteins. In general, proteins and peptides that have been modified to form S-nitrosothiol bonds are involved in soluble guanylate cyclase (sGC) -independent signaling by nitrogen oxides; though S-nitrosylation also affects sGC-dependent processes [5; Figure 1]. S-Nitrosylation can occur downstream of cellular NO synthase (NOS) activity, and can also be caused by extracellular sources of nitrogen oxides. Disorders of protein S-nitrosylation are relevant to the pathophysiology of many diseases [1–4, 6–12].
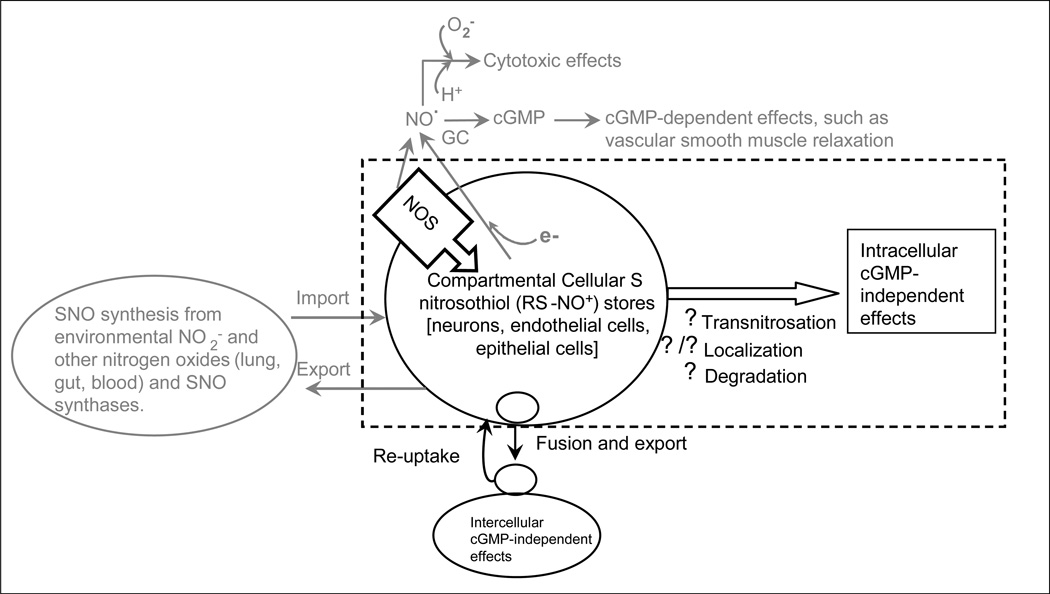
Figure 1. Scientific overview: An emerging paradigm of nitrogen oxide signaling.
Nitric oxide synthase (NOS) forms S-nitrosothiols (RS˙-˙NO, or, more commonly, RS−−NO+). Note there is also regulated cellular import of extracellular S-nitrosothiols. NOS-derived NO and NO reduced from S-nitrosothiols and NO2− can exert classical cytotoxic and cyclic GMP (cGMP)-dependent effects, the latter through activation of guanylyl cyclase (GC). Intracellular S-nitrosothiols can include protein and low-mass species, and are generally in sequestered locations in the cell, such as membranes and vesicles. These S-nitrosothiols can transfer NO+ equivalents to target proteins through transnitrosylation to cause cGMP-independent effects; this signaling can be regulated by movement of the S-nitrosothiols in the cell to target locations, and by degradation. Dr. Lewis’ recent data suggest that S-nitrosothiols can also be secreted into the extracellular space to signal intercellular, cGMP-independent effects—particularly in the autonomic nervous system—through extrusion from S-nitrosothiol-containing vesicles.
There is cross-talk between S-nitrosylation, phosphorylation and other post-translational signaling mechanisms that affect protein interactions. This is relevant to a spectrum of disease processes ranging from asthma to cancer. As an illustration, S-nitrosylation of wild-type (wt) Ras by endothelial NOS (eNOS) is required for tumor growth in a signaling pathway that also involves phosphorylation [11]. Specifically, oncogenic K-Ras activates proteins to initiate tumor growth. Of these proteins, only PI3 kinase, through activation of Akt, must remain activated by oncogenic K-Ras to maintain tumor growth [11]. The essential Akt phosphorylation substrate for this process is eNOS. Endothelial NOS activation, in turn, S-nitrosylates and activates wt H-Ras and N-Ras proteins at cysteine 118. Knockdown of eNOS or mutation of wt Ras cysteine 118 prevents tumor formation [11]. This signaling crosstalk appears to be relevant to the development of lung cancer [12].
S-Nitrosylation often acts through effects on protein-protein interactions. Of many examples, three are provided in this introduction. First, S-nitrosylation of procaspase-3 by NOS isoforms promotes procaspase-3 interaction with acid sphingomyelinase (ASM) and with NOS itself [13]. The interaction with ASM prevents apoptosis. Second, S-nitrosylation of apolipoprotein E (ApoE) isoforms at cysteine 112 (for ApoE3) by co-scaffolded nNOS prevents its interaction with low-density hypoprotein (LDL) receptor in the brain. This effect may contribute to the progression of Alzheimer’s disease (14; Figure 2). Third, S-nitrosylation of G protein receptor kinase 2 (GRK2) prevents interaction of GRK2 with the β2 adrenergic receptor (β2AR); this prevents β2AR phosphorylation and internalization in myocytes [15], preventing tachyphylaxis to β2 adrenergic agonists. Additional examples of regulation of protein-protein interactions by S-nitrosylation will be provided below.
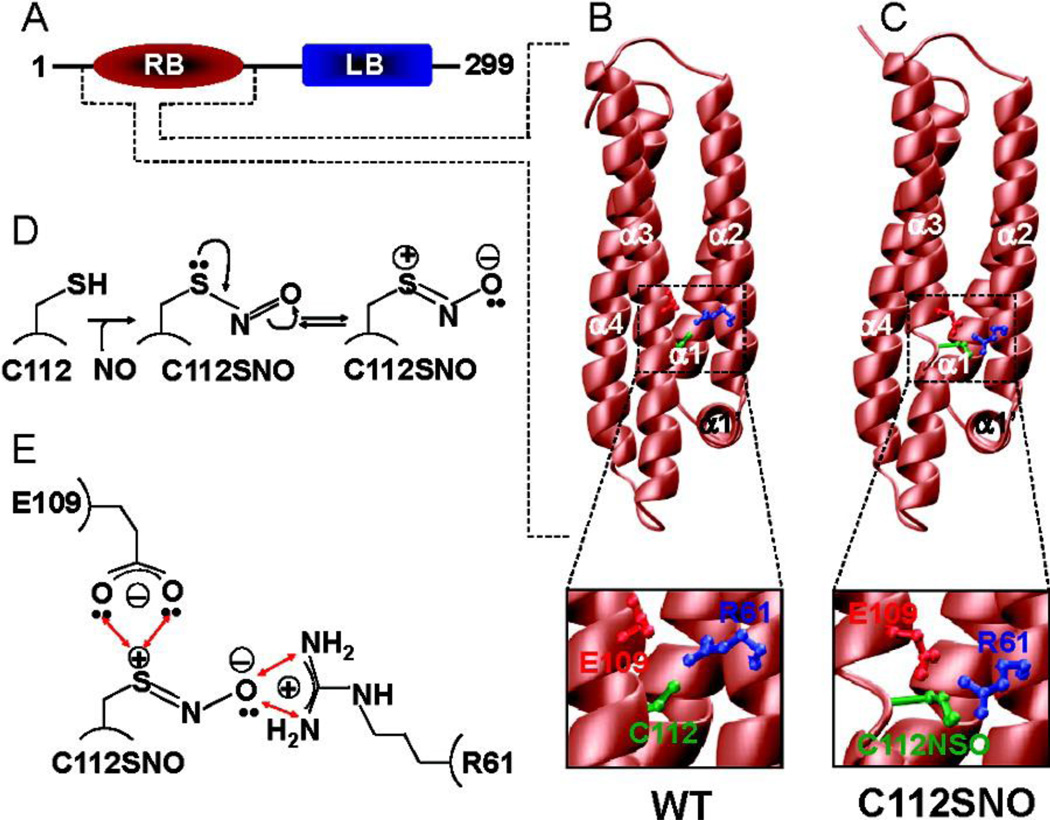
Figure 2. Effect of S-nitrosylation on the 3D structure of human ApoE3.
(A) Fully processed ApoE3, without the N-terminal signal peptide sequence (18 residues), is comprised of an N-terminal LDL receptor binding (RB) domain and a C-terminal lipid binding (LB) domain. Note that all the amino acid numbering used here is based on the amino acid sequence of the fully processed ApoE (residues 1−299). (B) 3D atomic model of the WT RB domain of ApoE. (C) 3D atomic model of the S-nitrosothiol derivative (C112SNO) of the RB domain of ApoE. Note that in both panels B and C, the RB domains are colored brown while the side chain moieties of R61, E109, and C112/C112SNO are colored blue, red, and green, respectively. Insets show close-ups of intramolecular interactions of C112/C112SNO with R61 and E109. (D) Schematic showing the S-nitrosylation of C112 within the RB domain of ApoE. Note that the resulting C112SNO S-nitrosothiol derivative may undergo resonance arrangement to form a zwitterion with an internal dipole characterized by the separation of a positive charge and a negative charge on sulfur and oxygen atoms, respectively. (E) Schematic showing a plausible hydrogen bonding and/or ion pairing network of the polarized S-nitrosothiol moiety of C112SNO, the guanidino group of R61, and the side chain carboxylate of E109. The double-headed red arrows indicate potential hydrogen bonding and/or ion pairing contacts. (from reference 14)
As these observations imply, the products of NOS activation can be highly localized to proteins co-scaffolded with NOS in specific cellular locations. These NOS products participate in covalent chemistry. This model contrasts with one in which NO simply diffuses randomly around the cell as a dissolved gas [Figure 1]. Though many observations over the years have suggested this covalent chemistry paradigm, it has only recently begun to be commonly recognized as an alternative view of NOS biochemistry [1–4,16]. S-nitrosylation is now a growing field of substantial importance to mammalian biology, human disease, antimicrobial therapy, plant biology, ecology and many other biological disciplines. Here, however, we will focus on S-nitrosylation signaling as it affects protein signaling in normal mammalian cell biology.
2. Biochemistry
a. General: Consensus motifs for NO addition to protein thiols
Consensus motifs have been identified for S-nitrosylation, including those in which the Cys is adjacent to basic, acidic or aromatic residues in the primary sequence or tertiary protein structure [4,17–19]. The chemistry is affected not only by the protein, but by the reactivity of the relevant nitrogen oxide signaling molecules. Specifically, S-nitrosothiol species (RS−−NO+), nitrous acid (HO−−NO+) and other NO+ donors can signal through NO+ (transnitrosylation) reactions [1,2,19–24] according to X− − NO+ + Y− − H+ ⇌ X− − H+ + Y− − NO+. In the case of protein S-nitrosothiols, these transnitrosylation reactions are favored or disfavored by changes in cellular location of the reactions and by specific protein conformations [1–4,17–19,22,23]. Though cysteines that are targets for transnitrosylation could theoretically be predicted to have relatively low pKa sulfhydryl groups, this is not a consistent finding [18]. Note that acidic conditions—such as those in protein microdomains, in lysosomes or in the mitochondrial inner space—may also promote SNO formation from nitrite protonation to form nitrous acid. Covalent chemistry is also relevant to reactions between nitroxyl (NO−/HNO) and thiol sites [26].
An NO equivalent bound covalently to a thiol is typically the NOS product that initiates S-nitrosylation cascades [7,15,27]. For example, following endothelial NOS (eNOS) activation by calcium ionophore in endothelial cells, eNOS itself is S-nitrosylated (NO+ attachment to an R-S thiol) followed by transnitrosylation (NO+ transfer) to downstream targets such as NOSiP, tubulin and heat shock protein (Hsp) 90 (Figure 3). These targets interact with one another: NO+ appears to be transferred to co-scaffolded interacting proteins.
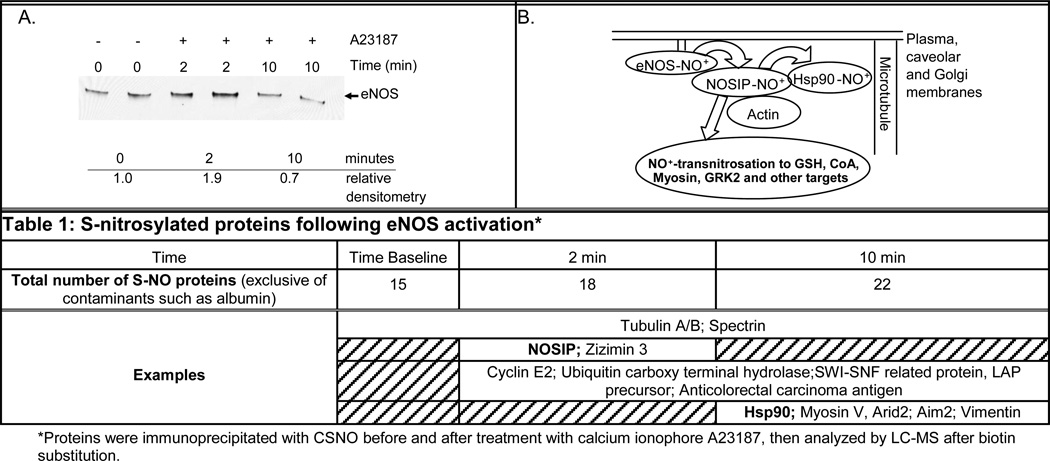
Figure 3. eNOS as a Nitrosonium Synthase.
A. Proteins were immunoprecipitated with anti-CSNO antibody before, and two and 10 min after, cell treatment with calcium ionophore A23187; they were then immunoblotted with anti-eNOS. B. Proposed general schematic of nitrosonium (NO+) transfer following eNOS activation, based on our preliminary data. Arrows represent transnitrosylation (NO+ transfer between cysteines). Protein-scaffolding scheme adapted from Su, Kondrikov, and Block (ref. 25).
Of note, however, NO radical can also be relevant to protein thiol modification in cells. Oxidation of NO to an NO+ equivalent may be favored by transition metals, oxygen or other oxidizing agents. For example, NO and O2 concentrate in hydrophobic compartments, which facilitates S-nitrosylation by N2O3 and protects the SNO from degradation by cytosolic reducing compounds [16,28]. Nitric oxide can also be oxidized by metalloproteins such as ceruloplasmin and hemoglobin to result in NO+ equivalents; these, in turn, modify cysteine thiolate sites [28,29]. In the case of ceruloplasmin, NO radical reacts with Cu2+ to form a NO+−Cu+ complex; after which NO+ is transferred to GSH with loss of a proton. The electron on Cu+ is transferred through the copper systems of the protein to oxygen, forming water and regenerating Cu2+ [28].
Note that cysteine S-nitrosylation contrasts with nitration reactions in which tryosine is modified by NO2+ addition. Nitration reactions are involved in cytotoxicity and tissue pathology. Unlike S-nitrosylation reactions, nitration reactions are not enzymatically reversed in cell biology: they do not normally represent reversible, physiological cell signaling reactions. Nitrosylation signaling is also contrasted with unregulated, inorganic NO addition reactions, or nitrosation.
b. Formation of the S-NO bond. How are proteins S-nitrosylated in cells?
Various proteins have been identified that catalyze formation of S-nitrosothiol bonds in cells. As noted above, these include all NOS isoforms, hemoglobin and ceruloplasmin. Activation of iNOS, eNOS and nNOS result in localized S-nitrosylation and functional cysteine modification in coscaffolded proteins, conventionally at cysteine S-nitrosylation motifs. For example, activation of iNOS—co-scaffolded with cyclooxygenase (Cox)2—activates Cox2 by S-nitrosylation where it is in macrophages [27]. Activation of eNOS leads to S-nitrosylation of several downstream proteins, depending on cellular location [see, for example, Figure 3], one of which is GRK2 [15]. In the heart, S-nitrosylation by nNOS can activate colocalized ryanodine-receptor to increase Ca2+ flux, while S-nitrosylation of the eNO-Scolocalized L-type Ca2+ channel, inhibits Ca2+ flux [7]. Biochemical evidence suggests the existence of additional SNO-synthases [S.J. Lewis, personal communication]. Note that different mechanisms have been identified by which NOS’s can be S-nitrosothiol synthases. In the case of iNOS, for example, the formation of one R-N-NO intermediate on the pterin has been identified near a GSH binding site (30).
Protein S-nitrosylation by nitrite can also be catalyzed. For example, cysteines in protein tyrosine-Xn-cysteine (YXnC) motifs are S-nitrosylated by myeloperoxidase in the presence of nitrite and hydrogen peroxide [31]; and hemoglobin can serve an S-nitrosothiol synthase through formation of HbFe (III)-NO intermediates in equilibrium with Fe(II)-NO+ [29]. S-nitrosylation can also result from nitrite protonation in relatively acidic cell compartments, such as the mitochondrial intermembrane space [32]; and S-nitrosothiols can be formed from inorganic reactions in acidic conditions in the lung and gut [Figure 1;20,33,34]. Indeed, the regulated formation, storage and transport of S-nitrosylated proteins and peptides can help to rationalize a wide range of paradoxical observations made in physiology, including tachyphylaxis of the endothelium-derived relaxing factor (EDRF) response to repeated endothelial stimulation with acetylcholine; nitrogen oxide bioactivity in the presence of mM concentrations of hemoglobin in blood; and the relatively benign phenotypes of multiple NOS knockout mice (see, for example, Figure 1).
c. Denitrosylation
The stability of cellular S-nitrosylated proteins varies substantially because of both enzymatic and non-enzymatic denitrosylation. S-Nitrosylation is balanced by denitrosylation much as phosphorylation is balanced by dephosphorylation [1]. Several enzymes and enzyme systems have been identified that serve as denitrosylases or transnitrosylases, including S-nitrosoglutathione (GSNO) reductase [36,37; Figure 4], thioredoxin reductase (with thioredoxin) [38–41], xanthine/xanthine oxidase [42], Cu/Zn superoxide dismutase (SOD) [43–45], carbonyl reductase [46] and protein disulphide isomerases [47,48]. Products vary according to the enzyme; they include alternate S-nitrosothiols, NO, peroxynitrite, hydroxylamine and ammonia. Thus, catabolism can result in inactivation or bioactivation, depending on location and upstream/downstream biochemistry. Transnitrosylation is in many cases permissive for enzymatic denitrosylation [36]. For example, transnitrosylation in the S-nitrosylated proteins to GSH, forming GSNO, permits GSNO reductase to serve as a protein denitrosylase (see, for example, 36). Note that the cellular location of these enzymes is critical, as discussed below: specific proteins are denitrosylated in specific cellular locations/organelles. Further, different denitrosylases serve in varying cell signaling pathways. For example, thioredoxin/thioredoxin reductase is critical in denitrosylating caspase 3 during cellular apoptosis [39]; while it is GSNO reductase that denitrosylates GPCR GRK2 during cell-membrane trafficking [15]. Kinetic parameters are as important as histologic localization. For example, denitrosylation downstream of GPCR or growth factor receptor activation occurs rapidly after receptor activation, whereas other S-nitrosylated proteins are more stable [4].
With regard to non-enzymatic decomposition of S-nitrosylated proteins, denitrosylation is generally favored by free copper and iron ions, by light, by heat, and by reducing agents such as ascorbate [35,49–52]. Copper and iron ions, however, are bound proteins; and most cells in the body are dark. Thus, thermal decomposition and reduction reactions are central to cellular inorganic S-nitrosothiol decomposition. Different S-nitrosothiols vary in stability [4,49,51]; and transnitrosylation can convert stable S-nitrosothiol species—particularly including stable S-nitrosoproteins—to less stable species—including S-nitrosocysteine (CSNO) [4,49,53]. This promotes non-enzymatic decomposition to cysteine and NO radical [34,49,51]. In biological systems, reduced thiols such as glutathione are the most abundant nucleophilic compounds that participate in both transnitrosylation reactions; but other nucleophilic compounds interact with S-nitrosothiols, including ascorbate, bilirubin and sulphite [35,54,55].
d. Transmembrane S-Nitrosothiol Trafficking
i. Extracellular NOS-independent sources
S-Nitrosothiols are present in the airway lumen, the gut, and other extracellular spaces [1,20,56] (Figure 2). As noted above, they can be formed in these spaces by protein-catalyzed reactions, such as hemoglobin-catalyzed S-nitrosothiol formation in the lung and elsewhere [1,27–31], or by inorganic reactions such as nitrite ingestion/inhalation in the context of the relatively low pH [20,56,57]. Relevant pH values are observed in the gut and distal airway, locations where thiol concentrations are also high. Note in this regard that both the gut and distal airway have high levels of GSNO reductase and other S-nitrosothiol catabolic enzymes to regulate S-nitrosothiol concentrations formed both organically and inorganically [57–60]; indeed, enzymes such as GSNO reductase can be upregulated by S-nitrosylation in the context of increased GSNO levels [60]. Transnitrosylation reactions can then form membrane-associated S-nitrosothiols that can transfer in and to erythrocytes, endothelial cells and other cells [27,56,61,62]. These cellular stores of S-nitrosothiols serve as NOS-independent sources of bioactive nitrogen oxides, regulated both by cell-cell interactions [1,61–64] and by transmembrane transport regulators (see below).
ii. Regulation of transmembrane S-nitrosothiol transport
S-nitrosoglutathione does not readily cross from the outside to the inside of the plasma membrane. However, γ-glutamyl transpeptidase (GGT) cleaves GSNO to form S-nitrosocysteinyl glycine which [65,66], in turn, is cleaved by dipeptidases to form S-nitroso-L-cysteine (L-CSNO). In this regard, the convergence between GSNO metabolism and cysteine leukotriene metabolism is striking; and there may be other novel isoforms of GGT appear to have differential reactivity towards GSNO and cysteinyl leukotrienes. L-CSNO transport across the plasma membrane, in turn, is regulated by the LAT transporter [66].
Additional transmembrane S-nitrosothiol transport proteins have been demonstrated. Protein disulfide isomerase isoforms are S-nitrosylated, serve as denitrosylases and can transfer NO from the extracellular protein, albumin, to the intracellular protein metallothionein [47,67]. Erythrocytic anion exchange protein 1 (AE 1) also serves a transmembrane transport role, providing a mechanism by which membrane-associated deoxyhemoglobin can signal from inside out to present S-nitrosothiols to vascular structures that can respond, in turn, to hemoglobin R:T conformation changes [61,62]. That said, the science of transmembrane interactions signaling by and through S-nitrosylation is in its infancy.
e. Intracellular stabilization of S-nitrosothiol bonds
As noted above, low-mass S-nitrosothiols are labile in the reducing environment of the cytosol. The intracellular S-nitrosothiol bond is stabilized by steric sequestration in S-nitrosylated proteins [4,19,62,63] and by localization in membranes and vesicles. Evidence from the Lewis group suggests that specific S-nitrosothiols are sequestered in cytosolic vesicles that have vesicular fusion and reuptake pathways reminiscent of other transporters involved in cell-cell interactions [Figure 1; 68]. Recent data from Straub, et al. are consistent with this paradigm [69]. Similarly, S-nitrosothiol bonds are stabilized in membranes and in the mitochondrial inter-membrane space [32]. In a number of situations (hemoglobin to AE1, for example [62]) this transfer of NO+ from one protein to a co-scaffolded protein both protects the S- nitrosothiol bond from non-specific reduction and localizes its activity to the target protein. Indeed, eNOS activation appears to transfer NO+ equivalents to downstream, coscaffolded proteins in a time-dependent, bucket-brigade process [Figure 3]. It is anticipated that proteins such as LAT in the plasma membrane (see above) will transfer L-CSNO to specific vesicles or other carrier protein to cause and/or preserve bioactivity; however, the nature of these stabilization transport systems require a substantial amount of additional study.
A word about concentration is in order. As implied from the discussion above, S-nitrosothiols do not conventionally serve simply as NO radical donors. Therefore, it is a mistake to perform experiments using high µM or mM concentrations of S-nitrosothiol with the idea that these will only generate nM concentrations of NO radical. The rate at which—and extent to which—these compounds generate NO radical normally reflects their catabolic inactivation: NO radical is often not the active moiety. Physiologically relevant cellular S-nitrosothiol concentrations involved in covalent chemistry are typically in the mid-high nM range [20,61]. They are somewhat higher in some tissues such as the brain [70], but, it is also a bit of a misconception to consider the global tissue concentration of S-nitrosothiols without considering the local cellular concentration of each specific low-mass S-nitrosothiol or S-nitrosylated protein of interest.
3. Subcellular localization of S-nitrosylated proteins and their metabolic enzymes
Specific S-nitrosylated proteins have been localized to the cell membrane [62], the endoplasmic reticulum [71], the Golgi [51,71], the mitochondria [32] and the nucleus [73]. Transfer of proteins from one space to another following S-nitrosylation is an important cell signaling mechanism. For example, Fas-/Fas-ligand binding at the cell membrane results in release of S-nitrosylated caspase 3 and caspase 9 from the mitochondrial intermembrane space [32]. When released from this sequestered location, these caspases are de-nitrosylated by the thioredoxin/thioredoxin reductase system [39], interacting with cytochrome C in the apoptosome and promoting cellular apoptosis. Many examples are now emerging regarding the role of S-nitrosylation in regulating transport between organelles (as reviewed below with regard to regulation of protein expression).
Note in this regard that S-nitrosothiol metabolic enzymes appear to traffic in the cell with their S-nitrosothiol products/substrates to subserve specific cellular functions [12]. For example, eNOS as an NO+ S-nitrosothiol synthase appears to localize in the base of cilia to form airway epithelial S-nitrosothiols [unpublished observation]. GSNO reductase traffics from cytosolic structures to the nucleus in anaphase and telaphase of mitosis following, along with mitotic spindles, those processes inhibited by colchicine [12]. Thus, S-nitrosothiols and their metabolic enzymes traffic with specific organelles and other structures in the cell in a regulated fashion to subserve specific functions.
4. S-Nitrosylation regulates protein interactions to affect a broad range of cell signaling functions
a. Epigenetic regulation
Regulation of protein interactions with chromatin can involve S-nitrosylation. For example, in neuronal development, brain-derived neurotrophic factor activates nNOS, which S-nitrosylates histone deacetylase 2 (HDAC2; cysteines 262 and 274), causing HDAC2 to dissociate from chromatin [74]. This increases histone acetylation, permitting transcription of beneficial target genes regulating dendritic growth (74); but it can also have potentially adverse effects, including increased expression of metastatic tumor antigen 1 [75]. Additionally, cell cycle regulation appears to involve S-nitrosylation and denitrosylation of critical proteins such as nuclear Ran-binding protein 3 [76] and, through E-box enhancer dependent effects, the clock protein, Period [77]. Localization of denitrosylating enzymes to the mitotic spindle also appears to be important to mitosis [12]. As with other cellular effects, however, the role of S-nitrosylation signaling in epigenetics and cell-cycle regulation is only beginning to be understood.
b. Transcriptional regulation
A number of nuclear regulatory protein interactions are modified by S-nitrosylation. For example, expression and DNA-binding of the nuclear regulatory specificity protein (Sp) 3 is increased, and those effects of Sp1 are decreased, by low GSNO concentrations; whereas the reverse is true at higher concentrations [78]. Supraphysiological (nitrosative stress) level effects of SNO or Sp’s are relevant to downstream regulation of the expression eNOS and cftr genes, as well as genes encoding 5-lipoxygenase and a variety of other critical cell regulatory proteins [78–80]. Another example is the regulation of Hypoxia-inducible factor 1 α (Hif1-α) expression, which is stabilized by physiological S-nitrosothiol levels through S-nitrosylation of protein von Hippel Lindau (C162). This prevents HIF1-α degradation [62], permitting HIF1-α interaction with HIF1-β. The resultant HIF1 heterodimer, in turn, binds to hypoxia-responsive elements in gene promotor regions to result in transcription of genes such as vascular endothelial growth factor (VEGF). This appears to be one mechanism by which oxyhemoglobin desaturation-derived S-nitrosothiols can signal gene regulatory effects in hypoxemia in vivo [64]. Of note, S-nitrosylation has a variety of other effects on HIF proteins, including direct effects on the protein itself [81,82]. S-Nitrosylation also has a variety of effects to alter NFκB activity, including S-nitrosylation of both NFκB subunits p50 (C 62) and p65 (C 38); as well as IκB kinase [82–88]. The net affect of these S-nitrosylation reactions is generally to increase cytosolic NFκB-IκB interaction, and/or preventing nuclear translocation of NFκB; these effects prevent interaction of NFκB with inflammatory gene promoters, inhibiting inflammation. Through these mechanisms, extracellular S-nitrosothiols (for example, in the context of oxyhemoglobin desaturation) or intracellularly generated S-nitrosothiols (for example, in the context of NOS upregulation) can signal in a targeted way to cause specific changes in gene expression.
c. Regulation of the activity of proteins involved in cellular energy metabolism
Many metabolic enzymes are now appreciated to be regulated by S-nitrosylation and/or interact with proteins that are affected by S-nitrosylation, including those in the mitochondria. Processes ranging from the S-nitrosylation-induced activation of glucokinase to S-nitrosylation-induced inactivation of the insulin receptor have been proposed to be related to the pathophysiology of diabetes [86–87]. The common glycolytic enzyme, glyceraldehyde-3-phosphate dehydrogenase (GAPDH), provides an example of the complexity of S-nitrosylation signaling as it relates to metabolism. While GAPDH S-nitrosylation reversibly inhibits the enzyme's glycolytic activity, the same reaction also protects it from irreversible inactivation by oxidation [88]. Strikingly, GAPDH S-nitrosylation also leads to its binding to the E-3 ubiquitin ligase, Siah, which—bound to GAPDH—is transported to the nucleus and is critically involved in apoptosis [89]. Surprisingly, GAPDH also stabilizes iron in cells, delivering it to the iNOS heme; this reaction is inhibited by GAPDH S-nitrosylation at C152 [90].
d. Regulation of cellular metal transport and homeostasis
S-Nitrosylated proteins regulate cellular proteins to affect metal transport and homeostasis. Several examples have been reported recently, including cellular regulation of zinc by metallothionein [91] and, as noted above, iron by GAPDH [90].
e. Regulation of protein translation and post-translational modifications
S-Nitrosylation can regulate both the folding and trafficking of normal proteins and the degradation of misfolded or unwanted proteins. It does this through modifying protein expression and protein/protein interactions in the endoplasmic reticulum, Golgi, submembrane vesicles and cytosol. An example is the regulation of cystic fibrosis transmembrane regulatory protein (CFTR), an epithelial Cl− channel. Only 30% of normally translated CFTR gene product is expressed and functional on the normal cell surface of epithelial cells in the airways, gut and reproductive organs. Genetic mutations in the cftr gene commonly result in misfolded proteins [92]. Nearly 100% of these misfolded proteins are targeted for degradation. The most common mutation associated with misfolded CFTR is delF508. This is a slightly abnormal gene product (missing a single amino acid) that, if expressed on the cell surface, can function normally. However, delF508 CFTR is targeted for degradation because of its misfolding: less than 1% is normally expressed on the cell surface. We and others have shown that S-nitrosylating agents augment the expression and cell-surface maturation of both wt and delF508 CFTR [51,71,93–95]. A variety of chaperones and co-chaperones escort CFTR as it is being translated, folded in the ER, glycosylated in the ER and Golgi, expressed on the cell surface and recycled in sub-plasma membrane vesicles [96]. Recent siRNA screening experiments confirm that heat shock protein (Hsp) 70 and Hsp90 are critical co-chaperones for successful CFTR maturation; and that heat shock cognate (Hsc) 70 is involved in targeting CFTR for degradation through CHIP and other cysteine-containing ubiquitin ligase systems [92]. There are several targets for S-nitrosylation in this system. These S-nitrosylated proteins track through the ER and Golgi at baseline (depending on NOS activity) and following S-nitrosothiol exposure. One of these targets is Hsc 70, inhibition of which by S-nitrosylation permits CFTR maturation [51]. Another critical target is Hsp70/Hsp90 organizing protein (Hop; or Stip1). It is S-nitrosylated on cysteine 403, targeting it for ubiquitination and degradation [71]. This loss of Hop prevents CFTR/Hop interaction, permitting not only CFTR glycosylation and maturation, but preventing loss of CFTR from the cell surface through recycling. The loss of Hop/CFTR interaction can be tracked through the ER and Golgi as CFTR is glycosylated and permitted to be expressed on the cell surface.
f. Regulation of protein degradation
As noted above, S-Nitrosylation affects the degradation of a number of proteins. The principal mechanism appears to be S-nitrosylation of the active site cysteine on specific E3 ubiquitin ligases preventing enzyme-target protein interaction. An example is the inhibition of Hif-1α degradation by S-nitrosylation of protein von Hippel Lindau (C162) mentioned in the section on gene regulation [64].
g. Regulation of the activities of membrane-associated proteins
Expression of membrane-associated proteins can be regulated through effects on transcription, translation, trafficking and degradation discussed above. Additionally, however, the activity of membrane-associated proteins such as ion channels can be affected directly by S-nitrosylation. S-Nitrosylation affects a broad spectrum of receptors including, for example, NMDA receptors [97]. It also affects many different channel ions. These range from the stereoselective effect of L-CSNO—but not D-CSNO—to inhibit the T-type Ca2+ channel in the thalamus [98] to the effect of NOS activation to modify K+ and Ca2+ channels in the heart. In the cardiac myocyte, for example, eNOS regulates plasma membrane K+ and Ca2+ through S-nitrosylation of other K+ channels and L-type Ca2+ channels; while nNOS regulates Ca2+ release from the endoplasmic reticulum through S-nitrosylation of the responsive receptors [1,7]. These effects often are initially dependent on protein-protein interactions. For example, interaction of ryanodine receptor type 1 (RyR1) with nNOS provides specificity for RyR1 S-nitrosylate during skeletal muscle disuse (99). Indeed, there are now many examples of how S-nitrosylation affects membrane protein-protein interactions. Several were discussed in previous sections of this paper, 1) including the inhibition of GRK2-βr AR interaction at the plasma membrane by eNOS-induced S-nitrosylation of GRK2; 2) augmentation of mitochondrial membrane procaspase-3-ASM interaction by eNOS-induced S-nitrosylation of procaspase-3; and 3) inhibition of LDL receptor binding of ApoE by nNOS-induced S-nitrosylation of ApoE.
Disorders of S-nitrosylation
Disorders of each of the cellular processes described above are observed in a variety of pathophysiological processes. These range from asthma to Parkinson’s disease. They include major causes of morbidity, mortality and increased health care costs world-wide [1–3,7–12,14,43,50–52,55–58,61,63,67,71,95,100]. Our review, however, have focused on the role of S-nitrosothiols in normal cell signaling. We anticipate that information regarding S-nitrosylation in the normal cell will serve as a basis for studies regarding abnormal S-nitrosylation signaling in a broad range of diseases. Some progress has already been made in the use of S-nitrosothiols for treating pulmonary diseases such as asthma, cystic fibrosis and pulmonary arterial hypertension [2].
Summary
Nitric oxide radical signals in conventional pathways represented as random gas diffusion through membranes to activate soluble guanylate cyclase. It also signals as a free radical through interaction with superoxide to form antimicrobial intermediates. Additionally, it is increasingly appreciated that cellular bioactivities caused by NOS activation and environmental nitrogen oxides can be regulated by reactions that signal specific, localized cellular effects through covalent transnitrosylation reactions that are independent of NO radical. These reactions are conventionally viewed as NO+ transfer reactions between proteins that affect—and are affected by—cellular protein interactions. They are observed in every part of the eukaryotic cell and are involved in the regulation of nearly every aspect of cell biology. Understanding the localized, covalent nature of this biochemistry represents a paradigm shift in the study of nitrogen oxides in cell biology. This new perspective, in turn, explains a number of paradoxes in cellular biochemistry of nitrogen oxides. It also opens doors for innovation in many areas of biology and medicine.
Research Highlights.
Nitric oxide synthase activation forms S-nitrosothiols
S-nitrosylation signaling is regulated
Virtually every aspect of cell biology is impacted S-nitrosylation signaling
Acknowledgments
We would like to thank Reid Townsend and Cheryl Lichti in the Washington University, St. Louis, Proteomics Core for their assistance with analysis of S-nitrosylated proteins downstream of eNOS activation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Nadzeya V. Marozkina, Email: nvm6v@virginia.edu.
Benjamin Gaston, Email: bmg3g@virginia.
References
- 1.Foster MW, Hess DT, Stamler JS. Protein S-nitrosylation in health and disease: a current perspective. Trends Mol. Med. 2009;15:391–404. doi: 10.1016/j.molmed.2009.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gaston B, Doctor A, Singel D, Stamler JS. S-Nitrosothiol signaling in respiratory biology. Am. J. Respir. Crit. Care Med. 2006;173:1186–1193. doi: 10.1164/rccm.200510-1584PP. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gow AJ, Chen Q, Hess DT, Day BJ, Ischiropoulos H, Stamler JS. Basal and stimulated protein S-nitrosylation in multiple cell types and tissues. J. Biol. Chem. 2002;277:9637–9640. doi: 10.1074/jbc.C100746200. [DOI] [PubMed] [Google Scholar]
- 4.Paige JS, Xu G, Stancevic B, Jaffrey SR. Nitrosothiol reactivity profiling identifies S-nitrosylated proteins with unexpected stability. Chem. Biol. 2008;15:1307–1316. doi: 10.1016/j.chembiol.2008.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mayer B, Pfeiffer S, Schrammel A, Koesling D, Schmidt K, Brunner F. A new pathway of nitric oxide/cyclic GMP signaling involving S-nitrosoglutathione. J. Biol. Chem. 1998;273:3264–3270. doi: 10.1074/jbc.273.6.3264. [DOI] [PubMed] [Google Scholar]
- 6.Altucci A, Mai A, Capogrossi MC, Puri PL, Gaetano C. HDAC2 blockade by nitric oxide and histone deacetylase inhibitors reveals a common target in Duchenne muscular dystrophy treatment. Proc. Nat. Acad. Sci. U.S.A. 2008;105:19183–19187. doi: 10.1073/pnas.0805514105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gonzalez DR, Treuer A, Sun Q-A, Stamler JS, Hare JM. S-nitrosylation of cardiac ion channels. J. Cardiovasc. Pharmacol. 2009;54:188–195. doi: 10.1097/FJC.0b013e3181b72c9f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ozawa K, Whalen EJ, Nelson CD, Mu Y, Hess DT, Lefkowitz RJ, Stamler JS. S-nitrosylation of beta-arrestin regulates beta-adrenergic receptor trafficking. Mol. Cell. 2008;31:395–405. doi: 10.1016/j.molcel.2008.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Moya MP, Gow AJ, Califf RM, Goldberg RN, Stamler JS. Inhaled ethyl nitrite gas for persistent pulmonary hypertension of the newborn. Lancet. 2002;360:141–143. doi: 10.1016/S0140-6736(02)09385-6. [DOI] [PubMed] [Google Scholar]
- 10.Snyder A, McPherson ME, Hunt JF, Johnson M, Stamler JS, Gaston B. Acute effects of aerosolized S-nitrosoglutathione in cystic fibrosis. Am. J. Respir. Crit. Care Med. 2002;165:922–926. doi: 10.1164/ajrccm.165.7.2105032. [DOI] [PubMed] [Google Scholar]
- 11.Lim KH, Ancrile BB, Kashatus DF, Counter CM. Tumour maintenance is mediated by eNOS. Nature. 2008;452:646–649. doi: 10.1038/nature06778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marozkina NV, Wei C, Yemen S, Wallrabe H, Nagji AS, Morozkina T, Jones DR, Gaston B. S-Nitrosoglutathione reductase in human lung cancer. J Respir Cell Molec Biol. 2011 doi: 10.1165/rcmb.2011-0147OC. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Matsumoto A, Comatas KE, Liu L, Stamler JS. Screening for nitric oxide-dependent protein-protein interactions. Science. 2003;301:657–661. doi: 10.1126/science.1079319. [DOI] [PubMed] [Google Scholar]
- 14.Abrams AB, Farooq A, Wang G. S-Nitrosylation of ApoE in Alzheimer’s disease. Biochem. 2011;50:3405–3407. doi: 10.1021/bi200266v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Whalen EJ, Foster MW, Matsumoto A, Ozawa K, Violin JD, Que LG, Nelson CC, Benhar M, Keys JR, Rockman HA, Koch WJ, Daaka Y, Lefkowitz RJ, Stamler JS. Regulation of beta-adrenergic receptor signaling by S-nitrosylation of G-protein-coupled receptor kinase 2. Cell. 2007;129:511–522. doi: 10.1016/j.cell.2007.02.046. [DOI] [PubMed] [Google Scholar]
- 16.Lancaster J, Gaston B. NO and nitrosothiols: spatial confinement and free diffusion. Am. J. Physiol. Lung Cell Mol. Physiol. 2004;287:465–466. doi: 10.1152/ajplung.00151.2004. [DOI] [PubMed] [Google Scholar]
- 17.Stamler JS, Toone EJ, Lipton SA, Sucher NS. (S)NO signals: translocation, regulation, and a consensus motif. Neuron. 1997;18:691–696. doi: 10.1016/s0896-6273(00)80310-4. [DOI] [PubMed] [Google Scholar]
- 18.Marino SM, Gladyshev VN. Structural analysis of cysteine S-nitrosylation: a modified acid-based motif and the emerging role of trans-nitrosylation. J. Molec. Biol. 2010;395:844–859. doi: 10.1016/j.jmb.2009.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Perez-Mato I, Castro C, Ruiz FA, Corrales FJ, Mato JM. Methionine adenosyltransferase S-nitrosylation is regulated by the basic and acidic amino acids surrounding the target thiol. J. Biol. Chem. 1999;274:17075–17079. doi: 10.1074/jbc.274.24.17075. [DOI] [PubMed] [Google Scholar]
- 20.Gaston B, Reilly J, Drazen JM, Fackler J, Ramdev P, Arnelle D, Mullins M, Sugarbaker D, Chee C, Singel D D, Loscalzo J, Stamler JS. Endogenous nitrogen oxides and bronchodilator S-nitrosothiols in human airways. Proc. Natl. Acad. Sci. U.S.A. 1993;90:10957–10961. doi: 10.1073/pnas.90.23.10957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bryan NS, Calvert JW, Gundewar S, Lefer DJ. Dietary nitrite restores NO homeostasis and is cardioprotective in endothelial nitric oxide synthase-deficient mice. Free Rad. Biol. Med. 2008;45:468–474. doi: 10.1016/j.freeradbiomed.2008.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Iwakiri Y, Satoh A, Chatterjee S, Toomre DK, Chalouni CM, Fulton D, Groxzmann RJ, Shah VH, Sessa WC. Nitric oxide synthase generates nitric oxide locally to regulate compartmentalized protein S-nitrosylation and protein trafficking. Proc. Natl. Acad. Sci. U.S.A. 2006;103:19777–19782. doi: 10.1073/pnas.0605907103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Matsushita K, Morrell CN, Cambien B, Yang SX, Yamakuchi M, Bao C, Hara MR, Quick RA, Cao W, O'Rourke B, Lowenstein JM, Pevsner J, Wagner DD, Lowenstein CJ. Nitric oxide regulates exocytosis by S-nitrosylation of N-ethylmaleimide-sensitive factor. Cell. 2003;115:127–129. doi: 10.1016/s0092-8674(03)00803-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Relph RA, Guasco TL, Elliott BM, Kamrath MZ, McCoy AB, Steele RP, Schofield DP, Jordan KD, Viggiano AA, Ferguson EE, Johnson MA. How the shape of an H-bonded network controls proton-coupled water activation in HONO formation. Science. 2010;327:308–312. doi: 10.1126/science.1177118. [DOI] [PubMed] [Google Scholar]
- 25.Su Y, Kondrikov D, Block ER. Cytoskeletal regulation of nitric oxide synthase. Cell Biochem. Biophys. 2005;43:439–449. doi: 10.1385/CBB:43:3:439. [DOI] [PubMed] [Google Scholar]
- 26.Miller TW, Cherney MM, Lee AJ, Francoleon NE, Farmer PJ, King SB, Hobbs AJ, Miranda KM, Burstyn JN, Fukuto JM. The effects of nitroxyl (HNO) on soluble guanylate cyclase activity: interactions at ferrous heme and cysteine thiols. J. Biol. Chem. 2009;284:21788–21796. doi: 10.1074/jbc.M109.014282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim SF, Huri DA, Snyder SH. Inducible nitric oxide synthase binds, S-nitrosylates and activates cyclooxygenase-2. Science. 2005;310:1966–1970. doi: 10.1126/science.1119407. [DOI] [PubMed] [Google Scholar]
- 28.Inoue K, Akaike T, Iyamoto Y, Okamoto T, Sawa T, Otagiri M, Suzuki S, Yoshimura T, Maeda H. Nitrosothiol formation catalyzed by ceruloplasmin. Implication for cytoprottective mechanism in vivo. J. Biol. Chem. 1999;274:27069–27075. doi: 10.1074/jbc.274.38.27069. [DOI] [PubMed] [Google Scholar]
- 29.Angelo M, Singel DJ, Stamler JS. An S-nitrosothiol (SNO) synthase function of hemoglobin that utilizes nitrite as a substrate. Proc. Natl. Acad. Sci. U.S.A. 2006;103:8366–8371. doi: 10.1073/pnas.0600942103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rosenfeld RJ, Bonaventura J, Szymczyna BR, MacCoss MJ, Arvai AS, Yates JR, III, Tainer JA, Getzoff ED. Nitric-oxide synthase forms N-NO-pterin and S-NO-Cys. J. Biol. Chem. 2010;285:31581–31589. doi: 10.1074/jbc.M109.072496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang H, Xu Y, Joseph J, Kalyanaraman B. Intramolecular electron transfer between tyrosyl radical and cysteine residue inhibits tyrosine nitration and induces thiyl radical formation in model peptides treated with myeloperoxidase, H2O2, and NO2−. J. Biol. Chem. 2005;280:40684–40698. doi: 10.1074/jbc.M504503200. [DOI] [PubMed] [Google Scholar]
- 32.Mannick J, Schonhoff C, Papeta N, Ghafourifar P, Szibor M, Fang K, Gaston B. S-Nitrosylation of mitochondrial caspases. J. Cell Biol. 2001;154:1111–1116. doi: 10.1083/jcb.200104008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ng ESM, Jourd’Heuil D, McCord JM, Hernandez D, Yasui M, Knight D, Kubes P. Enhanced S-nitroso-albumin formation from inhaled NO during ischemia/reperfusion. Circ. Res. 2004;94:559–565. doi: 10.1161/01.RES.0000117771.63140.D6. [DOI] [PubMed] [Google Scholar]
- 34.Bryan NS, Calvert JW, Elrod JW, Gundewar S, Ji SY, Lefer DJ. Dietary nitrite supplementation protects against myocardial ischemia-reperflusion injury. Proc. Natl. Acad. Sci. U.S.A. 2007;104:19144–19149. doi: 10.1073/pnas.0706579104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jaffrey SR, Erdjument-Bromage H, Ferris CD, Tempst P, Snyder SH. Detection and characterization of protein nitrosothiols. Nat. Cell Biol. 2001;3:193–197. doi: 10.1038/35055104. [DOI] [PubMed] [Google Scholar]
- 36.Liu L, Hausladen A, Zeng M, Que L, Heitman J, Stamler JS. A metabolic enzyme for S-nitrosothiol conserved from bacteria to humans. Nature. 2001;410:490–494. doi: 10.1038/35068596. [DOI] [PubMed] [Google Scholar]
- 37.Fang K, Johns R, Macdonald T, Kinter M, Gaston B. S-Nitrosoglutathione breakdown prevents airway smooth muscle relaxation in the guinea-pig. Am. J. Physiol. Lung Cell Molec. Biol. 2000;279:L716–L721. doi: 10.1152/ajplung.2000.279.4.L716. [DOI] [PubMed] [Google Scholar]
- 38.Nikitovic D, Holmgren A. S-Nitrosoglutathione is cleaved by the thioredoxin system with liberation of glutathione and redox regulating nitric oxide. J. Biol. Chem. 1996;271:19180–19185. doi: 10.1074/jbc.271.32.19180. [DOI] [PubMed] [Google Scholar]
- 39.Benhar M, Forrester MT, Hess DT, Stamler JS. Regulated protein denitrosylation by cytosolic and mitochondrial thioredoxins. Science. 2008;320:1050–1054. doi: 10.1126/science.1158265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Forrester MT, Seth D, Hausladen A, Cyler CE, Foster MW, Matsumoto A, Benhar M, Marshall HE, Stamler JS. Thioredoxin-interacting protein (Txnip) is a feedback regulator of S-nitrosylation. J. Biol. Chem. 2009;284:36160–36166. doi: 10.1074/jbc.M109.057729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Haendeler J, Hoffmann J, Tischler V, Berk BC, Zeiher AM, Dimmeler S. Redox regulatory and anti-apoptotic functions of thioredoxin depend on S-nitrosylation at cysteine 69. Nature Cell Biol. 2002;4:743–749. doi: 10.1038/ncb851. [DOI] [PubMed] [Google Scholar]
- 42.Trujillo M, Alvarez MN, Peluffo G, Freeman BA, Radi R. Xanthine oxidase-mediated decomposition of S-nitrosothiols. J. Biol. Chem. 1998;273:7828–7834. doi: 10.1074/jbc.273.14.7828. [DOI] [PubMed] [Google Scholar]
- 43.Johnson MA, Macdonald TL, Mannick JB, Conaway MR, Gaston B. Accelerated S-nitrosothiol breakdown by amyotrophic lateral sclerosis mutant copper, zinc-superoxide dismutase. J. Biol. Chem. 2001;276:39872–39878. doi: 10.1074/jbc.M102781200. [DOI] [PubMed] [Google Scholar]
- 44.Schonhoff CM, Matsuoka M, Tummala H, Johnson MA, Estevez AG, Kamaid A, Ricart KC, Hashimoto Y, Gaston B, Macdonald TL, Xu Z, Mannick JB. S-nitrosothiol depletion in amyotropic lateral sclerosis. Proc. Natl. Acad. Sci. USA. 2006;103:2404–2409. doi: 10.1073/pnas.0507243103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Romeo AA, Capobianco JA, English AM. Superoxide dismutase targets NO from GSNO to Cysbeta93 of oxyhemoglobin in concentrated but not dilute solutions of the protein. J. Am. Chem. Soc. 2003;125:14370–14278. doi: 10.1021/ja0289752. [DOI] [PubMed] [Google Scholar]
- 46.Bateman RL, Rauh D, Tavshanjian B, Shokat KM. Human carbonyl reductase 1 is an S-nitrosoglutathione reductase. J. Biol. Chem. 2008;283:35756–35762. doi: 10.1074/jbc.M807125200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sliskovic I, Raturi A, Mutus B. Characterization of the S-denitrosation activity of protein disulfide isomerase. J. Biol. Chem. 2005;280:8733–8741. doi: 10.1074/jbc.M408080200. [DOI] [PubMed] [Google Scholar]
- 48.Ramachandran N, Root P, Jiang XM, Hogg PJ, Mutus B. Mechanism of transfer of NO from extracellular S-nitrosothiols into the cytosol by cell-surface protein disulfide isomerase. Proc. Natl. Acad. Sci. U.S.A. 2001;98:9539–9544. doi: 10.1073/pnas.171180998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gaston B, Drazen JM, Jansen A, Sugarbaker DA, Loscalzo J, Stamler JS. Relaxation of human bronchial smooth muscle by S-nitrosothiols in vitro. J. Pharmacol. Exp. Ther. 1994;268:978–984. [PubMed] [Google Scholar]
- 50.Gaston B, Stamler JS. Nitrogen oxides and lung function. In: Crystal R, West J, Weibel E, Barnes P, editors. The Lung: Scientific Foundations. Philadelphia: Lippincott Raven; 1997. pp. 239–253. [Google Scholar]
- 51.Zaman K, Carraro L, Doherty J, Henderson E, Lendermon E, Liu L, Verghese G, Zigler M, Moss M, Park E, Palmer L, Doctor A, Stamler JS, Gaston B. A novel class of compounds that increase CFTR expression and maturation in epithelial cells. Mol. Pharmacol. 2006;70:1435–1442. doi: 10.1124/mol.106.023242. [DOI] [PubMed] [Google Scholar]
- 52.Askew SC, Butler AR, Flitney FW, Kemp GD, Megson IL. Chemical mechanisms underlying the vasodilator and platelet anti-aggregating properties of S-nitroso-N-acetyl-DL-penicillamine and S-nitrosoglutathione. Bioorgan. Med. Chemi. 1995;3:1–9. doi: 10.1016/0968-0896(94)00139-t. [DOI] [PubMed] [Google Scholar]
- 53.Hogg N, Singh R, Konorev E, Joseph J, Kalyanaraman B. S- Nitrosoglutathione as a substrate for γ-glutamyl transpeptidase. Biochem. J. 1997;323:477–481. doi: 10.1042/bj3230477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mancuso C, Bonsignore A, DiStasio E, Mordente A, Motterline R. Bilirubin and S-nitrosothiols interaction: evidence for a possible role of bilirubin as a scavenger of nitric oxide. Biochem. Pharmacol. 2003;66:2355–2363. doi: 10.1016/j.bcp.2003.08.022. [DOI] [PubMed] [Google Scholar]
- 55.Holder AA, Marshall SC, Wang PG, Kwak C-H. The mechanism of the decomposition of a bronchodilator, S-nitroso-N-acetyl-D,L-penicillamine (SNAP), by a bronchoconstrictor, Aqueous sulfite: Detection of the N-nitrosohydroxylamine-N-sulfonate ion. Bull. Korean Chem. Soc. 2003;24:350–356. [Google Scholar]
- 56.Marozkina N, Stsiapura V, Morozkina T, Stepuro I, Gaston B. S-Nitrosoglutathione formation in human gastric secretions is not prevented by ascorbic acid and antioxidant vitamin complex “resiston,” International Scientific Conference, “Reactive Oxygen and Nitrogen Species, Antioxidants and Human Health” 2011; Smolensk, Russia. [Google Scholar]
- 57.Gaston B, Kelly R, Urban P, Liu L, Henderson EM, Doctor A, Teague WG, Fitzpatrick A, Erzurum S, Hunt JF. Buffering airway acid decreases exhaled nitric oxide in asthma. J. Allergy Clin. Immunol. 2006;118:817–822. doi: 10.1016/j.jaci.2006.06.040. [DOI] [PubMed] [Google Scholar]
- 58.Que LG, Yang Z, Stamler JS, Lugogo NL, Kraft M. S-nitrosoglutathione reductase: an important regulator in human asthma. Am. J. Respir. Crit. Care Med. 2009;180:226–231. doi: 10.1164/rccm.200901-0158OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Baraona E, Abittan CS, Dohmen K, Moretti M, Pozzato G, Chayes ZW, Schaefer C, Lieber CS. Gender differences in pharmacokinetics of alcohol. Alcohol Clin. Exp. Res. 2001;25:502–507. [PubMed] [Google Scholar]
- 60.Brown-Steinke K, deRonde K, Yemen S, Palmer LA. Gender differences in S-nitrosoglutathione reductase activity in the lung. PLoS One. 2010;5:e14007. doi: 10.1371/journal.pone.0014007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Doctor A, Platt R, Sheram MD, Eischeid A, McMahon T, Maxey T, Doherty J, Axelrod M, Kline J, Gurka M, Gow A, Gaston B. Hemoglobin conformation couples S-nitrosothiol content in erythrocytes to oxygen gradients. Proc. Natl. Acad. Sci. U.S.A. 2005;102:5709–5714. doi: 10.1073/pnas.0407490102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pawloski JR, Hess DT, Stamler JS. Export by red blood cells of nitric oxide bioactivity. Nature. 2001;409:622–626. doi: 10.1038/35054560. [DOI] [PubMed] [Google Scholar]
- 63.Singel DJ, Stamler JS. Chemical physiology of blood flow regulation by red blood cells: the role of nitric oxide and S-nitrosohemoglobin. Ann. Rev. Physiol. 2005;67:99–145. doi: 10.1146/annurev.physiol.67.060603.090918. [DOI] [PubMed] [Google Scholar]
- 64.Palmer L, Doctor A, Chhabra P, Sheram ML, Laubach V, Karlinsey MZ, Forbes M, Macdonald T, Gaston B. S-Nitrosothiols signal hypoxia-mimetic vascular pathology. J. Clin. Invest. 2007;117:2592–2601. doi: 10.1172/JCI29444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhang Y, Hogg N. The mechanism of transmembrane S-nitrosothiol transport. Proc. Natl. Acad. Sci. U.S.A. 2004;101:7891–7896. doi: 10.1073/pnas.0401167101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lipton A, Johnson M, Macdonald T, Lieberman M, Gozal D, Gaston B. S-Nitrosothiols signal the ventilatory response to hypoxia. Nature. 2001;413:171–174. doi: 10.1038/35093117. [DOI] [PubMed] [Google Scholar]
- 67.Zhang LM, St Croix C, Cao R, Wasserloos K, Watkins SC, Stevens T, Li S, Tyurin V, Kagan VE, Pitt BR. Cell-surface protein disulfide isomerase is required for transnitrosylation of metallothionein by S-nitroso-albumin in intact rat pulmonary vascular endothelial cells. Exp. Biol. Med. 2006;231:1507–1515. doi: 10.1177/153537020623100909. [DOI] [PubMed] [Google Scholar]
- 68.Davisson RL, Shaffer RA, Johnson AK, Lewis SJ. Stimulation of lumbar sympathetic nerves may produce hindlimb vasodilation via the release of pre-formed stores of nitrosyl factors. Neurosci. 1996;72:881–887. doi: 10.1016/0306-4522(96)00090-5. [DOI] [PubMed] [Google Scholar]
- 69.Straub AC, Billaud M, Johnstone SR, Best AK, Yemen S, Dwyer ST, Looft-Wilson R, Lysiak JJ, Gaston B, Palmer L, Isakson BE. Compartmentalized connexin 43 S-nitrosylation/denitrosylation regulates heterocellular communication in the vessel wall. Arterioscler. Thromb. Vasc. Biol. 2010 Nov 11; doi: 10.1161/ATVBAHA.110.215939. [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kluge I, Gutteck-Amsler U, Zollinger M, Do KQ. S-nitrosoglutathione in rat cerebellum: identification and quantification by liquid chromatography-mass spectrometry. J. Neurochem. 1997;69:2599–2607. doi: 10.1046/j.1471-4159.1997.69062599.x. [DOI] [PubMed] [Google Scholar]
- 71.Marozkina NV, Borowitz M, Yemen S, Liu L, Sun F, Islam R, Erdmann-Gilmore P, Townsend RR, Lichti CF, Mantri S, Gaston B, Zaman K. S-Nitrosoglutathione targets Hsp 70/Hsp 90 organizing protein as a corrector therapy for cystic fibrosis. Proc. Natl. Acad. Sci. U.S.A. 2010;107:11393–11398. doi: 10.1073/pnas.0909128107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tranguch S, Huet-Hudson Y. Decreased viability of nitric oxide synthase double knockout mice. Mol. Reproduc. Dev. 2003;65:175–179. doi: 10.1002/mrd.10274. [DOI] [PubMed] [Google Scholar]
- 73.Ckless K, Reynaert NL, Taatjes DJ, Lounsbury KM, van Vliet A, Janssen-Heininger Y. In situ detection and visualization of S-nitrosylated proteins following chemical derivatization: identification of Ran GTPase as a target for S-nitrosylation. Nitric Oxide. 2004;11:216–227. doi: 10.1016/j.niox.2004.06.002. [DOI] [PubMed] [Google Scholar]
- 74.Nott A, Watson PM, Robinson JD, Crepaldi L, Riccio A. S-nitrosylation of histone deacetylase 2 induces chromatin remodeling in neurons. Nature. 2008;455:411–416. doi: 10.1038/nature07238. [DOI] [PubMed] [Google Scholar]
- 75.Pakala SB, Bui-Nguyen TM, Reddy SDN, Li D-Q, Peng S, Rayala SK, Behringer RR, Kumar R. Regulation of NF-kappaB circuitry by a component of the nucleosome remodeling and deacetylase complex controls inflammatory response homeostatis. J. Biol. Chem. 2010;285:23590–23597. doi: 10.1074/jbc.M110.139469. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 76.Marozkina NV, Gaston B. Eur. Respir. Soc. Meeting Abstract. Vienna, Austria: 2009. S-nitrosylation of Ran-binding protein 3 in human airway epithelial smooth muscle cells: identification using a novel, double-read protein microarray technology. [Google Scholar]
- 77.Kunieda T, Minamino T, Miura K, Katsuno T, Tateno K, Miyauchi H, Kaneko S, Bradfield CA, Fitzgerald GA, Komuro I. Reduced nitric oxide causes age-associated impairment of circadian rhythmicity. Circ. Res. 2008;102:607–614. doi: 10.1161/CIRCRESAHA.107.162230. [DOI] [PubMed] [Google Scholar]
- 78.Zaman K, Palmer LA, Doctor A, Hunt JF, Gaston B. Concentration-dependent effects of endogenous S-nitrosoglutathione on gene regulation by specificity proteins Sp3 and Sp1. Biochem. J. 2004;380:67–74. doi: 10.1042/BJ20031687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhang J, Wang S, Wesley RA, Danner RL. Adjacent sequence controls the response polarity of nitric oxide-sensitive Sp factor binding sites. J. Biol. Chem. 2003;278:29192–29200. doi: 10.1074/jbc.M213043200. [DOI] [PubMed] [Google Scholar]
- 80.Zaman K, Hanigan MH, Smith A, Vaughan J, Macdonald T, Jones Dr, Hunt JF, Gaston B. Endogenous S-nitrosoglutathione modifies 5-lipoxygenase expression in airway epithelial cells. Am. J. Respir. Cell Molec. Biol. 2006;34:387–393. doi: 10.1165/rcmb.2005-0336RC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sumbayev VV, Budde A, Zhou J, Brune B. HIF-1 alpha protein as a target for S-nitrosation. FEBS Lett. 2003;535:106–112. doi: 10.1016/s0014-5793(02)03887-5. [DOI] [PubMed] [Google Scholar]
- 82.Marshall HE, Hess DT, Stamler JS. S-nitrosylation: physiological regulation of NF-kappaB. Proc. Natl. Acad. Sci. U.S.A. 2004;101:8841–8842. doi: 10.1073/pnas.0403034101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Reynaert NL, Ckless K, Korn SH, Vox H, Guala AS, Wouters FM, Van der Vliea A, Janssen-Heininger YMWYM. Nitric oxide represses inhibitory IκB kinase through S-nitrosylation. Proc. Natl. Acad. Sci. U.S.A. 2004;101:8945–8950. doi: 10.1073/pnas.0400588101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Olson N, Kasahara DI, Hristova M, Bernstein R, Janssen-Heininger Y, van der Vliet A. Modulation of NFκB and HIF1 by S-nitrosoglutathione does not alter allergic inflammation in mice. Am. J. Respir. Cell Mol. Biol. 2010 doi: 10.1165/rcmb.2010-0035OC. [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kelleher ZT, Matsumoto A, Stamler JS, Marshall HE. NOS2 regulation of NF-κB by Snitrosylation of p65. J. Biol. Chem. 2007;282:30667–30672. doi: 10.1074/jbc.M705929200. [DOI] [PubMed] [Google Scholar]
- 86.Ding SY, Tribble ND, Kraft CA, Markwardt M, Gloyn AL, Rizzo MA. Naturally occurring glucokinase mutations are associated with defects in posttranslational S-nitrosylation. Mol. Endocrinol. 2010;24:171–177. doi: 10.1210/me.2009-0138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kaneki M, Shimizu N, Yamada D, Chang K. Nitrosative stress and pathogenesis of insulin resistance. Antioxid. Redox Signal. 2007;9:319–329. doi: 10.1089/ars.2006.1464. [DOI] [PubMed] [Google Scholar]
- 88.Mohr S, Stamler JS, Brüne B. Posttranslational modification of glyceraldehyde-3-phosphate dehydrogenase by S-nitrosylation and subsequent NADH attachment. J. Biol. Chem. 1996;271:4209–4214. doi: 10.1074/jbc.271.8.4209. [DOI] [PubMed] [Google Scholar]
- 89.Hara MR, Snyder SH. Nitric oxide-GAPDH-Siah: a novel cell death cascade. Cell. Mol. Neurobiol. 2006;26:527–538. doi: 10.1007/s10571-006-9011-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Chakravarti R, Aulak KS, Fox PL, Stuehr DJ. GAPDH regulates cellular heme insertion into inducible nitric oxide synthase. Proc. Natl. Acad. Sci. U.S.A. 2010;107:18004–18009. doi: 10.1073/pnas.1008133107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.St Croix CM, Wasserloos KJ, Dineley KE, Reynolds IJ, Levitan ES, Pitt BR. Nitric oxide-induced changes in intracellular zinc homeostatis are mediated by metallothionein/thionein. Am. J. Physiol. 2002;282:185–192. doi: 10.1152/ajplung.00267.2001. [DOI] [PubMed] [Google Scholar]
- 92.Collawn JF, Bebok Z, Matalon S. Search and rescue: finding ways to correct ΔF508 CFTR. Am. J. Respir. Cell Mol. Biol. 2009;40:385–387. doi: 10.1165/rcmb.2008-0006ED. [DOI] [PubMed] [Google Scholar]
- 93.Zaman K, McPherson M, Vaughan J, Hunt J, Mendes F, Gaston B, Palmer L. S-Nitrosoglutathione increases cystic fibrosis transmembrane regulator maturation. Biochem. Biophys. Res. Commun. 2001;284:65–70. doi: 10.1006/bbrc.2001.4935. [DOI] [PubMed] [Google Scholar]
- 94.Howard M, Fischer H, Roux J, Santos BC, Gullans SR, Yancey PH, Welch WJ. Mammalian osmolytes and S-nitrosoglutathione promote ΔF508 cystic fibrosis transmembrane conductance regulator (CFTR) protein maturation and function. J. Biol. Chem. 2003;278:35159–35167. doi: 10.1074/jbc.M301924200. [DOI] [PubMed] [Google Scholar]
- 95.Andersson C, Gaston B, Roomans GM. S-nitrosoglutathione induces functional ΔF508-CFTR in airway epithelial cells. Biochem. Biophys. Res. Commun. 2002;297:552–557. doi: 10.1016/s0006-291x(02)02245-3. [DOI] [PubMed] [Google Scholar]
- 96.Okiyoneda T, Barriere H, Bagdany M, Rabeh WM, Du K, Hohfeld J, Young JC, Lukacs GL. Peripheral protein quality control removes unfolded CFTR from the plasma membrane. Science. 2010;329:805–810. doi: 10.1126/science.1191542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Choi YB, Tenneti L, Le DA, Ortiz J, Bai G, Chen HS, Lipton SA. Molecular basis of NMDA receptor-coupled ion channel modulation by S-nitrosylation. Nature Neurosci. 2000;3:15–21. doi: 10.1038/71090. [DOI] [PubMed] [Google Scholar]
- 98.Joksovic PM, Doctor A, Gaston B, Todorovic SM. Functional modulation of T-type calcium channels by S-nitrosothiols in the rat thalamus. J. Neurophysiol. 2007;97:2712–2721. doi: 10.1152/jn.00926.2006. [DOI] [PubMed] [Google Scholar]
- 99.Salanova M, Schiffl G, Rittweger J, Felsenberg D, Blottner D. Ryanodine receptor type-1 (RyR1) expression and protein S-nitrosylation pattern in human soleus myofibres following bed rest and exercise countermeasure. Histochem. Cell Biol. 2008;130:105–118. doi: 10.1007/s00418-008-0399-6. [DOI] [PubMed] [Google Scholar]
- 100.Gaston B, Sears S, Woods J, Hunt J, Ponaman M, McMahon T, Stamler JS. Bronchodilator S-nitrosothiol deficiency in asthmatic respiratory failure. Lancet. 1998;351:1317–1319. doi: 10.1016/S0140-6736(97)07485-0. [DOI] [PubMed] [Google Scholar]